Open Access Article
Open Access ArticleSensitive detection of circulating methylated SEPT9 in hepatocellular carcinoma patients using a novel quantitative PCR assay†
Yen Hai
Tran
a,
Trang Thuy
Dao
b,
Ung Dinh
Nguyen
b,
Thien Ba
Tran
 c,
Loi Phuc
Luu
d,
Huy Quang
Duong
a and
Tho H.
Ho
c,
Loi Phuc
Luu
d,
Huy Quang
Duong
a and
Tho H.
Ho
 *be
*be
aDepartment of Gastroenterology and Hepatology, 103 Military Hospital, Vietnam Military Medical University, Hanoi, Vietnam. E-mail: tranhaiyen1812@gmail.com; huyduonghvqy@gmail.com; Tel: +84-988474574 Tel: +84-912626081
bDepartment of Genomics and Cytogenetics, Institute of Biomedicine and Pharmacy (IBP), Vietnam Military Medical University, Hanoi, Vietnam. E-mail: hohuutho@vmmu.edu.vn; daothuytrang392@gmail.com; dr.ungd4.vmmu@gmail.com; Tel: +84-916-9327-68 Tel: +84-326628152 Tel: +84-984078979
cOxford University Clinical Research Unit, Ho Chi Minh city, Vietnam. E-mail: thientb@oucru.org; Tel: +84-81-545-1019
dInstitute for Applied Research in Health Sciences and Aging (ARiHA), Thong Nhat Hospital, Ho Chi Minh City 70000, Vietnam. E-mail: luu.p.loi@googlemail.com
eDepartment of Microbiology, 103 Military Hospital, Vietnam Military Medical University, Hanoi, Vietnam. E-mail: hohuutho@vmmu.edu.vn; Tel: +84-916932768
First published on 14th February 2025
Abstract
Hepatocellular carcinoma (HCC) is a leading cause of cancer-related deaths worldwide. Early detection is crucial, yet reliable biomarkers are limited. Methylated SEPT9 (mSEPT9) has emerged as a promising biomarker for HCC. Building upon previous ExBP technology, we enhanced the semi-nested realtime PCR assay by integrating TaqMan probes, enabling quantitative detection of mSEPT9 in plasma samples of HCC patients. The assay was validated using synthetic DNA standards and plasma samples from 49 HCC patients, 20 chronic liver disease (CLD) patients, and 32 healthy donors (HDs). Our assay demonstrated sensitivity in detecting methylation ratios as low as 1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 100
100![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 000. The assay showed a strong linear correlation between Ct values and methylation levels over four orders of magnitude (R2 = 0.96178), indicating robust quantification. Clinically, the assay revealed significant differences in ΔCt values between HCC patients (median ΔCt = 19.55) and controls (CLD: 29.32 and HDs: 26.19, p < 0.005). ROC analysis for HCC vs. controls yielded an AUC of 0.729, with 77.55% sensitivity and 59.62% specificity at the optimal cutoff (≤25.98). Notably, the assay identified 72.73% of HCC cases with AFP levels below 20 ng mL−1, underscoring its potential in detecting AFP-negative cases. These findings suggest that the novel mSEPT9 assay is a sensitive and specific tool for early HCC detection, offering prognostic value for clinical monitoring.
000. The assay showed a strong linear correlation between Ct values and methylation levels over four orders of magnitude (R2 = 0.96178), indicating robust quantification. Clinically, the assay revealed significant differences in ΔCt values between HCC patients (median ΔCt = 19.55) and controls (CLD: 29.32 and HDs: 26.19, p < 0.005). ROC analysis for HCC vs. controls yielded an AUC of 0.729, with 77.55% sensitivity and 59.62% specificity at the optimal cutoff (≤25.98). Notably, the assay identified 72.73% of HCC cases with AFP levels below 20 ng mL−1, underscoring its potential in detecting AFP-negative cases. These findings suggest that the novel mSEPT9 assay is a sensitive and specific tool for early HCC detection, offering prognostic value for clinical monitoring.
1 Introduction
Hepatocellular carcinoma (HCC) is a prevalent malignant disease, ranking as the fifth most common cancer and the third leading cause of cancer-related deaths worldwide in both men and women.1 The absolute number of primary cancer cases, with HCC accounting for 75% to 85%, is expected to increase in 30 countries under investigation worldwide by 2030.2 Major risk factors for HCC include cirrhosis, hepatitis B virus (HBV), hepatitis C virus (HCV) infections, alcoholic liver disease, and non-alcoholic fatty liver disease.3 Early detection of HCC is crucial for improving patient survival, as international guidelines recommend regular surveillance of at-risk populations.4 Alpha-fetoprotein (AFP) is one of the most widely used serum biomarkers for HCC diagnosis and prognosis, particularly in cases with elevated levels. However, its limited sensitivity and specificity reduce its effectiveness for detecting early-stage HCC, highlighting the pressing need for reliable biomarkers and novel screening methods to improve early detection.5Epigenetic changes are a defining feature of human cancers, including HCC.6,7 One such alteration is the hypermethylation of the septin 9 (SEPT9) gene, which plays a crucial role in cell division regulation and acts as a tumor suppressor. This epigenetic modification has been associated with the development of liver cancer.8–10 In normal tissues, SEPT9 is widely expressed, but in liver cancer, its expression is often reduced or completely silenced due to abnormal promoter hypermethylation.11,12 Research focusing on the epigenome has pinpointed SEPT9 as an important epigenetic contributor to liver carcinogenesis, largely due to promoter hypermethylation.10
Liquid biopsy, a non-invasive diagnostic tool, has emerged as a promising method for detecting circulating tumor DNA (ctDNA) in patients with cancer. Methylated ctDNA, in particular, is one of the most intensively investigated targets for liquid biopsy due to its early occurrence in carcinogenesis. Multiple studies have demonstrated the potential of methylated gene markers in the diagnosis of HCC, offering new avenues for early detection.13,14 In the context of CRC, the US Food and Drug Administration (FDA)-approved Epi proColon assay, which targets methylated SEPT9, has demonstrated utility in CRC screening. However, the application of this technology to HCC is relatively novel. Furthermore, current methods, including Epi proColon, rely on complex assay designs involving non-extendable blocking probes with costly modified nucleotides, limiting their practical use for widespread screening. Additionally, these approaches often lack the sensitivity needed for detecting low levels of circulating tumor DNA in early-stage cancers and fail to provide quantitative measurements, which are critical for monitoring disease progression.15
In our previous study, we introduced ExBP technology16 for qualitative detection of mSEPT9 in colorectal cancer.17 However, this approach lacked the capacity for quantitative analysis, which is critical for monitoring tumor burden and disease progression. In this study, we advance this technology by integrating TaqMan probes, enabling robust and reliable quantification of mSEPT9 levels in HCC plasma samples.
