 Open Access Article
Open Access ArticleCreative Commons Attribution 3.0 Unported Licence
Casein microparticles filled with cellulase to enzymatically degrade nanocellulose for cell growth†
Céline
Bastard
 abc,
Jann Carl-Theodor
Schulte
d,
Md
Asaduzzaman
abc,
Jann Carl-Theodor
Schulte
d,
Md
Asaduzzaman
 d,
Calvin
Hohn
d,
Yonca
Kittel
d,
Calvin
Hohn
d,
Yonca
Kittel
 ab,
Laura
De Laporte
ab,
Laura
De Laporte
 *abc and
Ronald
Gebhardt
*abc and
Ronald
Gebhardt
 *d
*d
aInstitute for Technical and Macromolecular Chemistry, RWTH Aachen University, Worringerweg 1-2, 52074 Aachen, Germany. E-mail: delaporte@dwi.rwth-aachen.de
bDWI – Leibniz-Institute for Interactive Materials, Forckenbeckstr. 50, 52074 Aachen, Germany
cInstitute of Applied Medical Engineering (AME), Department of Advanced Materials for Biomedicine (AMB), University Hospital RWTH Aachen, Center for Biohybrid Medical Systems (CMBS), Forckenbeckstr. 55, 52074 Aachen, Germany
dChair of Soft Matter Process Engineering (AVT.SMP), RWTH Aachen University, Forckenbeckstr. 51, 52074 Aachen, Germany. E-mail: ronald.gebhardt@avt.rwth-aachen.de
First published on 13th March 2025
Abstract
For tissue engineering, nanocellulose-based three-dimensional hydrogel structures hold potential as biocompatible support materials for biomimetic scaffolds to regenerate damaged tissues. One challenge of this material is that nanocellulose does not degrade in the human body. Therefore, different carriers are needed to locally deliver cellulase in a controlled manner to degrade the scaffold at the same time the cells grow and proliferate. To achieve this, we developed casein microparticles (CMPs) as delivery vehicles as they are non-toxic and have high porosity with a stable structure at physiological pH values. The porosity of the CMPs was first tested by diffusion experiments with fluorescently labelled dextrans of different sizes as model molecules, demonstrating inward diffusion of dextrans up to 500 kDa. The CMPs continuously release active cellulase, resulting in the degradation of the nanocellulose hydrogel over a time of 21 days, supporting 3D cell growth.
Introduction
Cellulose nanofibrils (CNFs) have the potential to work as 3D scaffold materials in the biomaterials field for tissue engineering, 3D printing, wound dressing, or medical implants.1–3 CNFs are normally isolated from the cell walls of wood and plants and are a renewable feedstock, which makes them more economical than other materials.4,5 In general, nanocellulose materials are biocompatible,5,6 non-cytotoxic,7 and support cell growth.8–10 They also have low immunogenicity in vivo.11 However, CNFs do not degrade in the human body or under standard cell culture conditions in vitro. Therefore, in vitro, CNF hydrogels with a nanomesh structure have been crushed with a grid in pieces after which the CNF parts reannealed into a 3D construct resulting in macropores.9 These larger pores then supported cell growth which was not possible in the initial CNF hydrogels.Another way to create space inside CNF hydrogels for cells to grow is to degrade the structure in a controlled manner. The cellulase enzyme degrades cellulose in an orthogonal manner without affecting mammalian cell processes as cellulase is not toxic and not naturally present in the body.12,13 This is in contrast to the degradation mechanism usually used in 3D natural or synthetic hydrogels for cell growth, which mainly relies on matrix metalloprotease (MMP)-induced degradation.14 It is known that natural hydrogels like fibrin or collagen may degrade too rapidly in vivo to support tissue formation,15 while synthetic materials lead to synthetic degradation products. The degradation products of CNFs on the other hand are non-toxic sugars.
In particular, cellulase from Trichoderma reesei is stable for a long time and shows degradation of nanocellulose in a physiological pH range from 6 to 7.4. This enzyme has been used to degrade CNF structures after a cell monolayer was formed to achieve free-standing cell layers.8 Still, in vivo, the temperature and pH sensitivity of cellulase could be a challenge. One possibility would be to pre-treat a cellulose-based scaffold before implantation in vivo.16 Another possibility would be the integration of an enzyme carrier system in the nanocellulose biomaterial, which releases the enzyme over the long term.17 This encapsulation or immobilization could protect the cellulase.
