Open Access Article
Open Access ArticleCreative Commons Attribution 3.0 Unported Licence
Modular photoorigami-based 4D manufacturing of vascular junction elements†
Arpan
Biswas
 d,
Indra
Apsite
d,
Indra
Apsite
 d,
Sabine
Rosenfeldt
b,
Ivita
Bite
d,
Sabine
Rosenfeldt
b,
Ivita
Bite
 c,
Virginija
Vitola
c,
Virginija
Vitola
 c and
Leonid
Ionov
c and
Leonid
Ionov
 *a
*a
aFaculty of Engineering Sciences and Bavarian Polymer Institute, University of Bayreuth, Bayreuth 95447, Germany. E-mail: leonid.ionov@uni-bayreuth.de
bFaculty of Biology, Chemistry and Earth Sciences and Bavarian Polymer Institute, University of Bayreuth, Bayreuth 95447, Germany
cInstitute of Solid State Physics, University of Latvia, Kengaraga St. 8, Riga, LV-1063, Latvia
dFaculty of Engineering, University of Bayreuth, Bayreuth 95447, Germany
First published on 22nd April 2024
Abstract
Four-dimensional (4D) printing, combining three-dimensional (3D) printing with time-dependent stimuli-responsive shape transformation, eliminates the limitations of the conventional 3D printing technique for the fabrication of complex hollow constructs. However, existing 4D printing techniques have limitations in terms of the shapes that can be created using a single shape-changing object. In this paper, we report an advanced 4D fabrication approach for vascular junctions, particularly T-junctions, using the 4D printing technique based on coordinated sequential folding of two or more specially designed shape-changing elements. In our approach, the T-junction is split into two components, and each component is 4D printed using different synthesized shape memory polyurethanes and their nanohybrids, which have been synthesized with varying hard segment contents and by incorporating different weight percentages of photo-responsive copper sulfide-polyvinyl pyrrolidone nanoparticles. The formation of a T-junction is demonstrated by assigning different shape memory behaviors to each component of the T-junction. A cell culture study with human umbilical vein endothelial cells reveals that the cells proliferate over time, and almost 90% of cells remain viable on day 7. Finally, the formation of the T-junction in the presence of near-infrared light has been demonstrated after seeding the endothelial cells on the programmed flat surface of the two components and fluorescence microscopy at day 3 and 7 reveals that the cells adhered well and continue to proliferate over time. Hence, the proposed alternative approach has huge potential and can be used to fabricate vascular junctions in the future.
Introduction
Tissue engineering and regenerative medicine are continuously evolving to fulfill the increasing demand for organ transplantation worldwide.1 The potential of different types of biomaterials, cells, and fabrication techniques, individually or collectively, is being studied to repair damaged tissues or to regenerate the structure or/and function of tissues.2,3 Among different factors, the development of fabrication methods plays an essential role in tissue engineering and regenerative medicine. Usually, additive, subtractive, and formative manufacturing techniques are used to fabricate structures with or without cells in tissue engineering.4,5 However, all of them have certain limitations that restrict their preferential application within the field.6,7 Two strategies are commonly used to fabricate structures with cells: (i) fabrication of a structure incorporating the cells with the materials (cell–laden constructs) enables the construction of multicellular complex structures, but the issue with the viability of the incorporated cells restricts it to a few possible fabrication methods, while (ii) fabrication of a structure followed by cell seeding allows the use of almost all possible fabrication methods.7,8Additive manufacturing (AM) or three-dimensional (3D)9 printing has emerged as the most advanced and widely used technique for fabricating not only any complex structure in tissue engineering10,11 but also electronic devices and high-performance metamaterials.12,13 It enables the development of any 3D structure with good resolution, which helps to create new functionality or improves optimal performance. Recent advancements in 3D printing allow for the printing of multi-materials, which helps to develop heterogeneous structures or hybrids.14,15 Although these additional features of 3D printing have resulted in new innovative applications, they are limited. In 3D bioprinting, biological structures are fabricated layer-by-layer with accurate positioning of bioinks (biomaterials, cell, and bioactive molecules) through spatial control over the functional component placement.11 The most commonly used 3D printing techniques in tissue engineering or organ regeneration are inkjet printing,16,17 extrusion-based 3D printing,18,19 stereolithography,20 digital light processing (DLP),21etc. Despite recent achievements and the enormous future potential of 3D bioprinting in the fabrication of complex biological constructs with specific geometries and heterogeneities, it is still struggling with issues like appropriate cell orientation or achieving high resolution of constructs with good cell viability during the fabrication of hollow structures like blood vessels.22–25
Four-dimensional (4D) printing, an advanced extension of 3D printing, has attracted significant attention in various research fields including, but not limited to, smart materials and biomedical research.26–28 4D printing allows shape transformation within the material or structural design of any 3D printed structure with time in the presence of a predetermined stimulus, including osmotic pressure, light, heat, current, magnetic field, etc.29–32 4D printing, being a new sophisticated technique, not only provides tremendous opportunities for designing and fabricating smart or active structures efficiently and easily using shape-changing polymers (e.g. shape memory polymers, shape-changing hydrogels and liquid crystal elastomers) but also allows printing multiple materials (such as hydrogel(active)/polymer(passive) or polymer(active)/polymer(passive)) layer by layer, resulting in the much-desired shape transformation in the developed structure.33–36 The main advantage of 4D printing over 3D printing is the possibility of fabrication of hollow structures without the need for supporting materials. For example, the challenges associated with removing rod-like supports after 3D printing of tubular structures using a rotating rod or the issues with the attachment of the cells during seeding inside the fabricated tubes, are eliminated using the 4D printing technique.37 In the 4D printing approach, cells are seeded on a flat surface, and cells have time to attach and proliferate. Finally, the shape transformation enables the formation of tubular or scroll structures with uniformly distributed cells in the inner wall of the tube.37,38 Moreover, 4D printing successfully eliminates the requirement for 3D printing with high resolution that generates enormous shear stress during printing and negatively impacts the viability of the incorporated cells.38–40 Current 4D printing also has limitations regarding the shapes that can be created using a single shape-changing object. While a tube can easily be formed by rolling a rectangular film, achieving more complex shapes like, for example, T- and Y-junctions, which are essential elements of the vascular network, by shape-transformation of a single 2D object (film) is not possible.
To successfully execute 4D printing, the selection of materials is very crucial. Usually, shape-changing polymers (SCPs) are used for this type of approach, and among different SCPs, hydrogels, and shape memory polymers (SMPs) are very common.41–47 Hydrogels generally exhibit shape-changing behavior utilizing their swelling behavior, while SMPs can recover their shapes from a temporarily programmed shape in the presence of stimuli. Despite the numerous advantages of hydrogels (like tunable swelling behavior, porous structure, and excellent biocompatibility), their prolonged response rate (from a few minutes to a few days depending on the size), low mechanical strength (ranging from ∼1 kPa to ∼100 kPa) and fast degradation rate limit their applications in 4D printing for biomedical applications.48,49 In that scenario, the fast response time (from a few s to a few minutes), better mechanical properties (in the range of ∼MPa to ∼GPa), and the slow degradation rate of SMPs make them a better choice as compared to hydrogels.50,51 Among different available environmental stimuli, light as a stimulus has distinct advantages, including (i) precise application, (ii) remote applicability, and (iii) rapid switching abilities.52 Generally, one can introduce photo-responsiveness in shape memory polymers in two ways: (i) using photo-responsive dyes or chromophores or (ii) using photo-responsive fillers.53 Furthermore, photo-responsive fillers not only produces photo-responsiveness in SMP but will also help improve the viscoelastic properties during printing.2,54
Here, we introduce an advanced approach for fabricating vascular junctions, particularly T-junctions, using the sophisticated 4D printing technique based on coordinated sequential folding of two (and possibly more) shape-changing objects. While folding of single shape-changing objects can be compared with origami, coordinated folding of two or more shape-changing objects can be compared with the extension of traditional origami – modular origami. In this approach, the T-junction of the vascular network is split into two hollow structures and is 4D printed using different shape memory polymers to introduce different responsive behaviors to the two hollow structures. This paper demonstrates the proof of concept of this approach, which can be extended to more than two objects.
