Open Access Article
Open Access ArticleFluorescent nanodiamond immunosensors for clinical diagnostics of tuberculosis†
Trong-Nghia
Le
 a,
Mhikee Janella N.
Descanzo
b,
Wesley W.-W.
Hsiao
a,
Mhikee Janella N.
Descanzo
b,
Wesley W.-W.
Hsiao
 c,
Po-Chi
Soo
de,
Wen-Ping
Peng
c,
Po-Chi
Soo
de,
Wen-Ping
Peng
 b and
Huan-Cheng
Chang
b and
Huan-Cheng
Chang
 *acf
*acf
aInstitute of Atomic and Molecular Sciences, Academia Sinica, Taipei 106, Taiwan. E-mail: hchang@gate.sinica.edu.tw
bDepartment of Physics, National Dong Hwa University, Shoufeng, Hualien 97401, Taiwan
cDepartment of Chemical Engineering, National Taiwan University of Science and Technology, Taipei 106, Taiwan
dDepartment of Laboratory Medicine and Biotechnology, Tzu Chi University, Hualien 97004, Taiwan
eDepartment of Internal Medicine, Buddhist Tzu Chi General Hospital and Tzu Chi University, Hualien 97004, Taiwan
fDepartment of Chemistry, National Taiwan Normal University, Taipei 106, Taiwan
First published on 13th March 2024
Abstract
Fluorescent nanodiamonds (FNDs) are carbon nanoparticles containing a dense ensemble of nitrogen-vacancy defects as color centers. These centers have exceptional photostability and unique quantum properties, making them useful for ultrasensitive biosensing applications. This work employed FNDs conjugated with antibodies as magneto-optical immunosensors for tuberculosis (TB) diagnostics using competitive spin-enhanced lateral flow immunoassay (SELFIA). ESAT6 (6-kDa early secretory antigenic target) of Mycobacterium tuberculosis is a clinical marker of TB. We evaluated the assay's performance using the recombinant ESAT6 antigen and its antibodies noncovalently coated on FNDs. A detection limit of ∼0.02 ng mL−1 was achieved with the lateral flow membrane strip pre-structured with a narrow channel of 1 mm width. Adopting a cut-off value of 24.0 ng mm−1 for 100-nm FNDs on the strips, the method detected 49 out of 50 clinical samples with Mycobacterium tuberculosis complexes. In contrast, none of the assays for 10 clinical samples with non-tuberculous mycobacteria (NTM) isolates exhibited the presence of ESAT6. These results suggest that the SELFIA platform is applicable for TB detection and can differentiate TB from NTM infections, which also affect the human respiratory system. The FND-enabled immunosensing techniques are versatile and promising for early detection of TB and other diseases, opening a new avenue for biomedical applications of carbon-based nanomaterials.
Introduction
The negatively charged nitrogen-vacancy (NV−) defect in diamond has emerged as one of the most important quantum systems in the solid state since its identification in the 1970s.1 It was first applied to biology in 2005 by Yu et al. who produced fluorescent nanodiamonds (FNDs) containing a dense ensemble of NV− centers by proton irradiation of synthetic type-Ib diamond powders.2 When exposed to green light, these particles emit bright far-red fluorescence (ca. 600–800 nm wavelength). The fluorescence is exceptionally photostable (without photobleaching and blinking), making FNDs well-suited as biolabels for long-term imaging and tracking in cells and organisms.3 Another noticeable feature of the NV− centers is that their ground-state electron spins can be optically polarized, generating “bright” and “dark” states.4 This unique magneto-optical property has allowed selective detection of FNDs by applying a periodic, time-varying magnetic field to induce spin state mixing and thus modulate their fluorescence intensities, followed by lock-in demodulation.5 Similar background-free detection is achievable by microwave modulation of the spin resonances of the NV− centers, which has enabled ultrasensitive diagnosis of disease markers at the subfemtomolar level, as demonstrated by Miller et al.6Recently, by taking advantage of the unique properties of this nanomaterial, we have developed FNDs as high-sensitivity reporters for immunoassays and applied them for rapid antigen testing of infectious diseases such as COVID-19.7,8 The platform, called spin-enhanced lateral flow immunoassay (SELFIA),5,9 selectively detected antibody-conjugated FNDs captured on nitrocellulose membranes. Compared with conventional gold nanoparticle-based lateral flow immunoassays (LFIAs) using naked-eye detection, the SELFIA platform is 100-fold more sensitive and provides more objective and quantitative findings. These advantages have permitted high-sensitivity detection of all four serotypes of Dengue virus nonstructural protein NS1 as well as the nucleocapsid and spike proteins from the SARS-CoV-2 wild-type, Alpha, Delta, and Omicron variants.8,9 This study demonstrates the first application of FND immunosensors to clinical diagnostics of diseases like tuberculosis (TB) using SELFIA.
TB is a highly contagious disease caused by Mycobacterium tuberculosis (M. tuberculosis). While TB primarily affects the lungs (pulmonary TB), it can also impact other body parts (extrapulmonary TB) including the kidneys, bones, and brain.10,11 TB spreads through airborne transmission when an infected person coughs, sneezes or speaks. In 2021, approximately 10.6 million individuals developed TB, compared to 10.1 million in 2020. The number of TB-related deaths in 2021 reached 1.6 million, whereas it was 1.5 million in 2020. The incidence rate of TB increased by 3.6% from 2020 to 2021, marking a reversal from the nearly 2% annual decrease observed over the past two decades.10,12 TB is treatable with medication, but it is challenging to manage, and the emergence of drug-resistant strains is a growing concern. Although it was difficult to estimate the burden of TB during the COVID-19 pandemic, the number of undiagnosed and untreated cases increased, leading to a rise in community transmission and TB-related deaths.10 Vaccines for TB are available; however, effective public health measures like early diagnosis and proper treatment are crucial in controlling the disease spread.
