Organelle-resolved imaging of formaldehyde reveals its spatiotemporal dynamics†
Lei
Zhou
ac,
Yuan
Pan
a,
Xiaozhuan
Li
b,
Tingmin
Fan
b,
Xingguang
Liang
*b and
Xin
Li
 *a
*a
aCollege of Pharmaceutical Sciences, Zhejiang University, Hangzhou 310058, China. E-mail: lixin81@zju.edu.cn
bDepartment of Clinical Pharmacy, First Affiliated Hospital, School of Medicine, Zhejiang University, Hangzhou 310003, China. E-mail: lrvin@zju.edu.cn
cCollaborative Innovation Center of Yangtze River Delta Region Green Pharmaceuticals, Zhejiang University of Technology, Hangzhou 310014, China
First published on 3rd September 2024
Abstract
Understanding the spatiotemporal dynamics of formaldehyde (FA) is crucial for elucidating its pathophysiology. In this study, we designed a series of organelle-resolved probes to investigate FA dynamics. By incorporating various organelle anchors into a coumarin hydrazonate, we developed a series of probes with excellent organelle selectivity and FA specificity, enabling rapid and precise sensing of FA in an organelle-resolved way. Leveraging these probes, we captured the spatiotemporal dynamics of native FA in response to exogenous FA or oxidative stress challenges. In particular, we unveiled the distinct responses of various organelles to identical cellular stressors. Moreover, we observed the dynamic response within individual organelles when cells were exposed to stressors for varying durations. We envision these probes will serve as versatile tools for probing FA pathophysiology.
1. Introduction
Formaldehyde (FA), recognized as a notorious environmental carcinogen, has recently gained attention as an endogenous metabolic intermediate widely distributed in mammals at a relatively high concentration of approximately 87–400 μM,1 implying its essential role in cellular functions.2 FA is a key metabolite in one-carbon metabolism,3 predominantly generated as an intermediate during the conversion of serine to glycine catalyzed by serine hydroxymethyl transferases (SHMT).4,5 Additionally, FA is produced during the demethylation of methylated DNA or RNA bases (e.g. m6A),6N-methylated amino acid residues (e.g. lysine, arginine),7 or any other N-methylated amines (e.g. methylamine, adrenaline).8 Typically, FA readily condenses with tetrahydrofolate (THF) into 5,10-methylene-THF.9 Both 5,10-methylene-THF and THF are susceptible to decomposition, releasing free FA.3,10To counteract the toxic nature of FA, enzymatic mechanisms for its detoxification exist, with alcohol dehydrogenase 5 (ADH5) and aldehyde dehydrogenase 2 (ALDH2) serving as primary metabolic enzymes.11–13 Lethal FA accumulation due to deficiency of both ADH5 and ALDH2 leads to partial synthetic lethality,14 multisystem disorders,15 leukemia,16etc. Our previous work has revealed an increase in endogenous FA levels in the brains of aging mice, with a more pronounced tendency observed in Alzheimer's disease (AD) transgenetic mice,17,18 suggesting the involvement of FA in AD pathology. However, the specific contributions of FA to AD pathology remain unknown. Understanding its intracellular generation and organelle distribution should provide the most direct insights into FA-related pathophysiology. Nevertheless, the spatial and temporal information of FA remains largely uncharacterized due to the lack of effective methods for tracking subcellular FA in real time.
The volatile and highly electrophilic nature of FA results in a very short half-life of approximately 1–1.5 min,1 posing a significant challenge for its detection. Among the various available methods, activity-based sensing utilizing FA-specific fluorescent probes enables direct visualization of FA in live cells,19,20 offering a straightforward approach to detect its production, cellular trafficking, and metabolism. Currently, two categories of chemistry-based probes are developed for FA sensing: the first utilizes the azo-Cope rearrangement,21–24 while the second employs hydrazine-aldehyde condensation.25 These probes, characterized by their biocompatibility and selectivity, have shed light on the presence of endogenous FA in various pathologies, significantly enhancing understanding of FA-related pathophysiology.19,20 However, most of these probes suffer from slow sensing kinetics and irreversible signals, hindering real-time monitoring of FA dynamics in live cells. Furthermore, the development of organelle-specific probes remains an ongoing challenge.
