 Open Access Article
Open Access ArticleNovel photocrosslinking chemical probes utilized for high-resolution spatial transcriptomics†
Leslie
Spitalny
a,
Natalie
Falco
 a,
Whitney
England
a,
Tyler
Allred
a and
Robert C.
Spitale
a,
Whitney
England
a,
Tyler
Allred
a and
Robert C.
Spitale
 *abc
*abc
aDepartment of Pharmaceutical Sciences, University of California, Irvine, California 92697, USA. E-mail: rspitale@uci.edu
bDepartment of Chemistry, University of California, Irvine, California 92697, USA
cDepartment of Molecular Biology & Biochemistry, University of California, Irvine, California 92697, USA
First published on 20th January 2025
Abstract
The architecture of cells and the tissue they form within multicellular organisms are highly complex and dynamic. Cells optimize their function within tissue microenvironments by expressing specific subsets of RNAs. Advances in cell tagging methods enable spatial understanding of RNA expression when merged with transcriptomics. However, these techniques are currently limited by the spatial resolution of the tagging, the number of RNAs that can be sequenced, and multiplexing to isolate spatially-distinct cells within the same tissue landscape. To address these limitations, we developed CrossSeq, which employs photocrosslinking fluorescent probes and confocal microscopy activation to demarcate user-defined regions of interest on fixed cells for multiplexed spatial transcriptomic analysis. We investigate phenyl azide and diazirine crosslinking scaffolds and define their photoactivity profiles. We then deploy the aryl azide scaffold with three fluorophores for multiplexing on glyoxal fixed cells and analyze the defined populations using flow cytometry. Finally, we apply CrossSeq to investigate an in vitro MDA-MB-231-LM2 metastatic cancer migration model to evaluate changes in gene expression at the migratory cell front versus the exterior population. We anticipate this new technology will be a valuable tool addition as it will enable easier access to spatial transcriptomic analysis for the scientific community using conventional microscopy and analysis techniques.
Introduction
Multicellular organisms are composed of highly complex and heterogeneous three-dimensional cellular structures that evolve through molecular signaling within their microenvironment. Each cell's dynamic transcriptome collectively defines normal tissue homeostasis as well as the aberrant formation of disease.1 Understanding how the spatial organization of cells defines RNA expression is essential for elucidations into the fields of oncology, immunology, and neurobiology.2 Advances in next-generation sequencing (NGS) such as the development of single-cell RNA sequencing (scRNA-seq) have allowed for unbiased, transcriptome-wide analysis of thousands of individual cells from a single sample.3 Although scRNA-seq has proven to be a very powerful technique, any spatial information is lost once the multicellular sample is dissociated for analysis. To overcome this limitation, there has been an emergence in the field of spatial transcriptomics to develop methods for in situ genetic profiling.4Current in situ spatial transcriptomic techniques incorporate NGS or imaging-based approaches, each with its own advantages and limitations.5In situ capture techniques6,7 position a permeabilized tissue slice over mRNA capture probes printed onto a microarray slide, each containing a barcoded reverse transcription (RT) primer with unique positional tags. Upon capture, mRNA is reverse transcribed into cDNA containing the spatial barcode, which can then be sequenced and mapped back to its original location creating a gene expression map. These methods provide unbiased transcriptome-wide analysis but are limited in their detection efficiency due to the size of the capture array. Fluorescence in situ hybridization (FISH) methods8,9 use targeted fluorescent probes to identify thousands of pre-selected mRNA sequences through rounds of multiplex imaging to generate a gene abundance map. Although these methods have high detection efficiency and spatial resolution, they require costly, specialized equipment and have low sample throughput.
Our lab has been focused on developing chemical approaches to characterize the transcriptome. It would be advantageous to develop a more streamlined approach to tag cells with high spatial resolution and enable isolation through conventional cell sorting techniques. Photoactivatable probes can be used to selectively tag cells with fluorescent labeling to record their spatial information, and samples can then be dissociated and analyzed with applicable NGS technologies to generate RNA expression profiles. Such a method employs conventional microscopy imaging to enable user-defined activation of cell-permeable photocaged dyes10 or biotin-containing photocrosslinking probes11 to tag live cells or tissues for downstream analyses. Cellular tagging is an exciting alternative as it can be applied to any cell of interest. Additionally, conventional photo sources and microscopes can be employed for tagging, and post-tagging processes used are already adopted by many labs focused on understanding gene expression and cellular heterogeneity.