Cellulase has a total enzyme length of 18–21.5 nm and a diameter of 4–6.5 nm,18,19 thus larger pores are needed to release it. Different materials have been investigated for cellulase immobilization, such as mesoporous silica,20,21 Au-doped magnetic silica nanoparticles,22 and superparamagnetic chitosan microspheres,23,24 or alginate for encapsulation.17,25 Still, these materials have some drawbacks, for example, they need a combination of different materials or high production temperature resulting in high cost, have high variability in their particle sizes, are not stable over long time frames, or have not yet been tested for their biocompatibility. In this report, we test CMPs for cellulase encapsulation and delivery to degrade CNF hydrogels over time. These microparticles can be produced chemically,26 by enzymatic crosslinking27 or via spray drying.28 If produced from the main milk component by the use of pectin polysaccharides, the casein microparticles are especially interesting due to their production under gentle conditions at pH 6.8![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 29 and their sponge-like structure with a raspberry-like surface morphology that resembles casein micelles.30–32 The main ingredients in the preparation – casein and pectin – are also used in the pharmaceutical and food industries.33,34 The caseins are organized into casein micelles in aqueous solutions and, with the addition of pectin, they undergo phase separation and depletion flocculation.29,35 CMPs are produced via film drying followed by enzymatic hydrolysis of the pectin film matrix. They are especially interesting for drug delivery due to their volume change caused by swelling at different pH values,36 which has been demonstrated with α-tocopherol by mixing it into the solution during production.35 As their synthesis process does not require enzymatic crosslinking, thermal treatment, or high pressure, we hypothesize that they are also suitable for encapsulating enzymes. CMPs are spherical with diameters between 5 and 50 μm and are stable at 4 °C for up to 21 days in a Bis-Tris buffer solution.35,36 They do not show any significant structural changes when stored for up to 72 hours, retaining stability without visible degradation.37 Their stability increases under acidic conditions but the CMPs swell and disintegrate at highly alkaline pH values. The latter process takes seconds at pH 14, minutes at pH 11, and hours at pH 8.36,38 Another way to stabilize CMPs is the addition of calcium ions in the solution. 0.1 mM CaCl2 stabilizes the CMPs at a high alkali pH value of 11 for around 50 minutes, while without it, they disintegrated within 15 minutes. Higher concentrations of >10 mM CaCl2 resulted in no swelling of the CMPs at pH 11 over a time course of >1 h.31
29 and their sponge-like structure with a raspberry-like surface morphology that resembles casein micelles.30–32 The main ingredients in the preparation – casein and pectin – are also used in the pharmaceutical and food industries.33,34 The caseins are organized into casein micelles in aqueous solutions and, with the addition of pectin, they undergo phase separation and depletion flocculation.29,35 CMPs are produced via film drying followed by enzymatic hydrolysis of the pectin film matrix. They are especially interesting for drug delivery due to their volume change caused by swelling at different pH values,36 which has been demonstrated with α-tocopherol by mixing it into the solution during production.35 As their synthesis process does not require enzymatic crosslinking, thermal treatment, or high pressure, we hypothesize that they are also suitable for encapsulating enzymes. CMPs are spherical with diameters between 5 and 50 μm and are stable at 4 °C for up to 21 days in a Bis-Tris buffer solution.35,36 They do not show any significant structural changes when stored for up to 72 hours, retaining stability without visible degradation.37 Their stability increases under acidic conditions but the CMPs swell and disintegrate at highly alkaline pH values. The latter process takes seconds at pH 14, minutes at pH 11, and hours at pH 8.36,38 Another way to stabilize CMPs is the addition of calcium ions in the solution. 0.1 mM CaCl2 stabilizes the CMPs at a high alkali pH value of 11 for around 50 minutes, while without it, they disintegrated within 15 minutes. Higher concentrations of >10 mM CaCl2 resulted in no swelling of the CMPs at pH 11 over a time course of >1 h.31
In this study, we test the potential of using CMPs to encapsulate and release cellulase to degrade CNF hydrogel scaffolds during 3D cell growth (Fig. 1). We first investigate these processes with dextrans of different sizes as a model molecule and determine different diffusion coefficients. We then embedded CMPs inside a CNF hydrogel to examine whether the CMPs are able to release active cellulase by observing CNF degradation and calculating the decrease in the hydrogel size. Furthermore, we confirm that the produced cellulase-containing CMPs are not toxic for cells and add them to a 3D CNF hydrogel containing cells. This demonstrates that cellulase release from the CMPs enables L929 fibroblast cells to grow in 3D over a culture time of 21 days as the CNF hydrogel is slowly degraded.