Experimental section
Materials
Polycaprolactone diol (PCL-diol, Mn ∼ 2000 g mol−1), 1,6-hexamethylene diisocyanate (HDI), 1,4-butane diol (BD), dibutyltin laurate (DBTDL), and N,N-dimethyl formamide (DMF) were purchased from Merck and were used as received. Copper sulfide-poly vinyl pyrrolidone (CuS-PVP) nanoparticles were synthesized as discussed in ref. 55. Dulbecco's phosphate buffer solution (PBS) (Merck), calcein AM (Thermo Fisher Scientific), ethidium homodimer (EthD-1) 4,6-diamidino-2-phenylindole (DAPI) (Thermo Fischer Scientific), Triton X-100 (Merck), albumin fraction V (BSA) (Roth), phalloidin Dylight 488 (Thermo Fischer Scientific), Dulbecco's modified Eagle's medium (DMEM) (Merck), t-butyl alcohols (Merck), penicillin–streptomycin (Pen/Strep) (Gibco), fetal bovine serum (FBS) (Merck) were used.Synthesis
Different shape memory polyurethanes (SMPUs) were synthesized through a two-step polymerization process in a three-neck round bottom flask. In the 1st step, the prepolymer was synthesized by adding a predetermined amount of HDI into molten PCL-diol under an inert atmosphere (N2 atmosphere). In the 2nd step, the chain extender (BD) and catalyst (DBTDL: 0.1 ml of 1 wt% toluene solution) were added to complete the polymerization process (Fig. S1a, ESI†). The temperature was maintained at 70 °C throughout the polymerization. Different molar ratios of PCL-diol![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) HDI
HDI![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) BD were used to prepare polyurethanes with varying hard segment contents (HSCs) (Table 1). The chain extension was performed for 24 hours to ensure complete polymerization and to obtain a high molecular weight polymer. The synthesized polymers were recovered through precipitation in double distilled water (non-solvent) and dried in a vacuum oven under reduced pressure at 60 °C for 72 hours. The molecular weight of different synthesized SMPUs was determined through gel permeation chromatography (GPC), which is summarized in Table 1. The nanohybrids of different polyurethanes were prepared by dispersing CuS-PVP nanoparticles homogeneously into the DMF solution of SMPUs and drying the casted solution at 80 °C under reduced pressure in a vacuum oven. Different nanohybrids were prepared with varying wt% of nanoparticles in SMPU.
BD were used to prepare polyurethanes with varying hard segment contents (HSCs) (Table 1). The chain extension was performed for 24 hours to ensure complete polymerization and to obtain a high molecular weight polymer. The synthesized polymers were recovered through precipitation in double distilled water (non-solvent) and dried in a vacuum oven under reduced pressure at 60 °C for 72 hours. The molecular weight of different synthesized SMPUs was determined through gel permeation chromatography (GPC), which is summarized in Table 1. The nanohybrids of different polyurethanes were prepared by dispersing CuS-PVP nanoparticles homogeneously into the DMF solution of SMPUs and drying the casted solution at 80 °C under reduced pressure in a vacuum oven. Different nanohybrids were prepared with varying wt% of nanoparticles in SMPU.
| Polymers | Molar ratio (HMDI![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) : :![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) PCL PCL![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) : :![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) BD) BD) |
Hard segment content (HSC) | Polystyrene standard | ||
|---|---|---|---|---|---|
| M n | M w | PDI | |||
| HPB15 | 1.7![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) : :![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1 1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) : :![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 0.7 0.7 |
15% | 33![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 549 549 |
68![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 444 444 |
2.04 |
| HPB20 | 2.4![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) : :![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1 1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) : :![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1.4 1.4 |
20% | 58![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 562 562 |
93![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 919 919 |
1.4 |
| HPB30 | 3.7![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) : :![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1 1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) : :![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 2.7 2.7 |
30% | 24![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 728 728 |
53![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 070 070 |
2.15 |
Rheological properties
The rheological behavior of pure SMPUs and their nanohybrids was investigated using a multidrive rheometer of Anton Paar (MCR 702 MultiDrive). The samples were placed between two parallel plate–plate geometries with a diameter of 25 mm diameter. The samples' complex viscosity, storage modulus, and loss modulus were recorded by sweeping the angular frequency from 0.1 to 100 rad s−1 and varying the temperature from 200 °C to 100 °C.Dynamic mechanical analysis (DMA) of pure SMPUs and nanohybrids
The mechanical properties of 3D printed samples (dimension: 0.30 × 4 × 7 mm3) of different shape memory polyurethanes and their nanohybrids were investigated using Anton Paar MCR 702 MultiDrive. The change in elongational storage and loss modulus was recorded with increasing temperature from 20 to 50 °C at a constant frequency of 1 Hz and at various frequencies from 0.1 Hz to 10 Hz at a constant temperature of 40 °C. During the measurement, samples were stretched by 5% and stress was maintained at 0.5 MPa.Differential scanning calorimetry (DSC)
The melting temperature, crystallization temperature, and heat of fusion value (ΔH) of 3D printed samples of different SMPUs and their nanohybrids were investigated using a Mettler differential scanning calorimeter. The samples were scanned in the temperature range of −40 to 200 °C keeping the heating and cooling rate 10° min−1 and 5° min−1, respectively.Thermogravimetric analysis (TGA)
The thermal stability of different synthesized SMPUs and their nanohybrids was investigated using a Netzsch TG 209 thermogravimetric analyzer (TGA) in the temperature range of 25 to 600 °C at a heating rate of 20 °C min−1 under a nitrogen atmosphere.3D printing of pure SMPUs and their nanohybrids: For fabricating the desired geometries with different pure SMPUs and their nanohybrids, a pneumatic pressure-assisted extrusion-based 3D printer from RegenHU (3D Discovery) was used. The samples were extruded above their melting temperature (Tm) and under sufficient pneumatic pressure to ensure continuous extrusion of the samples from the nozzle during printing. A nozzle of ∼350 μm diameter, a layer thickness of 0.1 mm, and a feed rate of 6–10 mm s−1 were used to print the desired geometries. The desired 3D geometries were created using Fusion 360 CAD software, and slicing was carried out using Bio-CAD software provided by the RegenHU 3D printer.
Shape memory behavior of pure SMPUs and their nanohybrid: The shape memory behavior of different pure polymers and their nanohybrids was examined using 4D printed geometries. First, the 3D printed geometries (permanent shape) of different samples were deformed into temporary geometries (temporary shape) above their transition temperature (Tr ∼ melting temperature of soft segments of SMPUs, ∼40 °C), and the temporary shapes were fixed by lowering the temperature (Tf ∼ 10 °C) below the transition temperature (Tr). This process is called “programming”; the temporary shape is called the programmed shape. Furthermore, by increasing the temperature above Tr or using near-infrared (NIR) light (Philips R95 IR 100 W near-infrared light), recovery of the permanent shape from the temporary shape of different pure SMPUs and their nanohybrids was investigated. The shape fixity and shape recovery efficacy of different SMPUs and their nanohybrids were calculated using eqn (1) and (2).
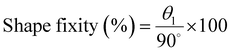 | (1) |
 | (2) |
Small-angle X-ray scattering (SAXS) measurement of SMPUs and their nanohybrids: For SAXS analysis, the SMPUs and their nanohybrids were used as obtained, where the 3D printed samples had a permanent and the programmed ones had a temporary shape. The measurements were performed under ambient conditions using a Double Ganesha AIR system (SAXSLAB/Xenocs), providing monochromatic radiation with a wavelength of λ = 1.54 Å (produced by a rotating Cu anode, MicroMax 007HF, Rigaku corporation) on the lateral area of the samples. The position-sensitive detector (Pilatus 300 K, Dectris) was placed at different distances from the sample to the detector to cover a wide range of scattering vectors q ( ; θ scattering angle). The 1-dimensional intensity I(q) was acquired through radial averaging and then normalized to the intensity of the incident beam, the sample thickness (1.5 mm), and the accumulation time.
; θ scattering angle). The 1-dimensional intensity I(q) was acquired through radial averaging and then normalized to the intensity of the incident beam, the sample thickness (1.5 mm), and the accumulation time.
