Open Access Article
Open Access ArticleCreative Commons Attribution 3.0 Unported Licence
Trehalose-polyamine/DNA nanocomplexes: impact of vector architecture on cell and organ transfection selectivity†
Fernando
Ortega-Caballero
 ab,
María L.
Santana-Armas
c,
Conchita
Tros de Ilarduya
ab,
María L.
Santana-Armas
c,
Conchita
Tros de Ilarduya
 c,
Christophe
Di Giorgio
d,
Raphäel
Tripier
c,
Christophe
Di Giorgio
d,
Raphäel
Tripier
 e,
Nathalie
Le Bris
e,
Nathalie
Le Bris
 e,
Cedric
Ollier
e,
Cedric
Ollier
 e,
Carmen
Ortiz Mellet
e,
Carmen
Ortiz Mellet
 a,
José M.
García Fernández
a,
José M.
García Fernández
 f,
José L.
Jiménez Blanco
f,
José L.
Jiménez Blanco
 *a and
Alejandro
Méndez-Ardoy
*a and
Alejandro
Méndez-Ardoy
 *af
*af
aDepartment of Organic Chemistry, Faculty of Chemistry, University of Seville, c/Profesor García González 1, Sevilla 41012, Spain. E-mail: jljb@us.es alejandroma@us.es
bDepartment of Organic Chemistry, Higher Polytechnic School, University of Seville, c/Virgen de África 7, Sevilla 41011, Spain
cDepartment of Pharmaceutical Sciences, School of Pharmacy and Nutrition, University of Navarra, Pamplona 31080, Spain
dInstitut de Chimie Nice, UMR 7272, Université Côte d’Azur, 28 Avenue de Valrose, Nice 06108, France
eUniversité de Brest, UMR CNRS 6521 CEMCA, 6 Avenue Victor le Gorgeu, Brest 29238, France
fInstituto de Investigaciones Químicas (IIQ), CSIC –, Universidad de Sevilla, Avda. Américo Vespucio 49, Sevilla 41092, Spain
First published on 5th March 2024
Abstract
A novel family of precision-engineered gene vectors with well-defined structures built on trehalose and trehalose-based macrocycles (cyclotrehalans) comprising linear or cyclic polyamine heads have been synthesized through procedures that exploit click chemistry reactions. The strategy was conceived to enable systematic structural variations and, at the same time, ensuring that enantiomerically pure vectors are obtained. Notably, changes in the molecular architecture translated into topological differences at the nanoscale upon co-assembly with plasmid DNA, especially regarding the presence of regions with short- or long-range internal order as observed by TEM. In vitro and in vivo experiments further evidenced a significant impact on cell and organ transfection selectivity. Altogether, the results highlight the potential of trehalose-polyamine/pDNA nanocomplex monoformulations to achieve targeting transfection without the need for any additional cell- or organ-sorting component.
Introduction
A range of new therapies relay on the safe delivery of nucleic acids into cells.1,2 One of the main hurdles is developing carrier systems (vectors) with the ability to transport the therapeutic gene material with high efficiency and selectivity to the target site of action, avoiding off-target effects. While viruses are well-suited for this purpose, viral systems still bring concerns about immunogenicity, random integration in the host genome, limited DNA packaging capacity and the cost of clinical grade production of large batches.3 Non-viral vector systems are an attractive alternative to bypass these issues.4 Currently approved non-viral gene therapies include the siRNA drug Patisiran (Onpattro),5 or the mRNA-based vaccines6 that played an essential role in global prophylaxis during the SARS-COV-2 pandemic,7 with a number of clinical trials ongoing.8Most of the nonviral vectors commonly used in nucleic acid therapeutics are intrinsically polydisperse in nature (e.g., cationic polymers),9–11 lack conformational definition (e.g., cationic dendrimers) and/or require multicomponent formulations (e.g. cationic lipids),9 which represents a major limitation for conducting structure–activity relationship (SAR) studies in view of vector optimization.12–15 Consequently, an urgent need remains in the field for well-defined molecular vectors susceptible to systematic chemical modifications through procedures warranting enantiomeric purity and well-suited for diversity-oriented