Unlike the previous qualitative assay, this newly developed method allows for relative quantification of mSEPT9 levels based on delta Ct values, which is critical for assessing tumor burden and monitoring disease progression in HCC. The assay also utilizes TaqMan probes, which improve both sensitivity and specificity, particularly when detecting low concentrations of cfDNA in liquid biopsy samples. This represents a significant advancement not only in the ability to detect mSEPT9 but also in providing a quantitative approach that can better support clinical decision-making. Additionally, the cost-effective nature of the assay and its application to HCC expand the versatility of ExBP technology beyond colorectal cancer, demonstrating its broader utility across different cancer types and marking a critical step forward in non-invasive cancer diagnostics.
2 Materials and methods
2.1. Study participants
A total of 101 participants were enrolled in this study between July 2021 and April 2024 at the 103 Military Hospital. Blood plasma samples were collected from 49 patients diagnosed with hepatocellular carcinoma (HCC) and 52 control individuals (Table 1). The control group comprised 32 healthy donors (HDs) and 20 patients with chronic liver diseases (CLDs), such as cirrhosis and hepatitis. The diagnosis of HCC followed the guidelines established by the American Association for the Study of Liver Diseases (AASLD),4,18 with liver biopsies performed to confirm any inconclusive cases. The Barcelona Clinic Liver Cancer (BCLC) staging system was used to evaluate the tumor stage of HCC patients.5 All control individuals were confirmed to be free of liver cancer following a thorough physical examination. CLD patients needed a confirmed diagnosis of chronic liver disease such as cirrhosis or hepatitis and were monitored for 6 months with liver ultrasound to rule out HCC. Healthy donors had no history of liver disease as confirmed by a thorough physical examination. Exclusion criteria included a history of other malignancies, prior treatment for HCC or other liver diseases that might interfere with the study results, pregnant or breastfeeding women, and any other condition that, in the opinion of the investigator, could interfere with the results of the study or the safety of the participants.| Variable | HCC (n = 49) | CLD (n = 20) | HD (n = 32) |
|---|---|---|---|
| a N/A: not applicable. | |||
| Age median (IQR) | 62 (50.0–69.5) | 45 (41.5–59.5) | 48.5 (37.25–60.5) |
![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) |
|||
| Gender | |||
| Male | 48 (98%) | 19 (95%) | 29 (90.6%) |
| Female | 1 (2%) | 1 (5%) | 3 (9.4%) |
![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) |
|||
| Etiology | |||
| HBV | 40 (81.6%) | 20 (100%) | N/A |
| HCV | 7 (14.3%) | N/A | |
| Alcoholic | 11 (22.4%) | N/A | |
![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) |
|||
| Liver enzymes | |||
| AST (U/L) median (IQR) | 49.76 (35.41–96.97) | 29.97 (25.77–50.91) | 25.48 (21.6–21.62) |
| ALT (U/L) median (IQR) | 43.44 (32.88–65.50) | 34.15 (25.2–46.43) | 24 (17.3–26.9) |
| Total bilirubin (mmol l−1) median (IQR) | 15.5 (11.97–21.35) | N/A | N/A |
| Albumin (g l−1) median (IQR) (g l−1) | 38 (35.87–40.82) | N/A | N/A |
| PLT (G/L)median (IQR) | 161 (114–191) | 173 (133.75–210.75) | N/A |
![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) |
|||
| Cirrhosis | |||
| Yes | 22 (44.9%) | 4 (20%) | N/A |
| No | 27 (55.1%) | 16 (80%) | N/A |
![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) |
|||
| Child pugh | |||
| A | 40 (81.6%) | N/A | N/A |
| B | 7 (14.3%) | N/A | N/A |
| C | 2 (4.1%) | N/A | N/A |
| Tumor size (mm) median (IQR) | 42 (27.5–71.5) | N/A | N/A |
![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) |
|||
| Tumor number | |||
| Single | 27 (55.1%) | N/A | N/A |
| Multi | 22 (44.9%) | N/A | N/A |
| AFP (ng ml−1) median (IQR) | 26.59 (9.2–1125.32) | 3.3 (3.31–4.28) | N/A |
![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) |
|||
| BCLC stage | |||
| 0-A | 27 (55.1%) | N/A | N/A |
| B | 8 (16.3%) | N/A | N/A |
| C | 12 (24.6%) | N/A | N/A |
| D | 2 (4.1%) | N/A | N/A |
All participants were thoroughly informed about the objectives and nature of the research, and they voluntarily agreed to participate. This informed consent process ensured a robust framework for analyzing the results, thereby supporting the validity and reliability of the study findings.
