Open Access Article
Open Access ArticleCreative Commons Attribution 3.0 Unported Licence
A semi-quantitative visual lateral flow immunoassay for SARS-CoV-2 antibody detection for the follow-up of immune response to vaccination or recovery†
Simone
Cavalera
 a,
Fabio
Di Nardo
a,
Fabio
Di Nardo
 *a,
Thea
Serra
*a,
Thea
Serra
 a,
Valentina
Testa
a,
Claudio
Baggiani
a,
Valentina
Testa
a,
Claudio
Baggiani
 a,
Sergio
Rosati
a,
Sergio
Rosati
 b,
Barbara
Colitti
b,
Ludovica
Brienza
b,
Irene
Colasanto
b,
Chiara
Nogarol
c,
Domenico
Cosseddu
d,
Cristina
Guiotto
d and
Laura
Anfossi
b,
Barbara
Colitti
b,
Ludovica
Brienza
b,
Irene
Colasanto
b,
Chiara
Nogarol
c,
Domenico
Cosseddu
d,
Cristina
Guiotto
d and
Laura
Anfossi
 a
a
aDepartment of Chemistry, University of Turin, Via Pietro Giuria 7, Turin, Italy. E-mail: fabio.dinardo@unito.it
bDepartment of Veterinary Science, University of Turin, Largo Braccini 2, Grugliasco (TO), Italy
cIn3diagnostic srl, Largo Braccini 2, Grugliasco (TO), Italy
dA.O. Ordine Mauriziano, Ospedale Umberto I di Torino, Via Magellano 1, Turin, Italy
First published on 26th January 2024
Abstract
The lateral flow immunoassay (LFIA) technique is largely employed for the point-of-care detection of antibodies especially for revealing the immune response in serum. Visual LFIAs usually provide the qualitative yes/no detection of antibodies, while quantification requires some equipment, making the assay more expensive and complicated. To achieve visual semi-quantification, the alignment of several lines (made of the same antigen) along a LFIA strip has been proposed. The numbering of the reacting lines has been used to correlate with the quantity of some biomarkers in serum. Here, we designed the first semiquantitative LFIA for detecting antibodies and applied it to classify the immune response to SARS-CoV-2 raised by vaccination or natural infection. We used a recombinant spike receptor-binding domain (RBD) as the specific capture reagent to draw two test lines. The detection reagent was selected among three possible ligands that are able to bind to anti-spike human antibodies: the same RBD, staphylococcal protein A, and anti-human immunoglobulin G antibodies. The most convenient detector, adsorbed on gold nanoparticles, was chosen based on the highest correlation with an antibody titre of 171 human sera, measured by a reference serological method, and was the RBD (Spearman's rho = 0.84). Incorporated into the semiquantitative LFIA, it confirmed the ability to discriminate high- and low-titre samples and to classify them into two classes (Dunn's test, P < 0.05). The proposed approach enabled the semiquantification of the immune response to SARS-CoV-2 by the unaided eye observation, thus overcoming the requirement of costly and complicated equipment, and represents a general strategy for the development of semiquantitative serological LFIAs.
Introduction
The detection of specific antibodies has been playing a crucial role in many aspects of the clinical practice.1 It is widely employed for the indirect diagnosis of infectious diseases (e.g., HIV,2,3 HCV,4 syphilis,5 Zika virus6 and many others) and autoimmune diseases (e.g., hepatitis,7 post-transfusion purpura and neonatal alloimmune thrombocytopenia,8 systemic autoimmune rheumatic disease cardiac autoimmunity,9,10 encephalitis,11 and haemolytic anaemia12), as well as for the monitoring of immunosuppression in organ transplants,13,14 or other clinical immune-related pathologies.15 It is also employed for the analysis of the acquired immunity from neutralizing antibodies,16 for therapy monitoring5 and for the extrapolation of information for vaccine design.17 Immunoassays, based on the specific interaction between antigens and specific antibodies, are the most widely and routinely employed analytical techniques for this purpose in clinical laboratories.18–20 Laboratory methods perform quantitative analyses and provide accurate results. The detection techniques include colorimetry, fluorescence, chemiluminescence, electrochemistry, and bioluminescence.18 Nevertheless, in circumstances where a considerable number of analyses is required, or in low-resource settings, low cost and portable screening techniques should be preferred. The lateral flow immunoassay (LFIA) is widely employed as a point-of-care test (POCT) for antibody testing. LFIA devices are portable, low cost and easy to use and these are the reasons for their massive employment in the last few decades and for the crucial role played during the SARS-CoV-2 pandemic.21–24 A typical LFIA device is a portable cassette including a strip composed of overlayered materials, which contains all the reagents required for the test. The device allows for the detection of the target molecule by exploiting the affinity and specificity of the antibody–antigen interaction. The LFIA can be completed in 5–15 min, and the results, especially with the typical colorimetric (visual) format, are simple to read, speeding up decision making and intervention.25 The major drawback of the LFIA is represented by the qualitative nature of the information provided. Quantitative detection capability has been introduced as well, through the integration of the LFIA with transducers, readers, smartphone-based apps and so on.26 This upgrade, however, reduces the portability, increases the cost, and requires an energy source, thus lowering the affordability of the devices. In fact, the qualitative LFIA, based on the visual inspection of results, still represents more than 80% of the employed LFIA devices.27 Ideally, the unaided eye readout of the colorimetric LFIA should also be maintained in the (semi)quantitative LFIA without the need for any external tool or complicated technology. Few works have been reported on visual semiquantitative LFIAs (sqLFIA). Hong and collaborators, in 2012, reported the detection of a cartilage oligomeric matrix protein using a LFIA equipped with a multi-spot pattern in the test zone for the semi-quantification.28 Anyway, the device required a commercial scanner and an imaging software program for the interpretation of results. The first truly stand-alone sqLFIA was proposed in 2019 by Oh et al., who developed a multi-test line LFIA for the semi-quantification of transferrin glycoforms in serum.29 The semi-quantification was achieved by counting the number of coloured test lines, which was directly proportional to the amount of the target protein. Similarly, Serebrennikova and collaborators developed a sqLFIA for procalcitonin.30 In 2020, Lee and collaborators included semi-quantification of microalbumin in urine through naked eye interpretation, based on printing the test zone as multiple spots; the quantitative information was achieved simply by counting the number of coloured spots.31In this work, we developed a visual (colorimetric) LFIA for the semi-quantitation of specific antibodies in serum. As a case of study, we considered the immune response to Covid-19 vaccination also because of the importance of monitoring the efficacy of the vaccination and the duration of the immunity towards SARS-CoV-2 over time. SARS-CoV-2 rapidly spread worldwide after December 2019 and caused 6.5 million confirmed deaths in the following two years of world pandemic, according to WHO reports.32 The disclosure of the sequence and structure of proteins encoded by this virus was crucial both for diagnostic purposes and for the management of the emergence through the development of vaccines.32,33 From this point of view, the most relevant SARS-CoV-2 proteins are the spike (Sp) and the nucleocapsid (Np) proteins. The Sp is a trimeric protein, located on the membrane, and used by the virus for the anchoring to the receptor (ACE-2) of target cells through a receptor binding domain (RBD). The Np is a RNA-binding protein necessary for the conservation of the folding of the viral genome. Both proteins can elicit an immune response in the host,34 and this aspect has been massively exploited. On the one hand, the immune response against Sp, and even more against Np, was used for the indirect diagnosis of infection by serological testing. In the very first phase of the pandemic, serology was employed as a screening tool to help diagnosis along with a laboratory testing based on molecular methods (RT-PCR).35 Most importantly, the spike protein was exploited to produce vaccines. In fact, the antibodies stimulated by the immune system against Sp have a neutralizing power, representing a temporary barrier against new infections. The first vaccines, therefore, contained the spike protein or mRNA encoding information for spike protein production.36–38 Undoubtedly, vaccination programs (first dose and boosters) allowed for a relatively rapid overcoming of the emergency state.39,40 Nevertheless, the immunity provided by different vaccines, as well as the one acquired after the infection, is a matter of discussion.41–46 The appearance of variants of SARS-CoV-2, differing mostly in the Sp than in other proteins, questioned the efficacy of the vaccines.47–50 In addition, the concurrence of natural infections, and the severity of symptoms, primed differently the immune response and the proneness to reinfection.51 The vaccination efficacy and persistency are not univocal in cases of vaccination, natural (from infection), and hybrid immuneresponse.52–56 Moreover, even if it is strongly recommended, the management and scheduling of booster doses is critical for susceptible individuals, such as elderly or immunocompromised population,57–59 patients with haematological malignancies,60,61 cancer,62,63 multiple sclerosis64 and other pathological conditions.65,66 These populations, in addition, should be monitored after vaccination for the average lower immunity compared to the rest of the population. The aspect of planning booster doses, future immunization policies and disease prevention strategies in highly vulnerable population led to the development of many serological tests for detecting antibodies to SARS-CoV-2. Both laboratory techniques67,68 and POCTs69,70 have been made available so far, including LFIA devices.71–74 These systems provide qualitative results and, in some cases, quantitative estimation with the introduction of equipment as reading systems.