M. tuberculosis secretes ESAT6 (early secretory antigenic target, 6 kDa) and CFP-10 (culture filtrate antigen, 10 kDa),13–15 two critical virulence factors that play vital roles in TB infection and immunity. Both proteins (ESAT6 and CFP-10) are considered ideal biomarkers, as their detection in clinical patient samples can be used to determine if a patient has TB.16 Additionally, they are not present in the Bacillus Calmette-Guérin (BCG) vaccine, which is widely used to prevent TB.17 There are two kinds of tests to detect TB infection: the TB skin test and TB blood tests.18 In the tuberculin skin test, a small amount of purified proteins from dead TB bacteria is first injected into the upper layer of the skin, and measurements are later made for the size of the lump that forms within 48–72 hours after the injection whereas for the blood tests, ESAT6, CFP10, and other TB antigens are detected through lymphocyte stimulation, and the levels of interferon-gamma (IFN-γ) released from the lymphocytes in patients’ blood are measured. Although both tests are highly sensitive, they demand specialized laboratory facilities and personnel with sufficient training. This prevents their accessibility in resource-limited settings where TB is common. Therefore, developing a rapid, cost-effective, and reliable method for point-of-care diagnostics of TB remains a formidable challenge. Quantitative LFIAs using FNDs as photostable reporters offer a promising and effective solution.
The FNDs used in this work were ∼100 nm in size and contained ∼10 ppm NV centers. We first demonstrated that the fluorescence properties of FNDs are perfectly stable, unaffected by the coating of antibodies on their surface. We next optimized the immunosensor production, assay buffer conditions, and incubation times and investigated the effect of membrane configurations on the assays. Finally, a performance evaluation of the SELFIA platform for TB diagnostics was conducted using clinical samples. The clinical samples comprised 50 culture supernatants with M. tuberculosis complexes (MTC) and 10 culture supernatants with non-tuberculous mycobacteria (NTM) isolates.19,20 By utilizing the excellent photostability of the FND reporters, we established a cut-off value of 24.0 ng mm−1 to categorize the result as “positive” or “negative” for TB screening or diagnostics using competitive SELFIA for the first time. Fig. 1 illustrates the working principle of the assay, which is rapid, simple, sensitive, specific, and quantitative. It adds a new dimension to the clinical detection of TB using carbon nanoparticles (such as carbon nanotubes and graphene oxides) targeting different immunogens.21,22
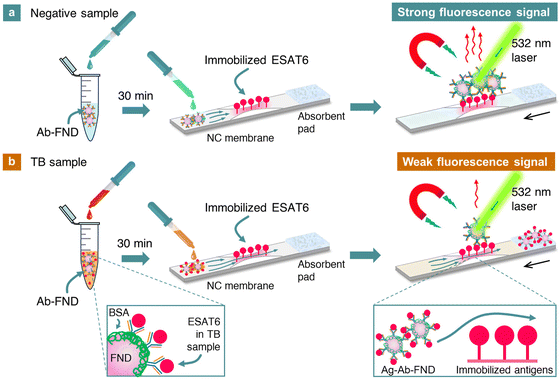 | ||
| Fig. 1 (a) and (b) Illustration of the detection of ESAT6 from M. tuberculosis in clinical samples by competitive SELFIA. The nitrocellulose membrane strip is scanned through the laser beam with a diameter of ∼0.6 mm using a motorized translation stage to obtain its fluorescence intensity line profile (Fig. S1, ESI†). Background-free detection is achieved through magnetic modulation of the fluorescence signals. The black arrows indicate the direction of the strip movement. The measured fluorescence intensity is expressed in terms of FND weight per unit area (ng mm−2). Abbreviations: Ab: antibody, Ag: antigen, NC: nitrocellulose. The principle of this competitive assay is based on the competition of ESAT6 in the sample and ESAT6 on the strip for a limited number of Ab binding sites on FNDs in the buffer. | ||
Results and discussion
FND immunosensors
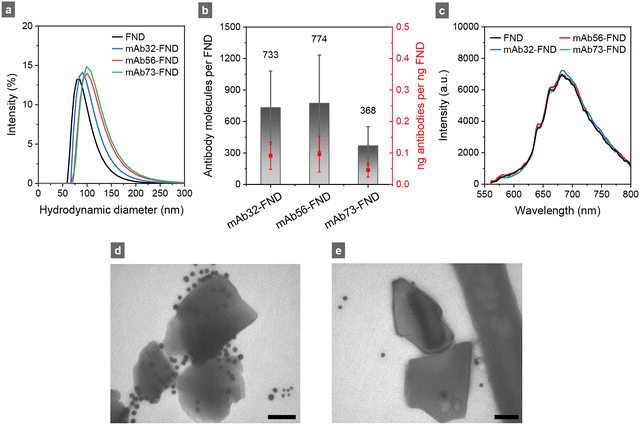
Fig. 2a shows the size distribution of FNDs after surface modifications by air oxidation and oxidizing acid washes.5,8,9 As is well documented in the literature,23–25 these nanodiamond particles possess a high binding affinity for proteins due to the abundance of various oxygen-containing groups on their surface, including –COOH, –COH, and –COOC–,26 which can interact noncovalently with the neutral, acidic, and basic moieties of antibodies at the interface to form stable antibody-FND complexes. In this study, three mouse monoclonal antibodies (mAb32, mAb56, and mAb73) were coated separately onto FNDs for testing. The goal was to find the antibodies with the highest possible specificity for ESAT6 detection. This was achieved by incubating FNDs with anti-ESAT6 at the weight ratio 5![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1 for 20 min, then covering the uncoated sites with bovine serum albumin (BSA) and washing them with phosphate-buffered saline (PBS). As shown in Fig. 2a, the average sizes of these particles increased by approximately 20 nm after the antibody conjugation, and their zeta potentials changed by approximately 15 mV, both suggesting successful antibody conjugation on the FND surface. Fourier-transform infrared spectra of bare FNDs and these antibody-conjugated FNDs supported the suggestion (Fig. S2 of the ESI†).
1 for 20 min, then covering the uncoated sites with bovine serum albumin (BSA) and washing them with phosphate-buffered saline (PBS). As shown in Fig. 2a, the average sizes of these particles increased by approximately 20 nm after the antibody conjugation, and their zeta potentials changed by approximately 15 mV, both suggesting successful antibody conjugation on the FND surface. Fourier-transform infrared spectra of bare FNDs and these antibody-conjugated FNDs supported the suggestion (Fig. S2 of the ESI†).
