Open Access Article
Open Access ArticleProduction of biohythane from cow dung using novel microbial synthetic consortia designed by heat-treated and acclimatized combined wastes†
Rashmi
Ira
a,
Vikas
Sharma
a,
Shrawan
Kumar
a,
Mira
Koul
a,
Lalita
Sharma
 b,
Aditi
Halder
b,
Aditi
Halder
 b and
Tulika
Prakash
b and
Tulika
Prakash
 *a
*a
aSchool of Biosciences and Bioengineering, Indian Institute of Technology, Mandi, Kamand, Mandi, Himachal Pradesh 175005, India. E-mail: tulika@iitmandi.ac.in; Tel: +91-1905-267140
bSchool of Chemical Sciences, Indian Institute of Technology, Mandi, Kamand, Mandi, Himachal Pradesh-175005, India
First published on 29th July 2024
Abstract
The world's increasing dependency on fossil fuels has become a significant energy and environmental concern as they contribute 83% of the global energy supply and produce large amounts of carbon dioxide. Biohythane, a blend of biomethane (5–10%) and biohydrogen (50–60%), is emerging as a promising and environmentally friendly alternative fuel derived from organic wastes and offers a sustainable solution. The existing methods of biohythane production suffer from major limitations of being cost- and labor-intensive due to adopting bulk substrate pretreatment to enhance biohythane yield thereby limiting their industrial applications. In this study, we have developed a synthetic microbial consortium (E(C2)Tx) for anaerobic digestion by combining various organic wastes and subjecting them to heat pre-treatment and acclimatization to enrich biohydrogen producers and methanogens, respectively. Raw cow dung was anaerobically digested as the substrate with E(C2)Tx and this resulted in the production of biohythane with 3% biohydrogen and 36% biomethane. The consortia designing strategy avoided any bulk substrate pretreatment and only included the pretreatment of the inoculum which is used in four times less volume than the substrate. A 16S rRNA gene based metagenomic analysis revealed that the CD samples treated with E(C2)Tx were enriched in cellulolytic and hydrogen-producing Firmicutes, along with methylotrophic and hydrogenotrophic methanogens. The developed technology offers promising commercial benefits by requiring less energy for biohythane production. In addition, it offers environmental advantages by providing an efficient CD waste management alternative and reducing climatic impact by lowering greenhouse gas emissions associated with fossil fuel burning. Using a waste complementarity approach for consortia designing aligns with the principles of circular economy and presents a sustainable, scalable energy solution. The developed method can support the growing energy market by increasing biohythane yield and lowering its production cost.
Environmental significanceBiohythane is a blend of CH4 and H2 and is an excellent replacement for non-renewable energy sources. The current biohythane production method involves the pre-treatment of a bulk substrate, which is a cost, energy, and labor-intensive process. Therefore, there is a need for the development of an improved synthetic microbial consortium for this issue, especially in single-stage anaerobic digestion. In this work, we have developed a synthetic microbial consortium using various organic wastes and adopted a heat pre-treatment strategy to enrich H2-producing microbes combined with the acclimatization process to augment methanogens. Following this, we tested the performance of two types of designed consortia (E(C2)Tx and E(C2)UTx) for biohythane production via anaerobic digestion of raw cow dung as a substrate. In addition, we explored the role of different microbial taxa in the anaerobic digestion process, revealing that our synthetic microbial consortium can overcome extreme environmental conditions and work more robustly. The organic composition of the substrate and inoculum allowed the dilution and counteracting of the various toxic compounds or acids from the substrate and inoculum blend, making suitable conditions for microbes' growth, degrading the substrate, and producing biogas/methane. Additionally, our consortia designing approach and findings would stimulate further research in understanding the waste complementarity concept at the substrate level and with various wastes as potential inoculum. |
Introduction
Due to the current global environmental and energy scenarios, the world's increasing dependency on fossil fuels has become a major concern. The global energy demand rose 2.9% in 2018 and is expected to reach 740 million terajoules by 2040, with an expansion of 30%.1 Currently, 83% of this energy is obtained from fossil fuels (mainly coal, oil, and natural gas) which substantially raises greenhouse gas (GHG) emissions (especially carbon dioxide (CO2)) and is leading to changing climatic conditions.2 Renewable energy sources such as biogas, wind, solar energy, etc., are emerging as better alternate options in terms of being more sustainable and generating a lower carbon footprint.3 In addition, implementing circular economy principles (namely reusing, reducing, and recycling wastes) helps lower the carbon footprints by reducing resource extraction and generation of waste.4,5 Following appropriate carbon pricing mechanisms, renewable energy policies are expected to reduce greenhouse gas emissions and lead to a lower carbon price level.5 As a result, renewable energy sources, including biogas, are forming a relatively significant part of primary energy sources in several countries worldwide.4The transition to renewable energy sources is not very swift and profitable and suffers from its own challenges including lower production and reduced supply of inputs.4 For any technology to be sustainable it must be profitable and able to attract investors' interest. From an investor's perspective, investing in funds with guaranteed returns may be a preferred choice,6 although a technology shock is shown to be associated with a growth in private investments. In addition to this, government policies to promote a circular economy via adopting renewable energy as a mechanism are also likely to play a significant role in investments in this direction.7 Another bottleneck in the utilisation of renewable energy sources is cost competitiveness. For example, microalgae emerged as a promising potential feedstock for biodiesel production while treating wastewater. However, the commercial and economic aspects of the microalgae biodiesel business depend on microalgae production cost, which itself suffers from several limitations.8,9
Recently, the blend of biomethane (CH4) with biohydrogen (H2) (concentration between 10% and 30% v/v), known as biohythane10 or H2-enriched compressed natural gas (HCNG), has emerged as another promising clean fuel. CH4 and H2 are widely used by chemical industries due to their high calorific values of 143 kJ g−1 and 55 kJ g−1, respectively.11 Biohythane offers a strong potential to enhance energy recovery by ∼8–43% along with a significant improvement in economic viability.12 As biohythane can reduce carbon monoxide (CO) emissions by up to 50%, it has been used directly as a commercial vehicle fuel in various countries including India, California, China (Beijing Hythane Bus Project), Canada (Montreal Hythane Bus Project),13,14 and the well-established automobile companies such as Volvo, Fiat, etc.15,16
The valorization of biohythane from biomass feedstocks or its microflora is emerging as a promising renewable energy generation technology. Biohythane fuel is derived from biomass feedstocks via the anaerobic digestion (AD) of organic waste, for example, food, microalgae, sewage sludge, and animal manure, including cow dung (CD).17 H2 production in a single-stage dark fermentation AD process yields biohythane with 5–10% H2, 50–60% CH4, and 30–40% carbon dioxide (CO2).10,18 It is estimated that animal manure produces 25 million tons of pollutants annually throughout the world.19 If untreated, these pollutants lead to the emission of noxious gases and repugnant odor, thereby necessitating the development of efficient waste management strategies. CD is an organic waste, which is rich in crude proteins, minerals, fibers, lignocellulosic components, and a diverse microbial community.20 Some of the CD waste management methods include the production of biogas (via anaerobic fermentation) and biochar (through pyrolysis) and recovery of plant nutrients21–23 among others. The utilization of CD as an independent substrate or in co-digestion with other organic wastes for biogas production through the AD process has been very widely explored.19,24–26 Cow manure of 1 kg can produce 35–40 L of biogas when mixed with water for a hydraulic retention time (HRT) of 55–60 days at an ambient temperature of 24–26 °C.27 However, there are only a limited number of studies that have focussed on using CD as a substrate for biohythane production.
The type of feedstock substrate, inoculum, and preprocessing strategies that are used during the AD process significantly affect the yield and quality of biohythane. Various studies have explored different approaches to optimize H2 and CH4 production to meet the requirements of standard biohythane composition.1,12,28 Many recent studies employing various different pretreatment strategies on different types of substrates (e.g., food waste, municipal waste, sludge, algal biomass, animal waste, and agricultural waste) and inoculums (pure or mixed cultures) to enhance H2 yield during biohythane production have been reviewed.1,12,13,29 It is clearly apparent that, first, there are very few studies that have utilised CD as a substrate for biohythane production. Second, in some of the studies simple and easily consumable carbon sources such as glucose, glycerine, cellulose, fruit juices, etc., which themselves are obtained by preprocessing, have been used as substrates.13
Third, in order to facilitate the hydrolysis of lignocellulosic components of a substrate such as CD and make them available for microbial digestion to improve biohythane (or biogas) production several pre-treatment strategies of the substrate have been adopted. These pre-treatment strategies utilize various physical (heat, sonication, etc.) and chemical methods (treatment using acid, alkali, etc.) or a combination of both.30–33 Biological pre-treatment is another alternative method of inoculating industrial cellulolytic enzymes or microbes into the substrate to degrade lignocellulosic components under mild and controlled environmental conditions. However, a major limitation of these substrate pre-treatment procedures is the increased consumption of energy and chemicals, which can lead to an extreme pH or temperature change and the formation of toxic by-products.34 Besides, these substrate pre-treatment procedures are labor- and/or energy-intensive which makes them economically not preferable for managing large quantities of the substrate in real-life applications.35
Fourth, a large majority of studies have adopted bulk substrate pre-treatment instead of pre-treating inoculum, which is used in much lower quantities as compared to the substrate. Some of the adopted pre-treatment methods of inoculum include heat treatment,30 sequential re-inoculation for enrichment,36 blending of different manures,37 acclimatizations,38etc., however, they have been employed independently of each other. Furthermore, some studies have used pure cultures and synthetic media to maintain them for enhancing H2 production. Pure cultures, although effective, require highly sterile operating conditions and extremely pure substrates, making them non-economic and impractical for industrial use. Mixed cultures, while economically viable, face challenges from non-hydrogen-producing organisms like methanogens, lactic acid bacteria, and homoacetogenic bacteria.13 Finally, some studies have performed two-stage AD or continuous AD under different organic load rates and HRT. Maintaining such a setup at the industrial scale requires additional costs limiting their economic viability.
Taken together, the existing methods of biohythane production suffer from several limitations including being labor-, energy-, and cost-intensive which make their real-life application very challenging. This indicates a need for the development of technologies for biohythane production which are environment friendly and free from bulk substrate treatment, use readily available feedstock, contribute towards waste management and thereby the circular economy, and most importantly are cost-effective. Towards this, in the present study, we have developed a technology for the preparation of novel synthetic microbial consortia by combining various types of wastes, namely, aged sludge (AS), paper mill sludge (PM), sewage wastewater, and CD, which is capable of producing biohythane from the CD substrate. Our approach mainly relies on pre-treating the synthetic inoculum, which is used in four times less quantity as compared to the bulk substrate in a single-stage dark fermentation reactor. This microbe-based AD process entails a complex and interconnected anaerobic microbial consortium that performs hydrolysis, acidogenesis, anaerobic/syntrophic oxidation, and methanogenesis in synergy. In such a process, the heat treatment method is known to inhibit the H2 consumers and the methanogenic activity, whereas the spore-forming H2-producing bacteria survive under such stressful conditions. Similarly, a pH range of 5.0 to 6.5 is known to favor the growth of H2-producing microbes.29,39,40 We adopted the heat pre-treatment and acclimatization strategies during our synthetic microbial consortia designing and compared the biohythane production among the treated and untreated inoculum with respect to the control. Furthermore, in order to understand the role of the microbial community dynamics during the AD processes, we have performed a 16S ribosomal RNA metagenomic sequencing analysis of the samples inoculated with heat-pre-treated (E(C2)Tx), untreated (E(C2)UTx), and control samples at the start of the AD process and after 40 days in a single-stage anaerobic digester at the lab-scale.
