Supramolecular DNA nanogels through host–guest interaction for targeted drug delivery†
Zongze
Duan
a,
Guizhi
Dong
b,
Hai
Yang
 a,
Zhengwei
Yan
a,
Simin
Liu
a,
Zhengwei
Yan
a,
Simin
Liu
 a,
Yuanchen
Dong
a,
Yuanchen
Dong
 *b and
Zhiyong
Zhao
*b and
Zhiyong
Zhao
 *a
*a
aKey Laboratory of Hubei Province for Coal Conversion and New Carbon Materials, School of Chemistry and Chemical Engineering, Wuhan University of Science and Technology, Wuhan 430081, China. E-mail: zhaozhiyong@wust.edu.cn
bCAS Key Laboratory of Colloid, Interface and Chemical Thermodynamics, Institute of Chemistry, Chinese Academy of Sciences, Beijing, 100190, China. E-mail: dongyc@iccas.ac.cn
First published on 22nd May 2024
Abstract
DNA hydrogels have been demonstrated with the advantages of good stability, easy modification, and extraordinary biocompatibility, which enables their great application prospects in biosensing, tissue engineering, and biomedicine. Based on the host–guest recognition properties of cucurbit[8]uril (CB[8]), we proposed a general method for constructing functional supramolecular DNA nanogels. Guest molecules have been conjugated into the DNA building units, which could be further crosslinked with CB[8] to construct supramolecular DNA nanogels. At the same time, the aptamer has also been modified into the hydrogel network to achieve cell targeting. These supramolecular DNA nanogels have been demonstrated with a controllable size and multiple stimuli responses, in addition to the excellent biocompatibility, stability and good targeting drug transport ability. Such a host–guest based strategy will provide a molecular library as a “toolbox” for the functionalization of DNA nanogels.
1. Introduction
DNA hydrogels are emerging biomaterials with good stability, flexibility, precise programmability and biocompatibility.1–5 During recent decades, while the functional nucleic acids such as aptamers,6i-motif structures,7 and CpG oligodeoxynucleotides8 have been introduced into DNA hydrogels to provide multiple functions,9 chemical functional groups or functional nanoparticles10,11 have also been integrated into DNA hydrogels to widen their biological applications.12–16 Recently, nanoscale DNA hydrogels, known as DNA nanogels, have been precisely prepared.17,18 Due to both the nano-size and the specific properties of DNA hydrogels, DNA nanogels from different skeletons including the Y-shaped skeleton,19 tetrahedron DNA skeleton20–22 and DNA dendrimer skeleton23 could be fabricated and utilized as vehicles for the targeted delivery of drug molecules in bioanalysis and biomedicine.Recently, supramolecular host–guest recognition has provided a new potential development strategy for the construction of functional DNA nanogels. In the last few decades, a variety of host molecules have been developed, such as crown ethers, cyclodextrins, calix[n]arenes, cucurbit[n]urils (CB[n]s) and pillar[n]arenes.24 Among these, the CB[n] (n = 5–8, 10, 13–15) family, as the fourth generation supramolecular macrocycle host molecules, has the following advantages in the construction of supramolecular hydrogels: (1) low cytotoxicity;25–28 (2) with different cavity sizes of CB[n]s, they can form complexes with controllable numbers of guest molecules;29 (3) the high affinity between CB[n]s and guest molecules30 guarantees the stability of the guest molecule, especially hydrophobic drugs;22 (4) the host–guest interaction is dynamically reversible by introducing external stimuli such as light,31 pH,32 FGG peptide,33etc. (5) the guest molecule could be functional, which introduces more functionality in the hydrogels. All these advantages indicate that the host–guest interaction between CB[n]s and guest molecules will be an effective strategy to construct supramolecular DNA nanogels.
Herein, we propose a strategy for the construction of photoresponsive supramolecular DNA nanogels (SDNs) through host–guest interaction between CB[8] and two different guest molecules (Scheme 1). In our design, we have prepared two DNA building units, termed as Y scaffold and the linker. Two guest molecules including azobenzene (Azo) and methyl violet (MV), have been chemically conjugated into the building blocks, respectively. It is known that the Azo molecule undergoes configuration inversion from trans to cis under UV irradiation, thus it could break out of the CB[8] cavity and destroy the 1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1
1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1 host–guest complex.34 This allows SDN photo-responsiveness and controls its disaggregation. In addition, the MUC1 aptamer35 was also introduced into SDNs to realize the target recognition. By controlling the concentration of the DNA building units, SDNs with different sizes can be obtained, which were verified by DLS and TEM. Then, the photo responsiveness of SDNs was investigated. Subsequently, the in vitro drug encapsulation and release experiments of SDNs were investigated. Finally, the targeted drug delivery and cytotoxicity of SDNs were revealed by cytotoxicity experiments, confocal laser scanning microscopy (CLSM) and flow cytometry.
1 host–guest complex.34 This allows SDN photo-responsiveness and controls its disaggregation. In addition, the MUC1 aptamer35 was also introduced into SDNs to realize the target recognition. By controlling the concentration of the DNA building units, SDNs with different sizes can be obtained, which were verified by DLS and TEM. Then, the photo responsiveness of SDNs was investigated. Subsequently, the in vitro drug encapsulation and release experiments of SDNs were investigated. Finally, the targeted drug delivery and cytotoxicity of SDNs were revealed by cytotoxicity experiments, confocal laser scanning microscopy (CLSM) and flow cytometry.
