Open Access Article
Open Access ArticleProtein aggregation monitoring in cells under oxidative stress: a novel fluorescent probe based on a 7-azaindole-BODIPY derivative†
Diego
Herrera-Ochoa
a,
Iván
Llano
b,
Consuelo
Ripoll
a,
Pierre
Cybulski
c,
Martin
Kreuzer
d,
Susana
Rocha
 *c,
Eva M.
García-Frutos
*c,
Eva M.
García-Frutos
 *be,
Iván
Bravo
*be,
Iván
Bravo
 a and
Andrés
Garzón-Ruiz
a and
Andrés
Garzón-Ruiz
 *a
*a
aDepartamento de Química Física, Facultad de Farmacia, Universidad de Castilla-La Mancha, Av. Dr José María Sánchez Ibáñez, s/n, 02071 Albacete, Spain. E-mail: andres.garzon@uclm.es
bInstituto de Ciencia de Materiales de Madrid (ICMM), CSIC, Cantoblanco, 28049 Madrid, Spain. E-mail: emgfrutos@icmm.csic.es
cMolecular Imaging and Photonics, Department of Chemistry, KU Leuven, Leuven Chem&Tech, Celestijnenlaan 200F, Leuven, 3001, Belgium. E-mail: susana.rocha@kuleuven.be
dALBA Synchrotron Light Source, Carrer de la Llum 2-26, Cerdanyola del Valles, 08290 Cerdanyola Del Vallès, Barcelona, Spain
eDepartamento de Química Orgánica y Química Inorgánica, Universidad de Alcalá, 28805 Alcalá de Henares, Madrid, Spain
First published on 10th July 2024
Abstract
The development of new fluorescent probes as molecular sensors is a critical step for the understanding of molecular mechanisms. BODIPY-based probes offer versatility due to their high fluorescence quantum yields, photostability, and tunable absorption/emission wavelengths. Here, we report the synthesis and evaluation of a novel 7-azaindole-BODIPY derivative to probe hydrophobic proteins as well as protein misfolding and aggregation. In organic solvents, this compound shows two efficiently interconverting emissive excited states. In aqueous environments, it forms molecular aggregates with unique photophysical properties. The complex photophysics of the 7-azaindole-BODIPY derivative was explored for sensing applications. In the presence of albumin, the compound is stabilized in hydrophobic protein regions, significantly increasing its fluorescence emission intensity and lifetime. Similar effects occur in the presence of protein aggregates but not with other macromolecules like pepsin, DNA, Ficoll 40, and coconut oil. Fluorescence lifetime imaging microscopy (FLIM) and two-photon fluorescence microscopy on breast (MCF-7) and lung (A549) cancer cells incubated with this compound display longer fluorescence lifetimes and higher emission intensity under oxidative stress. Synchrotron FTIR micro spectroscopy confirmed that the photophysical changes observed were due to protein misfolding and aggregation caused by the oxidative stress. These findings demonstrate that this compound can serve as a fluorescent probe to monitor protein misfolding and aggregation triggered by oxidative stress.
Introduction
The combination of boron dipyrromethene (BODIPY) and indole has been widely explored to develop fluorescent probes for biological applications.1–3 On the one hand, BODIPY is a well-known fluorescent dye and its derivatives are universally used in fluorescence imaging.1–5 On the other hand, indole is an electron-rich aromatic heterocyclic organic compound used for sensing a wide range of analytes, particularly anions.6–9 Despite the promising results reported for indole-BODIPY derivatives in fluorescence sensing applications, the combination of azaindole and BODIPY has received less attention. Azaindole is an aromatic heterocycle consisting of a pyridine ring fused to a pyrrole ring and, hence, electron-richer than indole. Many azaindoles exhibit biological activity and pharmacological properties including antitumor activity.10 Nevertheless, only three azaindole-BODIPY derivatives have been characterized to date (compounds 2–4 in Fig. 1).11,12 Compound 2 showed good performance as a ratiometric probe for the detection of F− ions in human colorectal carcinoma cells (HCT 116) by Mahapatra et al.11 Later, the sensitivity to HSO4− of compounds 3 and 4 was studied by Keşan et al.12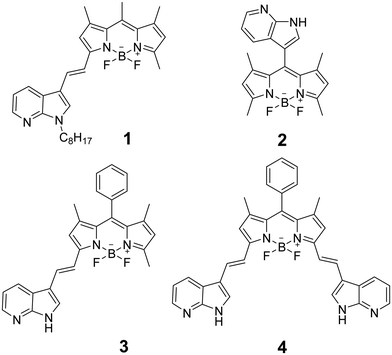 | ||
| Fig. 1 Chemical structure of the synthetized compound (1) and other reported 7-azaindole-BODIPY derivatives (2–4).11,12 | ||
Here, we present the synthesis of a novel 7-azaindole-BODIPY derivative (compound 1), the study of its photophysical properties and the evaluation of its use as a fluorescent probe for hydrophobic proteins as well as protein misfolding and aggregation associated with oxidative stress. It is well-known that the oxidative stress leads to accumulation and aggregation of misfolded proteins in cells due to diverse processes such as modification of chaperone protein activity, amino acid carboxylation, and cysteine oxidation, among others.13,14 The oxidative stress and protein aggregation are involved in biological aging and diverse diseases.13,15,16 In the present work, we demonstrate that the photophysical properties of compound 1 are sensitive to hydrophobic proteins as well as the hydrophobicity changes associated with the formation of protein aggregates. The response of compound 1 to cellular stress events was monitored by fluorescence lifetime imaging microscopy (FLIM) and two-photon fluorescence microscopy in different cell lines. Protein structural changes of cells under oxidative stress conditions were studied by synchrotron FTIR microscopy.