Materials and methods
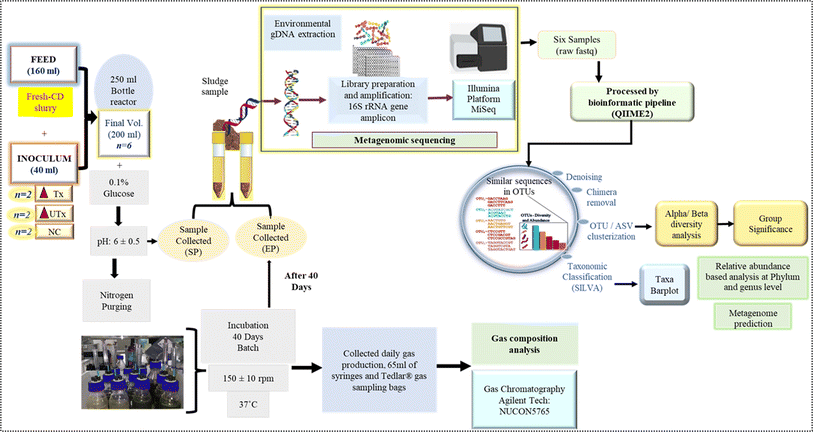
In this study, we have used fresh CD as a feed to evaluate the potential of novel synthetic microbial consortia developed by us to produce biohythane. The complete workflow and study design are given in Fig. 1.Novel synthetic microbial consortia as the inoculum
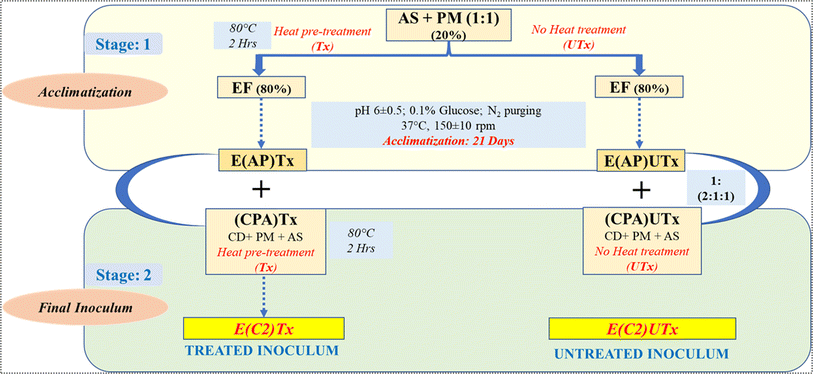
The novel synthetic microbial consortia used in this study consist of four different waste products as the natural mixed cultures: aged sludge (AS) obtained from the local sewage treatment plant (STP) at IIT Mandi (Kamand, Mandi, Himachal Pradesh, India), paper mill sludge (PM) obtained from Shreyans Paper Mill Ltd, Banah (Punjab, India), flocks of electro-flocculated (EF) wastewater were freshly obtained after performing the electro-flocculation technique on freshly collected sewage wastewater at IIT Mandi, as mentioned by Lalita et al. 2021,41 and CD obtained from local cowshed at Salgi, near IIT Mandi. The detailed method for the preparation of the inoculum is described in Fig. 2.Two types of synthetic microbial consortia were used in this study, namely, heat-pre-treated E(C2)Tx and untreated E(C2)UTx, and CD was used as a control. The compositions of both inocula were the same, however, the processing of the constituents to prepare the final inoculum differed on the basis of the heat pre-treatment step in the case of E(C2)Tx in order to retain the H2-producing microbial community. Heat-pre-treatment was performed by boiling the constituents at 80 °C for 2 hours with continuous magnetic stirring.
The preparation of the synthetic inoculum was carried out in two stages. In stage 1, acclimatization of the primary constituents (AS + PM) was performed for 21 days using EF as a carbon source for the microbial community at a pH of 6 and mesophilic temperature of 37 °C. 0.1% glucose (HiMedia) was added during this step to facilitate microbial growth and purging of N2 gas for 5 minutes to maintain an anaerobic environment. In stage 2, the acclimatized sludge prepared by using heat-pre-treated primary constituents was mixed with the heat-pre-treated CPA (CD + AS + PM) component in order to retain the effect of heat treatment throughout the process of inoculum designing for preparing the final treated inoculum (E(C2)Tx). The initial parametric conditions such as total solids (TS), volatile solids (VS), chemical oxygen demand (COD), and pH have been listed for the inoculum E(C2)Tx, in the recently published study by Thakur et al. (2023).24 However, for preparing the final untreated inoculum (E(C2)UTx), we mixed the acclimatized sludge prepared by using untreated primary constituents with untreated CPA components.
Feed preparation
Fresh CD was collected from a local cowshed at Salgi, near North Campus, IIT Mandi, (Kamand, Mandi, Himachal Pradesh, India). The cow was feeding on local vegetation, plants, and grass. Manual cleaning of the sample was performed by removing long grass. CD slurry was prepared by adding tap water in a proportion of 1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1 (v/v). The initial parameters including VS and TS (g VSadded and g TSadded, respectively) for CD as a substrate were calculated as previously described.24
1 (v/v). The initial parameters including VS and TS (g VSadded and g TSadded, respectively) for CD as a substrate were calculated as previously described.24
Experimental conditions
A mini-scale dark-fermentative single-stage bottle bioreactor was prepared in two replicates by using transparent 250 ml Duran® glass bottles that were sealed tightly with rubber corks having four septa, which were fitted with a needle of 65 ml syringes (Dispo Van®) for the collection of gas (Fig. 1). 160 ml of CD slurry as feed and 40 ml of synthetic microbial inoculum were added to prepare a final working volume of 200 ml in these 250 ml mini bottle reactors. This resulted in a substrate and inoculum composition of 4![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1 (v/v) i.e., 20% of synthetic microbial inoculum and 80% of the feed substrate. The final mixture was supplemented with 0.1% of glucose (HiMedia) and the pH was adjusted to 6 ± 0.5. To maintain the anaerobic conditions, the small-scale bottle bioreactor was purged with nitrogen gas. The bottle bioreactors were placed in an incubator in the dark at a temperature of 37 °C ± 1 °C at 150 ± 10 rpm for a period of 40 days (Fig. 1). Analysis of the gaseous mixture was carried out with the help of a gas chromatograph (Agilent, 7890B, Santa Clara, California, United States) which was equipped with two TCDs (Thermal Conductivity Detectors) and one FID (Flame Ionization Detector). All hydrocarbons were detected at the FID and hydrogen was detected at the TCD also known as auxiliary detectors where N2 was used as a reference gas. The injector and detector were maintained at 60 °C.
1 (v/v) i.e., 20% of synthetic microbial inoculum and 80% of the feed substrate. The final mixture was supplemented with 0.1% of glucose (HiMedia) and the pH was adjusted to 6 ± 0.5. To maintain the anaerobic conditions, the small-scale bottle bioreactor was purged with nitrogen gas. The bottle bioreactors were placed in an incubator in the dark at a temperature of 37 °C ± 1 °C at 150 ± 10 rpm for a period of 40 days (Fig. 1). Analysis of the gaseous mixture was carried out with the help of a gas chromatograph (Agilent, 7890B, Santa Clara, California, United States) which was equipped with two TCDs (Thermal Conductivity Detectors) and one FID (Flame Ionization Detector). All hydrocarbons were detected at the FID and hydrogen was detected at the TCD also known as auxiliary detectors where N2 was used as a reference gas. The injector and detector were maintained at 60 °C.
DNA extraction and metagenomic sequencing
The forward (5′TCGTCGGCAGCGTCAGATGTGTATAAGAGACAGCCTACGGGNGGCWGCAG3′) and reverse (5′GTCTCGTGGGCTCGGAGATGTGTATAAGAGACAGGACTACHVGGGTATCTAATCC3′) primers with overhang adapters were used for the amplification of the V3–V4 hypervariable regions of the microbial 16S rRNA gene by a polymerase chain reaction (PCR) (ProFlex PCR, Thermo Fisher Scientific, United States). PCR clean-up was performed using AMPure-beads (Beckman Coulter, California, United States) to purify the amplicons to remove free primers and primer dimers. Subsequently, barcoding for pooling multiple samples was performed using an index PCR with Illumina sequencing dual-index primers provided in Nextera XT Index Kit v2 (Illumina, San Diego, CA, United States) followed by PCR clean-up to generate the final library (4 nM). Spiking in with 25% Phi-X was used as a sequencing control. Equimolar amounts of the respective 16S rRNA gene amplicon pools were sequenced by using a MiSeq platform (Illumina, San Diego, CA, United States) available in the in-house NGS facility of IIT Mandi, by using MiSeq v3 reagents with paired-end 300-bp read chemistry (2 × 300 bp). After trimming the adapters and primer sequences, raw fastq sequence files were acquired and subjected to further bioinformatics analysis.
Post-sequencing bioinformatics analysis
Sequenced reads in the FASTQ format were subjected to a comprehensive bioinformatics processing and analysis using the QIIME2, version 2021.8. (Quantitative Insights into Microbial Ecology) software suite.42 Raw reads were imported into QIIME2 with the “Casava 1.8 paired end demultiplexed fastq” method (Fig. 1). The raw file was filtered and trimmed first to remove all the sequences with an average Phred score or quality value of below 20 or size less than 20 bp. The DADA2 (Divisive Amplicon Denoising Algorithm 2) plugin was used to denoise and filter the reads, which includes trimming and truncating low-quality regions, read dereplication, chimera filtering, and singleton removal.43 After denoising, open-reference clustering was performed with the VSEARCH plugin at 99% identity to generate operational taxonomic units (OTUs),44 where reads were organized in features, and a feature table was generated with a feature-table plugin (https://github.com/qiime2/q2-feature-Table). In the feature table, exactly one sequence represents each feature. The Amplicon Sequence Variants (ASVs) that have a frequency below 0.1% of the mean depth were excluded from further analysis. The feature-classifier plugin was used to assign taxonomy to OTUs against a pre-trained Sk-learn classifier45 based on the SILVA 138 database46 with a 99% similarity clustering threshold to generate taxonomy tables. Taxa filtering for eukaryota, mitochondria, and chloroplast was performed, and a final representative sequence file was generated for filtered taxonomy. The taxa bar plots depicting taxonomic hierarchy were generated using the QIIME2 taxa plugin.Alpha rarefaction for the phylogenetic diversity was performed by alpha_rarefaction.py script using the minimum and maximum depth parameters as 10 and 2227, respectively. The analysis of both alpha and beta diversity was performed by the q2-diversity plugin in QIIME2. The alpha diversity metrics to the evenness, observed OTUs, and Shannon were measured based on ASV levels. The alpha diversity metric dissimilarities were tested by the non-parametric Kruskal-Wallis test. Alpha-diversity-group significance was determined from community richness in addition to the Shannon index.47 Beta diversity metrics were measured by the Bray–Curtis distance, and the structure of the microbial community in all samples was explored with PERMANOVA48 by using the beta-group-significance command.