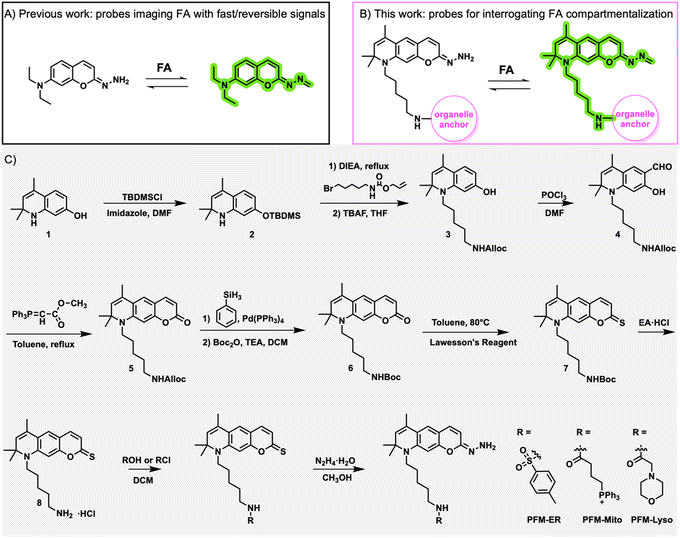
Our group pioneered the use of coumarin hydrazonate-based probes for detecting FA in live cells (Fig. 1A).17,18 These probes, characterized by their reduced nucleophilicity compared to their hydrazine counterparts, exhibited high selectivity toward FA while remaining inert toward other carbonyls, including acetaldehyde. Additionally, they demonstrated ultrafast detection kinetics and a reversible response profile to FA in both aqueous solutions and live cells. These advantages make them suitable for tracking cellular FA dynamics with high temporal resolution. Herein, by incorporating organelle anchors into the coumarin hydrazonate probe, we developed a series of organelle-specific probes for tracking the subcellular distribution of native FA (Fig. 1B). These probes retained the sensitivity and selectivity towards FA of the parent probe while acquiring the ability to target specific organelles. Enabled by these probes, we have uncovered the temporal dynamics of native FA in different organelles, including endoplasmic reticulum (ER), mitochondria, and lysosomes. These findings highlight the efficacy of our organelle-anchoring FA probes and underscore their potential for elucidating FA pathophysiology in the future.
2. Experimental section
2.1. Synthesis and characterization
The synthesis and structure characterization of the probes and intermediates were described in detail in the ESI.†2.2. Fluorescent spectra analysis methods
All the photophysical characterization experiments were carried out at ambient temperature. Fluorescence measurements were performed on an Agilent Cary Eclipse Fluorescence Spectrophotometer. Slit widths were set at 5 nm and 5 nm for excitement and emission respectively, and the sensitivity of the detector was kept at medium. Probes were dissolved in DMSO (5 mM) to make a stock solution and were diluted to the indicated concentrations for measurement. All spectra were measured in PBS buffer (10 mM, pH 7.4, 20% acetonitrile) solution.2.3. Cell culture
The HeLa cell line was purchased from ATCC. Briefly, the cells were cultured in Eagle's minimal essential medium (EMEM, Thermo Fisher Scientific) for HeLa cells, supplemented with 10% heat-inactivated fatal bovine serum (Thermo Fisher Scientific), penicillin (100 U mL−1, Thermo Fisher Scientific), and streptomycin (100 U mL−1, Thermo Fisher Scientific). The cultures were maintained at 37 °C in a 95% humidified atmosphere with 5% CO2.2.4. Confocal fluorescence imaging
For confocal microscopy imaging of the subcellular distribution of PFM-Mito, PFM-ER and PFM-Lyso in living cells, HeLa cells were incubated with ER-Tracker (1 μM), Mito-Tracker (0.5 μM) or Lyso-Tracker (1 μM) for 30 min. Then the residual tracker was washed three times with PBS (pH 7.4), followed by staining the cells with 1 μM PFM-Mito, PFM-ER and PFM-Lyso for 10 min. Cells were then imaged.For confocal fluorescent imaging experiments to evaluate the response of PFM-Mito, PFM-ER and PFM-Lyso in live system upon FA and H2O2 treatment, HeLa cells were first stained with 1 μM PFM-Mito, PFM-ER or PFM-Lyso at 37 °C for 15 min. The residual probe was washed three times with PBS. Cells were then treated with H2O2 or FA of indicated concentrations for 1 h, 2 h, 3 h, or 6 h. Cells were then imaged.