Spectroscopic measurement
The synthesized polymers’ proton nuclear magnetic resonance (1H-NMR) spectra were recorded using a Bruker Avance 500 spectrometer (500 MHz). The samples were completely dissolved in DMSO-d6 solvent and equilibrated in the magnetic field for 10 minutes before recording the spectrum. The chemical shifts were reported in ppm units relative to tetramethylsilane (TMS). Infrared spectra of solid films of different SMPUs and their nanohybrids were recorded using a Fourier transform infrared (FT-IR) spectrometer from Bruker Tensor 27, USA. The spectra were taken in the spectral range of 800 to 4000 cm−1 with a spectral resolution of 4 cm−1 at room temperature.Morphological investigation
The morphologies of the 3D printed structures of different SMPUs and their nanohybrids were investigated using a scanning electron microscope (SEM) from Thermo Fisher Scientific, USA. The samples were coated with platinum before measurement.Cell culture
Human umbilical vein endothelial cells (HUVECs) were seeded on the flat surfaces of different pure SMPUs and nanohybrids. The samples were 3D printed and prepared under aseptic conditions. Before culturing the cells, the surface of the samples was coated with a laminin solution to support the adhesion and growth of endothelial cells. The excess laminin solution was removed, and the sample was allowed to dry under the hood for 15 minutes. 2 × 105 cells per square centimeter were seeded on the 3D printed samples to assess cell adhesion, viability, and metabolic activity.Cell proliferation and cell viability
The proliferation of the cells on the samples' surface was investigated by measuring the metabolic activity of the seeded endothelial cells at predetermined time intervals using an alamarBlue assay. A 10% (v/v) solution of the alamarBlue reagent in HUVEC cell culture media was prepared and added to 48 well plates to cover the cells on the 3D-printed samples. After 90 minutes of incubation in a CO2 incubator with gentle shaking in all directions to ensure a homogeneous distribution, the media containing reduced almarBlue were taken out and kept under dark and icy conditions. After that, 100 μl aliquots were pipetted into 96 well plates, and the fluorescence intensity was measured at 600 nm after excitation at 535 nm using a plate reader (TriStar2 S LB 942 Multimode Microplate Reader, Berthold Technologies GmbH & Co. KG, Germany). The negative and positive controls were non-reduced and 100% reduced alamarBlue without cells, respectively.A live–dead assay was performed to investigate the viability of cells with time on the surface of the samples. In live–dead assay, cells were strained at a regular time interval (e.g., after 1, 3, and 7 days) with a mixed solution of calcein AM and ethidium homodimer-1 after removing the culture media from the 48 well plates. Then, the well plate was incubated under dark conditions for 30 minutes at room temperature. Finally, fluorescence images were taken using a Nikon Ti2 microscope. The mixed solution was prepared by adding 3.6 μl of calcein AM and 4 μl of ethidium homodimer-1 in 2 ml of DPBS.
Staining of actin filaments (F-actin) and nucleus
The adhesion and monolayer formation of endothelial cells over the surface of the samples were investigated by staining the fibronectin and nucleus of endothelial cells with Phalloidin Dylight 488 conjugated 300 units (prod # 21833 ThermoFisher) and 4′,6-diamidino-2-phenylindole (DAPI) at day 7. The staining solution was prepared by adding 500 μl of DAPI (concentration: 0.1 mg ml−1) and 250 ml of Phalloidin (200 units per ml) stock solution in 10 ml of DPBS. Before staining the cells, the media were aspirated from the well plate, and cells were washed with DPBS twice. Then, cells were treated with freshly prepared 3.7% formaldehyde for 5 minutes to fix the cells. After fixing the cells, the cell membrane was permeabilized by treating the cells with 0.1% Triton 100× solution for 5 min under ambient conditions. Finally, cells were washed twice with DPBS and stained with DAPI/phalloidin solution.Statistical analysis
The obtained data were presented as the mean ± standard deviation (SD) (3–7 replicates were used). Student t-test and one-way analysis of variance (ANOVA) were performed to analyze the differences between the experimental groups. A value of P ≤ 0.05 was considered statistically significant.Result and discussion
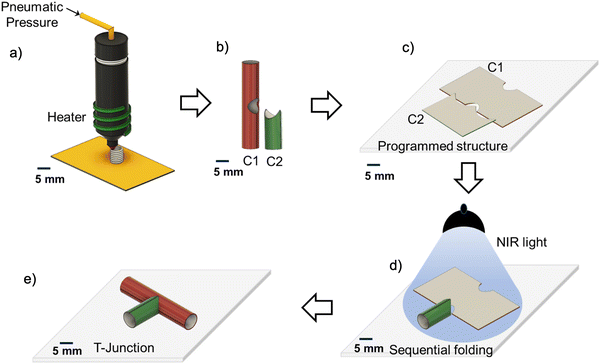
The approach of fabricating vascular junctions utilizing the sequential folding of the 4D printed structure is illustrated in Fig. 1. The fabrication involves three steps. The first step is the 3D printing of the components of vascular junctions individually using an extrusion-based 3D printer. In the second step, the 3D structures are programmed into flat structures. In the third step, the sequential folding of the arranged flat structures with or without seeded cells produces a vascular junction (T-junction). Both flat objects must fold at different times because simultaneous folding results in their collapse. To achieve the separation of object actuation over time, the materials used for their fabrication must possess either (i) different sensitivities, allowing the use of different signals to trigger their shape transformation or (ii) uniform sensitivity but different sensitivity thresholds, allowing triggering with one signal but at different time points. We have explored both possibilities using different synthesized thermoresponsive shape memory polyurethanes (SMPUs), which can be mixed with different amounts of light-sensitive nanoparticles.The SMPUs are synthesized through a two-step condensation polymerization process by mixing hexamethylene diisocyanate (HDI), polycaprolactone diol (PCL-diol), and 1,4-butane-diol (BD), and varying hard segment contents (Fig. S1a, ESI†). The synthesized SMPUs are designated as HPB15, HPB20, and HPB30; as shown in Table 1; the number means the mass percentage of the hard segment. The molecular weight of the synthesized SMPUs is measured using gel permeable chromatography (GPC) with respect to the polystyrene reference that shows a unimodal distribution of the molecular weight for all synthesized SMPUs (Fig. S1b, ESI†). The weight average molecular weights (Mw) of HPB15, HPB20, and HPB30 are 68, 94, and 53 kDa, respectively (Table 1). The chemical structure of the synthesized polyurethanes is characterized using 1H NMR (Fig. S1c, ESI†). The features of the spectra of three SMPUs are in good agreement with each component of polyurethane chains, and the peaks are assigned according to the literature.56 Generally, the peaks that appear between 1 to 2 ppm belong to the methylene groups present in HDI, PCL, and BD. The peak appearing at 7 ppm corresponds to >N–H protons of the hard segment, while the peak appearing at 2.92 ppm, belongs to >N–CH2 protons of the hard segment. The intensities of these peaks increase from HPB15 to HPB30, supporting the increase in hard segment content in SMPUs. Different nanohybrids of SMPUs are prepared by dispersing 0.2, 0.5, 0.75, and 1 wt% of the CuS-PVP nanoparticles in the pure SMPU (HPB20) matrix, and the prepared nanohybrids are designated as HPB20_0.2NH, HPB20_0.5NH, HPB_0.75NH, and HPB20_1NH, respectively. The presence of nanoparticles (NPs) in the SMPU matrix and the interaction of NPs with the polymer matrix is confirmed through the investigation of shifting of the vibrational frequencies of different functional groups in nanohybrids using FT-IR spectroscopy (Fig. S1d, ESI†). The absorption bands appearing at 1731 and 1688 cm−1 are correspond to the stretching frequencies of free carbonyl groups (>C![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) O) of ester and urethane moieties present in SMPU, while the absorption bands appearing at 1717 and 1668 cm−1 are correspond to the stretching frequencies of hydrogen-bonded carbonyl groups of ester and urethane moieties present in SMPU.51 In the nanohybrids (HPB20_0.2NH and HPB20_0.5NH), a single band appears at 1726 cm−1 instead of two individual bands at 1731 and 1717 cm−1 for the ester carbonyl group (>C
O) of ester and urethane moieties present in SMPU, while the absorption bands appearing at 1717 and 1668 cm−1 are correspond to the stretching frequencies of hydrogen-bonded carbonyl groups of ester and urethane moieties present in SMPU.51 In the nanohybrids (HPB20_0.2NH and HPB20_0.5NH), a single band appears at 1726 cm−1 instead of two individual bands at 1731 and 1717 cm−1 for the ester carbonyl group (>C![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) O) and the bands for free and H-bonded urethane carbonyl groups (>C
O) and the bands for free and H-bonded urethane carbonyl groups (>C![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) O) shifts to lower stretching frequencies to 1684 and 1665 cm−1 from 1688 and 1665 cm−1, respectively. Furthermore, the deformation frequency band of the >N–H group of urethane linkage shifts to 1539 from 1546 cm−1 in nanohybrids. This suggests a strong interaction between the nanoparticles and polymer chains, leading to a reduction in the stretching and deformation frequencies of >C
O) shifts to lower stretching frequencies to 1684 and 1665 cm−1 from 1688 and 1665 cm−1, respectively. Furthermore, the deformation frequency band of the >N–H group of urethane linkage shifts to 1539 from 1546 cm−1 in nanohybrids. This suggests a strong interaction between the nanoparticles and polymer chains, leading to a reduction in the stretching and deformation frequencies of >C![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) O and >N–H groups in nanohybrids.57 Thus, the results confirmed the successful synthesis of polycaprolactone–polyurethane copolymers and their hybrids with the nanoparticles.