Results and discussion
Designing of synthetic microbial consortia for producing biohythane by anaerobic digestion of cow dung
Establishing an enriched and efficient inoculum of microbial members to produce H2 is crucial for the fermentation process during AD. For microbial enrichment, mixed cultures can be directly used from the natural environments. In various natural environments, a variety of mixed cultures are found that have the extensive ability for fermentative H2 generation. These include hot spring sediment, anaerobic sludge, compost, municipal sewage sludge, and soil. Such mixed cultures have an additional advantage due to the simplicity of their operation and control under non-sterile conditions which is not feasible while using pure cultures.40 For the development of efficient synthetic microbial consortia the principle of “division of labor” has been used. This allows the microbial community to complete the process more quickly than monocultures by assigning various tasks to different members of the community. This enables a coordinated environment to support each process, i.e., each consortium member produces a molecule that is then used by the following link in the supply chain.49We have designed our synthetic microbial consortia by combining four different natural mixed cultures, namely aged sludge (AS), paper mill sludge (PM), cow dung (CD), and electro-flocculated wastewater (EF) (Fig. 2). PM is rich in compounds of fiber wood (lignocellulosic), such as carbohydrate polymers, mainly cellulose and hemicellulose, lignin, lipids, and other extractives. Some members of Firmicutes (Bacillus sp., Clostridium sp., and Paenibacillus sp.), and Actinobacteria (Streptomyces, Microbacterium, and Cellulomonas xylanilytica) present in PM sludge showed cellulase activity, which may be important for the breakdown of feedstock.50 AS obtained from STPs is rich in microbial diversity and contains a high concentration of organic matter with various nutrients including N2, P, K, Ca, Fe, Mg, Cu, Zn, Mn, and Si.51 AS contains aerobic and anaerobic microbes like bacteria, archaea, fungi, and protists. The predominant bacterial phyla in AS include Proteobacteria, Bacteroidota, Firmicutes, and Actinobacteria. The most dominating genera include Clostridium, Treponema, Propionibacterium, Syntrophus, Desulfobulbus, Brevundimonas, Paludibacter, Cloaci-bacterium, and Methylobacterium.52 The CD is enriched in hydrolyzing and acidifying bacteria and methanogenic archaea, such as Bifidobacterium, Bacillus, and Lactobacillus.53 EF is obtained by the electro-flocculation of sewage raw wastewater from the STP of IIT Mandi and is rich in Fe content in the form of Fe2+ and Fe3+ ions,41 which play important roles in the AD process.54 The production of H2 and CH4 requires different kinds of metal ions as micronutrients, namely, Fe2+, Zn2+, Ni2+, Na+, Mg2+, and Co2+ as cofactors, facilitators of transport processes, and structural skeletons of numerous enzymes mainly for the growth and metabolism of microbes.40 Based on the form of Fe available, namely Fe2+ or Fe3+, the microbial community is shifted towards CH4 or H2 production, respectively.54
Another method for enriching a community is based on employing a selection mechanism to favor bacteria that produce H2 while eradicating those that consume it. The majority of H2-producing bacteria are spore-forming, whereas methanogens and H2-consuming bacteria are not, except for a few hydrogenotrophic methanogens.55 The production of endospores can be encouraged by providing a stressed environment to the microbes which will be favorable for the endospore formation of bacteria, such as the H2-producers while removing the others. In the present study, we used two types of synthetic microbial inoculum, namely, heat-pre-treated and untreated, and tested the biohythane efficiency with CD feedstock as compared to the control group. Bacillus and Clostridium are the two most popular sporulating bacteria that are known to be mainly responsible for producing H2. Similarly, a slightly acidic pH, to avoid methanogenesis and solventogenesis (5.0 to 6.5), is one of the favoring parameters to support the growth of H2-producers and enhance the H2-production rate,29,40 whereas an optimum pH for methanogenic bacteria ranges between 6.0 and 7.5 and anaerobic bacteria work well below pH 6.56 So, in order to favor both H2 and CH4 production, we performed our experiments at a pH of 6–6.5.
For the design of the synthetic microbial consortia, at first, we used AS + PM to prepare two types of mixed sludge with (EAPTx) or without (EAPUTx) heat pre-treatment (Fig. 2). These mixed sludges were independently acclimatized with EF for 21 days to allow the microbial community to adapt to the AD process. After 21 days of acclimatization under anaerobic conditions, the microbial community is likely to be shifted towards methanogenesis and H2 scavenging conditions which may impact H2 yield. In general, the methanogenic archaeal community is a slow grower as compared to the anaerobic bacteria. Therefore, it is important to overcome this risk of complete community shifting towards H2 consumption and methanogenesis. Towards this, in the next step, we have mixed CPA consisting of AS, PM, and CD in our acclimatized sludge (EAPTx or EAPUTx). The heat-pre-treated CPA (CPA(Tx)) is combined with EAPTx and the untreated CPA ((CPA)UTx) is mixed with EAPUTx to result in the final synthetic microbial consortia, namely, E(C2)Tx and E(C2)UTx (Fig. 2). The effect of heat pre-treatment is again to enhance the H2 producers, which are mixed with a methanogenic microbial community enriched in the acclimatized sludge (EAPTx). It is important to note that we have used CD as the substrate for AD in our analysis which is classified as a lignocellulosic waste due to the presence of a large proportion of lignocellulose (50% in dry matter).33 Therefore, it is important to enhance the degradation of recalcitrant lignocellulose in CD so that the overall yield of biohythane can be increased. The use of PM and CD in the last step (CPA) is also to provide a community rich in lignocellulose-degrading microbes to facilitate the process of hydrolysis and fermentation.
Gas production and composition
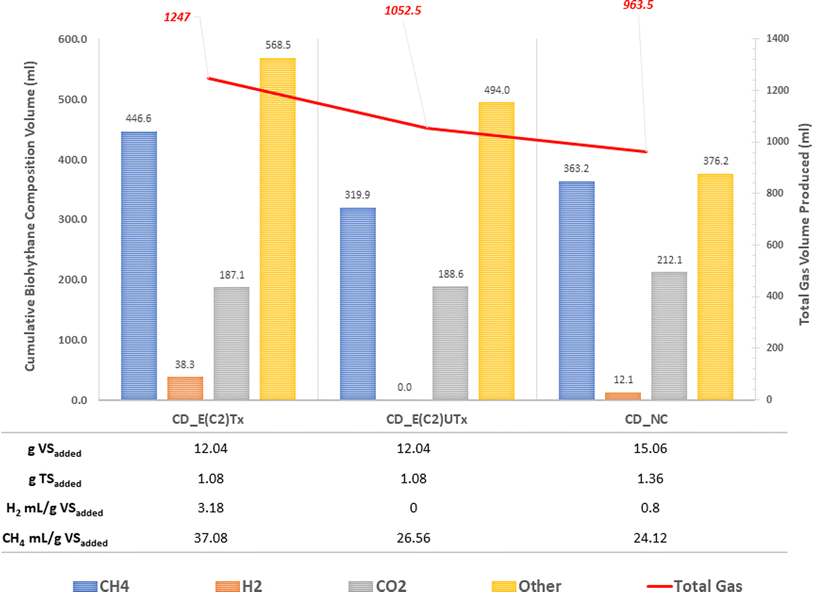
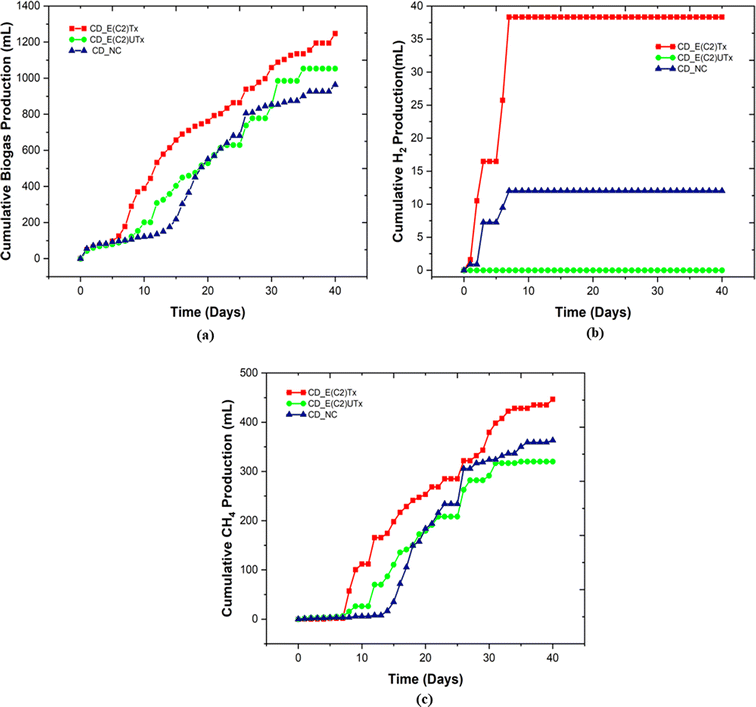
A mini-scale dark-fermentative single-stage bottle bioreactor system was used for biohythane production from CD as feed and synthetic microbial inoculum for a period of 40 days. The total gas production was measured under two experimental conditions, namely, the digestion of the CD substrate with E(C2)Tx and E(C2)UTx with respect to the control (only CD as feed without any inoculum). Fig. 3 represents the cumulative gas volumes for different gases produced in 40 days for this single-stage fermentation process. CD samples inoculated with E(C2)Tx showed the highest volume of total gas (1247 ml) produced followed by CD inoculated with E(C2)UTx (1052.5 ml) and CD control (963.5 ml).We also measured the biogas production daily to estimate the duration of a cycle for the production of H2 and CH4 (Fig. 4). In the case of E(C2)Tx, a drastic increase in gas production was observed on the 7th day of the experiment (Fig. 4(a)). In the case of E(C2)UTx samples, a rise in gas production was observed only after the 11th day, whereas for control samples this was observed after the 15th day (Fig. 4(a)). For the E(C2)Tx, the highest gas volume was obtained on the 8th day (112.5 ml) (Fig. 4(a)), whereas for E(C2)UTx and control samples, the highest gas volume was produced only on the 31st day (137.5 ml) and 26th day (125 ml), respectively. These observations show that there is a significant reduction in the cycle length of biogas production in the case of CD samples mixed with E(C2)Tx. A similar decrease in biogas production cycle length was observed previously while using E(C2)Tx inoculum for the anaerobic co-digestion of food waste (FW), CD, and sewage wastewater (FCR_E(C2)Tx) for 20 days.24
In the single-stage biohythane production, the rate-limiting step is H2 production, which ultimately also governs the composition of the biohythane. Cumulative H2 volume is 38.3 ml (3.17 ml H2 per g VSadded) for CD inoculated with E(C2)Tx and 12 ml (0.79 ml H2 per g VSadded) for the control, whereas the samples inoculated with E(C2)UTx do not show any H2 production (Fig. 3). There is a substantial increase in the H2 production in the E(C2)Tx sample on the 1st and 5th days and the maximum volume was obtained on the 7th day (12.6 ml) (Fig. 4(b)). For the control sample, H2 production started on the 2nd day and reached the maximum on the 3rd day (6.4 ml), although the total volume of H2 produced was less than that in the case of E(C2)Tx samples. However, no H2 was obtained in the E(C2)UTx sample till the end (Fig. 4(b)).
The production of H2 is followed by the production of CH4. The cumulative volume of CH4 is found to be the highest for CD inoculated with E(C2)Tx (446.6 ml, 37.08 ml g−1 VSadded) (Fig. 3). This is followed by the CH4 production in E(C2)UTx (319.9 ml, 26.56 ml g−1 VSadded) and control (363.2 ml, 24.12 ml g−1 VSadded) samples (Fig. 3). A gradual increase in CH4 production can be observed from the 7th day onwards for E(C2)Tx, with a maximum volume obtained on the 8th day (55.4 ml) (Fig. 4(c)). CH4 production continued to increase at the rate of approximately 16 ml per day up to day 32, after which only ∼5 ml per day of CH4 was obtained till the end of the experiment. While in the case of E(C2)UTx, a slight increase in CH4 production can be observed only from the 11th day onwards with a maximum CH4 volume of 54.7 ml obtained on day 25. In the case of the control samples, an increase in CH4 production was observed only by day 15, and the maximum volume was obtained on day 26 (71.7 ml).
These observations show that with E(C2)Tx, a higher proportion of H2 (3%) was obtained, whereas in control samples a lesser percentage of this gas was recovered (1.3%). In E(C2)UTx, no H2 production was observed. Additionally, with E(C2)Tx, the cycle of H2 production begins on the 1st day itself and that of CH4 production starts on the 7th day onwards, whereas in the control and E(C2)UTx samples a delayed CH4 production with less or no H2 production, respectively, was observed. This implies that the anaerobic digestion of the CD substrate with the E(C2)Tx inoculum is capable of generating biohythane, although with only 3% of H2 gas.