schemes.16–22 Molecular vectors fulfilling these postulates have become accessible by exploiting multifunctional platforms amenable to precision chemistry transformations.23–28 Carbohydrate-based scaffolds have proven particularly useful for such endeavor. Most of the work in this area has focused on the realm of cyclodextrins. Their axial symmetry, neat face differentiation and distinct hydroxyl reactivity allow the installation of multivalent displays of functional elements for nucleic acid recognition (i.e., cation ionizable groups and hydrogen-bonding centers) and self-organization (e.g., lipid tails or aromatic modules) with precise relative orientations.29–33 Interestingly, such favorable features are shared by the C2-symmetric disaccharide α,α’-trehalose. This was recently exploited to develop a novel family of gemini-type molecular vectors endowed with facial amphiphilicity, termed Siamese-twin surfactants that exhibited efficient DNA nanocomplexation and delivery in vitro and in vivo.34 Further on, α,α'-trehalose-based macrocycles (cyclotrehalans, CTs) were found to undergo co-assembly with plasmid DNA (pDNA) to afford transfectious nanocomplexes (CTplexes) with organ-selective gene delivery capabilities.35
The ensemble of the reported results on trehalose-based non-viral vectors evidences a remarkable influence of molecular architecture on the topological characteristics of the resulting vector-DNA nanocomplexes. The latter in turn translates into distinct cell selectivity and tissue tropism in mice models, probably due to shape and surface property-differentiated physiological barrier escape properties.36
The possibility of imparting targeting abilities by molecular vector design, without the need for installing a biorecognizable ligand or the coformulation with a third component, opens new avenues in non-viral gene delivery that remain largely unexplored. In this work we probe the effect of different linear and cyclic arrangements of polyamine heads displayed on trehalose or on a dimeric cyclotrehalan (Fig. 1) in the pDNA complexation and the transfection outcome in vitro and in vivo. We keep in mind reported molecular modelling studies on linear polyamines37–39 suggesting that their self-folding might hamper an optimal DNA complexation.40 Cyclic polyamines, such as cyclen (1,4,7,10-tetraazacyclododecane) and cyclam (1,4,8,11-tetraazacyclotetradecane), can mitigate this detrimental effect. Indeed, cyclen and cyclam have been proposed as delivery assistants after complexation with metallic cations, but less frequently as fundamental motifs to drive DNA binding.41–50
 | ||
| Fig. 1 Chemical structures of the trehalose-based molecular vector prototypes prepared and assayed in this work. All compounds 1–7 were isolated as the corresponding per-hydrochloride salts. | ||
Results and discussion
Synthesis of trehalose and cyclotrehalan gene vectors
The collection of trehalose-polyamine vectors prepared in this work is depicted in Fig. 1. They were conceived to assess the effect of linear versus cyclic polyamine arrangements, amphiphilic versus non amphiphilic character, macrocyclic dimers versus monomeric trehalose scaffolds and thiourea versus 1,2,3-triazole connectors in their pDNA complexation and delivery properties. The radial polycationic derivative 1 was prepared as reported previously (Fig. 1).32 The novel Janus-type amphiphilic derivatives 2–7 were accessed by “click” multiconjugation of cyclotrehalan or trehalose derivatives and suitably functionalized polyamine building blocks. Thus, a thiourea-forming reaction between cyclotrehalan hexaamine 8 and isothiocyanates 9 or 10 gave the corresponding Boc-protected adducts 11 and 12 (35% and 74% isolated yields). Subsequent acidic treatment afforded the target vectors 2 and 3 (Scheme 1). Similarly, the trehalose-cyclam thiourea conjugate 4 was obtained by coupling the trehalose diisothiocyanate 13 and the 2-aminoethyl cyclam derivative 1451 (→ 15), followed by Boc-deprotection (61% yield, Scheme 2). The triazole-linked vectors 5–7 were prepared by Cu(I)-catalyzed azide–alkyne coupling (CuAAC) between diazide 17 and alkynes 16–19 (→ 20-22) using resin-linked Cu(I)52 in H2O-tBuOH and final carbamate hydrolysis (38–71%; Scheme 3). A refined synthesis for compound 5 has been autonomously developed within the framework of a distinct project; comprehensive details are presented in a parallel paper.53pDNA complexation studies
