Open Access Article
Open Access ArticleCreative Commons Attribution 3.0 Unported Licence
Color-coded galectin fusion proteins as novel tools in biomaterial science†
Carina
Dey
 a,
Isabel K.
Sommerfeld
a,
Isabel K.
Sommerfeld
 bc,
Pavla
Bojarová
bc,
Pavla
Bojarová
 de,
Nikol
Kodra
a,
David
Vrbata
de,
Nikol
Kodra
a,
David
Vrbata
 d,
Miluše
Zimolová Vlachová
d,
Miluše
Zimolová Vlachová
 d,
Vladimír
Křen
d,
Vladimír
Křen
 d,
Andrij
Pich
d,
Andrij
Pich
 bcf and
Lothar
Elling
bcf and
Lothar
Elling
 *a
*a
aLaboratory for Biomaterials, Institute for Biotechnology and Helmholtz-Institute for Biomedical Engineering, RWTH Aachen University, Pauwelsstraße 20, 52074 Aachen, Germany. E-mail: l.elling@biotec.rwth-aachen.de; Tel: +49241 8028350
bDWI − Leibniz-Institute for Interactive Materials, e.V. Forckenbeckstr. 50, 52074 Aachen, Germany
cFunctional and Interactive Polymers, Institute of Technical and Macromolecular Chemistry, RWTH Aachen University, Worringerweg 2, 52074 Aachen, Germany
dLaboratory of Biotransformation, Institute of Microbiology of the Czech Academy of Sciences, Vídeňská 1083, Prague 4, 14200, Czech Republic
eDepartment of Health Care Disciplines and Population Protection, Faculty of Biomedical Engineering, Czech Technical University in Prague, nám. Sítná 3105, 27201 Kladno, Czech Republic
fAachen Maastricht Institute for Biobased Materials (AMIBM), Maastricht University, Brightlands Chemelot Campus, Urmonderbaan 22, 6167 RD Geleen, The Netherlands
First published on 29th January 2025
Abstract
The inherent carbohydrate-binding specificities of human galectins can serve as recognition elements in both biotechnological and biomedical applications. The combination of the carbohydrate-recognition domain (CRD) of galectins fused to peptides or proteins for purification, immobilization, and imaging enables multifunctional utilization within a single protein. We present here a library of color-coded galectin fusion proteins that incorporate a His6-tag, a fluorescent protein, and a SpyCatcher or SpyTag unit to enable immobilization procedures. These galectin fusion proteins exhibit similar binding properties to the non-fused galectins with micromolar apparent binding affinities. N- and C-terminal fusion partners do not interfere with the SpyCatcher/SpyTag immobilization. By applying SpyCatcher/SpyTag-mediated SC–ST-Gal-3 conjugates, we show the stepwise formation of a three-layer ECM-like structure in vitro. Additionally, we demonstrate the SpyCatcher/SpyTag-mediated immobilization of galectins in microgels, which can serve as a transport platform for localized targeting applications. The proof of concept is provided by the galectin-mediated binding of microgels to colorectal cancer cells.
Introduction
Galectins are a family of proteins binding to β-galactosyl-containing glycoproteins. The physiological significance of mammalian galectins is reflected in their diverse functions at the cellular level, such as the mediation of extracellular cross-linking events between cells and proteins or modulating apoptosis and inflammation processes by inducing intracellular cell signalling pathways. In addition to the research into galectin-inhibiting compounds, there is growing biomedical interest in the utilization of the intrinsic carbohydrate-binding properties of galectins. Galectins have already been used as therapeutics for various inflammatory diseases, cancer, and nephritis.1–3 The biotechnological application of galectin binding specificities has also been demonstrated in the galectin-mediated assembly of a biomimetic extracellular microenvironment.4 Galectin fusion proteins have been reported previously for prospective use as diagnostics and therapeutics. These include galectin chimeras,3,4 galectin-3 fusion proteins as in vivo reporters,5 cell-binding reporters6 as well as enzyme carriers.7 We have previously shown that galectin fusion proteins may serve as functional tools for glycobiological applications. These proteins contained several components, including a His6-tag for purification, a SNAP-tag for immobilization, a fluorescent protein for imaging applications, and either Gal-3 or Gal-1 CRD for carbohydrate binding. These proteins were characterized by in vitro binding assays and successfully applied in flow cytometric cell sorting or the preparation of Gal-3 affinity columns.8,9 After expanding our galectin fusion protein library, further analysis revealed that oligomerization of the SNAP-tag induced increased apparent binding signals in in vitro assays.10 This led us to reconsider the immobilization system for the application in follow-up studies. Beyond SNAP technology, the SpyCatcher/SpyTag (SC/ST) mechanism is a frequently used immobilization and crosslinking system for protein moieties.11 This system has been engineered by splitting a fibronectin-binding protein domain from Streptococcus pyogenes (S. pyogenes) into a peptide (SpyTag) and a protein (SpyCatcher), which spontaneously forms a stable amide bond between Asp117 and Lys31, respectively. The SC/ST system provides simple reaction conditions in terms of temperature, buffer system, and reaction time.12 Later on, various mutated versions of the SpyCatcher and SpyTag were developed for accelerated isopeptide formation.13,14 The versatility of the SC/ST immobilization technology has been demonstrated in various studies, including vaccine research,15,16 protein purification,11,17 design of artificial protein architectures,18 industrial applications,19 and production of protein-based hydrogel and biofilm materials.20–22The SC/ST technology is a promising tool for selective covalent immobilization of protein units in microgels. Microgels are soft polymer networks that are swollen in suitable solvents to form colloidal gels ranging from nanometers to micrometers in size.23 The uptake of proteins and other guest molecules either to the microgel shell or core is enabled by the porous structure of microgels.24 Different groups have previously used selective interactions for the immobilization of proteins in microgels. The selective eGFP (enhanced green fluorescent protein) immobilization through sortase A mediated binding between two peptide sequences was demonstrated.24 Sommerfeld et al. illustrated the SC/ST-mediated synthesis of glycosyltransferase-decorated microgels for glycan synthesis.25 The biocompatibility of PEG-based microgels is essential for bio-applications such as the development of new therapeutics,26,27 the delivery of antibodies,28 or the scavenging of toxins.29,30
The glycocalyx and extracellular matrix (ECM) form a complex network of glycoproteins, proteoglycans, water, and various macromolecules that enable cell–cell interactions and maintain structural integrity within different tissues.31,32 In a diseased state, such as in cancer and chronic inflammation, glycosylation patterns as well as the ECM microenvironment are altered, resulting in the development of glycan epitopes for galectin binding.33 Thus, galectin-functionalized microgels may be efficiently targeted to the cell surface by exploiting the β-galactosyl binding capabilities of galectins. In addition, galectin-directed cross-linking of glycoproteins can be used in chronic tissue inflammation to artificially reconstruct damaged extracellular networks.
In this study, we report for the first time the preparation of a library of color-coded galectin fusion proteins using the SpyCatcher/SpyTag immobilization system (Scheme 1). We have confirmed the functionality and characteristics of these galectin fusion proteins by comparing their binding behaviour with various glycoproteins through in vitro binding assays. The versatility of SpyCatcher- and SpyTag-bearing galectin fusion proteins was illustrated by the layer-by-layer assembly of an in vitro ECM-like structure We also demonstrate the fabrication of galectin-functionalized microgels and their binding to the surface of cancer cells, detected by flow cytometry. The presented targeting and transporting galectin-functionalized platforms are new promising tools for biomedical research.
Results and discussion
Production and characterization of SpyCatcher003 and SpyTag003-carrying galectin fusion proteins
The galectin fusion protein library was generated by cloning the SpyCatcher protein or SpyTag peptide sequence into the previously created fusion constructs.8–10 To construct the fusion proteins, we used the mutant versions (SpyCatcher003/SpyTag003) of the original sequence from S. pyogenes, which accelerate the formation of the conjugating isopeptide bond.13 The fusion constructs consisted of four protein subunits: a N-terminal His6-tag (H) for purification, SpyCatcher003 (SC003) protein/SpyTag003 (ST003) peptide for immobilization, the fluorescent protein for imaging, and the galectin carbohydrate recognition domain (CRD) for ligand binding (ESI, Scheme S1, for further abbreviations see Table S1†). The process of heterologous production of SpyCatcher/SpyTag-galectin fusion proteins is summarized in Tables S2 and S3 (ESI†). The fusion proteins were purified by IMAC, followed by ASF glycoprotein affinity chromatography as previously described.10 The resulting protein fractions were analysed by SDS-PAGE and western blot (ESI, Fig. S1–S8†).SDS-PAGE analysis and western blot confirmed pure protein fractions (ESI, Fig. S1†) which were further used in glycoprotein and cell binding assays. First, we compared the binding characteristics to asialofetuin (ASF) of the SpyCatcher003/SpyTag003-galectin-3 fusion proteins with those of the solely His6-tagged Gal-3 (HGal-3) (Fig. 1). HSC003eYFPGal-3 and HST003eYFPGal-3 displayed similar binding behaviour to HGal-3. The binding efficiency (BE) was calculated as a ratio of the maximum binding signal (Bmax) and half maximum binding value (KD) (Table S4†).34,35 A comparable KD value was reached in a single-digit micromolar range for galectin fusion proteins with SpyCatcher003 and SpyTag003 as well as for His6-tagged Gal-3 only (ESI, Table S4†).
In contrast to our previously reported SNAP-tag galectin fusion proteins, the SpyCatcher and SpyTag fusions did not induce aggregation of the galectin fusion proteins with an apparent higher binding signal (ESI, Fig. S9 and Table S4†).8,9
To provide a more in-depth analysis of the contribution of each fusion subunit to the binding to ASF, different SpyCatcher003 (SC003)- and SpyTag003 (ST003)-Gal-3 fusion proteins were generated including the complete fusion protein (HSC003eYFPGal-3, HST003eYFPGal-3), a fusion construct lacking the fluorescence protein (HSC003Gal-3, HST003Gal-3) or lacking SpyCatcher003 and SpyTag003 (HeYFPGal-3), respectively. Fusion proteins without the galectin CRD (HSC003eYFP, HST003eYFP) served as controls (ESI, Scheme S1†). Binding assays on ASF displayed a comparable binding behaviour of SC003-galectin fusion proteins (ESI, Fig. S10A†). The control protein HSC003eYFP did not bind to ASF confirming the absence of non-specific binding of the residual fusion components. These findings were additionally reflected by the submicromolar KD values (ESI, Table S5†). Based on these results, we concluded that the SpyCatcher does not interfere with the binding of the Gal-3 domain to ASF. Binding analysis of the corresponding SpyTag003-carrying Gal-3 fusion proteins revealed similar results (ESI, Fig. S10B and Table S5†).
The binding affinity of individual protein constructs of SpyCatcher- and SpyTag-galectins to ASF was verified to be stable over twelve days (ESI, Table S6†). We observed binding affinities in the micromolar range for both HSC003eYFPGal-3 and HST003eYFPGal-3. The binding efficiencies (BE) of HSC003eYFPGal-3 showed values ranging from 0.3–1.4 μM−1. For HST003eYFPGal-3, we observed binding efficiencies ranging from 0.7–2 μM−1. Remarkably, we observed an increase in binding efficiency for both proteins from the first to the second day, namely by a factor of 5 for HSC003eYFPGal-3 and by a factor of 2 for HST003eYFPGal-3. In the following days, the binding efficiencies readjusted to values between 0.5 and 1 μM−1. These time-dependent fluctuations could be due to the concentration-dependent tendency of eYFP to form weak dimers in solution36 or to Gal-3, which can form higher oligomers.37,38 Despite the occurrence of these fluctuations, we conclude that the proteins exhibit good stability under the given storage conditions (4 °C, PBS pH 7.5).