2.2. Sample collection, processing and preparation of standard DNA samples
Peripheral blood samples, approximately 10 mL each, were collected into K2 EDTA tubes to prevent clotting and processed within six hours to separate plasma. Plasma separation involved centrifugation at 120g for 20 minutes at 4 °C, after which the plasma was carefully transferred to clean 1.5 mL tubes to avoid contamination from cellular components. The plasma samples were stored at −80 °C until further analysis.For DNA extraction, 2 mL of plasma from each sample was processed using a QIAamp Circulating Nucleic Acid Kit (Qiagen, Germany). A total of 200 μL of Proteinase K solution and 1.6 mL of Buffer ACL containing carrier RNA were added to the plasma samples, followed by vortexing for 30 seconds. The lysate was incubated at 60 °C for 30 minutes to ensure efficient lysis. Afterward, 3.6 mL of Buffer ACB was added, and the mixture was incubated on ice for 5 minutes to facilitate DNA binding. The lysate was loaded onto binding columns, and DNA was purified through sequential washes with 600 μL of Buffer ACW1, 750 μL of Buffer ACW2, and 750 μL of 96–100% ethanol. DNA was eluted with 50 μL of Buffer AVE after incubation at room temperature for 3 minutes and centrifugation at 14![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 000 rpm for 1 minute. The extracted DNA was stored at −20 °C for subsequent bisulfite conversion.
000 rpm for 1 minute. The extracted DNA was stored at −20 °C for subsequent bisulfite conversion.
Bisulfite conversion of the extracted DNA was performed using an EZ DNA Methylation-Gold™ Kit (Zymo Research, Irvine, USA). The conversion reaction, in a total volume of 150 μL, included 20 μL of DNA sample and 130 μL of CT Conversion Reagent. Samples were subjected to thermal cycling conditions as follows: denaturation at 98 °C for 10 minutes, sulfonation at 64 °C for 150 minutes, and a final hold at 4 °C. The bisulfite-converted DNA was purified using spin columns provided in the kit and eluted in 25 μL of Buffer EB. This procedure converted unmethylated cytosines to uracil, enabling specific detection of methylated DNA during downstream methylation-specific analyses. Eluted DNA was stored at −80 °C until further use in quantitative PCR experiments.
For quantitative evaluation of the novel PCR assay's sensitivity to detect methylated SEPT9 (mSEPT9), synthetic DNA standards mimicking bisulfite-converted sequences of the SEPT9 gene were prepared. Methylated and unmethylated DNA calibrators were synthesized according to methods described previously.17 A series of standard samples was prepared by mixing a constant concentration of unmethylated SEPT9 DNA (105 copies per μL) with varying concentrations of methylated SEPT9 DNA to achieve methylated ratios ranging from 10![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1 to 1
1 to 1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 100
100![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 000. These standard samples allowed for the precise evaluation of the assay's responsiveness to different levels of DNA methylation, ensuring that it could detect even low levels of tumor-derived DNA in a clinical setting.
000. These standard samples allowed for the precise evaluation of the assay's responsiveness to different levels of DNA methylation, ensuring that it could detect even low levels of tumor-derived DNA in a clinical setting.
2.3. Quantitative PCR for detection of mSEPT9
This study differs from our earlier work17 by employing TaqMan probes alongside ExBP technology. The addition of TaqMan probes enables fluorescence-based quantification during realtime PCR cycles, which allows for precise measurement of mSEPT9 levels. This quantitative capability represents a key improvement over the qualitative detection method previously described.17The quantitative PCR assay for detecting methylated SEPT9 (mSEPT9) was designed as a two-round amplification process to achieve high specificity and sensitivity, particularly for low concentrations of circulating tumor DNA.
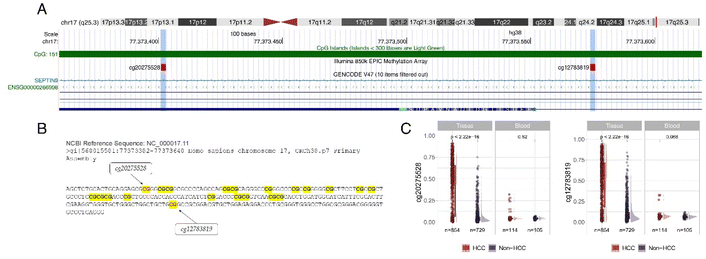
In silico validation of SEPT9 DNA methylation: To identify regions of the SEPT9 gene with differential methylation between HCC and non-HCC, an in silico analysis was conducted using available methylation databases. This analysis was performed using NCBI BLAST and the UCSC Genome Browser to examine existing data on SEPT9 methylation. The SEPT9 gene sequence, located on chromosome 17, was scrutinized, revealing specific CpG sites, cg20275528 and cg12783819 (Fig. 1A), situated within the 5′UTR, body, and first exon regions of the gene (chr17: 77373401–77373403 and chr17: 77373574–77373576, respectively). These CpG sites are located within a CpG island, indicating significant regulatory potential (Fig. 1B). The analysis identified these key CpG sites as being differentially methylated in HCC patients compared to controls (Fig. 1C).