Materials and methods
Materials
All chemicals were commercially available and used without further purification. 7-Azaindole, toluene, 1-iododoctane, phosphorus(V) oxychloride (POCl3), 5,5-difluoro-1,3,7,9,10-pentamethyl-5H-dipyrrolo[1,2-c:2′,1′-f][1,3,2]diazaborinin-4-ium-5-uide (boron dipyrromethene, BODIPY), acetic acid, piperidine and silica gel plates of type 60 F254 with a layer thickness of 0.2 mm were purchased from Sigma Aldrich. Acetone was purchased from VWR International. Tetrabutylammonium hydrogen sulfate [(Bu)4N(HSO4), a phase-transfer catalyst (PTC)] was provided by TCI. Potassium hydroxide (KOH) was acquired from Merck. Hexane, dichloromethane (CH2Cl2), magnesium sulfate anhydrous (MgSO4) and silica gel 60, 0.04–0.06 mm for flash chromatography and analytical thin layer chromatography (TLC) were purchased from Scharlab. Dimethylformamide (DMF) was provided by Fisher Chemicals and Panreac, respectively. Finally, ultrapure water (type I, 18.2 MΩ cm) was used throughout the work. 1H and 13C NMR spectra were recorded in CDCl3 using a BRUKER AVANCE 300 MHz instrument (300.1 and 75.5 MHz for 1H and 13C, respectively). Chemical shifts (δ) are quoted in parts per million (ppm), referenced to the residual solvent.Spectroscopic characterization experiments
UV-Vis absorption spectra were recorded using a V-650 (Jasco) spectrophotometer with a slit width of 0.4 nm and a scan rate of 600 nm min−1. A Peltier accessory was employed to control the temperature of the spectrophotometer's measuring cell. Steady-state fluorescence spectra were recorded using a FLS920 (Edinburgh Instruments) spectrofluorometer equipped with a 450 W Xe lamp as an excitation source, an MCP-PMT (microchannel plate-photomultiplier tube) detector (R3809 model) and a TCSPC (time-correlated single photon counting) data acquisition card (TCC900 model). An FS5 spectrofluorometer (Edinburgh Instruments) equipped with an integrating sphere, a 150 W Xe lamp as the light source and a PMT (photomultiplier tube) detector (R928P model) was employed to determine fluorescence quantum yields. A TLC 50 temperature-controlled cuvette holder (Quantum Northwest) was used for all the fluorescence experiments. The temperature was fixed at 20 °C.Time-resolved fluorescence emission was collected in a microscope (PicoQuant) equipped with a TCSPC card and two TAU-SPAD-100 avalanche photodiode detectors. Samples were excited with 511 and 637 nm diode pulse lasers with a repetition rate of 10 MHz. TCSPC resolution was 16 ps and the emission was recorded using 550/49 and 690/70 bandpass filters. The fluorescence intensity decay, I(t), was fitted to the following multiexponential function using an iterative least-square fit method:
 | (1) |
 | (2) |
Cell cultures
For FLIM experiments, human breast cancer MCF-7 cells (ATCC HTB-22) were cultured at 37 °C in a 5% CO2 humidified atmosphere, with DMEM (Dulbecco's Modified Eagle Medium) cell culture medium, 10% FBS, 1% penicillin–streptomycin and 1% glutamine. For two-photon fluorescence microscopy experiments, A549 cells (CCL-185) were cultured at 37 °C in a 5% CO2 humidified atmosphere, with DMEM cell culture medium, 10% FBS, 1% 1-glutamax and 0.1% gentamicin.The spheroid culture protocol was based on the study reported by Van Zundert et al.17 Briefly, A549 cells were seeded in previously prepared agarose microtissues and were kept at 37 °C in a 5% CO2 humidified incubator for 10 days until the spheroids reached the desired size or morphology. The culture medium was refreshed every 2–3 days. Then, cells were harvested from agarose microtissues and transferred to a 1.5 mL microcentrifuge tube.
Viability and reactive oxygen species (ROS) assays
The MTT (3-(4,5-dimethylthiazol-2-yl)-2,5-diphenyltetrazolium bromide) assay was performed on MCF-7 cells to assess the cytotoxicity of compound 1. A 96-well plate with 10![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 000 cells per well was cultured in DMEM without phenol red at 37 °C in a 5% CO2 humidified atmosphere for 24 hours. Cells were treated with different concentrations of 1, within a range of 25–500 nM, for 2 and 24 hours. The MTT assay was carried out according to the producer's instructions using a Multiskan Go plate reader spectrophotometer (Thermo Scientific). The cell viability was calculated based on the absorbance of the treated and untreated cells. The results were expressed as percent viability, considering the cell viability of the control cells (untreated) as 100%. Three independent measurements were carried out for each sample concentration and exposure time and the results were presented as the average value with ±2σ standard deviation.
000 cells per well was cultured in DMEM without phenol red at 37 °C in a 5% CO2 humidified atmosphere for 24 hours. Cells were treated with different concentrations of 1, within a range of 25–500 nM, for 2 and 24 hours. The MTT assay was carried out according to the producer's instructions using a Multiskan Go plate reader spectrophotometer (Thermo Scientific). The cell viability was calculated based on the absorbance of the treated and untreated cells. The results were expressed as percent viability, considering the cell viability of the control cells (untreated) as 100%. Three independent measurements were carried out for each sample concentration and exposure time and the results were presented as the average value with ±2σ standard deviation.
The fluorescence intensity related to the generation of ROS was measured using a FLUOstar OPTIMA microplate reader. 2′,7′-Dichlorodihydrofluorescein diacetate (H2DCFDA) from Sigma-Aldrich was used as an ROS indicator. MCF7 cell line was plated at 10![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 000 cells per well in 96-multiwell plates and incubated for 24 h at 37 °C and 5% of CO2. After that, the medium was aspirated, and the cells were incubated with H2DCFDA for 30 minutes at a final concentration of 10 μM in PBS. Then, the medium was removed, and the cells were washed with PBS two times. Next, cells were treated with cisplatin (10 μM), ethanol (1 M) and H2O2 (500 μM) for 3 hours. Finally, the fluorescence intensity was recorded at 520 nm after removing the treatment and washing twice with PBS.
000 cells per well in 96-multiwell plates and incubated for 24 h at 37 °C and 5% of CO2. After that, the medium was aspirated, and the cells were incubated with H2DCFDA for 30 minutes at a final concentration of 10 μM in PBS. Then, the medium was removed, and the cells were washed with PBS two times. Next, cells were treated with cisplatin (10 μM), ethanol (1 M) and H2O2 (500 μM) for 3 hours. Finally, the fluorescence intensity was recorded at 520 nm after removing the treatment and washing twice with PBS.
Fluorescence lifetime imaging of cells
MCF-7 cells were seeded onto 20 mm square glass cover slides into 6-well plates and cultured with DMEM medium without phenol red at 37 °C in a 5% CO2 humidified atmosphere until the cells reached 50–80% confluence. Cells were incubated with 0.1 μM of compound 1 in serum-free DMEM medium without phenol red for different times (0.5, 1 and 3 hours). To induce cellular stress, some cells were also incubated with ethanol, hydrogen peroxide or cisplatin, along with 1 for different times (0.5, 1 and 3 hours). Hydrogen peroxide is commonly used to induce oxidative stress.18–20 MCF-7 cells were exposed to a hydrogen peroxide dose of 0.5 mM, two-fold higher than the maximum concentration used by Mahalingaiah et al. to study the oxidative stress in the same type of cells.18 A concentration of 10 μM was chosen for cisplatin within the order of magnitude of IC50 values reported for MCF-7 cells in previous works.21–25After incubation, the cells were washed five times with PBS. After the treatment, FLIM images of the cells were acquired using a MicroTime 200 microscope (PicoQuant). A diode pulse laser of 511 nm (10 MHz and ∼0.9 μW) was chosen as an excitation source. TCSPC resolution was 16 ps and the emission was collected using a 690/70 bandpass filter. 80 × 80 μm regions were scanned using 156 nm per pixel spatial resolution and a dwell time of 2 ms. FLIM images were processed using SymphoTime 64 software (PicoQuant). The fluorescence intensity decays of each pixel were fitted to eqn (1) using an iterative least-squares fit method (all the fluorescence decays were best fitted to a three-exponential equation).