SpyCatcher- and SpyTag-mediated conjugation of galectin-3 fusion proteins
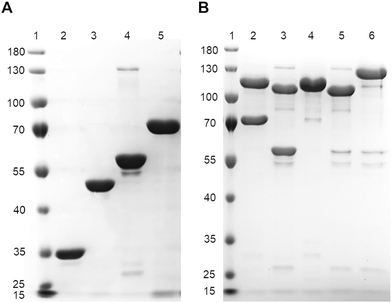
To confirm that the SpyCatcher003-Spytag003 interaction was preserved in the presence of N- and C-terminal fusion partners, we examined the isopeptide bond formation of HSC003eYFPGal-3 and HST003eYFPGal-3 with their respective reaction partners. Strep-SC003-sfGFP and strep-ST003-sfGFP served as controls. The proteins were purified to homogeneity by IMAC followed by SEC to ensure consistent band identification in the reducing SDS-PAGE gel and to prevent nonspecific interaction of protein fragments (Fig. 2A). The conjugation of SpyCatcher003 and SpyTag003 was carried out in PBS buffer at pH 7.5 for 1 h at 25 °C, according to the reaction conditions described previously.12 We observed the conjugation of HSC003eYFPGal-3 and strep-ST003-sfGFP at both equimolar and 2-fold excess molar ratios (Fig. 2B, lanes 2 and 4, respectively) of galectin fusion protein as a significant band corresponding to the molecular weight of the protein-conjugate (101 kDa, under denaturing conditions). The conjugation of HST003eYFPGal-3 and strep-SC003-sfGFP (101.5 kDa; Fig. 2B lanes 3 and 5, respectively) was also observed. Most importantly, the interaction of the two galectin-3 fusion proteins HSC003eYFPGal-3 and HST003eYFPGal-3 (denoted as SC–ST-Gal-3 conjugate) was detected as a divalent Gal-3 conjugate with two Gal-3 CRDs under the given denaturing conditions (124.7 kDa; Fig. 2B, lane 6). Based on these results, we concluded that SpyCatcher003–SpyTag003 interaction is not affected by the N-and C-terminal fusion partners. Our findings are consistent with other studies utilizing the SpyCatcher–SpyTag interaction for protein–protein conjugation15,16,18,19 and enables to utilize the potential of the SpyCatcher–SpyTagged galectin fusion proteins in further biomedical applications.After demonstrating SpyCatcher–SpyTag-driven assembly of two Gal-3 fusion proteins into an SC–ST-Gal-3 conjugate, we examined the binding behaviour of these conjugates in an in vitro binding assay on ASF (Fig. 3A). The SC–ST-Gal-3 conjugate exhibited a higher binding signal at lower protein concentrations (Fig. 3B). However, due to the high variability of the data points, the KD value and binding efficiency cannot be precisely calculated. At equivalent protein concentrations used in our previous binding experiments (Fig. 1 and Table S4†), the apparent KD of SC–ST-Gal-3 conjugate is estimated to be ≤0.3 μM, which is in the range of the analogous SNAP-tag construct (Table S4†). We hypothesized that instead of having a bivalent SC–ST-Gal-3 conjugate as expected (Fig. 3A), the formation of SC–ST-Gal-3 conjugate oligomers occurred upon binding, leading to higher a valency with a higher binding signal and an apparently higher affinity and binding efficiency.
Size exclusion chromatography (SEC) analysis of HST003eYFPGal-3 (57.5 kDa) revealed a single peak with a calculated molecular weight of 77.3 kDa (for 20 μM, ESI, Fig. S11A, S12A and Table S7†), indicating the monomeric form of the protein. The SEC profile of the SpyCatcher galectin fusion protein HSC003eYFPGal-3 (67.2 kDa) displayed a prominent peak corresponding to a molecular weight of 165.7 kDa, along with a smaller peak at 73.7 kDa (20 μM, ESI, Fig. S11B and Table S7†). This indicates that HSC003eYFPGal-3 exists primarily as a dimer and to a lesser extent as a monomer in solution. Similarly, the SC–ST-Gal-3 conjugate eluted as a prominent peak with a molecular weight corresponding to a dimer of the divalent conjugate (10 μM, ESI Fig. S11C†). In addition, a smaller peak with a molecular weight of 78.4 kDa was observed, probably representing the monomer of one of the single fusion proteins, although we did not observe any fragmentation of the SC–ST-Gal-3 conjugate in the SDS-PAGEs of the SEC samples. As the SEC results differed from the theoretical molecular weights, we conducted Dynamic Light Scattering (DLS) measurements of these samples and concluded that particles of (Dh) = 22.0 nm, 58.1 nm, and 11.0 nm were observed for HSC003eYFPGal-3, HST003eYFPGal-3, and the SC–ST-Gal-3 conjugate, respectively (ESI, Fig. S13 and Table S8†). We performed a prediction of each galectin fusion protein folding using the AlphaFold software and measured distances in PyMOL software resulting in average dimensions of 6–11 nm for HSC003YGal-3 and 6–9 nm for HST003YGal-3 (ESI, Fig. S14†) which is in good agreement with published Dh of respective single protein domains (3.3 ± 0.6 nm for the Gal-3CRD and 4.42 ± 0.1 nm for GFP, similar to YFP, Fig. S14†).39,40 These results are also in good agreement with the plot on the correlation of protein molecular weights vs. their Stokes radii published recently.41 Furthermore, the SEC-based calculation of Stokes diameters (DS) gave values of 7.9 nm, 10.2 nm and 12.2 nm for HST003YGal-3, HSC003YGal-3 and the SC–ST-Gal-3 conjugate (ESI Fig. S12B and Table S7†), which match with the results of the DLS measurements. The hydrodynamic diameter of the SpyCatcher003 protein and the SpyTag003 peptide is not reported in literature. Based on the DLS measurements, the AlphaFold models, and the reported Dh values, we conclude that the HSC003YGal-3 construct exists predominantly in a dimeric or trimeric form, which agrees with the obtained SEC results (ESI Fig. S11B and Table S7†). The HST003YGal-3 construct appears to be higher oligomers in contrast to the SEC results indicating the monomeric form (ESI Fig. S11A and Table S7†). These differences may be due to the changing Gal-3 monomer/oligomer equilibrium during the SEC separation. Considering the reported Dh values of the Gal-3 CRD and GFP, our DLS results clearly indicate that the SC–ST-Gal-3 (Dh 11 nm) conjugate exists in the bivalent form under the given conditions. For the SC–ST-Gal-3 conjugate, the discrepancy between SEC (dimer) and DLS (monomer) may be due to the rod-shaped structure giving rise to a different elution behaviour when compared to the globular standard calibration proteins. In this respect, the molecular weight indicating a dimer in SEC is overestimated. Importantly, these results cannot be related to the higher binding signal observed in the binding assays of the SC–ST-Gal-3 conjugate to ASF (Fig. 3B). A previous study demonstrated the concentration dependent aggregation of Gal-3 to higher oligomers (>5 Gal-3 units) triggered by the intermolecular interaction of the Gal-3 NT-domain with the CRD F-face after binding to a glycoprotein.38 However, it was also stated that fusion of proteins to the N-terminus (NT) of Gal-3 disturb oligomerization of Gal-3 via its NT-domain rendering preferentially monovalent carbohydrate binding.6 Our SEC results suggest that HSC003eYFPGal-3 forms a dimer at 10–20 μM concentrations. However, SpyCatcher-triggered dimerization has not been described so far. We assume that upon binding of the SC–ST-Gal-3 conjugate on ASF further oligomerization is induced. Based on these findings, we conclude that the increased binding signal of the SC–ST-Gal-3 conjugate to ASF in the ELISA is likely due to the formation of the SC–ST-Gal-3 conjugate clusters at the given concentration (Fig. 3).
Binding of galectin fusion proteins to ECM glycoproteins
The prepared SpyCatcher galectin fusion protein toolbox included CRD domains of Gal-1C2S, Gal-3 (incl. N-terminal tail), Gal-4NL, LGal-4C, Gal-8NL, and LGal-8C (incl. parts of the linker peptide (L)). Each galectin was color-coded by fusion with a different fluorescent protein (Scheme 2), facilitating fluorescence-based detection. | ||
| Scheme 2 Color-coded SpyCatcher galectin fusion proteins. A: construction of color-codes at the genetic level (created with BioRender.com); B: production of recombinant fluorescent fusion proteins in E. coli. Gal-1C2S: aa (amino acid) 1–135 (Uniprot entry: P09382); Gal-3: aa 1–250 (Uniprot entry: P17931); Gal-4NL: aa 1–160 (Uniprot entry: P 56470);1 LGal-4C: aa 169–323 (Uniprot entry: P 56470);1 Gal-8NL: aa 1–160 of Gal-8 isoform b;2 LGal-8C: aa 182–317 of Gal-8 isoform b.2 | ||
To ensure the functionality and targeting of the newly generated galectin fusion constructs, we conducted in vitro binding experiments to analyze their interaction with glycoproteins such as ASF, fetuin, and selected extracellular matrix (ECM) glycoproteins – laminin, fibronectin, and collagen IV, as well as the mucus-forming protein Muc 2 (porcine stomach) (Table 1, ESI, Fig. S15 and S16†). All galectin fusion proteins bound effectively to Muc 2 and exhibited affinities in the nano- to submicromolar range, highlighting this glycoprotein as a primary glycan target (Table 2). SpyCatcher fusion proteins of Gal-3, Gal-4NL, and LGal-4C exhibited the highest binding efficiencies (BE > 1.9 μM−1). Porcine stomach Muc 2 (Uniprot entry: A0A5G2QSD1) is a highly glycosylated protein (∼80%)42 that has a variety of glycan epitopes, including galactosyl-terminated O-glycans (core-1, -2, -3, and -4), blood group A antigens43 and a low degree of sialylation.44 Our results are consistent with previous studies on the interactions of Gal-3 with internal N-acetyllactosamine residues, core-1 structures, and blood group A antigens.45,46 Furthermore, the Gal-4 subunits were found to recognize both blood group A and B antigens,47,48 with the Gal-4N subunit showing a clear preference for blood group A.1
| ASF | Fetuin | Laminin | Fibronectin | Collagen IV | Muc 2 | ||
|---|---|---|---|---|---|---|---|
| Gal-3 | K D [μM] | 1.18 ± 0.33 | 2.17 ± 0.64 | 2.58 ± 0.49 | 7.52 ± 1.61 | 0.94 ± 0.15 | 0.23 ± 0.07 |
| B max [—] | 0.80 ± 0.05 | 0.83 ± 0.07 | 1.00 ± 0.06 | 1.08 ± 0.09 | 0.79 ± 0.03 | 0.75 ± 0.03 | |
| BE [μM−1] | 0.68 | 0.38 | 0.40 | 0.14 | 0.84 | 3.27 | |
| Gal-1C2S | K D [μM] | 1.57 ± 0.75 | 9.00 ± 3.30 | 9.10 ± 1.70 | 3.95 ± 2.15 | 0.95 ± 0.23 | 0.49 ± 0.06 |
| B max [—] | 0.68 ± 0.08 | 0.51 ± 0.08 | 0.96 ± 0.07 | 0.20 ± 0.04 | 0.68 ± 0.04 | 0.64 ± 0.01 | |
| BE [μM−1] | 0.43 | 0.06 | 0.11 | 0.05 | 0.72 | 1.31 | |
| Gal-4NL | K D [μM] | 4.50 ± 2.60 | n.d. | n.d. | 21.48 ± 1.78 | n.d. | 0.20 ± 0.07 |
| B max [—] | 0.14 ± 0.03 | n.d. | n.d. | 0.65 ± 0.19 | n.d. | 0.37 ± 0.01 | |
| BE [μM−1] | 0.03 | n.d. | n.d. | 0.03 | n.d. | 1.85 | |
| LGal-4C | K D [μM] | 3.52 ± 1.51 | 2.11 ± 1.47 | 21.22 ± 20.69 | 8.43 ± 3.47 | 7.55 ± 3.33 | 0.27 ± 0.07 |
| B max [—] | 0.29 ± 0.04 | 0.09 ± 0.02 | 0.18 ± 0.10 | 0.45 ± 0.08 | 0.35 ± 0.06 | 0.58 ± 0.07 | |
| BE [μM−1] | 0.08 | 0.04 | 0.01 | 0.05 | 0.05 | 2.12 | |
| Gal-8NL | K D [μM] | 0.64 ± 0.12 | 4.00 ± 1.58 | 37.60 ± 24.35 | 4.00 ± 1.58 | 0.04 ± 0.38 | 0.49 ± 0.06 |
| B max [—] | 0.65 ± 0.02 | 0.33 ± 0.04 | 1.32 ± 0.55 | 0.33 ± 0.04 | 0.08 ± 0.02 | 0.64 ± 0.01 | |
| BE [μM−1] | 1.01 | 0.08 | 0.04 | 0.08 | 2.00 | 1.30 | |
| LGal-8C | K D [μM] | 4.45 ± 1.62 | n.d. | 84.62 ± 117.81 | 4.12 ± 2.48 | n.d. | 0.50 ± 0.17 |
| B max [—] | 0.30 ± 0.04 | n.d. | 1.28 ± 1.39 | 0.28 ± 0.06 | n.d. | 0.52 ± 0.03 | |
| BE [μM−1] | 0.07 | n.d. | 0.02 | 0.07 | n.d. | 1.04 | |
| Reaction | Protein 1 | Conc. [μM] | Protein 2 | Conc. [μM] |
|---|---|---|---|---|
| 1 | Strep-ST003-sfGFP | 5 | HSC003eYFPGal-3 | 10 |
| 2 | Strep-SC003-sfGFP | 3.75 | HST003eYFPGal-3 | 7.5 |
| 3 | Strep-ST003-sfGFP | 7.5 | HSC003eYFPGal-3 | 7.5 |
| 4 | Strep-SC003-sfGFP | 5 | HST003eYFPGal-3 | 5 |
| 5 | HSC003eYFPGal-3 | 5 | HST003eYFPGal-3 | 5 |
Apart from Muc 2, HSC003eYFPGal-3 and HSC003DsRedMGal-1C2S exhibited binding to the ECM glycoproteins laminin, fibronectin, and collagen IV (Table 1). Laminin from EHS murine sarcoma, a high molecular weight heterotrimeric glycoprotein, contains up to 30% carbohydrate, predominantly in the form of N-glycans, featuring poly(N-acetyllactosamine) residues often terminated with α2,3-linked sialic acid or α-galactosyl moieties.49 In contrast to laminin, fibronectin is a dimeric glycoprotein with both O- and N-glycosylation sites, where N-glycans are primarily sialylated and the main O-glycoforms feature the sialyl-T-antigen.50 In contrast, human collagen IV is a major protein of the basement membrane. The alpha chains of collagen IV are O-glycosylated, composed mainly of galactosyl-hydroxylysines and glucosyl-galactosyl-hydroxylysines.51 Hence, the elevated results for HSC003eYFPGal-3 and HSC003DsRedMGal-1C2S probably arise from their interaction with internal and external galactosyl residues on the glycoproteins.52 The binding of HSC003mCherryGal-4NL to fibronectin and ASF was detected with binding efficiencies of less than 0.1 μM−1. Additionally, we could not observe any binding to fetuin and laminin (Table 1). Gal-4N preferentially recognizes sulfated and non-sialylated glycans.48,53 Furthermore, the presence of sialic acid residues is likely to hinder the binding of HSC003mCherryGal-4NL to fibronectin.54 In contrast, the glycoproteins ASF and fetuin as well as the ECM proteins laminin, fibronectin, and collagen IV were bound by HSC003mCherryLGal-4C (Table 1). However, they exhibited a low binding efficiency comparable to that observed for the N-terminal domain of Gal-4.