Imaging was performed on a laser scanning confocal microscope fluoview FV1000 (Olympus). Digital images were captured using the FV10-ASW 3.0 viewer software. The fluorescence of PFM-Mito and PFM-ER was monitored at λem = 500–550 nm (λex = 405 nm). The fluorescence of PFM-Lyso was monitored at λem = 500–550 nm (λex = 488 nm). Cell counts were performed using a 40× or 60× objective in at least five fields of view randomly selected from each coverslip. At least three independent experiments were counted. The fluorescence density was analyzed using ImageJ software (NIH, Bethesda, MD, USA).
2.5. Statistical analysis
The data were analyzed with ordinary one-way ANOVA. Pearson's correlation coefficient was applied to evaluate the correlation. All data are represented as the Mean ± S.E.M. A value of p < 0.05 was indicated as significant.3. Results and discussion
3.1. Probe design and synthesis
Recent years have witnessed significant advancements in strategies for targeting specific organelles. A wide array of organelle anchors is available, including biomolecules such as peptides, lipids, and oligosaccharides, and chemically designed moieties.26–28 Among the synthetic anchors, triphenylphosphonium is commonly used to target mitochondria due to its hydrophobic and cationic nature;29 the N-alkylmorpholine is frequently employed to anchor lysosomes owning to its reversible protonation at physiological pH;30 and phenyl sulfonamide is typically utilized for endoplasmic reticulum (ER) localization due to its affinity towards the sulphonylurea receptors of ATP-sensitive K+ channels on the ER.31 Consequently, we decided to conjugate these three anchors to our previously developed coumarin hydrazonate-based probes to create organelle-specific FA probes.To preserve the photophysical properties of the original coumarin hydrazonate probe, we opted to attach the anchors at the 7-amino terminal of the coumarin skeleton via an alkyl linker to minimize their impact on the electron push–pull effect of the fluorophore. Initially, we attempted to tether the anchors modularly using Cu(I)-catalyzed azide/alkyne click chemistry and synthesized 7-propargylamino-coumarin hydrazonate. However, we observed instability of the hydrazonate group in the presence of Cu(I). Consequently, we pursued tethering the anchors via a carbonyl acid-amine condensation reaction. After several attempts, we established the synthetic routes shown in Fig. 1C. Initially, an alkyl chain with a terminal allyloxycarbonyl (Alloc)-protected amino group was incorporated into the m-aminophenol starting material. Subsequently, the coumarin skeleton was constructed in three steps. The Alloc protection group was then replaced with t-butyloxy carbonyl (Boc), followed by thionation of the coumarin lactone. Deprotection of the terminal amino group allowed its condensation with various anchors. Subsequent hydrazinolysis yielded the desired probes.
3.2. Spectroscopic responses of the probes toward FA
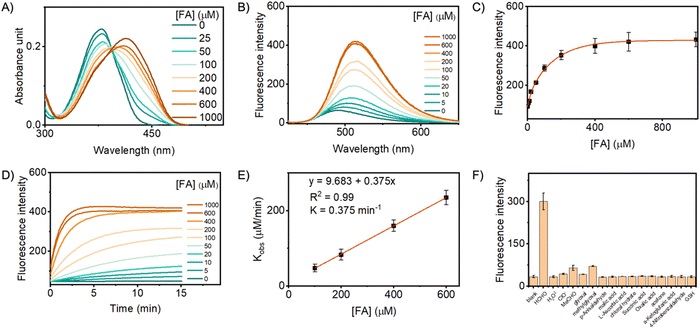
With the probes synthesized, we first assessed their spectroscopic responses to FA in a biologically relevant phosphate buffer solution (PBS, 10 mM, pH 7.4). The three probes exhibited similar responsive profiles towards FA, indicating that the organelle-anchoring moieties minimally influenced their FA sensing abilities. Taking probe PFM-ER as a representative, it exhibited maximum absorption at 380 nm (ε 12![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 220 M−1 cm−1). Treatment of PFM-ER (20 μM) with FA resulted in a dose-dependent bathochromic shift of the maximum absorption to 415 nm (ε 11
