Open Access Article
Open Access ArticleCreative Commons Attribution 3.0 Unported Licence
Sulfonium-based polymethacrylamides for antimicrobial use: influence of the structure and composition†
Sidra
Kanwal
a,
Umer Bin Abdul
Aziz
a,
Elisa
Quaas
b,
Katharina
Achazi
 b and
Daniel
Klinger
b and
Daniel
Klinger
 *a
*a
aInstitute of Pharmacy, Freie Universität Berlin, 14195 Berlin, Germany. E-mail: daniel.klinger@fu-berlin.de
bInstitute of Chemistry and Biochemistry, Freie Universität Berlin, 14195 Berlin, Germany
First published on 13th January 2025
Abstract
We are facing a shortage of new antibiotics to fight against increasingly resistant bacteria. As an alternative to conventional small molecule antibiotics, antimicrobial polymers (AMPs) have great potential. These polymers contain cationic and hydrophobic groups and disrupt bacterial cell membranes through a combination of electrostatic and hydrophobic interactions. While most examples focus on ammonium-based cations, sulfonium groups are recently emerging to broaden the scope of polymeric therapeutics. Here, main-chain sulfonium polymers exhibit good antimicrobial activity. In contrast, the potential of side-chain sulfonium polymers remains less explored with structure–activity relationships still being limited. To address this limitation, we thoroughly investigated key factors influencing antimicrobial activity in side-chain sulfonium-based AMPs. For this, we combined sulfonium cations with different hydrophobic (aliphatic/aromatic) and hydrophilic polyethylene glycol (PEG) groups to create a library of polymers with comparable chain lengths. For all compositions, we additionally examined the position of cationic and hydrophobic groups on the polymer backbone, i.e., we systematically compared same center and different center structures. Bactericidal tests against Gram-positive and Gram-negative bacteria suggest that same center polymers are more active than different center polymers of similar clog![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) P. Ultimately, sulfonium-based AMPs show superior bactericidal activity and selectivity when compared to their quaternary ammonium cationic analogues.
P. Ultimately, sulfonium-based AMPs show superior bactericidal activity and selectivity when compared to their quaternary ammonium cationic analogues.
Introduction
Antimicrobial resistance is a major threat to global human health that is often overlooked but requires immediate action.1 Over the last decades, overprescription and misuse of common first line antibiotics have caused a steep increase in bacterial resistance that drastically reduces our treatment options. In particular, for conventional small molecule antibiotics that target specific active sites in bacterial proteins, most pathogens are very quick to modify the target site, express efflux pumps, or develop drug degrading enzymes.2,3 Consequently, the development of new small molecule antibiotics is a never-ending race against the extremely rapid bacterial evolution. In contrast, polymeric antimicrobials can be more robust against the development of resistance due to their contrasting and simple mode of action.4–8 Inspired by natural host-defense peptides (HDPs), synthetic antimicrobial peptides and polymers (AMPs)9–11 combine two main active structural features: first, cationic residues promote electrostatic binding to anionic bacterial cell membranes. Second, hydrophobic moieties can insert into the non-polar membranes, thus destabilizing the membrane and causing bacterial death via lysis.12 Unlike the precise “lock and key” mechanism of many small molecule antibiotics, this membrane disruption pathway is less specific, making it more challenging for bacteria to develop resistance.13 In comparison with cationic small molecule amphiphiles, e.g., quaternary ammonium salts (QASs), the higher charge density of antimicrobial macromolecules also contributes to higher activity.14,15Despite these advantages, combining cationic and hydrophobic features in a polymer or peptide can cause toxicity in human cells as well, e.g., through hemolysis.16 Thus, the development of new AMPs requires a careful balance between maximized bactericidal activity and minimized systemic toxicity. This key challenge is addressed through adjusting the type, ratio, and spatial arrangement of these groups in the polymer chain.13 Such adjustments are easily performed in synthetic polymers due to the advancement of controlled polymerization methods that give access to well-defined random copolymers with tunable structures and compositions. In contrast to sequence-defined peptides that face high manufacturing costs, low stability due to proteolysis, and poor bioavailability, the production of AMPs is more cost-effective, scalable, and customizable.17
In such polymers, early optimization attempts focused on binary systems, where the ratio of randomly distributed cationic and hydrophobic groups was examined.18 By including neutral (non-ionic) hydrophilic units, the parameter room was later expanded to tertiary systems, which enabled a reduction in toxicity.19,20 Regarding structural features, diverse polymer architectures have been described.21 First, the position of the cationic, hydrophobic, and hydrophilic units on the polymer backbone was varied.17 Here, it can be distinguished between main-chain22 and side-chain23 AMPs. Second, the position of the cationic and hydrophobic groups on the polymers’ repeating units can be varied. In same center structures,24 both groups are side groups of the same monomer unit. In different center structures,25 cationic and hydrophobic moieties are each part of individual monomer units.17 Third, diverse polymer backbones26 have been examined that range from non-degradable poly(meth)acrylates16 and poly(meth)acrylamides27 to degradable aliphatic polycarbonates,28–30 polyesters,31,32 polypept(o)ides,33,34 and polyoxazolines.35 Fourth, the influence of polymer molecular weight was examined by adjusting the degree of polymerization through controlled polymerization techniques.14,36 Ultimately, the types of functional groups (hydrophobic,37 hydrophilic,20 or cationic38) can be varied. For hydrophobic and hydrophilic groups, a broad range of chemical structures have been examined. In contrast, the chemical room for the examined cations is much smaller and still mainly focuses on ammonium-based residues. The majority of available AMPs include either primary amines, quaternary ammonium salts (QAS), guanidinium-, or imidazolium groups.23,39
While such groups can successfully mimic the cations in naturally occurring HDPs, bacteria have already started to develop corresponding resistance, i.e., against QAS-based AMPs.40 The specific mechanisms can vary and remain under discussion. An important suggested pathway is based on the partial substitution of anionic cell surface constituents with cationic molecules.40–44 This decreases the affinity of the cell wall to AMPs. To address this reduced affinity, it is proposed to introduce other cation types into AMPs, thereby increasing their interaction with the bacterial cell membrane. Thus, exploring new cationic structures is crucial for retaining and expanding the therapeutic option of AMPs.45 In this context, trivalent sulfonium cations (SCs) are known to improve the activity of common antibiotics, e.g., vancomycin,46 and can be used as cations in amphiphilic antiseptics that exceed the antimicrobial activity of their QAS-based analogues, e.g., benzalkonium chloride (BKC) and cetylpyridinium chloride (CC).47 Thus, incorporating sulfonium cations into polymers is currently emerging as a new strategy to develop new AMPs.48 In comparison with quaternary ammonium- and phosphonium-polymers, the tertiary sulfonium analogues are more effective against bacteria.45 It is assumed that this stems from a combination of steric and electronic effects: as tertiary cations, steric hindrance for interactions with the bacterial cell wall is lower than that for quaternary ammonium and phosphonium cations. As large polarizable cations, they are more hydrophobic than their ammonium-based counterparts, which also enhances interactions with bacterial cell walls.45 While these factors suggest great potential, effective sulfonium-based AMPs are mostly realized through main-chain SC-containing polymers.25,49–52 In contrast, side-chain sulfonium-based polymers are less examined even though they show promise as AMPs.23,38 Thus, exploring the full potential of side-chain sulfonium-based antimicrobial polymers now requires systematic studies to develop structure–property relationships.
For addressing this need, we have prepared a polymer library to expand the state-of-the-art38 and systematically examine structural key parameters that influence antimicrobial activity in side-chain sulfonium-based AMPs. To be able to translate these findings into accurate structure–property relationships, we used a synthetic platform approach for accessing polymethacrylamides with the same chain lengths but different chemical compositions and structures. This was achieved by using one master batch of poly(pentafluorophenyl methacrylate) (P(PFPMA)) for a two-step post-polymerization functionalization. Here, we introduced varying combinations of pendant sulfonium cations, hydrophobic (aliphatic or aromatic), and hydrophilic (PEG) side groups. For all compositions, we additionally varied the position of cationic and hydrophobic groups on the polymer backbone, i.e., we systematically compared same center and different center structures. To benchmark the sulfonium chloride-based AMPs against established cationic AMPs, quaternary ammonium chloride-based analogues were prepared for the same center and different center polymers.
The bactericidal activity of polymers was quantified via broth dilution assays against Bacillus subtilis, Escherichia coli, Pseudomonas aeruginosa, and Staphylococcus aureus. Relating these results to the AMPs' hemolytic and cellular toxicity gave insights into their selectivity against bacteria over human cells. For all polymers, antimicrobial activity can be quantitatively linked to the copolymer hydrophobicity (as calculated by clog![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) P). Here, comparing AMP structures of similar hydrophobicity suggests that same center SC-polymers are more active than different center polymers. In addition, sulfonium-based AMPs show superior bactericidal activity and selectivity when compared to their quaternary ammonium-based analogues. Ultimately, additional hydrophilic polyethylene glycol (PEG) groups can shift the amphiphilic balance of the AMPs towards higher selectivity while retaining their antimicrobial activity. As a result, our findings contribute to a deeper understanding of the structural parameters that govern antimicrobial activity and selectivity in sulfonium-based side-chain AMPs. Thus, we propose that these structure–property relationships can guide the design of new polymeric antimicrobials.
P). Here, comparing AMP structures of similar hydrophobicity suggests that same center SC-polymers are more active than different center polymers. In addition, sulfonium-based AMPs show superior bactericidal activity and selectivity when compared to their quaternary ammonium-based analogues. Ultimately, additional hydrophilic polyethylene glycol (PEG) groups can shift the amphiphilic balance of the AMPs towards higher selectivity while retaining their antimicrobial activity. As a result, our findings contribute to a deeper understanding of the structural parameters that govern antimicrobial activity and selectivity in sulfonium-based side-chain AMPs. Thus, we propose that these structure–property relationships can guide the design of new polymeric antimicrobials.