Two-photon fluorescence imaging of 2D cell cultures. A549 cells were seeded in 29 mm, glass-bottom dishes (Cellvis, Mountain View, CA, USA) and grown until ∼70% confluency before adding compound 1. Cells were incubated with 0.1 μM of 1 in DMEM medium without phenol red for different times (0.5, 1 and 3 hours). To induce cellular stress, cells were incubated with ethanol (1 M) along with 1 for different times (0.5, 1 and 3 hours).
After the treatment, fluorescent images were recorded using a Leica TCS SP8 DIVE microscope (Leica microsystems) using a multi-photon Insight X3 laser and an HC PL IRAPO 40× water objective (NA 1.1). Two-photon pulse laser excitation of 1022 nm was chosen as an excitation source. Scanning speed and the image format were set at 400 Hz and 1024 × 1024 pixels. The emission was recorded at two different wavelength ranges (515–575 nm and 650–725 nm) using two HyD hybrid detectors.
Two-photon fluorescence imaging of cell spheroids. A549 cell spheroids were cultured as described above and were grown until they reached the required size. Then, the spheroids were incubated with 0.1 μM of compound 1 in DMEM medium without phenol red for 24 hours. After the treatment, fluorescent images were recorded using a Leica TCS SP8 DIVE microscope (Leica microsystems) using a multi-photon Insight X3 laser and FLUOTAR VISIR 25× water objective (NA 0.95). Two-photon pulse laser excitation, scanning speed and image format were identical to 2D cell fluorescence imaging described above. The emission was recorded by a HyD hybrid detector between 650–725 nm.
Synchrotron-based FTIR microspectroscopy
The SR-FTIR measurements were conducted at the MIRAS beamline at the ALBA synchrotron, Barcelona (Spain), the synchrotron light was used as the infrared light source, coupling with the 3000 Hyperion microscope coupled to a Vertex 70v spectrometer and a liquid nitrogen-cooled mercury cadmium telluride (MCT) detector.26 The spectroscopic data were collected in transmission mode using the 36× Schwarzschild objective and condenser and an aperture size of 10 × 10 μm.Two samples of control MCF-7 cells and two samples of MCF-7 cells treated with cisplatin were measured. Cells were seeded onto 13 mm diameter and 0.5 mm thick CaF2 glass into 6-well plates and cultured with DMEM medium at 37 °C in a 5% CO2 humidified atmosphere until the cells reached 50–80% confluence. Cells were incubated with 10 μM of cisplatin in serum-free DMEM medium for 3 hours. Cells were fixed with 4% formaldehyde for 15 min at room temperature. A total number of 193 control cells and 194 cisplatin-treated cells were measured, and the average spectra are presented. The data were collected in the 4000–800 cm−1 mid-infrared range at a spectral resolution of 4 cm−1 with 256 co-added scans per spectrum. The OPUS 8.2 (Bruker, Ettlingen, Germany) software package was used for data acquisition. Rubber-band baseline correction and vector normalization were performed for each single spectrum. The ratios of absorbances at different wavenumbers and the second derivatives were carried out using the Quasar software.27
Computational details
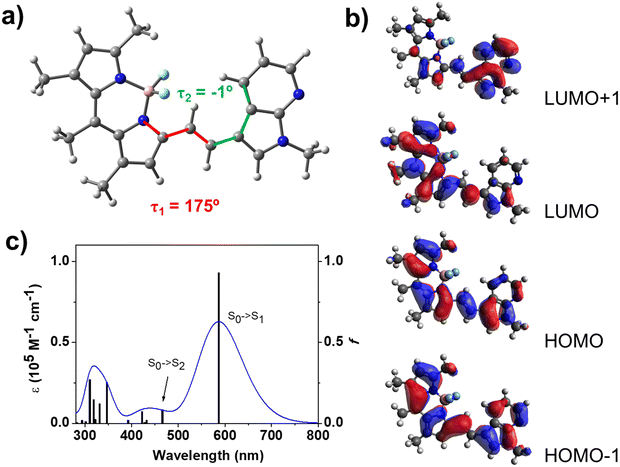
Full geometry optimizations were performed using the Gaussian16 (revision C.01)28 suite of programs at the BLYP/6-31G* level of theory29–31 (including solvent effects). Polarizable continuum model (PCM) was employed to include the solvent (water) effect.32,33 The electronic vertical transitions were calculated at the time-dependent (TD)-BLYP/6-31G* level (including solvent effects). In general, the most popular DFT functionals show a poor performance in the calculation of electronic transitions of BODIPY derivatives.34,35 Surprisingly, some pure generalized gradient approximation (GGA) functionals as BLYP lead to small deviations between calculated electronic transition wavelengths and absorption maximum wavelengths in BODIPY derivatives.34Results and discussion
Preparation of compound 1
The synthesis pathway for the preparation of 7-azaindole-BODIPY 1 is shown in Scheme 1 and ESI.† The incorporation of the alkyl chain from commercially available 7-azaindole (7AI) was carried out by using 1-iodooctane, KOH, and a phase-transfer catalyst (PTC) in acetone at 75 °C for 24 hours to give compound 7AI-5 (65%). Compound 7AI-6 was obtained by the Vilsmeier–Haack reaction of 7AI-5 with POCl3 in DMF to yield yellow oil with 68%. Finally, the Knoevenagel reaction method was used for the preparation of the novel 7-azaindole-BODIPY derivative, 1, by treating compound 7AI-6 with BODIPY in the presence of piperidine, acetic acid in a Dean–Stark apparatus with toluene for 48 hours. The new 7-azaindole-BODIPY derivative, 1, was isolated by column chromatography to afford the new monostyryl compound in a 5% yield as a pink powder. All compounds were identified through 1H, 13C NMR and mass spectrometry (see the ESI†).Spectroscopic characterization
An extended spectroscopic characterization was carried out to evaluate the suitability of compound 1 as a fluorescent probe. The absorption spectrum of 1 in THF shows a vibronic structure with an absorption maximum wavelength at 584 nm (λmaxab) and a shoulder at about 536 nm (Fig. 2a and Table S1, ESI†). A similar vibronic structure was found for 1 in other organic solvents such as toluene, ethanol and DMSO. The structured absorption bands observed in the absorption spectrum of 1 suggests a planar and highly conjugated structure, as confirmed by DFT calculations (Fig. 3a). Compound 3, the previously reported molecule with the most closely related structure, displayed a similar absorption spectrum with a λmaxab of 574 nm in acetonitrile solution.12 Compound 2, on the other hand, exhibited a significantly blue-shifted absorption maximum (λmaxab = 501 nm in an acetonitrile–water mixture)11 revealing the role of the ethylene bridge in the extension of the electronic conjugation in 7-azaindole-BODIPY system. Additionally, the presence of a second 7-azaindole group led to a substantial red-shift of the absorption band of compound 4 (λmaxab = 655 nm in acetonitrile solution).12 TD-DFT calculations revealed that the band centered at 584 nm corresponded to the S0 → S1 transition involving the HOMO and LUMO (Fig. 3c and Table S3, ESI†).The excitation and emission spectra of 1 follow the mirror rule (Fig. 2b, c and Fig. S2, ESI†), with excitation and emission maxima (λmaxex and λmaxem) in THF at 576 and 598 nm, respectively (see Table S1 for other solvents, ESI†). When dissolved in DMSO, a red-shift in the emission band was observed (λmaxem = 611 nm) due to the intramolecular charge-transfer character of the S0 → S1 transition. As shown in Fig. 3b, the HOMO is delocalized across the whole molecule, while the LUMO is mainly localized within the BODIPY moiety (similar findings were reported for compound 3).12 The fluorescence emission decay from the S1 state follows a mono-exponential profile with a