First round of amplification: the reaction mixture for the first round consisted of 1× HTOne MaX qPCR Green Master Mix (HT Biotec, Vietnam) with forward and reverse primers at a 0.1 μM concentration. The design of these primers and thermocycling conditions follows the methodology previously described in our previous study,17 targeting non-CpG regions in the SEPT9 gene to allow amplification of both methylated and unmethylated DNA. The amplified products were diluted 20-fold and used for the second round of amplification.
Second round of amplification: the second round of amplification used a semi-nested realtime PCR, incorporating the forward primer (SEPT9/Fi) and reverse primer (SEPT9/Ri), both previously described in our colorectal cancer detection work.17 However, unlike the previous method, which relied on DNA melting curve analysis for qualitative detection, this assay further introduces a TaqMan probe specific to the methylated SEPT9 sequence (Table 2). The TaqMan probe allows for relative quantification of mSEPT9, representing a significant advancement over the earlier qualitative approach. The reaction mixture included 1× HTOne MaX qPCR Green Master Mix, a forward primer (0.1 μM), a reverse primer (0.2 μM), and a TaqMan probe (0.2 μM). Amplification was performed using a Rotor-Gene Q instrument (Qiagen, Germany) with the following conditions: initial denaturation at 95 °C for 15 minutes, followed by one cycle at 94 °C for 15 seconds, 56 °C for 30 seconds, and 72 °C for 30 seconds and 45 cycles of 94 °C for 15 seconds, 60 °C for 30 seconds, and 72 °C for 30 seconds. Fluorescence detection occurred at 94 °C to avoid interference from non-specific SYBR Green signals.
| Amplification round | Primers/probe | Sequence (5’–3′) |
|---|---|---|
| 2nd PCR round | TaqMan probe (SEPT9_P) | FAM-TTAACCGCGAAATCCGAC-BHQ1 |
| Total SEPT9 quantification | Forward primer (SEPT9_Ft) | AATCCGAAATAATCCCATCCAACTA |
| Reverse primer (SEPT9_Ri) | CACACAGGAAACAGCTATGACCATG |
Quantification of total SEPT9 levels: to ensure accurate interpretation of the methylation analysis, total SEPT9 gene levels are quantified concurrently with the methylation-specific testing. This is done in a parallel PCR reaction using the same reverse primer SEPT9_Ri and a specific forward primer (SEPT9_Ft), following the thermocycling conditions of the second round. Fluorescence signals, emitted from the SYBR Green-like dye binding to double-stranded DNA, are collected at 72 °C during the elongation step of the thermocycling conditions in the second round of PCR, ensuring reliable quantification of total SEPT9 levels.
The relative methylation level of SEPT9 was calculated using the ΔCt method, which provides a comparative measure of the methylated SEPT9 (mSEPT9) fraction relative to the total SEPT9 gene. For each plasma sample, the Ct value obtained from the mSEPT9-specific reaction (amplifying only methylated DNA) was subtracted from the Ct value of the total SEPT9 reaction (amplifying both methylated and unmethylated DNA). This ΔCt value reflects the proportion of mSEPT9 present, where a lower ΔCt value indicates a higher relative methylation level due to earlier amplification of mSEPT9-specific sequences.
To ensure accuracy, each sample was analyzed in duplicate, and the average ΔCt value was calculated for final analysis. For samples where no amplification signal was detected, a default Ct value of 45 cycles was assigned, representing the maximum cycle number used in the assay.
2.4. Statistical analysis
Statistical analyses for this study were conducted using MedCalc software version 20.019 (MedCalc Software Ltd, Ostend, Belgium). Sensitivity and specificity were calculated for the quantitative PCR assay based on the plasma sample results from HCC patients and control individuals (including CLD and HD groups). Differences between groups were analyzed using the Mann–Whitney U test for non-parametric data.To evaluate the diagnostic accuracy of the mSEPT9 assay, receiver operating characteristic (ROC) curve analysis was performed. The area under the ROC curve (AUC) was calculated to determine the assay's effectiveness in distinguishing HCC patients from controls. The optimal cutoff value for mSEPT9 detection was identified using the Youden index, which maximizes the sum of sensitivity and specificity. Statistical significance was set at p < 0.05.
3 Results & discussion
3.1. Overview of the novel realtime PCR assay for sensitive mSEPT9 quantification
In this study, we developed an enhanced semi-nested realtime PCR assay for the quantitative detection of mSEPT9 in hepatocellular carcinoma (HCC). The assay builds on our previous work, which introduced the use of extendable blocking probe (ExBP) technology to improve the detection of methylated SEPT9 (mSEPT9) in colorectal cancer (CRC).17 However, while the previous assay was qualitative, the present work focuses on achieving quantitative detection using a TaqMan probe, allowing for relative quantification of mSEPT9 levels based on delta Ct values, which is crucial for assessing tumor burden and monitoring disease progression.In contrast to our earlier work, the second round of amplification employs a TaqMan probe specific to the methylated SEPT9 sequence, enabling precise detection and relative quantification of mSEPT9 in plasma samples. Fluorescence is measured during each cycle of amplification, and the Ct values are used to quantify mSEPT9 levels in relation to total SEPT9 levels (Fig. 2). To eliminate interference from SYBR Green fluorescence originating from the master mix, the fluorescence signal from the TaqMan probe is collected at 94 °C. This ensures reliable detection by isolating the signal specific to the TaqMan probe.