220 M−1 cm−1). Treatment of PFM-ER (20 μM) with FA resulted in a dose-dependent bathochromic shift of the maximum absorption to 415 nm (ε 11![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 010 M−1 cm−1 when FA was 1 mM) (Fig. 2A), indicating the extension of its π-conjugation system upon FA sensing. PFM-ER (5 μM) in PBS displayed a weak emission band centered at 490 nm. However, FA induced a redshift to 515 nm, which intensified in an FA dose-dependent manner (Fig. 2B). Plotting PFM-ER emission intensity at 515 nm against FA concentrations yielded an exponential regression, with FA concentrations in the range of 0–400 μM eliciting the most sensitive responses (Fig. 2C). A plateau effect was achieved when FA was dosed at 600 μM, suggesting the complete transformation of the probe under this condition. Given that cellular FA concentrations are reported in the range of 200–400 μM, this result suggests the potential of PFM-ER to quantify native FA in biological systems. We also recorded the sensing kinetics of PFM-ER. Aliquots of PFM-ER (5 μM) were treated with various concentrations of FA, and the emission intensity at 515 nm was recorded in a time-lapsed way (Fig. 2D). The sensing reaction proceeded rapidly, completing within a few minutes. Under pseudo-first-order conditions, an exceptionally fast rate constant (k) of 0.375 min−1 was determined (Fig. 2E), indicating that PFM-ER is sufficiently fast to capture transit FA signals. To investigate potential interference from other biomolecules, PFM-ER was exposed to various species commonly found in biological systems. Among the tested species, only FA induced a fluorescence response (Fig. 2F), indicating its specificity. Similar results were observed for PFM-Mito and PFM-Lyso (Fig. S1 and S2, ESI†). All these results suggest the rapid, sensitive, and selective response of the probes towards FA.
010 M−1 cm−1 when FA was 1 mM) (Fig. 2A), indicating the extension of its π-conjugation system upon FA sensing. PFM-ER (5 μM) in PBS displayed a weak emission band centered at 490 nm. However, FA induced a redshift to 515 nm, which intensified in an FA dose-dependent manner (Fig. 2B). Plotting PFM-ER emission intensity at 515 nm against FA concentrations yielded an exponential regression, with FA concentrations in the range of 0–400 μM eliciting the most sensitive responses (Fig. 2C). A plateau effect was achieved when FA was dosed at 600 μM, suggesting the complete transformation of the probe under this condition. Given that cellular FA concentrations are reported in the range of 200–400 μM, this result suggests the potential of PFM-ER to quantify native FA in biological systems. We also recorded the sensing kinetics of PFM-ER. Aliquots of PFM-ER (5 μM) were treated with various concentrations of FA, and the emission intensity at 515 nm was recorded in a time-lapsed way (Fig. 2D). The sensing reaction proceeded rapidly, completing within a few minutes. Under pseudo-first-order conditions, an exceptionally fast rate constant (k) of 0.375 min−1 was determined (Fig. 2E), indicating that PFM-ER is sufficiently fast to capture transit FA signals. To investigate potential interference from other biomolecules, PFM-ER was exposed to various species commonly found in biological systems. Among the tested species, only FA induced a fluorescence response (Fig. 2F), indicating its specificity. Similar results were observed for PFM-Mito and PFM-Lyso (Fig. S1 and S2, ESI†). All these results suggest the rapid, sensitive, and selective response of the probes towards FA.