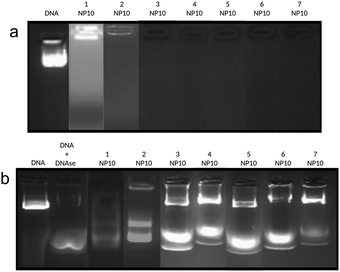
In order to determine the pDNA complexation abilities of 1–7, we mixed the vectors with a plasmid encoding luciferase (pCMV-Luc VR1216, 6934 bp) at different nitrogen (N) to phosphorous (P) stoichiometric ratios (N/P) in BHG buffer (HEPES 10 mM, pH 7.4, glucose 5% w/v). The pDNA protection efficacy and nanoparticle characteristics (size, surface potential, and morphology) were determined by electrophoretic mobility shift assay (EMSA) in agarose gel, dynamic light scattering (DLS), ζ-potential measurements and transmission electron microscopy (TEM).Gel retardation experiments using GelRed® as a visualization agent revealed that all vectors were able to inhibit DNA migration to positive potentials at N/P 10, therefore supporting that the negative charges of the DNA phosphate backbone are effectively screened by the vector cationic clusters. Except for 1, pDNA staining is prevented in the presence of the vector, suggesting limited accessibility of the plasmid in the nanocomplex to the intercalating probe (Fig. 2a). To test the protection capabilities of the vectors towards enzymatic degradation, the nanocomplexes were incubated in the presence of DNAse, then treated with sodium dodecylsulfate (SDS) to provoke disruption and pDNA release and tested by EMSA (Fig. 2b). pDNA degradation results in total fading of the fluorescence bands in the gel, as observed for the control.
The relatively weak fluorescence intensity in the case of complexes prepared from the cationic vector 1 points to incomplete protection. On the other hand, complexes prepared from 3–7 showed the presence of both pristine DNA and partially degraded DNA. This indicates that the protection abilities of vectors 3–7 were superior to those of 1–2.
The nanoparticle size distribution and surface charge were determined in solution by DLS measurements. The hydrodynamic diameter (Dh) values and ζ-potential of the formulations at N/P 10 and 20 are shown in Table 1.
| Vector | N/P 10 | N/P 20 | ||
|---|---|---|---|---|
| D h (nm) | ζ-potential (mV) | Size (nm) | ζ-potential (mV) | |
| a Data are from ref. 32 and are here included for comparative purposes. | ||||
| 1 | 133 ± 6 | +24 ± 3 | 107 ± 6 | +25 ± 1 |
| 2 | 166 ± 1 | +17 ± 2 | 103 ± 3 | +18 ± 1 |
| 3 | 111 ± 9 | +21 ± 1 | 91 ± 3 | +16 ± 3 |
| 4 | 101 ± 11 | +19 ± 5 | 90 ± 2 | +13 ± 1 |
| 5 | 157 ± 12 | +10 ± 5 | 122 ± 26 | +25 ± 10 |
| 6 | 173 ± 4 | +20 ± 4 | 139 ± 7 | +24 ± 5 |
| 7 | 161 ± 9 | +12 ± 3 | 132 ± 5 | +21 ± 5 |
The Dh values ranged from 90 to 173 nm, and surface charges were positive in all cases, ranging from +13 to +25 mV. Generally, the higher the N/P ratio, the smaller the nanoparticle size, which is compatible with a larger fraction of phosphates screened by the protonated amines. The incorporation of cyclic polyamines (e.g.2vs.3) had a positive impact on the ability of the vectors to generate smaller nanoparticles. In addition, the use of the thiourea linkers (e.g.4vs.5 and 6) also yielded smaller nanoparticles, suggesting that the cationic and hydrogen bond donor centers are optimally arranged to promote pDNA condensation upon co-assembly. At higher N/P ratios (20), cyclotrehalan-based DNA complexes (2 and 3) showed generally smaller hydrodynamic diameters than complexes prepared with trehalose derivatives 4, 5, 6 and 7 (91–103 nm vs. 90–139 nm). This could be related to entropic effects arising from the multivalent exposure of a higher number of cationic arms in the cyclotrehalan-based vectors.