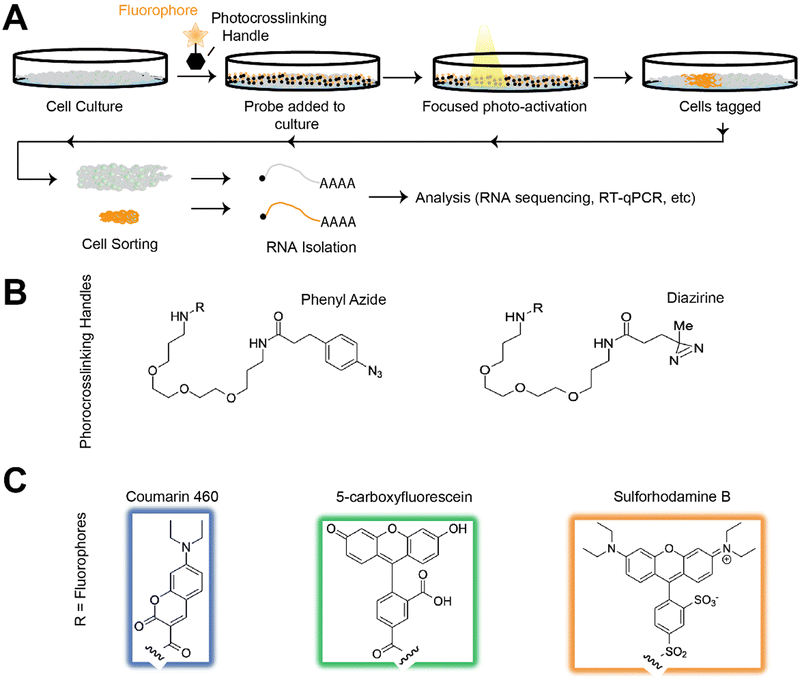
Herein, we aimed to develop and improve methods of tagging and analyzing cells with high spatial resolution. Our approach allows the user to define regions of interest (ROIs) on cells and crosslink a chemical probe with a fluorophore of choice to the ROI via near UV-light activation using confocal microscopy. This technique ensures that ROIs are defined with high resolution and does not require pre-target selection, mRNA capture arrays, or cellular engineering. Once ROIs are tagged, the sample is dissociated, sorted into the defined regions using flow cytometry, and then processed for whole transcriptome analysis with RNA-seq, as shown in (Fig. 1(A)). We demonstrate our platform's high-resolution multiplex capabilities with various irradiated ROIs in different shapes and sizes on assorted cell types. We also establish a simple, single-day workflow to map gene expression in situ on glyoxal fixed metastatic triple-negative breast cancer MDA-MB-231-LM2 cells in an in vitro wound healing model. We validated this system on glyoxal fixed cells to enable a less toxic fixation method, preserve samples for downstream processing, and produce high-integrity extractable RNA for a more user-friendly workflow.12 We anticipate our approach, and its optimized RNA-friendly workflow, will be adopted by many labs aiming to better understand RNA expression with spatial resolution.
Results
We aimed to develop a set of photo-crosslinkers that were relatively easy to synthesize, could be produced in high yield and abundance, and would give us access to different fluorophore wavelengths for cell sorting after tagging. Toward this goal, we designed our bi-functional CrossSeq probes with common photocrosslinking handles attached to fluorophores with varying emission wavelengths for multiplexing.13 We chose both the phenyl azide14 and diazirine15 crosslinking handles, which have different excitation maxima, to compare which would excite and react more quickly when exposed to light in our system. The three attached fluorophore options, shown in Fig. 1(C), were chosen based on their high quantum yield16–18 and distinct emission colors.For ease of use, we aimed to synthesize all probes in scalable parallel synthetic steps. Their syntheses are outlined in the ESI.† Briefly, commercially-available (ESI†) carboxy-fluorophores were first crosslinked to bi-functional amine linkers using carbodiimide amide coupling. The BOC-protected fluorophores were subsequently deprotected and coupled with a phenyl azide or diazirine to install UV-active cross-linkers. This straightforward synthetic scheme with minimal steps enables easy access to our CrossTag probes, furthering the user-friendliness of this platform.