Biohythane production, targeting more than 5% H2, has been explored in several studies previously.57–63 For example, Nguyen et al., (2022)64 used a mixture of swine manure and pineapple waste for the production of biohythane in a single-stage two-chamber digester with varying HRT (96 h to 6 h) and observed 60.5% of peak H2 content in biohythane at an HRT of 6 h. Ta et al., (2020)65 performed biohythane production in a single-stage anaerobic digester using synthetic nutrient media as feed and separately entrapped H2- and CH4-producing bacteria and obtained a peak of H2 (64.6 ml/L-d, 15%) and CH4 (395 ml/L-d). In another study, biohythane with a maximum of 3.21 mol H2 per mol hexose and 3.63 mol CH4 per mol hexose from acetic acid and glucose (synthetic wastewater) in a continuous stirred tank reactor (CSTR) was obtained.66 The AD of CD for biohythane production has also been explored, however, utilizing other co-digestion substrates.67,68 For example, Sufyan et al., (2023)68 performed a two-step AD using a mixture of domestic wastewater and CD with varying pH ranges and maintaining thermophilic conditions (55 °C) for H2 production and mesophilic conditions (35 °C) for CH4 production. They observed the highest yield of H2 (108.04 ml H2 per g VS) and CH4 (768.54 ml CH4 per g VS) at pH 5.5 and pH 7.5, respectively, and the highest cumulative biohythane (811.12 ml g−1 VS) was obtained at a pH of 7.5.68 As is evident, most of the studies are based on either the two-stage AD process or with a variety of different substrates (such as wastewater, palm oil mill effluent, food waste, distillery spent-wash, swine manure, fruit waste, algae, etc.) under energy or labor-intensive conditions, thereby leading to limited real life applications.
Taxonomy analysis of CD treated with different inocula
In order to estimate the microbial diversity dynamics before the start (SP) and end (EP) of AD, we performed the 16S rRNA-based metagenomic analysis. The amplicon sequencing yielded a total of 131![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 121 raw reads and 128
121 raw reads and 128![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 051 quality-filtered reads for all 6 samples (Table 1). The quality-filtered reads resulted in a total of 779 Operational Taxonomic Units (OTUs). After removing singletons and contaminants 756 OTUs were finally retained which were distributed among 30 phyla and 365 genera. A varying sampling depth across the samples is known to introduce bias in the estimation of rare taxa. A rarefaction analysis to estimate the minimum number of reads required to capture the complete diversity of each sample (ESI Fig. 1†) indicated that the sampling across all samples was found to be sufficient to capture the complete diversity of each dataset. The CD samples with E(C2)UTx exhibited the highest microbial diversity as they harbor the maximum number of unique OTUs followed by the samples with E(C2)Tx and controls (Table 1 and ESI Table 1†). This could be due to the loss of heat-intolerant microbial community during the heat pretreatment step of E(C2)Tx inoculum preparation. During heat pretreatment, the high temperature could lead to the disruption of chemical bonds of the microbial cells leading to a loss of heat labile community.69 This indicates that the type of inoculum pretreatment causes different microbial communities which can affect the overall H2 and CH4 production during the AD process. The beta-diversity analysis demonstrated no significant clustering of samples.
051 quality-filtered reads for all 6 samples (Table 1). The quality-filtered reads resulted in a total of 779 Operational Taxonomic Units (OTUs). After removing singletons and contaminants 756 OTUs were finally retained which were distributed among 30 phyla and 365 genera. A varying sampling depth across the samples is known to introduce bias in the estimation of rare taxa. A rarefaction analysis to estimate the minimum number of reads required to capture the complete diversity of each sample (ESI Fig. 1†) indicated that the sampling across all samples was found to be sufficient to capture the complete diversity of each dataset. The CD samples with E(C2)UTx exhibited the highest microbial diversity as they harbor the maximum number of unique OTUs followed by the samples with E(C2)Tx and controls (Table 1 and ESI Table 1†). This could be due to the loss of heat-intolerant microbial community during the heat pretreatment step of E(C2)Tx inoculum preparation. During heat pretreatment, the high temperature could lead to the disruption of chemical bonds of the microbial cells leading to a loss of heat labile community.69 This indicates that the type of inoculum pretreatment causes different microbial communities which can affect the overall H2 and CH4 production during the AD process. The beta-diversity analysis demonstrated no significant clustering of samples.
| Sr. no. | Sample ID | Total reads | Read length | Reads passing quality filtering | % reads passing quality filtering | Number of observed OTUs/feature count | Number of unique OTUs |
|---|---|---|---|---|---|---|---|
| 1 | CD_E(C2)Tx_SP | 24![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 416 416 |
301 | 23![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 801 801 |
97.50% | 3140 | 98 |
| 2 | CD_E(C2)Tx_EP | 25![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 539 539 |
301 | 24![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 905 905 |
97.50% | 3403 | 96 |
| 3 | CD_E(C2)UTx_SP | 21![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 991 991 |
301 | 21![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 505 505 |
97.80% | 4275 | 124 |
| 4 | CD_E(C2)UTx_EP | 23![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 072 072 |
301 | 22![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 401 401 |
97.10% | 3474 | 111 |
| 5 | CD_NC_SP | 22![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 272 272 |
301 | 21![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 847 847 |
98.10% | 2912 | 74 |
| 6 | CD_NC_EP | 13![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 831 831 |
301 | 13![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 592 592 |
98.30% | 2227 | 58 |
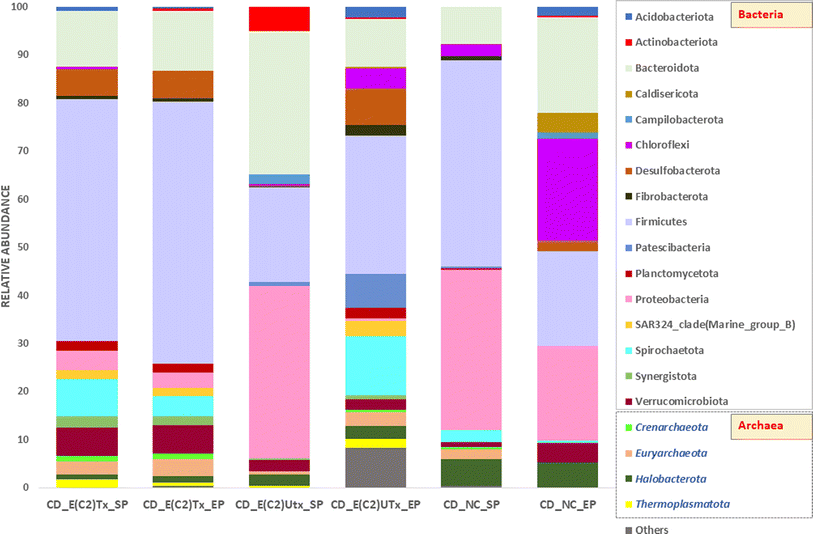
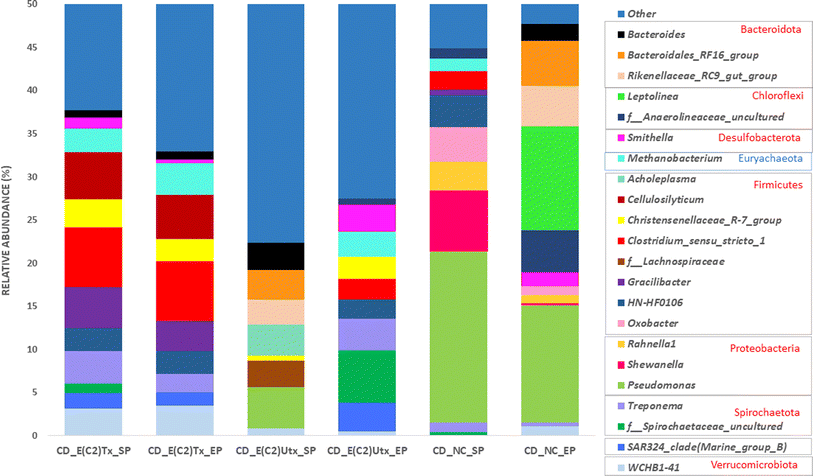
The top ten most abundant bacterial or archaeal phyla from each sample are shown in Fig. 5. The microbial community of the E(C2)Tx samples was mainly enriched with phylum Firmicutes, whereas in all other samples, the relative abundance of this phylum was comparatively less. The higher abundance of Firmicutes in E(C2)Tx samples throughout the AD process may be responsible for a higher hydrolysis rate and enhanced H2 and biohythane production.70 The members of this phylum participate in the hydrolysis, acidogenesis, and acetogenesis steps of AD and are known to degrade a wide range of substrates (protein, lipids, and carbohydrates).54,71 The abundances of Proteobacteria and Bacteroidota were the highest in the SP of E(C2)UTx samples. Proteobacteria include important acetogenesis bacteria and are well known for degrading or utilizing glucose, proteins, and various kinds of VFAs in manure and other lignocellulosic wastes.71 The phylum Bacteroidota can hydrolyze carbohydrates or proteins and transform organic substances to produce VFAs and is considered to participate in the hydrolysis and acidogenesis steps of AD.70 Interestingly, the EP of control samples showed the highest abundance of phylum Chloroflexi, which is found to be negligible in all other samples. The members of phylum Chloroflexi are often treated as primary fermenters and are ubiquitous in various AD systems including cattle manure, although their metabolism is not well defined though they are assumed to utilize glucose and break down tough-to-degrade organics.71
Among the archaeal taxa, Euryarchaeota is found to be enriched in samples with E(C2)Tx as compared to E(C2)UTx and controls, whereas Halobacterota is present in least abundance. A majority of the members of phylum Euryarchaeota utilize the hydrogenotrophic and methylotrophic pathways for methanogenesis.54Halobacterota can act on a wide range of substrates and produces CH4 using any of the methanogenesis pathways (acetoclastic, hydrogenotrophic, and methylotrophic).54,72
Roles of microbial community during anaerobic digestion of cow dung to produce biohythane
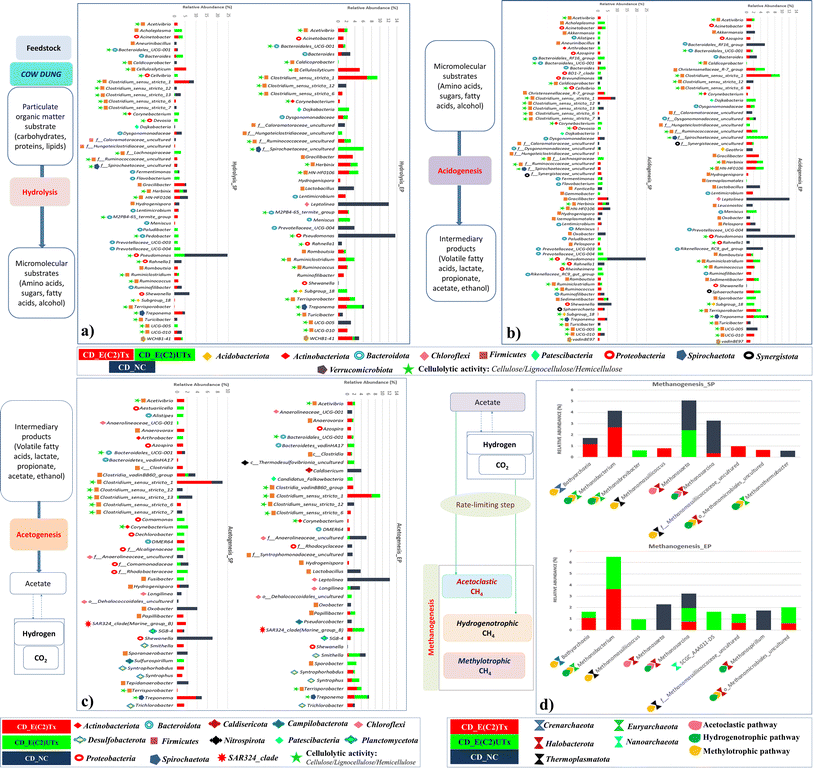
The process of microbial AD consists of four phases, namely, hydrolysis, acidogenesis, acetogenesis, and methanogenesis, where bacteria mediate the first three stages and archaea carry out the final one (Fig. 7).There are other prominent differences in the dominant microbes involved in the hydrolysis process in the E(C2)Tx and E(C2)UTx samples as compared to the control (Fig. 7). At the genus level, the cellulolytic activity to degrade the lignocellulosic feedstock is mainly carried out by the genus Clostridium and several other members of Firmicutes in E(C2)Tx samples. In the E(C2)UTx samples, the cellulolytic and hydrolytic activities are carried out by the members of the genus Pseudomonas and several other members of Bacteroidota. In the control samples, this is mainly carried out by Pseudomonas and Shewanella. A few studies have previously reported the microbial community of the AD process inoculated with heat-shock pre-treated inoculum on various substrates.39,69,76,77 Corroborating with our results, a dominance of H2 producers belonging to Clostridium sp. mainly Clostridium_sensu_stricto_1 and other Firmicutes (Prevotella) and Bacteroidota (Romboutsia), etc., have been observed in those studies. In addition, they also observed Enterococcus, Pseudomonas, and a few less explored taxa including Paraclostridium, Exiguobacterium, Aneurinibacillus, Ureibacillus, Xanthomonas, and Paenibacillus which could be due to the use of other substrates.39,69,76,77
The H2-production process can be hindered by several microbes including hydrogenotrophic methanogens, homoacetogens, nitrate-reducing, sulfate-reducing, iron-reducing, propionic acid-producing, and lactic acid bacteria. Propionogens or homoacetogens can produce propionic acid, which inhibits the dark fermentation process and reduces H2 yield.78Akkermansia and Alistipes are the two propionate producers and Candidatus_Falkowbact, Fusibacter, Caldisericum, and Sulfurospirillum are sulfate and thiosulfate reducing bacteria which are mainly present in E(C2)UTx and control samples which can impact H2 production adversely. Consequently, a reduction in H2 production has also been observed in these samples as compared to E(C2)Tx (ESI Table 2(A)†). Another important bacterial genera, namely Acinetobacter (Proteobacteria) showed a slightly higher abundance in E(C2)Tx than E(C2)UTx and was completely absent in control samples (ESI Table 2(A)†). Most of the members of this genera are pathogenic but play very important roles in hydrolysis, acidogenesis, and H2S removal. In addition, Acinetobacter is also an effective phosphate solubilizer which makes it an important taxa for phosphorus nutrient enrichment in E(C2)Tx.