3.3. Evaluating the organelle-specificity of the probes
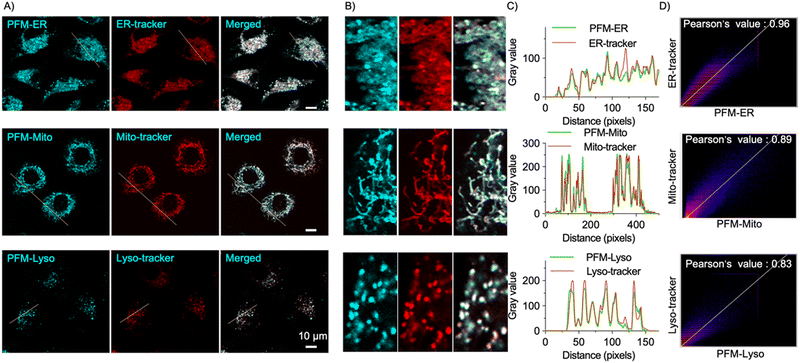
After confirming the sensitivity and specificity of the probes towards FA in solution-based experiments, we assessed their cytotoxicity using the CCK8 assay. The results demonstrated that PFM-Mito, PFM-ER and PFM-Lyso exhibited minimal toxicity in HeLa cells even at concentrations up to 50 μM with prolonged incubation time of 24 h (Fig. S3, ESI†), indicating their excellent biocompatibility.Subsequently, we evaluated their photostability for live cell imaging. Little change in cellular probe fluorescence intensity was observed upon continuous scan (Fig. S4, ESI†), suggesting their desirable photostability. Then, we moved to test their organelle specificity. HeLa cells were first stained with commercially available organelle-specific dye, loaded with the corresponding probe, and then imaged under confocal microscopy. All three probes exhibited significant cellular fluorescence (Fig. 3A), suggesting the presence of basal FA in live HeLa cells, consistent with previous reports. Importantly, the probe fluorescence colocalized well with the corresponding organelle-trackers (Fig. 3B). Intensity profile analysis of regions of interest across cells further confirmed their desirable spatial correlation (Fig. 3C). Co-localization areas were quantified using Pearson's coefficient. PFM-ER exhibited a Pearson's correlation coefficient of 0.96 with ER-tracker Red; PFM-Mito exhibited a Pearson's correlation coefficient of 0.89 with Mito-tracker red; and PFM-Lyso exhibited a Pearson's correlation coefficient of 0.83 with Lyso-tracker red (Fig. 3D). These results validated the organelle specificity of the probes and underscored their potential for spatially-resolved sensing of cellular FA.
3.4. Imaging exogenous FA-induced local FA disturbance in a spatiotemporally resolved manner
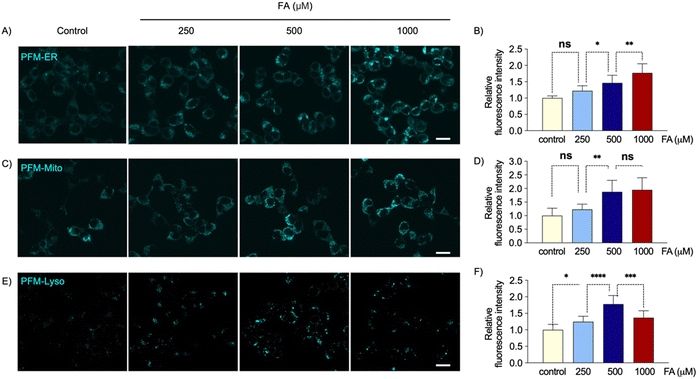
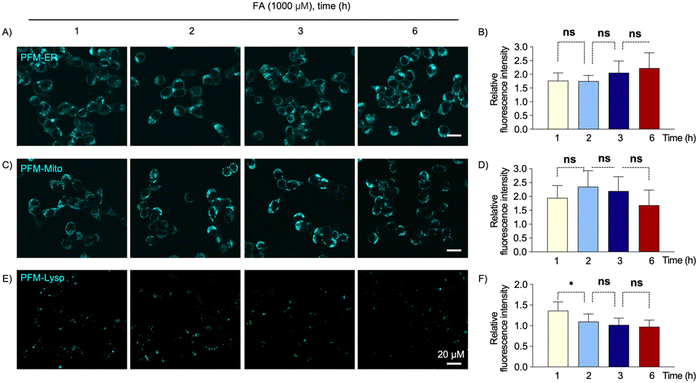
Environmental FA is a widely-present toxin, and understanding its impact on different organelles may illuminate its detailed toxic mechanisms. To investigate this, we utilized these organelle-specific probes to examine how different organelles respond to exogenous FA stimulation.HeLa cells were initially loaded with either PFM-ER, PFM-Mito, or PFM-Lyso. Subsequently, the cells were treated with various doses of exogenous FA. Following the indicated incubation period, cells were imaged using confocal microscopy. The cellular probe fluorescence and quantified intensity data revealed distinct responses of the ER, mitochondria, and lysosomes to exogenous FA stimulation (Fig. 4 and 5). Upon challenging cells with FA for 1 h, local FA concentrations in ER and mitochondria increased with escalating exogenous FA doses from 0 to 1000 μM, whereas lysosomes exhibited an initial increase followed by a decrease in local FA concentrations (Fig. 4). Notably, 250 μM FA in the culture medium caused no significant effect on local FA levels in ER and mitochondria; while significantly increased local FA levels in lysosomes (Fig. 4).