First, we determined each probe's photo-reactivity profile with bovine serum albumin (BSA) as a model protein to evaluate the required time and energy for binding.19 We incubated BSA with each CrossSeq probe individually at 100 μM concentration and then performed an irradiation time course with a 368 nm UV Crosslinker. The tagged BSA was then precipitated in methanol to remove the unbound probe. Next, crosslinking was resolved using denaturing gel electrophoresis, representing fluorescent signal accumulation per probe. As shown in Fig. 2(A), the phenyl azide probes react and bind more quickly than the diazirine scaffolds. This was unexpected as alkyl diazirines are known to excite with a longer UV wavelength, 350 nm, as compared to simple phenyl azide, 254–300 nm.20 A low level of background labeling of BSA in the absence of light was observed with the diazirine-containing probes conjugated to both 5-carboxyfluorescein and sulforhodamine B. Photoactivatable probes can produce spurious background due to ambient light exposure which can be difficult to completely exclude during sample processing.15 The quantitative representation shown in Fig. 2(B) confirms this result for each photoactivatable handle-fluorophore pair.
After validation that our CrossSeq probes crosslink to protein, we moved forward with evaluating our system on cells with a confocal microscope. Most confocal microscopes only operate in the visible light range, so we began testing to determine if the shortest wavelength option would activate our probes with the same efficiency as the UV light source. Using the 405 nm laser, we evaluated both the laser power and probe concentration required to successfully activate a ROI on in vitro cultured cells. In order to capture the RNA expression levels at a specific biological state, we decided to work with fixed cells to retain the biological relevance and limit the possibility of RNA alteration due to stress or degradation. Fixed cells are also more clinically relevant as pathologists will fix tissue for long-term storage and evaluation.2 Most researchers use a form of formaldehyde for tissue fixation, but this reagent is known to be detrimental to RNA retrieval and integrity as nucleic acids become crosslinked to proteins through Schiff base adduct formation and are then lost during RNA isolation or become degraded through de-crosslinking thermal treatment.12 Therefore, we decided to utilize glyoxal as a less toxic fixative that does not crosslink nucleic acids to proteins. An adduct can be formed on the Watson–Crick face of guanine at the acidic pH required for glyoxal fixation, but this adduct is unstable at neutral pH or higher and will readily reverse to preserve nucleosides.12
We optimized probe binding at low micromolar concentrations to minimize background signal and with low laser power to attenuate cellular degradation. Through this testing process we determined that even if the probe's crosslinking handle was the same, the conjugated fluorophore was the determining factor for how long and how much laser power was required to activate and bind each probe successfully. We postulate this may be due to the selected fluorophores' xanthene scaffold and their ability to aggregate, dimerize, and absorb the 405 nm activation energy due to their blue-shifted absorption properties in these states.16,21,22 Thus, the more aromatic and electron-accepting the fluorophores were, the more energy was required to efficiently activate the crosslinking azide handle. As shown in Fig. 3(A), the time and laser power required to activate a single ROI varies greatly per probe with 100% 405 nm laser for almost 3 minutes required for the sulforhodamine compared to 1% 405 nm laser for 30 seconds for the coumarin probe. Additionally, the concentration of the probes depended on the fluorophores’ photostability. For example, 5-carboxyfluorescein photobleaches readily23 and therefore requires the highest concentration of the three probes, 200 μM, compared to sulforhodamine b which is the most stable and only required 1 μM. Once the activation parameters were determined, it was further confirmed that a longer excitation wavelength did not diminish the binding of the phenyl azide probes, and tagged the cells more quickly than the diazirine. Consequently, we moved forward with the phenyl azide probes for multiplexing and flow cytometry evaluation.
In vitro cultured, glyoxal fixed HeLa cells were exposed to all three fluorophore-phenyl azide probes sequentially and were activated to achieve multiplexed ROIs, as shown in Fig. 3(A). We next used these same activation parameters to tag equal-sized ROIs within a single well on an ibidi 8-well optical μ-slide confluent with fixed HeLa cells. We defined ROIs in the navigator mode which creates a map of the sample with square tiles. This sample tile map is shown in Fig. 3(B), where each highlighted ROI occupies about 3% of the overall well surface area. Using the 20× water objective, each ROI was excited sequentially with each phenyl azide-fluorophore probe. The sample was then incubated in the gentle cell detachment solution, Accumax, for 10 minutes at 37 °C, resuspended in the same solution, and flown on an Agilent Novocyte Quanteon flow cytometer containing violet, blue, yellow, and red lasers. As seen in Fig. 3(C), each CrossSeq probe-tagged population was clearly defined and detected. The gated, fluorescent populations compose slightly higher than 2% of the overall sample population per probe type showing consistent tagging capability with minimal fluorescent cross talk.