In our study, we pursued the realization of the visual sqLFIA for specific antibody quantification. To this aim, we first compared three assay formats for antibody detection by the LFIA and then evaluated the conversion into sqLFIA (Fig. 1). The same specific capture reagent (RBD) was used to design the LFIA formats, while the detection of the captured antibodies was accomplished using three different probes, comprising gold nanoparticles adsorbed with (1) again RBD, (2) staphylococcal protein A (SpA), and (3) anti-human immunoglobulin G antibody (anti-hIgG). The three formats aimed at detecting specific anti-spike antibodies, but with some substantial differences in terms of proneness to matrix effects and biological variability (interference or enhancing effects caused by other non-specific immunoglobulins present in the serum). The SpA, for example, can react with immunoglobulins (Igs) and create lattices of gold-conjugates crosslinked by non-specific immunoglobulins, increasing the signal and, ultimately, increasing the sensitivity. The same is true for the anti-human immunoglobulin G antibody, however with different affinities and with the exception of Igs of A and M classes. The double antigen approach could be more prone to the saturating effect,75,76 as, in this case, the binding of the target antibodies to the labelled antigen may prevent the interaction with the antigen deposed onto the membrane. On the other hand, it should not suffer interference from the non-specific Igs. Each format was evaluated by testing 171 human sera (92 collected pre-covid and 79 belonging to vaccinated individuals) previously characterized by their antibody content by a reference serological method (SARS-CoV-2 IgG II Quant, Abbott Diagnostics, USA). The performances of the three prototypal devices were compared in terms of diagnostic sensitivity and correlation of the colour intensity of the test line with the specific antibody titre, as measured by the reference assay. Once selected, the best format was exploited to design the semi-quantitative device for the discrimination between different populations of ‘positive’ individuals (classified according to their anti-Sp antibody titre).
The principle of the semi-quantification consisted of using the same RBD spotted on two following test lines. In the case of high-titre sera, the second test line was supposed to capture the excess of anti-spike antibodies that were not retained by the first test line. In this way, two coloured lines became visible for high positive samples. On the other hand, low positive samples were supposed to provide just one coloured test line, since the few antibodies present were efficiently captured by the first test line (Fig. 1). The 171 samples were assayed once again by the sqLFIA and classified according to the number of coloured lines formed. The results were correlated with the amount of the antibody, and boxplots, one-way ANOVA on Ranks (Kruskal–Wallis), statistical pairwise comparisons (Dunn's test), and ROC curves were made to characterise the discrimination capacity of the sqLFIA.
We also produced a recombinant RBD via mammalian expression strategy, using a high-yield transient expression system based on high density, suspension adapted, HEK cells, to obtain a highly concentrated antigen. Protein purification was achieved directly from culture supernatants, using the twin-Strep-Tag/Strep-Tactin XT® affinity matrix, combining high specificity, mild conditions, and low temperature. This strategy enabled us to circumvent the limited stability of the commercial recombinant RBD, which is a major challenge in the development and production of devices including this bioreagent. The new RBD was incorporated into the LFIA devices and compared to the commercially available analogue.
Experimental
Chemicals and materials
Gold(III) chloride trihydrate (ACS reagent), staphylococcal protein A (SpA), bovine serum albumin (BSA), casein sodium salt from milk, avidin, and sucrose were obtained from Sigma–Aldrich (St. Louis, MO, USA). Sulfo–NHS–LC-Biotin was obtained from Thermo Scientific (Waltham, MA, USA). The commercial recombinant spike protein (cSp) was purchased from Rekom Biotech (Granada, España). Materials for the synthesis of the new recombinant spike protein (nSp) production were purchased from Genscript (Leiden, Netherland), Thermo Fisher Scientific (Waltham, MA, USA) and IBA-lifesciences (GÖttingen, Germany). Tween20 and other chemicals were purchased from VWR International (Milan, Italy). The anti-human IgG antibody (anti-hIgG) produced in the mouse was purchased from Medix Biochemica (Espoo, Finland). Nitro-cellulose membranes, cellulose absorbent pads, and blood separator sample pads were purchased from MDI membrane technologies (Ambala, India). Glass fibre conjugate pads were obtained from Merck Millipore (Billerica, MA, USA).Serum samples
Whole blood from donors was collected by venous puncture, after collecting informed consensus. Serum was obtained on the same day of collection, immediately heat inactivated at 56 °C for 30 min and stored at −20 °C until analysis. Samples were transported and handled in compliance with international standards for biosecurity and biocontainment. A total of 79 serum samples belonging to vaccinated/recovered individuals were collected, and 76 were diagnosed as positive for the antibodies directed to the spike protein from SARS-CoV-2 (Fig. S1, ESI†). The serological reference method was performed at the Biochemical Laboratory of A.O. Ordine Mauriziano, Ospedale Umberto I by means of SARS-CoV-2 IgG II Quant (Abbott Diagnostics, USA) coupled with an Alinity analyser detecting antibodies towards the RBD of the spike protein from SARS-CoV-2 and the positivity was intended for values of antibody titre >15 AU mL−1.77 To assess diagnostic specificity, 92 pre-covid serum samples were tested in this work. All serum samples used in this work were collected in accordance with the approval of the ethical committee Prot. n. 0432967 del 21/07/2023 – [UOR: SI000045 – Classif. III/11].Production of the recombinant RBD spike subunit
The spike gene subunit encoding for the receptor binding domain (RBD) was PCR amplified and cloned into the pBRICO eukaryotic expression vector in a frame with a secretory leader peptide and twin-strep-tag peptide, enabling efficient purification from the culture supernatant. The Expi293 expression system was employed according to the guideline and reagents were provided by the manufacturer. The culture supernatant after transfection was directly used for protein purification using Strep-Tactin XT 4Flow and using a buffer set and protocol recommended by IBA biosciences. The purity and yield of the recombinant antigen were evaluated by SDS-PAGE and Bradford methods, respectively.Labelling functional proteins with gold nanoparticles