O and >N–H groups in nanohybrids.57 Thus, the results confirmed the successful synthesis of polycaprolactone–polyurethane copolymers and their hybrids with the nanoparticles.
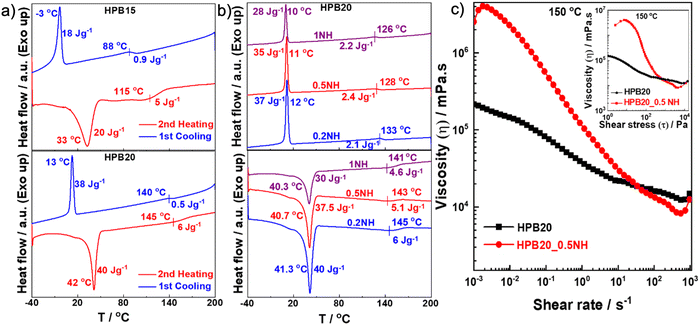
Information such as the precise melting temperature (Tm) of the polymer, the flow and relaxation behavior, moduli near the printing temperature, and the thermal stability or degradation temperature of the polymer are essential for 3D printing of polymers using a melt extrusion-based 3D printer. The melting and crystallization temperatures of different synthesized SMPUs and their nanohybrids are measured through differential calorimetric analysis (DSC) (Fig. 2a, b and Fig. S2, ESI†). It is observed that the melting temperature (and the corresponding heat of fusion value (ΔH)) of the hard segment increases from 101 °C (5 J g−1) to 150 °C (10 J g−1) with increasing hard segment content (%). At the same time, the melting temperature for the soft segment (and the corresponding heat of fusion value (ΔH)) increases from 33 °C (20 J g−1) for HPB15 to 42 °C (40 J g−1) for HPB20 and then decreases slightly to 41 °C (35 J g−1) for HPB30. Hence, the melting temperature increases with an increase in the hard segment content (HSC) of polyurethane. A similar trend is also observed in the case of crystallization temperature and the corresponding heat of fusion (ΔH) of both soft and hard segments. The H-bonding between urethane linkages of the hard segments as well as between the ester >C![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) O group of the soft segment and the amide >N–H group of the hard segment of the polyurethane chains increases with increasing HSC, which helps to improve the melting temperature SMPUs with increasing HSC.56 However, incorporating the NPs in the polymer matrix decreases both the melting and crystallization temperatures for the soft and hard segments of SMPU nanohybrids. The interaction of the polymer chains with dispersed CuS-PVP NPs (cf. evident from FT-IR measurement) is responsible for the decrease in the melting temperature (Tm) in nanohybrids.51,57 Hence, synthesized SMPUs and nanohybrids have two melting and crystallization temperatures, and the value of melting and crystallization temperature increases with increasing hard segment content and decreases with increasing concentration of NPs. The melting of soft segments in the 30–40 °C temperature range will allow the SMPUs to show shape memory behaviors in the range of human body temperatures. In contrast, melting the hard segment at high temperatures facilitates the 3D printing of both SMPUs and nanohybrids.
O group of the soft segment and the amide >N–H group of the hard segment of the polyurethane chains increases with increasing HSC, which helps to improve the melting temperature SMPUs with increasing HSC.56 However, incorporating the NPs in the polymer matrix decreases both the melting and crystallization temperatures for the soft and hard segments of SMPU nanohybrids. The interaction of the polymer chains with dispersed CuS-PVP NPs (cf. evident from FT-IR measurement) is responsible for the decrease in the melting temperature (Tm) in nanohybrids.51,57 Hence, synthesized SMPUs and nanohybrids have two melting and crystallization temperatures, and the value of melting and crystallization temperature increases with increasing hard segment content and decreases with increasing concentration of NPs. The melting of soft segments in the 30–40 °C temperature range will allow the SMPUs to show shape memory behaviors in the range of human body temperatures. In contrast, melting the hard segment at high temperatures facilitates the 3D printing of both SMPUs and nanohybrids.
The flow behavior and moduli of different SMPUs and nanohybrids, which are relevant for processing via 3D printing, are investigated at different temperatures using a rheometer by varying the strain rate (![[small gamma, Greek, dot above]](https://www.rsc.org/images/entities/i_char_e0a2.gif) ), the amplitude of strain (γ), and angular frequency (ω) (Fig. 2c and Fig. S3, ESI†). The investigation of the effect of shear rate on the viscosity of pure HPB20 and its nanohybrid HPB20_0.5NH at 150 °C shows a continuous decrease in viscosity with the increasing shear rate for HPB20, suggesting typical shear thinning behavior due to the breaking of hydrogen bonds between polymer chains and contacts between particles.58,59 Hence, the nanohybrid needs a critical level of stress (yield stress, which is around 50 Pa) to start the flow, and below this critical stress (τy), it behaves like a solid (inset of Fig. 2b) because of the interaction between nanoparticles. This type of flow behavior (high viscosity at low stress) helps the extruded materials retain their shape during printing and increases the resolution of the printed structure.60 The investigation of the effect of amplitude sweeps on the moduli of both the pure polymer and its nanohybrids at 150 °C shows that the storage and loss moduli of pure HPB20 and HPB20_1NH remained linear with an increase in the amplitude of the shear strain and started to decrease after reaching 13% of the shear strain (Fig. S3b, ESI†). Furthermore, in frequency sweep measurements at different temperatures, the storage (G′) and loss (G′′) modulus increases for all pure SMPUs and nanohybrids with increasing angular frequency (ω) as well as with decreasing temperature (Fig. S3c, ESI†). The HPB15 and HPB30 polymers show terminal flow behavior above 130 °C. HPB20 shows a more complex rheological behavior: either G′ or G′′ dominate depending on the frequency due to different kinds of relaxation processes. Interestingly, the increase in G′ was observed upon a decrease in frequency. Any model of viscoelastic behavior cannot explain this behavior. We believe that the origin of this behavior is the re-establishment of hydrogen bonds between polymer chains which occur over time at low frequencies that are measured at the end of the measurement and takes longer to occur. However, the most important observation is that the rheological properties of HPB20 are different from those of HPB15 and HPB30: the last two polymers flow (nearly pure viscous behavior), while HPB20 is viscoelastic. The reason for this difference is the high molecular weight of HPB20, resulting in a large relaxation time of polymer chains and the observation of viscoelastic behavior on the time scale studied by rheology. However, in nanohybrids, especially with a higher concentration of nanoparticles (0.75 and 1 wt%), G′′ remains higher than G′ at 160 and 150 °C within the whole frequency range (0.1 to 100 rad s−1). Crossing between G′ and G′′ happens at lower temperatures (140 °C and 130 °C) because of the lowering of the melting temperature (Tm) in the presence of NPs (cf. evident from the DSC thermogram). It is worth mentioning that all the frequency sweep measurements have been conducted within the linear region of the modulus.
), the amplitude of strain (γ), and angular frequency (ω) (Fig. 2c and Fig. S3, ESI†). The investigation of the effect of shear rate on the viscosity of pure HPB20 and its nanohybrid HPB20_0.5NH at 150 °C shows a continuous decrease in viscosity with the increasing shear rate for HPB20, suggesting typical shear thinning behavior due to the breaking of hydrogen bonds between polymer chains and contacts between particles.58,59 Hence, the nanohybrid needs a critical level of stress (yield stress, which is around 50 Pa) to start the flow, and below this critical stress (τy), it behaves like a solid (inset of Fig. 2b) because of the interaction between nanoparticles. This type of flow behavior (high viscosity at low stress) helps the extruded materials retain their shape during printing and increases the resolution of the printed structure.60 The investigation of the effect of amplitude sweeps on the moduli of both the pure polymer and its nanohybrids at 150 °C shows that the storage and loss moduli of pure HPB20 and HPB20_1NH remained linear with an increase in the amplitude of the shear strain and started to decrease after reaching 13% of the shear strain (Fig. S3b, ESI†). Furthermore, in frequency sweep measurements at different temperatures, the storage (G′) and loss (G′′) modulus increases for all pure SMPUs and nanohybrids with increasing angular frequency (ω) as well as with decreasing temperature (Fig. S3c, ESI†). The HPB15 and HPB30 polymers show terminal flow behavior above 130 °C. HPB20 shows a more complex rheological behavior: either G′ or G′′ dominate depending on the frequency due to different kinds of relaxation processes. Interestingly, the increase in G′ was observed upon a decrease in frequency. Any model of viscoelastic behavior cannot explain this behavior. We believe that the origin of this behavior is the re-establishment of hydrogen bonds between polymer chains which occur over time at low frequencies that are measured at the end of the measurement and takes longer to occur. However, the most important observation is that the rheological properties of HPB20 are different from those of HPB15 and HPB30: the last two polymers flow (nearly pure viscous behavior), while HPB20 is viscoelastic. The reason for this difference is the high molecular weight of HPB20, resulting in a large relaxation time of polymer chains and the observation of viscoelastic behavior on the time scale studied by rheology. However, in nanohybrids, especially with a higher concentration of nanoparticles (0.75 and 1 wt%), G′′ remains higher than G′ at 160 and 150 °C within the whole frequency range (0.1 to 100 rad s−1). Crossing between G′ and G′′ happens at lower temperatures (140 °C and 130 °C) because of the lowering of the melting temperature (Tm) in the presence of NPs (cf. evident from the DSC thermogram). It is worth mentioning that all the frequency sweep measurements have been conducted within the linear region of the modulus.