The morphology of the nanocomplexes was investigated by TEM after uranyl acetate staining (Fig. 3).
 | ||
| Fig. 3 Representative TEM micrographs of the DNA-vector complexes at N/P 20 in HEPES 20 mM, pH 7.4, DNA 303 μM phosphate. | ||
The non-amphiphilic derivative 1 (Fig. 3a and Fig. S1, ESI†) co-assembled with pDNA into networked blobs of about 200 nm with no observable internal structure. Differently, the micrographs of the nanocomplexes formulated from the amphiphilic vectors 2–7 showed well-defined quasi-spherical or pear-shaped nanoparticles with different ultrathin organizations. Thus, for compounds 2, 5 and 7 onion-like long-range multilamellar arrangements of dark (high electron density) and light regions are apparent (Fig. 3b, e, and g and Fig. S1, ESI†), in agreement with alternate dispositions of pDNA and cationic lipid bilayers (Fig. 3h and Fig. S1, ESI†).30 Nanocomplexes obtained from 3 and 4 also showed alternating dark and light regions, but in a highly sinusoidal short-range arrangement with multiple arcs (Fig. 3c, d and Fig. S1, ESI†). Compound 6 nanocomplexes had mixed characteristics, with areas displaying long- and short-range order (Fig. 3f and Fig. S1, ESI†).
In vitro gene delivery and toxicity studies
The pDNA delivery efficiencies of vectors 1–7 were first evaluated in vitro in African green monkey kidney fibroblast (COS-7), human cervix cancer (HeLa), human cellular hepatocarcinoma (HepG2) and mouse ascites macrophage (RAW264.7) cells. After 4 h incubation, the medium was replaced and the cells were incubated for 48 h, then lysed and the luciferase expression measured and standardised against the total amount of protein per well. The results are shown in Fig. 4.In general, increasing the N/P ratio from 10 to 20 led to higher transfection levels (Fig. 4b), though at the expense of a toxicity penalty (see below). The tetraethyleneimine-decorated vectors showed the highest efficacy across all cell lines, overpassing poly(ethyleneimine)-formulated polyplexes (N/P 10) used as a positive control (Fig. 4b). They behave as broad range transfection agents, performing especially well in HeLa and HepG2 cells. Trehalose-cyclam and -cyclen-based vectors generally reached lower expression levels but displayed higher cell selectivity. Thus, compounds 3 and 4 mediated transfection preferentially in RAW264.7 cells, 5 and 7 in HeLa cells and 6 in HepG2, while they all lacked substantial expression levels in COS-7 cells. Ascribing these differences in relative cell transfection performances to the above commented dissimilarities in the internal order in the corresponding nanocomplexes is appealing. Indeed, a direct relationship between the presence of short-range order, long-range order, or mixed areas, in the nanocomplexes and the transfection outcome can be drawn. Such generalization must be taken with care, however. The diversity of transfection efficiency across cell lines can reflect intrinsic differences in endocytic processes or endosomal escape mechanisms that depend on an ensemble of cell and vector factor.52
Toxicity of the formulations prepared in this work was assessed by the MTT assay. The data revealed low toxicity in all cases (Fig. S2, ESI†). At N/P 10, cell viabilities were above 90%. Increasing the N/P ratio to 20 led to an increase in toxicity. Nevertheless, it remained above 80% with an average cell viability of 90%. Anyway, all formulations with the synthesized vectors display lower toxicity than PEI polyplexes (50–75% cell viability across the different cell lines; ESI,† Fig. S2).