AuNPs with a localized surface plasmon resonance (LSPR) of 525.5 nm (a mean diameter of ca. 32 nm)78 were prepared by tetrachloroauric acid reduction with sodium citrate79 as reported in previous works80,81 (see the ESI† for details). Characterization of the AuNPs was performed by means of transmission electron microscopy (TEM, Fig. S2, ESI†) and visible spectrophotometry (Table S1 and Fig. S3, ESI†). Three gold conjugates were synthesized by adsorbing the following proteins onto the surface of AuNPs: (i) a commercial recombinant spike RBD protein from SARS-CoV-2 (AuNP-cSp); (ii) staphylococcal protein A (AuNP-SpA); (iii) anti-human G immunoglobulin antibody produced in the goat (AuNP-anti-hIgG). In addition, a new recombinant spike RBD protein from SARS-CoV-2 was produced and adsorbed onto the gold nanoparticle surface (AuNP-nSp). Gold conjugates were obtained by passive adsorption of bioreagents (cSp, nSp, SpA, and anti-human IgG antibody) onto the surface of the AuNPs, by mixing different amounts of the protein (1–2–4 μg of cSp, nSp, SpA, and 5–10–15 μg of anti-hIgG) to 1 mL of gold nanoparticles at an optical density of 1. Details for the preparation and characterisation of the gold conjugates included in this work are reported in the ESI.† To incorporate the control line into the LFIA device including AuNPs adsorbed with spike RBD proteins, an additional gold conjugate was produced by adsorbing biotinylated bovine beta casein on AuNPs (10 μg mL−1). The bovine beta casein was linked to Sulfo–NHS–LC-Biotin following the protocol recommended by the manufacturer and the gold conjugate AuNP-biotin was prepared as reported in a previous work.82Design of experiments for optimizing AuNP–protein conjugates
The best performing gold conjugate for each serological LFIA format was defined based on the best compromise between the amount of a functional protein adsorbed on the AuNPs (protein-to-AuNP ratio, μg OD−1) and the amount of a gold conjugate in the single strip (optical density, OD), as previously described.83 Designing of experiment sets was performed to individuate the conditions providing the highest analytical signal (colour intensity on the test line). The levels of the protein-to-AuNP ratio were chosen upon the minimum stabilising amount, defined through the salt-induced aggregation test (Fig. S4, ESI†). The salt-induced aggregation test was performed on AuNPs adjusted to pH 8, except for SpA (pH 6, see the ESI† for further details). The test defines the minimum amount of protein needed for stabilising the AuNPs from aggregation caused by a saline shock. The levels used in the optimisation were factors of this amount (0.5×–1.0×–2.0×). As a positive control, a high titre human serum (#1001) was used for the full set of experiments and a pre-covid serum (negative control) was used for confirming the absence of the non-specific binding signal (false positive). Details on the experimental designs are reported in the ESI.†Production of the qualitative LFIA strips
The recombinant RBD of the spike protein was applied to the nitrocellulose (NC) membrane to form the test lines (the RBD from the spike protein 1.0 mg mL−1). Avidin (1.0 mg mL−1) was used in the control line. Reagents were dotted at 1 μL cm−1 by means of a XYZ3050 platform (Biodot, Irvine, CA, USA), equipped with a BioJetQuanti™ 3000 Line Dispenser for non-contact dispensing, keeping 3 mm between the lines. The gold conjugates were absorbed onto the glass fibre conjugate pad previously saturated with the AuNP storage buffer (see the ESI† for details on the buffer composition). The conjugates were mixed with a ratio of X/1 (AuNP-protein/AuNP-biotin, where X is the optimal OD of the specific gold conjugate) and diluted with the AuNP storage buffer. The pad was dipped in the gold conjugate mix solution and dried for 4 h at room temperature. NC membranes were dried at 37 °C for 60 min under vacuum, layered with samples, conjugates and absorbent pads, cut into strips (4.2 mm width) by means of a CM4000 guillotine (Biodot, Irvine, CA, USA) and inserted into plastic cassettes (Kinbio, Shangai, China) to fabricate the ready-to-use LFIA devices. Cassettes were stored in the dark, in plastic bags containing silica at room temperature until use.Testing serum samples with the LFIA
The three qualitative LFIA formats were evaluated with 76 positive, 3 negative, and 92 pre-covid samples.On the day of the analysis, sera were thawed for 30 min at room temperature, gently shaken and diluted 1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 10 by using the running buffer (34 mM tris/80 mM glycine buffer pH 8.2, 1% BSA, 1% Tween 20, and 0.05% sodium azide). Assays to detect SARS-CoV-2 antibodies were conducted at room temperature, by adding 80 μL of the diluted serum to the sample well. Qualitative results were judged by the naked eye after 20 min from the sample application. Samples were analysed in duplicate, and results were observed by three operators. The test line intensities were acquired by means of a portable scanner (OpticSlim 550 scanner, Plustek Technology GmbH, Norderstedt, Germany) and the area of the coloured lines was quantified using a QuantiScan 3.0 software (BioSoft, China). Values below 15 arbitrary units (a.u.) corresponded to no signal detected by the naked eye and were then set at zero.
10 by using the running buffer (34 mM tris/80 mM glycine buffer pH 8.2, 1% BSA, 1% Tween 20, and 0.05% sodium azide). Assays to detect SARS-CoV-2 antibodies were conducted at room temperature, by adding 80 μL of the diluted serum to the sample well. Qualitative results were judged by the naked eye after 20 min from the sample application. Samples were analysed in duplicate, and results were observed by three operators. The test line intensities were acquired by means of a portable scanner (OpticSlim 550 scanner, Plustek Technology GmbH, Norderstedt, Germany) and the area of the coloured lines was quantified using a QuantiScan 3.0 software (BioSoft, China). Values below 15 arbitrary units (a.u.) corresponded to no signal detected by the naked eye and were then set at zero.
The sqLFIA device to visually discriminate high/low positive samples
The conversion to the sqLFIA was achieved by interposing a second test line between the first test and the control lines. The concentration of the second test line was varied (1.0–0.5–0.25–0.1 mg mL−1) and the discrimination power was checked by using 4 serum samples representing a high titre (#1001, 5680 AU mL−1), a medium-high titre (#1010, 3238 AU mL−1), a medium-low titre (#1048, 417 AU mL−1), and a low titre (#1050, 90.5 AU mL−1).Both the RBD and SpA detectors were considered in the study. Finally, the sqLFIA was fabricated by drawing two test lines (1.0 and 0.1 mg mL−1, respectively) and using the AuNP-cSp detector.
In addition, another prototype was produced by substituting the commercial RBD (cSp) with the in-house produced recombinant RBD (nSp). The nSp was applied at 1 mg mL−1 and 0.25 mg mL−1 to form the first and second test lines, respectively.