Next, to demonstrate the utility of the CrossSeq platform to characterize gene expression within a specific population, we conducted an in vitro cell migration assay to investigate the highly metastatic triple-negative breast cancer cell line, MDA-MB-231-LM2 (MDA-LM2). This wound-healing model provides a standardized technique to measure cellular migration in a two-dimensional in vitro environment.24 The MDA-LM2 cell line was derived from the parent human triple-negative breast cancer line, MDA-MB-231, by selecting for cancer cell populations that successfully migrated from the primary tumor to form lung metastases in a xenograft mouse model.25 We specifically aimed to elucidate changes in gene expression for the cells that migrated into the wound gap versus the exterior static cell population over 48 hours, just before the complete closure of an inflicted wound (Fig. S3, ESI†).
A 920 μm wound with clearly marked boundaries was created on confluent MDA-LM2 cultures. Then the growth media was replaced with a low percent serum to promote migration, instead of proliferation, into the gap. The cells were then allowed to migrate for 0, 24, and 48 hours before being fixed with glyoxal, and were then examined via confocal microscopy. A map of each sample was then created in navigator mode (Fig. 4(A)) and the cells along the border and within the wound were tagged by 405 nm laser activation of the coumarin CrossTag probe with a 10× air objective (Fig. 4(B)). The coumarin CrossTag probe was chosen for this tagging application as it has a quick activation time and requires the least laser power. After the cells were dissociated with Accumax, samples were sorted by fluorescence-activated cell sorting (FACS) by their coumarin-tagged fluorescent label (Fig. 4(C)) and subjected to SMARTer Stranded Total RNA-Seq.26
Our overall goal was to develop a workflow for the analysis of RNA isolated from tagged cells. We aimed to test this directly by using probes in a cell-based workflow for RNA isolation and analysis. The methods used for the analysis are described in the supplementary information file. Briefly, cDNA reads were pseudoaligned using kallisto 0.48.027 to a GENCODE v29. Following alignment, differentially expressed RNAs were determined using DESeq2 1.31.16.28 GO term analysis was performed using statistical overrepresentation tests against Biological Process GO terms through PANTHER 17.0.29 GO term network analysis was performed using the EnrichmentMap 3.3.4 plugin30 in Cytoscape 3.8.2.31
Analysis of the differentially expressed transcripts revealed changes in gene expression when comparing the migrated cell front at 24 and 48 hours post wounding to the migration front at 0 hours when the cells had not yet migrated and the wound was first inflicted. Some key upregulated genes were identified as having known roles in proliferation and tumor cell migration. For example, Small Cajal body-associated RNA 2 (scaRNA2) regulates DNA repair pathway choice by inhibiting DNA-PK and increased expression of scaRNA2 also promotes cell proliferation, migration, and invasion in cancer cells.32,33 Another example, tumor necrosis factor receptor superfamily member 6b (TNFRSF6B or DcR3) plays an important role in cancer progression and immunosuppression.34 TNFRSF6B has also been demonstrated to be highly overexpressed in more aggressive metastatic cancers and also is a strong prognosticator for cancer metastasis when taken into account with other features of human sera.35 Also, chondroadherin (CHAD), an extracellular matrix protein, showed increased expression and is known to be correlated with increased tumor size and metastasis.36–38 Additionally, several long non-coding RNA genes, MIR210HG and PPP1R26-AS1, were upregulated which have been proven to be oncogenic in breast cancer.39–42 Conversely, a down-regulation in known tumor suppressors, sodium/calcium exchanger 2 (SLC8A2) and aryl hydrocarbon receptor repressor (AHRR), were observed in cells at the migration front, aiding in invasion.43–46 These changes in gene expression were seen in both the 24 and 48-hour migration front cells. Interestingly, expression of C-X-C chemokine receptor type 5 (CXCR5) was shown to be downregulated at 24 hours but upregulated at 48 hours. This receptor is known to aid in signaling for tumor cell development and migration, suggesting a longer timeline from wounding is required for increased expression.47–49 Overall, these data support the development of a robust protocol to tag cells with high resolution, sort the cells, and isolate the RNA for downstream bioinformatic analyses.