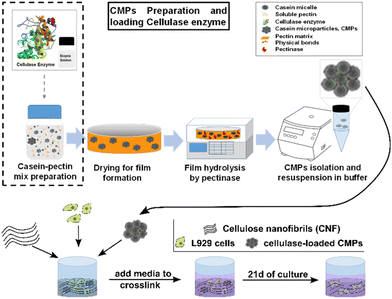 | ||
| Fig. 1 A schematic illustration of the production of cellulase-containing CMPs to locally deliver cellulase inside a CNF hydrogel to degrade the structure and support 3D cell growth. | ||
Materials and methods
CMPs preparation
The CMPs are prepared according to Zhuang et al.29 and optimized according to Schulte et al.31 In short, a dispersion of casein micelles (10% (w/w)) was prepared by dissolving casein micelle concentrate powder MC 80 (Milei GMBH, Leutkirch im Allgäu, Germany) in simulated milk ultra-filtrate solution (SMUF). The SMUF solution was prepared based on the protocol of Dumpler et al.39 The casein micelle solution was stirred for 1 h at room temperature (RT), 4 h at 4 °C, and 1 h at 37 °C. The pectin solution (2%) was prepared by adding pectin powder into a BisTris + CaCl2 buffer (pH 6.8) and stirring the mixture at 80 °C for 2–3 h until it became a clear solution. To prepare the casein–pectin solution (10 g), BisTris + CaCl2 buffer (pH 6.8, 4.28 g) was mixed at room temperature with the casein solution (4.07 g), the pectin solution (1.5 g) and, if needed, the cellulase enzyme (0.15 g) to obtain a 3% (w/w) casein and 0.3% pectin solution. A 70 mm glass Petri dish was filled with 3.9 g of the casein–pectin solution and placed under a drying hood overnight for film formation. Enzymatic hydrolysis with a pectinase solution was performed at 47 °C for 2 h in an automatic shaker (160 rpm). After cooling down, the clear solution was collected and centrifuged (22 °C, 1500 RCF, 10 min). The supernatant was removed, PBS was added at a concentration of 0.5 g L−1, and the CMPs were stored in the fridge (4 °C) for further use.Characterization of the diffusion properties of CMPs
The diffusion properties of the CMPs were characterized with FITC-dextran molecules (40 kDa, 70 kDa, 500 kDa, and 2000 kDa) via the fluorescence recovery after photobleaching (FRAP) technique using confocal laser scanning microscopy (CLSM) on a Leica TCS SP8 microscope (Leica Microsystems, Germany). The CMPs were incubated in an aqueous solution of FITC (0.5 mg mL−1) overnight. The bleaching experiments were performed using a 63× glycerol objective and the 488 nm line of an argon-ion laser operating at 80% output power. A region of interest (ROI) was set on the microgel and a time series of digital images (512 × 512 pixels) were recorded with an interval of 1.292 s using a highly attenuated laser beam (1.3% transmission). After recording 30 pre-bleach images, a circular area with a diameter of ca. 10 μm was bleached at maximum laser intensity (100% transmission) for 1.292 s. Other time series of 100 post-bleach images with the same settings were recorded. The FRAP data analysis was performed according to Kittel et al.40SDS-PAGE, bicinchoninic acid (BCA), and EnzCheck assays for indirect cellulase loading measurement
The encapsulated cellulase (Sigma c2730) was indirectly measured using SDS-PAGE, BCA, and an EnzCheck assay.For the SDS, samples were mixed with the loading buffer and incubated for 10 min at 95 °C. Then, different volumes were loaded onto the 4–20% gradient gel (140 V for 70 min) and the proteins were visualized by Coomassie staining. For the SDS, 1 mL CMPs solution was spun down and resuspended in 250 μl to increase the concentration (4× concentrated). The CMPs did break apart during heating.
The total soluble protein concentration of the cellulase and the intact CMPs samples was determined by the BCA assay (Pierce BCA Protein Assay Kit, 23225) following the manufacturer's instruction for the MTP format (Greiner BIO-ONE, Microplate, 96-well, PS, F-Bottom, Clear, 655101). Bovine serum albumin (BSA) was used to generate a standard curve. Absorbance was measured using a CLARIOstar Plus (BMG Labtech) plate reader.
The activity of cellulase in the intact CMPs samples with and without cellulase loading was determined using the EnzCheck cellulase substrate (Invitrogen, E33953) following the manufacturer's instructions at pH 5. The assay was performed in the MTP format (Greiner BIO-ONE, Microplate, 96-well, PP, F-Bottom, Black, 655209). A serial dilution of cellulase was used to generate a standard curve ranging from 2.67–2734 ng mL−1 (protein concentration based on the BCA assay). After incubation for 30 min at RT, the fluorescence was measured using the CLARIOstarPlus (BMG Labtech) plate reader at an excitation of 339/10 nm and an emission of 452/10 nm.
Preparation of the CNF suspension and hydrogel formation
A 1.0 wt% suspension of CNF was prepared by the group of Prof. Walther according to Isogai et al.4 by the TEMPO-mediated oxidation under alkaline conditions (pH = 10.5) for 30 min of a softwood kraft pulp and subsequent homogenization at pH 8.5 with an MRT model CR5 pressure microfluidizer, applying 2 passes at 1400 bar and 2 passes at 1000 bar. The resulting apparent viscosity degree of polymerization (DPv) is 600 and the content of carboxyl groups is 0.85 mmol g−1. Due to the TEMPO-mediated oxidation, the carboxyl groups of the CNF are deprotonated and therefore negatively charged under physiologically mimetic conditions, i.e., PBS or media at pH 7.4. When in contact with cations, the anionic CNF interfibrillar network stabilizes, forming an integer hydrogel that no longer exhibits shear-thinning behavior.9 The hydrogel dimensions formed here are around 5 mm in diameter and 0.5 mm in height per well.Hydrogel degradation comparing free cellulase with cellulase encapsulated in CMPs
For the degradation experiment, cellulase was encapsulated in the CMPs as described above. A solution with the same stock concentration of free cellulase, empty CMPs, and CMPs containing cellulase was added to 10 μL of nanocellulose and incubated in PBS or DMEM over 21 days while constantly re-adding PBS or DMEM to avoid evaporation. It was imaged every 2–3 days using an Opera Phenix Plus High-Content Screening System (PerkinElmer, USA) with a 10× air objective in brightfield mode and Z-stacks of 300–350 μm thickness.Cell culture