A quantification for the number of antibodies attached to the FND surface involved measuring the remaining antibodies in the supernatant through ultraviolet absorbance at 280 nm, assuming a molar extinction coefficient of 2.1 × 105 cm−1 M−1 for IgG antibodies.27,28 The results in Fig. 2b demonstrated the efficient binding of all three antibodies (mAb32, mAb56, and mAb73) to FNDs through noncovalent interactions. The antibody loadings were about 0.04–0.1 ng ng−1 FND or 300–700 molecules per FND, assuming a spherical shape for the nanoparticles. Fig. 2c compares the fluorescence spectra between bare and antibody-conjugated FNDs suspended in distilled deionized water at the same concentration (0.1 mg mL−1). Interestingly and importantly, no significant changes in the fluorescence intensity of the particles before and after coating with the three antibodies were found. This is because most (∼75%) of the fluorescent NV centers are deeply embedded in the diamond matrix and are separated from the surface by more than 5 nm. Therefore, the light-emitting properties of FNDs are not really affected by environmental changes, such as the changes in composition of the culture medium.
To further confirm the attachment of antibodies onto the FND surface, we employed an immunogold staining technique to directly visualize the events using goat anti-mouse IgG-10 nm colloidal gold as the label. Using a transmission electron microscope (TEM) operating at 25 kV, we unambiguously identified the 10 nm gold nanoparticles surrounding the mAb56-FND complexes (Fig. 2d). In contrast, no significant colloidal gold attachment was found for FNDs mixed with BSA without mAb56 (Fig. 2e). These results indicated that the antibodies could bind spontaneously with surface-oxidized and carboxylated FNDs via noncovalent interactions. Moreover, BSA is highly effective in preventing undesired non-specific interactions from occurring.
Direct and competitive SELFIA
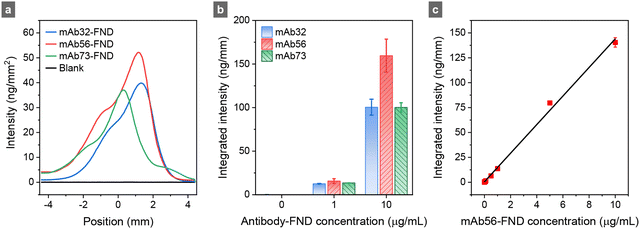
Following material characterization, we searched for the optimal monoclonal antibodies against ESAT6 using direct SELFIA (Fig. S3, ESI†), analogous to the direct enzyme-linked immunosorbent assay (ELISA). The experimental setup is shown in Fig. S1 (ESI†) and has been detailed in ref. 5. The assay was performed by immobilizing an excess amount (0.75 μg) of recombinant monomeric ESAT6 on the nitrocellulose membrane, followed by adding a known amount of antibody-conjugated FNDs to the strip without antigens. Fig. 3a illustrates three representative fluorescence intensity line profiles of antibody-conjugated FNDs captured by the immobilized ESAT6. The similarity in profile implies that the affinity of these three antibodies (mAb32, mAb56, and mAb73) for the antigen is comparable. Their integrated fluorescence intensities increased as more antibody-conjugated FNDs were added to the strips (Fig. 3b). Notably, mAb56-FND exhibited the highest fluorescence intensity among all conjugates tested, showing a linear response over a wide concentration range of 0–10 μg mL−1 (Fig. 3c). We chose it as the immunoreagent in subsequent experiments with competitive SELFIA (Fig. 1).The competitive SELFIA for ESAT6 involved incubating the sample solution (50 μL) with mAb56-FNDs prior to adding the mixture to the test strip. For the ESAT6-negative samples, all the antibodies attached to FNDs are available for capturing by antigens immobilized on the strip, giving rise to the highest possible fluorescence signal. However, if the sample contained ESAT6, which could bind with antibodies on the FND surface, the capture efficiency of mAb56-FNDs by ESAT6 on the strip was reduced, yielding a diminished or no fluorescence signal. The magnitude of the signal reduction (not increase) is directly correlated with the concentration of ESAT6 in the sample. To optimize the assay performance, we varied the experimental conditions, including the weight ratios of antibodies to FNDs, the incubation times of mAb56-FNDs with the samples, and the types of running and washing buffers. Results are presented in the ESI† (Fig. S4 and S5), which led us to adopt the following conditions in the ensuing experiments: 100 ng mAb56-FNDs prepared at the 1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 5 weight ratio as the immunoreagents, an incubation time of 30 min between mAb56-FNDs and the sample, 3% BSA/PBS as the running buffer, and 0.2% BSA + 0.2% Tween 20 (wt%) in acetate buffer pH 5 as the washing buffer. Compared with the commonly used sandwich immunoassay for rapid antigen testing, the competitive assay does not need a highly specific antibody pair; moreover, it has a wider dynamic detection range and no Hook effect at high antigen concentrations.29
5 weight ratio as the immunoreagents, an incubation time of 30 min between mAb56-FNDs and the sample, 3% BSA/PBS as the running buffer, and 0.2% BSA + 0.2% Tween 20 (wt%) in acetate buffer pH 5 as the washing buffer. Compared with the commonly used sandwich immunoassay for rapid antigen testing, the competitive assay does not need a highly specific antibody pair; moreover, it has a wider dynamic detection range and no Hook effect at high antigen concentrations.29
Sensitivity enhancement
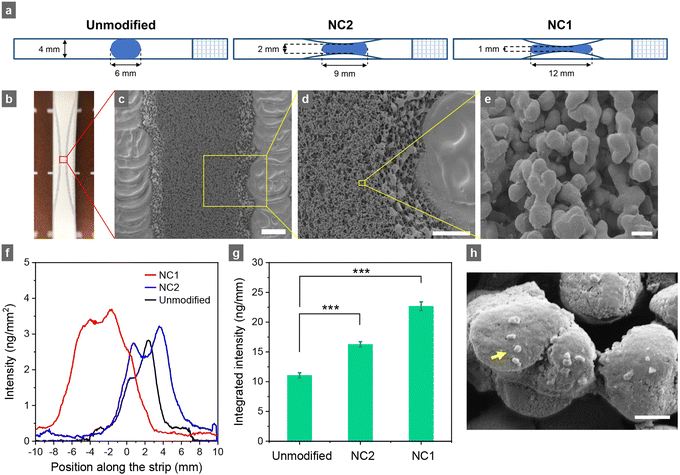