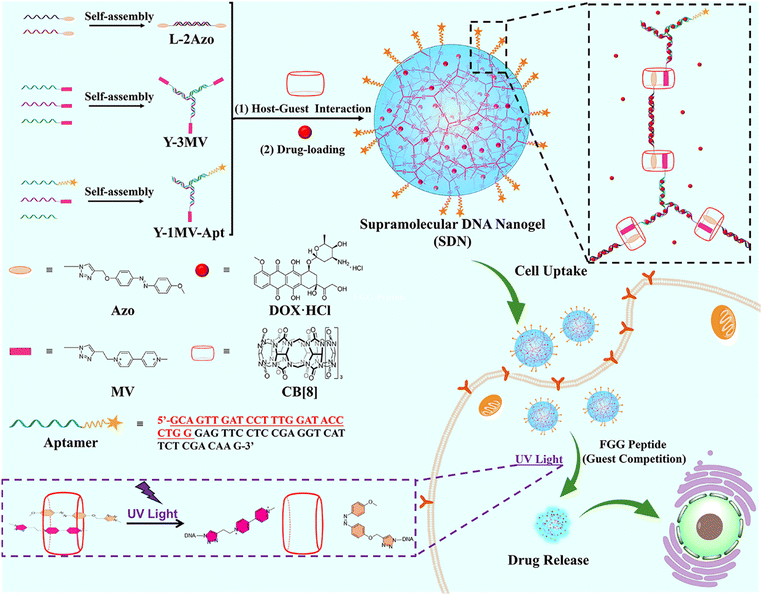 | ||
| Scheme 1 The formation of photo-responsive supramolecular DNA nanogels and their targeted drug delivery. | ||
2. Experimental section
2.1 Materials and instruments
The chemicals and reagents used in the experiments were commercially obtained and used without further purification, unless otherwise stated. All the DNA sequences were sourced from hippobio (Zhejiang, China). The water used in all experiments was Milli-Q deionized water (18.2 MΩ cm).The concentration of DNA and the functional DNA strands were determined by UV-Vis (Shimadzu, Japan) absorption at 260 nm. Structures of compounds were confirmed by 1H NMR spectroscopy (Agilent Technologies, China) and MALDI-TOF mass spectrometry (Shimadzu, Japan). The polyacrylamide gel electrophoresis (PAGE) analysis was carried out on an electrophoresis analyser (Jun Yi, Beijing) and imaged on a Gel Imager (Bio-Rad, USA). The size and morphology were determined by dynamic light scattering (DLS, PSS, USA), and transmission electron microscopy (TEM, JEOL, USA). A UV-vis spectrophotometer (UV-3600, shimadzu, Japan), a fluorescence spectrophotometer (LS 55, PerkinElmer, UK), and a flow cytometer (FACSCelesta, BD/Becton Dickinson, US) were also used in this study.
2.2 Synthesis of the two guest molecules
The synthesis steps of the two guest molecules Azo and MV are detailed in the ESI† (Schemes S1 and S2).2.3 Modification of DNA by guest molecules and construction of DNA building units
Specifically, 20 μL ssDNA-N3 (0.5 mM, 10 nmol), 2 μL copper sulfate (20 mM, 40 nmol), 2 μL sodium ascorbate (100 mM, 200 nmol), and 2 μL Azo (20 mM, 40 nmol) were mixed together. 5 μL DMF was added to the reaction system to increase the solubility of Azo, with the reaction temperature set to 55 °C and the reaction time to 6 h. After the reaction, it was purified by 20% polyacrylamide gel electrophoresis (PAGE).The reaction of MV is roughly the same; no additional DMF is needed, and the reaction time is shortened to be within 2 h, because the extension of this reaction time will cause the decomposition of DNA.
Two ssDNA modified with Azo molecules form a straight double stranded DNA with Azo groups at both ends (L-2Azo) through base complementary pairing. Three ssDNA modified with MV molecules form a Y-shaped DNA structure (Y-3M) in the same way. Meanwhile, Y-1M with only one MV molecule and Y-1M-Apt with an MUC1 aptamer were prepared as end cappers. These DNA assemblies will be used as building units in the construction of supramolecular DNA nanogels. Reaction conditions: 95 °C for 5 min; after annealing, they were placed at room temperature for 12 h, and the required DNA building units were obtained. L-2Azo is composed of ssDNA1-Azo and ssDNA2-Azo, Y-3M is composed of ssDNA3-MV, ssDNA4-MV and ssDNA5-MV, Y-1M is composed of ssDNA3, ssDNA4-MV and ssDNA5, and Y-1M-Apt is composed of ssDNA3-Apt, ssDNA4-MV and ssDNA5.
2.4 Construction of supramolecular DNA nanogels through host–guest interaction
Aqueous solutions were prepared with 50 μM L-2A, 30 μM Y-3M, 10 μM Y-1M and 10 μM Y-1M-Apt. For CB[8] solution concentrations refer to previous studies.36 As shown in Table S1 (ESI†), the corresponding DNA building units were mixed with CB[8] solution, shaken well, and then sonicated for 1 to 2 min to make CB[8] fully recognize the two guest molecules. Finally, the mixture was placed at room temperature for 24 h to obtain SDNs.2.5 Drug loading experiment of supramolecular DNA nanogels
Firstly, we plotted the DOX·HCl concentration dependent UV standard curve. Then, DOX·HCl (the total volume of solution is 100 μL, and the final concentration of DOX is 50 μg ml−1) was added for co incubation during the preparation of SDN-2. After incubation, SDN-2 was separated from the solution by high-speed centrifugation (9000 rpm) for 10 min. The concentration of unencapsulated DOX was determined by measuring the UV absorption intensity of DOX in its supernatant. The calculation formulas for drug loading and encapsulation efficiency are as follows:| Drug loading rate = (WT − WF)/WSDN-2 × 100% |
| Encapsulation rate = (WT − WF)/WT × 100% |
2.6 Stimulation response experiment of supramolecular DNA nanogels
Photo response: The SDN-2 was irradiated for 10, 30 and 60 min under 90 mW cm−2 UV light, and then prepared into TEM samples for observation.FGG peptide response: 100 equivalent FGG peptide and SDN-2 were incubated in PBS solution for 1 h. After incubation, DNA was prepared into TEM samples for observation.