The thermal stability of polymers is a very important factor for melt extrusion-based 3D printing. The thermal stability of pure shape memory polyurethanes and nanohybrids is investigated using thermogravimetric analysis (TGA) (Fig. S4, ESI†). It is observed that the thermal stability slightly decreases with the increasing concentration of the nanoparticles in the SMPUs. The degradation temperature at 5% degradation is 315 °C for HPB20, which decreases to 304 °C for HPB20_1NH. The thermal degradation of CuS-PVP nanoparticles dispersed in chloroform shows a three-stage degradation profile in which 1st weight loss is responsible for the evaporation of the solvent. In contrast, the 2nd weight loss is responsible for PVP degradation (∼300–400 °C), and the final weight loss is for CuS nanoparticles (∼700 °C). Hence, the SMPUs and nanohybrids are printable around their melting temperature without any thermal degradation.
Dynamical mechanical analysis (DMA) has been performed with increasing temperature at a constant frequency of 1 Hz to understand the phase/relaxation/structural transitions of the programmed shapes of the 3D printed SMPUs and nanohybrids. Fig. 3a and Fig. S5a (ESI†) show changes in the storage (E′) and loss (E′′) modulus for all the SMPUs and nanohybrids with an increase in temperature from 20 °C to 50 °C. A significant lowering of the moduli has been observed within 30 to 40 °C for HPB15, HPB20, and HPB20_0.5NH with a plateau below and above this temperature range, while no significant changes in moduli have been observed for HPB30 within this temperature range (Fig. S5b, ESI†). Hence, a clear transition taking place for HPB15, HPB20, and HPB20_0.5NH within the temperature range of 30 to 40 °C, allowing them to demonstrate the shape memory behavior within the physiological temperature range of most mammals, including humans.51,61 Furthermore, it is also observed that the decrease in moduli in the case of HPB15 and HPB20_0.5NH takes place at a relatively lower temperature as compared to HPB20, supporting the early recovery of the programmed shape of HPB15 and HPB20_0.5NH as compared to HPB20 during the formation of T junctions through the sequential folding of the two components.
In general, in SMPUs, three types of blocks (diisocyanates, diols, and chain extenders) are linked together. These blocks are, however, able to undergo phase segregation. Small-angle X-ray scattering (SAXS) is well-suited for examining the individual small units inside the SMPUs and their hybrids, and it is important for understanding phase separation. For the SAXS study (Fig. 3b and c), samples that show the intended shape-changing behavior in a temperature range suitable for biomedical applications were chosen, specifically HTP15, HTP20, and HTP20_0.5NH. All samples are investigated in the permanent shape (after 3D printing) and in the temporary shape (after 3D printing and programming). An anisotropic scattering pattern is observed in the 2D SAXS diffraction pattern for the permanent shapes of pure SMPUs and nanohybrids, indicating the alignment of the lamellae in a particular direction during 3D printing.62,63 Except for HTP20 in its temporary shape, all samples show a relatively small spatial/angular distribution of lamellae, indicating a high degree of order compared to the mainly randomly dispersed ones of programmed HPB20. Interestingly, the presence of NPs improves the ordering of the lamellae after programming compared to others.64,65 This is to mention that Bragg reflections are predominantly visible at 90° to the print/programming direction. The corresponding 1-dimensional data (1D SAXS, Fig. 3c) show a correlation peak in the range of q ≈ 0.25–1.0 nm−1 (maximum around qmax ≈ 0.4 nm−1), characteristic of the polyurethanes' phase-segregated morphology. According to Bragg's law, the average size of the interdomains is ca. l ≈ 15 ± 3 nm  for all samples. Note that the correlation length (l) value is slightly smaller for the programmed samples compared to that of the corresponding 3D printed ones, indicating alignment of the actuating domain (soft segment) during programming. Comparing HPB20 in its permanent and temporary shape, the alignment of the lamellae is more regular in the permanent shape. Additionally, after adding 0.5 wt% CuS-PVP NPs as fillers to programmed HPB20, the regular alignment of interdomains is significantly enhanced compared to the data of programmed HPB20 (more pronounced Bragg peaks), allowing the formation of smaller but more easily orientable crystalline domains.
for all samples. Note that the correlation length (l) value is slightly smaller for the programmed samples compared to that of the corresponding 3D printed ones, indicating alignment of the actuating domain (soft segment) during programming. Comparing HPB20 in its permanent and temporary shape, the alignment of the lamellae is more regular in the permanent shape. Additionally, after adding 0.5 wt% CuS-PVP NPs as fillers to programmed HPB20, the regular alignment of interdomains is significantly enhanced compared to the data of programmed HPB20 (more pronounced Bragg peaks), allowing the formation of smaller but more easily orientable crystalline domains.
To fabricate T-junctions, individual components are 3D printed using a commercially available pneumatic pressure-assisted melt extrusion-based 3D printer (Fig. S6a and Video SV1, ESI†). The digital model of individual components is prepared using commercial CAD software (Autodesk Fusion360) and modified using the native printer software. The surrounding temperature of the extruder that is required to melt the polymer as well as to maintain the continuous flow of the polymers through the nozzle of melt extrusion-based 3D printing increases from 140 °C for HPB15 to 180 °C for HPB30 (Fig. S6b, ESI†). Hence, the printing temperature increases with the increasing HSC of SMPUs because of an increase in melting temperature from 105 to 150 °C with the increasing HSC of the SMPUs (cf. evident from the DSC thermogram). Furthermore, the thickness of the wall of the 3D printed components increases with the increase in the printing temperature as well as with an increase in the pneumatic pressure (Fig. S6c and d, ESI†). For HPB20, the wall thickness increases from 0.3 mm to 1.1 mm with increasing temperature from 140 °C to 165 °C when pressure is constant. While the wall thickness increases from 0.25 mm to 1.5 mm with increasing pressure from 0.3 mm to 0.7 mm at a constant temperature. It is also observed that the concentration of the nanoparticles (up to 1 wt%) does not affect the printing conditions. The smallest achievable diameter of the components using a nozzle of 350 μm diameter is 2 mm. (Fig. S7a, ESI†). The morphological investigation of the printed structure using a scanning electron microscope shows a microgroove pattern on the surface of the printed structure. Interestingly, the diameter of this individual microgroove is higher (50 μm) in HPB20_0.5NH nanohybrids (Fig. S7b, ESI†) as compared to pure HPB20 (40 μm). The high yield stress in HPB20_0.5NH restricts the spreading of the nanohybrid compared to pure HPB20 after extrusion and helps to retain the diameter of the microgrooves.66 Fig. S6e (ESI†) shows two different 3D-printed components of the T junction, which are hollow tubular self-standing polymeric structures. The bigger component is component 1 (C1) and the smaller component is component 2 (C2). Appropriate arrangements of C1 and C2 can construct a T junction. Furthermore, it is also possible to construct the T junction of different diameters using two individual 3D printed components of different diameters.