After optimizing assay conditions, we examined whether detection sensitivity could be improved by altering the nitrocellulose membrane configuration (such as the width).30,31 In conventional studies and commercial setups, a nitrocellulose membrane strip of 4 mm wide is typically used (Fig. 4a). However, as the diameter of the laser beam to probe FND reporters on the strips in our home-built SELFIA reader is only about 0.6 mm (Fig. 1), the region of interest can be reduced to 1 mm wide to enhance the sensitivity. Fig. 4a depicts the configurations of three strips: an unmodified strip, a 2 mm-wide narrow-channel (NC2) strip, and a 1 mm-wide narrow-channel (NC1) strip, all created on 4 mm strips using laser engraving. By precisely regulating the intensity of a CO2 laser and its engraving speed, parts of the nitrocellulose membrane were removed without harming the active region. Fig. 4b shows a photograph of the pre-structured NC1 strip. Scanning electron microscopy (SEM) imaging of the strip revealed a highly porous architecture of the nitrocellulose membrane in the narrow channel post-laser engraving (Fig. 4c–e), with a distinct boundary in which the nitrocellulose material was completely removed. Although there were slight structural changes at the edges of the narrow channels, the membrane at the center maintained its original structure and porosity, affirming the preservation of its functionality.In Fig. 4f and g, we display the fluorescence intensity line profiles and integrated fluorescence intensities of mAb56-FNDs captured by ESAT6 immobilized on the strips with different formats by direct SELFIA. SEM also verified the capture of these antibody-conjugated FNDs on the strips (Fig. 4h). Although the heights of the line profiles in Fig. 4f did not change substantially across the unmodified, NC2, and NC1 strips, the bands were broader as the channels became narrower (from 4 mm to 1 mm wide). Consequently, the total signal intensity increased when the channel width decreased. The result is attributed to a concentration effect, where more mAb56-FNDs are forced to pass through the narrow channel at the strip center. Therefore, they can be more readily captured by the immobilized ESAT6 and also excited by the laser beam when the strip is scanned over to obtain the fluorescence intensity line profile (Fig. 1). However, the signal enhancement was less than a factor of 4. This is because the flow rate increased roughly 2-fold in the narrow channel region (Video S1, ESI†),30 which decreased the binding efficiency between mAb56-FNDs and the immobilized ESAT6. Such a signal enhancement effect was further verified by using other biomarkers, e.g., the non-structural protein NS1 of the Dengue virus (Fig. S6, ESI†), suggesting the potential use of these pre-structured membrane strips to enhance the performance of LFIA.
Clinical detection
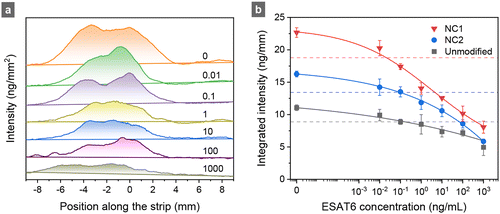
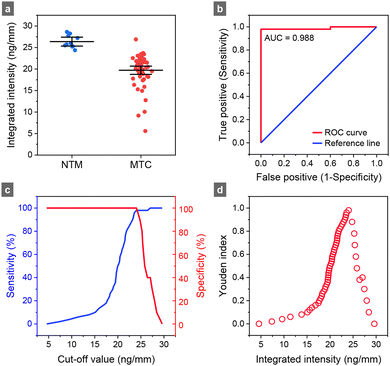
To quantitatively assess the performance of competitive SELFIA of clinical samples, calibration curves were first constructed for the recombinant ESAT6 using unmodified, NC2, and NC1 test strips. As shown in Fig. 5a for the NC1 strips, the fluorescence intensity gradually decreased as the ESAT6 concentration increased from 0 to 1000 ng mL−1. Consistently across all samples, NC1 strips exhibited a higher fluorescence intensity than the NC2 and the original 4 mm wide strips (Fig. 5b). Each experimental data set could fit well with a four-factor logistic function. Following the definitions of LOB (limit of blank) = mean + 1.645 × (SD of blank), n = 10, and LOD (limit of detection) = LOB + 1.645 × (SD of the low concentration sample, 0.01 ng mL−1), n = 3,32 we obtained LOD = 0.19, 0.10, and 0.02 ng mL−1 of the competitive SELFIA for ESAT6 using unmodified, NC2, and NC1 strips, respectively.Next, we applied competitive SELFIA to detect ESAT6 in 50 MTC culture-positive and 10 NTM samples using the NC1 strips. Procedures for sputum sample collection and bacterial identification have been previously reported19,20 and are described briefly in the Materials and methods section. Fig. 6 and Fig. S7 (ESI†) show the results of the assays and associated data analysis. The average values of the integrated intensities of mAb56-FNDs captured on the strips were 26.4 ng mm−1 (95% confidence interval: 25.3–27.4 ng mm−1) for the NTM samples and 19.7 ng mm−1 (95% confidence interval: 18.8–20.7 ng mm−1) for the MTC samples (Fig. 6a). The apparent positive detection rate was 98% (i.e., 49/50). Various analytical methods can be applied to assess the assay's efficacy, including receiver operating characteristic (ROC) analysis, area under the curve (AUC), optimal cut-off value determination, and Youden index calculation. The ROC curve plots the true positive rate (sensitivity) against the false positive rate (1 − specificity) at various threshold settings (Fig. 6b). The curve provides a visual tool to assess the trade-off between sensitivity and specificity and helps determine the optimal cut-off value for classification. A higher AUC value indicates a better discriminatory power of the test, with an AUC of 1 representing a perfect classifier and 0.5 for a random classifier. The Youden index, calculated as J = sensitivity + specificity − 1, is also derived from the ROC curve. It quantifies the effectiveness of a diagnostic test or biomarker by maximizing the difference between the true positive rate and the false positive rate. A higher Youden index value (ranging from 0 to 1) indicates a better overall diagnostic test performance, with 1 representing a perfect test. Fig. 6c and d show that the AUC of the present assays was 0.988, and the corresponding Youden index was 0.98 at the cut-off value of 24.0 ng mm−1. Considering that FND is perfectly photostable and its fluorescence intensity is a reliable quantitative measure, this cut-off value (e.g., 24.0 ng mm−1 for 100-nm FNDs) may be generally applicable for TB screening or diagnostic tests using competitive SELFIA. To validate the method further, a systematic investigation of the inter-assay and intra-assay coefficients of variation using more clinical samples is currently in progress.