Conclusion
Herein we report the development of novel chemical reagents that enable high-resolution imaging and cell sorting of distinct populations of cells in 2-dimensional culture conditions. We demonstrate the utility of an efficient synthetic scheme to derive three-colored crosslinking reagents which can be activated by exposure to UV light.In vitro crosslinking with BSA demonstrated similar yield and kinetics of UV-induced adduct formation. We also reveal that phenyl azide crosslinking reagents have higher yield of crosslinking than diazirine-containing reagents in our hands. Using a confocal microscope, we were able to use our novel reagents to rapidly and clearly define cellular boundaries for cell tagging with fluorophores representing three different and distinguishable wavelengths. Cells tagged with these probes were efficiently sorted with high accuracy, demonstrating the ease of our approach to analyze specific cell populations after tagging.
We finally demonstrated the utility of our approach in a metastatic MDA-LM2 cell migration assay where we used one of our probes to tag migrating cells for cell sorting and analysis by RNA sequencing. To enable RNA sequencing experiments we optimized a glyoxal preservation protocol, moving away from paraformaldehyde (PFA) and formalin-fixed paraffin-embedded (FFPE) tissues additives, which are known to reduce RNA integrity and introduce issues with ‘omic measurements of RNA expression, such as RNA sequencing. Using our approach we were able to profile changes in gene expression of migrating cells in the migration assay and identify differences in migrating cells by RNA sequencing. This overall demonstrates a full workflow for analysis of cells by gene expression using photo-activated cell tagging with high resolution.
We anticipate that the application of the CrossSeq platform will be very useful for identifying differences in RNA expression depending on cellular orientation or organization in space. Such processes as development, organ organization, or conditions where cells are moving through structured tissues (in vivo migration and metastasis) would benefit from spatial resolution and analysis to better understand how cell organization contributes to RNA expression programs. With improved RNA integrity, glyoxal fixation allows for theoretical compatibility with many next-generation sequencing (NGS) methods, such as scRNA-seq3 and slam-seq-total,50 which provide RNAome analysis from a single cell. This stringent sequencing coupled with three-dimensional tissue profiling can help to further elucidate the biological contribution of RNA, both coding and non-coding, to disease development. We envision that the introduction of this innovative technology will enhance the accessibility of spatial transcriptomic analysis for the scientific community by integrating seamlessly with traditional microscopy and analysis methods. These approaches and extending this method to tissue samples is currently underway in our lab and will be reported in due course.
Data availability
The experimental section, figures and tables; NMR spectra; and other details are in the ESI† and available upon request.Conflicts of interest
The authors declare no competing financial interests.Acknowledgements
We thank members of the Spitale lab for their critical reading of the manuscript. This work is supported by the Ono Pharma Foundation and R. C. S. is a Pew Biomedical Scholar.References
- T. A. Cooper, L. Wan and G. Dreyfuss, RNA and Disease, Cell, 2009, 136, 777–793 CrossRef.
- M. Asp, J. Bergenstråhle and J. Lundeberg, Spatially Resolved Transcriptomes—Next Generation Tools for Tissue Exploration, BioEssays, 2020, 42, 1900221 CrossRef PubMed.
- O. Stegle, S. A. Teichmann and J. C. Marioni, Computational and analytical challenges in single-cell transcriptomics, Nat. Rev. Genet., 2015, 16, 133–145 CrossRef.
- S. K. Longo, M. G. Guo, A. L. Ji and P. A. Khavari, Integrating single-cell and spatial transcriptomics to elucidate intercellular tissue dynamics, Nat. Rev. Genet., 2021, 22, 627–644 CrossRef.
- L. Yue, F. Liu, J. Hu, P. Yang, Y. Wang, J. Dong, W. Shu, X. Huang and S. Wang, A guidebook of spatial transcriptomic technologies, data resources and analysis approaches, Comput. Struct. Biotechnol. J., 2023, 21, 940–955 CrossRef.
- P. L. Ståhl, F. Salmén, S. Vickovic, A. Lundmark, J. F. Navarro, J. Magnusson, S. Giacomello, M. Asp, J. O. Westholm and M. Huss, et al., Visualization and analysis of gene expression in tissue sections by spatial transcriptomics, Science, 2016, 353, 78–82 CrossRef PubMed.