Data analysis
The D-optimal design of experiments and elaboration of the results were performed using a Chemometric Agile Tool. Statistical calculations were carried out using a SigmaPlot 14.0 software (SyStat Software Inc, Palo Alto, CA, USA). The distribution of serological titres of the positive samples was evaluated according to the Shapiro–Wilk normality test. The number of positive results from the three qualitative LFIAs was used to define the diagnostic sensitivity (Se%). The Spearman correlation (Spearman's rho, rS) for the non-Gaussian distribution (according to Shapiro–Wilk test <0.05%) was calculated by plotting the colour intensity on the test line against the anti-spike antibody titre obtained from the reference method. ROC curves were made to define the titre cut-off level for different sqLFIA prototypes. The Mann–Whitney test was run for determining the statistical significance of differences between the high-titre and low-titre groups.Results and discussion
Selection of the serological LFIA format
The strategy for the development of the semiquantitative LFIA for monitoring the antibody response to SARS-CoV-2 included, as the first step, the individuation of the LFIA format most convenient for the purpose of quantitation. For this reason, three qualitative (single line) LFIA formats were studied. The capture reagent was kept constant (RBD spike protein) while different detectors were investigated (Fig. 1). One format (LFIA-1) was based on the ‘double antigen’ approach, and the detector was made by absorbing the commercial spike RBD to the AuNP (AuNP-cSp). The second format (LFIA-2) employed the bacterial ligand SpA as the specific ligand adsorbed to the AuNP (AuNP-SpA). SpA was considered because of its high binding capacity towards immunoglobulins. The third (LFIA-3) was a typical serological format, including a secondary antibody directed towards human immunoglobulin G as the detector (AuNP-anti-hIgG). First, the three AuNP–protein conjugates were optimized by means of an experimental design by varying the levels of the protein adsorbed (protein-to-AuNP ratio) and the amount of the detector (optical density of the conjugate solution) included in the LFIA device (Table S2 and Fig. S5, ESI†). The combinations were studied by applying a negative sample and a positive sample and recording the signal formed in the test line. The optimal detector was defined according to maximizing the signal for the positive sample and showing no false positivity for the negative sample. For the LFIA-1, the best combination was characterised by 1 μg of the recombinant RBD absorbed for each optical density unit of the AuNP, and an optical density of 4 in the assay. The optimal combination for the LFIA-2 was characterised by 4 μg of SpA absorbed for each optical density unit of the AuNP, and an optical density of 9 in the assay, while, for the LFIA-3, the best detector was characterised by 15 μg of the anti-hIgG antibody absorbed for each optical density unit of the AuNP, and an optical density of 6 in the assay.The three optimized qualitative LFIA devices were applied to detect the presence of anti-spike antibodies in 171 human sera. The rate of true positive results (diagnostic sensitivity) is reported in Table 1, along with the correlation of the measured signal intensity with the results of the serological reference assay. Among the three, the LFIA-2 approach showed the highest diagnostic sensitivity (93.8%, C.I.95%: 88.6–99.1), while the LFIA-3 resulted as the less sensitive (85.2%, C.I.95%: 77.4–92.9). We hypothesized that non-SARS specific immunoglobulins reacted differently with the probes, and therefore, the inter-individual variability impacted differently on the assay formats. Some samples probably had a high concentration of non-specific Igs so preventing the probes from binding to the specific antibodies. In general, the highest values of the test line intensity were recorded with the LFIA-2. This result may be explained by the valency of SpA, which has up to five binding domains for IgGs, and likely rendered it less prone to saturation. In addition, SpA also enabled enhancing the specific signal via cross-linking of several AuNPs mediated by non-specific immunoglobulins.
| Format | Protein | Sensitivity % (C.I.95%) | Correlationa, rS |
|---|---|---|---|
| a Spearman's rho towards the serological reference method, calculated for not normally distributed data (according to Shapiro–Wilk one-way ANOVA). | |||
| LFIA-1 | cSp | 86.4 (79.0–93.8) | 0.84 |
| LFIA-2 | SpA | 93.8 (88.6–99.1) | 0.47 |
| LFIA-3 | anti-hIgG | 85.2 (77.4–92.9) | 0.48 |
Concerning the correlation with the reference serological test, the highest Spearman's rho value was recorded for the LFIA-1 format (0.84 for cSp). This was explained by considering the double specificity of the antigen–antibody recognition both in capture and in detection, which resulted in the absence of interference from non-specific immunoglobulins. In contrast, the correlation for the LFIA-2 (0.48) and LFIA-3 (0.47) was significantly lower probably because the gold labelled proteins interacted with both specific and non-specific antibodies, increasing the dispersion of data (Fig. 2).
Conversion of LFIA-1 into sqLFIA
Among the three qualitative formats used to detect anti-spike antibodies in the serum, the LFIA-1 was the most performing one in terms of correlation with quantitative measurements.Thus, we investigated further this approach for the development of a (semi)quantitative visual LFIA. To this aim, we added an additional test line comprising the same capture protein used in the previous experiments. The test lines were located as close as possible to the sample well to minimize the contact time between the target antibodies and the detector before they reached the capture reagent. Actually, when the same reagent was used for capturing and detection, a competition for the binding to target antibodies occurred, analogously to what happens for the single-epitope sandwich in the antigenic LFIA75,83,84. The effect of the competition was a reduction of the sensitivity, which could be counteracted efficiently by the precaution of narrowing the distance between the test lines and the sample application point.75,83,84
The concentration of the second test line (T2) was defined according to the ability of correctly classifying four samples chosen to have a high titre (#1001, 5680 AU mL−1), a medium-high titre (#1010, 3238 AU mL−1), a medium-high titre (#1048, 417 AU mL−1), and a low titre (#1050, 90.5 AU mL−1). In particular, the interpretation rule was defined as follows: the high-titre sample should provide two coloured test lines, the low-titre sample should provide one coloured test line, and the negative control should provide no coloured test lines. Examples of results are shown in Fig. S6 and are summarised in Table S3 (ESI†).
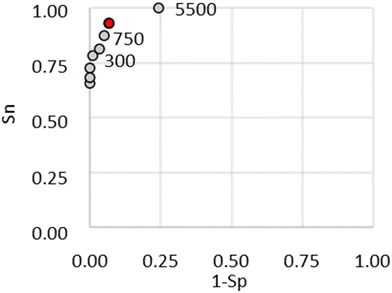
The sqLFIA was able to discriminate high and low titre samples by using a concentration of the RBD to form the second test line T2 of 0.1 mg mL−1 (Fig. S6a, ESI†). Under this condition, the LFIA device allowed us to discriminate also between the #1048 (medium-low-titre) and #1050 (low-titre) samples (Fig. 3a). To confirm the results obtained by exploiting the LFIA-1 format, a second prototype sqLFIA was prepared, based on the LFIA-2 format. This last was characterized by higher sensitivity, but a lower correlation with quantitative measurements. The prototype derived from LFIA-2 resulted less efficient in discriminating among high and low positive samples, despite the ability to detect extremely low titre sera. In this format, the second test line showed a parallel behaviour to the first, without adding information compared to the device with a single test line (Fig. S6b, ESI†). Then, a total of 171 samples, including negative and positive sera, were tested with the sqLFIA device based on the double antigen format (LFIA-1). The classification ability and the discrimination cut-off, in terms of antibody titre, were determined. According to the Shapiro–Wilk normality test, the set of positive sera was not normally distributed; therefore, non-parametric statistics were used. Samples that gave two test lines (2xT), one test line (1xT), and no test line (neg) responses were clustered and analysed for their distribution by means of a Kruskal–Wallis one-way analysis of variance on ranks, and the significance of the differences was determined by Dunn's method for all pairwise multiple comparison procedure (Table 2). The 2 x T cluster enclosed 48 samples with a median antibody titre of 977 AU mL−1 (25–75%: 797–3548 AU mL−1), 1 x T contained 21 samples with a median of 238 AU mL−1 (25–75%: 0–552 AU mL−1), and negative responses were given by 102 samples, including the pre-covid ones. The results are also reported as box plots in Fig. 3b. The boxes are separated among the three clusters and Dunn's method confirmed the significant difference between the clusters (Table 2). The discrimination level between the 2 x T and 1 x T groups was defined through the ROC curve and resulted in 750 AU mL−1 with a sensitivity (Se, probability to correctly classify the 2 x T) of 93% and a specificity (Sp, probability to correctly classify the 1 x T) of 93% (Fig. 4).