Comparison with other assays
TB is highly contagious, primarily affecting the lungs and other body parts. The global burden of TB is increasing, with millions of new cases and deaths reported annually. Drug resistance complicates treatment efforts. In this context, the detection of specific biomarkers like CFP10 and ESAT6 is critical for facilitating the diagnostics of active TB. Recently, various types of assays such as immuno-polymerase chain reaction,33 ELISA,34,35 LFIA,36,37 mass spectrometry,19,20,38 electrochemiluminescence,39 and electrochemical immunosensing40–44 have been developed for ESAT6/CFP10 detection. Specifically, Omar et al. demonstrated the use of a Ni/polyaniline/reduced graphene oxide electrode in electrochemical immunosensors, achieving an LOD of 1.0 ng mL−1 for ESAT6.40 Mohd Bakhori et al. utilized quantum dot-silica nanoparticles on a screen-printed carbon electrode, achieving a LOD value of 0.15 ng mL−1 for ESAT6/CFP10 detection.43 Although these electrochemical immunoassays are highly sensitive, clinical validation is needed prior to their applications in practical settings. Other assays, such as the aptamer-based quantitative polymerase chain reaction by Kil et al., showed 50.0% sensitivity, 91.8% specificity, and 78.1% accuracy, comparable to that of sputum smear microscopy.33 An ESAT6-specific ELISA of culture supernatants also enabled a sensitivity of 95.4% and a specificity of 100%.34 However, these assays are more time-consuming and require more specific equipment than LFIAs. In this work, the LOD measured for ESAT6 with the improved SELFIA platform is 0.02 ng mL−1, approximately 300-fold lower than the reported value of LOD = 6.0 ng mL−1 with a colloidal gold-based LFIA.36 Table S1 (ESI†) compares the performance of competitive SELFIA with other reported assays for EAST6.The NTM family comprises approximately 170 species of mycobacteria, including M. avium complex, M. kansasii, and M. abscessus, as the causative agents of pulmonary diseases in humans.45 NTM infections are primarily acquired from the environment, although the exact transmission mode remains unclear. In addition to affecting the lungs, NTM infections commonly involve the lymphatic system, skin, and soft tissues.46 Distinguishing between TB and NTM species in clinical specimens is challenging as both show positive results in conventional smear acid-fast staining.47 Consequently, the incidence of NTM has often been underestimated in many countries with a high burden of TB. In this work, with the SELFIA platform operated in a competitive mode and a cut-off value set at 24.0 ng mm−1, 49 out of 50 MTC samples were successfully detected as ESAT6-positive. In contrast, none of the assays for 10 NTM samples exhibited the presence of ESAT6. The clinical sensitivity and specificity were 98% and 100%, respectively.
Conclusions
We have demonstrated the potential use of antibody-conjugated FNDs as immunosensors for detecting disease markers (such as ESAT6) in clinical samples. Our findings strongly suggest that the nanomaterial-based SELFIA platform is a promising analytical tool for differentiating between TB and NTM infections. Notably, the observed high sensitivity and low LOD (∼0.02 ng mL−1) prove the assay's capacity to detect ESAT6 even at extremely low concentrations, which is of utmost importance in facilitating early diagnosis. The SELFIA reader is an easy-to-use device that requires no special training and skill. With the help of further technological advancements, the FND-enabled platform is expected to find practical applications in the field of in vitro diagnostics and can be used to monitor a wide range of diseases in point-of-care settings and clinics.Experimental section
Chemicals and materials
Recombinant M. tuberculosis ESAT6 protein (GTX57459-pro) and M. tuberculosis ESAT6 mouse monoclonal antibodies, GTX40232 (mAb32) and GTX36773 (mAb73), were purchased from GeneTex. M. tuberculosis ESAT6 mouse monoclonal antibody MBS312556 (mAb56) was obtained from MyBioSource. Goat anti-mouse IgG-10 nm colloidal gold (A-31561) was purchased from Thermo Fisher Scientific. BSA, PBS, and all other chemicals were from Sigma Aldrich.Preparation of antibody-conjugated FNDs
FNDs were produced by ion irradiation of synthetic type-Ib diamond powders, followed by vacuum annealing at 800 °C, air oxidation at 450 °C, and concentrated sulfuric-nitric acid washes at 100 °C, as detailed previously.48,49 Through physical adsorption, they were conjugated separately with three antibodies, including mAb32, mAb56, and mAb73. Each conjugation was made by mixing FNDs (1 mg mL−1) and the antibodies of interest at various weight ratios (Ab![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) FND = 1
FND = 1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 5 to 1
5 to 1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 15) and then incubating the mixture at room temperature for 20 min. Afterward, an aqueous solution containing 3% BSA was added to block empty sites on the FND surface without washing away unbound antibodies. Following centrifugal isolation of the antibody-FND complexes at 20
15) and then incubating the mixture at room temperature for 20 min. Afterward, an aqueous solution containing 3% BSA was added to block empty sites on the FND surface without washing away unbound antibodies. Following centrifugal isolation of the antibody-FND complexes at 20![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 000 × g for 5 min, the pellets were dispersed in 3% BSA/PBS to a final concentration of 100 μg mL−1. A microplate reader (Spark, Tecan) operating at 280 nm measured the changes in absorbance of free antibody molecules in distilled deionized water before and after mixing with FNDs to determine the antibody loading on the solid-phase supports. The same reader was applied to measure the changes in the fluorescence intensity of FNDs with or without antibody conjugation by photoexcitation at 530 nm.