Experimental section
Materials
All starting materials and chemical and biological reagents were purchased from commercial vendors and used as received, unless noted otherwise (see the ESI† for further details). Concentrated human red blood cells for research purposes were purchased from DRK Blutspendedienst Nordost. Some starting materials such as pentafluorophenylmethacrylate (PFPMA), 1,2-epoxy nonane, epoxy PEG and 1-bromo-2-hexane were synthesized according to published procedures (see the ESI† for detailed experimental procedures).Polymer synthesis
The master batch of the P(PFPMA) precursor polymer was prepared via a modified established protocol53 (see the ESI† for a detailed experimental procedure). This single master batch was further used to prepare two libraries of cationic polymers, i.e. same center and different center polymers.Synthesis of same center polymers
P(PFPMA) was functionalized via a 2-step functionalization strategy to synthesize sulfonium and ammonium-based polymers. Exemplary protocols are briefly described here for polymer synthesis; detailed procedures on synthesis and purification can be found in the ESI.†For the synthesis of sulfonium-based polymers, firstly, P(PFPMA) (7 g, 0.0278 mol of PFPMA units) was reacted with 2-(methylthio)ethylamine (12.65 g, 0.1388 mol, 5.0 mol eq. w.r.t. PFPMA units) in the presence of triethylamine (14.05 g, 0.1388 mol) at 50 °C in 200 mL anhydrous DMF for 5 days. After dialyzing the reaction mixture in DMF and Milli-Q water, and consequently lyophilizing, the thioether group containing polymer, i.e., poly[(N-(2-(methylthio)ethyl)methacrylamide)] P(MTEMAA), was obtained (see the ESI† for the detailed experimental procedure). In the second step, P(MTEMAA) was reacted with various hydrophobic and hydrophilic epoxide moieties to introduce sulfonium cations in the side chain to generate the library of same center cationic sulfonium polymers. In a representative reaction, P(MTEMAA) (200 mg, 1.25 × 10−3 mol, 1 mol eq.) was dissolved in DCM and 1,2-epoxyhexane (0.721 g, 7.5 × 10−3 mol, 6.0 mol eq. to thioether groups) and trifluoroacetic acid (0.86 g, 7.5 × 10−3 mol) was added and stirred at 35 °C for 5 days. During the reaction two phases formed and the DCM phase could be decanted off afterwards. The other phase was dissolved in MeOH and precipitated twice in Et2O. The product was obtained as a slightly yellow/brown solid (see the ESI† for further purification and characterization).
For the synthesis of ammonium-based polymers in the first step, poly[(N-(2-(dimethylamino)ethyl)methacrylamide)] P(DMAEMAA) was obtained by reacting P(PFPMA) (10 g, 0.0396 mol of PFPMA units) with N,N-dimethylethylendiamine (17.45 g, 0.198 mol, 5.0 mol eq. w.r.t. PFPMA units) in the presence of triethylamine (20.04 g, 0.198 mol) at 50 °C in 200 mL anhydrous DMF for 5 days. In the second step, P(DMAEMAA) was reacted with different 1-bromo-2-hydroxyalkanes. As an example, P(DMAEMAA) (112 mg, 1.0 eq. of amine groups, 7.18 × 10−4 mol of amine groups) was dissolved in DMF (10 mL) followed by the addition of 12 equivalents (w.r.t. amines) of 1-bromo-2-butanol (1.89 g, 8.62 × 10−3 mol) and the reaction mixture was stirred at 70 °C for 10 days. For purification, the reaction mixture was dialyzed against DMF for 3 days followed by extensive dialysis against water (see the ESI† for further purification and characterization).
Synthesis of different center polymers
Poly(PFPMA) was functionalized via a 2-step functionalization strategy to synthesize sulfonium and ammonium-based different center polymers. An exemplary protocol for the synthesis of PS+-prdc is briefly described here. Detailed procedures for the synthesis and purification can be found in the ESI.†In a representative reaction for the synthesis of PS+-prdc, [poly(PFPMA)] (0.5 g, 1.984 × 10−3 mol of PFPMA units) was reacted with propylamine (0.078 g, 1.32 × 10−3 mol), 2-(methylthio)ethylamine (0.12 g, 1.32 × 10−3 mol), and HPA (0.099 g, 1.32 × 10−3 mol) in 10 mL DMF at 50 °C for 3 days, which resulted in the synthesis of thioether-based copolymers, P[(MTEMAA)-co-n(propyl)MAA-co-HPMAA], which was methylated further by reacting with 10 eq. of CH3I in DMF to generate PS+-prdc (see the ESI† for further experimental details and for the synthesis of PN+-Xdc).
All polymers from same and different center libraries were dialyzed against NaCl to exchange the counter ion with chloride ions for all polymers. Finally, the dialyzed aqueous solutions of polymers were run through a Sephadex G-10 column to remove low molecular weight impurities. The purified samples were then freeze dried and characterized through 1H NMR spectroscopy and GPC (see the ESI† for details).
Methods
ATR-FTIR spectroscopy
Attenuated total reflectance Fourier transformed infrared (ATR-FTIR) spectroscopy was used to characterize the first functionalization of precursor polymer, poly(PFPMA) to thioether and tert-amine group containing polymers in same and different center polymers (see the ESI† for further details).Nuclear magnetic resonance (NMR) spectroscopy
NMR spectroscopy was used to characterize the synthesis of starting materials and polymers and their functionalization reactions at individual steps. All NMR (1H, 19F and 13C) spectra were recorded at 300 K on either a Jeol Eclipse 600 MHz (Tokyo, Japan) or a Bruker AVANCE 600 MHz spectrometer (Billerica, MA, USA). Chemical shifts δ were documented in ppm and the deuterated solvent peaks were used as a standard. All data were processed using MestReNova software (v6.2.1-7569). All the samples were measured at concentrations of 10–20 mg mL−1.Gel permeation chromatography (GPC)
GPC was used to determine the relative polymer sizes by using a customized chromatography system (PSS Polymer Standards Services GmbH, Mainz, Germany). DMF + 10 mM LiBr was used as the mobile phase to characterize the synthesis and first functionalization of PPFMPA using a concentration of 1.5 mg mL−1. However, all cationic polymers were measured in H2O at 3.0 mg mL−1 by keeping the flow rate of 1.0 mL min−1 (see the ESI† for further instrument and setup details).Dynamic light scattering (DLS)
The polymer–protein complex (PPC) formation of polymers in the LB medium was characterized by dynamic light scattering at 90° in multiangle round cell glass cuvettes using a NICOMP nano Z3000 system (Particle Sizing Systems, USA). For each measurement, the polymer sample was prepared by dissolving it at a concentration of 1 mg mL−1 in Milli-Q water and LB medium at 23 °C in triplicate.Zeta potential
Zeta potentials of the cationic polymers were determined using a Zetasizer Nano (Malvern Instruments) at a concentration of 1 mg mL−1 in Milli Q water and LB medium in DTS1060 folded capillary cells at 25 °C in triplicate.MIC determination via broth dilution assays
To determine the susceptibility of bacteria to a compound, the broth dilution assays were performed using a 96-well plate format with the modified protocol from the literature.25 For this purpose, two Gram-negative bacterial strains (E. coli and P. aeruginosa) and two Gram-positive bacterial strains (B. subtilis and S. aureus) were used. For each strain, a preculture in 5 mL of Luria broth (LB) medium was incubated overnight at 37 °C with shaking at 140 rpm. The OD600 value of the bacterial culture was diluted to 0.02 before adding to the 96-well plate. The stock solutions of the polymer (1024 μg mL−1 each) were prepared in sterile Milli-Q water. These were then used to prepare a series of different polymer concentrations by 2-fold dilutions (from 512 to 0.5 μg mL−1; each 100 μL in 96-well plates). Subsequently, 100 μL of the diluted bacterial suspension (OD ∼0.02) was added to each well giving final concentrations of 256, 128, 64, 32, 16, 8, 4, 2, 1, 0.5, and 0.25 μg mL−1. The inoculated LB medium (OD ∼0.02) without any polymer solution was used as a positive control. On the other hand, sterile LB medium without any polymer or bacteria was used as a negative control. Samples in all experiments were tested in triplicate. All samples and controls were incubated overnight at 37 °C. MIC is defined as the lowest polymer concentration at which more than 90% bacterial growth (MIC90) was inhibited. For the same center polymers, narrower concentration ranges were tested to get more detailed information about the MIC90 (see the ESI† for further experimental details).Hemolysis assay
The hemolytic activity of the synthesized polymers was investigated on human erythrocytes. The erythrocytes were isolated from fresh human blood and washed 3 times by sterile PBS. The assays were performed by following the reported protocol with little modification.54 Each polymer was dissolved in PBS except PS+-he, PS+-he-PEG, PS+-hedc and PN+-hedc where a mixture of PBS and DMSO (less than 5%) was required to dissolve the polymers based on their relatively low solubility in aqueous solution. In conical bottom 96-well plates, multiple concentrations (5 mg mL−1–50 μg mL−1) of the polymers were added (100 μL), followed by the addition of 150 μL of 5% v/v erythrocyte suspension in PBS to each well. 10% v/v solution of Triton X-100 in PBS and PBS alone were used as positive and negative controls, respectively. The microplates were incubated for 1 h at 37 °C and then centrifuged at 1500 rpm for 10 minutes at 4 °C. Finally, 150 μL of supernatants were transferred to empty 96-well plates and the absorbance was measured at 543 nm. The percentage hemolysis was calculated using the provided equation.Each experiment was conducted in quadruplicate. Data are presented as HC50 which means that the concentration of the polymer at which 50% hemolysis occurred (see the ESI† for further experimental details).