2.7 Drug release of supramolecular DNA nanogels in vitro
First, the fluorescence curves of a series of DOX·HCl drugs with gradient concentration were configured and tested, and the drug release concentration was calculated using the linear regression equation obtained from the fluorescence emission at 590 nm. Then, the SDN-2 collected by centrifugation was carefully dispersed into PBS buffer solution with a pH of 7.4 and the control group was set (group 1: no treatment; group 2: UV irradiation for 30 min; group 3: add DNase I, the final concentration is 5 U ml−1; group 4: heat to 70 °C for 30 min; group 5: add DNase I, the final concentration is 50 U ml−1). The SDN-2 of each group was incubated at 37 °C, and the drug release concentration was determined by measuring the DOX·HCl fluorescence intensity in the supernatant after centrifugation in a high-speed centrifuge at intervals. After that, SDNs were re-dispersed into PBS buffer solution.2.8 Cell culture
MCF-7 cells were cultured in Dulbecco's modified Eagle medium (DMEM, Gibco, USA) containing 10% FBS and 1% penicillin/streptomycin at 37 °C and 5% CO2.MDA-MB-231 cells were cultured in the RMPI medium 1640 containing 10% FBS and 1% penicillin/streptomycin at 37 °C and 5% CO2.
2.9 Cells uptake experiment
The proliferative MCF-7 was inoculated with 1 × 105 cells per well in a confocal dish and incubated for 24 h under standard conditions. The diluted DMEM culture medium containing free DOX, SDN-2@DOX and SDN-2-Apt@DOX (DOX concentration is 5 μg mL−1) was incubated for 4 h, the waste liquid was taken out, PBS (PH = 7.4) was used to clean three times, and then DAPI was added to incubate in the incubator for 15 min to dye the nucleus. PBS was used to clean three times, and then 200 μL of paraformaldehyde tissue fixative was added. Finally, a confocal microscope was used to obtain all the cell uptake pictures at 60× magnification.The proliferative MCF-7 was seeded into a 6-well plate with 5 × 104 cells per well and incubated for 24 h under standard conditions. All wells were divided into four groups (blank control, free DOX, SDN-2@DOX and SDN-2-Apt@DOX). The blank control group did not undergo any treatment. The concentration of DOX in the other three groups was 5 μg mL−1. After adding the sample, they were incubated for 4 h, the old culture medium was taken out, PBS was used to wash three times to remove the sample that has not entered the cell, and then trypsin digestion solution was used to digest and collect the cells in each group in a 1.5 mL centrifuge tube, and the concentration of cells in each group was adjusted to 1 × 106 cells per mL. The cell suspension was collected and centrifuged at 1500 rpm at 4 °C for 3 min with a low temperature, the waste liquid was discarded, 1 mL of PBS was added, blown gently to resuspend the cells, and centrifuged again, and this step was repeated twice. For the last time, the waste liquid was discarded and 200 μL PBS was added, which was mixed evenly, and then the membrane was passed through for observation and detection by flow cytometry. The operation and treatment of 231 cells remained the same.
2.10 MTT assay
The cytotoxicity of free DOX, SDN-2@DOX and SDN-2-Apt@DOX to cells in vitro was determined by the MTT method. MCF-7 cells were seeded into 96 well plates with 6 × 103 cells per well and incubated for 24 h. Then the old medium was taken out, and 100 μL of the prepared fresh medium containing free DOX, SDN-2@DOX and SDN-2-Apt@DOX with different concentrations of DOX was added to a 96-well plate. After incubation, MTT (5 mg mL−1) was added, incubation was continued for 4 h, then the medium containing MTT was taken out, and 150 μL of DMSO was added, and the OD value at 492 nm was measured after the vibration is uniform.The biocompatibility of each building unit and SDN-2 without drug loading was determined using the same steps above.
The operation and treatment of 231 cells remained the same.
2.11 Cell apoptosis assay
The proliferative MCF-7 cells were seeded into a 6-well plate with 5 × 104 cells per well and incubated for 24 h. The cells were incubated for 24 h in a medium including free Dox, SDN-2@DOX and SDN-2-Apt@DOX, respectively. The concentration of DOX in the other three groups was 5 μg mL−1. Then the cells were rinsed three times with PBS and harvested, centrifuged at 1500 × g for 3 min, and Annexin V-FITC/propidium iodide (PI) staining agents were added. Cells were harvested for fluorescence-activated cell sorting analysis by flow cytometry.3. Results and discussion
3.1 Synthesis and characterization of supramolecular DNA nanogels
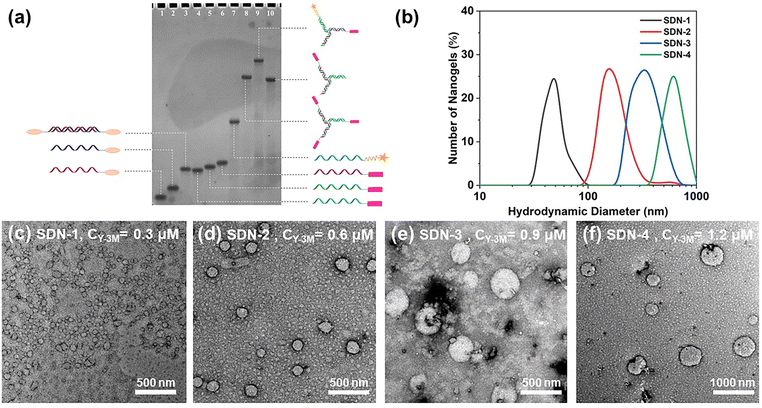
Azo and MV were modified with alkynyl groups (Fig. S1, ESI†), and then the two molecules can be conjugated to the single strand DNA (ssDNA) with azide groups at the 5 end (Fig. S2a, ESI†) by a click reaction.37 The DNA sequence and specific structure are detailed in Fig. S2b (ESI†). The modified DNA was purified by polyacrylamide gel electrophoresis (PAGE) and further characterized by mass spectrometry (Fig. S3 to S4, ESI†). Linear double strand DNA (dsDNA) with Azo (L-2Azo) at both ends was obtained by annealing treatment. By the same operation, Y-type dsDNA with one MV at the end (Y-1MV) and Y-type dsDNA with three MV at the end (Y-3MV) were also obtained. As shown in Fig. 1a, these dsDNAs will be used as building units to construct SDNs.In previous studies,6,7,35 DNA nanogels of different sizes were prepared by changing the concentration of the DNA building units. Here, we also studied the relationship between the DNA building unit concentration and the size of SDNs. We have precisely controlled the concentration and proportion of each DNA building unit, which makes the total amount of these two guest molecules at the sticky end the same (Table S1, ESI†). As expected, after controlling the initial concentration of DNA building units, we obtained SDNs with different sizes. Through DLS and TEM analysis shown in Fig. 1, it can be seen that the SDN is spherical and its size ranges from about 50 nm to 600 nm, and it has good particle dispersion (PI of good samples can be close to 0.1). The controllable size of SDNs allows more choices when dealing with the complex internal environment. Considering the stability and EPR effect,38 we chose SDN-2 with a size of about 160 nm for subsequent experiments.