In E(C2)UTx samples, the methanogenic community is mainly dominated by SCGC_AAA011-D5, Methanobacterium, and Methanosaeta among others, whereas the control samples exhibit a higher abundance of Methanosaeta, Methanosarcina, and Methanospirillum. Methanosarcina and Methanosaeta are expert taxa in the acetate cleavage process that produces methane, however, Methanosarcina grows best in environments with high acetate concentrations and encompasses both acetoclastic and hydrogenotrophic pathways for methane production.82,83Methanospirillum sp. is a member of the Methanomicrobiales of Halobacterota and is crucial for methanogenesis.84
Methylotrophic microbes are known to play a lesser role in methanogenesis as compared to hydrogenotrophic and acetoclastic ones.74 The known substrates for the methylotrophic pathway are methyl compounds, alcohols, and acetate. In addition, some of the methylotrophic microbes are strictly dependent on H2 for their activity. At the same time, the available alcohol and acetate produced during acidogenesis might be consumed via the methylotrophic pathway for CH4 production. These indicate the possibility of an active participation of the methylotrophic pathway also for methanogenesis in E(C2)Tx. It is known that acetoclastic processes contribute more towards CH4 production.85 Towards this, some acetoclastic microbes are observed in E(C2)Tx samples, such as Methanosarcina. However, a higher abundance of methylotrophic and hydrogenotrophic microbes are found for CH4 production in E(C2)Tx samples (ESI Fig. 2†). This implies that due to the effect of heat treatment the methylotrophic and hydrogenotrophic pathways might be favored for methane production in E(C2)Tx. Corroborating with our results, in another study, acetoclastic and methylotrophic methanogens have been observed while performing thermal hydrolysis pre-treatment of protein rich tofu as the substrate.86
Proposed mechanism of biohythane production via anaerobic digestion of CD using E(C2)Tx inoculum
The E(C2)Tx samples are found to be enriched with the H2-producing and cellulolytic microbial communities throughout the experiment duration as compared to E(C2)UTx and controls. Due to this, an enhanced substrate degradation is expected to take place and subsequently, more H2 will be produced till the end of the experimental cycle. A major hindrance in the retrieval of H2 in AD is caused by the presence of H2-consuming methanogens, as they use up all the available H2 to form CH4. It is important to note that the indigenous microbial community of the substrate CD is rich in methanogens. However, the fact that methanogens are very slow-growing microbes74 and the microbial community in the E(C2)Tx samples is enriched in cellulolytic bacteria suggests that there may be a faster degradation of a substrate and subsequent H2 production but a relatively slower consumption of H2 for CH4 formation in the initial phase of the experiment. Therefore, due to the high cellulolytic activity, the abundance of H2 may be so much that even after the production of CH4 by consuming the available H2, sufficient H2 is left which can be detected and recovered in E(C2)Tx samples. Subsequently, methanogens will start to adapt to the compounds available in the environment which include H2, acetic acid, alcoholic compounds, methyl compounds, and others, and will start growing. This will lead to the enrichment of hydrogenotrophic, acetoclastic, and methylotrophic microbes during the AD process for CH4 production. Interestingly, in the E(C2)Tx samples, a dominance of methylotrophic and hydrogenotrophic communities was observed throughout the experiment (ESI Table 2(B)†).The overall stable community composition throughout the AD process of 40 days in the E(C2)Tx samples also offers other advantages. For example, the microbial community in these samples has a negligible abundance of nitrifying and denitrifying microbes and sulfate- and thiosulfate-reducing microbes throughout the experiment. This implies that the process of ammonification and sulfur reduction is not predominant in this case. These processes are known to increase the alkalinity of the system, leading to a hindrance to the growth of H2-producing microbes and an overall disruption of the AD process, as well as the production of biogas.74 Another factor that is mainly responsible for the cessation of the AD process is the accumulation of VFAs and CO2, which leads to an acidic environment and is known to hinder the growth of methanogenic and H2-producing microbial communities.54 The methylotrophic pathway already plays an important role in the metabolization of alcohol and acetic acid. These observations imply that there exists an optimal balance between the shifting of the environment to excessively acidic or alkaline conditions, which can adversely affect the microbial community dynamics and prevent the community from shifting to the extreme.
The main driving mechanism behind efficient biohythane production from CD in this study is the optimized designing and pre-treatment of the synthetic microbial consortia – E(C2)Tx. For the designing of E(C2)Tx consortia, heat pre-treatment was used to selectively enrich the community with spore-forming H2-producers. Heat treatment can increase the activity of hydrolytic enzymes in the inoculum and can accelerate the degradation of complex organic compounds into simpler molecules, which can be more easily utilized by microbes for H2 and CH4 production.87 Acclimatization of the inoculum's components under AD conditions allowed the microbial community to adapt to the new components such as CD, AS, PM, EF, and environmental conditions. Acclimatization promotes the development of synergistic interactions between various microbial species, improving the stability and overall metabolic performance of microbial consortia. During the acclimatization period, the microbial community becomes more tolerant towards AD inhibitors such as VFAs leading to more consistent biogas production.88–90 There are some studies that have adopted heat pretreatment and acclimatization of inoculum to enrich biohythane with H2. For instance, Rafieenia et al. (2018)91 used heat pre-treatment of inoculum as one of the methods to enrich H2-producing bacteria and performed the two-stage AD of food waste. However, food waste (FW) as a substrate is known to be rich in organic content (lipids, proteins, carbohydrates, starch, cellulose) which is very different from CD composition. In another study, Shi et al., (2023)86 demonstrated the use of heat pretreatment in combination with acclimatization, but they performed thermal hydrolysis pretreatment on a bulk substrate and acclimatization of inoculum. Besides, the substrate used by them was tofu, which is a highly protein-rich food product.
Suitability of the cow dung substrate for biohythane production
Among different substrates, carbohydrate-rich substrates are best suited for biohythane production, followed by those rich in proteins, while fatty acid-rich substrates are least suited.29 For example, the H2 production from FW is reported to yield 205 L H2 per kg VS, whereas cellulosic wastes such as hyacinth yielded 51.7 L H2 per kg VS.92 However, water hyacinth plants need harvesting, cutting/chopping, and pretreatment before AD and for FW continuous pH maintenance is required during the AD process to get H2 and CH4 because FW contains various inhibitory components which can cause instability of the system.24The substrates with a carbon/nitrogen (C/N) ratio of more than 20 are ideal for stable H2 production, whereas a C/N ratio between 20 and 30 is observed as suitable for efficient CH4 yield.92,93 The C/N ratio of CD is in the range of 16–25,94 which means it is the best-suited feedstock for the production of both H2 and CH4 or even biohythane. Furthermore, in the biohydrogen production system, H2-producing bacteria need vital nutrients such as carbon (C), nitrogen (N), and phosphorus (P). The cellulose component of CD can act as a very good C source. In contrast, N can be supplied by proteins, nitrates, nitrites, and ammonium salts (also function as a buffer for organic acids) from the undigested organic matter of CD.
Phosphorus concentration plays a crucial role in the DF process by maintaining the buffer capacity of the system during any reaction.92 Although P is an extremely important nutrient for various life forms, including plants and animals, its ready availability is a great challenge.23,95 Recovery of phosphorus from CD is often limited by the extremely small size of phosphorus-containing struvite crystals.96 Phosphate solubilizing microbes can hydrolyze organic and inorganic phosphorus into soluble forms.97 Thus, phosphorus is likely provided in the form of phosphates in the AD process,98 and its concentration is positively associated with H2 production. In addition to this, CD from indigenous Indian cows is also enriched with certain inorganic metals, including zinc (Zn), copper (Cu), cobalt (Co), manganese (Mn), and traces of sulfur (S), iron (Fe), and magnesium (Mg), which are all required for microbial growth.22
Economic perspective for biohythane production
A techno-economic analysis for the production of biohythane from wheat straw estimated the production cost as 23.04 $ per kg biohythane.99 The nutrient cost involved in this process was found to be the major cost driver. It was additionally highlighted that a reduction in the nutrient cost by 80% could lead to a decrease of 44% in the total production costs of the process. The production cost of biohythane was estimated previously as 7.20–48.96 $ per kg biohythane using a two-stage dark fermentation and AD of potato steam starch.100 The nutrients (yeast extract) required for both dark fermentation and AD were found to contribute the highest towards the total production cost. Subsequently, it was highlighted that using fewer or less costly nutrients for the AD process for single- or two-stage- DF and AD process would be critical for lowering the production costs. Furthermore, it was also noted that the two-stage systems for biohythane production demonstrated a net positive energy balance than the single-stage H2/CH4 reactors while also considering the energy required for pretreatment and reactor heating.101,102For exploring the industrial applications of any technology, the capital investment estimates need to be explored. For setting up a two-stage AD process for biohythane production, an approximate amount of 12![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 687.7 € has been estimated for treating 13.4 tons of waste biomass (i.e., 946.84 € per ton).100 In Europe, a pilot experiment was set up in a two-stage thermophilic AD on food waste, which was found to generate an annual income of 540
687.7 € has been estimated for treating 13.4 tons of waste biomass (i.e., 946.84 € per ton).100 In Europe, a pilot experiment was set up in a two-stage thermophilic AD on food waste, which was found to generate an annual income of 540![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 874 € per year from 27 tons of daily waste feed (i.e., 54.88 € of income per ton).103 For any investments in this direction, a payback period varying from 2 to 6 years has been reported, which also depends upon the nature of feedstocks.104 Governmental schemes and policies under the waste-to-energy theme have been known to play major roles in the proliferation and implementation of novel technologies for biohythane production in the market.