Cytotoxicity assay
Cell viability assays were performed on HaCat and L929 cells by using Cell Counting Kit-8 (CCK-8).55 HaCat cells were cultured in Roswell Park Memorial Institute (RPMI) 1640 medium, and L929 cells were cultured in Dulbecco's modified Eagle's medium (DMEM), both supplemented with 10% (v/v) fetal bovine serum (FBS), 100 U mL−1 penicillin and 100 μg mL−1 streptomycin. HaCat and L929 cells were seeded in a 96-well plate at a density of 5 × 104 cells per mL in 90 μL of RPMI and DMEM, respectively, per well overnight at 37 °C and 5% CO2. 10 μL of sample solutions (dissolved in sterile deionized water) were added in serial dilutions to the wells including positive (1% SDS) and negative (culture medium and H2O) controls and incubated for another 24 h at 37 °C with 5% CO2. Moreover, the wells containing no cells but only the polymer samples were used for background subtraction. After the incubation period (24 h), 10 μL per well of the CCK8 solution was added, and absorbance (450 nm/650 nm) of the dye was measured consequently after incubating for approximately another 3 h. The cell viability for individual concentrations was calculated by setting the non-treated control as a reference to 100% and the non-cell control to 0% after subtraction of the background signal. Each experiment was conducted in triplicate and repeated 3 times (see the ESI† for further experimental details).Results and discussion
To develop structure–property relationships in side-chain sulfonium-based AMPs, we have designed a polymer library that consists of 4 sets of polymers (see Scheme 1). In this library, all polymers contain a polymethacrylamide backbone with different pendant groups. To examine the influence of the chemical composition and polymer structure, 3 different parameters were varied. (1) Chemical composition of the side chains: These contain cations and hydrophobic groups. We used different hydrophobic groups to vary the carbon-to-cation ratio and control the overall hydrophobicity of the polymer. Moreover, additional neutral hydrophilic PEG side chains were introduced to balance the hydrophobic influences of alkyl and aromatic groups. (2) Structural location of cationic and hydrophobic groups: we prepared same center and different center side-chain AMPs. In the same center polymers, the hydrophobic groups are connected to the cations through a hydrophilic hydroxyl-containing spacer. Thus, starting from the backbone, the side chain structure of these polymers can be divided into the following parts: cation, hydroxyl-based spacer and hydrophobic group. To ensure comparability between the same center and different center polymers, we used these parts as individual side groups in the different center polymers. With this, we aim for similarity in chemical composition but variation in functional group distribution (Scheme 1). (3) Type of cation: Focusing on sulfonium-based AMPs, we used trivalent sulfonium cations as the main type of cation in same center and different center AMPs. To compare their activity and selectivity to established systems, we used structural analogues that contained quaternary ammonium cations. In all systems, chloride counterions were used to ensure comparability.Synthesis and characterization of antimicrobial polymers
All polymers of the library were prepared by post-polymerization functionalization from one batch of reactive precursor polymers.56 This enabled us to circumvent copolymerization of different monomers which could lead to batch-to-batch inconsistencies in the degree of polymerization and dispersity. In contrast, the polymer functionalization approach ensured the same backbone length for all polymers (same center and different center). As a reactive precursor polymer, we chose poly(pentafluorophenyl methacrylate) (P(PFPMA)) since these polymers are known for their facile and efficient functionalization with primary amines. To adjust the degree of polymerization (DP) and ensure low dispersity (Đ), we used reversible addition–fragmentation transfer (RAFT) polymerization for the preparation of a well-defined precursor polymer (30 g, Mn,GPC = 17![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 800 g mol−1, Đ = 1.24; see the ESI† for synthetic details). Here, we targeted a DP of 70 since it is reported for AMPs that such chain lengths show optimum antibacterial activity.23,25
800 g mol−1, Đ = 1.24; see the ESI† for synthetic details). Here, we targeted a DP of 70 since it is reported for AMPs that such chain lengths show optimum antibacterial activity.23,25
Sulfonium-based same center AMPs (PS+-X) were synthesized by first substituting all reactive ester groups in PPFPAM with 2-methylthioethylamine to give the thioether-based homopolymer, i.e., P(MTEMAA). Quantitative functionalization was demonstrated via ATR-FTIR, where the complete disappearance of the ester band and the appearance of an amide band were observed (ESI, Fig. S1†). In addition, the disappearance of all fluorine signals in 19F-NMR supported this assumption (ESI, Fig. S2†). Finally, quantitative analysis of 1H-NMR spectra revealed complete conversion of the PFP-ester groups, thus suggesting the successful formation of the thioether-containing homopolymer (ESI, Fig. S3†). Overall, the first functionalization step gave access to well-defined homopolymers with a DP and molecular weight distribution that is determined by the precursor polymer (see ESI Fig. S4† for GPC traces of P(MTEMAA) in comparison with P(PFPMA)).
In the second step, the homopolymer P(MTEMAA) was reacted with various hydrophobic epoxide moieties to simultaneously generate the cationic moieties and install hydrophobic side groups. Here, the acid-catalyzed nucleophilic attack of the thioether to the epoxide led to the formation of the tertiary sulfonium cation by ring opening of the epoxide. To adjust AMP hydrophobicity and systematically investigate the effect of hydrophobic groups, we choose three different epoxides with varying aliphatic alkyl chains, i.e., methyl (me), propyl (pr), hexyl (he), and two aromatic functional groups, i.e., benzyl ether (be), and benzyl (bz). These hydrophobic groups were selected due to their reported antimicrobial activity4,6 and gave five hydrophobic homopolymers (PS+-me, PS+-pr, PS+-he, PS+-be, and PS+-bz) that covered a range of calculated clog![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) P from −0.37 to 21.73 (Fig. 1b and ESI, Fig. S24, Table S3,† for details of calculations). By combining two different hydrophobic groups in a 50
P from −0.37 to 21.73 (Fig. 1b and ESI, Fig. S24, Table S3,† for details of calculations). By combining two different hydrophobic groups in a 50![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 50 molar ratio, binary hydrophobic copolymers could be prepared that show clog
50 molar ratio, binary hydrophobic copolymers could be prepared that show clog![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) P values between the respective homopolymers. Thus, by the formation of PS+-me-be and PS+-me-he, a set of SC-AMPs was generated that covered a wide range of different hydrophobicities (Fig. 1b). Ultimately, the combination of a hydrophobic group with a hydrophilic PEG group (70
P values between the respective homopolymers. Thus, by the formation of PS+-me-be and PS+-me-he, a set of SC-AMPs was generated that covered a wide range of different hydrophobicities (Fig. 1b). Ultimately, the combination of a hydrophobic group with a hydrophilic PEG group (70![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 30 molar ratio) led to the formation of binary amphiphilic copolymers (PS+-me-PEG, PS+-pr-PEG, PS+-he-PEG). Here, the introduction of the additional hydrophilic group aimed to balance the influence of hydrophobic groups, thus reducing potential toxic effects.
30 molar ratio) led to the formation of binary amphiphilic copolymers (PS+-me-PEG, PS+-pr-PEG, PS+-he-PEG). Here, the introduction of the additional hydrophilic group aimed to balance the influence of hydrophobic groups, thus reducing potential toxic effects.
All same center PS+-X polymers were purified by size exclusion chromatography (SEC) over a Sephadex column to remove any unreacted hydrophobic epoxides (see ESI Fig. S5†). Afterwards, the counterions of the sulfonium cations were exchanged. Since the acid-catalyzed functionalization reaction was performed with trifluoroacetic acid (TFA), the crude sulfonium polymers contained the respective TFA counterions. To ensure comparability to established AMPs with halogen counterions, we exchanged the TFA anions with chloride anions by extensive dialysis of the polymers against NaCl solution (see the ESI for details, Fig. S6†). The successful formation of all sulfonium-based homo- and copolymers was demonstrated by 1H-NMR spectroscopy. Here, quantitative peak analysis revealed a conversion of thioethers to sulfoniums that exceeded 95% for all polymers (ESI, Fig. S7–17, and Table S1†). As shown by GPC traces, the introduction of pendant hydrophobic and hydrophilic groups caused a slight increase in molecular weight from the thioether precursors to the final polymers (ESI, Fig. S18†). However, the dispersity remained similar to the precursors, thus suggesting the successful generation of well-defined sulfonium-based AMPs with different side chain compositions.
Quaternary ammonium-based polymers (PN+-X) were prepared as benchmark polymers. Thus, to ensure comparability to the sulfonium analogues, the synthesis of PN+-X started with the functionalization of the same master batch of P(PFPMA) precursor polymers. In the first step, this polymer was reacted with excess N,N-dimethylethylenediamine (DMEDA) to substitute all PFP ester groups. Hereby, the resulting amine-functionalized homopolymer P(DMAEMAA) exhibited a DP and dispersity similar to the PS+-X polymers. Quantitative functionalization was confirmed by a combination of ATR-FTIR, 19F-NMR, and 1H-NMR analyses (ESI, Fig. S1–S3†). This initial step yielded well-defined homopolymers with a molecular weight distribution determined by the precursor polymer (ESI, Fig. S19† for GPC traces of P(DMAEMAA) vs. P(PFPMA)).
In the second step, P(DMAEMAA) was further functionalized to generate the cations and introduce hydrophobic side groups in the same reaction. To ensure structural similarity to the sulfonium polymer, we attempted functionalization with the same epoxide moieties that were used for the generation of the PS+-X polymers. However, in comparison with the thioether moieties, the tertiary amines exhibit lower nucleophilicity and higher steric hindrance which hinders the opening of the epoxide ring. As a result, no quantitative conversion could be achieved even by examining various reaction conditions. As an alternative, 1-bromo-2-hydroxy alkanes were employed to functionalize P(DMAEMAA). These molecules were chosen to give quaternary ammonium polymers with functional groups that are analogous to those in the sulfonium polymers, i.e., a β-hydroxy group next to the cation (Fig. 1a). Using this strategy, we prepared two benchmark polymers that contain either a methyl (PN+-me) or propyl (PN+-pr) group attached to the β-carbon.
Purification of both QAS polymers was achieved via size exclusion chromatography over a Sephadex column (ESI, Fig. S20†). Afterwards, the bromide counterions were exchanged for chlorides via extensive dialysis against NaCl solution. Hereby, comparability to the PS+-X polymers was ensured. The successful synthesis of both quaternary ammonium control polymers is demonstrated by 1H-NMR which shows quantitative conversion of all tertiary amines (ESI, Fig. S21, S22, and Table S2†). GPC traces illustrate that sulfonium and ammonium polymers exhibit comparable molecular distributions (Fig. S23 and S24†). Here, DP and dispersity are determined by the P(PFPMA) precursor.