To test the stability of SDN-2, we measured the size of the SDN-2 stored in TM buffer solution through DLS for 7 consecutive days and took TEM images of the final sample (Fig. S5, ESI†). After analysing DLS, we observed that after long-term storage, the size of SDN-2 was gradually decreasing, but still within an acceptable range. It can be seen from TEM images that the samples stored for 7 days still have very clear outlines. These results indicate that our supramolecular DNA nanogels constructed via host–guest interaction have relatively good stability.
3.2 Stimulation responsiveness of supramolecular DNA nanogels
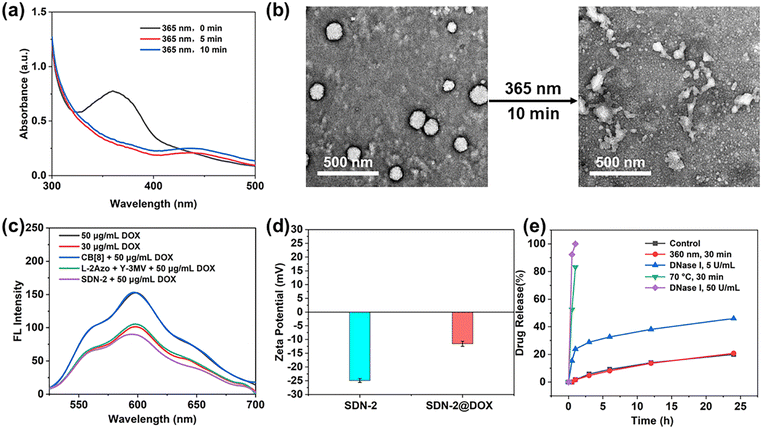
In previous studies, pure DNA hydrogels were observed to respond to pH,39 nucleic acid,40,41 ATP41,42 and enzymes.43 It can be anticipated that the introduction of supramolecular host–guest systems will bring more advantages in the case of coexistence. In our study, the stimuli response of the supramolecular DNA nanogels is divided into two parts: photoresponsiveness and biomolecule response such as FGG peptide or spermine.First of all, the configuration of the Azo molecule will change under ultraviolet light irradiation, and its ultraviolet characteristic absorption peak at 365 nm will decrease, while its ultraviolet characteristic absorption peak at 450 nm will increase. Since the UV characteristic absorption peak of DNA is about 260 nm, after nanogels are assembled, the UV characteristic absorption peak of high concentration DNA seriously affects the characteristic peak of Azo at 365 nm. Therefore, it is not possible to observe the changes in the UV characteristic absorption peak of SDN-2 under ultraviolet radiation. Therefore, the UV characteristic absorption peak of L-2Azo was directly detected. As shown in Fig. 2a, L-2Azo exhibits a distinct 365 nm UV characteristic peak when not illuminated by ultraviolet radiation. When exposed to UV light for 5 min, the peak at 365 nm disappeared, while the peak at 450 nm appeared. When the light prolonged for 10 min, the change was not significant, indicating that Azo modified on L-2Azo reacted quickly to UV and completely transformed at 5 min. When SDN-2 is exposed to ultraviolet radiation, it can be clearly seen from Fig. 2b that SDN-2 undergoes partial dissociation and becomes loose from its original regular structure. Due to the relatively dense structure after assembly into SDN-2, the photoresponsiveness speed of SDN-2 is not as fast as that of L-2Azo. As the UV irradiation time continues to extend, SDN-2 gradually dissociates completely (Fig. S6a and b, ESI†). The above results show that the photoresponsiveness of Azo is successfully endowed to supramolecular DNA nanogels, which enables the nanogels to undergo structural disintegration under ultraviolet irradiation, and also shows that it is completely feasible to realize the function of guest molecules in the DNA nanogel system.