L929 mouse fibroblasts (passage 8–12, obtained from Deutsche Sammlung von Mikroorganismen und Zellenkulturen GmbH, DSMZ ACC-2) were cultured in tissue culture flasks with Dulbecco's modified Eagle's medium (DMEM, Gibco) containing 10% fetal bovine serum (FBS, Biowest or Gibco) and 1% antibiotic/antimycotic solution (AMB, Gibco). The cells were cultured at 37 °C in 5% CO2 in a humidified environment.Live-dead assay
To determine the influence of CMPs on cells, L929 fibroblasts were seeded in 24-well plates and incubated for 1 or 5 days with serial dilution of 0.5 g L−1 cellulase-loaded CMPs in DMEM. Cell viability was assessed by live/dead staining according to the manufacturer's instructions (Invitrogen, Thermo Fisher) and images were recorded using the Opera system. To analyze the percentage of living cells, the green cells are compared to the total number of cells in brightfield mode.Proliferation assay (MTS assay)
For the MTS assay, around 1 × 104 cells are seeded in each hydrogel in a 24-well plate with a serial dilution of 0.5 g L−1 cellulase-loaded CMPs and cultivated for 1 day. The MTS working solution is prepared with cell culture media (1000 μL) and MTS reagent (200 μL). The solution containing growth media is removed and replaced with the MTS working solution. The sample is incubated in the dark for 2–4 h at 37 °C. 100 μL of the solution is added to a 96-well plate (3× for each well). Then it is measured using a microplate reader (Synergy HT from BioTek) at 490 nm.Nanocellulose–cell–CMPs model
To add the CMPs together with the cells into a nanocellulose gel, the CMPs were first centrifuged at 1500 rpm for 10 min, and the PBS was removed. 1 × 104 cells were stained with Vybrant DiO (488, Thermo Fisher) solution, added to the CMPs without mixing, and then centrifuged at 300 rpm for 5 min. After removing the media, the CMPs and cells were pipetted with 10 μL nanocellulose to form a mixture. The same was done with CMPs or just cells as controls. The nanocellulose–cell–CMPs mixture was pipetted into a well plate and medium was added. The cells were imaged for 0 d to 21 d on the Opera Phenix Plus High-Content Screening System (PerkinElmer, USA) with a 10× air objective in brightfield mode, a 488 nm laser, and Z-stacks of 300–350 μm thickness.Data analysis
To visualize the effect of the free or encapsulated cellulase on the nanocellulose gel, a mathematical model for the size loss over time was established assuming that nanocellulose gel size decreases monoexponentially (eqn (1)).| Size(t) = a·e−b·t, | (1) |
| Volume(t) = volumet=0 + a(1 − e−b·t), | (2) |
A linear model is fitted viaeqn (3) and shows that the cell volume correlates with the degradation of nanocellulose.
Cell volume = −144.88 × size of nanocellulose + 23![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 483.9. 483.9.
| (3) |
Image processing and analysis
The samples are imaged using a PerkinElmer Opera Phenix Plus system with a 10× air objective. Z-stacks of 300–350 μm thickness are acquired for each sample. These images are captured using the appropriate excitation wavelengths and the emission signals are captured using suitable detection sCMOS sensor cameras. The obtained z-stacks are evaluated using the Harmony analyzing program or ImageJ-Fiji.Statistical analysis
Statistical data analysis was performed using OriginPro 2022 using a one-way ANOVA with pair comparisons using Tukey's methods. The p-value below 0.05 is considered a significant difference (p* < 0.05; p** < 0.01; p*** < 0.001).Results and discussion
FITC-dextran release from CMPs
As cellulase is a large enzyme,18,19 it needs to be encapsulated in capsules that over time provide sufficiently large pores in the range of 40–60 nm so as not to hinder the diffusion and release of the cellulase.18The synthesized CMPs, produced via a previously described method,29,31 are almost perfectly spherical with diameters in the range of 5–50 μm and are known to swell depending on the pH level. Inside, they have a granular, porous structure, which is formed by a gel network of μm-sized building blocks, interspersed with water channels of the same size (Fig. 2A and ESI Fig. 1A–D†). As CMPs are quite porous with a sponge-like structure,41 the diffusion of different-sized fluorescein isothiocyanate (FITC)-dextran molecules (40–2000 kDa) through the pores of the CMPs is first analyzed. The prepared CMPs are incubated in solutions with different-sized FITC-dextran molecules overnight and images are taken using confocal laser scanning microscopy. This demonstrates that the FITC dextran molecules of 40–70 kDa completely fill the particles, those of 500 kDa partially, and those of 2000 kDa much less (Fig. 2B–E). After incubation, we performed fluorescence recovery after photobleaching (FRAP) to determine the diffusion coefficients. In the case of FITC-dextran of 40–70 kDa, the molecules directly diffused back inside the spot after bleaching, so their diffusion coefficients were not detectable due to this fast recovery. With Stokes radii between 4.5 and 5.8 nm, these dextran molecules are also much smaller than the cellulase to be encapsulated. For the 500 and 2000 kDa FITC-dextran molecules with dimensions of approximately 14.7 and 27.0 nm, the diffusion coefficients are calculated to be 164 ± 28 μm2 s−1 and 28 ± 2 μm2 s−1, respectively (Fig. 2F). These diffusion coefficients are higher than those measured for dextran molecules inside PEG microgels of much smaller mesh size than the CMPs (e.g. diffusion coefficients in the range of 4–10 μm2 s−1 for 40–70 kDa FITC-dextran).40 The results suggest that cellulase should be trapped inside the CMPs during synthesis, with the ability to diffuse out when incorporated into CNF hydrogels.