000 × g for 5 min, the pellets were dispersed in 3% BSA/PBS to a final concentration of 100 μg mL−1. A microplate reader (Spark, Tecan) operating at 280 nm measured the changes in absorbance of free antibody molecules in distilled deionized water before and after mixing with FNDs to determine the antibody loading on the solid-phase supports. The same reader was applied to measure the changes in the fluorescence intensity of FNDs with or without antibody conjugation by photoexcitation at 530 nm.
Characterization of antibody-conjugated FNDs
Immunogold staining was used to validate the noncovalent conjugation between antibodies and FNDs. Briefly, goat anti-mouse IgG-10 nm colloidal gold (5 μL) was mixed with the mAb56-FND suspension (5 μL, 200 μg mL−1) and then incubated for 60 min at room temperature. Subsequently, the mixture was dropped on a copper grid with an ultrathin carbon film supported by a Lacey carbon film (Ted Pella) and incubated for 20 min. The liquid was then removed, and the grid was washed twice with distilled deionized water and dried in a vacuum. The same procedures were applied to BSA-conjugated FNDs, which served as the control. TEM images of the nanoparticle bioconjugates were acquired using a low-voltage electron microscope (LVEM25, Delong) operating at 25 kV.Fabrication of pre-structured membrane strips
Narrow channels on nitrocellulose membranes (FF120HP plus, Millipore) were configured with a 30 W CO2 laser engraving platform (Beamo, Flux). Parameters such as laser power and engraving speed were carefully optimized to minimize membrane damage while exclusively removing unwanted nitrocellulose materials. The optimal laser power was 18%, and the engraving speed was 120 mm s−1. After the configuration, an absorption pad was assembled at the far end of the laser-etched membrane. The resulting membranes were cut into 4 mm wide strips and stored in a desiccator at room temperature until use. A field-emission scanning electron microscope (JSM-7800F, JEOL) operating at an accelerating voltage of 5 kV examined the changes in the membrane's architecture before and after laser engraving as well as the capture of antibody-conjugated FNDs on the strips.Direct and competitive SELFIA
Test strips, each consisting of a modified or unmodified nitrocellulose membrane and an absorbent pad, were assembled in plastic cassettes. To carry out direct SELFIA, a drop of the ESAT6 solution (1.5 μL and 0.5 mg mL−1) was first deposited at the center of the membrane strip and then air-dried at room temperature. Following pre-wetting of the test strip with 3% BSA/PBS (30 μL), antibody-conjugated FND solution (50 μL, 2 μg mL−1) was suspended at the front end of the membrane. The strip was then left in air at room temperature for 10 min and added with 0.2% BSA + 0.2% Tween 20 in acetate buffer pH 5 (50 μL) as the washing buffer to complete the assay. The strips (with their cassette caps removed) were finally dried in a microwave oven for 1 min before fluorescence measurements were taken using a home-built SEFLIA reader, as described in previous reports.5,8,9 The same protocols were applied for competitive SELFIA, except that the antibody-conjugated FND solution (10 μL, 10 μg mL−1) was first mixed with the ESAT6 solution (50 μL) prepared at different concentrations (0–1000 ng mL−1) in 3% BSA/PBS for 30 min at room temperature before each assay.Clinical samples
Clinical sputum samples were originally collected at Buddhist Tzu Chi General Hospital for nucleic acid amplification tests on reducing delayed diagnosis and misdiagnosis of TB, as previously reported.50 These samples were used again in this study with the dataset containing no information that could be connected to the identity of the patients. In brief, sputum samples were collected, liquefied, and decontaminated using the standard N-acetyl-L-cysteine-sodium method, followed by inoculation of the specimens in an automated liquid culture system (BACTEC MGIT 960, Becton Dickinson).19,20,50 The samples were incubated at 37 °C for 5–14 days, and the growth of bacteria was automatically monitored until a positive signal was shown. The supernatant of the culture medium was then filtered. An aliquot of the filtrate was examined by using conventional methods such as microscopy, subcultures, and biochemical identification, including niacin, nitrate reduction, and 68 °C catalase tests.51 Another aliquot of the same filtrate was tested with competitive SELFIA.Protocols for clinical detection
Clinical samples, including MTC and NTM, were employed to investigate the sensitivity and specificity of the FND-based competitive SELFIA for TB. First, the sample solution (50 μL) was incubated with antibody-conjugated FNDs (10 μL and 10 μg mL−1) for 30 min. Subsequently, 3% BSA/PBS (40 μL) was added to the mixed solution. Prior to the assays, test strips immobilized with ESAT6 were pre-wetted with 3% BSA/PBS (30 μL). The sample/antibody-conjugated FND mixtures were then added to the test strips and allowed to run for 10 min, followed by washing the strips with the acetate buffer containing 0.2% BSA and 0.2% Tween 20 (pH 5, 50 μL) to complete the assay. After drying the test strips in a microwave oven, a home-built SELFIA reader obtained the fluorescence intensity line profiles of FNDs on the nitrocellulose membranes.Data analysis
SPSS 20.0 software was used to determine statistical significance via one-way analysis of variance (ANOVA). All the values were presented as means ± standard deviations (SDs). Additionally, the same method was applied to assess the accuracy of the assay for clinical samples by receiver operating characteristic (ROC) curve analysis.Author contributions
Trong-Nghia Le: conceptualization, methodology, formal analysis, investigation, writing – original draft, writing – review & editing, visualization. Mhikee Janella N. Descanzo: investigation, resources, writing – review & editing. Wesley W.-W. Hsiao: writing – review & editing. Po-Chi Soo: resources, writing – review & editing. Wen-Ping Peng: resources, writing – review & editing. Huan-Cheng Chang: conceptualization, methodology, formal analysis, writing – original draft, writing – review & editing, supervision, funding acquisition.Conflicts of interest
There are no conflicts to declare.Acknowledgements
This work was supported by Academia Sinica and the Ministry of Science and Technology, Taiwan, under Grant No. 108-2923-B-001-001-MY3 and 110-2113-M-001-046-MY3. We thank Dr Chih-Bin Lin and Dr Jen-Jyh Lee at Buddhist Tzu Chi General Hospital for providing clinical samples.Notes and references
- G. Davies and M. F. Hamer, Proc. R. Soc. London, Ser. A, 1976, 348, 285 CAS.