ASF and collagen IV emerged as the most effective ligands for HSC003eGFPGal-8NL, displaying BE greater than 1.0 μM−1. Gal-8N is reported to prefer sialylated glycan residues,55 but weak binding was detected with fetuin. In contrast, Gal-8N displayed a higher affinity to ASF (0.64 ± 0.12 μM), which is in line with the studies reporting on its high affinity to N-acetyllactosamine residues.56 In contrast, only weak binding was observed for laminin and fibronectin, with BE lower than 0.1 μM−1. The SpyCatcher galectin fusion protein, featuring the C-terminal domain of Gal-8, demonstrated only weak binding to ASF, fibronectin, and laminin (BE < 0.1 μM−1), while no binding was observed for collagen IV and fetuin. This lack of LGal-8C binding may be attributed to the preference of the Gal-8C subdomain for (poly)N-acetyllactosamine and blood group antigens.46,56,57 Inhibition assays with lactose confirmed the glycan-derived binding of the SpyCatcher003-galectin fusion proteins (ESI, Fig. S17†). Here, the highest binding signal was compared with the signal observed in the presence of lactose. Lactose significantly inhibited glycan binding of all galectins.
These results have confirmed the functionality of SpyCatcher003-galectin fusion proteins and demonstrated their ability to bind to various glycoprotein targets. In particular, HSC003eYFPGal-3 proved to be a potent binder to all tested ECM glycoproteins, including Muc 2 proteins with the highest binding efficiencies. Considering the high efficiencies of binding of the SC–ST-Gal-3 conjugates to ASF (Fig. 3), we next investigated the SC–ST-Gal-3 conjugate for building an ECM-like network using a layer-by-layer mode in a microtiter plate assay.
Application of galectin-3 fusion proteins for layering ECM glycoproteins
Galectins mediate cell-matrix and cell–cell interactions in the external microenvironment. Their oligomerization, from dimers (e.g., Gal-1) to oligomers (e.g., Gal-3), enables high-affinity recognition and crosslinking of galactose-containing glycoconjugates.38,58–61 In biomaterial science, the lectin-mediated in vitro mimicking of extracellular structures facilitates the reconstitution of natural microenvironment.62 In this context, we exploited the ability of the generated SC–ST-Gal-3 conjugates to bind ECM glycoproteins for the layer-by-layer assembly of a flexible in vitro ECM-like layer, incorporating laminin, fibronectin, and collagen IV. The optimal concentration of each glycoprotein layer and crosslinking by the SC–ST-Gal-3 conjugate was determined using in vitro binding assays on the respective protein. Since laminin is a major component of the basement membrane, it was chosen as the first layer. Initially, laminin-coated microtiter well plates were incubated with SC–ST-Gal-3 conjugate in concentrations of 0–5 μM (Fig. 4A). A concentration of 2.5 μM SC–ST-Gal-3 conjugate was chosen to optimally occupy all glycan-binding sites of the laminin layer. In the next step, the laminin layer was saturated with 2.5 μM SC–ST-Gal-3 conjugate, and fibronectin was added at a concentration of 0–20 μg mL−1 (Fig. 4B). The results displayed a saturated fibronectin layer at concentrations above 5 μg mL−1. To ensure maximum occupancy of glycan binding sites, the subsequent experiment was conducted with fibronectin-coated well plates to determine the optimal concentration of SC–ST-Gal-3 conjugate, which crosslinks the second fibronectin layer with the third layer composed of collagen IV, revealing a maximum binding at 5 μM SC–ST-Gal-3 conjugate (Fig. 4C). Lastly, the stepwise construction of 5 μg mL−1 laminin, 2.5 μM SC–ST-Gal-3 conjugate, 10 μg mL−1 fibronectin, and 5 μM SC–ST-Gal-3 conjugate enabled the binding of collagen IV serving as the third layer (Fig. 4D). Here, the layer density was achieved at concentrations exceeding 200 μg mL−1, which can be attributed to the limited glycosylation of human collagen IV and the presence of mono-galactosyl residues.51,54 Herein, we demonstrated the SC–ST-Gal-3 conjugate-mediated stepwise formation of an in vitro ECM-like layer composed of three glycoprotein layers. The formation of a Muc2 layer could not be demonstrated due to the cross-reactions of the available antibodies with the underlying ECM glycoproteins. In summary, the covalent interaction between SpyCatcher and SpyTag with galectins facilitates the assembly of different galectin recognition domains with unique glycan specificities, enabling the construction of customized ECM protein layers. Since HSC003eYFPGal-3 also binds Muc 2 (Table 2 and Fig. S16C†), the formation of a galectin-mediated mucin layer could help the reconstruction of the mucus layer. This is particularly important in the context of chronic inflammatory diseases such as inflammatory bowel disease (IBD), where the integrity of the mucus layer is compromised, leading to increased inflammation and disease progression. Reconstruction of this layer could help to restore barrier function and reduce inflammation in affected tissues.Binding of galectin fusion proteins to adenocarcinoma cell line DLD-1
In line with our objective to utilize SpyCatcher galectin fusion proteins as targeting or transport tools in an IBD-related context, we focused on exploring the binding interaction of these fusion proteins with human colorectal adenocarcinoma DLD-1 cells. Typically, colon adenocarcinoma cell lines are employed for investigating the galectin role in chronic inflammation63,64 or to appropriately simulate IBD conditions.65 DLD-1 is a human colorectal cancer cell line originally isolated from the large intestine of a colon adenocarcinoma patient,66 employed in studies investigating IBD-correlated tumorigenesis.67,68 The previous cells were shown to express all investigated galectins69,70 (ESI, Fig. S18†) and present glycan ligands on their surface. The N-glycome predominantly comprises glycans of the high mannose and (di- or tri-antennary) complex type. The latter are characterized by approximately 30% fucosylation and 40% sialylation.71,72 We tested the ability of SpyCatcher galectin fusion proteins of Gal-1C2S, Gal-3, and the subdomains Gal-4 and Gal-8 in varying concentrations to bind to the glycocalyx surface of DLD-1 cells by exploiting the color code in flow cytometry (Fig. 5 and ESI Fig. S19†). Unfortunately, the binding of Gal-8 subdomains to DLD-1 cells interferes with the autofluorescence of the cells,73 which hampered proper data evaluation (results not shown). Galectin fusion protein constructs of Gal-1, Gal-3, and Gal-4NL bound to the surface of DLD-1 cells in a dose-dependent manner (Fig. 5). Gal-1 and Gal-3 showed a distinct interaction with the DLD-1 cell surface. Notably, concentrations of HSC003eYFPGal-3 higher than 5 μM resulted in a fluorescence intensity that was out of the detection limit of the laser/photodetector (Fig. 5B). We tried to reclone this construct with DsRedM fluorescence to verify that the binding is not influenced by the type of the fluorescence part but the result was the same – the results were out of the detection limit. Apparently, irrespective of the type of the fluorescence label, the Gal-3 protein constructs bound so strongly and in such high amounts to the cell surface that the fluorescence exceeded the detection limit. We conclude that the binding of HSC003eYFPGal-3/HSC003eDsRedMGal-3 to the DLD-1 cells is very robust regardless of the fluorescent label. Moreover, Gal-3 caused a strong aggregation of cancer cells (in a concentration higher than 17 μM) as also reported in the literature.74Due to the different binding behaviour of Gal-3 compared to the other constructs, we verified that it is not cytotoxic for DLD1 cancer cells (tested on the construct HGal-3 without fusion partner due to the interference of the fluorescent label; ESI, Fig. S20†). The binding of Gal-4 subunits varied. While HSC003mCherryGal4NL bound similarly to HSC003DsRedMGal-1C2S (Fig. 5C), HSC003mCherryLGal4C did not bind to the cells whatsoever, even at 80 μM concentration (Fig. 5D). In the available range of concentrations, saturated binding was reached with neither of the three proteins, which indicates a high number of available glycan ligands on the cell surface. The extracellular binding of Gal-3 to DLD-1 has been previously reported69,75 and corroborates our findings. However, the binding of the SpyCatcher fusion protein of Gal-1C2S is not in accordance with previous studies indicating that Gal-1 does not bind to this cell line.76 However, we observed binding of Gal-1 even without the SpyCatcher fusion (data unpublished). We assume that HSC003DsRedMGal-1C2S may also occur as synthetic dimer of the dimeric Gal-1 by the SC fusion as discussed for the SC-Gal-3 fusion proteins (generating tetravalent Gal-1). Tetravalency of the SC-fused Gal-1 may support binding to low affinity glycan ligands. As stated above, Gal-4 is expressed by DLD-1 cells, but there is no evidence in the literature for binding to DLD-1. The binding of SpyCatcher galectin fusion proteins to the cell surface was inhibited by the addition of lactose in a dose-dependent manner (Fig. 6). For HSC003eYFPGal-3 (2.5 μM), lactose was a stronger inhibitor than for HSC003DsRedMGal-1C2S (2.5 μM); in the former case 100 mM lactose decreased the binding to 5%, while binding of HSC003DsRedMGal-1C2S was reduced only to 27%. The results were confirmed with 1 μM Gal-1 and Gal-3 and reflect previous inhibition studies with both galectins.77 The binding of 18 μM Gal-4NL was inhibited by only 53% with 100 mM lactose.