![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1/3
1/3![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1/3 mixture of primary aminoalkanes that contain the respective functional groups. Following this approach, a set of thioether-based ternary copolymers P[(MTEMAA)-co-n(alkyl)MAA-co-HPMAA] and a set of tert-amine containing ternary copolymers P[(DMAEMAA)-co-n(alkyl)MAA-co-HPMAA] were prepared. Each set consisted of three copolymers with varying hydrophobicity, i.e., varying alkyl chains of propyl (pr), butyl (bu), and hexyl (he) to generate AMPs with clog
1/3 mixture of primary aminoalkanes that contain the respective functional groups. Following this approach, a set of thioether-based ternary copolymers P[(MTEMAA)-co-n(alkyl)MAA-co-HPMAA] and a set of tert-amine containing ternary copolymers P[(DMAEMAA)-co-n(alkyl)MAA-co-HPMAA] were prepared. Each set consisted of three copolymers with varying hydrophobicity, i.e., varying alkyl chains of propyl (pr), butyl (bu), and hexyl (he) to generate AMPs with clog![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) P values that resemble those of their same center analogues (see the ESI for the calculation of clog
P values that resemble those of their same center analogues (see the ESI for the calculation of clog![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) P, Fig. S38, and Table S6†). In addition, the neutral hydrophilic group 2-hydroxy propylamine (HPA) was used in both sets to mimic the hydroxyl-containing spacer that was generated during functionalization of the same center polymers. After synthesis, ATR-FTIR and 19F-NMR showed quantitative conversion of the PFP esters to the respective amides (ESI, Fig. S26 and S27†). In addition, 1H-NMR analysis was used to determine the terpolymer composition, i.e., the incorporation ratio of the three different functional groups (ESI, Fig. S28 and S29†). It was found that the incorporation ratio between the groups closely resembled the targeted ratio as programmed by the feed ratio (ESI, Tables S4 and S5†).
P, Fig. S38, and Table S6†). In addition, the neutral hydrophilic group 2-hydroxy propylamine (HPA) was used in both sets to mimic the hydroxyl-containing spacer that was generated during functionalization of the same center polymers. After synthesis, ATR-FTIR and 19F-NMR showed quantitative conversion of the PFP esters to the respective amides (ESI, Fig. S26 and S27†). In addition, 1H-NMR analysis was used to determine the terpolymer composition, i.e., the incorporation ratio of the three different functional groups (ESI, Fig. S28 and S29†). It was found that the incorporation ratio between the groups closely resembled the targeted ratio as programmed by the feed ratio (ESI, Tables S4 and S5†).
In the second step, the nucleophilic thioether groups of P[(MTEMAA)-co-n(alkyl)MAA-co-HPMAA] and the tertiary amine groups of P[(DMAEMAA)-co-n(alkyl)MAA-co-HPMAA] were methylated with methyl iodide to give the tertiary sulfonium and quaternary ammonium-based terpolymers, respectively (see ESI Fig. S30†). To ensure comparability to the same center polymers, the iodide counterions were exchanged for chlorides and the polymers were purified by SEC over a Sephadex column. The final AMPs were examined via1H-NMR, which demonstrated successful quantitative methylation (ESI, Fig. S31–S36†). In addition, GPC analysis revealed comparable molecular weight distributions to their same center analogues, thus demonstrating comparability (ESI, Fig. S24 and S37†).
Polymers in biological media: polymer–protein complex (PPC) formation and stability
Accurately assessing the AMPs' biological effect requires good solubility in the respective aqueous medium. In deionized (DI) water, all polymers were readily soluble, and no aggregates were detected by dynamic light scattering (DLS) at a concentration of 1 mg mL−1. However, in the bacterial growth medium, an increase in turbidity and hydrodynamic diameter (dh) was observed for polymers with high hydrophobicity directly after sample preparation (ESI, Tables S7 and S8†). We suggest that this could be attributed to the formation of polymer–protein complexes (PPCs), i.e., colloidal assemblies that originate from electrostatic and hydrophobic interactions between cationic AMPs and proteins in biological media.37,38 Since the formation of PPCs can hinder the cationic groups from interacting with the anionic bacterial membranes, the antimicrobial activity of the AMPs can be reduced. Furthermore, the resulting turbidity of the PPC dispersion can bias the optical density values during the determination of minimum inhibitory concentration (MIC). Thus, to accurately test the biological effect, large aggregates should be prevented or dissolved. We found that this can be achieved by simply incubating the polymers for a longer time in the respective medium. For polymers that did show aggregation, time-dependent DLS measurements revealed that the PPC formation is reversible and that the initial large aggregates dissolved after 2 hours.To further examine the potential interaction between AMPs and proteins in solution, we determined the zeta potential (ζ) of all polymers. Here, a positive ζ-potential represents a net cationic charge of the polymer and thereby governs the interaction with the negatively charged bacteria.57 In DI water, all polymers exhibit positive ζ-potentials ranging from +11 to +50 mV (ESI, Fig. S39–S42†). This can be attributed to the free sulfonium and ammonium cationic groups. Notably, PEG-containing polymers possess reduced potentials when compared to their non-PEGylated counterparts. This reduction stems from the negative contribution of PEG, which can be attributed to the affinity of hydroxide ions (asymmetric adsorption of water ions).58 In comparison with these values from DI water, the ζ-potentials in the LB medium are reduced but still overall positive (Fig. S39–S42†). We suggest that this is the result of electrostatic interactions between the AMPs and negative charges of proteins in the medium. However, DLS did not reveal large aggregates in these samples. Thus, we suggest that the polymers in our library can interact with proteins but not to an extent that causes the formation of large aggregates which could hinder interactions with bacteria.
Bioactivity: antimicrobial activity, hemolytic activity, and cellular toxicity of polymers
Estimating the therapeutic potential of AMPs requires quantification of two key properties: (i) their antimicrobial activity and (ii) their hemolytic activity. The balance between these properties determines how efficient the polymers are in inhibiting bacterial growth while avoiding toxic side effects. To examine the influence of the polymer structure and composition on these parameters, the following sets of experiments were conducted.First, the antimicrobial activity of the polymer library was tested against Gram-positive and Gram-negative bacteria to determine the susceptibility of different bacterial cell walls. As representative strains for Gram-positive bacteria, we selected B. subtilis and S. aureus. As representative strains for Gram-negative bacteria, E. coli and P. aeruginosa were selected. All tested strains were non-resistant to avoid undefined and varying influences of different resistance mechanisms that can occur in resistant strains, especially in clinical isolates.59–61 Thus, focusing on non-resistant strains allows accurate comparisons that are needed to develop the required structure–property relationships. To quantitatively examine the inhibitory effect of the polymers, a standard broth micro-dilution method was conducted. Here, the optical density of the bacterial broth was measured with respect to its dependence on the polymer concentration at fixed time points. First, polymer concentrations in 2-fold dilution steps from 256 to 0.25 μg mL−1 were tested in triplicate to give a first estimate of the minimum inhibitory concentration, i.e., the lowest polymer concentration at which more than 90% bacterial growth was inhibited (MIC90). For same center polymers, additional tests were performed in smaller dilution steps in a narrower concentration range. With this, we aimed to get more detailed information about the MIC90 values (see the ESI† for experimental details).
Second, hemolytic activity of the polymers was tested against isolated fresh human erythrocytes. A standard hemoglobin release mediated assay was used to determine the influence of polymer concentration and gave access to the hemolytic concentration at which 50% hemolysis occurred (HC50) (see the ESI† for experimental details).
Finally, cell cytotoxicity of the polymers was tested on L929 and HaCat cell lines. The Cell Counting Kit-8 (CCK-8) assay was performed to test the influence of polymers on human cells and this gave access to the percentage cell viability of polymers at different concentrations (0.1–1 mg mL−1) tested (see the ESI† for experimental details).
![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) P of the polymers allowed a systematic correlation between biological activity and polymer hydrophobicity, i.e., chemical composition.
P of the polymers allowed a systematic correlation between biological activity and polymer hydrophobicity, i.e., chemical composition.
Regarding the antimicrobial activity, we found that the same center sulfonium-based polymers were active against all tested strains. In all cases, a clear dependency on the clog![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) P was observed. This means that MIC90 decreased with increasing polymer hydrophobicity until the best activity was reached for clog
P was observed. This means that MIC90 decreased with increasing polymer hydrophobicity until the best activity was reached for clog![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) P values between 8 and 10 (Fig. 3c–f). The most hydrophilic PS+-me showed the highest MIC90: around 24–32 μg mL−1 against B. subtilis and E. coli and around 70 μg mL−1 against S. aureus and P. aeruginosa. For all strains, the lowest MIC90 was observed for PS+-me-he polymers with a clog
P values between 8 and 10 (Fig. 3c–f). The most hydrophilic PS+-me showed the highest MIC90: around 24–32 μg mL−1 against B. subtilis and E. coli and around 70 μg mL−1 against S. aureus and P. aeruginosa. For all strains, the lowest MIC90 was observed for PS+-me-he polymers with a clog![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) P value of around 11. This corresponds to 12 μg mL−1 against B. subtilis, 20 μg mL−1 against E. Coli, 26 μg mL−1 against S. aureus and 26 μg mL−1 against P. aeruginosa. A further increase in hydrophobicity did not decrease the MIC90 value anymore. In contrast, with large hydrophobic groups, i.e., hexyl groups in PS+-he, the MIC90 increased again. In analogy to observations by other groups, we assume that this effect can be caused by the reduced aqueous solubility of the polymer. This can cause the assembly of hydrophobic groups, thus hindering their interaction with the bacterial cell membrane.21,62,63
P value of around 11. This corresponds to 12 μg mL−1 against B. subtilis, 20 μg mL−1 against E. Coli, 26 μg mL−1 against S. aureus and 26 μg mL−1 against P. aeruginosa. A further increase in hydrophobicity did not decrease the MIC90 value anymore. In contrast, with large hydrophobic groups, i.e., hexyl groups in PS+-he, the MIC90 increased again. In analogy to observations by other groups, we assume that this effect can be caused by the reduced aqueous solubility of the polymer. This can cause the assembly of hydrophobic groups, thus hindering their interaction with the bacterial cell membrane.21,62,63
While an increase in polymer hydrophobicity increased the antibacterial activity, it also increased the hemolytic activity. As shown in Fig. 3f, the HC50 value decreased with an increasing clog![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) P value of the polymers. For clog
P value of the polymers. For clog![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) P ≥ 7, penetration of hydrophobic groups into the mammalian cell membranes causes severe hemolysis, i.e., an HC50 value below 30 μg mL−1. Thus, only same center sulfonium-based polymers with small hydrophobic side groups (me, me-be, and be) and a corresponding clog
P ≥ 7, penetration of hydrophobic groups into the mammalian cell membranes causes severe hemolysis, i.e., an HC50 value below 30 μg mL−1. Thus, only same center sulfonium-based polymers with small hydrophobic side groups (me, me-be, and be) and a corresponding clog![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) P below 7 were suitable to prevent toxic side effects.