874 € per year from 27 tons of daily waste feed (i.e., 54.88 € of income per ton).103 For any investments in this direction, a payback period varying from 2 to 6 years has been reported, which also depends upon the nature of feedstocks.104 Governmental schemes and policies under the waste-to-energy theme have been known to play major roles in the proliferation and implementation of novel technologies for biohythane production in the market.
The economic viability and competitiveness of renewable energy sources, including biohythane, are significantly influenced by the cost of traditional fossil fuels, particularly crude oil.105 Any fluctuations in the oil or fossil fuel prices on a global scale impact the economic sustainability of renewable energy sources by influencing the currency exchange rates, which will further affect the cost of energy import and export.106 For an effective competition of biohythane as a future energy source with fossil fuels, despite the fluctuations in their costs, biohythane production technology needs to be economically viable. This emphasizes the need for more innovation to develop cost-effective strategies for the production of biohythane to ensure its financial sustainability.
Limitations of the study and future prospects
There are a few limitations of this study that may be addressed to enhance the commercial potential of the technology developed. The lab-scale experimental set-up with small working volumes, small study sample size, and the purity of reactants used limits the direct transferability of the findings of the study to the industrial settings. Another important challenge is a lower than required (>5–10%) amount of H2 gas (3%) in biohythane composition recovered during the AD process. Furthermore, a parametric analysis was outside the main frame of this study, which might be one of the major factors for not attaining the appropriate range of biohythane composition.Keeping these limitations in mind, a comprehensive standardization of the bioprocess and substrate parameters (such as COD, TS, VS, etc.) can be taken into consideration in future studies. Second, lab-based or small-scale experiments may not accurately represent the challenges and complexity of commercial-scale operations. Therefore, future research should focus on scaling up the experiments to increase working volumes, which may match the industrial demands. Third, co-digestion of the CD substrate with other wastes can be attempted to enhance the proportion of H2 to meet the standard biohythane composition. Previously, we have attempted co-digesting food waste, CD, and sewage wastewater mixed in an appropriate proportion by using the E(C2)Tx inoculum for enhancing the H2 production.24 However, the AD process requires continuous pH maintenance during the entire process, which is likely to increase production costs if applied at the commercial scale.
The cost-effective utilization of fermentative or AD residue is crucial for improving the financial performance of biohythane production. For future studies, the use of anaerobic digestate for generating various cost-effective and sustainable products, such as effective biofertilizers, biochar, bio-cement, bio-bricks, etc., may be targeted. Converting anaerobic digestate into biochar or bio-cement not only addresses waste management issues but can sequester carbon and provide an alternative solution for soil health and fertility with new market opportunities.107,108 The future research may also focus on enhancing the efficacy of the system by providing key elements (C, H, N, and P) in addition to essential micronutrients (Fe, Ni, Co, Mo, Cu) in the form of nanoparticles for the growth of anaerobic bacteria.109
Conclusion
The current study investigated the potential of synthetically designed microbial consortia for biohythane production using single-stage anaerobic digestion of CD waste. The (E(C2)Tx) inoculum generated biohythane with 3% H2 and 36% CH4 without requiring any pretreatment of the bulk substrate, thereby, proving to be more economically viable. In contrast the existing approaches heavily rely on cost- and labor-intensive bulk substrate pretreatment strategies limiting their commercial potential. The presented approach involved a pretreatment of the inoculum which is used in four times less volume than the bulk substrate and is made up of different wastes as well. The strategy for the pretreatment of inoculum included heat treatment and acclimatization, which resulted in a stable microbial community enriched in hydrolytic and cellulose degrading microbes as well as hydrogenotrophic and methylotrophic methanogens, which have their own advantages in increasing H2 and CH4 yields by also reducing the cycle length of obtaining these gases. The presented technology shows promising cost-, resource- and energy-effectiveness, which can offer biohythane as a competitive alternative renewable energy source over the traditional fossil fuels, which also have their own environmental impacts. In addition to this, the developed technology for generating biohythane from CD presents a very efficient CD waste management strategy and a good example of waste to energy conversion supporting circular economy and sustainable development initiatives. Future research in this direction should focus on developing economic models for quantifying financial benefits and scaling up the process with comprehensive process parameters and open the avenues for its commercial adoption worldwide.Data availability
Raw sequencing reads have been submitted to the NCBI Short Reads Archive (SRA) under BioProject accession number PRJNA828359, with respective SRA IDs (viz., SRR18827896, SRR18906513, SRR18827912, SRR18828078, SRR18828097 and SRR18828098). These data can be accessed using the following link: https://www.ncbi.nlm.nih.gov/sra/PRJNA828359.Author contributions
Rashmi Ira and Vikas Sharma designed the experimental setup, and performed sample collection, DNA extraction, and metagenomic analysis. Shrawan Kumar and Tulika Prakash performed NGS library preparation and sequencing. Mira Koul assisted in metagenomic data analysis. Lalita Sharma carried out EF synthesis and Aditi Halder provided inputs and leads for EF production. Tulika Prakash conceptualized the problem, arranged resources, managed the project, and provided guidance for all experiments and computational analysis. Rashmi Ira and Tulika Prakash wrote the original draft and final manuscript. All authors have read and approved the final version of the manuscript.Conflicts of interest
The authors have declared no conflict of interests.Acknowledgements
The work was supported by grant number IITM/MHRD/IMPRINT/AD/169 from Ministry of New and Renewable Energy (MNRE) and the Ministry of Education, formerly known as the Ministry of Human Resource Development (MHRD), Govt of India for Project No. 7801, entitled “Sustainable wastewater treatment through bio-photoelectrocatalysis and biofuel production” under scheme IMPacting Research INnovation and Technology-1 or IMPRINT-1. Rashmi Ira acknowledges the Ministry of Education or MHRD, India for providing the research fellowships. We acknowledge the help of Dr Shyam K. Masakapalli for discussions towards experimental design.References
- J. Sivanesan, A. Vijayalakshmi, B. Sivaprakash and N. Rajamohan, Biohythane as a sustainable fuel – A review on prospective synthesis based on feedstock preprocessing, optimization approach and circular economy concept, Process Saf. Environ. Prot., 2024, 185, 739–753 CrossRef CAS.
- F. Crotogino, in Storing Energy, Elsevier, 2022, pp. 633–649 Search PubMed.
- Q. Hassan, S. Algburi, A. Z. Sameen, T. J. Al-Musawi, A. K. Al-Jiboory, H. M. Salman, B. M. Ali and M. Jaszczur, A comprehensive review of international renewable energy growth, Energy Built Environ., 2024 DOI:10.1016/j.enbenv.2023.12.002.
- B. Bencoova, R. Grosos, M. Gomory, K. Bacova and S. Michalkova, Use of biogas plants on a national and international scale, Acta Montan. Slovaca, 2021, 139–148 Search PubMed.
- Y. Guo, X. Gou, Z. Xu and M. Skare, Carbon Pricing Mechanism for the Energy Industry: A Bibliometric Study of Optimal Pricing Policies, Acta Montan. Slovaca, 2022, 49–69 Search PubMed.
- H. Pavolova, T. Bakalari, K. Kyšeľa, M. Klimek, Z. Hajduova and M. Zawada, The analysis of investment into industries based on portfolio managers, Acta Montan. Slovaca, 2021, 161–170 Search PubMed.
- M. Akbari, N. Loganathan, H. Tavakolian, A. Mardani and D. Streimikiene, The Dynamic Effect of Micro-Structural Shocks on Private Investment Behavior, Acta Montan. Slovaca, 2021, 1–17 Search PubMed.
- J. Maroušek, A. Maroušková, B. Gavurová, D. Tuček and O. Strunecký, Competitive algae biodiesel depends on advances in mass algae cultivation, Bioresour. Technol., 2023, 374, 128802 CrossRef PubMed.
- J. Maroušek, B. Gavurová, O. Strunecký, A. Maroušková, M. Sekar and V. Marek, Techno-economic identification of production factors threatening the competitiveness of algae biodiesel, Fuel, 2023, 344, 128056 CrossRef.
- S. Roy and D. Das, Biohythane production from organic wastes: present state of art, Environ. Sci. Pollut. Res., 2016, 23, 9391–9410 CrossRef CAS PubMed.
- D. Bolzonella, F. Battista, C. Cavinato, M. Gottardo, F. Micolucci, G. Lyberatos and P. Pavan, Recent developments in biohythane production from household food wastes: A review, Bioresour. Technol., 2018, 257, 311–319 CrossRef CAS PubMed.
- S. Shanmugam, T. Mathimani, E. R. Rene, V. Edwin Geo, A. Arun, K. Brindhadevi and A. Pugazhendhi, Biohythane production from organic waste: Recent advancements, technical bottlenecks and prospects, Int. J. Hydrogen Energy, 2021, 46, 11201–11216 CrossRef CAS.
- K. B. Sasidhar, P. S. Kumar and L. Xiao, A critical review on the two-stage biohythane production and its viability as a renewable fuel, Fuel, 2022, 317, 123449 CrossRef CAS.
- O. Sarkar, J. A. Modestra, U. Rova, P. Christakopoulos and L. Matsakas, Waste-Derived Renewable Hydrogen and Methane: Towards a Potential Energy Transition Solution, Fermentation, 2023, 9, 368 CrossRef CAS.
- L. M. Das, R. Gulati and P. K. Gupta, A comparative evaluation of the performance characteristics of a spark ignition engine using hydrogen and compressed natural gas as alternative fuels, Int. J. Hydrogen Energy, 2000, 25, 783–793 CrossRef CAS.
- S. A. Abdur Rawoof, P. S. Kumar, D.-V. N. Vo, T. Devaraj and S. Subramanian, Biohythane as a high potential fuel from anaerobic digestion of organic waste: A review, Renew. Sustain. Energy Rev., 2021, 152, 111700 CrossRef CAS.
- C. Mamimin, A. Singkhala, P. Kongjan, B. Suraraksa, P. Prasertsan, T. Imai and S. O-Thong, Two-stage thermophilic fermentation and mesophilic methanogen process for biohythane production from palm oil mill effluent, Int. J. Hydrogen Energy, 2015, 40, 6319–6328 CrossRef CAS.
- S. Sevda, V. K. Garlapati, S. Sharma and T. R. Sreekrishnan, in Delivering Low-Carbon Biofuels with Bioproduct Recovery, ed. L. Singh and D. M. Mahapatra, Elsevier, 2021, pp. 165–176 Search PubMed.
- M. Langone, M. Soldano, F. Claudio, F. Pirozzi and G. Andreottola, Anaerobic Digestion of Cattle Manure Influenced by Swirling Jet Induced Hydrodynamic Cavitation, Appl. Biochem. Biotechnol., 2018, 184(4), 1200–1218 CrossRef CAS PubMed.
- G. Randhawa and D. J. Kullar, Bioremediation of Pharmaceuticals, Pesticides, and Petrochemicals with Gomeya/Cow Dung, ISRN Pharmacol., 2011, 2011, 362459 Search PubMed.
- J. Maroušek, S. Hašková, R. Zeman, J. Váchal and R. Vaníčková, Processing of residues from biogas plants for energy purposes, Clean Technol. Environ. Policy, 2015, 17, 797–801 CrossRef.
- K. K. Gupta, K. R. Aneja and D. Rana, Current status of cow dung as a bioresource for sustainable development, Bioresour. Bioprocess., 2016, 3, 28 CrossRef.
- J. Maroušek and B. Gavurová, Recovering phosphorous from biogas fermentation residues indicates promising economic results, Chemosphere, 2022, 291, 133008 CrossRef PubMed.
- H. Thakur, R. Ira, N. K. Verma, V. Sharma, S. Kumar, A. Dhar, T. Prakash and S. Powar, Anaerobic co-digestion of food waste, bio-flocculated sewage sludge, and cow dung in CSTR using E(C2)Tx synthetic consortia, Environ. Technol. Innovat., 2023, 32, 103263 CrossRef CAS.