To summarize, we have verified the glycan-driven interaction of SpyCatcher galectin fusion constructs with human intestinal DLD-1 cells and demonstrated the applicability of the generated color-coding system in flow cytometry. Our model human cancer cell line DLD-1 does not accurately reflect the glycosylation pattern of intestinal epithelial cells. The use of primary cells would more accurately reflect natural conditions-but is associated with complex cultivation conditions.78 Nevertheless, the usability of SpyCatcher galectin fusion constructs with living biomaterials enables the investigation of the targeting and transport capability of galectins.
Galectin-functionalized microgels as a targeting tool for biomedical applications
We further exploited the specific binding properties of SpyCatcher galectin fusion proteins to target cells in a biomedical context. For this aim, we have implemented galectin-functionalized microgels as a targeting platform using the SpyCatcher/SpyTag technology that combines the glycan-targeting capabilities of galectins with the properties of microgels. SpyTag003-presenting microgels were synthesized by droplet microfluidics as previously shown for enzyme-decorated microgels.25 The visualization of microgels through optical microscopy (ESI, Fig. S21A and B†) and the manual measurement of the microgel diameters confirmed similar sizes, with diameters of 146 ± 5 nm (non-functionalized) and 142 ± 4 nm (SpyTag003-functionalized). The microgels were then subjected to a fluorescamine assay, which we have previously used for the visualization of peptides and enzymes in microgels.25,79 While staining of the non-functionalized microgels did not deliver a fluorescent product (ESI, Fig. S21C†), the presence of primary amines in the SpyTag003-microgels was confirmed, demonstrating a uniform distribution of the SpyTag003 throughout the entire microgel (ESI, Fig. S21D†). The permeability80 was investigated by dispersing the microgels in FITC-dextran solutions with varying molecular weights (4, 40, 150, and 500 kDa), followed by confocal analysis of the fluorescence intensity profiles (ESI, Fig. S22†). Comparison of normalized fluorescence intensities of microgels with and without SpyTag003 showed a reduced permeability for higher molecular weight molecules (∼15% of 500 kDa dextran fluorescence intensity was detected in the microgels), indicating that the fluorescent molecules are too large to fit through microgel pores. We observed a higher diffusion of 4 kDa FITC-dextran into the microgels (>75%). Here, SpyTag003-microgels displayed a lower permeability (75% of 4 kDa FITC-dextran in the microgel) compared to non-functionalized microgels (80%). The use of 8-arm PEG-VS for SpyTag incorporation (instead of 4-arm PEG-VS), along with the incorporation of the SpyTag itself, probably contributed to the reduced permeability. The lower permeability of the SpyTag003-microgels was also reflected in the fluorescence intensity of the 4 kDa and 40 kDa FITC-dextrans.Attachment of SpyCatcher galectin fusion proteins to SpyTag003-microgels was accomplished via post-attachment. SpyCatcher003-carrying fusion proteins of each galectin class were incubated with SpyTag003 microgels and non-functionalized microgels as controls, including HSC003DsRedMGal-1C2S, HSC003eYFPGal-3, HSC003mCherryGal-4NL and HSC003eGFPGal-8NL. Subsequent visualization of the microgels with confocal microscopy revealed that non-functionalized microgels did not display significant fluorescence (Fig. 7A–D), indicating that only minimal non-specific interactions of the galectin fusion proteins with the microgels exists. In contrast, we expected considerable fluorescence of the SpyTag003-microgels due to the attachment of SpyCatcher003-carrying fluorescent galectin fusion proteins, which was confirmed for all tested galectins (Fig. 7E–H). We observed microgels that exhibited fluorescence throughout the microgel, indicating diffusion of galectin fusion proteins into the microgel mass. On the other hand, some microgels exhibited fluorescence only in the outer shell of the microgel. We conclude that an increasing attachment of galectins to the microgel shell may lead to the blockage of the microgel pores, which prevents galectins from further diffusing into the microgel due to steric hindrance. Galectin fusion proteins, including Gal-1, Gal-3, Gal-8N, and Gal-4N, can form concentration-dependent oligomers, ranging from dimers to oligomers.1,37,38,46,81,82 Our study demonstrates that SpyCatcher-fused Gal-3 forms dimers, which may reduce the diffusion into the microgels. Additionally, fluorescent proteins originating from A. Victoria tend to form weak dimers, such as eGFP and eYFP.36,83 We expect that the overall diffusion of galectin multimers into microgels is lower than that of galectin monomers due to their increased size under the given conditions. The increased concentration of larger galectin clusters in the outer shell of the microgel may further reduce the permeability of the microgel and hinder the ability of the galectins to react with the SpyTag003 units in the core of the microgel. The permeability studies indicated that the galectins (sized from 54.6 to 67.2 kDa; Table S3†), can partially diffuse into the microgels. Importantly, the results cannot be directly compared on a one-to-one basis with the molecular weights of galectins, which arises from the differences in the molecular structure and associated interactions with the polymer network. Furthermore, the permeability is not only influenced by the molecular weight-but also by the different three-dimensional structures of the SpyCatcher galectin fusion proteins. Even so, the results indicate that the diffusion of galectins into microgels would be much less probable when clusters are formed: the permeability already decreased strongly when comparing 40 with 150 kDa FITC-dextran and was even lower for larger dextrans.
Quantification of microgel-bound galectin fusion proteins was performed using the Bradford method calibrated for BSA.84 Analysis of the supernatants from two independent galectin–microgel coupling experiments revealed significant SpyCatcher/SpyTag-mediated galectin attachment compared to non-functionalized microgels (Fig. 8). About 75% of the initially added amount of HSC003eYFPGal-3 and HSC003DsRedMGal-1C2S bound to SpyTag003 microgels corresponding to absolute values of 12.4 ± 2.3 nmol and 12.4 ± 1.8 nmol, respectively. In contrast, the binding efficiency of fusion proteins containing the N-terminal subdomains of Gal-4 and Gal-8 were more than 20% lower with 45% (7.6 ± 0.5 nmol) for HSC003eGFPGal-8NL and 54% (7.8 ± 0.6 nmol) for HSC003mCherryGal-4NL. The factors contributing to this reduced attachment could be the potential formation of oligomers between the single subdomains of Gal-4NL and Gal-8NL as previously reported.1,46 Even though the fluorescent fusion protein eGFP may theoretically enhance oligomerization effects due to its intrinsic tendency to form dimers,36,83 mCherry is not expected to cause problems as it mainly appears as a monomeric protein.85 The formation of large oligomer clusters at the microgel shell could hinder their diffusion into the microgel and reduce the amount of binding. We hypothesized that only a small quantity of galectins would be bound to the microgel network via non-specific interactions (1.4 to 2.7 nmol), which corresponds to 9–16% of the initial amount of galectin in the reaction (Fig. 8). These data suggest that galectins were mainly bound to microgels through specific SpyTag/SpyCatcher interactions.
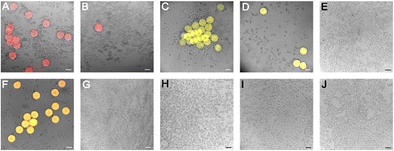
Following the establishment of microgel-galectin coupling conditions, we conjugated SpyCatcher fluorescent galectin constructs to the SpyTag003-microgels and assessed the binding of these galectin-functionalized microgels (Gal-MGs) in a culture with DLD-1 cells by fluorescent microscopy (Fig. 9). Gal-1C2S-DsRedM-MGs and Gal-3-eYFP-MGs bound strongly to the cells, even pulling adhered cells off the plate surface (Fig. 9A and C). Gal-4 subunits showed the same binding as observed in the flow cytometry experiment: Gal-4NL-mCherry-MGs bound to DLD-1 cells whereas the LGal-4C-mCherry-MGs did not bind (Fig. 9F and H). Lactose (200 mM) inhibited cell surface binding of the galectin fusion proteins (Fig. 9B, D, G and I), indicating a glycan-driven galectin interaction. Our findings demonstrate the potential usability of the galectin-loaded microgels as a targeting and transporting tool for biomedically relevant materials. Boesveld et al.30 previously established microgels as a platform for antibody-mediated cytokine scavenging in the context of chronic inflammation. Thus, the assembly of functional microgels with our galectin-targeting tools would enable the positioning of these microgels in the areas of inflammation. In addition, galectin-functionalized microgels can be interesting colloidal building blocks for the cell-triggered assembly and fabrication of hydrogel scaffolds for tissue engineering applications.86
Experimental
Protein constructs
The N-terminal His6-tagged (H) proteins utilized in this study, including HGal-3 and HeYFPGal-3, were generated as described elsewhere.1,8,10 Galectin fusion proteins incorporating SpyCatcher003 are based on the pET17b (Novagen, Germany) or pET28a (Novagen, Germany) vector (ESI, Table S2†). Restriction enzymes, Klenow-Polymerase I and DNA-ligases, were purchased from New England Biolabs (Germany). All genes were optimized for recombinant expression in E. coli, and successful cloning was confirmed through DNA sequencing. To create SpyCatcher003-carrying galectin fusion proteins, the established SNAP-tag vector DNA (pET17b::HSNAPeYFPGal-3, pET17::HSNAPDsRedMGal-1C2S, pET17b::HSNAPeGFPGal-8NL/LGal-8C) was employed as a template.10 The synthetic gene encoding SpyCatcher003 protein from S. pyogenes14 was introduced using NheI and AgeI restriction sites. pET17b::HSpyCatcher003Gal-4NL/LGal-4C was cloned in several successive steps. Here, the mCherry synthetic gene (Genbank: KJ567138.1 81) was introduced into the existing vector pET17b::HSNAPeYPGal-3 through AgeI and BsrGI restriction sites followed by the substitution of Gal-3 with subdomains of human Gal-4N (aa 1–150) and Gal-4C (aa 179–323), respectively. To enhance the binding activity, the subdomains were recloned including the segments of the natural linker peptide (L).1 For the amplification of Gal-4NL, a PCR with forward primer 5′-CTCTAGATGTACAAAATGGCATATGTTCCGGCACCGGG-3′ and reverse primer 5′-GATCGCGGCCGCTTAGGGTCCCTGGGGCCGGAGGGGCTGACCACCAATAAAATTAATGCTCTGC-3′ was employed. The subdomain LGal-4C was cloned using forward primer 5′-GATCTGTACAAACCCGGACATTGCCATCAACAGCTGAACAGCCTGCCTACCATGGAAGGCCCGC-3′ and reverse primer 5′-ATCCGCGGCCGCTTAAATCTGAACATAACTCAGGG-3′. Both genes were introduced through restriction sites BsrI and NotI. In the case of HSC003DsRedMGal-3, the Gal-1C2S domain from pET17b::HSC003DsRedMGal-1C2S was exchanged with the human Gal-3 domain by BsrGI and NotI. For the production of control proteins (HSC003eYFP, HSC003eGFP, HSC003DsRedM, HSC003mCherry), galectin domains were removed with BsrGI and NotI. The remaining overhanging DNA bases were treated with Klenow polymerase I, followed by blunt-end ligation. The synthesis of HSC003Gal-3 involved removing the fluorescence gene eYFP from pET17b::HSC003eYFPGal-3 with AgeI and BsrGI, followed by overhanging base digestion and blunt end ligation. Genes encoding for SpyTag003 (ST003)14 protein variants HST003eYFPGal-3 (pET28a), HST003eYFP (pET17b), HST003Gal-3 (pET17b) and strep-SC003/ST003-sfGFP (pETDuetI) were custom-synthesized (Biocat GmbH, Germany).Expression and purification of protein constructs
The protein constructs were expressed in E. coli BL21 (DE3), E. coli Rosetta (DE3) pLysS, or E. coli Rosetta 2 (DE3) pLysS (Merck, Germany) strains (ESI, Table S2†). Transformation, protein expression, cell disruption, and purification of His6-tagged proteins were performed as described previously.10 Strep-tagged proteins were isolated on 5 mL StrepTrap XT Sepharose (Cytiva GmbH, Germany) using strep binding buffer (100 mM Tris-HCl pH 8, 150 mM NaCl, 1 mM EDTA) and 50 mM desthiobiotin as an elution reagent. Purified proteins were dialyzed in PBS (50 mM NaH2PO4 pH 7.5, 150 mM NaCl) overnight at 4 °C (molecular weight cut off: 14 kDa).SDS-PAGE and immunoblot
Discontinuous SDS-PAGE (4% stacking gel, 10–14% separation gel) was used for the analysis of galectin expression and purification. Protein samples were treated with 50 mM dithiothreitol (Carl Roth GmbH, Germany) and NuPAGE™ LDS 1× sample buffer (Thermo Fisher Scientific, Germany) for 15 min at 95 °C. Gel electrophoresis was conducted at 200 V (const.) for 60 min, followed by Coomassie Blue staining (Serva Electrophoresis GmbH, Germany). Western blotting was carried out on PVDF membranes (Immobilon®-P, pore size 0.45 μm, Merck, Germany) for 60 min at 30 V (const.). His6-tagged proteins were detected with 100 mU mL−1 of peroxidase-conjugated anti-His6-antibody (sc-8036, Santa Cruz Biotechnology, Germany). The SpyCatcher incorporation was detected with primary rabbit anti-SpyCatcher antibody (200 ng mL−1, HCA-379, Bio-Rad Laboratories, Germany) and goat anti-rabbit peroxidase-conjugate at a 1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 4000 dilution (A9169, Sigma Aldrich, Germany). After washing with 0.05% v/v TBS-Tween 20, Pierce 1-step Ultra TMB substrate solution (Thermo Fisher Scientific, Germany) was utilized for calorimetric visualization.