P below 7 were suitable to prevent toxic side effects.
Regarding cell viability, only the most hydrophilic PS+-me showed 100% cell viability up to a concentration of 100 μg mL−1 (Fig. S44b†). For the other polymers, cell viability decreased with an increase in hydrophobicity (PS+-me-be, PS+-be, PS+-pr, PS+-bz, PS+-me-he, and PS+-he). However, for all polymers, cell viability was not significantly reduced in the concentration range that is needed for antimicrobial activity <100 μg mL−1. Only for PS+-he, viability was reduced to 50–60% even at 10 μg mL−1 (see Fig. S44a and b†). This trend was observed in both HaCat and L929 cell lines (see the ESI for further details, Fig. S44†).
![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) P < 7. However, in this hydrophilicity range, the antibacterial activity is also reduced. Thus, we aimed to examine whether a change to different center polymer structures can improve these biological properties. For this, we examined different center polymers (PS+-Xdc) with clog
P < 7. However, in this hydrophilicity range, the antibacterial activity is also reduced. Thus, we aimed to examine whether a change to different center polymer structures can improve these biological properties. For this, we examined different center polymers (PS+-Xdc) with clog![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) P values in a comparable hydrophilicity range (1 < clog
P values in a comparable hydrophilicity range (1 < clog![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) P < 5), i.e., PS+-prdc, PS+-budc, and PS+-hedc. These polymers contain similar functional groups as the same center polymers but distributed along different repeating units (see Fig. 4a). The hydrophobic side groups vary slightly from those in their same center analogues to ensure a similar clog
P < 5), i.e., PS+-prdc, PS+-budc, and PS+-hedc. These polymers contain similar functional groups as the same center polymers but distributed along different repeating units (see Fig. 4a). The hydrophobic side groups vary slightly from those in their same center analogues to ensure a similar clog![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) P value to the constant quantitative descriptor. With this, we aim to compensate for the inherent incomparability that would arise from identical hydrophobic groups on same center and different center polymers. For example, using hydrophobic hexyl side groups results in a same center polymer PS+-he with a clog
P value to the constant quantitative descriptor. With this, we aim to compensate for the inherent incomparability that would arise from identical hydrophobic groups on same center and different center polymers. For example, using hydrophobic hexyl side groups results in a same center polymer PS+-he with a clog![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) P > 20, whereas the same hexyl groups on a different center polymer (PS+-hedc) decrease the clog
P > 20, whereas the same hexyl groups on a different center polymer (PS+-hedc) decrease the clog![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) P to 5.03.
P to 5.03.
The antibacterial activity of these polymers followed a similar trend to that for the same center polymers and the MIC90 value decreased with increasing polymer hydrophobicity. For the Gram-negative strains, a change in polymer hydrophobicity from clog![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) P = 1.05 (PS+-prdc) to clog
P = 1.05 (PS+-prdc) to clog![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) P = 5.03 (PS+-hedc) decreased the MIC90 value from >256 μg mL−1 to 32 μg mL−1 against E. coli (Fig. 4e). In the case of P. aeruginosa, it decreased from >256 μg mL−1 to 64 μg mL−1 (Fig. 4f). For the Gram-positive strains, all polymers showed good activity against B. subtilis (MIC90 = 32–8 μg mL−1, see Fig. 4c), whereas none of the polymers was active against S. aureus (Fig. 4d).
P = 5.03 (PS+-hedc) decreased the MIC90 value from >256 μg mL−1 to 32 μg mL−1 against E. coli (Fig. 4e). In the case of P. aeruginosa, it decreased from >256 μg mL−1 to 64 μg mL−1 (Fig. 4f). For the Gram-positive strains, all polymers showed good activity against B. subtilis (MIC90 = 32–8 μg mL−1, see Fig. 4c), whereas none of the polymers was active against S. aureus (Fig. 4d).
Overall, the different center polymers' antimicrobial properties deviate from those of the same center polymers. To give a rough estimate of the influence of the polymer structure, we compared the MIC90 values of same center and different center polymers with similar hydrophobicity.
Here, PS+-me-be and PS+-budc both show similar hydrophobicity with a clog![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) P value of around 2 (1.88 and 2.38, respectively). In this direct comparison, the different center polymers show less antimicrobial activity, i.e., a higher MIC90, against all tested strains. However, the reduction of polymer structure to clog
P value of around 2 (1.88 and 2.38, respectively). In this direct comparison, the different center polymers show less antimicrobial activity, i.e., a higher MIC90, against all tested strains. However, the reduction of polymer structure to clog![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) P as a descriptor can only give a rough estimate of the structural effect since it omits the specific influence of polymer–membrane interactions. Here, we assume that the longer hexyl side groups in PS+-me-he can show a stronger interaction with the hydrophobic lipids in the bilayer membranes, thus reducing the MIC90.37,64 In addition, we suggest that electronic effects also need to be taken into consideration. In the same center polymers, the sulfonium cations are accompanied by β-hydroxy groups. These substituents are known to enhance the stability of sulfonium cations.25 As a result, interactions between the same center sulfoniums and the anionic bacterial cell membrane might be enhanced in comparison with the different center polymers without β-hydroxy substituents.
P as a descriptor can only give a rough estimate of the structural effect since it omits the specific influence of polymer–membrane interactions. Here, we assume that the longer hexyl side groups in PS+-me-he can show a stronger interaction with the hydrophobic lipids in the bilayer membranes, thus reducing the MIC90.37,64 In addition, we suggest that electronic effects also need to be taken into consideration. In the same center polymers, the sulfonium cations are accompanied by β-hydroxy groups. These substituents are known to enhance the stability of sulfonium cations.25 As a result, interactions between the same center sulfoniums and the anionic bacterial cell membrane might be enhanced in comparison with the different center polymers without β-hydroxy substituents.
The hemolytic activity of the different center polymers also increased with increasing polymer hydrophobicity. PS+-prdc and PS+-budc showed good compatibility with red blood cells with HC50 = 5000 and 4000 μg mL−1, respectively. However, for PS+-hedc, the hemolytic activity increased drastically, which corresponds to an HC50 value of 70 μg mL−1. Overall, these results show that for clog![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) P values below 5, the different center polymers show better compatibility with red blood cells than the same center polymers of comparable hydrophobicity.
P values below 5, the different center polymers show better compatibility with red blood cells than the same center polymers of comparable hydrophobicity.
Similarly, the viability of HaCat and L929 cells decreased with increasing polymer hydrophobicity, i.e., increasing alkyl chain length (pr > bu > he). Overall, PS+-prdc showed the best compatibility e.g., 100% viability for HaCat cells, even at 1000 μg mL−1 (Fig. S46b†). For the other polymers PS+-budc and PS+-hedc, cell viability was reduced at high concentrations. However, cytotoxic effects were negligible in the concentration range that is needed for antimicrobial activity <100 μg mL−1 (Fig. S46a and b†). In comparison with the same center sulfonium polymers, the different center analogues showed a reduced cytotoxic effect (see the ESI for further details, Fig. S44 and S46†).
![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) P values below 7, both polymer types show only limited hemolytic activity that would allow their use in a therapeutic application. In this hydrophobicity range, the same center structures show a higher antimicrobial activity than different center structures. However, different center polymers show better compatibility to red blood cells, i.e., a reduced hemolytic activity.
P values below 7, both polymer types show only limited hemolytic activity that would allow their use in a therapeutic application. In this hydrophobicity range, the same center structures show a higher antimicrobial activity than different center structures. However, different center polymers show better compatibility to red blood cells, i.e., a reduced hemolytic activity.
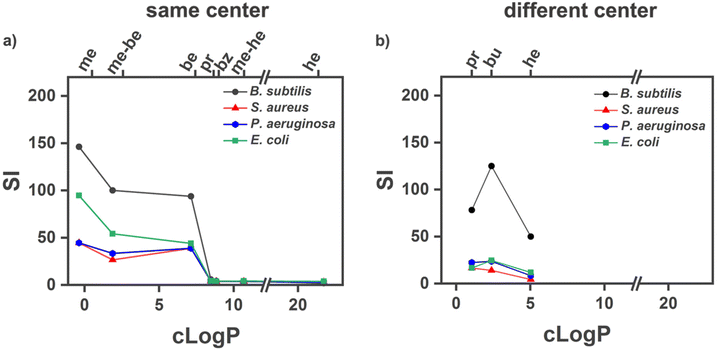
Thus, estimating a therapeutic potential requires weighing both effects against each other. For this, a selectivity index (SI) was used to represent the ratio of HC50 to MIC90.65 In the case of maximum values like MIC90 > 256 μg mL−1 and HC50 > 5000 μg mL−1, 256 and 5000 μg mL−1 were used for calculations, respectively. While this interpretation can introduce deviations from real values, we used the highest concentration tested as a published standard protocol,38 thus reducing these deviations to a systematic effect. In Fig. 4, we plotted the SI for the examined polymers against their clog![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) P values. For clog
P values. For clog![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) P values below 7, it can be seen that same center polymers were more selective than different center polymers of comparable hydrophobicity. In this hydrophobicity range, the higher antimicrobial activity of the same center structures can compensate for their slightly higher hemolytic activity. Above clog
P values below 7, it can be seen that same center polymers were more selective than different center polymers of comparable hydrophobicity. In this hydrophobicity range, the higher antimicrobial activity of the same center structures can compensate for their slightly higher hemolytic activity. Above clog![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) P values of 7, the hemolytic toxicity dominates, and selectivity is lost.