The integration of ExBP and TaqMan probe technology enhances the overall performance of the assay by improving specificity and enabling relative quantification of mSEPT9 levels. While the detection limit remains consistent with previous methods, the addition of TaqMan probes provides a fluorescence-based quantification capability, offering more precise analysis of methylation levels. This approach addresses the limitations of traditional qualitative assays by offering a cost-effective and reliable method for quantitative analysis, providing a valuable tool for both clinical diagnostics and research applications.
The relative quantification method in this study employs the ΔCt approach, comparing Ct values from methylated SEPT9 amplification to those from total SEPT9, ensuring proportionality and reliability by using primers targeting the same genomic region. This design minimizes variability, enhances amplification efficiency, and eliminates the need for external reference genes, enabling robust detection of methylation levels. By normalizing within the same gene, the assay provides a precise and sensitive measure, particularly valuable for identifying low-abundance methylation signals critical for early HCC diagnosis.
3.2. Assay validation with standard samples
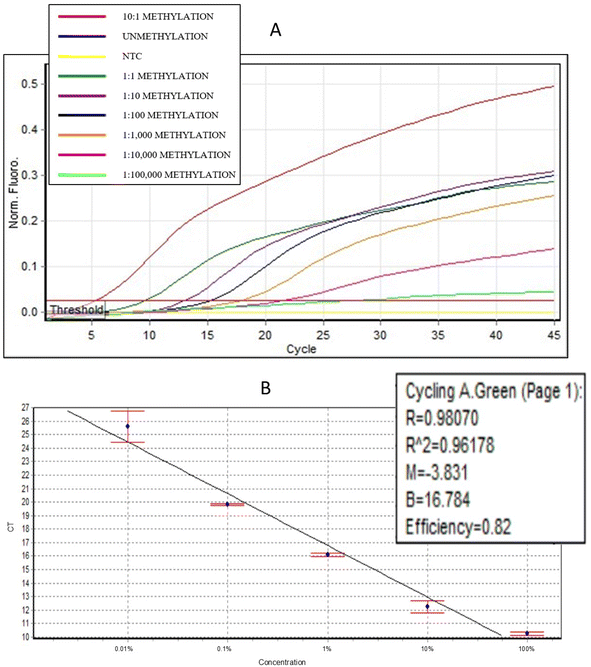
To validate the newly developed semi-nested PCR assay for detecting mSEPT9, we conducted a thorough evaluation using synthetic DNA standards (Fig. 3). These synthetic standards represented bisulfite-converted SEPT9 gene sequences, including fully methylated and a range of methylated-to-unmethylated mixtures. The mixtures maintained a constant concentration of unmethylated SEPT9 DNA (105 copies per μL) and varied concentrations of mSEPT9 DNA, creating methylation ratios from 10![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1 to 1
1 to 1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 100
100![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 000.
000.
The assay successfully detected mSEPT9 across the full range of methylated-to-unmethylated DNA ratios. For example, the 10![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1 methylated sample showed early amplification, indicating the assay's ability to detect relatively high methylation levels. Lower methylation ratios, such as 1
1 methylated sample showed early amplification, indicating the assay's ability to detect relatively high methylation levels. Lower methylation ratios, such as 1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 10, 1
10, 1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 100, and even 1
100, and even 1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 100
100![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 000, produced progressively higher Ct values, demonstrating the assay's sensitivity to minimal methylation. The fully unmethylated sample exhibited no amplification, confirming the high specificity of the assay in excluding non-methylated sequences (Fig. 3A).
000, produced progressively higher Ct values, demonstrating the assay's sensitivity to minimal methylation. The fully unmethylated sample exhibited no amplification, confirming the high specificity of the assay in excluding non-methylated sequences (Fig. 3A).
The results showed a strong linear correlation between the Ct values and methylation levels across a dynamic range of four orders of magnitude (R2 = 0.96178), underscoring the assay's robustness (Fig. 3B). For synthetic standard samples, the Ct values for total SEPT9 remained uniform across all tested methylation ratios, providing a stable baseline for calculating ΔCt values. This uniformity directly enabled the observed strong linear relationship between ΔCt values and methylation concentrations, as illustrated in supplementary Fig. 1S.† While both Ct and ΔCt values demonstrated linear relationships with methylation levels for synthetic standards, the ΔCt value is particularly valuable in clinical plasma samples, where variations in total SEPT9 levels are more likely due to individual differences in circulating DNA composition. By normalizing for these variations, ΔCt provides a more reliable measure of relative methylation levels in real-world clinical settings, ensuring higher accuracy in detecting mSEPT9 in plasma samples.
To further assess the reproducibility of the assay, we analyzed the coefficient of variation (CV) values across a range of methylation ratios, including both low and higher concentrations. At the lowest tested methylation ratio of 0.001% (1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 100
100![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 000), the assay achieved a CV of 12.37%, demonstrating acceptable reproducibility under challenging low-concentration conditions. For higher methylation ratios ranging from 0.01% to 10%, the CV values were consistently robust, ranging from 1.14% to 5.56%. These findings highlight the assay's reliability across a wide dynamic range, ensuring its applicability for detecting both trace and abundant levels of methylated SEPT9. This performance further reinforces the assay's potential as a sensitive and reliable tool for quantitative mSEPT9 analysis in clinical and research settings.