To investigate the time-dependent response, cells were exposed to the same doses of FA (1000 μM) for varying durations (1–6 h). Throughout this duration, local FA concentrations in ER and mitochondria remained relatively stable, while lysosomal FA concentrations decreased slightly from 1 h to 2 h, maintaining consistency over the subsequent 4 hours (Fig. 5). These findings indicate differing responses of various organelles to the same cell stress. We hypothesize that each organelle may possess a buffering capacity to tolerate FA stress, with different organelles exhibiting distinct thresholds for their buffering capabilities.
3.5. Imaging oxidative stress-induced endogenous FA in a spatiotemporally resolved way
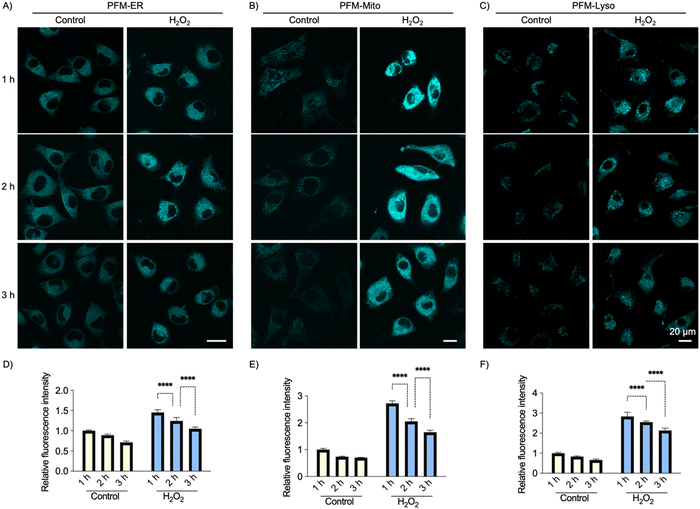
Previously, we uncovered a burst of native FA generation in live cells upon oxidative or ER stress challenge.18,32 However, the dynamic profile of this FA generation has not been recorded in an organelle-resolved manner. Leveraging the probes’ capability to image cellular FA in a spatiotemporally resolved manner, we investigated how various organelles respond to oxidative stress in terms of their local FA generation.HeLa cells were initially loaded with PFM-ER, PFM-Mito or PFM-Lyso (1 μM) at 37 °C for 15 min. Subsequently, the cells were treated with H2O2 (250 μM) for the indicated time and imaged. Cells stained with the corresponding probe but not challenged with H2O2 served as controls. As anticipated, H2O2 induced a dramatic upregulation of cellular FA, evident from the significantly increased cellular probe fluorescence in the H2O2-treated groups compared to the control groups (Fig. 6). Notably, extending the H2O2 incubation time from 1 h to 3 h led to a gradual decrease in local FA concentrations in ER, mitochondria, and lysosomes. This observation suggests that native FA generation upon oxidative stress stimulation is a rapid response. The decline in FA levels within the organelles is likely attributable to a greater metabolic rate than the generation rate.
4. Conclusion
In summary, we have developed chemical probes PFM-ER, PFM-Mito, and PFM-Lyso for imaging cellular FA in a spatiotemporally resolved manner. The probes exhibit high specificity towards FA, demonstrating rapid and FA-concentration-dependent fluorogenic response, making them sensitive tools for interrogating temporal fluctuations of native FA. Additionally, they display excellent organelle selectivity, as evidenced by their desirable co-localization with well-recognized organelle trackers, enabling the tracking of native FA within specific organelles. Utilizing these probes, we observed distinct organelle responses to exogenous FA stimulation in terms of native FA levels. Furthermore, we observed rapid upregulation of native FA in ER, mitochondria, and lysosomes upon H2O2 stimulation, followed by gradual decay. These findings highlight the reliability of the probes for imaging cellular FA in a spatiotemporally resolved manner and suggest their utility in facilitating pathophysiological studies on biological FA.Data availability
The authors confirm that the data supporting the findings of this study are available within the article and its ESI.†Conflicts of interest
The authors declare no competing financial interest.Acknowledgements
This work was supported by the National Natural Science Foundations of China (22377106, 22077112), and the Natural Science Foundation of Zhejiang Province (LZ23H300001, LY21H090012, LY19H030010). X. L. was supported by the National Program for Support of Top-notch Young Professionals (grant 2021). We thank the technical support of Ximin Jin from the microscopy core facility, Central Laboratory, the First Affiliated Hospital, Zhejiang University School of Medicine with the confocal microscopy.References
- E. F. S. Authority, EFSA J., 2014, 12, 3550–3560 Search PubMed.