Degrading the CNF hydrogel by embedding cellulase-loaded CMPs
Cellulase is encapsulated inside CMPs by mixing cellulase with casein–pectin inside a BisTris buffer. The resulting aggregates of casein micelles with entrapped cellulase are solidified into cellulase-loaded CMPs during film drying before they are isolated from the film matrix by enzymatic hydrolysis.The presence of cellulase in the CMPs formulations is indicated in SDS-PAGE (ESI Fig. 2A†). However, since cellulase (and pectinase as well) are not pure enzymes (both technical enzymes naturally produced by fungi), no single band corresponding to the cellulase at ∼68 kDa (calculated size of cellulase) is observed. Therefore, the cellulase loading capacity in the CMPs cannot be quantified by SDS-PAGE. In the future, cellulase could be produced recombinantly leading to higher purity and a defined activity profile. The supernatant fraction containing unincorporated cellulase also demonstrated a dominant band at ∼68 kDa, indicating that cellulase is used in excess during the fabrication process, which needs to be further optimized. To determine the amount of loaded cellulase in the CMPs, an indirect measurement was, therefore, performed. Directly after the fabrication procedure, the intact cellulase-loaded CMPs (total protein concentration of ∼0.5 mg mL−1) contained on average ∼35 μg mL−1 active cellulase as measured using a fluorescent cellulase substrate (ESI Fig. 2B†). Together with the measurements of the total protein amounts inside the empty and cellulase-loaded CMPs (ESI Fig. 2C†), this indicates a loading capacity of ∼20 weight/weight% of cellulase in the particles (Fig. 3B). The low level of enzyme activity in the empty CMPs is caused by the pectinase that is used to produce the CMPs (ESI Fig. 2D†). The Aspergillus niger organism, which produces pectinase also produces a cellulase with a molecular weight of ∼28 kDa,42 which was detected by the reactivity assay. The difference in total soluble protein concentration between the empty CMPs and cellulase-loaded CMPs agrees well with the quantified cellulase concentration based on the activity assay.
To investigate if active cellulase is released from the CMPs, cellulase-loaded CMPs (Fig. 3A) are embedded inside CNF hydrogels, and the hydrogel dimensions are observed over 21 days at 37 °C in PBS and DMEM. The hydrogel is marked with a black line and the black dots in the surrounding liquid are CMPs that were not completely embedded in the hydrogel as they immediately detached from the hydrogel after immersion in the liquid (Fig. 3C and D). The degradation kinetics with cellulase-loaded CMPs is compared with that of empty CMPs or free cellulase (cellulase added directly to PBS or DMEM at the same total concentration as the cellulase-loaded CMPs). It is apparent, after 21 days at 37 °C in DMEM, that the CNF gel shrinks following the addition of CMPs loaded with cellulase (Fig. 3C and D). If there are empty CMPs in the gel, the hydrogel does not degrade at all during 21 days neither in PBS nor DMEM. The free cellulase degrades the gel more rapidly compared with cellulase-loaded CMPs, as in this case the cellulase has to be released first. After its release, it is clear that the cellulase is active and degrades the hydrogels efficiently (Fig. 3E). The same trend is visible in PBS (Fig. 3F), with a slightly lower degradation rate compared to that in the DMEM (∼28% of the gel degraded in PBS after 21 days versus 53% in DMEM). This is unexpected as the addition of CaCl2 to the medium has previously been shown to decrease the swelling of the CMPs at pH 11 and prevent the swelling process completely at pH 8.31
The faster CNF hydrogel degradation is likely caused by the higher activity of cellulase at lower pH, which was demonstrated before.8 This is confirmed by the fact that free cellulase also degrades the CNF hydrogel faster in the medium compared to PBS (∼55% of the gel degraded in PBS after 21 days and 66% in DMEM). As cellulase is an enzyme and thus a catalyst, the hydrogels continue to degrade over time.
During enzymatic degradation, the size of the nanocellulose hydrogel initially decreases linearly after which degradation is observed to proceed more slowly (Fig. 3D and E). All kinetics with pure cellulase or CMPs-encapsulated cellulase fit a monoexponential function (see eqn (1) in the Materials and methods section). The fit parameters for degradation in DMEM or PBS are shown in Table 1.
| Parameter | Cellulase-loaded CMPs in DMEM | In PBS | Free cellulase in DMEM | In PBS |
|---|---|---|---|---|
| value | ||||
| a | 98.2% | 98.4% | 93.2% | 96.2% |
| b | 1.44 × 10−3 h−1 | 6.6 × 10−4 h−1 | 2.27 × 10−3 h−1 | 1.49 × 10−3 h−1 |
From the reciprocal values of the determined rates, b, characteristic times can be determined after which nanocellulose has shrunk to 37% of its initial value. This demonstrates that for both buffer systems, the encapsulation of the cellulase in CMPs can extend the duration of degradation by 200 h, i.e. approx. 8 days compared to the free enzyme.