After printing the components (C1 and C2) of the T-junction using different shape memory polyurethanes (SMPUs) and their nanohybrids, their shape-changing efficacy was investigated in terms of shape-changing temperature, shape fixity, shape recovery, and recovery kinetics for the successful construction of the T-junction through the sequential folding of the two components. To check the shape-changing behavior of different SMPUs and their nanohybrids, the 3D printed cylindrical hollow components were cut manually and made flat at the abovementioned transition temperature of polymers, and the flat shapes were immobilized by decreasing the temperature below the transition temperature. This whole process is called programming.67 The programmed or temporary shapes started to recover their permanent shape with time in the presence of an appropriate stimulus. Here, the temperature is used as a stimulus for pure SMPUs, while near-infrared light is used as a stimulus in nanohybrids. It is observed that the shape recovery temperature (Tr) for pure SMPUs increases from 33 °C for HPB15 to 70 °C for HPB30 (Fig. S8a, ESI†). Hence, the shape recovery temperature increases with an increase the hard segment content (HSC) of SMPU. Furthermore, the shape recovery efficacy also increases initially from 81% for HPB15 to 90% for HPB20 and then decreases to 87% for HPB30 (Fig. S8b, ESI†). Similarly, the shape fixity increases initially from 84% for HPB15 to 97% for HPB20 and then decreases to 90% for HPB30. Although the shape fixing of HPB20 is also possible at 20 °C, the shape fixing of all the SMPUs has been conducted at 10 °C to achieve maximum shape fixity. Hence, HPB20 shows better shape memory behavior in terms of better shape fixity and a higher recovery ratio as compared to HPB15 and HPB30. The better temporary shape retention efficacy of HPB20 as compared to HPB15 and HPB30 is because of its higher molecular weight, which ensures elastic behavior in a broader time scale range. Furthermore, only HPB15 and HPB20 show shape recovery behavior within the physiological temperature range. In the case of nanohybrids, the recovery of the permanent shape occurs under illumination with light because of the photothermal effect of CuS-PVP nanoparticles. The CuS-PVP nanoparticles produce heat, absorbing near-infrared light (∼800 nm), helping in the melting of the crystalline soft segments (actuating segment) of the SMPUs. Fig. 4a shows that the shape recovery percentage increases from 90% for pure HPB20_0.2NH to 93% for HPB20_0.5NH, and then it decreases to 86% for HPB20_1NH, while the shape fixity continuously decreases from 99% for HPB20_0.2NH to 86% for HPB20_1NH. Hence, the temporary shape retention efficiency of SMPU decreases with the increasing concentration of CuS-PVP NPs. In contrast, the shape recovery efficiency is high at lower concentrations of CuS-PVP NPs, but at high concentrations of NPs, it decreases even more than with pure HPB20. The reason for this trend is the formation of particle network elements with yield behavior – structures formed by particles slow down (high viscosity) and even oppose the recovery of the polymer (the particle network has elastic properties below the yield stress).
The shape recovery kinetics of different nanohybrids in the presence of light is investigated by plotting the recovery percentage of shapes at different time intervals during the recovery process. The shape recovery profile of different nanohybrids with time (Fig. 4b and Fig. S8c, ESI†) shows that HPB20_0.2NH takes 33 seconds to recover the shape while HPB20_1NH takes 12 seconds to recover the shape. Hence, the shape recovery rate increases with the increasing concentration of CuS-PVP NPs in the SMPU matrix – the higher concentration of CuS-PVP NPs produces more heat by absorbing light, thereby helping in the faster recovery process. Furthermore, it is also observed that the shape recovery time increases from 12 seconds to 17 seconds for HPB20_1NH as the distance between the near IR light source and samples increases from 50 cm to 60 cm because of the decrease of the intensity of light with increasing height (Fig. S8d, ESI†). Thus, variations in the amount of nanoparticles and intensity of light can be used to tune the actuation behavior of SMPs.
The optimization of the shape recovery behavior of different SMPUs and nanohybrids offers three possible combinations of SMPUs and nanohybrids to achieve the T shape through the sequential folding of SMPUs. They are as follows: (i) a combination of two pure SMPUs with different transition temperatures; (ii) a combination of one pure SMPU and one nanohybrid, and (iii) a combination of two nanohybrids with different shape recovery kinetics. For all the combinations, the cylindrical hollow components are programmed into flat shapes and arranged perpendicularly, and kept in contact with each other using adhesive at the bottom of the two components before being exposed to heat or light. Fig. S8e (ESI†) shows the flat shape of component 1, which is made of HPB20, while component 2 is made of HPB15, arranged perpendicularly at room temperature (first combination) (Video SV2, ESI†). The gradual increase in temperature from room temperature (22 °C) triggers the folding of component 2, followed by component 1, finally forming the T shape. The fast recovery kinetics of component 2 (made of HPB15) as compared to component 1 is due to the lower melting temperature of the actuating segment (33 °C) of HPB15 as compared to HPB20 (40 °C). However, the limited shape recovery efficiency of C2 (HPB15) results in an incomplete cylindrical shape. In the second combination (Fig. S8f, ESI†), component 1 is made of pure HPB20, and component 2 is made of HPB20_0.5NH. The flat structure of component 2 starts to fold back to its permanent shape on exposure to light, while component 1 completes its folding upon exposure to a hot water bath (40 °C). Hence, the sequential folding of two components on exposure to different stimuli results in a T shape in the end. In the third combination (Fig. 4c and Video SV3, ESI†), on exposure to infrared light, both components' programmed and arranged structures start to fold. The sequential folding of C2 and C1 finally results in a T shape. C2 responds immediately while C1 responds later to light because of the high shape recovery kinetics of HPB20_0.5NH as compared to HPB20_0.2NH in response to infrared (IR) light that helps them to fold sequentially without inhibiting the folding of the other component. Among the different combinations, the combination of two nanohybrids with different recovery kinetics can construct a T-shape more efficiently and completely. Furthermore, their folding behavior can also be controlled remotely. These advantages make the combination of two nanohybrids the best option among three different combinations for constructing T-junctions with cells.
For future biomedical applications, it is very important to investigate the cell-material interactions in terms of cell adhesion, cell proliferation, and cell viability on top of the developed materials. As endothelial cells are responsible for the interior of blood vessels,68 we performed all the in vitro cell culture studies using endothelial cells (HUVECs). Two sets of samples were prepared for cell culture studies: 3D printed flat structures of HPB20 and HPB20_0.5NH were prepared for investigating the cell proliferation and viability, while a 3D printed T-junction was used for investigating the adhesion and monolayer formation of cells inside the T-junction after folding. The proliferation of the endothelial cells with time on the 3D printed flat surface has been investigated using an alamarBlue reduction assay. The intercellular metabolic reduction of resazurin produces a highly fluorescent form of resazurin, and the fluorescence intensity is directly proportional to the number of living cells respiring.69Fig. 5a shows that the fluorescence intensities on day 1 are 43 and 51% for HPB20 and HPB_0.5NH, respectively, increasing to 62 and 67% on day 7. Hence, the metabolic rate of the endothelial cells increases with time for both pure SMPUs and nanohybrids, indicating that cells are proliferating on the surface of both materials. Furthermore, the viability of the endothelial cells on the surface of the pure SMPU and nanohybrid has been investigated using a live–dead assay in which calcein AM is used to stain the live cells. In contrast, ethidium bromide-1 is used to stain the dead cells only. The cell viability (%) has been determined using the number density of live cells with respect to total live and dead cells. Typically, calcein AM is hydrolyzed by the esterase of living cells only, resulting in green fluorescence retained within the cytoplasm of live cells, while ethidium homodimer-1 binds with the nucleic acid of dead cells, producing red fluorescence. Fig. 5b and c show high viability of the endothelial cells on day 7 with slightly higher viability of cells on HPB20_0:5NH (90%) compared to pure HPB20 (85%).
To prove the folding behavior of T-junctions with cells (Fig. 5d), followed by the proliferation of the cells on the inner side of the tube, a 3D printed T-junction was programmed into a flat structure and placed in a Petri dish. It is worth mentioning that for this study, T-junctions are 3D printed using HPB20_0.2NH (C1) and HPB20_0.5NH (C2). Then, HUVACs were seeded only on the top of the flat surface and transferred inside a CO2 incubator for 6 hours to confirm the cells' attachment on the programmed structure's surface. After that, the folding of the programmed flat structure was carried out in the presence of NIR light as stimuli inside a biosafety cabinet. After completion of the folding, the Petri dish is filled with media and placed inside the CO2 incubator. The proliferation of the HUVECs and the monolayer formation inside the T-junction were analyzed by staining the actin filaments and the cell nuclei (Fig. 5e) after culturing the cells on the T-junction for 3 and 7 days. Microscopic imaging of the T-junction reveals the cell-to-cell interaction, and the density of the adherent cells increases with time which is essential for the formation of the HUVEC monolayer.70 Furthermore, scanning the whole T-junction after seven days reveals that cells are distributed homogeneously throughout the tubular T-junction (Fig. 5f). Therefore, the proposed fabrication technique has huge potential and could be used to fabricate an artificial T-junction to replace damaged vascular junctions.