- S. G. Rodriques, R. R. Stickels, A. Goeva, C. A. Martin, E. Murray, C. R. Vanderburg, J. Welch, L. M. Chen, F. Chen and E. Z. Macosko, Slide-seq: A scalable technology for measuring genome-wide expression at high spatial resolution, Science, 2019, 363, 1463–1467 CrossRef PubMed.
- K. H. Chen, A. N. Boettiger, J. R. Moffitt, S. Wang and X. Zhuang, Spatially resolved, highly multiplexed RNA profiling in single cells, Science, 2015, 348, aaa6090 CrossRef PubMed.
- C.-H. L. Eng, M. Lawson, Q. Zhu, R. Dries, N. Koulena, Y. Takei, J. Yun, C. Cronin, C. Karp and G.-C. Yuan, et al., Transcriptome-scale super-resolved imaging in tissues by RNA seqFISH+, Nature, 2019, 568, 235–239 CrossRef.
- A. S. Genshaft, C. G. K. Ziegler, C. N. Tzouanas, B. E. Mead, A. M. Jaeger, A. W. Navia, R. P. King, M. D. Mana, S. Huang and V. Mitsialis, et al., Live cell tagging tracking and isolation for spatial transcriptomics using photoactivatable cell dyes, Nat. Commun., 2021, 12, 4995 CrossRef PubMed.
- Q. Tang, L. Liu, Y. Guo, X. Zhang, S. Zhang, Y. Jia, Y. Du, B. Cheng, L. Yang and Y. Huang, et al., Optical Cell Tagging for Spatially Resolved Single-Cell RNA Sequencing, Angew. Chem., Int. Ed., 2022, 61, e202113929 CrossRef PubMed.
- P. Channathodiyil and J. Houseley, Glyoxal fixation facilitates transcriptome analysis after antigen staining and cell sorting by flow cytometry, PLoS One, 2021, 16, e0240769 CrossRef PubMed.
- G. T. Hermanson, Bioconjugate Techniques, 3rd edn, 2013, pp. 229–58 Search PubMed.
- S. Reddington, P. Watson, P. Rizkallah, E. Tippmann and D. D. Jones, Genetically encoding phenyl azide chemistry: new uses and ideas for classical biochemistry, Biochem. Soc. Trans., 2013, 41, 1177–1182 CrossRef PubMed.
- A. V. West, G. Muncipinto, H.-Y. Wu, A. C. Huang, M. T. Labenski, L. H. Jones and C. M. Woo, Labeling Preferences of Diazirines with Protein Biomolecules, J. Am. Chem. Soc., 2021, 143, 6691–6700 CrossRef PubMed.
- J. Q. Umberger and V. K. LaMer, The Kinetics of Diffusion Controlled Molecular and Ionic Reactions in Solution as Determined by Measurements of the Quenching of Fluorescence 1,2, J. Am. Chem. Soc., 1945, 67, 1099–1109 CrossRef.
- R. F. Kubin and A. N. Fletcher, Fluorescence quantum yields of some rhodamine dyes, J. Lumin., 1983, 27, 455–462 CrossRef.
- G. Jones, W. R. Jackson, C. Y. Choi and W. R. Bergmark, Solvent effects on emission yield and lifetime for coumarin laser dyes. Requirements for a rotatory decay mechanism, J. Phys. Chem., 1985, 89, 294–300 CrossRef.
- B. Liu, Y. Pang, R. Bouhenni, E. Duah, S. Paruchuri and L. McDonald, A step toward simplified detection of serum albumin on SDS-PAGE using an environment-sensitive flavone sensor, Chem. Commun., 2015, 51, 11060–11063 RSC.
- G. T. Hermanson, Bioconjugate Techniques, 3rd edn, 2013, pp. 275–98 Search PubMed.
- O. Valdes-Aguilera and D. C. Neckers, Aggregation phenomena in xanthene dyes, Acc. Chem. Res., 1989, 22, 171–177 CrossRef.
- Y. E. Litman, H. B. Rodríguez, S. E. Braslavsky and E. S. Román, Photophysics of Xanthene Dyes at High Concentrations in Solid Environments: Charge Transfer Assisted Triplet Formation, Photochem. Photobiol., 2018, 94, 865–874 CrossRef PubMed.
- L. Song, E. J. Hennink, I. T. Young and H. J. Tanke, Photobleaching kinetics of fluorescein in quantitative fluorescence microscopy, Biophys. J., 1995, 68, 2588–2600 CrossRef.