- S.-J. Yu, M.-W. Kang, H.-C. Chang, K.-M. Chen and Y.-C. Yu, J. Am. Chem. Soc., 2005, 127, 17604 CrossRef CAS PubMed.
- Y. Y. Hui, W. W.-W. Hsiao, S. Haziza, M. Simonneau, F. Treussart and H.-C. Chang, Curr. Opin. Solid State Mater. Sci., 2017, 21, 35 CrossRef CAS.
- R. Schirhagl, K. Chang, M. Loretz and C. L. Degen, Annu. Rev. Phys. Chem., 2014, 65, 83 CrossRef CAS PubMed.
- Y. Y. Hui, O. J. Chen, H.-H. Lin, Y.-K. Su, K. Y. Chen, C.-Y. Wang, W. W.-W. Hsiao and H.-C. Chang, Anal. Chem., 2021, 93, 7140 CrossRef CAS PubMed.
- B. S. Miller, L. Bezinge, H. D. Gliddon, D. Huang, G. Dold, E. R. Gray, J. Heaney, P. J. Dobson, E. Nastouli, J. J. L. Morton and R. A. McKendry, Nature, 2020, 587, 588 CrossRef CAS PubMed.
- W. W.-W. Hsiao, T.-N. Le, D. M. Pham, H.-H. Ko, H.-C. Chang, C.-C. Lee, N. Sharma, C.-K. Lee and W.-H. Chiang, Biosensors, 2021, 11, 295 CrossRef CAS PubMed.
- W. W.-W. Hsiao, N. Sharma, T.-N. Le, Y.-Y. Cheng, C.-C. Lee, D.-T. Vo, Y. Y. Hui, H.-C. Chang and W.-H. Chiang, Anal. Chim. Acta, 2022, 1230, 340389 CrossRef PubMed.
- T.-N. Le, W. W.-W. Hsiao, Y.-Y. Cheng, C.-C. Lee, T.-T. Huynh, D. M. Pham, M. Chen, M.-W. Jen, H.-C. Chang and W.-H. Chiang, Anal. Chem., 2022, 94, 17819 CrossRef CAS PubMed.
- World Health Organization, Global Tuberculosis Report 2022, World Health Organization, 2022, p. 68 Search PubMed.
- A. H. Baykan, H. S. Sayiner, E. Aydin, M. Koc, I. Inan and S. M. Erturk, Insights Imaging, 2022, 13, 39 CrossRef PubMed.
- S. Bagcchi, Lancet Microbe, 2023, 4, e20 CrossRef PubMed.
- A. L. Sørensen, S. Nagai, G. Houen, P. Andersen and A. B. Andersen, Infect. Immun., 1995, 63, 1710 CrossRef PubMed.
- F.-X. Berthet, P. B. Rasmussen, I. Rosenkrands, P. Andersen and B. Gicquel, Microbiology, 1998, 144, 3195 CrossRef CAS PubMed.
- P. N. Brusasca, R. Colangeli, K. P. Lyashchenko, X. Zhao, M. Vogelstein, J. S. Spencer, D. N. Mcmurray and M. L. Gennaro, Scand. J. Immunol., 2001, 54, 448 CrossRef CAS PubMed.
- L. A. H. v Pinxteren, P. Ravn, E. M. Agger, J. Pollock and P. Andersen, Clin. Diagn. Lab. Immunol., 2000, 7, 155 CrossRef PubMed.
- M. Harboe, T. Oettinger, H. G. Wiker, I. Rosenkrands and P. Andersen, Infect. Immun., 1996, 64, 16 CrossRef CAS PubMed.
- Centers for Disease Control and Prevention, Tuberculosis Testing & Diagnosis, https://www.cdc.gov/tb/topic/testing/default.htm (accessed 18 November 2023).
- P.-C. Soo, C.-J. Kung, Y.-T. Horng, K.-C. Chang, J.-J. Lee and W.-P. Peng, Anal. Chem., 2012, 84, 7972 CrossRef CAS PubMed.
- P.-C. Soo, Y.-T. Horng, A.-T. Chen, S.-C. Yang, K.-C. Chang, J.-J. Lee and W.-P. Peng, Tuberculosis, 2015, 95, 620 CrossRef CAS PubMed.
- S.-J. Kahng, S. D. Soelberg, F. Fondjo, J.-H. Kim, C. E. Furlong and J.-H. Chung, Biomed. Microdevices, 2020, 22, 50 CrossRef CAS PubMed.
- J. Jeon, J. Lee, J. So, J. B. Lee, H. Lee, Y. Chang, S. Shin, J. Jo and C. Ban, Sens. Actuators, B, 2020, 317, 128126 CrossRef CAS.
- X. L. Kong, L. C.-L. Huang, C.-M. Hsu, W.-H. Chen, C.-C. Han and H.-C. Chang, Anal. Chem., 2005, 77, 259 CrossRef CAS PubMed.
- T. T.-B. Nguyen, H.-C. Chang and V. W.-K. Wu, Diamond Relat. Mater., 2007, 16, 872 CrossRef CAS.
- L.-J. Su, H.-H. Lin, M.-S. Wu, L. Pan, K. Yadav, H.-H. Hsu, T.-Y. Ling, Y.-T. Chen and H.-C. Chang, Bioconjugate Chem., 2019, 30, 2228 CrossRef CAS PubMed.
- A. Krueger and D. Wang, Adv. Funct. Mater., 2012, 22, 890 CrossRef CAS.
- T.-N. Le, H.-Y. Chen, X. M. Lam, C.-C. Wang and H.-C. Chang, Anal. Chem., 2023, 95, 12080 CrossRef CAS PubMed.