Conclusions
In this paper, we have reported a novel approach for fabricating vascular junctions, particularly the T-junction of the vascular network, using the advanced extension 4D printing technique that resembles modular origami. Our approach permits the construction of a T-junction through coordinated sequential folding of two shape-changing elements with different shape-changing behaviors. To successfully execute our objective, the T-junction was split into two tubular hollow components and 4D printed using synthesized shape-memory polyurethanes (SMPUs) and their blends with nanoparticles. We extensively characterized the properties of the polymers and blends, enabling the identification of optimal conditions to achieve the best printability and appropriate stimuli-responsive properties, which allow coordinated shape transformation, as well as shape fixation and recovery. Here, we demonstrated three approaches to achieve the T-junction through coordinated sequential folding of two components by combining differential shape memory behaviors of different synthesized SMPUs and nanohybrids. Among these approaches, the combination of two different nanohybrids with different shape recovery responses proves to be the most effective and precise in constructing the T-junction compared to other combinations. The used polymers, blends and printed structures were found to be biocompatible, as evidenced by the high cell viability observed. Finally, the T-junction with attached cells has been fabricated through coordinated sequential folding in the presence of NIR light. The histochemical analysis reveals that the cells adhere nicely to the surface of the T-junction and proliferate with time. Hence, the proposed modular-origami-inspired 4D printing approach will open new possibilities for the fabrication of complex structures, which are important not only for tissue engineering in general but also for other technical fields.Author contributions
Arpan Biswas: methodology, formal analysis, investigation, data curation, and writing – original draft. Indra Aspite assisted in cell culture studies. Sabine Rosenfeldt performed and analyzed the X-ray scattering data. Ivita Bite synthesized CuS-PVP nanoparticles. Virginija Vitola: synthesized CuS-PVP nanoparticles. Leonid Ionov: review and editing, project administration, and supervision.Conflicts of interest
There are no conflicts of interest to declare.Acknowledgements
This work was financially supported by TRR 225 project A08, project “Light-activated 4D printed materials” of the Baltic-German University Liaison Office, which is supported by the German Academic Exchange Service (DAAD) with funds from the Foreign Office of the Federal Republic of Germany, DFG IO 68/17-1, DFG IO 68/16-1, IO 68/11-1, IO 68/10-1, and VW Experiment, and lzp-2023/1-0521 project. Institute of Solid State Physics, University of Latvia as the Center of Excellence has received funding from the European Union’s Horizon 2020 Framework Programme H2020-WIDESPREAD-01-2016-2017-TeamingPhase2 under grant agreement No.739508, project CAMART2.References
- M. E. Gomes, M. T. Rodrigues, R. M. A. Domingues and R. L. Reis, Tissue Eng., Part B, 2017, 23, 211–224 CrossRef PubMed.
- I. Apsite, A. Biswas, Y. Li, L. Ionov, I. Apsite, A. Biswas, Y. Li and L. Ionov, Adv. Funct. Mater., 2020, 30, 1908028 CrossRef CAS.
- L. Moroni, J. A. Burdick, C. Highley, S. J. Lee, Y. Morimoto, S. Takeuchi and J. J. Yoo, Nat. Rev. Mater., 2018, 3, 21–37 CrossRef CAS PubMed.
- P. Bajaj, R. M. Schweller, A. Khademhosseini, J. L. West and R. Bashir, Annu. Rev. Biomed. Eng., 2014, 16, 247 CrossRef CAS PubMed.
- C. J. Ferris, K. G. Gilmore, G. G. Wallace and M. in het Panhuis, Appl. Microbiol. Biotechnol., 2013, 97, 4243–4258 CrossRef CAS PubMed.
- G. Constante, I. Apsite, H. Alkhamis, M. Dulle, M. Schwarzer, A. Caspari, A. Synytska, S. Salehi and L. Ionov, ACS Appl. Mater. Interfaces, 2021, 13, 12767–12776 CrossRef CAS PubMed.
- L. Ionov, Adv. Healthcare Mater., 2018, 7, 1800412 CrossRef PubMed.
- P. Zhao, H. Gu, H. Mi, C. Rao, J. Fu and L. Sheng Turng, Front. Mech. Eng., 2017, 13, 107–119 CrossRef.
- J. W. Stansbury and M. J. Idacavage, Dent. Mater., 2016, 32, 54–64 CrossRef CAS PubMed.
- B. Derby, Science, 2012, 338, 921–926 CrossRef CAS PubMed.
- S. V. Murphy and A. Atala, Nat. Biotechnol., 2014, 32, 773–785 CrossRef CAS PubMed.
- X. Zheng, H. Lee, T. H. Weisgraber, M. Shusteff, J. DeOtte, E. B. Duoss, J. D. Kuntz, M. M. Biener, Q. Ge, J. A. Jackson, S. O. Kucheyev, N. X. Fang and C. M. Spadaccini, Science, 2014, 344, 1373–1377 CrossRef CAS PubMed.
- Z. C. Eckel, C. Zhou, J. H. Martin, A. J. Jacobsen, W. B. Carter and T. A. Schaedler, Science, 2016, 351, 58–62 CrossRef CAS PubMed.
- N. W. Bartlett, M. T. Tolley, J. T. B. Overvelde, J. C. Weaver, B. Mosadegh, K. Bertoldi, G. M. Whitesides and R. J. Wood, Science, 2015, 349, 161–165 CrossRef CAS PubMed.
- G. Gao, W. Park, B. S. Kim, M. Ahn, S. Chae, W. W. Cho, J. Kim, J. Y. Lee, J. Jang and D. W. Cho, Adv. Funct. Mater., 2021, 31, 2008878 CrossRef CAS.
- E. Masaeli, V. Forster, S. Picaud, F. Karamali, M. H. Nasr-Esfahani and C. Marquette, Biofabrication, 2020, 12, 025006 CrossRef CAS PubMed.
- L. H. Solis, Y. Ayala, S. Portillo, A. Varela-Ramirez, R. Aguilera and T. Boland, Biofabrication, 2019, 11, 045005 CrossRef CAS PubMed.
- J. H. Kim, I. Kim, Y. J. Seol, I. K. Ko, J. J. Yoo, A. Atala and S. J. Lee, Nat. Commun., 2020, 11, 1–12 CrossRef PubMed.
- A. Kirillova, R. Maxson, G. Stoychev, C. T. Gomillion and L. Ionov, Adv. Mater., 2017, 29, 1703443 CrossRef PubMed.
- H. G. Yi, H. Lee and D. W. Cho, Bioengineering, 2017, 4, 10 CrossRef PubMed.
- Y. Sun, K. Yu, J. Nie, M. Sun, J. Fu, H. Wang and Y. He, Biofabrication, 2021, 13, 035032 CrossRef PubMed.
- M. A. Heinrich, W. Liu, A. Jimenez, J. Yang, A. Akpek, X. Liu, Q. Pi, X. Mu, N. Hu, R. M. Schiffelers, J. Prakash, J. Xie and Y. S. Zhang, Small, 2019, 15, 1805510 CrossRef PubMed.
- N. Hong, G. H. Yang, J. H. Lee and G. H. Kim, J. Biomed. Mater. Res., Part B, 2018, 106, 444–459 CrossRef CAS PubMed.
- B. K. Gu, D. J. Choi, S. J. Park, M. S. Kim, C. M. Kang and C. H. Kim, Biomater. Res., 2016, 20, 1–8 CrossRef PubMed.
- I. Cicha, R. Detsch, R. Singh, S. Reakasame, C. Alexiou and A. R. Boccaccini, Curr. Opin. Biomed. Eng., 2017, 2, 83–89 CrossRef.
- S. Miao, N. Castro, M. Nowicki, L. Xia, H. Cui, X. Zhou, W. Zhu, S. Jun Lee, K. Sarkar, G. Vozzi, Y. Tabata, J. Fisher and L. G. Zhang, Mater. Today, 2017, 20, 577–591 CrossRef CAS PubMed.
- R. L. Truby and J. A. Lewis, Nature, 2016, 540, 371–378 CrossRef CAS PubMed.
- A. Sydney Gladman, E. A. Matsumoto, R. G. Nuzzo, L. Mahadevan and J. A. Lewis, Nat. Mater., 2016, 15, 413–418 CrossRef CAS PubMed.
- The next wave: 4D printing – Atlantic Council, https://www.atlanticcouncil.org/in-depth-research-reports/report/the-next-wave-4d-printing-and-programming-the-material-world/, (accessed 30 May 2022).
- J. Meng, F. Chu, J. Hu and C. Li, Adv. Funct. Mater., 2019, 29, 1902220 CrossRef.