- J. E. N. Jonkman, J. A. Cathcart, F. Xu, M. E. Bartolini, J. E. Amon, K. M. Stevens and P. Colarusso, An introduction to the wound healing assay using live-cell microscopy, Cell Adhes. Migr., 2015, 8, 440–451 CrossRef PubMed.
- A. J. Minn, G. P. Gupta, P. M. Siegel, P. D. Bos, W. Shu, D. D. Giri, A. Viale, A. B. Olshen, W. L. Gerald and J. Massagué, Genes that mediate breast cancer metastasis to lung, Nature, 2005, 436, 518–524 CrossRef.
- S. Picelli, Å. K. Björklund, O. R. Faridani, S. Sagasser, G. Winberg and R. Sandberg, Smart-seq2 for sensitive full-length transcriptome profiling in single cells, Nat. Methods, 2013, 10, 1096–1098 CrossRef.
- N. L. Bray, H. Pimentel, P. Melsted and L. Pachter, Near-optimal probabilistic RNA-seq quantification, Nat. Biotechnol., 2016, 34, 525–527 CrossRef PubMed.
- M. I. Love, W. Huber and S. Anders, Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2, Genome Biol., 2014, 15, 550 CrossRef PubMed.
- P. D. Thomas, D. Ebert, A. Muruganujan, T. Mushayahama, L. Albou and H. Mi, PANTHER: Making genome-scale phylogenetics accessible to all, Protein Sci., 2022, 31, 8–22 CrossRef PubMed.
- D. Merico, R. Isserlin, O. Stueker, A. Emili and G. D. Bader, Enrichment Map: A Network-Based Method for Gene-Set Enrichment Visualization and Interpretation, PLoS One, 2010, 5, e13984 CrossRef PubMed.
- P. Shannon, A. Markiel, O. Ozier, N. S. Baliga, J. T. Wang, D. Ramage, N. Amin, B. Schwikowski and T. Ideker, Cytoscape: A Software Environment for Integrated Models of Biomolecular Interaction Networks, Genome Res., 2003, 13, 2498–2504 CrossRef.
- B. Nsengimana, F. A. Khan, U. A. Awan, D. Wang, N. Fang, W. Wei, W. Zhang and S. Ji, Pseudogenes and Liquid Phase Separation in Epigenetic Expression, Front. Oncol., 2022, 12, 912282 CrossRef PubMed.
- S. Bergstrand, E. M. O’Brien, C. Coucoravas, D. Hrossova, D. Peirasmaki, S. Schmidli, S. Dhanjal, C. Pederiva, L. Siggens and O. Mortusewicz, et al., Small Cajal body-associated RNA 2 (scaRNA2) regulates DNA repair pathway choice by inhibiting DNA-PK, Nat. Commun., 2022, 13, 1015 CrossRef.
- C. Zhang, H. Li, Y. Huang, Y. Tang, J. Wang, Y. Cheng, Y. Wei, D. Zhu, Z. Cao and J. Zhou, Integrative analysis of TNFRSF6B as a potential therapeutic target for pancreatic cancer, J. Gastrointest. Oncol., 2021, 12, 1673–1690 CrossRef PubMed.
- J. Li, N. Xie, J. Yuan, L. Liu, Q. Zhou, X. Ren, Q. Chen, G. Zhang, Q. Ruan and Y. H. Chen, et al., DcR3 combined with hematological traits serves as a valuable biomarker for the diagnosis of cancer metastasis, Oncotarget, 2017, 8, 107612 CrossRef PubMed.
- J. P. Rennhack, B. To, M. Swiatnicki, C. Dulak, M. P. Ogrodzinski, Y. Zhang, C. Li, E. Bylett, C. Ross and K. Szczepanek, et al., Integrated analyses of murine breast cancer models reveal critical parallels with human disease, Nat. Commun., 2019, 10, 3261 CrossRef PubMed.
- S. Gokce, D. Herkiloglu, E. İ. Kaygusuz, O. Cevik and S. Ahmad, Association of chondroadherin with leiomyosarcoma, Gynecol. Oncol. Rep., 2023, 46, 101144 CrossRef.