000), the assay achieved a CV of 12.37%, demonstrating acceptable reproducibility under challenging low-concentration conditions. For higher methylation ratios ranging from 0.01% to 10%, the CV values were consistently robust, ranging from 1.14% to 5.56%. These findings highlight the assay's reliability across a wide dynamic range, ensuring its applicability for detecting both trace and abundant levels of methylated SEPT9. This performance further reinforces the assay's potential as a sensitive and reliable tool for quantitative mSEPT9 analysis in clinical and research settings.
3.3. Clinical performance of novel realtime PCR assay using plasma samples
Following the promising results from the initial evaluation using synthetic DNA standards, the assay was further validated on blood plasma samples from patients with hepatocellular carcinoma (HCC) and control groups, including those with chronic liver disease (CLD) and healthy donors (HDs). This comprehensive analysis aimed to assess the assay's diagnostic utility in a clinical setting, comparing ΔCt values between the groups and evaluating the assay's performance using ROC curve analysis.The ΔCt values showed significant differences among the hepatocellular carcinoma (HCC), chronic liver disease (CLD), and healthy donor (HD) groups (Fig. 4A). HCC patients had a median ΔCt of 19.55, significantly lower than the CLD group's median of 29.32 (p = 0.0004) and the HD group's median of 26.19 (p = 0.0024). The 95% confidence intervals (CIs) for the medians were 13.87 to 25.82 for HCC, 21.50 to 31.24 for CLD, and 20.07 to 29.16 for HDs. These differences highlight the lower ΔCt values in HCC patients compared to the control groups.
The ROC curve analysis for ΔCt values in HCC vs. control groups (CLD + HD) yielded an AUC of 0.729 (p < 0.0001), indicating acceptable diagnostic accuracy (Fig. 4B). The optimal cutoff value was ≤25.98, providing a sensitivity of 77.55% and a specificity of 59.62%. When analyzed separately, the ROC curve for HCC vs. CLD yielded an AUC of 0.774, suggesting acceptable diagnostic accuracy, with the optimal cutoff value of ≤26.15, providing a sensitivity of 79.59% and a specificity of 65.00% (Fig. 4C). For HCC vs. HD, the ROC curve analysis yielded an AUC of 0.700, indicating acceptable diagnostic accuracy, with an optimal cutoff value of ≤25.77, providing a sensitivity of 75.51% and a specificity of 59.38% (Fig. 4D). These results suggest that the novel assay is effective in identifying HCC cases, with its relatively high sensitivity making it a reliable screening tool for early detection. Among HCC patients with AFP levels below 20 ng mL−1, the novel assay detected 16 out of 22 cases (72.73%), using the optimal cutoff value of ≤25.98. This highlights the assay's potential to complement traditional AFP testing, particularly in cases where AFP is not elevated.
A significant strength of our assay is its ability to detect very low concentrations of mSEPT9. This was evidenced by our control experiments using synthetic DNA standards, where the assay could detect mSEPT9 at extremely low levels while completely ignoring unmethylated SEPT9 even at high concentrations (5 × 105 copies per reaction). This level of sensitivity and specificity highlights the assay's robustness and its potential utility in clinical settings where detecting minimal levels of methylated DNA is crucial.
When comparing our results with those of previously published studies, our assay demonstrated a sensitivity of 77.55% and specificity of 59.62% at an optimal cutoff value of ΔCt ≤ 25.98 for HCC detection. This performance is comparable to that in the study by Yurika Kotoh et al. (2020), which reported a sensitivity of 63.2% and specificity of 90.0% for detecting HCC using a sensitive methylated SEPT9 assay, with an AUC of 0.81.19 While our AUC was slightly lower at 0.729, the higher sensitivity observed in our assay indicates that it may identify a greater number of HCC cases, albeit with a trade-off in specificity.
The CORD approach used in Kotoh et al.'s study highlights the potential of digital PCR technologies for sensitive and specific detection of methylated DNA.19 However, the requirement for digital PCR equipment and methylation-sensitive restriction enzymes may limit its accessibility, particularly in resource-limited settings. Advanced digital PCR instruments are not universally available, and the additional steps involving three restriction enzymes introduce procedural complexity that could affect throughput and consistency. Nevertheless, digital PCR offers unparalleled precision and robustness in absolute quantification, which remains a significant advantage for applications requiring detailed quantitative insights.
In comparison, our semi-nested realtime PCR assay presents a practical alternative by leveraging widely available equipment and a simplified workflow. This assay achieves high sensitivity for mSEPT9 detection while maintaining cost-effectiveness and operational simplicity. These attributes make it an attractive option for broader clinical adoption, particularly in settings where resources and infrastructure may be constrained.
Furthermore, when compared to the study by Jörn Lewin et al. (2021), our assay demonstrated a similar level of clinical performance. Lewin's study reported a sensitivity of 76.7% and specificity of 64.1% for the HCCBloodTest (Epigenomics AG), which also targets mSEPT9.20 Overall, our findings suggest that the novel mSEPT9 assay is a valuable tool for the early detection of HCC.