4000 dilution (A9169, Sigma Aldrich, Germany). After washing with 0.05% v/v TBS-Tween 20, Pierce 1-step Ultra TMB substrate solution (Thermo Fisher Scientific, Germany) was utilized for calorimetric visualization.
Galectin binding and inhibition assays
Glycoproteins used in this study are asialofetuin (fetal calf serum; Sigma Aldrich, Germany), fetuin (fetal bovine serum; Sigma Aldrich, Germany), Muc 2 (porcine stomach; Sigma Aldrich, Germany), laminin (Engelbreth-Holm-Swarm murine sarcoma; Sigma Aldrich, Germany), fibronectin (human plasma; Sigma Aldrich, Germany), collagen IV (human placenta; Sigma Aldrich, Germany). The analysis of galectin binding to glycoproteins was conducted using established assays.8–10 Briefly, glycoproteins (1 μg per well in 100 mM Na2CO3 pH 9.6) were immobilized on adsorptive 96 well microtiter plates (Nunc Immuno Maxisorp; Thermo Scientific, Germany) overnight at 4 °C. After three washes with PBS-T (PBS pH 7.5 + 0.05% v/v Tween 20), the remaining active sites on the plates were blocked with 2% w/v bovine serum albumin in PBS-T. Subsequently, galectin dilutions of up to 10 μM (50 μL per well) were incubated for 1 h followed by incubation with a 50 μL of 200 ng μL−1 peroxidase-conjugated anti-His6-antibody (sc-8036, Santa Cruz Biotechnology, Germany). Galectin binding was detected with 50 μL per well 3,3′,5,5′-tetramethylbenzidine (TMB; Kementec, Denmark) and 3 M HCl (50 μL per well) as a stopping reagent. Photometric absorption at 450 nm was measured (BioTek Synergy™ 2; Agilent Technologies, USA) and the mean signal was determined based on 2–3 data points. The binding signals underwent normalization with the highest binding signal set to 1 followed by the subtraction of the background signal. Apparent KD values of galectins were computed (SigmaPlot 14.0 software) assuming one-site saturation with galectin concentration for half-maximum binding. The galectin binding efficiencies (μM−1) were then determined as the ratio of the maximum binding signal (Bmax) and the apparent KD value.10 Glycan-derived galectin-binding was confirmed by inhibition with lactose. For this, 30 μM galectin was incubated for 1 h, either with 200 mM lactose or without lactose. The highest binding signal for each galectin was normalized to 100% and inhibition was calculated as the residual binding signal [%] in relation to the non-inhibited binding.Conjugation of galectin fusion constructs through the interaction of SpyCatcher and SpyTag
The constructs strep-SC003-sfGFP, strep-ST003-sfGFP, HSC003eYFPGal-3, and HST003eYFPGal-3 were isolated from the bacterial crude extract following the described procedure (Section Expression and purification of protein constructs) and subsequently purified to homogeneity through SEC (HiPrep 26/60 Sephacryl-S200-HR; Cytiva GmbH, Germany). The reactions involving HSC003eYFPGal-3, HST003eYFPGal-3, and sfGFP proteins were conducted using both equimolar and 2-fold molar excess ratios. The Gal-3 conjugation of HSC003eYFPGal-3 and HST003eYFPGal-3 (referred to as SC–ST-Gal-3 conjugate) was conducted at an equimolar ratio of both fusion proteins (Table 2). All experiments were carried out in a total volume of 100 μL and incubated in PBS pH 7.5 for 1 h at 25 °C. Afterward, the protein samples were applied to a reducing gel as outlined (Section SDS-PAGE and Immunoblot).Dynamic light scattering (DLS) measurements
80 μL of each solution (1 mg mL−1 of HSC003eYFPGal3 and HST003eYFPGal-3; 10 μM of SC–ST-Gal-3 conjugate) was measured in a DynaPro® NanoStar® DLS Detector (Wyatt Technology, Germany) using disposable micro-cuvettes (MOS Plastics Inc., USA). Each sample was measured ten times under the conditions shown in Table 3. The data were analyzed with DYNAMIC software (Version 2.3.7.3).| Property | Value |
|---|---|
| Number of acquisition | 10 |
| Acquisition time | 5 s |
| Read interval | 1 |
| Temperature | 20 °C |
| Laser wavelength | 662.7 nm |
| DLS detector angle | 90° |
Size exclusion chromatography (SEC)
For size exclusion chromatography, we prepared 200 μL solutions of two-fold purified HSC003eYFPGal-3 (20 μM) and HST003eYFPGal-3 (20 μM), along with an SC–ST-Gal-3 conjugate solution (10 μM). These solutions were filtered through a 0.2 μM filter. The samples were applied onto a Superdex Increase 200 10/300 GL column (Cytiva, USA), using PBS pH 7.5 as the running buffer, with a flow rate set at 0.75 mL min−1, and a detection wavelength of 280 nm. To determine the molecular weight of the samples, the SEC column was equilibrated with protein standards from a Gel Filtration Calibration Kit (Cytiva, USA).10 Stokes calibration was calculated based on the Stokes radii of the standard proteins87–89 and an online protocol (https://www.phenomenex.com/documents/2022/10/18/16/04/white-paper-sec-for-the-determination-of-protein-hydrodynamic-radius).In vitro ECM-layer formation
The stepwise in vitro built-up of an ECM-like layer was constructed using laminin (Engelbreth-Holm-Swarm murine sarcoma, Sigma Aldrich, Germany), fibronectin (human plasma, Sigma Aldrich, Germany), and collagen IV (human placenta, Merck Millipore, USA). Interaction of these proteins was achieved using SpyCatcher and SpyTag-conjugated Gal-3 fusion proteins (SC–ST-Gal-3 conjugate). 10 μM solutions of HSC003eYFPGal-3 and HST003eYFPGal-3 were mixed in a rotary shaker for 1 h at 4 °C, resulting in a 5 μM conjugate solution which was further diluted (0–5 μM) in PBS at pH 7.5. In vitro binding assays were conducted to determine the saturating concentrations of the respective glycoprotein/galectin for each layer. Initially, 200 μL per well of a 5 μg mL−1 laminin solution was immobilized on adsorptive microtiter plates (Nunc Immuno Maxisorp, Thermo Scientific, Germany) overnight at 4 °C. Following three washes with PBS-T, non-occupied binding sites were blocked in PBS-T supplemented with 2% w/v BSA for 1 h at room temperature. Subsequently, freshly prepared dilutions of the SC–ST-Gal-3 conjugates (50 μL per well) were added and incubated for 1 h at room temperature. Aliquots of 50 μL per well of an anti-His6-antibody dilution (1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1000 in PBS pH 7.5; sc-8036, Santa Cruz Biotechnology, Germany) were added and incubated for 1 h at room temperature. Galectin binding was detected by adding 50 μL per well of TMB followed by 50 μL per well of 3 M HCl as a stopping reagent and photometrical measurement was conducted at 450 nm. In the following binding experiment, saturating amounts of SC–ST-Gal-3 conjugate (2.5 μM; 50 μL per well) were applied to the laminin layer for binding fibronectin. To achieve this, 50 μL per well of fibronectin dilutions (0–20 μg mL−1; in PBS pH 7.5) were added and incubated for 1 h at room temperature. Bound fibronectin was then detected by adding 50 μL per well of a rabbit anti-fibronectin antibody (1
1000 in PBS pH 7.5; sc-8036, Santa Cruz Biotechnology, Germany) were added and incubated for 1 h at room temperature. Galectin binding was detected by adding 50 μL per well of TMB followed by 50 μL per well of 3 M HCl as a stopping reagent and photometrical measurement was conducted at 450 nm. In the following binding experiment, saturating amounts of SC–ST-Gal-3 conjugate (2.5 μM; 50 μL per well) were applied to the laminin layer for binding fibronectin. To achieve this, 50 μL per well of fibronectin dilutions (0–20 μg mL−1; in PBS pH 7.5) were added and incubated for 1 h at room temperature. Bound fibronectin was then detected by adding 50 μL per well of a rabbit anti-fibronectin antibody (1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1000 in PBS pH 7.5; F3648, Sigma Aldrich, Germany) for 1 h at room temperature, followed by the addition of 50 μL per well of peroxidase-conjugated anti-rabbit-antibody from goat (A9169, Sigma Aldrich, Germany). The binding was detected photometrically after adding TMB and 3 M HCl, following the same procedure as described. The subsequent binding assay was conducted to determine the amount of SC–ST-Gal-3 conjugate necessary to crosslink fibronectin with collagen IV. Here, 200 μL per well of a 10 μg mL−1 fibronectin solution was immobilized on adsorptive plates and incubated with dilutions of SC–ST-Gal-3 conjugate, as described for the laminin assay. Finally, the whole layer was built up with 200 μL per well of 5 μg mL−1 laminin, 50 μL per well of 2.5 μM SC–ST-Gal-3 conjugate, 50 μL per well of 10 μg mL−1 fibronectin and 50 μL per well of 5 μM SC–ST-Gal-3 conjugate, each solution incubated for 1 h at room temperature with intermittent PBS-T washing steps. Subsequently, aliquots of 50 μL per well of collagen IV in the range of 0–200 μg mL−1 were incubated for 1 h at room temperature. The binding of collagen IV was detected by incubation with 50 μL per well of mouse anti-collagen IV-antibody (1
1000 in PBS pH 7.5; F3648, Sigma Aldrich, Germany) for 1 h at room temperature, followed by the addition of 50 μL per well of peroxidase-conjugated anti-rabbit-antibody from goat (A9169, Sigma Aldrich, Germany). The binding was detected photometrically after adding TMB and 3 M HCl, following the same procedure as described. The subsequent binding assay was conducted to determine the amount of SC–ST-Gal-3 conjugate necessary to crosslink fibronectin with collagen IV. Here, 200 μL per well of a 10 μg mL−1 fibronectin solution was immobilized on adsorptive plates and incubated with dilutions of SC–ST-Gal-3 conjugate, as described for the laminin assay. Finally, the whole layer was built up with 200 μL per well of 5 μg mL−1 laminin, 50 μL per well of 2.5 μM SC–ST-Gal-3 conjugate, 50 μL per well of 10 μg mL−1 fibronectin and 50 μL per well of 5 μM SC–ST-Gal-3 conjugate, each solution incubated for 1 h at room temperature with intermittent PBS-T washing steps. Subsequently, aliquots of 50 μL per well of collagen IV in the range of 0–200 μg mL−1 were incubated for 1 h at room temperature. The binding of collagen IV was detected by incubation with 50 μL per well of mouse anti-collagen IV-antibody (1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1000 in PBS pH 7.5, C1926, Sigma-Aldrich, Germany) for 1 h and incubation with goat anti-mouse antibody (1
1000 in PBS pH 7.5, C1926, Sigma-Aldrich, Germany) for 1 h and incubation with goat anti-mouse antibody (1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1000 in PBS pH 7.5, A4416, Sigma Aldrich, Germany) under the same conditions.