| Kruskal–Wallis one way analysis of variance on ranks | All pairwise multiple comparison procedures (Dunn's method) | ||||||
|---|---|---|---|---|---|---|---|
| N | Median | 25–75% | Diff of ranks | Q | P < 0.05 | ||
| 2xT | 48 | 977 | 797–3548 | 2xT vs. 1xT | 46.03 | 3.554 | Yes |
| 1xT | 21 | 238 | 0–552 | 2xT vs. neg | 89.086 | 10.28 | Yes |
| neg | 102 | 0 | 0 | 1xT vs. neg | 43.056 | 3.629 | Yes |
Number of test lines in the sqLFIA
Since adding a second test line enabled us to introduce semi-quantification ability to the LFIA, the addition of a third test line, again composed of the RBD spotted at 0.1 mg mL−1 was investigated to verify if the discrimination power could be improved. The 171 samples were tested using a three-test-line sqLFIA device (Fig. S7, ESI†). Despite the trends of responses agreed with serum titres, and 4 clusters of samples were obtained, with a distinct number of coloured test lines (3 x T, 2 x T, 1 x T, and neg), the number of samples in two of these clusters (2 x T and 1 x T) was very low. The Mann–Whitney pairwise comparison confirmed that there was no significant difference between the 2 x T and 1 x T clusters (Table S4, ESI†).Therefore, the following work was carried out by maintaining the 2-test-line configuration.
Production of the RBD
The gene subunit encoding RBD was successfully amplified by the PCR, digested with appropriate restriction enzymes, and cloned into a mammalian expression vector. Sequence analysis of at least two positive clones confirmed the authenticity and the correct frame of the gene of interest. Plasmid midiprep was then performed using the endotoxin free method and used for the transfection experiment. Expy293F cells were cultured in 25 mL culture volume in Erlenmeyer flasks (125 mL capacity), incubated at 37 °C with 80% humidity and 8%CO2 using an orbital shaker platform (120 rpm) until reaching a density of 3 × 106 viable cells/mL and transfected according to the expression system recommendation. Up to 5 flasks were transfected and the culture medium was harvested 5 days post transfection (p.i.). Purification with the Strep-Tactin® XT affinity matrix was conducted with 1 mL of the resin column by gravity flow and eluted in 3 fractions (0.6 mL E1; 1.6 mL E2; 0.8 mL E3), being fraction E2 expected to contain most of the eluted protein (Fig. S8, ESI†). The protein concentration was measured using the Bradford method and fraction E2 reached 3 mg mL−1 of the pure RBD (4.8 mg).Inclusion of the new RBD into the sqLFIA
The newly produced RBD was linked to the gold nanoparticles (AuNP-nSp) and was optimised as described above for the commercial protein. The gold-protein conjugates were spectrophotometrically characterised (Fig. S9 and Table S5, ESI†) and submitted to the experimental design to identify the combination providing the highest colour intensity of the test line for the positive sample. The optimized detector was the one including 2 μg with an optical density of 4 in the assay (Table S6 and Fig. S10, ESI†). It was used to prepare a new qualitative LFIA, which was tested again by applying the 171 serum samples. The analytical performances were comparable, or even superior, to the ones achieved by the commercial protein. In particular, the sensitivity was higher (Se%: 93.5, C.I.95%: 86.4–98.2) and the correlation with reference measurements was comparable (Spearman's rho: 0.87). Then, we converted it into a two line (sqLFIA) format using the same concentration to form the second test line of the LFIA-1 (0.1 mg mL−1). The classification ability was confirmed as the device discriminated between the #1010 (medium-high-titre) and #1050 (medium-low-titre) samples (Table S7, ESI†). The classification of the 171 serum samples furnished by the new sqLFIA was quite different compared to the one provided by the LFIA including the commercial RBD. The majority of the samples (59) resulted in a 1 x T response, with a median of 715 AU mL−1 (25–75%: 329–974 AU mL−1), while the 2 x T samples were 14, pooled in the highest values of the antibody titre corresponding to the maximum limit of the serological reference method, with a median of 5680 AU mL−1 (25–75%: 3017–5680 AU mL−1). Interestingly, the sqLFIA based on the new recombinant RBD furnished a lower number of false negative results (6), which, furthermore, were given by samples characterised by antibody titres <100 AU mL−1. The overlapping between the 2 x T and 1 x T clusters, due to the high dispersion of the antibody titres of the samples included in the 1 x T cluster, is appreciable from the boxplot (Fig. S11a, ESI†). The pairwise multiple comparison Dunn method confirmed that there was no significant difference between the 2 x T and 1 x T groups (Table S8, ESI†). Nevertheless, the ROC curve individuated a discrimination level (4500 AU mL−1), with a Se of 78% and a Sp of 97% (Fig. S12a, ESI†). Apparently, a higher amount of the new recombinant RBD on the second test line was needed to replicate the behaviour of the commercial one, in terms of classification power. In fact, the prototype employing 0.25 mg mL−1 of the RBD as the second test line allowed us to discriminate between the #1048 (medium-low-titre) and #1050 (low-titre) samples (Table S7 and Fig. S11b, ESI†). Samples were also clustered likewise: 54 samples with a median at 969 AU mL−1 (25–75%: 784–3253 AU mL−1) were included in the 2 x T cluster, 19 samples with a median of 238 AU/mL (25–75%: 90–341 AU mL−1) in the 1 x T cluster, and 98 were assigned as negative samples. The difference between clusters was significant using Dunn's method (Table S8, ESI†). The discriminating value between the 2 x T and 1 x T groups, individuated by the ROC curve, was between 400 AU mL−1 (Se: 93%; Sp: 97%) and 500 AU mL−1 (Se: 98%; Sp: 94%) (Fig. S12b, ESI†).Conclusions