Cell viability and proliferation assay
Before mixing the CMPs with cells in nanocellulose, they are first tested for their effect on cell viability and proliferation in 2D. For this, dilutions of CMPs from 0.5 g L−1 to 0.125 g L−1 are tested with L929 fibroblasts and cultivated for 1 day to analyze cell viability and proliferation and 5 days for viability. Fig. 4 shows that cell viability is not affected by the CMPs at any of the concentrations tested both on day 1 and day 5. The percentage of live cells is calculated as the ratio of green cells and cells counted in brightfield mode as the red BOBO-3-Iodide stain interacted with the CMPs (ESI Fig. 2A–D†). As the CMPs (without cells) interfere with the MTS proliferation assay, we only performed the proliferation assay after 24 h. The MTS levels indeed increase in the presence of more CMPs, which makes it difficult to interpret the data but when subtracting the particle background level, it looks like cell proliferation is not hindered (ESI Fig. 2E†). This fits well with the literature where casein micelles were also proven to be non-toxic to cells.33L929 cell growth in the nanocellulose gel in the presence of CMPs with encapsulated cellulase
As nanocellulose hydrogels do not degrade in water, cells embedded inside these gels do not have the space to spread or grow. Here, we study the use of CMPs as a local delivery system for cellulase to degrade the hydrogels and support cell growth inside a nanocellulose gel. L929 fibroblasts and cellulase-loaded CMPs are both mixed within the nanocellulose. With the addition of media, the nanocellulose crosslinks, and the cells are cultivated for 21 days while being live-imaged to analyze their growth. The L929 fibroblasts, stained with a Vybrant stain, are seeded in nanocellulose with cellulase-loaded CMPs, empty CMPs, or without CMPs. To analyze the L929 cell growth inside the nanocellulose hydrogels, the total cell volume is measured in 3D confocal image stacks. As shown in Fig. 5, only in the presence of cellulase-loaded CMPs do the cells spread and grow, as demonstrated by the increase in total cell volume (Fig. 5H). Cells with empty CMPs and without CMPs do not show any growth and remain round (Fig. 5C–F). Here, it is clear that the number of cells increases inside CNF with cellulase-loaded CMPs during the 21 days of culture (Fig. 5G). Compared to this, the number of cells stagnates and even decreases without the presence of CMPs or empty CMPs. As the amount of Vybrant staining decreases in the cells with each doubling, it could be that not all cells were detected during the counting. Also, the total cell volumes do not increase in these cases as the L929 cells alone are not able to degrade the nanocellulose (Fig. 5H). The cellulase released from the CMPs, therefore, is proven to be active, resulting in nanocellulose degradation, providing space for the cells to spread and increase their volume (Fig. 5A and B). To better analyze L929 cell growth inside the nanocellulose hydrogels in the presence of cellulase-loaded CMPs, a volume model is established (see eqn (2) in the Materials and methods section). The used parameters are shown in Table 2.| Parameter | Value |
|---|---|
| Volumet=0 | 8979.83 μm3 |
| a | 10![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 469.52 μm3 469.52 μm3 |
| b | 2.33 × 10−3 h−1 |
For t → ∞, the cell volume will approach a value of Volume0 + a = 19![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 449.35 μm3.
449.35 μm3.
Assuming the model is correct, the cells would reach 37% of this maximum cell number/expansion after  , i.e. around 18 days. When plotting the total cell volume as a function of the size of the nanocellulose hydrogels embedded with cellulase-loaded CMPs from Fig. 3, a linear model (Fig. 5I) is fitted viaeqn (3) (see Materials and methods) and shows that the cell volume correlates with the degradation of nanocellulose. It is important to mention that in the presence of the cells, the hydrogels did not shrink as much but became softer from the inside out, providing space for the cells to grow. This could be explained by the fact that in the presence of cells, the media, and thus the released cellulase, are replaced with fresh media every 3–4 days. In addition, it is known that cells locally produce acids and that the cellulase activity increases at lower pH values, which can enhance the activity of the enzyme inside the gel where the cells are located. Furthermore, the cells will produce extracellular matrix proteins, which can stabilize the 3D cell construct. As different tissues need different degradation rates, different concentrations of cellulase-loaded CMPs could be added to the CNF hydrogel, as it has been demonstrated before that the degradation kinetics of CNF hydrogels depends on the cellulase concentration.8
, i.e. around 18 days. When plotting the total cell volume as a function of the size of the nanocellulose hydrogels embedded with cellulase-loaded CMPs from Fig. 3, a linear model (Fig. 5I) is fitted viaeqn (3) (see Materials and methods) and shows that the cell volume correlates with the degradation of nanocellulose. It is important to mention that in the presence of the cells, the hydrogels did not shrink as much but became softer from the inside out, providing space for the cells to grow. This could be explained by the fact that in the presence of cells, the media, and thus the released cellulase, are replaced with fresh media every 3–4 days. In addition, it is known that cells locally produce acids and that the cellulase activity increases at lower pH values, which can enhance the activity of the enzyme inside the gel where the cells are located. Furthermore, the cells will produce extracellular matrix proteins, which can stabilize the 3D cell construct. As different tissues need different degradation rates, different concentrations of cellulase-loaded CMPs could be added to the CNF hydrogel, as it has been demonstrated before that the degradation kinetics of CNF hydrogels depends on the cellulase concentration.8