In vivo gene delivery
The in vivo delivery efficiency was evaluated after systemic injection of the nanocomplexes prepared at N/P 10 (HEPES 10 mM, pH 7.4, glucose 5% w/v) in the mouse tail vein. All experiments were carried out according to ethical guidelines established by Directive 86/609/EEC-RD 53/2013 February 1, and with the approval of the Committee on Animal Research at the University of Navarra (number of protocol 017-19). Twenty-four hours after injection the mice were sacrificed, and the luciferase expression measured in heart, kidneys, liver, lungs, and spleen (Fig. 5). Surprisingly, the formulation obtained from the non-amphiphilic derivative 1 showed negligible luciferase levels in all organs. This could arise from aggregation leading to premature clearance or because of degradation due to inefficient cargo protection, according to the EMSA data. On the opposite, compound 2 promoted substantial luciferase expression in all organs, with higher expression levels in the lungs and the liver. Paralleling the in vitro results, cyclam and cyclen derivatives 3–7 revealed a lower total expression of luciferase but a higher selectivity towards specific organs. Hence, vectors 3, 4 and 6 showed preferential expression in the lungs, the spleen, and the liver, respectively, whereas vectors 5 and 7 showed a marked tropism to the kidney (Fig. 5a and b).The data evidence a clear advantage of amphiphilic over non-amphiphilic molecular vectors for in vivo applications. They also support that restricting the flexibility of the cationic centres by using cyclic instead of linear polyamines as cationic heads enhances organ selectivity. This effect can only in part be rationalized in terms of the impact of changes at the molecular level on the organizational patterns at the nanoscale.
Other molecular and supramolecular features may be responsible for such observations. For instance, compounds 2, 3 and 4, the three forming spherical nanocomplexes with onion-line multilamellar internal structures, show a differential organ selectivity. Thus, polyethyleneimine amphiphile 2 behaves as a broad-range organ selective vector with preference towards liver and lungs. In contrast, cyclam derivatives 3 and 4 are more selective and exhibit differential preference for the lungs and the spleen, respectively. Nanocomplexes prepared from 5 and 7, which exhibit a characteristic sinusoidal internal structure, both transfect preferentially the kidney, but the cyclen derivative 7 is significantly more selective than the cyclam analogue 5. Finally, the enhanced tropism of 6 towards the liver as compared with 2 might be due to the partial loss long-range order of the corresponding nanocomplexes.
Conclusions
A novel collection of structurally defined gene delivery systems with varied molecular architectures have been prepared from trehalose and linear or cyclic polyamine precursors by a synthetic strategy based on click chemistry multiconjugation reactions. Co-formulation with pDNA rendered self-assembled transfectious nanocomplexes with topological characteristics and internal organizational patterns that are strongly dependent on the molecular features of the vector. A comparative evaluation of the in vitro and in vivo transfection capabilities evidenced remarkable increases in cell and organ selectivity for compounds bearing cyclic polyamine-type cationic heads. Depending on the overall architecture, different cells and organs could be targeted without the need for involving any biorecognition component. Collectively, these results reinforce the notion that subtle differences in the molecular vector design can have a meaningful impact at the nanoscale and strongly influence the ability of the resulting nanocomplexes to cross biological barriers. They further reveal a new strategy to finely customize the vector in order to program a precise destination in biological environments. Finally, they warrant additional investigations on molecular gene vector structure-nanocomplex property-transfection efficiency relationships in view of realizing the goal of ligand-free site-specific nucleic acid therapeutics.Author contributions
F. Ortega Caballero: investigation, supervision and writing-review & editing. María L. Santana-Armas: investigation. C. Tros de Ilarduya: investigation, Christophe Di Giorgio: investigation. Raphäel Tripier: resources. Nathalie Le Bris: investigation, resources. Cedric Ollier: investigation. C. Ortiz Mellet: project administration, funding acquisition and writing-review & editing. J. M. García Fernández: conceptualization, project administration and writing-review & editing. J. L. Jiménez Blanco: supervision, investigation and writing – original draft. Alejandro Méndez-Ardoy: investigation, writing – original draft.Conflicts of interest
There are no conflicts to declare.Acknowledgements
We acknowledge the Ministerio de Ciencia, Innovación y Universidades and the Agencia Estatal de Investigación (projects RTI2018-097609-B-C21 and RTI2018-097609-B-C22), AEI/10.13039/501100011033 and “ERDF A way of making Europe” (PID2021-124247OB-C21 and PID2022-141034OB-C22) and the COST action GLYCONanoPROBES (CM18132). J. M. G. F. acknowledges funding by the European Union's Horizon Europe research and innovation programme under the Marie Skłodowska-Curie grant agreement 101130235-Bicyclos. This work was supported by Grant RYC2021-034263-I (A. M.-A.), founded by the Ministry of Science and Innovation/State Research Agency/10.13039/501100011033 and the European Union “NextGenerationEU/Recovery, Transformation and Resilience Plan”. The CITIUS (University of Seville) is also acknowledged for technical support.Notes and references
- B. B. Mendes, J. Conniot, A. Avital, D. Yao, X. Jiang, X. Zhou, N. Sharf-Pauker, Y. Xiao, O. Adir, H. Liang, J. Shi, A. Schroeder and J. Conde, Nat. Rev. Methods Primers, 2022, 2, 24 CrossRef CAS PubMed.