To the best of our knowledge, in the present work, we report the development of the first semiquantitative serological LFIA based on visual interpretation. Based on our results, the double antigen format was the most suitable for pursuing quantitation by colorimetric LFIA. We hypothesized that the higher correlation with quantitative measurement of anti-spike antibodies reached by the double antigen format was due to the absence of interference by non-specific immunoglobulins present in the serum, which may vary largely among samples belonging to different subjects. We also suggested that the use of staphylococcal protein A as the detector enabled us to reach high diagnostic sensitivity (positive/negative discrimination); however, it was less suitable for developing quantitative serological LFIAs. In addition, two RBD spike subunits, a commercial and a newly produced one, were employed to set up sqLFIAs based on multiline interpretation, in which samples (containing high and low amounts of antibodies) were discriminated by numbering the coloured test lines formed upon the application to the device. Interestingly, the two recombinant RBD proteins clustered samples in distinct groups, which may be explained by a different affinity towards antibodies present in the samples. In both cases, the multiline strategy was effective for achieving semi-quantification through visual inspection of LFIA devices (allowing us to reach sensitivity and specificity above 90%) without requiring additional equipment or complicated, multi-step protocols.Author contributions
Simone Cavalera: conceptualization, methodology, and writing – original draft; Fabio Di Nardo: writing – original draft; Thea Serra: investigation and data analysis; Valentina Testa: investigation and data analysis; Claudio Baggiani: formal analysis and writing – review and editing; Barbara Colitti: investigation and resource (SARS-CoV-2 recombinant spike protein); Ludovica Brienza: investigation and resource (SARS-CoV-2 recombinant spike protein); Irene Colasanto: investigation and resource (SARS-CoV-2 recombinant spike protein); Laura Anfossi: funding acquisition, conceptualization, formal analysis, writing – review and editing, and project management; Segio Rosati: funding acquisition, project management, methodology, writing – review and editing, and resource (SARS-CoV-2 recombinant spike protein); Chiara Nogarol: funding acquisition and project management; Cristina Guiotto: resource (clinical samples and diagnosis); Domenico Cosseddu: resource (clinical samples and diagnosis). All authors have read and agreed to the published version of the manuscript.Conflicts of interest
There are no conflicts to declare.Acknowledgements
Authors gratefully thanks Dr Fabiana Marnetto from Neuroscience Institute Cavalieri Ottolenghi AOU San Luigi Gonzaga (Orbassano, Torino, Italy) for having provided pre-covid sera. This research acknowledges support from the Project CH4.0 under the MUR program “Dipartimenti di Eccellenza 2023–2027” (CUP:D13C22003520001) and the FSC Azione 173 Piano Riparti Piemonte INFRA-P2 COVID Linea B (ASF 378 – 64).Notes and references
- A. Lewandowicz-Uszyńska, P. Naporowski, G. Pasternak and D. Witkowska, Postepy Hig. Med. Dosw., 2018, 72, 1162–1178 CrossRef.
- R. J. Chappel, K. M. Wilson and E. M. Dax, Future Microbiol., 2009, 4, 963–982 CrossRef PubMed.
- S. G. Devare, J. Med. Virol., 2007, 79, 11–15 CrossRef.
- S. Chevaliez, Expert Rev. Anti-Infect. Ther., 2019, 17, 341–347 CrossRef CAS PubMed.
- E. W. Hook, Lancet, 2017, 389, 1550–1557 CrossRef PubMed.
- I. B. B. Silva, A. S. da Silva, M. S. Cunha, A. D. Cabral, K. C. A. de Oliveira, E. De Gaspari and C. R. Prudencio, J. Venomous Anim. Toxins Incl. Trop. Dis., 2020, 26, e20200019 Search PubMed.
- G. N. Dalekos and N. K. Gatselis, Eur. J. Intern. Med., 2023, 108, 9–17 CrossRef CAS PubMed.
- L. Porcelijn, E. Huiskes and M. de Haas, Transfus. Apher. Sci., 2020, 59, 102705 CrossRef CAS PubMed.
- A. Willitzki, R. Hiemann, V. Peters, U. Sack, P. Schierack, S. Rödiger, U. Anderer, K. Conrad, D. P. Bogdanos, D. Reinhold and D. Roggenbuck, Clin. Dev. Immunol., 2012, 12, 284740 Search PubMed.
- C. Wehlou and J. R. Delanghe, Clin. Chim. Acta, 2009, 408, 114–122 CrossRef CAS PubMed.
- H. Mojžišová, M. Elišák, J. Hanzalová, M. Petržalka and P. Marusič, Ceska Slov. Neurol. Neurochir., 2020, 83, 251–256 CrossRef.
- M. Raos, M. Lukic, D. Pulanic, M. Vodanovic and B. G. Cepulic, Blood Transfus., 2022, 20, 319–328 Search PubMed.
- M. Israeli, T. Klein, G. Brandhorst and M. Oellerich, Clin. Chim. Acta, 2012, 413, 1374–1378 CrossRef CAS PubMed.
- A. Picascia, T. Infante and C. Napoli, Clin. Exp. Nephrol., 2012, 16, 373–381 CrossRef CAS PubMed.
- M. W. Mather, S. Powell, B. Talks, C. Ward, C. D. Bingle, M. Haniffa and J. Powell, Expert Rev. Mol. Med., 2021, 23, 1–10 CrossRef PubMed.
- K. L. Mansfield, S. A. La Rocca, M. Khatri, N. Johnson, F. Steinbach and A. R. Fooks, J. Virol. Methods, 2013, 188, 139–144 CrossRef CAS PubMed.
- D. R. Burton and L. Hangartner, Annu. Rev. Immunol., 2016, 34, 635 CrossRef CAS PubMed.
- Y. F. W. Wang and M. Kobayashi, Advanced Techniques in Diagnostic Microbiology, Springer, US, 2013, pp. 53–73 Search PubMed.
- Y. F. Wang, Advanced Techniques in Diagnostic Microbiology: Volume 1: Techniques, Springer International Publishing, 3rd edn, 2018, pp. 127–147 Search PubMed.
- A. H. B. B. Wu, Clin. Chim. Acta, 2006, 369, 119–124 CrossRef CAS PubMed.
- C.-C. C. Lai, C.-Y. Y. Wang and P.-R. R. Hsueh, J. Microbiol., Immunol. Infect., 2020, 53, 505–512 CrossRef CAS PubMed.
- A. P. Espejo, Y. Akgun, A. F. Al Mana, Y. Tjendra, N. C. Millan, C. Gomez-Fernandez and C. Cray, Am. J. Clin. Pathol., 2020, 154, 293–304 CrossRef CAS PubMed.
- Y. Zhou, Y. Wu, L. Ding, X. Huang and Y. Xiong, Trends Anal. Chem., 2021, 145, 116452 CrossRef CAS PubMed.
- J. He, S. Zhu, J. Zhou, W. Jiang, L. Yin, L. Su, X. Zhang, Q. Chen and X. Li, Front. Bioeng. Biotechnol., 2022, 10, 1090281 CrossRef PubMed.
- M. Sajid, A.-N. Kawde and M. Daud, J. Saudi Chem. Soc., 2014, 19, 689–705 CrossRef.
- J. Park, Sensors, 2022, 22, 7398 CrossRef CAS PubMed.
- F. Di Nardo, M. Chiarello, S. Cavalera, C. Baggiani and L. Anfossi, Sensors, 2021, 21, 5185 CrossRef CAS PubMed.
- S. Y. Hong, Y. M. Park, Y. H. Jang, B. H. Min and H. C. Yoon, BioChip J., 2012, 6, 213–220 CrossRef CAS.
- J. Oh, S. J. Kwon, J. S. Dordick, W. J. Sonstein, R. J. Linhardt and M. G. Kim, Theranostics, 2019, 9, 4182–4191 CrossRef CAS PubMed.
- K. V. Serebrennikova, J. V. Samsonova and A. P. Osipov, Microchim. Acta, 2019, 186, 423 CrossRef PubMed.