On the other hand, CB[8] has a high affinity for peptide of sequences Phe-Gly-Gly (FGG peptide) (the binding constant Ka = 1.5 × 1011 M−2).44 Its binding constant is much larger than that of the host–guest complex between CB[8] and Azo, MV guests (the binding constant Ka = 1.58 × 109 M−2).45,46 This makes it possible for the FGG peptide to compete with guest molecules and induce the dissociation of DNA nanogels. Here, we use FGG peptides as competing guest molecules to conduct experiments. As competitive guests, the FGG peptide can replace Azo from the cavity of CB[8]. To investigate the effect of the FGG peptide on the dissociation of SDN-2, excess FGG peptide was co-incubated with SDN-2 at 37 °C for 1 h. As shown in Fig. S6c (ESI†), in the presence of FGG peptide, SDN-2 gradually disintegrates just like under ultraviolet radiation. This result indicates that supramolecular DNA nanogels can also undergo responsive dissociation under the influence of FGG peptides or other biomolecules.
3.3 In vitro drug release of supramolecular DNA nanogels
Next, we evaluated the drug loading and release ability of SDN-2 in vitro. By high-speed centrifugation, SDN-2 was separated from the solution (Fig. S7, ESI†).47 As shown in Fig. S8a (ESI†), after co-incubation with DOX-HCl for 24 h, the UV characteristic absorption peak of DOX at 490 nm significantly decreased, indicating that DOX was loaded onto the nanogel, which was also been supported by the fluorescence spectra and zeta potential (Fig. 2c and d). It is obvious that with the increased DOX concentration, the higher loading efficiency of nanogels can be achieved, up to about 58%. However, the encapsulation efficiency of DOX is not linearly related to the concentration of DOX. At low concentrations, the encapsulation efficiency may not be very high due to molecular diffusion. At high concentrations, due to the upper limit of drug loading of DNA nanogels, a considerable part of DOX cannot be loaded (Fig. S8c, ESI†).DOX-loaded SDN-2(SDN-2@DOX) was then incubated in PBS buffer solution (pH = 7.4) to evaluate its drug release ability (Fig. 2e). Without any treatment, the drug release of SDN-2@DOX at 24 h was less than 20%, which indicates that SDN can effectively encapsulate DOX and maintain certain stability. As a positive control, when SDN-2@DOX was heated at 70 °C for 30 min, its drug release reached 80%. In our study, we found that the drug release of SDN-2@DOX did not increase significantly under single UV irradiation, which can be explained by DOX interacting with the base double helix.48 However, when co-incubated with 5 U mL−1 DNase I for 24 h, SDN-2@DOX also has relatively good stability, and its drug release is about 50%. The possible reason is that the tight DNA structure and high-density surface charges hinder the interaction between DNase I and DNA.49 When the concentration of DNase I was increased to 50 U mL−1, 100% drug release was achieved within 1 h. These results demonstrate that our nanogels have good degradable ability, which could facilitate the drug release.
3.4 Cell experiments
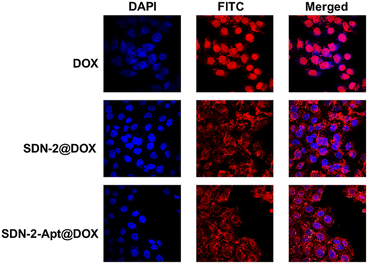
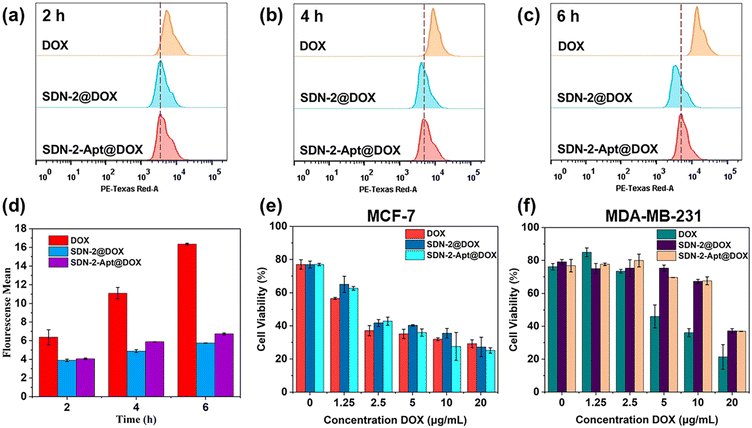
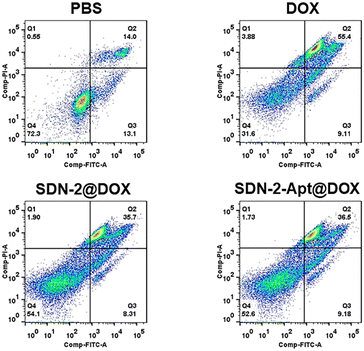
We continue to evaluate the potential of SDNs in cell drug delivery. As shown in Fig. 3, three groups of samples were incubated with MCF-7 cells for 6 h. It can be observed that most of the drugs are concentrated in the nuclear region of the cells treated with DOX directly, while most of the drug fluorescence is in the cytoplasmic region and a small part is in the nucleus of the other two groups of samples using SDN-2 as a drug delivery agent. Due to the water solubility of DOX hydrochloride, the absorption of drugs by cells is faster than that of carriers. However, it can also be seen that the nanogel successfully delivered the drug molecules into the cells.The targeted drug delivery ability was evaluated by flow cytometry. MCF-7 cells and MDA-MB-231 cells were selected for the experiment. MUC1 aptamers are loaded on the SDN-2(SDN-2-Apt). As shown in Fig. 4a–c, after incubation with cells for 2, 4, and 6 h, it can be observed that the cell uptake of the sample increases with time. Among the two types of cells, the pure drug treatment group had the highest cell uptake, which is consistent with the previous CLSM results. Compared to SDN-2@DOX, SDN-2-Apt@DOX has more cellular uptake in MCF-7 cells, about 20% (Fig. 4d), but the difference is not significant in 231 cells (Fig. S10, ESI†).