- A. Gupta, J. L. Andresen, R. S. Manan and R. Langer, Adv. Drug Delivery Rev., 2021, 178, 113834 CrossRef CAS PubMed.
- K. Kaygisiz and C. V. Synatschke, Biomater. Sci., 2020, 8, 6113–6156 RSC.
- I. Lostalé-Seijo and J. Montenegro, Nat. Rev. Chem., 2018, 2, 258–277 CrossRef.
- E. A. Narasipura, R. VanKeulen-Miller, Y. Ma and O. S. Fenton, Bioconjugate Chem., 2023, 34, 1177–1197 CrossRef CAS PubMed.
- U. Sahin, K. Karikó and Ö. Türeci, Nat. Rev. Drug Discovery, 2014, 13, 759–780 CrossRef CAS PubMed.
- M. D. Shin, S. Shukla, Y. H. Chung, V. Beiss, S. K. Chan, O. A. Ortega-Rivera, D. M. Wirth, A. Chen, M. Sack, J. K. Pokorski and N. F. Steinmetz, Nat. Nanotechnol., 2020, 15, 646–655 CrossRef CAS PubMed.
- C. Chen, Z. Yang and X. Tang, Med. Res. Rev., 2018, 38, 829–869 CrossRef PubMed.
- I. S. Pinto, R. A. Cordeiro and H. Faneca, J. Controlled Release, 2023, 353, 196–215 CrossRef CAS PubMed.
- T. T. Smith, S. B. Stephan, H. F. Moffett, L. E. McKnight, W. Ji, D. Reiman, E. Bonagofski, M. E. Wohlfahrt, S. P. S. Pillai and M. T. Stephan, Nat. Nanotechnol., 2017, 12, 813–820 CrossRef CAS PubMed.
- J. M. Priegue, D. N. Crisan, J. Martínez-Costas, J. R. Granja, F. Fernandez-Trillo and J. Montenegro, Angew. Chem., Int. Ed., 2016, 55, 7492–7495 CrossRef CAS PubMed.
- P. Huang, H. Deng, Y. Zhou and X. Chen, Matter, 2022, 5, 1670–1699 CrossRef CAS.
- Z. P. Tolstyka, H. Phillips, M. Cortez, Y. Wu, N. Ingle, J. B. Bell, P. B. Hackett and T. M. Reineke, ACS Biomater. Sci. Eng., 2015, 2, 43–55 CrossRef PubMed.
- A. Sizovs, L. Xue, Z. P. Tolstyka, N. P. Ingle, Y. Wu, M. Cortez and T. M. Reineke, J. Am. Chem. Soc., 2013, 135, 15417–15424 CrossRef CAS PubMed.
- W. S. Boyle, K. Senger, J. Tolar and T. M. Reineke, Biomacromolecules, 2016, 18, 56–67 CrossRef PubMed.
- X. Zhao, J. Chen, M. Qiu, Y. Li, Z. Glass and Q. Xu, Angew. Chem., Int. Ed., 2020, 59, 20083–20089 CrossRef CAS PubMed.
- M. Qiu, Y. Li, H. Bloomer and Q. Xu, Acc. Chem. Res., 2021, 54, 4001–4011 CrossRef CAS PubMed.
- L. Miao, L. Li, Y. Huang, D. Delcassian, J. Chahal, J. Han, Y. Shi, K. Sadtler, W. Gao, J. Lin, J. C. Doloff, R. Langer and D. G. Anderson, Nat. Biotechnol., 2019, 37, 1174–1185 CrossRef CAS PubMed.
- K. A. Hajj, R. L. Ball, S. B. Deluty, S. R. Singh, D. Strelkova, C. M. Knapp and K. A. Whitehead, Small, 2019, 15, 1805097 CrossRef PubMed.