lifetime, τF, of 3.2 ns and a fluorescence quantum yield, ΦF, of 58% in THF (Fig. S3 and Table S2, ESI†). These results are consistent with those previously reported for compound 3 (τF = 4.2 ns and ΦF = 42% in acetonitrile solution).12 Furthermore, additional minor excitation bands were detected at shorter wavelengths (around 530 nm and 350–400 nm), which were also observed in the absorption spectrum (Fig. 2c). The excitation band observed at approximately 530 nm was attributed to the S0 → S2 transition involving the HOMO−1, HOMO, LUMO and LUMO+1. The resolution of this band in the absorption spectrum was hindered by its overlap with the tail of the most intense absorption band (assigned to the S0 → S1 transition). Interestingly, when 1 was excited at 465 nm, two emission bands emerged, centered at 525 and 598 nm. This excitation wavelength avoided the direct excitation of the S1 state but allowed the excitation of the S2 state (Fig. 2d and Table S1, ESI†). Thus, the first emission band (525 nm) corresponds to the fluorescence emission from the S2 state while the second emission band (598 nm) is associated with the emission from the S1 state preceding internal conversion (IC) from the S2 state (hereafter noted as S2/S1→ S0). Of note, the fluorescence intensity in both emission maxima showed a linear correlation with the fluorophore concentration, ruling out the formation of aggregates (Fig. S4, ESI†). The similar intensity of both emission bands suggests that S2/S1→ S0 is an efficient IC process that competes against the fluorescence emission from the S2 state. Accordingly, comparable fluorescence lifetimes were found for both processes (4.3 ns for S2 → S0 and 3.9 ns for S2/S1 → S0; see Table S2, ESI†). Note that the fluorescence emission decay from the S2/S1 → S0 process also follows a mono-exponential profile, with a lifetime of 0.7 ns longer than for the S1 → S0 process. This increase in the fluorescence lifetime indicates the association with an IC process.
As far as we know, the existence of two emissive states in BODIPY derivatives has been scarcely reported. Cho et al. observed that some BODIPY derivatives in THF solution led to ‘anomalous’ emission from the S2 excited state,36 while Schäfer et al. reported the existence of two emissive states in BODIPY dye films.37 The possible combined use of both emissive states could offer unique advantages in bioimaging applications. It is possible to excite the S2 state using blue light (465 nm) and collect the fluorescence emission in the red spectral range from the S1 state (600–750 nm) mediating an IC process. The large difference between the excitation and collection wavelengths reduces the impact of the background fluorescence in microscopy images of biological samples. Noteworthy, red fluorescence emission was observed even when exciting 1 with ultraviolet light (at 350 nm), indicating additional IC processes between high-energy excited states and S1. The presence of the unusual S2/S1 → S0 process prompted the investigation of the sensing potential of compound 1.
In aqueous solution, a quite different photophysical behavior was observed compared to organic solvents. As shown in Fig. 2a, a new and intense absorption band appears at 625 nm, while the peak associated with the S0 → S1 transition remains at 580 nm. The new red-shifted band was attributed to the presence of molecular aggregates.38Fig. 4a shows the effect of the temperature on the stability of aggregates. The ratio of the absorbances at 580 and 630 nm increased with the temperature increase in aqueous solution, while no spectral changes were observed in THF (Fig. S1, ESI†). Our data show that aggregation of compound 1 leads to fluorescence quenching of both S1 → S0 and S2 → S0 transitions (a faint and broad band is recorded between 600 and 750 nm, alongside a weak band at 515 nm). pH changes from 5 to 9 did not affect photophysical properties of the aggregates (Fig. S5 shows that emission spectrum is not sensitive to pH, ESI†). On the contrary, the addition of bovine serum albumin (BSA), while exciting at 560 nm led to an emission band centered at 590 nm (S1 → S0 transition), suggesting a shift in the equilibrium toward the non-aggregated form of 1 (Fig. 4b). Exciting at 465 nm, the intensity of both the band centered at 530 nm (S2 → S0 transition) and the tail extending beyond 575 nm (S2/S1 → S0 transition) also increases upon addition of BSA. Interestingly, the fluorescence intensity associated with the S2/S1 → S0 process shows a higher increase than the fluorescence intensity from the S2 → S0 transition, as evident in the normalized emission spectrum (Fig. S6, ESI†). BSA is frequently used as a model to test new fluorophores as probes for protein surface hydrophobicity,39 in that protein contains two hydrophobic pockets (binding sites I and II)40 along with diverse superficial hydrophobic areas.39 The non-aggregated form of 1 (emissive) could be stabilized in the hydrophobic regions of the protein recovering the fluorescence emission. In accordance with this hypothesis, the fluorescence enhancement is reversed by adding a specific site I binder such as warfarin, showing that 1 is displaced from the site I and the equilibrium is shifted toward the aggregated form of 1 (non-emissive) (Fig. 4c).41 Nevertheless, the fluorescence emission is not fully quenched when the concentration of warfarin (1.25 mM) becomes 25-times higher than that of 1 (50 μM) and hence the fluorophore can have more than one binding site. An increase in the fluorescence intensity akin to BSA was not observed in the presence of pepsin, in accordance with a previous report of He et al.42 who showed that BSA had higher affinity for hydrophobic phenolic compounds such as gallotannins than pepsin. In this sense, pepsin has multiple binding sites for a hydrophilic antioxidant such as glutathione in contrast with the single binding site found for a hydrophobic antioxidant such as melatonin.43 Compound 1 did not show sensitivity to coconut oil or macromolecules such as DNA and Ficoll 40 (Fig. S7, ESI†) but was sensitive to the formation of BSA aggregates induced by heating and addition of NaCl to the protein solution.44,45 The fluorescence intensity of the two emission bands of 1 increased with the incubation time of the protein at 80 °C (Fig. 4d). The misfolding of BSA leads to the exposure of hydrophobic amino acid residues on the surface of the protein and the aggregation of the protein.46,47 As previously mentioned, protein misfolding and aggregation are physiological processes associated with oxidative stress and are linked to various diseases.13,15,16 Congo red, Nile red, and Thioflavin T are dyes commonly used for staining protein aggregates, particularly used for monitoring amyloid fibril assembly.48–51 The potential use of compound 1 as a fluorescent probe for detecting protein misfolding and aggregation induced by oxidative stress is discussed below.