1000 in PBS pH 7.5, A4416, Sigma Aldrich, Germany) under the same conditions.
Microgel synthesis and attachment of galectin fusion proteins
![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 000 dpi) dark-field photomask. An epoxy-based photoresist (SU-8) was then patterned onto a silicon wafer through photolithography of the printed photomask to form the used master molds. To produce PDMS-based microfluidic devices, soft lithography was used. The Sylgard 184 Elastomer Kit (Mavom), was used to fill the master molds, more specifically a two-component mixture of 10 parts siloxane and 1 part curing agent (crosslinker). After filling the master molds, the mixture underwent overnight curing at 60 °C, forming a fully cross-linked polydimethylsiloxane (PDMS) device. The device was carefully removed, and equipped with tube connections using a 0.75 mm biopsy punch. The device and glass slide, cleaned with isopropyl alcohol and water, and dried at 60 °C for 1 h, underwent oxygen plasma treatment. The PDMS and glass slide were bonded to form irreversible bonds, facilitated by the introduction of polar Si–Ox, Si–OH, and C–OH groups during plasma treatment. To minimize wetting in microfluidic channels, an Aquapel® (PWG Auto Glass, LLC) hydrophobic film was applied, and the channels were flushed with air after a 2-minute exposure.
000 dpi) dark-field photomask. An epoxy-based photoresist (SU-8) was then patterned onto a silicon wafer through photolithography of the printed photomask to form the used master molds. To produce PDMS-based microfluidic devices, soft lithography was used. The Sylgard 184 Elastomer Kit (Mavom), was used to fill the master molds, more specifically a two-component mixture of 10 parts siloxane and 1 part curing agent (crosslinker). After filling the master molds, the mixture underwent overnight curing at 60 °C, forming a fully cross-linked polydimethylsiloxane (PDMS) device. The device was carefully removed, and equipped with tube connections using a 0.75 mm biopsy punch. The device and glass slide, cleaned with isopropyl alcohol and water, and dried at 60 °C for 1 h, underwent oxygen plasma treatment. The PDMS and glass slide were bonded to form irreversible bonds, facilitated by the introduction of polar Si–Ox, Si–OH, and C–OH groups during plasma treatment. To minimize wetting in microfluidic channels, an Aquapel® (PWG Auto Glass, LLC) hydrophobic film was applied, and the channels were flushed with air after a 2-minute exposure.
For microfluidic experiments, a microfluidic station comprising three syringe pumps (PHD Ultra, Harvard Apparatus, Holliston, USA) was used. GASTIGHT® Syringes (Hamilton, USA) were placed in each syringe pump to accurately control the flow rates on the microfluidic chip. Through fine-bore PE tubing with an inner diameter of 0.38 mm, the syringes were attached to a PDMS microfluidic device with an 80 μm channel diameter. During the synthesis, an inverted microscope (Motic AE2000, TED PELLA, INC., Redding, CA) with a camera (Flea3, Point Grey, Richmond, CA) enabled real-time observation of microfluidic flow and droplet formation.
Two distinct aqueous phases, each at a flow rate of 150 μL h−1, were introduced separately into the microfluidic device. These phases were merged into a single channel, maintaining laminar flow conditions. Following the combination, an immiscible oil phase (Novec™ HFE-7500 with 2% w/w FluoSurf-C surfactant, Emulseo) was used to transform the aqueous stream into water-in-oil (W/O) droplets at 600 μL h−1 in a flow-focusing microfluidic channel. The pre-polymers in the droplets underwent a Michael-type addition reaction, resulting in the formation of monodisperse, spherical microgels. For their purification, microgels were washed three times with HFE-7500 (Emulseo), once with n-heptane, and five times with PBS.
The two aqueous phases consist of the SH-phase containing PEG thiol (PEG-SH, Biopharma PEG Scientific Inc.) and the VS-phase containing PEG vinyl sulfone (PEG-VS, Creative PEGWorks). To prepare SpyTag-functionalized microgels, the VS-phase was prepared from 5% w/v 8-arm PEG-VS (20 kDa) in 100 mM HEPES pH 8. The SH-phase contained 2.5% w/v 4-arm PEG-SH solution (10 kDa) and 1% w/v SpyTag in H2O due to the low solubility of the used peptide in the buffer.25 The peptide C-SpyTag003 (BioCat GmbH, 98% purity), designated as SpyTag003, was custom-synthesized with the sequence CRGVPHIVMVDAYKRYK (2034.06 g mol−1).14 The microfluidic synthesis was designed according to our previous work where an excess of VS functional groups was used for the attachment of the Spy-Tag through an incorporated cysteine.25 For the preparation of non-functionalized microgels, a 2.5% w/v 4-arm PEG-VS solution (10 kDa) in 100 mM HEPES pH 8 formed the VS-phase. The SH-phase was prepared with 2.5% w/v 4-arm PEG-SH (10 kDa) in 100 mM HEPES pH 8. Using Olympus cellSens Standard 3.2 software, microscopy images of microgels were captured using an Olympus CKX53 equipped with a camera (Olympus DP23). Microgel sizes were determined using Fiji (ImageJ), with a minimum of 20 particles measured for analysis.
Confocal microscopy
A fluorescamine assay was used for the detection of the SpyTag peptide in microgels.25,79 Therefore, microgels (40 μL in 100 mM PBS pH 7.5) were mixed vigorously with a solution of 4′-phenylspiro[2-benzofuran-3,2′-furan]-1,3′-dione (fluorescamine, 10 μL, 3 mg mL−1 in acetone) as described previously.91 The product was then examined using confocal microscopy and possesses an excitation wavelength λex of 390 nm and an emission wavelength λem of 475 nm.75Permeability studies were carried out according to Bulut et al.80 Microgels were mixed with 1 mg mL−1 FITC-dextran solutions of varying molecular weights (4, 40, 150, and 500 kDa). The fluorescence intensity profile was analyzed by confocal microscopy.
For the visualization of fluorescent microgels, confocal laser scanning microscopy (SP8 Tandem Confocal, Leica Microsystems Inc.) was used. A photodiode with 405 nm was used for the analysis of fluorescamine derivates (455–500 nm emission; blue channel), an argon laser with 476 nm for eYFP (500–550 nm emission; yellow channel), and an argon laser with 488 nm for eGFP (500–550 nm emission; green channel). A diode-pumped solid-state laser with 561 nm was used to analyze samples with DsRedM (575–650 nm emission) and mCherry (600–650 nm emission; red channel). Image processing was performed with LasX software.
Cell culture
The human adenocarcinoma DLD-1 cell line was kindly provided by Prof. P. Šebo (Institute of Microbiology of the Czech Academy of Sciences, Prague). Cells were grown in RPMI supplemented with 10% v/v fetal bovine serum and 1% v/v antibiotic mixture (penicillin, 100 U mL−1, and streptomycin, 100 g mL−1; all Gibco, Thermo Fisher Scientific, Waltman, MA, USA). Cells were incubated in a humidified atmosphere containing 5% CO2 at 37 °C.Western blot for the detection of galectins in DLD-1 cells
The detection of all galectins was done by western blot as described earlier with Gal-3,69 or Gal-8![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 70 in DLD-1. We used primary rabbit antibodies (1
70 in DLD-1. We used primary rabbit antibodies (1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1000) against human Gal-1 (D608 T, Cell Signaling Technology, Danvers, MA, USA), against human Gal-3 (D4I2, Cell Signaling Technology, USA), against human Gal-4 (PA5-95353, Thermo Fisher Scientific, Waltman, MA, USA), or against human Gal-8 (JB85-35, Novus Biologicals, USA), respectively. Goat anti-rabbit secondary antibody conjugated with horseradish peroxidase (31460, ThermoFisher Scientific, USA) with SuperSignal™ West Femto Maximum Sensitivity substrate (Thermofisher, USA) were used for chemiluminescence detection.
1000) against human Gal-1 (D608 T, Cell Signaling Technology, Danvers, MA, USA), against human Gal-3 (D4I2, Cell Signaling Technology, USA), against human Gal-4 (PA5-95353, Thermo Fisher Scientific, Waltman, MA, USA), or against human Gal-8 (JB85-35, Novus Biologicals, USA), respectively. Goat anti-rabbit secondary antibody conjugated with horseradish peroxidase (31460, ThermoFisher Scientific, USA) with SuperSignal™ West Femto Maximum Sensitivity substrate (Thermofisher, USA) were used for chemiluminescence detection.
Binding of galectin fusion proteins to colorectal cancer cells
The serially diluted galectin fusion proteins in PBS/BSA buffer (50 mM NaH2PO4/150 mM NaCl/1% w/v bovine serum albumin, pH 7.5) were mixed with the suspension of DLD-1 cells (1 × 105 in 100 μL final volume) in U-bottom 96-well tissue culture test plates and incubated for 60 min on ice. The carbohydrate-driven galectin binding was verified by inhibition with lactose (200 and 100 mM). The cells were washed with PBS/BSA buffer and centrifuged (200g, 3 min, 4 °C). The cells (resuspended in PBS/BSA buffer) were analyzed by flow cytometry with Hoechst 33258 stain (1 mg mL−1). Two independent experiments were performed in duplicate. In data analysis using FlowJo software (Tree Star, Ashland, OR, USA), cell aggregates and dead cells were excluded by appropriate gating. Values are given as mean and SEM.Binding of galectin-functionalized microgels to the cell surface
DLD-1 cells were seeded in the complete medium into a 24-well plate at 12 × 105 in 500 μL final volume. After 40 h of incubation (ca. 90% confluence of cells), the cells were washed with Hanks’ balanced salt solution (HBSS). Following the coupling, the galectin-functionalized microgels were dissolved in 200 μL of HBSS or 200 mM lactose (in HBSS), added to the cells, and incubated on ice for 1 h. After washing in HBSS, the samples were examined using an Olympus IX83 fluorescence microscope equipped with a 10× 2PH-UPLFLN/0.3 N.A. objective. An appropriate filter set was used for the excitation/emission of the fluorescently labeled proteins. The resulting images consist of a fluorescence and bright-field image using FiJi software.92Cytotoxicity
The cytotoxicity of HGal-3 was tested using CellCounting Kit-8 (CCK-8; Dojindo Laboratories, Kumamoto, Japan) according to the manufacturer's protocol. Briefly, DLD-1 cells (20![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 000 cells per well) were seeded in 100 μL of standard growth medium in 96-well plates. After overnight stabilization, the used medium was removed and the cells were treated with serially diluted HGal-3 (final concentration 0.5–30 μM) and cultured for 24 h. Then, 10 μL of CCK-8 solution was added to each well. After 2 h, absorbance was measured at 450 nm using Sunrise Absorbance Microplate Reader (Tecan Group Ltd, Maennedorf, Switzerland). All samples were tested at least twice. The relative viability of cells was determined, with the negative control (without substances) set at 100%.
000 cells per well) were seeded in 100 μL of standard growth medium in 96-well plates. After overnight stabilization, the used medium was removed and the cells were treated with serially diluted HGal-3 (final concentration 0.5–30 μM) and cultured for 24 h. Then, 10 μL of CCK-8 solution was added to each well. After 2 h, absorbance was measured at 450 nm using Sunrise Absorbance Microplate Reader (Tecan Group Ltd, Maennedorf, Switzerland). All samples were tested at least twice. The relative viability of cells was determined, with the negative control (without substances) set at 100%.