- K. Won Lee, Y. Chan Yu, H. Jin Chun, Y. Han Jang, Y. Duk Han and H. C. Yoon, Biosensors, 2020, 10, 87 CrossRef PubMed.
- R. M. El-Shabasy, M. A. Nayel, M. M. Taher, R. Abdelmonem, K. R. Shoueir and R. Kenawy, Int. J. Biol. Macromol., 2022, 204, 161–168 CrossRef CAS PubMed.
- M. Karamese, Eurasian J. Med., 2022, 54, S106–S114 CrossRef CAS PubMed.
- D. P. Maison, Y. Deng and M. Gerschenson, Front. Immunol., 2023, 14, 1195871 CrossRef CAS PubMed.
- G. Alhamid, H. Tombuloglu, A. A. Rabaan and E. Al-Suhaimi, Saudi J. Biol. Sci., 2022, 29, 103465 CrossRef CAS PubMed.
- R. Patel, M. Kaki, V. S. Potluri, P. Kahar and D. Khanna, Hum. Vaccines Immunother., 2022, 18, 1–12 Search PubMed.
- M. J. Hogan and N. Pardi, Annu. Rev. Med., 2022, 73, 17–39 CrossRef CAS PubMed.
- M. Li, H. Wang, L. Tian, Z. Pang, Q. Yang, T. Huang, J. Fan, L. Song, Y. Tong and H. Fan, Signal Transduction Targeted Ther., 2022, 7, 146 CrossRef CAS PubMed.
- Y. Shao, Y. Wu, Y. Feng, W. Xu, F. Xiong and X. Zhang, Front. Med., 2022, 16, 185–195 CrossRef PubMed.
- C. Zheng, W. Shao, X. Chen, B. Zhang, G. Wang and W. Zhang, Int. J. Infect. Dis., 2022, 114, 252–260 CrossRef CAS PubMed.
- H. Ward, M. Whitaker, B. Flower, S. N. Tang, C. Atchison, A. Darzi, C. A. Donnelly, A. Cann, P. J. Diggle, D. Ashby, S. Riley, W. S. Barclay, P. Elliott and G. S. Cooke, Nat. Commun., 2022, 13, 907 CrossRef CAS PubMed.
- T. A. Bates, S. K. McBride, H. C. Leier, G. Guzman, Z. L. Lyski, D. Schoen, B. Winders, J. Y. Lee, D. X. Lee, W. B. Messer, M. E. Curlin and F. G. Tafesse, Sci. Immunol., 2022, 7, 1–8 Search PubMed.
- L. Coppeta, C. Ferrari, G. Somma, A. Mazza, U. D’ancona, F. Marcuccilli, S. Grelli, M. T. Aurilio, A. Pietroiusti, A. Magrini and S. Rizza, Vaccines, 2022, 10, 141 CrossRef CAS PubMed.
- M. I. Samanovic, A. R. Cornelius, S. L. Gray-Gaillard, J. R. Allen, T. Karmacharya, J. P. Wilson, S. W. Hyman, M. Tuen, S. B. Koralov, M. J. Mulligan and R. S. Herati, Sci. Transl. Med., 2022, 14, 8961 CrossRef PubMed.
- T. Gebrecherkos, Y. K. Kiros, F. Challa, S. Abdella, A. Gebreegzabher, D. Leta, A. Desta, A. Hailu, G. Tasew, M. Abdulkader, M. Tessema, G. Tollera, T. Kifle, Z. G. Arefaine, H. H. D. F. Schallig, E. R. Adams, B. C. Urban, T. F. Rinke de Wit and D. Wolday, PLoS One, 2022, 17, 1–18 CrossRef PubMed.
- Q. Fernandes, V. P. Inchakalody, M. Merhi, S. Mestiri, N. Taib, D. Moustafa Abo El-Ella, T. Bedhiafi, A. Raza, L. Al-Zaidan, M. O. Mohsen, M. A. Yousuf Al-Nesf, A. A. Hssain, H. M. Yassine, M. F. Bachmann, S. Uddin and S. Dermime, Ann. Med., 2022, 54, 524–540 CrossRef CAS PubMed.
- G. McLean, J. Kamil, B. Lee, P. Moore, T. F. Schulz, A. Muik, U. Sahin, Ö. Türeci and S. Pather, mBio, 2022, 13, 1–24 CrossRef CAS PubMed.
- K. J. Bruxvoort, L. S. Sy, L. Qian, B. K. Ackerson, Y. Luo, G. S. Lee, Y. Tian, A. Florea, H. S. Takhar, J. E. Tubert, C. A. Talarico and H. F. Tseng, Lancet Reg. Heal. Am., 2022, 6, 100134 Search PubMed.
- P. Mistry, F. Barmania, J. Mellet, K. Peta, A. Strydom, I. M. Viljoen, W. James, S. Gordon and M. S. Pepper, Front. Immunol., 2022, 12, 809244 CrossRef PubMed.
- B. Zeng, L. Gao, Q. Zhou, K. Yu and F. Sun, BMC Med., 2022, 20, 200 CrossRef CAS PubMed.
- K. A. Holder, D. P. Ings, D. O. A. Harnum, R. S. Russell and M. D. Grant, NPJ vaccines, 2022, 7, 122 CrossRef CAS PubMed.
- S. Pilz, V. Theiler-Schwetz, C. Trummer, R. Krause and J. P. A. Ioannidis, Environ. Res., 2022, 209, 112911 CrossRef CAS PubMed.
- D. R. Feikin, M. M. Higdon, L. J. Abu-Raddad, N. Andrews, R. Araos, Y. Goldberg, M. J. Groome, A. Huppert, K. L. O’Brien, P. G. Smith, A. Wilder-Smith, S. Zeger, M. Deloria Knoll and M. K. Patel, Lancet, 2022, 399, 924–944 CrossRef CAS PubMed.
- Y. Mao, W. Wang, J. Ma, S. Wu and F. Sun, Chin. Med. J., 2022, 135, 145 CrossRef PubMed.
- P. Ssentongo, A. E. Ssentongo, N. Voleti, D. Groff, A. Sun, D. M. Ba, J. Nunez, L. J. Parent, V. M. Chinchilli and C. I. Paules, BMC Infect. Dis., 2022, 22, 1–12 CrossRef PubMed.
- C. Stein, H. Nassereldine, R. J. D. Sorensen, J. O. Amlag, C. Bisignano, S. Byrne, E. Castro, K. Coberly, J. K. Collins, J. Dalos, F. Daoud, A. Deen, E. Gakidou, J. R. Giles, E. N. Hulland, B. M. Huntley, K. E. Kinzel, R. Lozano, A. H. Mokdad, T. Pham, D. M. Pigott, R. C. Reiner, T. Vos, S. I. Hay, C. J. L. Murray and S. S. Lim, Lancet, 2023, 401, 833–842 CrossRef PubMed.
- S. Galmiche, L. B. Luong Nguyen, E. Tartour, X. de Lamballerie, L. Wittkop, P. Loubet and O. Launay, Clin. Microbiol. Infect., 2022, 28, 163–177 CrossRef CAS PubMed.
- H. Meng, J. Mao and Q. Ye, J. Med. Virol., 2022, 94, 2369–2375 CrossRef CAS PubMed.