P values of 7, the hemolytic toxicity dominates, and selectivity is lost.
In general, selectivity is based on differences in the composition and structure of outer membranes from bacterial and mammalian cells: While bacterial membranes have a strong negative charge due to anionic phospholipids, mammalian cell membranes are mostly made of zwitterionic phospholipids, which result in a significantly reduced net charge.17,66 Thus, cationic polymers are more prone to interacting with bacterial cells than mammalian cells. However, to exhibit antimicrobial activity, hydrophobic groups are also required to disrupt the membrane. These interactions are not selective. Thus, optimizing the SI requires balancing the cation-based selectivity with antimicrobial activity and hemolytic toxicity.
![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) P ≥ 7, hemolytic toxicity dominates over the good antimicrobial activity (Fig. 3 and Fig. 5). In contrast, for hydrophilic polymers with clog
P ≥ 7, hemolytic toxicity dominates over the good antimicrobial activity (Fig. 3 and Fig. 5). In contrast, for hydrophilic polymers with clog![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) P ∼ 0, hemolytic activity is negligible. But antimicrobial activity is reduced, too. This problem of balancing competing biological responses in copolymer structures is well-known for ammonium-based copolymers.67–69 Thus, we aimed to compare our sulfonium polymers to their ammonium analogues to determine the effect of the cation type. For this, we focused on two exemplary polymers: First, PS+-me was chosen as a representative for a hydrophilic polymer structure (clog
P ∼ 0, hemolytic activity is negligible. But antimicrobial activity is reduced, too. This problem of balancing competing biological responses in copolymer structures is well-known for ammonium-based copolymers.67–69 Thus, we aimed to compare our sulfonium polymers to their ammonium analogues to determine the effect of the cation type. For this, we focused on two exemplary polymers: First, PS+-me was chosen as a representative for a hydrophilic polymer structure (clog![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) P ∼ 0) that shows good compatibility with blood cells but moderate antimicrobial activity. Second, PS+-pr was chosen as a representative of a moderately hydrophobic polymer structure (clog
P ∼ 0) that shows good compatibility with blood cells but moderate antimicrobial activity. Second, PS+-pr was chosen as a representative of a moderately hydrophobic polymer structure (clog![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) P = 7) where the hemolytic toxicity starts to dominate over good antimicrobial activity (Fig. 6a). The biological properties (HC50 and MIC90) of these polymers were compared to their ammonium-based structural analogues PN+-me and PN+-pr (see Fig. 1).
P = 7) where the hemolytic toxicity starts to dominate over good antimicrobial activity (Fig. 6a). The biological properties (HC50 and MIC90) of these polymers were compared to their ammonium-based structural analogues PN+-me and PN+-pr (see Fig. 1).
For all tested bacterial strains, the sulfonium polymers (PS+-me, PS+-pr) showed higher antimicrobial activity than their ammonium analogues (PN+-me, PN+-pr) (Fig. 6a). Importantly, the ammonium-based polymers only showed activity against B. subtilis.
Regarding the hemolytic activity of both cation types, PS+-me was less hemolytic than its ammonium counterpart, PN+-me (Fig. 6a). In contrast, PS+-he showed high hemolytic activity with an HC50 value of 50 μg mL−1. Here, its counterpart PN+-pr caused comparably low hemolytic effects even up to a concentration of 2000 μg mL−1 (Fig. 6b).
Cell viability tests showed that the ammonium polymers are generally less cytotoxic than their sulfonium counterparts (Fig. S45a and b†). Here, PN+-me was found to be the least cytotoxic polymer with 70–80% cell viability even at 1000 μg mL−1. While this demonstrates good cellular compatibility of ammonium-based polymers, their antimicrobial activity is also drastically reduced (see Fig. 6b).
In summary, these results demonstrate that the methyl-functionalized same center polymers showed a good balance between antimicrobial activity, low hemolytic toxicity, and high cell compatibility. Here, PS+-me was more selective against all tested strains in comparison with its ammonium analogue PN+-me (Fig. 6c).
In general, sulfonium polymers PS+-budc and PS+-hedc were more active against the tested strains than their corresponding ammonium analogues (PN+-budc, PN+-hedc) (Fig. 7a). Here, it is noteworthy that the ammonium polymers were active only against B. subtilis and that all different center polymers showed very low antimicrobial activity against S. aureus.
Regarding the hemolytic activity, the ammonium polymers did not show significant hemolytic activity (only 1% hemolysis is observed at 5000 μg mL−1). In particular, for the hexyl-functionalized polymers, this is in direct contrast to their sulfonium counterparts where PS+-hedc showed 50% hemolysis at a concentration of 400 μg mL−1 (Fig. 7b).
Cell viability of different center sulfonium and ammonium-based polymers decreased with increasing hydrophobicity of hydrophobic side groups (pr, bu, he). This trend was observed equally in both sulfonium and ammonium-based polymers for the first two hydrophobic side groups (pr, bu) (Fig. S46a and b†). However, for the hexyl group, PN+-hedc showed better cell compatibility up to 100 μg mL−1 in comparison with its sulfonium analogue PS+-hedc, which is cytotoxic at this particular concentration (see the ESI for further details, Fig. S46a and b†).
Overall, these results suggest that for different center polymers, the cation type has a pronounced effect on biological activity. Here, the ammonium polymers did not show any significant activity which also translates to limited selectivity. In contrast, the sulfonium polymers are still active but show less activity and selectivity than their same center counterparts.
![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) P value of 8.47. It is assumed that the respective large hydrophobic side groups (pr, he) can effectively penetrate the lipid bilayers of cell walls, which leads to high antimicrobial activity with an MIC90 value below 16 μg mL−1 but also an HC50 value of only 50 μg mL−1 (Fig. 3a and b). Thus, to exploit this high antimicrobial activity, the polymers' hemolytic toxicity needs to be reduced. In this context, it has been shown that incorporating PEG (polyethylene glycol) oligomers into AMPs can increase their selectivity towards microbes by enhancing compatibility with erythrocytes and various mammalian cell lines.70–73 Based on these findings, we aimed to further increase the performance of hydrophobic same center PS+-X polymers by adding oligomeric PEG chains as neutral hydrophilic groups into the polymer structure.
P value of 8.47. It is assumed that the respective large hydrophobic side groups (pr, he) can effectively penetrate the lipid bilayers of cell walls, which leads to high antimicrobial activity with an MIC90 value below 16 μg mL−1 but also an HC50 value of only 50 μg mL−1 (Fig. 3a and b). Thus, to exploit this high antimicrobial activity, the polymers' hemolytic toxicity needs to be reduced. In this context, it has been shown that incorporating PEG (polyethylene glycol) oligomers into AMPs can increase their selectivity towards microbes by enhancing compatibility with erythrocytes and various mammalian cell lines.70–73 Based on these findings, we aimed to further increase the performance of hydrophobic same center PS+-X polymers by adding oligomeric PEG chains as neutral hydrophilic groups into the polymer structure.
For this, we prepared random same center copolymers where half of the sulfonium groups were functionalized with a hydrophobic alkyl chain and the other half of the sulfonium groups contained PEG oligomers (Mn = 400 g mol−1, n = 8) (see Fig. 1). These copolymers are denoted as PS+-me-PEG, PS+-pr-PEG, and PS+-he-PEG. Moreover, a control polymer (PS+-PEG) containing only PEG side chains was also prepared. All polymers were examined with respect to their antimicrobial activity and their hemolytic toxicity. To determine the influence of the neutral hydrophilic side chains, the results were compared to the respective homopolymers that contained the same hydrophobic groups but no PEG, i.e., PS+-me, PS+-pr, and PS+-he. As illustrated in Fig. 8, PEGylation increased the MIC90 against all strains. In particular, for the methyl-functionalized polymers, this led to a loss of antimicrobial activity. Here, the PEG groups outweighed the short methyl groups, thus resulting in activity that resembles the fully PEGylated control polymer, PS+-PEG, without any hydrophobic groups (see ESI Fig. S43†). However, for the propyl- and hexyl-functionalized polymers, the increase in MIC90 was less pronounced. In particular, for the propyl-containing polymers the antimicrobial activity of the PEGylated copolymers PS+-pr-PEG closely resembled the activity of the non-PEGylated homopolymer PS+-pr.
Regarding the hemolytic activity, PEGylation can lead to good cell compatibility, i.e., the PS+-PEG control without any hydrophobic groups showed only 1% hemolysis at a concentration of 7000 μg mL−1 (ESI, Fig. S43†). For all amphiphilic copolymers PS+-X-PEG, PEGylation increased the HC50 value in comparison with their homopolymer counterparts PS+-X.
Moreover, PEGylation led to an increase in cell viability too; for instance, the PS+-PEG control showed 100% cell viability even at a concentration of 1000 μg mL−1 (the highest tested concentration, ESI, Fig. S47a and b†). For all amphiphilic copolymers PS+-X-PEG, PEGylation increased the cell viability in comparison with their homopolymer counterparts PS+-X. All PS+-X-PEG polymers showed no pronounced cytotoxic effects up to 100 μg mL−1 (ESI, Fig. S47a and b†), whereas non-PEGylated counter polymers, particularly PS+-pr and PS+-he, showed clear toxicity at this concentration (see ESI Fig. S44†).