Conclusions
This study showcased the versatile potential of galectin fusion proteins as innovative glyco-tools for biomedically relevant applications, particularly in the context of chronic inflammation. We established a color-coded SpyCatcher galectin fusion protein library and performed in vitro binding assays on glycoproteins, ECM glycoconjugates, and the cell surfaces, which revealed their functionalities and binding affinities towards the respective ligands. SpyCatcher/SpyTag immobilization technology enabled the in vitro layer-by-layer construction of an ECM-like network and the synthesis of galectin-functionalized microgels, which were targeted to DLD-1 glycocalyx through the interaction between galectin and glycocalyx carbohydrates. These findings show the perspective for the transport of functional and biocompatible materials, such as microgels, to the desired locations. Future research should explore conjugating different galectin-binding domains with each other to elucidate the properties of newly combined constructs. Additionally, utilizing primary cells from IBD patients would provide a more accurate picture of natural cell surface glycosylation in this disease and will be the subject of the future research. In conclusion, this work demonstrates the potential use of galectin fusion proteins as novel tools in biomaterial science.Author contributions
Conceptualization, C. D., L. E., V. K., and P. B.; methodology, C. D., I. S., N. K., D. V., and M. Z. V.; writing – draft preparation, C. D., I. S., D. V., and M. Z. V.; writing – review and editing, L. E., V. K., P. B., and A. P. All authors have read and agreed to the published version of the manuscript.Data availability
The data supporting this article have been included as part of the ESI.†Conflicts of interest
The authors declare no conflicts of interest.Acknowledgements
The authors thank Nikol Kodra, Niels Hoffmann, and Truc Pham for their support in carrying out the experiments.The financial support of the project MiRAGE from the BMBF (Federal Ministry of Education and Research, Germany) within the project cluster “Future technologies for the industrial bioeconomy: focus on biohybrid technology” is greatly acknowledged. Parts of the analytical investigations were performed at the Center for Chemical Polymer Technology CPT, which is supported by the European Commission and the federal state of North Rhine-Westphalia (No. 300088302). The authors would like to thank Selin Bulut for performing microgel permeability studies, and Selin Bulut and Luis Paulo Busca Guerzoni for supplying the master molds for microfluidic microgel synthesis. Furthermore, the support of Tudor Lile as a research assistant is sincerely appreciated.
C. D. and L. E. acknowledge support from the Collaborative Research Center (CRC) grant SFB 985 project C3 from DFG (Deutsche Forschungsgemeinschaft). I. S. and A. P. thank the Federal Ministry of Education and Research (BMBF) for financial support for the MiRAGE project (Microgel Countercurrent Flow Reactor for Automated Glycan Synthesis – 031B1116B). L. E. and V. K. gratefully acknowledge the financial support from the German-Czech joint project by the German and Czech Science Foundations (DFG, project no. EL 135/19-1, and GAČR 22-00197K). P. B. gratefully acknowledges the financial support from the Ministry of Health of the Czech Republic, project NU-23-08-00307, and from the Ministry of Education, Youth and Sports of the Czech Republic, mobility project no. LUC23148 related to COST Action CA21135 - Modelling immunotherapy response and toxicity in cancer (IMMUNO-model).
References
- K. Slámová, J. Červený, Z. Mészáros, T. Friede, D. Vrbata, V. Křen and P. Bojarová, Molecules, 2023, 28(10), 4039 CrossRef PubMed.
- S. K. Patnaik, B. Potvin, S. Carlsson, D. Sturm, H. Leffler and P. Stanley, Glycobiology, 2006, 16, 305–317 CrossRef CAS PubMed.
- A. K. Ludwig, M. Michalak, Q. Xiao, U. Gilles, F. J. Medrano, H. Ma, F. G. FitzGerald, W. D. Hasley, A. Melendez-Davila, M. Liu, K. Rahimi, N. Y. Kostina, C. Rodriguez-Emmenegger, M. Moller, I. Lindner, H. Kaltner, M. Cudic, D. Reusch, J. Kopitz, A. Romero, S. Oscarson, M. L. Klein, H. J. Gabius and V. Percec, Proc. Natl. Acad. Sci. U. S. A., 2019, 116, 2837–2842 CrossRef CAS PubMed.
- M. M. Fettis, S. A. Farhadi and G. A. Hudalla, Biomater. Sci., 2019, 7, 1852–1862 RSC.
- S. A. Farhadi, E. Bracho-Sanchez, M. M. Fettis, D. T. Seroski, S. L. Freeman, A. Restuccia, B. G. Keselowsky and G. A. Hudalla, Nat. Commun., 2018, 9, 4943 CrossRef PubMed.
- S. A. Farhadi, R. Liu, M. W. Becker, E. A. Phelps and G. A. Hudalla, Proc. Natl. Acad. Sci. U. S. A., 2021, 118(19), e2024117118 CrossRef CAS PubMed.
- E. Bracho-Sanchez, F. G. Rocha, S. K. Bedingfield, B. D. Partain, S. L. Macias, M. A. Brusko, J. M. Colazo, M. M. Fettis, S. A. Farhadi, E. Y. Helm, K. Koenders, A. J. Kwiatkowski, A. Restuccia, B. S. Morales, A. Wanchoo, D. Avram, K. D. Allen, C. L. Duvall, S. M. Wallet, G. A. Hudalla and B. G. Keselowsky, Nat. Biomed. Eng., 2023, 7, 1156–1169 CrossRef CAS PubMed.
- S. Böcker and L. Elling, Glycobiology, 2017, 27, 457–468 Search PubMed.
- C. E. Kupper, S. Böcker, H. Liu, C. Adamzyk, J. van de Kamp, T. Recker, B. Lethaus, W. Jahnen-Dechent, S. Neuss, G. Muller-Newen and L. Elling, Curr. Pharm. Des., 2013, 19, 5457–5467 CrossRef CAS PubMed.
- C. Dey, P. Palm and L. Elling, Molecules, 2023, 28, 1054 CrossRef CAS PubMed.
- A. H. Keeble and M. Howarth, Chem. Sci., 2020, 11, 7281–7291 RSC.
- B. Zakeri, J. O. Fierer, E. Celik, E. C. Chittock, U. Schwarz-Linek, V. T. Moy and M. Howarth, Proc. Natl. Acad. Sci. U. S. A., 2012, 109, E690–E697 CrossRef CAS PubMed.
- A. H. Keeble, A. Banerjee, M. P. Ferla, S. C. Reddington, I. Anuar and M. Howarth, Angew. Chem., Int. Ed., 2017, 56, 16521–16525 CrossRef CAS PubMed.
- A. H. Keeble, P. Turkki, S. Stokes, I. N. A. Khairil Anuar, R. Rahikainen, V. P. Hytönen and M. Howarth, Proc. Natl. Acad. Sci. U. S. A., 2019, 116(52), 26523–26533 CrossRef CAS PubMed.
- Z. Liu, H. Zhou, W. Wang, W. Tan, Y.-X. Fu and M. Zhu, Sci. Rep., 2014, 4, 7266 CrossRef CAS PubMed.
- S. C. Reddington and M. Howarth, Curr. Opin. Chem. Biol., 2015, 29, 94–99 CrossRef CAS PubMed.
- I. N. A. Khairil Anuar, A. Banerjee, A. H. Keeble, A. Carella, G. I. Nikov and M. Howarth, Nat. Commun., 2019, 10(1), 1734 CrossRef PubMed.
- M. Fairhead, G. Veggiani, M. Lever, J. Yan, D. Mesner, C. V. Robinson, O. Dushek, P. A. van der Merwe and M. Howarth, J. Am. Chem. Soc., 2014, 136, 12355–12363 CrossRef CAS PubMed.
- C. Schoene, J. O. Fierer, S. P. Bennett and M. Howarth, Angew. Chem., Int. Ed., 2014, 53, 6101–6104 CrossRef CAS PubMed.
- Z. Botyanszki, P. K. R. Tay, P. Q. Nguyen, M. G. Nussbaumer and N. S. Joshi, Biotechnol. Bioeng., 2015, 112, 2016–2024 CrossRef CAS PubMed.
- X. Gao, J. Fang, B. Xue, L. Fu and H. Li, Biomacromolecules, 2016, 17, 2812–2819 CrossRef CAS PubMed.
- F. Sun, W. B. Zhang, A. Mahdavi, F. H. Arnold and D. A. Tirrell, Proc. Natl. Acad. Sci. U. S. A., 2014, 111, 11269–11274 CrossRef CAS PubMed.
- F. A. Plamper and W. Richtering, Acc. Chem. Res., 2017, 50, 131–140 CrossRef CAS PubMed.
- E. Gau, D. M. Mate, Z. Zou, A. Oppermann, A. Töpel, F. Jakob, D. Wöll, U. Schwaneberg and A. Pich, Biomacromolecules, 2017, 18, 2789–2798 CrossRef CAS PubMed.
- I. K. Sommerfeld, P. Palm, K. P. Hussnaetter, M. I. Pieper, S. Bulut, T. Lile, R. Wagner, J. J. Walkowiak, L. Elling and A. Pich, Biomacromolecules, 2024, 25, 3807–3822 CrossRef CAS PubMed.
- Y. Kittel, A. J. C. Kuehne and L. De Laporte, Adv. Healthcare Mater., 2022, 11, 2101989 CrossRef CAS PubMed.
- J. J. Rice, M. M. Martino, L. De Laporte, F. Tortelli, P. S. Briquez and J. A. Hubbell, Adv. Healthcare Mater., 2013, 2, 57–71 CrossRef CAS PubMed.
- A. Erfani, A. Hanna, P. Zarrintaj, S. Manouchehri, K. Weigandt, C. P. Aichele and J. D. Ramsey, Soft Matter, 2021, 17, 5349–5361 RSC.
- S. Boesveld, A. Jans, D. Rommel, M. Bartneck, M. Möller, L. Elling, C. Trautwein, P. Strnad and A. J. C. Kuehne, ACS Appl. Mater. Interfaces, 2019, 11, 25017–25023 CrossRef CAS PubMed.
- S. Boesveld, Y. Kittel, Y. Luo, A. Jans, B. Oezcifci, M. Bartneck, C. Preisinger, D. Rommel, T. Haraszti, S. P. Centeno, A. J. Boersma, L. De Laporte, C. Trautwein, A. J. C. Kuehne and P. Strnad, Adv. Healthcare Mater., 2023, 12, e2300695 CrossRef PubMed.
- C. Frantz, K. M. Stewart and V. M. Weaver, J. Cell Sci., 2010, 123, 4195–4200 CrossRef CAS PubMed.
- J. K. Kular, S. Basu and R. I. Sharma, J. Tissue Eng., 2014, 5, 2041731414557112 CrossRef PubMed.
- K. V. Mariño, A. J. Cagnoni, D. O. Croci and G. A. Rabinovich, Nat. Rev. Drug Discovery, 2023, 22, 295–316 CrossRef PubMed.
- D. Laaf, P. Bojarová, H. Pelantová, V. Křen and L. Elling, Bioconjugate Chem., 2017, 28, 2832–2840 CrossRef CAS PubMed.
- T. Fischöder, D. Laaf, C. Dey and L. Elling, Molecules, 2017, 22(8), 1320 CrossRef PubMed.
- D. A. Zacharias, J. D. Violin, A. C. Newton and R. Y. Tsien, Science, 2002, 296, 913–916 CrossRef CAS PubMed.
- N. Ahmad, H.-J. Gabius, S. André, H. Kaltner, S. Sabesan, R. Roy, B. Liu, F. Macaluso and C. F. Brewer, J. Biol. Chem., 2004, 279, 10841–10847 CrossRef CAS PubMed.
- Z. Zhao, X. Xu, H. Cheng, M. C. Miller, Z. He, H. Gu, Z. Zhang, A. Raz, K. H. Mayo, G. Tai and Y. Zhou, Proc. Natl. Acad. Sci. U. S. A., 2021, 118(19), e2021074118 CrossRef CAS PubMed.