- R. Nicolle, M. Ayadi, A. Gomez-Brouchet, L. Armenoult, G. Banneau, N. Elarouci, M. Tallegas, A.-V. Decouvelaere, S. Aubert and F. Rédini, et al., Integrated molecular characterization of chondrosarcoma reveals critical determinants of disease progression, Nat. Commun., 2019, 10, 4622 CrossRef PubMed.
- W. Shi, Y. Tang, J. Lu, Y. Zhuang and J. Wang, MIR210HG promotes breast cancer progression by IGF2BP1 mediated m6A modification, Cell Biosci., 2022, 12, 38 CrossRef PubMed.
- Y. Du, N. Wei, R. Ma, S.-H. Jiang and D. Song, Long Noncoding RNA MIR210HG Promotes the Warburg Effect and Tumor Growth by Enhancing HIF-1α Translation in Triple-Negative Breast Cancer, Front. Oncol., 2020, 10, 580176 CrossRef PubMed.
- S. Xu, D. Kong, Q. Chen, Y. Ping and D. Pang, Oncogenic long noncoding RNA landscape in breast cancer, Mol. Cancer, 2017, 16, 129 CrossRef.
- S. Zhou, S. Zhang, H. Zhang, J. Ma, H. Dai, L. Qu and M. Zhou, Clinical Potential of lncRNA PPP1R26-AS1 in Breast Cancer and Its Contribution to Cancer Progression, Mol. Biotechnol., 2022, 64, 660–669 CrossRef PubMed.
- A. S. Beltran, A. Russo, H. Lara, C. Fan, P. M. Lizardi and P. Blancafort, Suppression of Breast Tumor Growth and Metastasis by an Engineered Transcription Factor, PLoS One, 2011, 6, e24595 CrossRef.
- M. Qu, J. Yu, H. Liu, Y. Ren, C. Ma, X. Bu and Q. Lan, The Candidate Tumor Suppressor Gene SLC8A2 Inhibits Invasion, Angiogenesis and Growth of Glioblastoma, Mol. Cells, 2017, 40, 761–772 CrossRef CAS.
- C. F. A. Vogel and T. Haarmann-Stemmann, The aryl hydrocarbon receptor repressor – More than a simple feedback inhibitor of AhR signaling: Clues for its role in inflammation and cancer, Curr. Opin. Toxicol., 2017, 2, 109–119 CrossRef PubMed.
- C. F. A. Vogel, G. Lazennec, S. Y. Kado, C. Dahlem, Y. He, A. Castaneda, Y. Ishihara, C. Vogeley, A. Rossi and T. Haarmann-Stemmann, et al., Targeting the Aryl Hydrocarbon Receptor Signaling Pathway in Breast Cancer Development, Front. Immunol., 2021, 12, 625346 CrossRef CAS.
- N. A. Mitkin, C. D. Hook, A. M. Schwartz, S. Biswas, D. V. Kochetkov, A. M. Muratova, M. A. Afanasyeva, J. E. Kravchenko, A. Bhattacharyya and D. V. Kuprash, p53-dependent expression of CXCR5 chemokine receptor in MCF-7 breast cancer cells, Sci. Rep., 2015, 5, 9330 CrossRef CAS PubMed.
- D. Zhu, W. Ye and J. Jiang, Clinical significance of CXCL13/CXCR5 axis in human cancers, Transl. Cancer Res., 2018, 7, 1737–1742 CrossRef.
- M. G. Kazanietz, M. Durando and M. Cooke, CXCL13 and Its Receptor CXCR5 in Cancer: Inflammation, Immune Response, and Beyond, Front. Endocrinol., 2019, 10, 471 CrossRef.
- A. Isakova, N. Neff and S. R. Quake, Single cell profiling of total RNA using Smart-seq-total, bioRxiv, 2020, preprint DOI:10.1101/2020.06.02.131060.
Footnote |
| † Electronic supplementary information (ESI) available. See DOI: https://doi.org/10.1039/d4cb00262h |
| This journal is © The Royal Society of Chemistry 2025 |




![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 000 events were recorded at the beginning of sorting. (D) and (E) RNA-seq analysis of differentially expressed genes per time point: Volcano plot analyses comparing the genes that were statistically up- or down-regulated at the tagged migration front for samples 0
000 events were recorded at the beginning of sorting. (D) and (E) RNA-seq analysis of differentially expressed genes per time point: Volcano plot analyses comparing the genes that were statistically up- or down-regulated at the tagged migration front for samples 0