Different examples of fluorescent probes for hydrophobic proteins39,52,53 and protein aggregation54,55 have been reported recently. Some of them are based in BODIPY derivates39,55 as compound 1. The applications of these probes are mainly based on measurements of fluorescence intensity. It is known that fluorescence-lifetime probes have significant advantages over fluorescence-intensity probes because the fluorescence lifetime is not sensitive to either the fluorophore concentration, excitation source intensity or duration of light exposure.56–58 Therefore, we are also interested in knowing if compound 1 could be also used as a fluorescence-lifetime probe for hydrophobic proteins and protein aggregation. For that, the effect of BSA on the fluorescence decay profile of the unusual S2/S1 → S0 photophysical process was studied using FLIM (Fig. 5a). This specific transition was chosen for two reasons: (i) its fluorescence intensity is very sensitive to albumin; (ii) the fluorescence background effects are minimized due to the large difference between the excitation wavelength (511 nm) and the acquired wavelengths (655–725 nm region). As showed in Fig. 5b, the average fluorescence lifetime of the S2/S1 → S0 transition increased from 3.4 ns to 3.6–4.0 ns in the presence of BSA (0.05–0.25 mM), while the opposite effect was observed in the presence of other macromolecules such as Ficoll 40 and coconut oil. The average fluorescence lifetime of 1 also increased in the presence of BSA when the aggregation of the protein was induced by heating (80 °C) and adding NaCl.44,45Fig. 5c shows how the fluorescence lifetime of the probe increases with the denaturation time of BSA. The fluorescence lifetime of 1 reached approximately 5.0 ns after an incubation period of 45 minutes with BSA under heating. Fig. 5d shows a FLIM image of BSA aggregates of tenths of micrometer in aqueous solution using compound 1 as a probe. In contrast, no fluorescent aggregates are observed in the absence of BSA or the presence of non-denatured BSA (Fig. 5d).
Fluorescence imaging in live cells
The potential use of compound 1 as a fluorescent probe for detecting protein misfolding and aggregation was tested on two cancer cell lines (human breast cancer MCF-7 cells and human lung cancer A549 cells) under oxidative stress conditions. Fluorescence imaging experiments were carried out using two fluorescence microscopy approaches: one based on time-resolved fluorescence measurements (FLIMs), and another based on fluorescence intensity measurements (two-photon fluorescence microscopy). FLIM experiments showed that compound 1 was efficiently taken up by MCF-7 cells (Fig. 6). Cell viability remained high, above 93% for concentrations up to 0.5 mM after 24-hour incubation with 1 (Fig. S8, ESI†). In a similar way, a moderate impact on cell viability was reported for the homologous 7-azaindole-BODIPY derivative 3 after 48 hours of incubation in lymphoblast cells (K562 cell line, cell viability of about 70%).12 Oxidative stress was induced in MCF-7 cells by adding ethanol (1 M),59 hydrogen peroxide (0.5 mM),18–20 or cisplatin (10 μM).60,61 It was confirmed that the ROS level increased significantly in MCF-7 cells to 22% and 260% after cisplatin and hydrogen peroxide treatments (for 3 hours), respectively, with respect to the control cells (Fig. S9, ESI†). The ethanol treatment for 3 hours only induced a slight increase in the ROS level (3%). FLIM images (Fig. 6 and Fig. S10–S12, ESI†) show intracellular regions with longer fluorescence lifetimes for oxidant-treated MCF-7 cells (colored in green and red), contrasting to the shorter fluorescence lifetimes found in control cells (colored in blue). This fact is particularly evident for MCF-7 cells exposed to cisplatin and hydrogen peroxide, for incubation periods of 1 and 3 hours. Considering the photophysical behavior of compound 1, the regions with longer lifetimes in the vicinity of the nucleus were attributed to the accumulation of misfolded proteins or aggregates. Consequently, a broader distribution of (intracellular) fluorescence lifetimes (and a shift to longer lifetimes) was observed for the oxidant-treated MCF-7 cells than for the untreated cells (Fig. 7). Fig. 8 shows a quantitative analysis of the evolution of the average fluorescence lifetimes determined for single MCF-7 cells treated with ethanol, hydrogen peroxide, or cisplatin (see also Table S4, ESI†). In general, the average fluorescence lifetimes are higher, and the distribution is broader, for the oxidant-treated MCF-7 cells than for the untreated cells. For instance, the average fluorescence lifetimes of the control cells are within the range of 2–4 ns, while a significant proportion of cells treated with hydrogen peroxide or cisplatin for 3 hours showed average lifetimes within 2–6 ns. The response of compound 1 to protein misfolding and aggregation associated with oxidative stress seems to be consistent with previous studies. For instance, the formation of protein aggregates and the increase of ROS are concomitant processes observed in kidney tubular cells after cisplatin administration.62,63 The endoplasmic reticulum stress and unfolded protein aggregation also seems to be related to the cardiotoxicity of cisplatin in mice.64 | ||
| Fig. 7 Fluorescence lifetime distribution histograms corresponding to the FLIM images of Fig. 6. The histograms were fitted to a Gaussian function. | ||
 | ||
| Fig. 8 Box plots of average fluorescence lifetimes (τav) determined for single MCF-7 cells treated with 1 (0.1 μM) along with ethanol (1 M), hydrogen peroxide (0.5 mM), or cisplatin (10 μM). The incubation times were (a) 0.5 hours, (b) 1 hour, and (c) 3 hours. Raw data are collected in Table S4 (ESI†). | ||
The sensitivity of compound 1 to protein misfolding and aggregation was also evaluated in A549 cells using a two-photon fluorescence microscope. An excitation wavelength of 1022 nm was chosen in order to reach the S2 state of compound 1 and the fluorescence emission was recorded in wavelength windows: 515–575 nm (green channel) and 650–750 nm (red channel) to monitor the florescence emission associated with the S2 → S0 and S2/S1 → S0 transitions, respectively. Two-photon fluorescence microscopy images showed that compound 1 was efficiently taken up by control A549 cells (Fig. 9 and Fig. S14, ESI†). For the untreated cells, the fluorescence emission recorded in the green channel is more intense compared to that in the red channel. This scenario is modified in cells treated with ethanol, where the fluorescence intensity from the red channel significantly increases with respect to the green channel. The fluorescence emission collected in the red channel also increases with the incubation time (quantitative data are shown in Fig. S15 and Table S5, ESI†). Again, the notable increase of fluorescence intensity associated with the S2/S1 → S0 transition (red channel) seems to be related to protein misfolding and aggregation induced by oxidative stress. These results were in accordance with the findings of Kim et al., who showed that metabolic stress induced the formation of protein aggregates in A549 cells.65 Additionally, two-photon fluorescence microscopy allows a deeper penetration of the excitation laser beam into tissues and cell spheroids. Fig. 10 shows that compound 1 can be taken up by cells in spheroids. After an incubation period of 24 hours, compound 1 stains the periphery of A549 cell spheroids and penetrates about 50 micrometers inside. These results open the door to the future use of compound 1 as a fluorescent probe in more complex systems such as tissues and even in vivo models.