Conclusions and outlook
This report demonstrates the efficient production and use of CMPs to locally deliver active cellulase inside nanocellulose hydrogels to trigger degradation and support 3D cell growth. The CMPs are porous and facilitate the release of dextran molecules < 2000 kDa as a model molecule of cellulase. When cellulase-loaded CMPs are embedded inside nanocellulose hydrogels, the hydrogels degrade gradually over a time course of 21 days, demonstrating that the enzyme is successfully encapsulated in the microparticles and released over time. The enzymatic degradation of the nanocellulose can be delayed by approximately 8 days through the use of cellulase-loaded CMPs compared to cellulase added to the buffer or media via the CMP structure-related release process. CMPs added to the media are non-toxic and do not hinder cell proliferation. The degradation of nanocellulose with embedded cellulase-loaded CMPs and the increase of total cell volume of growing L929 cells can both be analyzed by a mono-exponential function. The characteristic times for both processes estimated by the model fits are very close. The process variables of total cell volume and size of the nanocellulose hydrogel are negatively correlated with each other, i.e. the more the nanocellulose is degraded, the more the cells can grow. This provides a system that can be adapted to a wide range of applications. Past research demonstrated easy non-toxic methods to further stabilize the particles via heat treatment or the addition of calcium,37 but further studies are needed to understand the changes in particle morphology and the mechanisms that affect structural integrity to evaluate possible limitations of this delivery system. We demonstrated that confocal microscopy is a suitable method for this structural analysis. In addition, simulations can be performed to predict the release kinetics at different pH values, such as for the human digestion system. In this study, we demonstrate that the CMPs can be used as a delivery system with potential applications in food, drugs, or biomedicine to encapsulate, protect, and release bioactive compounds. In future studies, CNF hydrogels incorporated with the enzyme-loaded CMPs will be tested in vivo to analyze cell infiltration into these 3D hydrogels.Author contributions
L. D. L., R. G., C. B., and J. C. T. S. conceived the initial idea. C. B. and L. D. L. designed all CNF and cell experiments, which were performed by C. B., while J. C. T. S, M. A., and C. H. produced the CMPs. R. G. performed the model fittings. Y. K. and C. B. performed the FRAP experiments, which were analyzed by Y. K. All authors read and provided feedback on the manuscript.Data availability
The data that support the findings of this study are available from the corresponding author upon reasonable request.Conflicts of interest
The authors declare no conflict of interest.Acknowledgements
The authors gratefully acknowledge funding from the German Research Foundation (DFG) within the project LA 3606/3-1. The authors gratefully acknowledge financial support from the Exploratory Research Space of RWTH Aachen University (project OPSF580).References
- A. Ghilan, R. Nicu, D. E. Ciolacu and F. Ciolacu, Materials, 2023, 16, 4447 CAS.
- A. K. Tamo, J. Mater. Chem. B, 2024, 12, 7692–7759 RSC.
- S. S. Athukoralalage, R. Balu, N. K. Dutta and N. R. Choudhury, Polymers, 2019, 11, 898 CrossRef CAS PubMed.
- T. Saito, Y. Nishiyama, J. L. Putaux, M. Vignon and A. Isogai, Biomacromolecules, 2006, 7, 1687–1691 CrossRef CAS PubMed.
- Y. R. Lou, L. Kanninen, T. Kuisma, J. Niklander, L. A. Noon, D. Burks, A. Urtti and M. Yliperttula, Stem Cells Dev., 2014, 23, 380 CrossRef CAS PubMed.
- H. Cai, S. Sharma, W. Liu, W. Mu, W. Liu, X. Zhang and Y. Deng, Biomacromolecules, 2014, 15, 2540–2547 CrossRef CAS PubMed.
- J. Vartiainen, T. Pöhler, K. Sirola, L. Pylkkänen, H. Alenius, J. Hokkinen, U. Tapper, P. Lahtinen, A. Kapanen, K. Putkisto, P. Hiekkataipale, P. Eronen, J. Ruokolainen and A. Laukkanen, Cellulose, 2011, 18, 775–786 CrossRef CAS.
- J. G. Torres-Rendon, M. Köpf, D. Gehlen, A. Blaeser, H. Fischer, L. De Laporte and A. Walther, Biomacromolecules, 2016, 17, 905–913 CrossRef CAS PubMed.
- D. B. Gehlen, N. Jürgens, A. Omidinia-Anarkoli, T. Haraszti, J. George, A. Walther, H. Ye and L. De Laporte, Macromol. Rapid Commun., 2020, 41, 1–10 CrossRef PubMed.
- Y. Hu and J. M. Catchmark, J. Biomed. Mater. Res., Part B, 2011, 97B, 114–123 CrossRef CAS PubMed.
- M. T. Farcas, E. R. Kisin, A. L. Menas, D. W. Gutkin, A. Star, R. S. Reiner, N. Yanamala, K. Savolainen and A. A. Shvedova, J. Toxicol. Environ. Health, Part A, 2016, 79, 984 CrossRef CAS PubMed.
- M. Tenkanen, J. Puls and K. Poutanen, Enzyme Microb. Technol., 1992, 14, 566–574 CrossRef CAS.
- A. Suurnäkki, M. Tenkanen, M. Siika-Aho, M. L. Niku-Paavola, L. Viikari and J. Buchert, Cellular, 2000, 7, 189–209 CrossRef.