- V. N. Pham, K. J. Bruemmer, J. D. W. Toh, E. J. Ge, L. Tenney, C. C. Ward, F. A. Dingler, C. L. Millington, C. A. Garcia-Prieto, M. C. Pulos-Holmes, N. T. Ingolia, L. B. Pontel, M. Esteller, K. J. Patel, D. K. Nomura and C. J. Chang, Science, 2023, 382, eabp9201 CrossRef CAS PubMed.
- G. Burgos-Barragan, N. Wit, J. Meiser, F. A. Dingler, M. Pietzke, L. Mulderrig, L. B. Pontel, I. V. Rosado, T. F. Brewer, R. L. Cordell, P. S. Monks, C. J. Chang, A. Vazquez and K. J. Patel, Nature, 2017, 548, 549–554 CrossRef CAS PubMed.
- V. Schirch and D. M. E. Szebenyi, Curr. Opin. Chem. Biol., 2005, 9, 482–487 CrossRef CAS PubMed.
- H. S. Fernandes, M. J. Ramos and N. M. F. S. A. Cerqueira, ACS Catal., 2018, 8, 10096–10110 CrossRef CAS.
- J. D. W. Toh, S. W. M. Crossley, K. J. Bruemmer, E. J. Ge, D. He, D. A. Iovan and C. J. Chang, Proc. Natl. Acad. Sci. U. S. A., 2020, 117, 25284–25292 CrossRef CAS PubMed.
- Y. Shi, F. Lan, C. Matson, P. Mulligan, J. R. Whetstine, P. A. Cole, R. A. Casero and Y. Shi, Cell, 2004, 119, 941–953 CrossRef CAS PubMed.
- P. J. Boor, M. B. Trent, G. A. Lyles, M. Tao and G. A. S. Ansari, Toxicology, 1992, 73, 251–258 CrossRef CAS PubMed.
- R. G. Kallen and W. P. Jencks, J. Biol. Chem., 1966, 241, 5851–5863 CrossRef CAS PubMed.
- X. L. Chen, S. Y. Chothia, J. Basran and R. J. Hopkinson, Chem. Commun., 2021, 57, 5778–5781 RSC.
- I. V. Rosado, F. Langevin, G. P. Crossan, M. Takata and K. J. Patel, Nat. Struct. Mol. Biol., 2011, 18, 1432–1434 CrossRef CAS PubMed.
- L. B. Pontel, I. V. Rosado, G. Burgos-Barragan, J. I. Garaycoechea, R. Yu, M. J. Arends, G. Chandrasekaran, V. Broecker, W. Wei, L. M. Liu, J. A. Swenberg, G. P. Crossan and K. J. Patel, Mol. Cell, 2015, 60, 177–188 CrossRef CAS PubMed.
- F. A. Dingler, M. Wang, A. F. Mu, C. L. Millington, N. Oberbeck, S. Watcham, L. B. Pontel, A. N. Kamimae-Lanning, F. Langevin, C. Nadler, R. L. Cordell, P. S. Monks, R. Yu, N. K. Wilson, A. Hira, K. Yoshida, M. Mori, Y. Okamoto, Y. Okuno, H. Muramatsu, Y. Shiraishi, M. Kobayashi, T. Moriguchi, T. Osumi, M. Kato, S. Miyano, E. Ito, S. Kojima, H. Yabe, M. Yabe, K. Matsuo, S. Ogawa, B. Göttgens, M. R. G. Hodskinson, M. Takata and K. J. Patel, Mol. Cell, 2020, 80, 996–1012 CrossRef CAS PubMed.
- J. Nakamura, D. W. Holley, T. Kawamoto and S. J. Bultman, Genes Environ., 2020, 42, 21 CrossRef CAS PubMed.
- Y. Oka, M. Hamada, Y. Nakazawa, H. Muramatsu, Y. Okuno, K. Higasa, M. Shimada, H. Takeshima, K. Hanada, T. Hirano, T. Kawakita, H. Sakaguchi, T. Ichimura, S. Ozono, K. Yuge, Y. Watanabe, Y. Kotani, M. Yamane, Y. Kasugai, M. Tanaka, T. Suganami, S. Nakada, N. Mitsutake, Y. Hara, K. Kato, S. Mizuno, N. Miyake, Y. Kawai, K. Tokunaga, M. Nagasaki, S. Kito, K. Isoyama, M. Onodera, H. Kaneko, N. Matsumoto, F. Matsuda, K. Matsuo, Y. Takahashi, T. Mashimo, S. Kojima and T. Ogi, Sci. Adv., 2020, 6, eabd7197 CrossRef CAS PubMed.