As a result, PEGylation of the propyl-functionalized polymers showed an optimal balance of retained antimicrobial activity but reduced hemolytic toxicity, i.e., an HC50 value of 1500 μg mL−1. Thus, PS+-pr-PEG combined good selectivity with reasonable MIC90 values against each tested strain. This demonstrates the successful balance between the structure of alkyl side chains and the addition of neutral hydrophilic groups.
Conclusion
In this work, we systematically examined the potential of sulfonium-based side chain polymers as antimicrobial agents. For these polymers, we systematically varied the polymer structure and composition to investigate the influence of these parameters on antimicrobial and hemolytic activities. For accurate comparison between the biological activities of polymers, all polymers were synthesized from a master batch of PPFPAM precursor polymers through a 2-step functionalization strategy. This ensured the same degree of polymerization and the same dispersity in all polymer samples.First, we examined the influence of the polymer structure. For this, cationic sulfonium polymers with same center and different center structures were compared. For both polymer structures, different hydrophobic side groups were used to change the overall hydrophobicity of the polymers. Independent of the polymer structure (same center or different center), antimicrobial activity increased with polymer hydrophobicity, i.e., clog![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) P. However, cytotoxicity against human cells also increased with clog
P. However, cytotoxicity against human cells also increased with clog![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) P, thus drastically reducing selectivity for polymers with clog
P, thus drastically reducing selectivity for polymers with clog![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) P > 7. For more hydrophilic polymers (clog
P > 7. For more hydrophilic polymers (clog![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) P < 7), a direct comparison of polymers with similar clog
P < 7), a direct comparison of polymers with similar clog![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) P values showed that the same center polymers were more active than their different center analogues.
P values showed that the same center polymers were more active than their different center analogues.
Second, the influence of the cation type was investigated by comparing two polymers from the same and different center libraries (PS+-me and PS+-pr) with their comparable QAC analogues (PN+-me and PN+-pr). Our findings demonstrate that sulfonium polymers show superior antimicrobial activity compared to the ammonium ones. Nevertheless, the ammonium-based polymers show slightly reduced toxicity.
Based on these tests, we established two key structure–property relationships for sulfonium-based side chain polymers: (i) same center polymers show higher selectivity than their different center analogues and (ii) sulfonium-based polymers show higher antimicrobial activity than their ammonium-based analogues but show a slightly increased hemolytic activity.
Thus, to reduce the cellular toxicity of same center sulfonium-based polymers, we introduced additional neutral hydrophilic PEG side groups into the most active polymers. We then compared the biological activity of PS+-me-PEG, PS+-pr-PEG, and PS+-he-PEG with their corresponding non-PEGylated counterparts PS+-me, PS+-pr, and PS+-he. These tests revealed that PEG moieties reduced the toxic effects of PS+-pr while retaining its antimicrobial activity. As a result, this systematic optimization based on structure–property relationships gave access to a promising candidate, i.e., PS+-pr-PEG, which showed good selectivity against all tested bacterial strains.
Overall, the development of sulfonium polymers as potential new therapeutics is still in its infancy. At this point, it was crucial to determine the general potential of these side chain sulfonium polymers as antimicrobial agents. For this, it was important to thoroughly understand the impact of structural and chemical variations on their activity, toxicity, and selectivity. Thus, in this work, we developed structure–property relationships that complement existing studies on main-chain sulfonium polymers. The resulting more fundamental understanding of this AMP class can guide the development of promising candidates for future tests and applications.
Translating these polymers to clinical applications is envisioned to be versatile. Potential applications range from parenteral administration, over topical administration in wounds, to polymer brushes or films as coatings for implants. The actual field of application depends on future studies. These would start with testing the most promising sulfonium polymers (PS+-me and PS+-pr-PEG) against resistant strains and examining the potential formation of resistance against these polymers. Here, sulfonium polymers are suggested to reduce the potential of resistance formation due to the unique properties of the sulfonium cations. Then, next steps would include testing these polymers in in vitro and in vivo infection models. In such examinations, the AMPs should then be tested against relevant antibiotic/antimicrobial benchmarks.
Author contributions
D.K. contributed to the concept of this work, participated in the thorough discussion of the experimental results, and provided all necessary reagents/materials/analysis tools to perform this research work. S.K conceived and designed the experiments, performed the synthesis and characterization of the materials, and analyzed the data. U.B.A.A performed the hemolysis assays and trained S.K to carry out MIC assays. E.Q. and K.A. performed the cytotoxicity studies. All authors have participated in the writing and given approval to the final version of the manuscript.Data availability
The data supporting this article have been included as part of the ESI.† Raw data are available via Zenodo, DOI: 10.5281/zenodo.14577780.Conflicts of interest
The authors declare no conflicts of interest.Acknowledgements
Sidra Kanwal acknowledges the German academic exchange program (DAAD) for the scholarship granted for her PhD studies (Funding programme number: 57507871, personal reference number: 91765401). Umer B. A. Aziz is grateful to Investitionsbank Berlin for a postdoc funding (VAL130/2023). We thank Prof. Charlotte Kloft for kindly providing us the bacterial strains. We thank Dr Stefanie Wedepohl for helping with the hemolytic assays. We also extend our gratitude to Ms Sijie Liu for kindly calculating the clog![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) P of our polymers.
P of our polymers.
References
- D. M. Brogan and E. Mossialos, Global Health J., 2016, 12, 8 CrossRef PubMed.
- P. A. Lambert, Adv. Drug Delivery Rev., 2005, 57, 1471–1485 CrossRef CAS PubMed.
- F. C. Tenover, Am. J. Infect. Control, 2006, 34, S3–S10 CrossRef PubMed.
- P. Fernandes, Nat. Biotechnol., 2006, 24, 1497–1503 CrossRef CAS PubMed.
- S. B. Levy and M. Bonnie, Nat. Med., 2004, 10, S122–S129 CrossRef PubMed.
- C. Nathan, Nature, 2004, 431, 899–902 CrossRef PubMed.
- M. A. Fischbach and C. T. Walsh, Science, 2009, 325, 1089 CrossRef PubMed.
- H. F. Chambers and F. R. DeLeo, Nat. Rev. Microbiol., 2009, 7, 629–641 CrossRef.
- M. Zasloff, Nature, 2002, 415, 389–395 CrossRef.
- K. A. Brogden, Nat. Rev. Microbiol., 2005, 3, 238–250 CrossRef.
- K. Kuroda and G. A. Caputo, Wiley Interdiscip. Rev.:Nanomed. Nanobiotechnol., 2013, 5, 49–66 Search PubMed.
- J. P. S. Powers and R. E. W. Hancock, Peptides, 2003, 24, 1681–1691 CrossRef PubMed.
- C. Ergene, K. Yasuhara and E. F. Palermo, Polym. Chem., 2018, 9, 2407–2427 RSC.
- E. R. Kenawy, S. D. Worley and R. Broughton, Biomacromolecules, 2007, 8, 1359–1384 CrossRef PubMed.
- B. Dizman, M. O. Elasri and L. J. Mathias, J. Appl. Polym. Sci., 2004, 94, 635–642 CrossRef.
- K. Kuroda, G. A. Caputo and W. F. DeGrado, Chem. – Eur. J., 2009, 15, 1123 CrossRef PubMed.
- P. Pham, S. Oliver and C. Boyer, Macromol. Chem. Phys., 2023, 224, 1–28 CrossRef.
- K. Hu, N. W. Schmidt, R. Zhu, Y. Jiang, G. H. Lai, G. Wei, E. F. Palermo, K. Kuroda, G. C. L. Wong and L. Yang, Macromolecules, 2013, 46, 1908–1915 CrossRef PubMed.
- B. C. Allison, B. M. Applegate and J. P. Youngblood, Biomacromolecules, 2007, 8, 2995–2999 CrossRef PubMed.
- P. Pham, S. Oliver, E. H. H. Wong and C. Boyer, Polym. Chem., 2021, 12, 5689–5703 RSC.
- S. Laroque, M. Reifarth, M. Sperling, S. Kersting, S. Klöpzig, P. Budach, J. Storsberg and M. Hartlieb, ACS Appl. Mater. Interfaces, 2020, 12, 30052–30065 CrossRef.
- Q. Wei, T. Becherer, S. Angioletti-Uberti, J. Dzubiella, C. Wischke, A. T. Neffe, A. Lendlein, M. Ballauff and R. Haag, Angew. Chem., Int. Ed., 2014, 53, 8004–8031 CrossRef PubMed.
- J. Guo, J. Qin, Y. Ren, B. Wang, H. Cui, Y. Ding, H. Mao and F. Yan, Polym. Chem., 2018, 9, 4611–4616 RSC.
- V. Sambhy, B. R. Peterson and A. Sen, Angew. Chem., Int. Ed., 2008, 47, 1250–1254 CrossRef PubMed.
- J. Oh and A. Khan, Biomacromolecules, 2021, 22, 3534–3542 CrossRef PubMed.
- M. Hartlieb, E. G. L. Williams, A. Kuroki, S. Perrier and K. E. S. Locock, Curr. Med. Chem., 2017, 24, 2115–2140 CrossRef.
- S. E. Exley, L. C. Paslay, G. S. Sahukhal, B. A. Abel, T. D. Brown, C. L. McCormick, S. Heinhorst, V. Koul, V. Choudhary, M. O. Elasri and S. E. Morgan, Biomacromolecules, 2015, 16, 3845–3852 CrossRef PubMed.
- K. Punia, A. Punia, K. Chatterjee, S. Mukherjee, J. Fata, P. Banerjee, K. Raja and N. L. Yang, RSC Adv., 2017, 7, 10192–10199 RSC.
- C. Ergene, K. Yasuhara and E. F. Palermo, Polym. Chem., 2018, 9, 2407–2427 RSC.
- J. Tan, Y. Zhao, J. L. Hedrick and Y. Y. Yang, Adv. Healthcare Mater., 2022, 11, 1–10 Search PubMed.