Synchrotron FTIR microspectroscopy
FTIR spectra of MCF-7 cells were recorded to provide additional information on the sensitivity of compound 1 to protein misfolding events. These spectra can inform about the protein structural changes in the cells under oxidative stress conditions. Average FTIR spectra of MCF-7 cells untreated and treated with cisplatin (10 μM for 3 hours) were acquired using a synchrotron light source. Fig. 11a shows the average FTIR spectra of amide I and II regions for control and treated cells. Amide I band is shifted toward higher wavenumbers in the treated cells (1654 cm−1) with respect to the control cells (1653 cm−1). In contrast, amide II band is shifted toward lower wavenumbers (from 1541 cm−1 for control cells to 1539 cm−1 for treated cells). The shifts of the amide I and II bands are typically associated with changes in secondary structure during misfolding and aggregation events.66–70 The second derivate of the average FTIR spectra shows a minimum at 1655 cm−1 and a shoulder at 1635 cm−1 typically attributed to α-helix and β-sheet structures, respectively (Fig. 11b).66–70 As shown in Fig. 11c, a decrease in the β-sheet/α-helix ratio (ratio of absorbances at 1635 and 1655 cm−1) was observed in cells treated with cisplatin with respect to the control cells. Maps of β-sheet/α-helix ratio acquired for groups of control and treated cells are shown in Fig. 11d and e, respectively. The blue color is predominant in Fig. 11e indicating the decrease of the β-sheet/α-helix ratio with respect to the control cells (in green and yellow colors). The protein structural changes observed in MCF-7 cells under oxidative stress conditions are consistent with previous studies. For instance, an increase in α-helix structure was observed in gastric adenocarcinoma AGS cells treated with a brominated derivative of coelenterazine, which has anticancer activity.66 Altered protein α-helix structures were also detected in human embryonic liver WRL68 cells treated with doxorubicin.69 Here, we have found that cisplatin, under similar conditions as the previous FLIM experiments, induces conformational changes in the proteins of MCF-7 cells. The observation of these changes is additional evidence for the sensitivity of compound 1 to protein misfolding and aggregation events associated with cellular stress responses.Conclusions
The photophysical properties and sensing applications of a novel 7-azaindole-BODYPY derivative 1 have been studied here. Two emissive excited states (S1 and S2) were found for 1, as well as an efficient IC process between both states. In an aqueous solution, the fluorescence emission intensity of 1 is strongly quenched because of the formation of molecular aggregates. The fluorescence emission of 1 significantly increases upon the addition of albumin and formation of protein aggregates because the monomeric form of 1 is stabilized in the hydrophobic environments. Compound 1 showed less sensitivity to other macromolecules such as pepsin, DNA and Ficoll 40 as well as coconut oil. The fluorescence lifetime of 1 (particularly that coming from the transition S2/S1 → S0, which involves the internal conversion between S2 and S1 states) is highly sensitive to the presence of protein aggregates.Different microscopy techniques, in two different cell lines, demonstrated that the S2/S1 → S0 transition of compound 1 can be used to monitor protein misfolding and aggregation associated with cellular stress responses. Firstly, FLIM images showed subcellular regions where the fluorescence lifetime significantly increased in breast cancer MCF-7 cells treated with ethanol, hydrogen peroxide or cisplatin. These regions were attributed to the accumulation of misfolded proteins or aggregates in response to the oxidative stress. Synchrotron FTIR microscopy showed that cisplatin induced conformational changes in the proteins of MCF-7 cells at short incubation periods (3 h) evidencing the sensitivity 1 to protein misfolding events associated with cellular stress responses. Secondly, two-photon fluorescence images showed that the emission intensity associated with the S2/S1 → S0 transition markedly increased when stress was induced in lung cancer A549 cells by adding ethanol. Compound 1 was also efficiently taken up and distributed by spheroids of A549 cells. These results encourage further research on the applications of 1 as a fluorescent probe in more complex systems such as tissues and even in vivo models.
Data availability
Data are available from the authors upon reasonable request.Author contributions
A. G. R., I. B., and E. M. G. F. conceived the idea. A. G. R., I. B., E. M. G. F., S. R., and M. K. designed and supervised the experiments. D. H. O. performed the measurements. I. L. carried out the synthesis of the studied compound. All authors contributed to the discussion of the results and agreed with the results. All the authors contributed to writing the manuscript.Conflicts of interest
There are no conflicts to declare.Acknowledgements
This work is supported by ‘Universidad de Castilla-La Mancha’ through Projects 2022-GRIN-34143 and 2019-GRIN-27175, by KU Leuven (C14/22/085, IDN/20/021, and KA/20/026), and by the ‘Junta de Comunidades de Castilla-La Mancha’ by the grants SBPLY/21/180501/000050, SBPLY/23/180502/000013 and SBPLY/21/180501/000042. The authors also acknowledge the support through the grants CPP2021-008597, and PID2021-128761OA-C22 and PID2020-117788RB-I00 funded by MCIN and Next Generation EU through Plan de Recuperación, Transformación y Resiliencia. P. C. and S. R. acknowledge the financial support from the SuperCol project that received funding from the European Union's Horizon 2020 research and innovation program under the Marie Skłodowska-Curie grant agreement No. 860914. D. H. O. also thanks the ‘Universidad de Castilla-La Mancha’ for his predoctoral fellowship (2019/5964). The computational study was performed in SSC (Servicio de Supercomputación) from Universidad de Castilla-La Mancha. The authors acknowledge the ALBA Synchrotron Facility for beamtime allocation proposal number 2023027543 (instrument BL01 – MIRAS). The authors thank the Agencia Estatal de Investigación, MCIN/AEI/10.13039/501100011033 (Spain, projects PID2019-105479RB-I00 and PID2022-137889OB-I00) and “ERDF A way of making Europe” (EU, project PID2022-137889OB-I00).References
- E. Ozcan, H. H. Kazan and B. Çoşut, Luminescence, 2020, 35, 168–177 CrossRef CAS PubMed.
- N. Boens, V. Leen and W. Dehaen, Chem. Soc. Rev., 2012, 41, 1130–1172 RSC.
- T. Kowada, H. Maeda and K. Kikuchi, Chem. Soc. Rev., 2015, 44, 4953–4972 RSC.
- C. Duan, Y. Zhou, G. G. Shan, Y. Chen, W. Zhao, D. Yuan, L. Zeng, X. Huang and G. Niu, J. Mater. Chem. C, 2019, 7, 3471–3478 RSC.
- B. Zhu, X. Xing, J. Kim, H. Rha, C. Liu, Q. Zhang, L. Zeng, M. Lan and J. S. Kim, Biomaterials, 2024, 304, 122419 CrossRef CAS PubMed.
- P. Jayasudha, R. Manivannan, S. Ciattini, L. Chelazzi and K. P. Elango, Sens. Actuators, B, 2017, 242, 736–745 CrossRef CAS.
- J. Shao, Y. Wang, H. Lin, J. Li and H. Lin, Sens. Actuators, B, 2008, 134, 849–853 CrossRef CAS.
- K. R. Rathikrishnan, V. K. Indirapriyadharshini, S. Ramakrishna and R. Murugan, Tetrahedron, 2011, 67, 4025–4030 CrossRef CAS.
- R. Manivannan, A. Satheshkumar, E.-S. H. El-Mossalamy, L. M. Al-Harbi, S. A. Kosa and K. P. Elango, New J. Chem., 2015, 39, 3936–3947 RSC.