- M. A. Hink, R. A. Griep, J. W. Borst, A. van Hoek, M. H. Eppink, A. Schots and A. J. Visser, J. Biol. Chem., 2000, 275, 17556–17560 CrossRef CAS PubMed.
- L. Pirone, M. P. Lenza, S. Di Gaetano, D. Capasso, M. Filocaso, R. Russo, C. Di Carluccio, M. Saviano, A. Silipo and E. Pedone, Int. J. Mol. Sci., 2024, 25, 2895 CrossRef CAS PubMed.
- E. Brookes and M. Rocco, Sci. Rep., 2022, 12, 7349 CrossRef CAS PubMed.
- G. J. Strous and J. Dekker, Crit. Rev. Biochem. Mol. Biol., 1992, 27, 57–92 CrossRef CAS PubMed.
- N. G. Karlsson, H. Nordman, H. Karlsson, I. Carlstedt and G. C. Hansson, Biochem. J., 1997, 326(Pt 3), 911–917 CrossRef CAS PubMed.
- M. P. Quintana-Hayashi, M. Padra, J. T. Padra, J. Benktander and S. K. Lindén, Microorganisms, 2018, 6, 55 CrossRef PubMed.
- M. Hoffmann, M. R. Hayes, J. Pietruszka and L. Elling, Glycoconjugate J., 2020, 37, 457–470 CrossRef CAS PubMed.
- S. R. Stowell, C. M. Arthur, P. Mehta, K. A. Slanina, O. Blixt, H. Leffler, D. F. Smith and R. D. Cummings, J. Biol. Chem., 2008, 283, 10109–10123 CrossRef CAS PubMed.
- J. I. Quintana, S. Delgado, R. Núñez-Franco, F. J. Cañada, G. Jiménez-Osés, J. Jiménez-Barbero and A. Ardá, Front. Chem., 2021, 9, 193 Search PubMed.
- O. A. Vokhmyanina, E. M. Rapoport, S. André, V. V. Severov, I. Ryzhov, G. V. Pazynina, E. Korchagina, H. J. Gabius and N. V. Bovin, Glycobiology, 2012, 22, 1207–1217 CrossRef CAS PubMed.
- R. N. Knibbs, F. Perini and I. J. Goldstein, Biochemistry, 1989, 28, 6379–6392 CrossRef CAS PubMed.
- D. Liu, S. Wang, J. Zhang, W. Xiao, C. H. Miao, B. A. Konkle, X.-F. Wan and L. Li, Front. Chem., 2021, 9(415), 691217 CrossRef CAS PubMed.
- T. Basak, L. Vega-Montoto, L. J. Zimmerman, D. L. Tabb, B. G. Hudson and R. M. Vanacore, J. Proteome Res., 2016, 15, 245–258 CrossRef CAS PubMed.
- M. F. Troncoso, M. T. Elola, A. G. Blidner, L. Sarrias, M. V. Espelt and G. A. Rabinovich, J. Biol. Chem., 2023, 299, 105400 CrossRef CAS PubMed.
- K. Bum-Erdene, H. Leffler, U. J. Nilsson and H. Blanchard, Sci. Rep., 2016, 6, 20289 CrossRef CAS PubMed.
- M. I. Nielsen, J. Stegmayr, O. C. Grant, Z. Yang, U. J. Nilsson, I. Boos, M. C. Carlsson, R. J. Woods, C. Unverzagt, H. Leffler and H. H. Wandall, J. Biol. Chem., 2018, 293, 20249–20262 CrossRef CAS PubMed.
- H. Ideo, A. Seko, I. Ishizuka and K. Yamashita, Glycobiology, 2003, 13, 713–723 CrossRef CAS PubMed.
- S. Carlsson, C. T. Oberg, M. C. Carlsson, A. Sundin, U. J. Nilsson, D. Smith, R. D. Cummings, J. Almkvist, A. Karlsson and H. Leffler, Glycobiology, 2007, 17, 663–676 CrossRef CAS PubMed.
- S. Kumar, M. Frank and R. Schwartz-Albiez, PLoS One, 2013, 8, e59761 CrossRef CAS PubMed.
- M. T. Elola, A. G. Blidner, F. Ferragut, C. Bracalente and G. A. Rabinovich, Biochem. J., 2015, 469, 1–16 CrossRef CAS PubMed.
- J. He and L. G. Baum, Methods Enzymol., 2006, 417, 247–256 CAS.
- I. R. Nabi, J. Shankar and J. W. Dennis, J. Cell Sci., 2015, 128, 2213–2219 CrossRef CAS PubMed.
- R. Y. Yang, G. A. Rabinovich and F. T. Liu, Expert Rev. Mol. Med., 2008, 10, e17 CrossRef PubMed.
- M. V. Beer, C. Rech, P. Gasteier, B. Sauerzapfe, J. Salber, A. Ewald, M. Möller, L. Elling and J. Groll, Adv. Healthcare Mater., 2013, 2, 306–311 CrossRef CAS PubMed.
- C. Muglia, N. Mercer, M. A. Toscano, M. Schattner, R. Pozner, J. P. Cerliani, R. P. Gobbi, G. A. Rabinovich and G. H. Docena, Cell Death Dis., 2011, 2, e163–e163 CrossRef CAS PubMed.
- M. Puthenedam, F. Wu, A. Shetye, A. Michaels, K.-J. Rhee and J. H. Kwon, Inflamm. Bowel Dis., 2010, 17, 260–267 CrossRef PubMed.
- A. Joshi, A. Soni and S. Acharya, In Vitro Models, 2022, 1, 213–227 CrossRef PubMed.
- D. Ahmed, P. W. Eide, I. A. Eilertsen, S. A. Danielsen, M. Eknæs, M. Hektoen, G. E. Lind and R. A. Lothe, Oncogenesis, 2013, 2, e71–e71 CrossRef CAS PubMed.
- M. Lee, Y.-S. Kim, S. Lim, S.-H. Shin, I. Kim, J. Kim, M. Choi, J. H. Kim, S.-J. Koh, J.-W. Park and H.-W. Shin, Nat. Commun., 2023, 14, 2363 CrossRef CAS PubMed.
- M. Romano, F. De Francesco, L. Zarantonello, C. Ruffolo, G. A. Ferraro, G. Zanus, A. Giordano, N. Bassi and U. Cillo, Anticancer Res., 2016, 36, 1447 CAS.
- V. Heine, M. Hovorková, M. Vlachová, M. Filipová, L. Bumba, O. Janoušková, M. Hubálek, J. Cvačka, L. Petrásková, H. Pelantová, V. Křen, L. Elling and P. Bojarová, Eur. J. Med. Chem., 2021, 220, 113500 CrossRef CAS PubMed.
- D. Vrbata, J. Červený, N. Kulik, M. Hovorková, S. Balogová, M. Vlachová, H. Pelantová, V. Křen and P. Bojarová, Bioorg. Chem., 2024, 145, 107231 CrossRef CAS PubMed.
- S. Holst, A. J. Deuss, G. W. van Pelt, S. J. van Vliet, J. J. Garcia-Vallejo, C. A. Koeleman, A. M. Deelder, W. E. Mesker, R. A. Tollenaar, Y. Rombouts and M. Wuhrer, Mol. Cell. Proteomics, 2016, 15, 124–140 CrossRef CAS PubMed.
- D. Wang, V. Kuzyk, K. Madunić, T. Zhang, O. A. Mayboroda, M. Wuhrer and G. S. M. Lageveen-Kammeijer, Int. J. Mol. Sci., 2023, 24, 4842 CrossRef CAS PubMed.
- N. Billinton and A. W. Knight, Anal. Biochem., 2001, 291, 175–197 CrossRef CAS PubMed.
- A. K. Michel, P. Nangia-Makker, A. Raz and M. J. Cloninger, ChemBioChem, 2014, 15, 2106–2112 CrossRef CAS PubMed.
- K. L. Wu, C. M. Kuo, E. Y. Huang, H. M. Pan, C. C. Huang, Y. F. Chen, C. C. Hsiao and K. D. Yang, Am. J. Transl. Res., 2018, 10, 2402–2412 CAS.
- A. Hittelet, H. Legendre, N. Nagy, Y. Bronckart, J.-C. Pector, I. Salmon, P. Yeaton, H.-J. Gabius, R. Kiss and I. Camby, Int. J. Cancer, 2003, 103, 370–379 CrossRef CAS PubMed.
- M. R. Tavares, M. Bláhová, L. Sedláková, L. Elling, H. Pelantová, R. Konefał, T. Etrych, V. Křen, P. Bojarová and P. Chytil, Biomacromolecules, 2020, 21, 641–652 CrossRef CAS PubMed.
- O. Piwocka, M. Musielak, K. Ampuła, I. Piotrowski, B. Adamczyk, M. Fundowicz, W. M. Suchorska and J. Malicki, Cancer Cell Int., 2024, 24, 28 CrossRef PubMed.
- S. Udenfriend, S. Stein, P. Böhlen, W. Dairman, W. Leimgruber and M. Weigele, Science, 1972, 178, 871–872 CrossRef CAS PubMed.
- S. Bulut, S. H. Jung, T. Bissing, F. Schmitt, M. Bund, S. Braun and A. Pich, Small, 2023, 19, e2303783 CrossRef PubMed.
- M. Cho and R. D. Cummings, Biochemistry, 1996, 35, 13081–13088 CrossRef CAS PubMed.
- H. M. Jan, S. C. Wu, C. J. Stowell, M. L. Vallecillo-Zúniga, A. Paul, K. R. Patel, S. Muthusamy, H. Y. Lin, D. Ayona, R. P. Jajosky, S. P. Varadkar, H. Nakahara, R. Chan, D. Bhave, W. J. Lane, M. Y. Yeung, M. A. Hollenhorst, S. Rakoff-Nahoum, R. D. Cummings, C. M. Arthur and S. R. Stowell, Mol. Cell. Proteomics, 2024, 23, 100747 CrossRef CAS PubMed.
- D. E. Strongin, B. Bevis, N. Khuong, M. E. Downing, R. L. Strack, K. Sundaram, B. S. Glick and R. J. Keenan, Protein Eng., Des. Sel., 2007, 20, 525–534 CrossRef CAS PubMed.
- M. M. Bradford, Anal. Biochem., 1976, 72, 248–254 CrossRef CAS PubMed.
- N. C. Shaner, R. E. Campbell, P. A. Steinbach, B. N. G. Giepmans, A. E. Palmer and R. Y. Tsien, Nat. Biotechnol., 2004, 22, 1567–1572 CrossRef CAS PubMed.
- S. Bulut, D. Günther, M. Bund, C. Haats, T. Bissing, C. Bastard, M. Wessling, L. De Laporte and A. Pich, Adv. Healthcare Mater., 2024, 13, 2302957 CAS.
- V. La Verde, P. Dominici and A. Astegno, Bio-Protoc., 2017, 7, e2230 Search PubMed.
- T. Scheidt, T. Kartanas, Q. Peter, M. M. Schneider, K. L. Saar, T. Müller, P. K. Challa, A. Levin, S. Devenish and T. P. J. Knowles, Lab Chip, 2020, 20, 2663–2673 RSC.
- T. F. O'Connor, P. G. Debenedetti and J. D. Carbeck, J. Am. Chem. Soc., 2004, 126, 11794–11795 CrossRef PubMed.
- S. H. Jung, S. Bulut, L. P. B. Busca Guerzoni, D. Günther, S. Braun, L. De Laporte and A. Pich, J. Colloid Interface Sci., 2022, 617, 409–421 CrossRef CAS PubMed.
- S. A. J. Watkin, A. Hashemi, D. R. Thomson, V. M. Nock, R. C. J. Dobson and F. G. Pearce, Methods Enzymol., 2023, 682, 429–464 CAS.
- M. Abramoff, P. Magalhães and S. J. Ram, Biophotonics Intern., 2003, 11, 36–42 Search PubMed.
Footnote |
| † Electronic supplementary information (ESI) available. See DOI: https://doi.org/10.1039/d4bm01148a |
| This journal is © The Royal Society of Chemistry 2025 |