Additionally, our assay demonstrated its potential as a complementary diagnostic tool by effectively detecting HCC cases in patients with low AFP levels, addressing a critical gap in traditional screening methods. This capability is further supported by Kotoh et al., who reported mSEPT9 detection in 49 out of 84 cases with AFP levels below 20 ng mL−1, emphasizing the biomarker's utility for early HCC diagnosis.19 These results highlight the potential of the mSEPT9 assay, especially for improving detection in cases where traditional AFP screening is insufficient.
These comparisons indicate that while our novel realtime PCR assay for mSEPT9 provides a robust and sensitive method for detecting HCC, the differences in patient populations and study designs can influence the observed diagnostic performance. Future studies should focus on further validating our assay in diverse patient cohorts and exploring its utility in conjunction with other biomarkers to enhance early HCC detection.
Evaluating the ΔCt values of methylated SEPT9 in HCC patients according to Barcelona Clinic Liver Cancer (BCLC) staging showed significant differences between early-stage (BCLC 0 and A) and advanced-stage (BCLC B, C, and D) patients. The ΔCt values were significantly higher in early-stage patients (median = 22.69) compared to advanced-stage patients (median = 15.68), with a p-value of 0.0027 (Fig. 5A). This suggests that higher ΔCt values are associated with earlier stages of HCC. The ROC curve analysis for ΔCt values in distinguishing between early and advanced stages of HCC yielded an AUC of 0.751 (p = 0.001), indicating acceptable diagnostic accuracy (Fig. 5B). The optimal cutoff value was ≤16.765, providing a sensitivity of 76.92% and a specificity of 65.22%. Using this cutoff, the odds ratio was 5.1852 (p = 0.0092), suggesting a significant association between lower ΔCt values and advanced HCC stages.
The significant correlation between mSEPT9 levels and BCLC staging indicates the potential of this biomarker in prognostication. Higher ΔCt values in early-stage HCC patients reflect lower methylation levels of SEPT9, which may be indicative of a better prognosis and potential for curative treatments. This is consistent with findings by Yurika Kotoh and colleagues, who reported that methylated SEPT9 levels increase with the progression of HCC.19 Therefore, the quantitative assessment of mSEPT9 using this novel assay could provide valuable prognostic information and aid in clinical decision-making.
4 Conclusion
In conclusion, our study introduces a novel semi-nested quantitative PCR assay that integrates ExBP technology with TaqMan probes for quantitative detection of circulating methylated SEPT9 in plasma samples from HCC patients. This approach demonstrates promising sensitivity and specificity, effectively distinguishing between HCC patients and control groups, including those with CLD and HDs. The assay's ability to detect mSEPT9 even in HCC patients with low AFP levels highlights its potential as a valuable tool for early cancer detection and monitoring. Furthermore, the significant correlation between mSEPT9 levels and BCLC staging underscores its prognostic utility, aiding in patient stratification and treatment decision-making. While these results are encouraging, further research is needed to validate the assay in larger and more diverse populations to confirm its clinical applicability. Emphasis should be placed on its cost-effectiveness and high sensitivity, particularly in resource-limited settings. Future studies should also explore the assay's utility in conjunction with other biomarkers to enhance early HCC detection and improve patient outcomes.Ethical statement
All procedures performed in studies involving human participants were in accordance with the ethical standards of the institutional research committee of 103 Military Hospital, Vietnam Military Medical University (reference number: 15/2021/CNChT-HĐĐĐ, dated 29/06/2021) and with the 1964 Helsinki Declaration and its later amendments or comparable ethical standards. Informed consent was obtained from all individual participants included in the study.Data availability
All data generated or analysed during this study are included in this published article.Author contributions
Conceptualization, Yen Tran, Huy Duong and Tho Ho; formal analysis, Yen Tran, Trang Dao, Ung Nguyen and Tho Ho; methodology, Yen Tran, Trang Dao, Ung Nguyen, Thien Tran, Loi Luu, Huy Duong and Tho Ho; validation, Yen Tran, Trang Dao, Ung Nguyen and Tho Ho; writing – original draft, Yen Tran, Trang Dao and Tho Ho; writing – review & editing, Yen Tran, Trang Dao, Ung Nguyen, Huy Duong and Tho Ho. All authors have read and agreed to the published version of the manuscript.Conflicts of interest
Yen Hai Tran declares that she has no conflict of interest. Trang Thuy Dao declares that she has no conflict of interest. Ung Dinh Nguyen declares that he has no conflict of interest. Thien Ba Tran declares that he has no conflict of interest. Loi Phuc Luu declares that he has no conflict of interest. Huy Quang Duong declares that he has no conflict of interest. Tho Huu Ho declares that he has no conflict of interest.Acknowledgements
This research received no external funding. The APC was funded by the 103 Military Hospital. We thank Trang Thu Hoang, Hoai Bui, Tu Cam Vu, Kim Ngan Thi Dinh, Quyen Van Pham, Tuan Minh Ngo, Quyen Thi Nguyen, and Thu Hang Pham for their exceptional technical assistance.References
- J. Ferlay, M. Ervik, F. Lam, M. Colombet, L. Mery, M. Piñeros, et al., Global Cancer Observatory: Cancer Today, international agency for research on cancer, Lyon, France, 2018, pp. 1–6 Search PubMed.
- P. C. Valery, M. Laversanne, P. J. Clark, J. L. Petrick, K. A. McGlynn and F. Bray, Projections of primary liver cancer to 2030 in 30 countries worldwide, Hepatology, 2018, 67(2), 600–611 Search PubMed.