- C. A. N. Xavier, V. Moset, R. Wahid and H. B. Møller, The efficiency of shredded and briquetted wheat straw in anaerobic co-digestion with dairy cattle manure, Biosyst. Eng., 2015, 139, 16–24 CrossRef.
- S. Simm, A. Orrico, M. Orrico, N. Sunada, A. Schwingel, W. Lopes, K. Whittinghill, F. Miranda de Vargas Junior and M. Costa, Contribute of crude glycerin to increase the efficiency of anaerobic digestion process of dairy cattle manure, Environ. Prog. Sustain. Energy, 2018, 37(4), 1305–1311 CrossRef CAS.
- A. Kalia and S. Singh, Development of a Biogas Plant, Energy Sources, 2004, 26, 707–714 CrossRef.
- J. Santhosh, O. Sarkar and S. Venkata Mohan, Green Hydrogen-Compressed natural gas (bio-H-CNG) production from food waste: Organic load influence on hydrogen and methane fusion, Bioresour. Technol., 2021, 340, 125643 CrossRef CAS PubMed.
- S. O-Thong, C. Mamimin and P. Prasertsan, in Advances in Biofuels and Bioenergy, InTech, 2018 Search PubMed.
- S. V. Mohan, Fermentative hydrogen production with simultaneous wastewater treatment: influence of pretreatment and system operating conditions, J. Sci. Ind. Res., 2008, 67, 950–961 CAS.
- N. M. C. Saady, F. Rezaeitavabe and J. E. Ruiz Espinoza, Chemical Methods for Hydrolyzing Dairy Manure Fiber: A Concise Review, Energies, 2021, 14, 6159 CrossRef CAS.
- M. Langone, M. Soldano, C. Fabbri, F. Pirozzi and G. Andreottola, Anaerobic Digestion of Cattle Manure Influenced by Swirling Jet Induced Hydrodynamic Cavitation, Appl. Biochem. Biotechnol., 2018, 184, 1200–1218 CrossRef CAS PubMed.
- Y. Li, J. Zhao, J. Krooneman and G. J. W. Euverink, Strategies to boost anaerobic digestion performance of cow manure: Laboratory achievements and their full-scale application potential, Sci. Total Environ., 2021, 755, 142940 CrossRef CAS PubMed.
- S. Mirmohamadsadeghi, K. Karimi, R. Azarbaijani, L. Parsa Yeganeh, I. Angelidaki, A.-S. Nizami, R. Bhat, K. Dashora, V. K. Vijay, M. Aghbashlo, V. K. Gupta and M. Tabatabaei, Pretreatment of lignocelluloses for enhanced biogas production: A review on influencing mechanisms and the importance of microbial diversity, Renew. Sustain. Energy Rev., 2021, 135, 110173 CrossRef CAS.
- L. Sun, P. B. Pope, V. G. H. Eijsink and A. Schnürer, Characterization of microbial community structure during continuous anaerobic digestion of straw and cow manure, Microb. Biotechnol., 2015, 8, 815–827 CrossRef PubMed.
- A. Ferraro, G. Massini, V. Mazzurco Miritana, S. Rosa, A. Signorini and M. Fabbricino, A novel enrichment approach for anaerobic digestion of lignocellulosic biomass: Process performance enhancement through an inoculum habitat selection, Bioresour. Technol., 2020, 313, 123703 CrossRef CAS PubMed.
- J. Castilla-Archilla, C. E. Thorn, S. Pau and P. N. L. Lens, Screening for suitable mixed microbial consortia from anaerobic sludge and animal dungs for biodegradation of brewery spent grain, Biomass Bioenergy, 2022, 159, 106396 CrossRef CAS.
- I. M. Sicchieri, T. C. F. de Quadros, M. A. Bortoloti, F. Fernandes and E. K. Kuroda, Selection, composition, and validation of standard inoculum for anaerobic digestion assays, Biomass Bioenergy, 2022, 164, 106558 CrossRef CAS.
- C. Hernández, Z. L. Alamilla-Ortiz, A. E. Escalante, M. Navarro-Díaz, J. Carrillo-Reyes, I. Moreno-Andrade and I. Valdez-Vazquez, Heat-shock treatment applied to inocula for H2 production decreases microbial diversities, interspecific interactions and performance using cellulose as substrate, Int. J. Hydrogen Energy, 2019, 44, 13126–13134 CrossRef.
- M. Nageswara-Rao, Advances in Biofuels and Bioenergy, InTech, Rijeka, 2018 Search PubMed.
- L. Sharma, S. Prabhakar, V. Tiwari, A. Dhar and A. Halder, Optimization of EC parameters using Fe and Al electrodes for hydrogen production and wastewater treatment, Environ. Adv., 2021, 3, 100029 CrossRef CAS.
- J. G. Caporaso, J. Kuczynski, J. Stombaugh, K. Bittinger, F. D. Bushman, E. K. Costello, N. Fierer, A. G. Peña, J. K. Goodrich, J. I. Gordon, G. A. Huttley, S. T. Kelley, D. Knights, J. E. Koenig, R. E. Ley, C. A. Lozupone, D. McDonald, B. D. Muegge, M. Pirrung, J. Reeder, J. R. Sevinsky, P. J. Turnbaugh, W. A. Walters, J. Widmann, T. Yatsunenko, J. Zaneveld and R. Knight, QIIME allows analysis of high-throughput community sequencing data, Nat. Methods, 2010, 7, 335–336 CrossRef CAS PubMed.
- B. J. Callahan, P. J. McMurdie, M. J. Rosen, A. W. Han, A. J. A. Johnson and S. P. Holmes, DADA2: High-resolution sample inference from Illumina amplicon data, Nat. Methods, 2016, 13, 581–583 CrossRef CAS PubMed.
- T. Rognes, T. Flouri, B. Nichols, C. Quince and F. Mahé, VSEARCH: a versatile open source tool for metagenomics, PeerJ, 2016, 4, e2584 CrossRef PubMed.
- G. Varoquaux, L. Buitinck, G. Louppe, O. Grisel, F. Pedregosa and A. Mueller, Scikit-learn, GetMobile Mob. Comput. Commun., 2015, 19, 29–33 CrossRef.
- C. Quast, E. Pruesse, P. Yilmaz, J. Gerken, T. Schweer, P. Yarza, J. Peplies and F. Glöckner, The SILVA ribosomal RNA gene database project: Improved data processing and web-based tools, Nucleic Acids Res., 2012, 41(D1), D590–D596 CrossRef PubMed.
- R. Wenlong, Y.-J. Wen, J. Dunwell and Y.-M. Zhang, pKWmEB: integration of Kruskal-Wallis test with empirical Bayes under polygenic background control for multi-locus genome-wide association study, Heredity, 2018, 120(3), 208–218 CrossRef PubMed.
- B. J. Kelly, R. Gross, K. Bittinger, S. Sherrill-Mix, J. D. Lewis, R. G. Collman, F. D. Bushman and H. Li, Power and sample-size estimation for microbiome studies using pairwise distances and PERMANOVA, Bioinformatics, 215, 31(15), 2461–2468 CrossRef PubMed.
- K. Zhou, K. Qiao, S. Edgar and G. Stephanopoulos, Distributing a metabolic pathway among a microbial consortium enhances production of natural products, Nat. Biotechnol., 2015, 33(4), 377–383 CrossRef CAS PubMed.
- M. L. Maki, M. Broere, K. T. Leung and W. Qin, Characterization of some efficient cellulase producing bacteria isolated from paper mill sludges and organic fertilizers, Int. J. Biochem. Mol. Biol., 2011, 2, 146–154 CAS.
- M. Jackson, M. Line, S. Wilson and S. Hetherington, Application of Composted Pulp and Paper Mill Sludge to a Young Pine Plantation, J. Environ. Qual., 2000, 29, 407–414 CrossRef CAS.
- A. Nascimento, A. de Souza, P. A. De Andrade, F. Andreote, A. Coscione, F. Oliveira and J. Regitano, Sewage sludge microbial structures and relations to their sources, treatments, and chemical attributes, Front. Microbiol., 2018, 9, 372394 Search PubMed.
- K. Gupta, K. Aneja and D. Rana, Current status of cow dung as a bioresource for sustainable development, Bioresour. Bioprocess., 2016, 3(1), 28 CrossRef.
- R. Xu, S. Fang, L. Zhang, W. Huang, Q. Shao, F. Fang, Q. Feng, J. Cao and J. Luo, Distribution patterns of functional microbial community in anaerobic digesters under different operational circumstances: A review, Bioresour. Technol., 2021, 341, 125823 CrossRef CAS PubMed.
- T. Duangmanee, S. I. Padmasiri, J. J. Simmons, L. Raskin and S. Sung, Hydrogen Production by Anaerobic Microbial Communities Exposed to Repeated Heat Treatments, Water Environ. Res., 2007, 79, 975–983 CrossRef CAS PubMed.
- S. W. Van Ginkel, S.-E. Oh and B. E. Logan, Biohydrogen gas production from food processing and domestic wastewaters, Int. J. Hydrogen Energy, 2005, 30, 1535–1542 CrossRef CAS.
- A. Ghimire, V. Luongo, L. Frunzo, P. N. L. Lens, F. Pirozzi and G. Esposito, Biohythane production from food waste in a two-stage process: assessing the energy recovery potential, Environ. Technol., 2022, 43, 2190–2196 CrossRef CAS PubMed.
- S. B. Pasupuleti and S. Venkata Mohan, Single-stage fermentation process for high-value biohythane production with the treatment of distillery spent-wash, Bioresour. Technol., 2015, 189, 177–185 CrossRef CAS PubMed.
- S. Lunprom, O. Phanduang, A. Salakkam, Q. Liao, T. Imai and A. Reungsang, Bio-hythane production from residual biomass of Chlorella sp. biomass through a two-stage anaerobic digestion, Int. J. Hydrogen Energy, 2019, 44, 3339–3346 CrossRef CAS.
- S. Aslanzadeh, K. Rajendran and M. J. Taherzadeh, A comparative study between single- and two-stage anaerobic digestion processes: Effects of organic loading rate and hydraulic retention time, Int. Biodeterior. Biodegrad., 2014, 95, 181–188 CrossRef CAS.
- J. Seengenyoung, C. Mamimin, P. Prasertsan and S. O-Thong, Pilot-scale of biohythane production from palm oil mill effluent by two-stage thermophilic anaerobic fermentation, Int. J. Hydrogen Energy, 2019, 44, 3347–3355 CrossRef CAS.
- M. Hans and S. Kumar, Biohythane production in two-stage anaerobic digestion system, Int. J. Hydrogen Energy, 2019, 44, 17363–17380 CrossRef CAS.
- M.-L. Thi Nguyen, P.-C. Hung, T.-P. Vo, C.-H. Lay and C.-Y. Lin, Effect of food to microorganisms (F/M) ratio on biohythane production via single-stage dark fermentation, Int. J. Hydrogen Energy, 2021, 46, 11313–11324 CrossRef CAS.
- T.-T. Nguyen, D.-T. Ta, C.-Y. Lin, C.-Y. Chu and T.-M.-N. Ta, Biohythane production from swine manure and pineapple waste in a single-stage two-chamber digester using gel-entrapped anaerobic microorganisms, Int. J. Hydrogen Energy, 2022, 47, 25245–25255 CrossRef CAS.
- D. T. Ta, C.-Y. Lin, T. M. N. Ta and C.-Y. Chu, Biohythane production via single-stage anaerobic fermentation using entrapped hydrogenic and methanogenic bacteria, Bioresour. Technol., 2020, 300, 122702 CrossRef CAS PubMed.
- H. Hafez, G. Nakhla and H. El Naggar, An integrated system for hydrogen and methane production during landfill leachate treatment, Int. J. Hydrogen Energy, 2010, 35, 5010–5014 CrossRef CAS.