The therapeutic effects of SDN-2@DOX and SDN-2-Apt@DOX on tumour cells were tested through MTT experiments. We first evaluated the cytotoxicity of various DNA building units and SDN-2 in MCF-7 cells without drug loading. As shown in Fig. S11 (ESI†), the cell survival rate was above 90%, indicating good biocompatibility of the nanogel. After drug loading, both SDN-2@DOX and SDN-2-Apt@DOX showed significant cytotoxicity to MCF-7 cells (Fig. 4e), which was similar to the results of free DOX. Compared with the results of SDN-2@DOX, SDN-2-Apt@DOX showed good cytotoxicity. In the 231 cell experimental group (Fig. 4f), the cytotoxicity of free DOX was significantly higher than that of SDN-2@DOX and SDN-2-Apt@DOX, and there was no difference in the results between the SDN-2@DOX and SDN-2-Apt@DOX groups. The above cytotoxic results demonstrate that nanogels have good drug delivery ability and tumour treatment efficacy.
From the apoptosis results of MCF-7 cells in Fig. 5, similar to the MTT results, the highest number of apoptotic cells was observed with direct treatment with DOX, approaching 80%. The number of apoptotic cells treated with SDN-2@DOX and SDN-2-Apt@DOX was relatively low, only about 50%. In Fig. S12 (ESI†), the results were similar, with the apoptosis rates for SDN-2@DOX and SDN-2-Apt@DOX being lower than that for DOX, but still relatively high. The above results suggest that SDN-2 can efficiently deliver DOX into cells to achieve its therapeutic effect.
 | ||
| Fig. 5 Apoptosis assay of the MCF-7 cells treated with DOX, SDN-2@DOX and SDN-2-Apt@DOX (DOX concentration: 5 μM). | ||
4. Conclusions
In this study, we proposed a simple and general method to construct functional supramolecular DNA nanogels through CB[8] and guest molecular recognition. Our supramolecular DNA nanogels have controllable size, good biocompatibility and targeted drug delivery ability. More importantly, the photoresponsive ability has been successfully endowed to supramolecular DNA nanogels by adopting Azo as a guest molecule. The competitive properties of the host–guest system can also be demonstrated in the supramolecular DNA nanogel system. These results indicate that the host–guest supramolecular system is compatible with the DNA nanogel system. It is worth noting that in the past decade, research on host–guest recognition has continued to advance, and the vast host–guest recognition molecular library has provided us with many choices and conveniences. Using this strategy, the excellent properties of guest molecules can be integrated into the DNA nanogel system through convenient modification methods. Overall, this strategy provides a good choice for the construction of multifunctional DNA nanogel platforms.Author contributions
Z. D.: methodology, investigation, formal analysis, and writing – original draft. G. D.: methodology and investigation. H. Y.: methodology and investigation. Z. Y.: methodology. S. L.: conceptualization, supervision, and funding acquisition. Z. Z.: conceptualization, writing – review and editing, supervision, and funding acquisition. Y. D.: conceptualization and writing – review and editing.Conflicts of interest
There are no conflicts to declare.Acknowledgements
This work was financially supported by the National Natural Science Foundation of China (No. 21604066, 21871216 and 21971248). The authors gratefully acknowledge the technical assistance from Binglin Sui and Professor Junlin Chen of Wuhan University of Science and Technology.Notes and references
- C. M. Niemeyer, DNA as a material for nanotechnology, Angew. Chem., Int. Ed. Engl., 1997, 36, 585–587 CrossRef CAS.
- J. Li, L. Mo, C. Lu, T. Fu, H. Yang and W. Tan, Functional nucleic acid-based hydrogels for bioanalytical and biomedical applications, Chem. Soc. Rev., 2016, 45, 1410–1431 RSC.
- F. Li, J. Tang, J. Geng, D. Luo and D. Yang, Polymeric DNA hydrogel: Design, synthesis and applications, Prog. Polym. Sci., 2019, 98, 101163 CrossRef CAS.
- F. Li, D. Lyu, S. Liu and W. Guo, DNA hydrogels and microgels for biosensing and biomedical applications, Adv. Mater., 2020, 32, 1806538–1806546 CrossRef CAS PubMed.
- X. Zhang, W. Yan, Z. Song, S. Asif, I. Hussain, C. Xiao and X. Chen, DNA nanogel for cancer therapy, Adv. Ther., 2023, 6, 2200287 CrossRef CAS.
- P. Song, D. Ye, X. Zuo, J. Li, J. Wang, H. Liu, Mt Hwang, J. Chao, S. Su and L. Wang, DNA hydrogel with aptamer-toehold-based recognition, cloaking, and decloaking of circulating tumor cells for live cell analysis, Nano Lett., 2017, 17, 5193–5198 CrossRef CAS PubMed.
- E. Cheng, Y. Xing, P. Chen, Y. Yang, Y. Sun, D. Zhou, L. Xu, Q. Fan and D. Liu, A pH-Triggered, fast-responding DNA hydrogel, Angew. Chem., Int. Ed., 2009, 121, 7796–7799 CrossRef.
- M. Nishikawa, Y. Mizuno, K. Mohri, N. Matsuoka, S. Rattanakiat, Y. Takahashi, H. Funabashi, D. Luo and Y. Takakura, Biodegradable CpG DNA hydrogels for sustained delivery of doxorubicin and immunostimulatory signals in tumor-bearing mice, Biomaterials, 2011, 32, 488–494 CrossRef CAS PubMed.
- Y. Yong and C. Fan, Nanocomposite DNA hydrogels emerging as programmable and bioinstructive materials systems, Chem, 2022, 8, 1554–1566 Search PubMed.
- Y. Dong, D. Liu and Z. Yang, A brief review of methods for terminal functionalization of DNA, Methods, 2014, 67, 116–122 CrossRef CAS PubMed.