- M. P. Lutolf, J. L. Lauer-Fields, H. G. Schmoekel, A. T. Metters, F. E. Weber, G. B. Fields and J. A. Hubbell, Proc. Natl. Acad. Sci. U. S. A., 2003, 100, 5413–5418 CAS.
- K. M. Lorentz, S. Kontos, P. Frey and J. A. Hubbell, Biomaterials, 2011, 32, 430–438 CrossRef CAS PubMed.
- E. Entcheva, H. Bien, L. Yin, C. Y. Chung, M. Farrell and Y. Kostov, Biomaterials, 2004, 25, 5753–5762 CAS.
- A. K. Tamo, I. Doench, A. M. Helguera, D. Hoenders, A. Walther and A. O. Madrazo, Polymers, 2020, 12, 1522 CAS.
- P. Bubner, J. Dohr, H. Plank, C. Mayrhofer and B. Nidetzky, J. Biol. Chem., 2012, 287, 2759 CAS.
- P. M. Abuja, I. Pilz, M. Claeyssens and P. Tomme, Biochem. Biophys. Res. Commun., 1988, 156, 180–185 CAS.
- R. H. Y. Chang, J. Jang and K. C. W. Wu, Green Chem., 2011, 13, 2844–2850 CAS.
- V. Lombardi, M. Trande, M. Back, S. V. Patwardhan and A. Benedetti, Nanomaterials, 2022, 12, 626 CAS.
- E. J. Cho, S. Jung, H. J. Kim, Y. G. Lee, K. C. Nam, H. J. Lee and H. J. Bae, Chem. Commun., 2011, 48, 886–888 Search PubMed.
- K. Khoshnevisan, A. K. Bordbar, D. Zare, D. Davoodi, M. Noruzi, M. Barkhi and M. Tabatabaei, Chem. Eng. J., 2011, 171, 669–673 CrossRef CAS.
- H. Peniche, A. Osorio, N. Acosta, A. De La Campa and C. Peniche, J. Appl. Polym. Sci., 2005, 98, 651–657 CAS.
- D. Çamurlu and S. Önal, Biocatal. Biotransform., 2021, 39, 292–301 Search PubMed.
- S. Milenkova, B. Pilicheva, Y. Uzunova, T. Yovcheva and M. Marudova, Materials, 2022, 15, 1333 CAS.
- T. Huppertz, M. A. Smiddy and C. G. de Kruif, Biomacromolecules, 2007, 8, 1300–1305 CAS.
- M. M. Baracat, A. M. Nakagawa, R. Casagrande, S. R. Georgetti, W. A. Verri and O. De Freitas, AAPS PharmSciTech, 2012, 13, 364–372 CAS.
- Y. Zhuang, J. Sterr, U. Kulozik and R. Gebhardt, Int. J. Biol. Macromol., 2015, 74, 44–48 CAS.
- M. Ouanezar, F. Guyomarc'h and A. Bouchoux, Langmuir, 2012, 28, 4915–4919 CrossRef CAS PubMed.
- J. Schulte, M. Stöckermann, S. Thill and R. Gebhardt, Int. Dairy J., 2020, 105, 104692 CrossRef CAS.
- A. Bouchoux, G. Gésan-Guiziou, J. Pérez and B. Cabane, Biophys. J., 2010, 99, 3754–3762 CrossRef CAS PubMed.
- U. Sadiq, H. Gill and J. Chandrapala, Foods, 2021, 10, 1965 CAS.
- Y. Luo, K. Pan and Q. Zhong, Int. J. Pharm., 2015, 486, 59–68 CrossRef CAS PubMed.
- Y. Zhuang, J. Sterr, A. Schulte, U. Kulozik and R. Gebhardt, Food Biophys., 2016, 11, 332–338 CrossRef.
- J. Schulte, M. Stöckermann and R. Gebhardt, Food Hydrocolloids, 2020, 105, 105741 CAS.
- J. C.-T. Schulte, R. Gebhardt and U. Kulozik, Diss. Rheinisch-Westfälische Tech. Hochschule Aachen, 2022 DOI:10.18154/RWTH-2022-09032.
- S. Khanna, J. Herzog and R. Gebhardt, Chem. Ing. Tech., 2020, 92, 1329–1329 CAS.
- J. Dumpler, I. Kieferle, H. Wohlschläger and U. Kulozik, Int. Dairy J., 2017, 68, 60–69 CAS.
- Y. Kittel, L. P. B. Guerzoni, C. Itzin, D. Rommel, M. Mork, C. Bastard, B. Häßel, A. Omidinia-Anarkoli, S. P. Centeno, T. Haraszti, K. Kim, J. Guck, A. J. C. Kuehne and L. De Laporte, Angew. Chem., Int. Ed., 2023, 135(44) DOI:10.1002/anie.202309779.
- R. Gebhardt, M. Burghammer, C. Riekel, S. V. Roth and P. Müller-Buschbaum, Macromol. Biosci., 2008, 8, 347–354 CAS.
- P. L. Hurst, J. Nielsen, P. A. Sullivan and M. G. Shepherd, Biochem. J., 1977, 165, 33–41 CAS.
Footnote |
| † Electronic supplementary information (ESI) available. See DOI: https://doi.org/10.1039/d4bm01508h |
| This journal is © The Royal Society of Chemistry 2025 |