- Y. Han, W. Dong, Q. Guo, X. Li and L. Huang, Eur. J. Med. Chem., 2020, 203, 112506 CrossRef CAS PubMed.
- A. K. Mahapatra, R. Maji, K. Maiti, S. S. Adhikari, C. Das Mukhopadhyay and D. Mandal, Analyst, 2014, 139, 309–317 RSC.
- G. Keşan, B. Topaloğlu, E. Özcan, H. H. Kazan, E. T. Eçik, E. Şenkuytu, I. F. Sengul, H. Kandemir and B. Çoşut, Spectrochim. Acta, Part A, 2019, 213, 73–82 CrossRef PubMed.
- S. Alberti and A. A. Hyman, Nat. Rev. Mol. Cell Biol., 2021, 22, 196–213 CrossRef CAS PubMed.
- E. Lévy, N. El Banna, D. Baïlle, A. Heneman-Masurel, S. Truchet, H. Rezaei, M. E. Huang, V. Béringue, D. Martin and L. Vernis, Int. J. Mol. Sci., 2019, 20, 3896 CrossRef PubMed.
- G. R. Mallucci, D. Klenerman and D. C. Rubinsztein, Annu. Rev. Cell Dev. Biol., 2020, 36, 165–189 CrossRef CAS PubMed.
- C. A. Ross and M. A. Poirier, Nat. Med., 2004, 10, S10 CrossRef PubMed.
- I. Van Zundert, N. Maenhoudt, S. De Vriendt, H. Vankelecom, B. Fortuni and S. Rocha, Bio. Protoc., 2022, 12, e4469 CAS.
- P. K. S. Mahalingaiah and K. P. Singh, PLoS One, 2014, 9, e87371 CrossRef PubMed.
- C. Ransy, C. Vaz, A. Lombès and F. Bouillaud, Int. J. Mol. Sci., 2020, 21, 1–14 CrossRef PubMed.
- J. J. P. Gille and H. Joenje, Mutat. Res., 1992, 275, 405–414 CAS.
- H. Ciftci, B. Sever, N. Bayrak, M. Yıldız, H. Yıldırım, H. Tateishi, M. Otsuka, M. Fujita and A. F. TuYuN, Pharmaceuticals, 2022, 15, 1266 CrossRef CAS PubMed.
- A. Ezzat, W. Fayad, A. Ibrahim, Z. Kamel, A. I. El-Diwany, K. H. Shaker and M. A. Esawy, Biocatal. Agric. Biotechnol., 2020, 24, 101526 CrossRef.
- A. Wawruszak, J. J. Luszczki, A. Grabarska, E. Gumbarewicz, M. Dmoszynska-Graniczka, K. Polberg and A. Stepulak, PLoS One, 2015, 10, e0143013 CrossRef PubMed.
- A. Wawruszak, J. Luszczki, M. Halasa, E. Okon, S. Landor, C. Sahlgren, A. Rivero-Muller and A. Stepulak, Int. J. Mol. Sci., 2021, 22, 5184 CrossRef CAS PubMed.
- R. Vrzal, P. Štarha, Z. Dvořák and Z. Trávníček, J. Inorg. Biochem., 2010, 104, 1130–1132 CrossRef CAS PubMed.
- I. Yousef, L. Ribó, A. Crisol, I. Šics, G. Ellis, T. Ducic, M. Kreuzer, N. Benseny-Cases, M. Quispe, P. Dumas, S. Lefrançois, T. Moreno, G. García, S. Ferrer, J. Nicolas and M. A. G. Aranda, Synchrotron Radiat. News, 2017, 30, 4–6 CrossRef.
- M. Toplak, G. Birarda, S. Read, C. Sandt, S. M. Rosendahl, L. Vaccari, J. Demšar and F. Borondics, Synchrotron Radiat. News, 2017, 30, 40–45 CrossRef.
- M. J. Frisch, G. W. Trucks, H. B. Schlegel, G. E. Scuseria, M. A. Robb, J. R. Cheeseman, G. Scalmani, V. Barone, G. A. Petersson, H. Nakatsuji, X. Li, M. Caricato, A. V. Marenich, J. Bloino, B. G. Janesko, R. Gomperts, B. Mennucci, H. P. Hratchian, J. V. Ortiz, A. F. Izmaylov, J. L. Sonnenberg, D. Williams-Young, F. Ding, F. Lipparini, F. Egidi, J. Goings, B. Peng, A. Petrone, T. Henderson, D. Ranasinghe, V. G. Zakrzewski, J. Gao, N. Rega, G. Zheng, W. Liang, M. Hada, M. Ehara, K. Toyota, R. Fukuda, J. Hasegawa, M. Ishida, T. Nakajima, Y. Honda, O. Kitao, H. Nakai, T. Vreven, K. Throssell, J. A. Montgomery, Jr., J. E. Peralta, F. Ogliaro, M. J. Bearpark, J. J. Heyd, E. N. Brothers, K. N. Kudin, V. N. Staroverov, T. A. Keith, R. Kobayashi, J. Normand, K. Raghavachari, A. P. Rendell, J. C. Burant, S. S. Iyengar, J. Tomasi, M. Cossi, J. M. Millam, M. Klene, C. Adamo, R. Cammi, J. W. Ochterski, R. L. Martin, K. Morokuma, O. Farkas, J. B. Foresman and D. J. Fox, Gaussian 16, Revision C.01, Gaussian, Inc., Wallingford CT, 2016 Search PubMed.
- C. Lee, W. Yang and R. G. Parr, Phys. Rev. B: Condens. Matter Mater. Phys., 1988, 37, 785 CrossRef CAS PubMed.
- B. Miehlich, A. Savin, H. Stoll and H. Preuss, Chem. Phys. Lett., 1989, 157, 200–206 CrossRef CAS.
- A. D. Becke, Phys. Rev. A: At., Mol., Opt. Phys., 1988, 38, 3098 CrossRef CAS PubMed.
- M. Cossi, N. Rega, G. Scalmani and V. Barone, J. Comput. Chem., 2003, 24, 669–681 CrossRef CAS PubMed.
- J. Tomasi, B. Mennucci and R. Cammi, Chem. Rev., 2005, 105, 2999–3093 CrossRef CAS PubMed.
- M. R. Momeni and A. Brown, J. Chem. Theory Comput., 2015, 11, 2619–2632 CrossRef CAS PubMed.
- Q. Alkhatib, W. Helal and A. Marashdeh, RSC Adv., 2022, 12, 1704–1717 RSC.
- D. W. Cho, M. Fujitsuka, J. H. Ryu, M. H. Lee, H. K. Kim, T. Majima and C. Im, Chem. Commun., 2012, 48, 3424–3426 RSC.
- C. Schäfer, J. Mony, T. Olsson and K. Börjesson, Chem. – Eur. J., 2020, 26, 14295–14299 CrossRef PubMed.
- A. B. Descalzo, P. Ashokkumar, Z. Shen and K. Rurack, ChemPhotoChem, 2020, 4, 120–131 CrossRef CAS.
- N. Dorh, S. Zhu, K. B. Dhungana, R. Pati, F. T. Luo, H. Liu and A. Tiwari, Sci. Rep., 2015, 5, 18337 CrossRef CAS PubMed.