- Z. Zhao, Y. Dong, Z. Duan, D. Jin, W. Yuan and D. Liu, DNA-organic molecular amphiphiles: Synthesis, self-assembly, and hierarchical aggregates, Aggregate, 2021, 2, e95 CrossRef CAS.
- L. Zhang, S. Jean, S. Ahmed, P. Aldridge, X. Li, F. Fan, E. Sargent and S. Kelley, Multifunctional quantum dot DNA hydrogels, Nat. Commun., 2017, 8, 381–389 CrossRef PubMed.
- W. Chen, W. Liao, Y. Sohn, M. Fadeev, A. Cecconello, R. Nechushtai and I. Willner, Stimuli-responsive nucleic acid-based polyacrylamide hydrogel-coated metal–organic framework nanoparticles for controlled drug release, Adv. Funct. Mater., 2018, 28, 1705137 CrossRef.
- W. C. Liao, S. Lilienthal, J. S. Kahn, M. Riutin, Y. S. Sohn and R. Nechushtai, et al., pH- and ligand-induced release of loads from DNA–acrylamide hydrogel microcapsules, Chem. Sci., 2017, 8, 3362–3373 RSC.
- J. Geng, C. Yao, X. Kou, J. Tang, D. Luo and D. Yang, A Fluorescent biofunctional DNA hydrogel prepared by enzymatic polymerization, Adv. Healthcare Mater., 2018, 7, 1700998 CrossRef PubMed.
- L. Zhang, R. Abdullah, X. Hu, H. Bai, H. Fan, L. He, H. Liang, J. Zou, Y. Liu, Y. Sun, X. Zhang and W. Tan, Engineering of bioinspired, size-controllable, self-degradable cancer-targeting DNA nanoflowers via the incorporation of an artificial sandwich Base, J. Am. Chem. Soc., 2019, 141, 4282–4290 CrossRef CAS PubMed.
- J. Li, C. Zheng, S. Cansiz, C. Wu, J. Xu, C. Cui, Y. Liu, W. Hou, Y. Wang, L. Zhang, I. Teng, H. Yang and W. Tan, Self-assembly of DNA nanohydrogels with controllable size and stimuli-responsive property for targeted gene regulation therapy, J. Am. Chem. Soc., 2015, 137, 1412–1415 CrossRef CAS PubMed.
- H. V. P. Thelu, S. K. Albert, M. Golla, N. Krishnan, D. Ram, S. M. Srinivasula and R. Varghese, Size controllable DNA nanogels from the self-assembly of DNA nanostructures through multivalent host-guest interactions, Nanoscale, 2018, 10, 222–230 RSC.
- X. Zhang, P. Zhang, C. Xiao and X. Chen, ROS-responsive self-degradable DNA nanogels for targeted anticancer drug delivery, ACS Macro Lett., 2023, 12, 1317–1323 CrossRef CAS PubMed.
- H. Xue, F. Ding, J. Zhang, Y. Guo, X. Gao, J. Feng, X. Zhu and C. Zhang, DNA tetrahedron-based nanogels for siRNA delivery and gene silencing, Chem. Commun., 2019, 55, 4222–4225 RSC.
- W. Tang, L. Han, S. Duan, X. Lu, Y. Wang, X. Wu, J. Liu and B. Ding, An aptamer-modified DNA tetrahedron-based nanogel for combined chemo/gene therapy of multidrug-resistant tumors, ACS Appl. Bio Mater., 2021, 4, 7701–7707 CrossRef CAS PubMed.
- H. Yang, Z. Duan, F. Liu, Z. Zhao and S. Liu, Cucurbit[7]uril-based supramolecular DNA nanogel for targeted codelivery of chemo/photodynamic drugs, ACS Macro Lett., 2023, 12, 295–301 CrossRef CAS PubMed.
- H. Zhu, J. Wu, J. Zhao, L. Yu, B. Liyarita, X. Xu, Y. Xiao, X. Hu, S. Shao, J. Liu, X. Wang and F. Shao, Dual-functional DNA nanogels for anticancer drug delivery, Acta Biomater., 2024, 175, 240–249 CrossRef CAS PubMed.
- Z. Liu, S. K. M. Nalluri and J. F. Stoddart, Surveying macrocyclic chemistry: from flexible crown ethers to rigid cyclophanes, Chem. Soc. Rev., 2017, 46, 2459–2478 RSC.
- V. D. Uzunova, C. Cullinane, K. Brix, W. M. Nau and A. I. Day, Toxicity of cucurbit[7]uril and cucurbit[8]uril: an Exploratory in vitro and in vivo study, Org. Biomol. Chem., 2010, 8, 2037–2042 RSC.
- G. Hettiarachchi, D. Nguyen, J. Wu, D. Lucas, D. Ma and L. Isaacs, et al., Toxicology and drug delivery by cucurbit[n]uril type molecular containers, PLoS One, 2010, 5, e10514 CrossRef PubMed.
- R. Oun, R. S. Floriano, L. Isaacs, E. G. Rowan and N. J. Wheate, The ex vivo neurotoxic myotoxic and cardiotoxic activity of cucurbituril-based macrocyclic drug delivery vehicles, Toxicol. Res., 2014, 3, 447–455 CrossRef CAS PubMed.
- H. Chen, J. Y. W. Chan, X. Yang, I. W. Wyman, D. Bardelang, D. H. Macartney, S. M. Y. Lee and R. Wang, Developmental and organ-specific toxicity of cucurbit[7]uril: in vivo study on zebrafish models, RSC Adv., 2015, 5, 30067–30074 RSC.
- S. J. Barrow, S. Kasera, M. J. Rowland, J. del Barrio and O. A. Scherman, Cucurbituril-based molecular recognition, Chem. Rev., 2015, 115, 12320–12406 CrossRef CAS PubMed.