- Y. Wang, H. Cui, T. Esworthy, D. Mei, Y. Wang and L. G. Zhang, Adv. Mater., 2022, 34, 2109198 CrossRef CAS PubMed.
- Q. Ge, H. J. Qi and M. L. Dunn, Appl. Phys. Lett., 2013, 103, 131901 CrossRef.
- V. Stroganov, J. Pant, G. Stoychev, A. Janke, D. Jehnichen, A. Fery, H. Handa and L. Ionov, Adv. Funct. Mater., 2018, 28, 1706248 CrossRef.
- Q. Ge, A. H. Sakhaei, H. Lee, C. K. Dunn, N. X. Fang and M. L. Dunn, Sci. Rep., 2016, 6, 1–11 CrossRef PubMed.
- J. Wu, C. Yuan, Z. Ding, M. Isakov, Y. Mao, T. Wang, M. L. Dunn and H. J. Qi, Sci. Rep., 2016, 6, 1–11 CrossRef PubMed.
- D. Raviv, W. Zhao, C. McKnelly, A. Papadopoulou, A. Kadambi, B. Shi, S. Hirsch, D. Dikovsky, M. Zyracki, C. Olguin, R. Raskar and S. Tibbits, Sci. Rep., 2014, 4, 1–8 Search PubMed.
- G. Constante, I. Apsite, H. Alkhamis, M. Dulle, M. Schwarzer, A. Caspari, A. Synytska, S. Salehi and L. Ionov, ACS Appl. Mater. Interfaces, 2021, 13, 12767–12776 CrossRef CAS PubMed.
- A. Kirillova, R. Maxson, G. Stoychev, C. T. Gomillion and L. Ionov, Adv. Mater., 2017, 29, 1703443 CrossRef PubMed.
- Y. Wang, H. Cui, T. Esworthy, D. Mei, Y. Wang and L. G. Zhang, Adv. Mater., 2022, 34, 2109198 CrossRef CAS PubMed.
- I. Apsite, G. Constante, M. Dulle, L. Vogt, A. Caspari, A. R. Boccaccini, A. Synytska, S. Salehi and L. Ionov, Biofabrication, 2020, 12, 035027 CrossRef CAS PubMed.
- X. Kuang, K. Chen, C. K. Dunn, J. Wu, V. C. F. Li and H. J. Qi, ACS Appl. Mater. Interfaces, 2018, 10, 7381–7388 CrossRef CAS PubMed.
- Y. Zhang, L. Huang, H. Song, C. Ni, J. Wu, Q. Zhao and T. Xie, ACS Appl. Mater. Interfaces, 2019, 11, 32408–32413 CrossRef CAS PubMed.
- S. E. Bakarich, R. Gorkin, M. in het Panhuis and G. M. Spinks, Macromol. Rapid Commun., 2015, 36, 1211–1217 CrossRef CAS PubMed.
- C. M. González-Henríquez, M. A. Sarabia-Vallejos and J. Rodriguez-Hernandez, Prog. Polym. Sci., 2019, 94, 57–116 CrossRef.
- M. Champeau, D. A. Heinze, T. N. Viana, E. R. de Souza, A. C. Chinellato and S. Titotto, Adv. Funct. Mater., 2020, 30, 1910606 CrossRef CAS.
- Y. Dong, S. Wang, Y. Ke, L. Ding, X. Zeng, S. Magdassi and Y. Long, Adv. Mater. Technol., 2020, 5, 2000034 CrossRef CAS.
- Y. Zhang, L. Huang, H. Song, C. Ni, J. Wu, Q. Zhao and T. Xie, ACS Appl. Mater. Interfaces, 2019, 11, 32408–32413 CrossRef CAS PubMed.
- Y. Mao, Z. Ding, C. Yuan, S. Ai, M. Isakov, J. Wu, T. Wang, M. L. Dunn and H. J. Qi, Sci. Rep., 2016, 6, 1–13 CrossRef PubMed.
- A. Sydney Gladman, E. A. Matsumoto, R. G. Nuzzo, L. Mahadevan and J. A. Lewis, Nat. Mater., 2016, 15, 413–418 CrossRef CAS PubMed.
- A. Lendlein and S. Kelch, Shape-memory Polymers, Encyclopedia of Polymer Science and Technology, 2002, vol. 4, pp. 125–136 DOI:10.1002/0471440264.PST336.
- A. Biswas, A. P. Singh, D. Rana, V. K. Aswal and P. Maiti, Nanoscale, 2018, 10, 9917–9934 RSC.
- G. Stoychev, A. Kirillova and L. Ionov, Adv. Opt. Mater., 2019, 7, 1900067 CrossRef.
- D. Hua, X. Zhang, Z. Ji, C. Yan, B. Yu, Y. Li, X. Wang and F. Zhou, J. Mater. Chem. C, 2018, 6, 2123–2131 RSC.
- X. Kuang, D. J. Roach, J. Wu, C. M. Hamel, Z. Ding, T. Wang, M. L. Dunn and H. J. Qi, Adv. Funct. Mater., 2019, 29, 1805290 CrossRef.
- V. Vitola, I. Bite, I. Apsite, A. Zolotarjovs and A. Biswas, J. Polym. Res., 2021, 1, 3, DOI:10.1007/s10965-020-02375-z.
- A. Biswas, V. K. Aswal, B. Ray and P. Maiti, J. Phys. Chem. C, 2018, 122, 11167–11176 CrossRef CAS.
- A. Biswas, V. K. Aswal and P. Maiti, J. Colloid Interface Sci., 2019, 556, 147–158 CrossRef CAS PubMed.
- A. Dodero, M. Castellano, P. Lova, M. Ottonelli, E. Brunengo, S. Vicini and M. Alloisio, Polymers, 2021, 13, 1604 CrossRef CAS PubMed.
- S. A. Wilson, L. M. Cross, C. W. Peak and A. K. Gaharwar DOI:10.1021/acsami.7b13602.
- D. Y. Shin, S. S. Yoo, H. E. Song, H. Tak and D. Byun, Sci. Rep., 2015, 5, 1–7 Search PubMed.
- M. Denda, T. Sokabe, T. Fukumi-Tominaga and M. Tominaga, J. Invest. Dermatol., 2007, 127, 654–659 CrossRef CAS PubMed.
- B. Peng, Y. Yang, K. A. Cavicchi, S. A. Ghodbane, S. Murthy, M. G. Dunn and J. Kohn, Biofabrication, 2019, 11, 045004 CrossRef PubMed.
- J. Liu, W. Li, Y. Guo, H. Zhang and Z. Zhang, Composites, Part A, 2019, 120, 140–146 CrossRef CAS.
- S. Shi, T. Xu, D. Wang and M. Oeser, Polymers, 2020, 12, 1994 CrossRef CAS PubMed.
- H. Koerner, J. J. Kelley and R. A. Vaia, Macromolecules, 2008, 41, 4709–4716 CrossRef CAS.
- Z. Jiang, B. Diggle, M. L. Tan, J. Viktorova, C. W. Bennett and L. A. Connal, Adv. Sci., 2020, 7, 2001379 CrossRef CAS PubMed.
- M. Behl, K. Kratz, J. Zotzmann, U. Nöchel and A. Lendlein, Adv. Mater., 2013, 25, 4466–4469 CrossRef CAS PubMed.
- Dorland's Illustrated Medical Dictionary – Google Books, https://books.google.com/books?hl=en&lr=&id=EGOHxtpN1VAC&oi=fnd&pg=PA20&ots=uuG-t1s0tU&sig=XnOpBueNkK4b5-V0QKKWlBGa1m8#v=onepage&q&f=false, (accessed 3 October 2022).
- J. O’Brien, I. Wilson, T. Orton and F. Pognan, Eur. J. Biochem., 2000, 267, 5421–5426 CrossRef PubMed.
- A. Krüger-Genge, S. Hauser, A. T. Neffe, Y. Liu, A. Lendlein, J. Pietzsch and F. Jung, ACS Biomater. Sci. Eng., 2021, 7, 527–540 CrossRef PubMed.
Footnote |
| † Electronic supplementary information (ESI) available: supporting information is available on the RSC Publications website or from the author. Schematic reaction scheme, GPC, 1H-NMR, FT-IR, of synthesized SMPUs (S1), thermal and viscoelastic properties of SMPUs (S2–S4), DMTA data (S5), 3D printing parameters and printed structures (S6 and S7), self-folding of SMPUs (S8) and videos (VS1 to VS3). See DOI: https://doi.org/10.1039/d4tb00236a |
| This journal is © The Royal Society of Chemistry 2024 |