- J. M. Llovet, R. K. Kelley, A. Villanueva, A. G. Singal, E. Pikarsky and S. Roayaie, et al., Hepatocellular carcinoma, Nat. Rev. Dis. Primers, 2021, 7(1), 6 Search PubMed.
- J. A. Marrero, L. M. Kulik, C. B. Sirlin, A. X. Zhu, R. S. Finn and M. M. Abecassis, et al., Diagnosis, Staging, and Management of Hepatocellular Carcinoma: 2018 Practice Guidance by the American Association for the Study of Liver Diseases, Hepatology, 2018, 68(2), 723–750 Search PubMed.
- European Association for the Study of the Liver, EASL Clinical Practice Guidelines: Management of hepatocellular carcinoma, J. Hepatol., 2018, 69(1), 182–236 Search PubMed.
- S. Moran, A. Martinez-Cardús, S. Boussios and M. Esteller, Precision medicine based on epigenomics: the paradigm of carcinoma of unknown primary, Nat. Rev. Clin. Oncol., 2017, 14(11), 682–694 CrossRef PubMed.
- X. Hao, H. Luo, M. Krawczyk, W. Wei, W. Wang and J. Wang, et al., DNA methylation markers for diagnosis and prognosis of common cancers, Proc. Natl. Acad. Sci. U. S. A., 2017, 114(28), 7414–7419 Search PubMed.
- M. Scott, P. L. Hyland, G. McGregor, K. J. Hillan, S. E. Russell and P. A. Hall, Multimodality expression profiling shows SEPT9 to be overexpressed in a wide range of human tumours, Oncogene, 2005, 24(29), 4688–4700 Search PubMed.
- A. Kakehashi, N. Ishii, T. Shibata, M. Wei, E. Okazaki and T. Tachibana, et al., Mitochondrial prohibitins and septin 9 are implicated in the onset of rat hepatocarcinogenesis, Toxicol. Sci., 2011, 119(1), 61–72 CrossRef CAS PubMed.
- A. Villanueva, A. Portela, S. Sayols, C. Battiston, Y. Hoshida and J. Méndez-González, et al., DNA methylation-based prognosis and epidrivers in hepatocellular carcinoma, Hepatology, 2015, 61(6), 1945–1956 Search PubMed.
- M. Uhlén, L. Fagerberg, B. M. Hallström, C. Lindskog, P. Oksvold and A. Mardinoglu, et al., Proteomics. Tissue-based map of the human proteome, Science, 2015, 347(6220), 1260419 Search PubMed.
- R. Wasserkort, A. Kalmar, G. Valcz, S. Spisak, M. Krispin and K. Toth, et al., Aberrant septin 9 DNA methylation in colorectal cancer is restricted to a single CpG island, BMC Cancer, 2013, 13, 398 Search PubMed.
- W. Okajima, S. Komatsu, D. Ichikawa, M. Miyamae, T. Kawaguchi and S. Hirajima, et al., Circulating microRNA profiles in plasma: identification of miR-224 as a novel diagnostic biomarker in hepatocellular carcinoma independent of hepatic function, Oncotarget, 2016, 7(33), 53820–53836 Search PubMed.
- Y. H. Su, A. K. Kim and S. Jain, Liquid biopsies for hepatocellular carcinoma, Transl. Res., 2018, 201, 84–97 CrossRef PubMed.
- D. Wu, G. Zhou, P. Jin, J. Zhu, S. Li and Q. Wu, et al., Detection of Colorectal Cancer Using a Simplified SEPT9 Gene Methylation Assay Is a Reliable Method for Opportunistic Screening, J. Mol. Diagn., 2016, 18(4), 535–545 CrossRef CAS PubMed.
- T. H. Ho, K. X. Dang, S. Lintula, K. Hotakainen, L. Feng and V. M. Olkkonen, et al., Extendable blocking probe in reverse transcription for analysis of RNA variants with superior selectivity, Nucleic Acids Res., 2014, 43(1), e4 Search PubMed.
- L. T. Duong, T. T. Dao, H. T. Bui, U. D. Nguyen, U. T. Hoang and D. V. Tran, et al., Innovative Semi-Nested Realtime PCR Assay with Extendable Blocking Probe for Enhanced Analysis of SEPT9 Methylation in Colorectal Cancer, Biomedicines, 2024, 12(7), 1458 CrossRef CAS PubMed.
- J. K. Heimbach, L. M. Kulik, R. S. Finn, C. B. Sirlin, M. M. Abecassis and L. R. Roberts, et al., AASLD guidelines for the treatment of hepatocellular carcinoma, Hepatology, 2018, 67(1), 358–380 Search PubMed.
- Y. Kotoh, Y. Suehiro, I. Saeki, T. Hoshida, M. Maeda and T. Iwamoto, et al., Novel Liquid Biopsy Test Based on a Sensitive Methylated SEPT9 Assay for Diagnosing Hepatocellular Carcinoma, Hepatol. Commun., 2020, 4(3), 461–470 CrossRef CAS PubMed.
- J. Lewin, D. Kottwitz, J. Aoyama, T. deVos, J. Garces and O. Hasinger, et al., Plasma cell free DNA methylation markers for hepatocellular carcinoma surveillance in patients with cirrhosis: a case control study, BMC Gastroenterol., 2021, 21(1), 136 CrossRef CAS PubMed.
Footnote |
| † Electronic supplementary information (ESI) available. See DOI: https://doi.org/10.1039/d4ay02168a |
| This journal is © The Royal Society of Chemistry 2025 |