- S. Perez-Amodio, P. Holownia, C. L. Davey and C. P. Price, Anal. Chem., 2001, 73, 3417 CrossRef CAS PubMed.
- J.-H. Bong, T.-H. Kim, J. Jung, S. J. Lee, J. S. Sung, C. K. Lee, M.-J. Kang, H. O. Kim and J.-C. Pyun, BioChip J., 2021, 15, 100 CrossRef CAS PubMed.
- E. Eriksson, J. Lysell, H. Larsson, K. Y. Cheung, D. Filippini and W. C. Mak, Research, 2019, 2019, 8079561 CrossRef CAS PubMed.
- S. K. Bikkarolla, S. E. McNamee, S. McGregor, P. Vance, H. McGhee, E. L. Marlow and J. McLaughlin, AIP Advances, 2020, 10, 125316 CrossRef CAS.
- D. A. Armbruster and T. Pry, Clin. Biochem. Rev., 2008, 29(Suppl 1), S49 Search PubMed.
- B. Kil, E. Kim, M. Kang, J. Jeon, Y. Chang, C. L. Chang and C. Ban, Sens. Actuators, B, 2023, 381, 133427 CrossRef CAS.
- T. T. Feng, C. M. Shou, L. Shen, Y. Qian, Z. G. Wu, J. Fan, Y. Z. Zhang, Y. W. Tang, N. P. Wu, H. Z. Lu and H. P. Yao, Int. J. Tuberc. Lung Dis., 2011, 15, 804 CrossRef PubMed.
- R. Keshavarz, N. Mosavari, K. Tadayon, F. Fardid, R. Ghaderi, K. S. Babadi, R. Arefpajoohi, S. H. Sajadi, M. M. Taheri, S. Dashtipour, M. Dehghanpour and S. G. Magami, Int. J. Mycobact., 2015, 4, 114 CrossRef.
- X. Wu, Y. Wang, T. Weng, C. Hu, F. X. C. Wang, Z. Wu, D. Yu, H. Lu and H. Yao, Medicine, 2017, 96, e9350 CrossRef CAS PubMed.
- N. Ariffin, N. A. Yusof, J. Abdullah, S. F. Abd Rahman, N. H. Ahmad Raston, N. Kusnin and S. Suraiya, J. Sens., 2020, 2020, 1365983 Search PubMed.
- C. Liu, C. J. Lyon, Y. Bu, Z. Deng, E. Walters, Y. Li, L. Zhang, A. C. Hesseling, E. A. Graviss and Y. Hu, Clin. Chem., 2018, 64, 791 CrossRef PubMed.
- T. Broger, M. Tsionksy, A. Mathew, T. L. Lowary, A. Pinter, T. Plisova, D. Bartlett, S. Barbero, C. M. Denkinger, E. Moreau, K. Katsuragi, M. Kawasaki, P. Nahid and G. B. Sigal, PLoS One, 2019, 14, e0215443 CrossRef CAS PubMed.
- R. A. Omar, N. Verma and P. K. Arora, Front. Immunol., 2021, 12, 653853 CrossRef CAS PubMed.
- U. Z. M. Azmi, N. A. Yusof, J. Abdullah, F. Mohammad, S. A. A. Ahmad, S. Suraiya, N. H. A. Raston, F. N. M. Faudzi, S. K. Khiste and H. A. Al-Lohedan, Nanomaterials, 2021, 11, 2446 CrossRef CAS PubMed.
- U. Z. Mohd Azmi, N. A. Yusof, J. Abdullah, S. A. Alang Ahmad, F. N. Mohd Faudzi, N. H. Ahmad Raston, S. Suraiya, P. S. Ong, D. Krishnan and N. K. Sahar, Microchim. Acta, 2021, 188, 20 CrossRef CAS PubMed.
- N. Mohd Bakhori, N. A. Yusof, J. Abdullah, H. Wasoh, S. K. Ab Rahman and S. F. Abd Rahman, Materials, 2020, 13, 149 CrossRef PubMed.
- Y. Xia, H. Zhu, R. Liu, X. Li, X. Xie, J. Zhao, J. Li, Z. Yang and X. Chen, Talanta, 2023, 253, 124052 CrossRef CAS.
- M. M. Johnson and J. A. Odell, J. Thorac. Dis., 2014, 6, 210 Search PubMed.
- D. E. Griffith, T. Aksamit, B. A. Brown-Elliott, A. Catanzaro, C. Daley, F. Gordin, S. M. Holland, R. Horsburgh, G. Huitt, M. F. Iademarco, M. Iseman, K. Olivier, S. Ruoss, C. F. V. Reyn, J. Richard, J. Wallace and K. Winthrop, Am. J. Respir. Crit. Care Med., 2007, 175, 367 CrossRef CAS PubMed.
- R. Gopalaswamy, S. Shanmugam, R. Mondal and S. Subbian, J. Biomed. Sci., 2020, 27, 74 CrossRef PubMed.
- Y.-R. Chang, H.-Y. Lee, K. Chen, C.-C. Chang, D.-S. Tsai, C.-C. Fu, T.-S. Lim, Y.-K. Tzeng, C.-Y. Fang and C.-C. Han, Nat. Nanotechnol., 2008, 3, 284 CrossRef CAS PubMed.
- L.-J. Su, C.-Y. Fang, Y.-T. Chang, K.-M. Chen, Y.-C. Yu, J.-H. Hsu and H.-C. Chang, Nanotechnology, 2013, 24, 315702 CrossRef PubMed.
- J.-Y. Feng, C.-J. Lin, J.-Y. Wang, S.-T. Chien, C.-B. Lin, W.-C. Huang, C.-H. Lee, C.-C. Shu, M.-C. Yu, J.-J. Lee and C.-Y. Chiang, Sci. Rep., 2022, 12, 12064 CrossRef CAS PubMed.
- F. Nolte, Mycobacterium, Man. Clin. Microbiol., 1995, 400–437 Search PubMed.
Footnote |
| † Electronic supplementary information (ESI) available. See DOI: https://doi.org/10.1039/d3tb03038e |
| This journal is © The Royal Society of Chemistry 2024 |