- M. Mizutani, E. F. Palermo, L. M. Thoma, K. Satoh, M. Kamigaito and K. Kuroda, Biomacromolecules, 2012, 13, 1554–1563 CrossRef PubMed.
- E. A. Chamsaz, S. Mankoci, H. A. Barton and A. Joy, ACS Appl. Mater. Interfaces, 2017, 9, 6704–6711 CrossRef.
- Q. Gao, P. Li, H. Zhao, Y. Chen, L. Jiang and P. X. Ma, Polym. Chem., 2017, 8, 6386–6397 RSC.
- K. Klinker and M. Barz, Macromol. Rapid Commun., 2015, 36, 1943–1957 CrossRef PubMed.
- M. Concilio, R. G. Maset, L. P. Lemonche, V. Kontrimas, J. I. Song, S. K. Rajendrakumar, F. Harrison, C. R. Becer and S. Perrier, Adv. Healthcare Mater., 2023, 12, 1–13 Search PubMed.
- K. Lienkamp, K. N. Kumar, A. Som, K. Nüsslein and G. N. Tew, Chem. – Eur. J., 2009, 15, 11710–11714 CrossRef PubMed.
- P. T. Phuong, S. Oliver, J. He, E. H. H. Wong, R. T. Mathers and C. Boyer, Biomacromolecules, 2020, 21, 5241–5255 CrossRef PubMed.
- P. Pham, S. Oliver, D. T. Nguyen and C. Boyer, Macromol. Rapid Commun., 2022, 43, 1–13 CrossRef.
- B. T. Benkhaled, S. Hadiouch, H. Olleik, J. Perrier, C. Ysacco, Y. Guillaneuf, D. Gigmes, M. Maresca and C. Lefay, Polym. Chem., 2018, 9, 3127–3141 RSC.
- W. Ji, R. R. Koepsel, H. Murata, S. Zadan, A. S. Campbell and A. J. Russell, Biomacromolecules, 2017, 18, 2583–2593 CrossRef PubMed.
- J. L. Anaya-López, J. E. López-Meza and A. Ochoa-Zarzosa, Crit. Rev. Microbiol., 2013, 39, 180–195 CrossRef PubMed.
- A. Peschel, Trends Microbiol., 2002, 10, 179–186 CrossRef PubMed.
- A. Peschel and H. G. Sahl, Nat. Rev. Microbiol., 2006, 4, 529–536 CrossRef PubMed.
- S. Maria-Neto, K. C. De Almeida, M. L. R. Macedo and O. L. Franco, Biochim. Biophys. Acta, Biomembr., 2015, 1848, 3078–3088 CrossRef PubMed.
- Y. Gong, X. Xu, M. Aquib, Y. Zhang, W. Yang, Y. Chang, H. Peng, C. Boyer, A. K. Whittaker and C. Fu, ACS Appl. Polym. Mater., 2024, 6, 6966–6975 CrossRef.
- D. Guan, F. Chen, Y. Qiu, B. Jiang, L. Gong, L. Lan and W. S. Huang, Angew. Chem., Int. Ed., 2019, 58, 6678 CrossRef PubMed.
- R. G. Carden, K. J. Sommers, C. L. Schrank, A. J. Leitgeb, J. A. Feliciano, W. M. Wuest and K. P. C. Minbiole, ChemMedChem, 2020, 15, 1974 CrossRef PubMed.
- A. Activity, Biocontrol Sci., 2011, 16, 23–31 CrossRef PubMed.
- Y. Hu, J. Zhao, J. Zhang, Z. Zhu and J. Rao, ACS Macro Lett., 2021, 10, 990–995 CrossRef PubMed.
- X. Wang, G. Wang, J. Zhao, Z. Zhu and J. Rao, ACS Macro Lett., 2021, 10, 1643–1649 Search PubMed.
- Y. Zhao, X. Wang, Y. Hu, J. Zhao, M. Sun, M. Yang, H. Xuan, X. Wang, J. Zhang, Z. Zhu and J. Rao, ACS Appl. Polym. Mater., 2023, 5, 4437–4447 CrossRef.
- Y. Hu, J. Zhao, M. Yang, X. Wang, H. Zhang, J. Zhang, Z. Zhu and J. Rao, ACS Appl. Polym. Mater., 2022, 4, 4868–4875 CrossRef.
- M. Eberhardt, R. Mruk, R. Zentel and P. Théato, Eur. Polym. J., 2005, 41, 1569–1575 CrossRef.
- U. B. A. Aziz, A. Saoud, M. Bermudez, M. Mieth, A. Atef, T. Rudolf, C. Arkona, T. Trenkner, C. Böttcher, K. Ludwig, A. Hoelzemer, A. C. Hocke, G. Wolber and J. Rademann, Nat. Commun., 2024, 15, 3537 CrossRef PubMed.
- T. M. P. Neumann-Tran, C. López-Iglesias, L. Navarro, E. Quaas, K. Achazi, C. Biglione and D. Klinger, ACS Appl. Polym. Mater., 2023, 5, 7718–7732 CrossRef.
- X. Ma, M. Lin, J. Sun and X. Chen, Adv. NanoBiomed Res., 2023, 3, 1–9 Search PubMed.
- R. J. Kopiasz, N. Kulbacka, K. Drężek, R. Podgórski, I. Łojszczyk, J. Mierzejewska, T. Ciach, E. Augustynowicz-Kopeć, A. Głogowska, A. Iwańska, W. Tomaszewski and D. Jańczewski, Macromol. Biosci., 2022, 22, 2200094 CrossRef PubMed.
- R. Zimmermann, U. Freudenberg, R. Schweiß, D. Küttner and C. Werner, Curr. Opin. Colloid Interface Sci., 2010, 15, 196–202 CrossRef.
- F. O. Olorunmola, D. O. Kolawole and A. Lamikanra, Afr. J. Infect. Dis., 2013, 7, 1–7 Search PubMed.
- K. J. Aroca Molina, S. J. Gutiérrez, N. Benítez-Campo and A. Correa, Microorganisms, 2024, 12, 1116 CrossRef PubMed.
- E. Peterson and P. Kaur, Front. Microbiol., 2018, 9, 1–21 CrossRef PubMed.
- J. Kim, H. Yu, E. Yang, Y. Choi and P. S. Chang, LWT–Food Sci. Technol., 2023, 174, 114421 CrossRef.
- M. Bustelo, A. Pinazo, M. A. Manresa, M. Mitjans, M. P. Vinardell and L. Pérez, Colloids Surf., A, 2017, 532, 501–509 CrossRef.
- W. Chin, G. Zhong, Q. Pu, C. Yang, W. Lou, P. F. De Sessions, B. Periaswamy, A. Lee, Z. C. Liang, X. Ding, S. Gao, C. W. Chu, S. Bianco, C. Bao, Y. W. Tong, W. Fan, M. Wu, J. L. Hedrick and Y. Y. Yang, Nat. Commun., 2018, 9, 1–14 CrossRef PubMed.
- L. Buzoglu Kurnaz, Y. Luo, X. Yang, A. Alabresm, R. Leighton, R. Kumar, J. H. Hwang, A. W. Decho, P. Nagarkatti, M. Nagarkatti and C. Tang, Bioact. Mater., 2023, 20, 519–527 Search PubMed.
- Z. Si, W. Zheng, D. Prananty, J. Li, C. H. Koh, E. T. Kang, K. Pethe and M. B. Chan-Park, Chem. Sci., 2022, 13, 345–364 RSC.
- M. Zasloff, Nature, 2002, 415, 389–395 CrossRef PubMed.
- M. Santos, A. Fonseca, P. Mendonça, R. Branco, A. Serra, P. Morais and J. Coelho, Materials, 2016, 9, 599 CrossRef.
- H. Takahashi, G. A. Caputo, S. Vemparala and K. Kuroda, Bioconjugate Chem., 2017, 28, 1340 CrossRef PubMed.
- S. Colak, C. F. Nelson, K. Nusslein and G. N. Tew, Biomacromolecules, 2009, 10, 353–359 CrossRef PubMed.
- C. A. Cho, C. Liang, J. Perera, M. A. Brimble, S. Swift and J. Jin, Macromol. Biosci., 2020, 20, 65 CrossRef.
- J. Morris, K. Beck, M. A. Fox, D. Ulaeto and M. G. G. C. Clark, Antimicrob. Agents Chemother., 2012, 56, 3298 CrossRef.
- A. K. Lam, E. L. Moen, J. Pusavat, C. L. Wouters, H. Panlilio, M. J. Ferrell, M. B. Houck, D. T. Glatzhofer and C. V. Rice, ACS Omega, 2020, 5(40), 26262–26270 CrossRef PubMed.
Footnote |
† Electronic supplementary information (ESI) available: FTIR spectra of the precursor polymer and its functionalization to thio and tert-amine containing polymers. GPC traces for the first and second functionalizations of poly(PFPMA) to synthesize cationic polymers in same center and different center libraries. 1H NMR spectra of precursor polymers and cationic polymers after functionalization. 19F NMR spectra confirming the transformation of poly(PFPMA) to thio and tert-amine containing precursor polymers. 1H NMR spectra of individual cationic polymers from each library with integration. clog![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) P calculation for exemplary polymers from same center and different center polymers. Confirmation of protein–polymer complex (PPC) formation in LB medium by DLS characterization. The presence of positive charges on sulfonium and ammonium cationic polymers as represented by ζ-potential values. HC50 and MIC90 data for the PS+-PEG control polymer. Cell viability assays for polymers from each library. See DOI: https://doi.org/10.1039/d4bm01247j P calculation for exemplary polymers from same center and different center polymers. Confirmation of protein–polymer complex (PPC) formation in LB medium by DLS characterization. The presence of positive charges on sulfonium and ammonium cationic polymers as represented by ζ-potential values. HC50 and MIC90 data for the PS+-PEG control polymer. Cell viability assays for polymers from each library. See DOI: https://doi.org/10.1039/d4bm01247j |
| This journal is © The Royal Society of Chemistry 2025 |