- A. F. Mu, A. Hira, A. Niwa, M. Osawa, K. Yoshida, M. Mori, Y. Okamoto, K. Inoue, K. Kondo, M. T. Kanemaki, T. Matsuda, E. Ito, S. Kojima, T. Nakahata, S. Ogawa, K. Tanaka, K. Matsuo, M. K. Saito and M. Takata, Blood, 2021, 137, 2021–2032 CrossRef CAS PubMed.
- R. R. Tao, M. H. Liao, Y. X. Wang, H. Wang, Y. H. Tan, S. Y. Qin, W. J. Wei, C. Z. Tang, X. G. Liang, Y. F. Han and X. Li, Anal. Chem., 2022, 94, 1308–1317 CrossRef CAS PubMed.
- X. G. Liang, B. Chen, L. X. Shao, J. Cheng, M. Z. Huang, Y. Chen, Y. Z. Hu, Y. F. Han, F. Han and X. Li, Theranostics, 2017, 7, 2305–2313 CrossRef CAS PubMed.
- J. Ohata, K. J. Bruemmer and C. J. Chang, Acc. Chem. Res., 2019, 52, 2841–2848 CrossRef CAS PubMed.
- Y. H. Tang, Y. Y. Ma, J. L. Yin and W. Y. Lin, Chem. Soc. Rev., 2019, 48, 4036–4048 RSC.
- A. Roth, H. Li, C. Anorma and J. Chan, J. Am. Chem. Soc., 2015, 137, 10890–10893 CrossRef CAS PubMed.
- T. F. Brewer and C. J. Chang, J. Am. Chem. Soc., 2015, 137, 10886–10889 CrossRef CAS PubMed.
- T. F. Brewer, G. Burgos-Barragan, N. Wit, K. J. Patel and C. J. Chang, Chem. Sci., 2017, 8, 4073–4081 RSC.
- K. J. Bruemmer, R. R. Walvoord, T. F. Brewer, G. Burgos-Barragan, N. Wit, L. B. Pontel, K. J. Patel and C. J. Chang, J. Am. Chem. Soc., 2017, 139, 5338–5350 CrossRef CAS PubMed.
- Y. H. Tang, X. Q. Kong, A. Xu, B. L. Dong and W. Y. Lin, Angew. Chem., Int. Ed., 2016, 55, 3356–3359 CrossRef CAS PubMed.
- H. Zhu, J. L. Fan, J. J. Du and X. J. Peng, Acc. Chem. Res., 2016, 49, 2115–2126 CrossRef CAS PubMed.
- M. K. Goshisht, N. Tripathi, G. K. Patra and M. Chaskar, Chem. Sci., 2023, 14, 5842–5871 RSC.
- J. J. Yang, A. Griffin, Z. Qiang and J. Ren, Signal Transduction Targeted Ther., 2022, 7, 379 CrossRef CAS PubMed.
- J. Zielonka, J. Joseph, A. Sikora, M. Hardy, O. Ouari, J. Vasquez-Vivar, G. Cheng, M. Lopez and B. Kalyanaraman, Chem. Rev., 2017, 117, 10043–10120 CrossRef CAS PubMed.
- V. Nguyen and H. D. Li, Molecules, 2023, 28, 6550 CrossRef PubMed.
- M. I. Thabrew and G. O. Emerole, Comp. Biochem. Physiol., Part C: Pharmacol., Toxicol. Endocrinol., 1983, 74, 473–476 CrossRef CAS PubMed.
- J. Cheng, Y. Ren, Y. J. Huang, X. Z. Li, M. Z. Huang, F. Han, X. G. Liang and X. Li, Anal. Chem., 2020, 92, 1409–1415 CrossRef CAS PubMed.
Footnote |
| † Electronic supplementary information (ESI) available: Probe synthetic methods and structure characterization data, probe absorption and emission measurement method, cell culture, cell proliferation assay, imaging method, and supplementary figures. See DOI: https://doi.org/10.1039/d4tb01317d |
| This journal is © The Royal Society of Chemistry 2024 |