- K. Manothummetha, N. Chuleerarux, A. Sanguankeo, O. S. Kates, N. Hirankarn, A. Thongkam, M. V. Dioverti-Prono, P. Torvorapanit, N. Langsiri, N. Worasilchai, C. Moonla, R. Plongla, W. M. Garneau, A. Chindamporn, P. Nissaisorakarn, T. Thaniyavarn, S. Nematollahi and N. Permpalung, JAMA Netw. open, 2022, 5, E226822 CrossRef PubMed.
- P. Langerbeins and M. Hallek, Blood, 2022, 140, 236–252 CrossRef CAS PubMed.
- N. Gagelmann, F. Passamonti, C. Wolschke, R. Massoud, C. Niederwieser, R. Adjalle, B. Mora, F. Ayuk and N. Kroger, Haematologica, 2022, 107, 1840–1849 CrossRef CAS PubMed.
- J. Barrière, M. Carles, C. Audigier-Valette, D. Re, Z. Adjtoutah, B. Seitz-Polski, V. Gounant, D. Descamps and G. Zalcman, Eur. J. Cancer, 2022, 162, 182–193 CrossRef PubMed.
- C. Corti, G. Antonarelli, F. Scotté, J. P. Spano, J. Barrière, J. M. Michot, F. André and G. Curigliano, Ann. Oncol., 2022, 33, 158–168 CrossRef CAS PubMed.
- F. Capone, M. Rossi, A. Cruciani, F. Motolese, F. Pilato and V. Di Lazzaro, Neural Regener. Res., 2023, 18, 284 CAS.
- A. Jena, D. James, A. K. Singh, U. Dutta, S. Sebastian and V. Sharma, Clin. Gastroenterol. Hepatol., 2022, 20, 1456–1479.e18 CrossRef CAS PubMed.
- A. Jena, S. Mishra, P. Deepak, P. Kumar-M, A. Sharma, Y. I. Patel, N. A. Kennedy, A. H. J. Kim, V. Sharma and S. Sebastian, Autoimmun. Rev., 2022, 21, 102927 CrossRef CAS PubMed.
- S. Dowlatshahi, E. Shabani and M. J. Abdekhodaie, Arch. Virol., 2021, 166, 715–731 CrossRef CAS PubMed.
- G. Fedele, A. Palmieri, C. Damiano, A. Di Lonardo, P. Leone, I. Schiavoni, C. Trevisan, A. M. Abbatecola, C. Cafariello, A. Malara, P. Minchella, G. Panduri, R. Antonelli Incalzi, A. T. Palamara, P. Stefanelli and G. Onder, Aging: Clin. Exp. Res., 2022, 34, 2577–2584 Search PubMed.
- R. Peng, Y. Pan, Z. Li, Z. Qin, J. M. Rini and X. Liu, Biosens. Bioelectron., 2022, 197, 113762 CrossRef CAS PubMed.
- A. K. M. Furuya, C. Wagner, J. Connors, J. Bodnar, T. Miller, N. L. Ryman, C. J. Warszycki, S. J. Wong, A. C. Walsh, K. A. McDonough and W. T. Lee, Diagn. Microbiol. Infect. Dis., 2021, 99, 115298 CrossRef CAS PubMed.
- C. Cervia, Y. Zurbuchen, P. Taeschler, T. Ballouz, D. Menges, S. Hasler, S. Adamo, M. E. Raeber, E. Bächli, A. Rudiger, M. Stüssi-Helbling, L. C. Huber, J. Nilsson, U. Held, M. A. Puhan and O. Boyman, Nat. Commun., 2022, 13, 1–12 Search PubMed.
- L. Spicuzza, D. Campagna, C. Di Maria, E. Sciacca, S. Mancuso, C. Vancheri and G. Sambataro, AIMS Microbiol., 2023, 9, 375 CAS.
- Y. Zhou, Y. Chen, W. Liu, H. Fang, X. Li, L. Hou, Y. Liu, W. Lai, X. Huang and Y. Xiong, Sens. Actuators, B, 2021, 343, 130139 CrossRef CAS PubMed.
- S. Medrano, M. Martínez-Rodríguez, L. Vallejo, E. Culebras and A. Delgado-Iribarren, Rev. Española Quimioter., 2022, 35, 538 CAS.
- S. Cavalera, E. Alladio, Efrem, E. A. Foglia, S. Grazioli, B. Colitti, S. Rosati, C. Nogarol, F. Di Nardo, T. Serra, V. Testa, C. Baggiani, G. Maccabiani, E. Brocchi and L. Anfossi, Microchim. Acta, 2024, 191, 9 CrossRef CAS PubMed.
- S. Cavalera, G. Pezzoni, S. Grazioli, E. Brocchi, S. Baselli, D. Lelli, B. Colitti, T. Serra, F. Di Nardo, M. Chiarello, V. Testa, S. Rosati, C. Baggiani and L. Anfossi, Biosensors, 2022, 12, 739 CrossRef CAS PubMed.
- M. Daperno, C. Guiotto, I. Casonato, G. Pagana, S. Micalizzi, M. C. R. Azzolina, C. Norbiato, D. Cosseddu and R. Rocca, Intern. Med. J., 2021, 51, 1049–1059 CrossRef CAS PubMed.
- N. G. Khlebtsov, Anal. Chem., 2008, 80, 6620–6625 CrossRef CAS PubMed.
- J. Turkevich, P. C. Stevenson and J. Hillier, Discuss. Faraday Soc., 1951, 11, 55–75 RSC.
- S. Cavalera, A. Russo, E. A. Foglia, S. Grazioli, B. Colitti, S. Rosati, C. Nogarol, F. Di Nardo, T. Serra, M. Chiarello, C. Baggiani, G. Pezzoni, E. Brocchi and L. Anfossi, Talanta, 2022, 240, 123–155 CrossRef PubMed.
- S. Cavalera, F. Di Nardo, L. Forte, F. Marinoni, M. Chiarello, C. Baggiani and L. Anfossi, Sensors, 2020, 20, E6609–E6609 CrossRef PubMed.
- S. Cavalera, B. Colitti, S. Rosati, G. Ferrara, L. Bertolotti, C. Nogarol, C. Guiotto, C. Cagnazzo, M. Denina, F. Fagioli, F. Di Nardo, M. Chiarello, C. Baggiani and L. Anfossi, Talanta, 2020, 223, 121737 CrossRef PubMed.
- S. Cavalera, G. Pezzoni, S. Grazioli, E. Brocchi, S. Baselli, D. Lelli, B. Colitti, T. Serra, F. Di Nardo, M. Chiarello, V. Testa, S. Rosati, C. Baggiani and L. Anfossi, Biosensors, 2022, 12, 739 CrossRef CAS PubMed.
- S. Cavalera, B. Colitti, G. M. De Mia, F. Feliziani, S. D. Giudici, P. P. Angioi, F. D’Errico, D. Scalas, A. Scollo, T. Serra, M. Chiarello, V. Testa, F. Di Nardo, C. Baggiani, A. Oggiano, S. Rosati and L. Anfossi, Talanta, 2023, 258, 124443 CrossRef CAS PubMed.
Footnote |
| † Electronic supplementary information (ESI) available: (S1) salt-induced aggregation test; (S2) gold conjugate synthesis; (S3) optimization of the three serological LFIA device for format selection by design of experiments; (S4) characterisation the AuNP conjugates. See DOI: https://doi.org/10.1039/d3tb02895j |
| This journal is © The Royal Society of Chemistry 2024 |