- J. Lagona, P. Mukhopadhyay, S. Chakrabarti and L. Isaacs, The Cucurbit[n]uril family, Angew. Chem., Int. Ed., 2005, 44, 4844–4870 CrossRef CAS PubMed.
- Y. Lan, Y. Wu, A. Karas and O. A. Scherman, Photoresponsive hybrid raspberry-like colloids based on cucurbit[8]uril host-guest interactions, Angew. Chem., Int. Ed., 2014, 53, 2166–2169 CrossRef CAS PubMed.
- G. Stephenson, R. M. Parker, Y. Lan, Z. Yu, O. A. Scherman and C. Abell, Supramolecular colloidosomes: Fabrication, characterisation and triggered release of cargo, Chem. Commun., 2014, 50, 7048–7051 RSC.
- W. Wang, B. Qi, X. Yu, W. Li, Z. Yang, H. Zhang, S. Liu, Y. Liu and X. Wang, Modular design of supramolecular organic frameworks for image-guided photodynamic therapy, Adv. Funct. Mater., 2020, 30, 2004452 CrossRef CAS.
- F. Tian, D. Jiao, F. Biedermann and O. A. Scherman, Orthogonal switching of a single supramolecular complex, Nat. Commun., 2012, 3, 1207–1214 CrossRef PubMed.
- H. Zhang, S. Ba, J. Y. Lee, J. Xie, T.-P. Loh and T. Li, Cancer biomarker-triggered disintegrable DNA nanogels for intelligent drug delivery, Nano Lett., 2020, 20, 8399–8407 CrossRef CAS PubMed.
- X. Yang, F. Liu, Z. Zhao, F. Liang, H. Zhang and S. Liu, Cucurbit[10]uril-based chemistry, Chin. Chem. Lett., 2018, 29, 1560–1566 CrossRef CAS.
- T. Du, W. Yuan, Z. Zhao and S. Liu, Reversible morphological tuning of DNA–perylenebisdiimide assemblies through host–guest interaction, Chem. Commun., 2019, 55, 3658–3661 RSC.
- A. Lacroix and H. F. Sleiman, DNA nanostructures: Current challenges and opportunities for cellular delivery, ACS Nano, 2021, 15, 3631–3645 CrossRef CAS PubMed.
- G. Pan, Q. Mou, Y. Ma, F. Ding, J. Zhang, Y. Guo, X. Huang, Q. Li, X. Zhu and C. Zhang, pH-responsive and gemcitabine-containing DNA nanogel to facilitate the chemodrug delivery, ACS Appl. Mater. Interfaces, 2019, 11, 41082–41090 CrossRef CAS PubMed.
- Q. Yang, Y. Wang, T. Liu, C. Wu, J. Li, J. Cheng, W. Wei, F. Yang, L. Zhou, Y. Zhang, S. Yang and H. Dong, Microneedle array encapsulated with programmed DNA hydrogels for rapidly sampling and sensitively sensing of specific microRNA in dermal interstitial fluid, ACS Nano, 2022, 16, 18366–18375 CrossRef CAS PubMed.
- J. Wang, J. Li, Y. Chen, R. Liu, Y. Wu, J. Liu, X. Yang, K. Wang and J. Huang, Size-controllable and self-assembled DNA nanosphere for amplified microRNA imaging through ATP-fueled cyclic dissociation, Nano Lett., 2022, 22, 8216–8223 CrossRef CAS PubMed.
- N. Li, X. Wang, M. Xiang, J. Liu, R. Yu and J. Jiang, Programmable self-assembly of protein-scaffolded DNA nanohydrogels for tumor-targeted imaging and therapy, Anal. Chem., 2019, 91, 2610–2614 CrossRef CAS PubMed.
- Y. Lin, Y. Huang, Y. Yang, L. Jiang, C. Xing, J. Li, C. Lu and H. Yang, Functional self-assembled DNA nanohydrogels for specific telomerase activity imaging and telomerase-activated antitumor gene therapy, Anal. Chem., 2020, 92, 15179–15186 CrossRef CAS PubMed.
- L. M. Heitmann, A. B. Taylor, P. J. Hart and A. R. Urbach, Sequence-specific recognition and cooperative dimerization of N-terminal aromatic peptides in aqueous solution by a synthetic host, J. Am. Chem. Soc., 2006, 128, 12574–12581 CrossRef CAS PubMed.
- W. Jeon, H. Kim, C. Lee and K. Kim, Control of the stoichiometry in host–guest complexation by redox chemistry of guests: Inclusion of methylviologen in cucurbit[8]uril, Chem. Commun., 2002, 1828–1829 RSC.
- F. Tian, D. Jiao, F. Biedermann and O. A. Scherman, Orthogonal switching of a single supramolecular complex, Nat. Commun., 2012, 3, 1207–1214 CrossRef PubMed.
- G. Zhu, R. Hu, Z. Zhao, Z. Chen, X. Zhang and W. Tan, Noncanonical self-assembly of multifunctional DNA nanoflowers for biomedical applications, J. Am. Chem. Soc., 2013, 135, 16438–16445 CrossRef CAS PubMed.
- L. Peng, M. You, Q. Yuan, C. Wu, D. Han, Y. Chen, Z. Zhong, J. Xue and W. Tan, Macroscopic volume change of dynamic hydrogels induced by reversible DNA hybridization, J. Am. Chem. Soc., 2012, 134, 12302–12307 CrossRef CAS PubMed.
- C. Li, S. Luo, J. Wang, Z. She and Z. Wu, Nuclease-resistant signaling nanostructures made entirely of DNA oligonucleotides, Nanoscale, 2021, 13, 7034–7051 RSC.
Footnote |
| † Electronic supplementary information (ESI) available. See DOI: https://doi.org/10.1039/d4tb00853g |
| This journal is © The Royal Society of Chemistry 2024 |