- J. Ghuman, P. A. Zunszain, I. Petitpas, A. A. Bhattacharya, M. Otagiri and S. Curry, J. Mol. Biol., 2005, 353, 38–52 CrossRef CAS PubMed.
- G. Rabbani and S. N. Ahn, Int. J. Biol. Macromol., 2019, 123, 979–990 CrossRef CAS PubMed.
- Q. He, B. Shi and K. Yao, Food Chem., 2006, 95, 250–254 CrossRef CAS.
- X. Li and T. Ni, Eur. Biophys. J., 2016, 45, 165–174 CrossRef CAS PubMed.
- J. H. Park, J. A. Jackman, A. R. Ferhan, G. J. Ma, B. K. Yoon and N. J. Cho, ACS Appl. Mater. Interfaces, 2018, 10, 32047–32057 CrossRef CAS PubMed.
- X. Liu, W. Zhang, J. Liu, R. Pearce, Y. Zhang, K. Zhang, Q. Ruan, Y. Yu and B. Liu, Food Hydrocolloids, 2020, 101, 105450 CrossRef CAS.
- N. S. Lebedeva, E. S. Yurina, Y. A. Gubarev and O. I. Koifman, Spectrochim. Acta, Part A, 2021, 246, 118975 CrossRef CAS PubMed.
- L. Wen, M. Lyu, H. Xiao, H. Lan, Z. Zuo and Z. Yin, Mol. Pharm., 2018, 15, 2257–2267 CrossRef CAS PubMed.
- C. Xue, T. Y. Lin, D. Chang and Z. Guo, R. Soc. Open. Sci., 2016, 4, 160696 CrossRef PubMed.
- E. I. Yakupova, L. G. Bobyleva, I. M. Vikhlyantsev and A. G. Bobylev, Biosci. Rep., 2019, 39, BSR20181415 CrossRef PubMed.
- P. Frid, S. V. Anisimov and N. Popovic, Brain Res. Rev., 2007, 53, 135–160 CrossRef CAS PubMed.
- F. Kundel, L. Tosatto, D. R. Whiten, D. C. Wirthensohn, M. H. Horrocks and D. Klenerman, FEBS J., 2018, 285, 3604–3630 CrossRef CAS PubMed.
- L. Peng, R. Wei, K. Li, Z. Zhou, P. Song and A. Tong, Analyst, 2013, 138, 2068–2072 RSC.
- Y. Chen, K. Li, S. Zhang, P. Xu and B. Song, Dyes Pigm., 2022, 202, 110267 CrossRef CAS.
- S. Yang, X. Liu, S. Wang, J. Song, J. Wu, B. Shen, H. Jia, S. Guo, Y. Wang, Y. Yang, Y. Jiang, H. Yang and J. Chang, Sens. Actuators, B, 2023, 395, 134514 CrossRef CAS.
- A. K. Mora, S. Murudkar, N. Shivran, S. Mula, S. Chattopadhyay and S. Nath, Int. J. Biol. Macromol., 2021, 166, 1121–1130 CrossRef CAS PubMed.
- C. Liu, X. Wang, Y. Zhou and Y. Liu, Sci. World J., 2013, 2013, 801901 CrossRef PubMed.
- M. Y. Berezin and S. Achilefu, Chem. Rev., 2010, 110, 2641–2684 CrossRef CAS PubMed.
- M. J. Ruedas-Rama, J. M. Alvarez-Pez, L. Crovetto, J. M. Paredes and A. Orte, FLIM Strategies for Intracellular Sensing, Advanced Photon Counting: Applications, Methods, Instrumentation, Springer, 2014, pp. 191–223 Search PubMed.
- M. Comporti, C. Signorini, S. Leoncini, C. Gardi, L. Ciccoli, A. Giardini, D. Vecchio and B. Arezzini, Genes Nutr., 2010, 5, 101–109 CrossRef CAS PubMed.
- W. Yu, Y. Chen, J. Dubrulle, F. Stossi, V. Putluri, A. Sreekumar, N. Putluri, D. Baluya, S. Y. Lai and V. C. Sandulache, Sci. Rep., 2018, 8, 4306 CrossRef PubMed.
- R. Pratibha, R. Sameer, P. V. Rataboli, D. A. Bhiwgade and C. Y. Dhume, Eur. J. Pharmacol., 2006, 532, 290–293 CrossRef CAS PubMed.
- A. Takahashi, T. Kimura, Y. Takabatake, T. Namba, J. Kaimori, H. Kitamura, I. Matsui, F. Niimura, T. Matsusaka, N. Fujita, T. Yoshimori, Y. Isaka and H. Rakugi, Am. J. Pathol., 2012, 180, 517–525 CrossRef CAS PubMed.
- S. M. Sears and L. J. Siskind, J. Am. Soc. Nephrol., 2021, 32, 1559–1567 CrossRef CAS PubMed.
- H. Ma, K. R. Jones, R. Guo, P. Xu, Y. Shen and J. Ren, Clin. Exp. Pharmacol. Physiol., 2010, 37, 460–465 CrossRef CAS PubMed.
- C. H. Kim, H. M. Jeon, S. Y. Lee, E. K. Jeong, M. K. Ju, B. J. Park, H. G. Park, S.-C. Lim, S. I. Han and H. S. Kang, Int. J. Oncol., 2010, 37, 97–102 CAS.
- C. M. Magalhaes, T. Dučić, R. B. Pereira, P. González-Berdullas, J. E. Rodríguez-Borges, D. M. Pereira, J. C. G. Esteves da Silva, M. Algarra and L. Pinto da Silva, Arch. Biochem. Biophys., 2023, 743, 109660 CrossRef CAS PubMed.
- T. Dučić and J. C. Koch, Cells, 2023, 12, 1451 CrossRef PubMed.
- A. Dróżdż, A. Sławińska-Brych, D. Kubera, M. Kimsa-Dudek, J. M. Gola, J. Adamska, C. Kruszniewska-Rajs, A. Matwijczuk, D. Karcz, W. Dąbrowski, A. Stepulak and M. Gagoś, Int. J. Mol. Sci., 2022, 23, 15260 CrossRef PubMed.
- X. Zhou, J. Zhong, W. Yu and Y. Tang, Spectrochim. Acta, Part A, 2022, 283, 121773 CrossRef CAS PubMed.
- T. Dučić, A. Sanchez-Mata, J. Castillo-Sanchez, M. Algarra and E. Gonzalez-Munoz, Spectrochim. Acta, Part A, 2023, 297, 122713 CrossRef PubMed.
Footnote |
| † Electronic supplementary information (ESI) available: Additional details on synthesis. Additional information on spectroscopic characterization (Fig. S1–S7). DFT calculations. Figures of cytotoxicity and ROS level assays (Fig. S8 and S9). Additional FLIM images (Fig. S10–S13) and two-photon fluorescence microscopy images and quantitative data (Fig. S14 and S15). See DOI: https://doi.org/10.1039/d4tb00567h |
| This journal is © The Royal Society of Chemistry 2024 |