- W. Prapinagsorn, S. Sittijunda and A. Reungsang, Co-Digestion of Napier Grass and Its Silage with Cow Dung for Bio-Hydrogen and Methane Production by Two-Stage Anaerobic Digestion Process, Energies, 2017, 11, 47 CrossRef.
- F. Sufyan, M. Ali, S. Khan and N. Hossain, Biohythane Production from Domestic Wastewater Sludge and Cow Dung Mixture Using Two-Step Anaerobic Fermentation Process, Sustainability, 2023, 15, 14417 CrossRef CAS.
- G. Yang, Y. Yin and J. Wang, Microbial community diversity during fermentative hydrogen production inoculating various pretreated cultures, Int. J. Hydrogen Energy, 2019, 44, 13147–13156 CrossRef CAS.
- X. Zheng and R. Li, Critical Review on Two-Stage Anaerobic Digestion with H2 and CH4 Production from Various Wastes, Water, 2024, 16, 1608 CrossRef CAS.
- M. Abid, J. Wu, M. Seyedsalehi, Y. Hu and G. Tian, Novel insights of impacts of solid content on high solid anaerobic digestion of cow manure: Kinetics and microbial community dynamics, Bioresour. Technol., 2021, 333, 125205 CrossRef CAS PubMed.
- S. L. Bräuer, N. Basiliko, H. M. P. Siljanen and S. H. Zinder, Methanogenic archaea in peatlands, FEMS Microbiol. Lett., 2020, 367(20) DOI:10.1093/femsle/fnaa172.
- M. S. Demissie, N. H. Legesse and A. A. Tesema, Isolation and characterization of cellulase producing bacteria from forest, cow dung, Dashen brewery and agro-industrial waste, PLoS One, 2024, 19, e0301607 CrossRef CAS PubMed.
- J. W. Lim, T. Park, Y. W. Tong and Z. Yu, The microbiome driving anaerobic digestion and microbial analysis, Adv. Bioenergy, 2020, 5, 1–61 CrossRef CAS.
- J. Struckmann Poulsen, N. de Jonge, W. Vieira Macêdo, F. Rask Dalby, A. Feilberg and J. Lund Nielsen, Characterisation of cellulose-degrading organisms in an anaerobic digester, Bioresour. Technol., 2022, 351, 126933 CrossRef CAS PubMed.
- K. Vamshi Krishna and S. Venkata Mohan, Selective enrichment of electrogenic bacteria for fuel cell application: Enumerating microbial dynamics using MiSeq platform, Bioresour. Technol., 2016, 213, 146–154 CrossRef CAS.
- G. Yang, J. Wang and Y. Shen, Antibiotic fermentation residue for biohydrogen production using different pretreated cultures: Performance evaluation and microbial community analysis, Bioresour. Technol., 2019, 292, 122012 CrossRef CAS PubMed.
- X. Qu, H. Zeng, Y. Gao, T. Mo and Y. Li, Bio-hydrogen production by dark anaerobic fermentation of organic wastewater, Front. Chem., 2022, 10 DOI:10.3389/fchem.2022.978907.
- H. Pasalari, M. Gholami, A. Rezaee, A. Esrafili and M. Farzadkia, Perspectives on microbial community in anaerobic digestion with emphasis on environmental parameters: A systematic review, Chemosphere, 2021, 270, 128618 CrossRef CAS PubMed.
- P. Chellapandi, M. Bharathi, C. Sangavai and R. Prathiviraj, Methanobacterium formicicum as a target rumen methanogen for the development of new methane mitigation interventions: A review, Anim. Vet. Sci., 2018, 6, 86–94 CrossRef CAS PubMed.
- L. Blasco, M. Kahala, E. Tampio, M. Vainio, S. Ervasti and S. Rasi, Effect of Inoculum Pretreatment on the Composition of Microbial Communities in Anaerobic Digesters Producing Volatile Fatty Acids, Microorganisms, 2020, 8, 581 CrossRef CAS PubMed.
- A. Li, Y. Chu, X. Wang, L. Ren, J. Yu, X. Liu, J. Yan, L. Zhang, S. Wu and S. Z. Li, A pyrosequencing-based metagenomic study of methane-producing microbial community in solid-state biogas reactor, Biotechnol. Biofuels, 2013, 6, 3 CrossRef CAS.
- Y. Liu and W. B. Whitman, Metabolic, Phylogenetic, and Ecological Diversity of the Methanogenic Archaea, Ann. N. Y. Acad. Sci., 2008, 1125(1), 171–189 CrossRef CAS PubMed.
- D.-M. Yin, M. Westerholm, W. Qiao, S.-J. Bi, S. M. Wandera, R. Fan, M.-M. Jiang and R.-J. Dong, An explanation of the methanogenic pathway for methane production in anaerobic digestion of nitrogen-rich materials under mesophilic and thermophilic conditions, Bioresour. Technol., 2018, 264, 42–50 CrossRef CAS PubMed.
- S. Harirchi, S. Wainaina, T. Sar, S. A. Nojoumi, M. Parchami, M. Parchami, S. Varjani, S. K. Khanal, J. Wong, M. K. Awasthi and M. J. Taherzadeh, Microbiological insights into anaerobic digestion for biogas, hydrogen or volatile fatty acids (VFAs): a review, Bioengineered, 2022, 13(3), 6521–6557 CrossRef CAS PubMed.
- J. Shi, G. Zhang, H. Zhang, F. Qiao, J. Fan, D. Bai and G. Xu, Effect of Thermal Hydrolysis Pretreatment on Anaerobic Digestion of Protein-Rich Biowaste: Process Performance and Microbial Community Structures Shift, Front. Environ. Sci., 2022, 9, 805078 CrossRef.
- J.-J. Lay, Y.-J. Lee and T. Noike, Feasibility of biological hydrogen production from organic fraction of municipal solid waste, Water Res., 1999, 33, 2579–2586 CrossRef CAS.
- J. A. Rackliffe, J. Ni and N. S. Mosier, Effect of acclimatization rate on biogas production from anaerobic digestion of biodiesel waste products, Biofuels, Bioprod. Biorefin., 2023, 17, 1541–1553 CrossRef CAS.
- J. V. Mercado, M. Koyama and K. Nakasaki, Complexity of acclimatization substrate affects anaerobic digester microbial community response to organic load shocks, Environ. Res., 2023, 216, 114722 CrossRef CAS PubMed.
- J. Zeng, Y. Dai, R. Xu, S. Cheng, R. Sun and S. Shi, Effect of acclimation on inoculum functioning and dynamics within a microbial community, Biomass Bioenergy, 2019, 128, 105312 CrossRef CAS.
- R. Rafieenia, A. Pivato and M. C. Lavagnolo, Effect of inoculum pre-treatment on mesophilic hydrogen and methane production from food waste using two-stage anaerobic digestion, Int. J. Hydrogen Energy, 2018, 43, 12013–12022 CrossRef CAS.
- V. Mozhiarasi, T. S. Natarajan and K. Dhamodharan, A high-value biohythane production: Feedstocks, reactor configurations, pathways, challenges, technoeconomics and applications, Environ. Res., 2023, 219, 115094 CrossRef CAS PubMed.
- J. Maroušek, O. Strunecký, L. Kolář, M. Vochozka, M. Kopecký, A. Maroušková, J. Batt, M. Poliak, M. Šoch, P. Bartoš, T. Klieštik, M. Filip, P. Konvalina, J. Moudrý, J. Peterka, K. Suchý, T. Zoubek and E. Cera, Advances in nutrient management make it possible to accelerate biogas production and thus improve the economy of food waste processing, Energy Sources, Part A Recovery, Util. Environ. Eff., 2020, 1–10 Search PubMed.
- K. Hagos, J. Zong, D. Li, C. Liu and X. Lu, Anaerobic co-digestion process for biogas production: Progress, challenges and perspectives, Renew. Sustain. Energy Rev., 2017, 76, 1485–1496 CrossRef CAS.
- J. Stávková and J. Maroušek, Novel sorbent shows promising financial results on P recovery from sludge water, Chemosphere, 2021, 276, 130097 CrossRef.
- C. Schott, L. Yan, U. Gimbutyte, J. R. Cunha, R. D. van der Weijden and C. Buisman, Enabling efficient phosphorus recovery from cow manure: Liberation of phosphorus through acidification and recovery of phosphorus as calcium phosphate granules, Chem. Eng. J., 2023, 460, 141695 CrossRef CAS.
- L. I. da Silva, M. C. Pereira, A. M. X. de Carvalho, V. H. Buttrós, M. Pasqual and J. Dória, Phosphorus-Solubilizing Microorganisms: A Key to Sustainable Agriculture, Agriculture, 2023, 13, 462 CrossRef.
- X. Qu, H. Zeng, Y. Gao, T. Mo and Y. Li, Bio-hydrogen production by dark anaerobic fermentation of organic wastewater, Front. Chem., 2022, 10, 978907 CrossRef CAS PubMed.
- K. Willquist, V. N. Nkemka, H. Svensson, S. Pawar, M. Ljunggren, H. Karlsson, M. Murto, C. Hulteberg, E. W. J. van Niel and G. Liden, Design of a novel biohythane process with high H2 and CH4 production rates, Int. J. Hydrogen Energy, 2012, 37, 17749–17762 CrossRef CAS.
- M. Ljunggren and G. Zacchi, Techno-economic analysis of a two-step biological process producing hydrogen and methane, Bioresour. Technol., 2010, 101, 7780–7788 CrossRef CAS PubMed.
- S. Shanmugam, M. Sekar, R. Sivaramakrishnan, T. Raj, E. S. Ong, A. H. Rabbani, E. R. Rene, T. Mathimani, K. Brindhadevi and A. Pugazhendhi, Pretreatment of second and third generation feedstock for enhanced biohythane production: Challenges, recent trends and perspectives, Int. J. Hydrogen Energy, 2021, 46, 11252–11268 CrossRef CAS.
- S. A. A. Rawoof, P. S. Kumar, D.-V. N. Vo and S. Subramanian, Sequential production of hydrogen and methane by anaerobic digestion of organic wastes: a review, Environ. Chem. Lett., 2021, 19, 1043–1063 CrossRef CAS.
- F. Micolucci, M. Gottardo, P. Pavan, C. Cavinato and D. Bolzonella, Pilot scale comparison of single and double-stage thermophilic anaerobic digestion of food waste, J. Clean. Prod., 2018, 171, 1376–1385 CrossRef CAS.
- Y. Wang, Y. Zhang, L. Meng, J. Wang and W. Zhang, Hydrogen–methane production from swine manure: Effect of pretreatment and VFAs accumulation on gas yield, Biomass Bioenergy, 2009, 33, 1131–1138 CrossRef CAS.
- M. Vochozka, J. Horák, T. Krulický and P. Pardal, Predicting future Brent oil price on global markets, Acta Montan. Slovaca, 2020, 25, 375–392 Search PubMed.
- M. Vochozka, Z. Rowland, P. Suler and J. Marousek, The Influence of the International Price of Oil on the Value of the EUR/USD Exchange Rate, J. Compet., 2020, 12, 167–190 Search PubMed.
- J. Maroušek, A. Maroušková, B. Gavurová and B. Minofar, Techno-economic considerations on cement substitute obtained from waste refining, J. Clean. Prod., 2023, 412, 137326 CrossRef.
- J. Maroušek, B. Minofar, A. Maroušková, O. Strunecký and B. Gavurová, Environmental and economic advantages of production and application of digestate biochar, Environ. Technol. Innovat., 2023, 30, 103109 CrossRef.
- J. Maroušek, Review: Nanoparticles can change (bio)hydrogen competitiveness, Fuel, 2022, 328, 125318 CrossRef.
Footnote |
| † Electronic supplementary information (ESI) available. See DOI: https://doi.org/10.1039/d4va00107a |
| This journal is © The Royal Society of Chemistry 2024 |