- I. Louzao, R. García-Fandiño and J. Montenegro, J. Mater. Chem. B, 2017, 5, 4426–4434 RSC.
- K. A. Whitehead, J. R. Dorkin, A. J. Vegas, P. H. Chang, O. Veiseh, J. Matthews, O. S. Fenton, Y. Zhang, K. T. Olejnik, V. Yesilyurt, D. Chen, S. Barros, B. Klebanov, T. Novobrantseva, R. Langer and D. G. Anderson, Nat. Commun., 2014, 5, 4277 CrossRef CAS PubMed.
- C. Gehin, J. Montenegro, E.-K. Bang, A. Cajaraville, S. Takayama, H. Hirose, S. Futaki, S. Matile and H. Riezman, J. Am. Chem. Soc., 2013, 135, 9295–9298 CrossRef CAS PubMed.
- L. Gallego-Yerga, I. Posadas, C. de la Torre, J. Ruiz-Almansa, F. Sansone, C. Ortiz Mellet, A. Casnati, J. M. García Fernández and V. Ceña, Front. Pharmacol., 2017, 8, 249 CrossRef PubMed.
- J. Gasparello, M. Lomazzi, C. Papi, E. D’Aversa, F. Sansone, A. Casnati, G. Donofrío, R. Gambari and A. Finotti, Mol. Ther.–Nucleic Acids, 2019, 18, 748–763 CrossRef CAS PubMed.
- L. Gallego-Yerga, J. M. Benito, L. Blanco-Fernández, M. Martínez-Negro, I. Vélaz, E. Aicart, E. Junquera, C. Ortiz Mellet, C. Trosde Ilarduya and J. M. García Fernández, Chem. – Eur. J., 2018, 24, 3825–3835 CrossRef CAS PubMed.
- D. Manzanares, I. Araya-Durán, L. Gallego-Yerga, P. Játiva, V. Márquez-Miranda, J. Canan, J. L. Jiménez Blanco, C. O. Mellet, F. D. González-Nilo, J. M. García Fernández and V. Ceña, Nanomedicine, 2017, 12, 1607–1621 CrossRef CAS PubMed.
- W.-B. Zhang, X. Yu, C.-L. Wang, H.-J. Sun, I.-F. Hsieh, Y. Li, X.-H. Dong, K. Yue, R. Van Horn and S. Z. D. Cheng, Macromolecules, 2014, 47, 1221–1239 CrossRef CAS.
- J. L. Jiménez Blanco, J. M. Benito, C. Ortiz Mellet and J. M. García Fernández, J. Drug Delivery Sci. Technol., 2017, 42, 18–37 CrossRef.
- F. Ortega-Caballero, C. Ortiz Mellet, L. Le Gourriérec, N. Guilloteau, C. Di Giorgio, P. Vierling, J. Defaye and J. M. García Fernández, Org. Lett., 2008, 10, 5143–5146 CrossRef CAS PubMed.
- A. Díaz-Moscoso, L. L. Gourriérec, M. Gómez-García, J. M. Benito, P. Balbuena, F. Ortega-Caballero, N. Guilloteau, C. D. Giorgio, P. Vierling, J. Defaye, C. Ortiz Mellet and J. M. García Fernández, Chem. – Eur. J., 2009, 15, 12871–12888 CrossRef PubMed.
- C. Ortiz Mellet, J. M. Benito and J. M. García Fernández, Chem. – Eur. J., 2010, 16, 6728–6742 CrossRef CAS PubMed.
- A. I. Carbajo-Gordillo, J. López-Fernández, J. M. Benito, J. L. Jiménez Blanco, M. L. Santana-Armas, G. Marcelo, C. Di Giorgio, C. Przybylski, C. Ortiz Mellet, C. Tros de Ilarduya, F. Mendicuti and J. M. García Fernández, Macromol. Rapid Commun., 2022, 43, 2200145 CrossRef CAS PubMed.
- A. I. Carbajo-Gordillo, J. L. Jiménez Blanco, J. M. Benito, H. Lana, G. Marcelo, C. Di Giorgio, C. Przybylski, H. Hinou, V. Ceña and C. Ortiz Mellet, et al. , Biomacromolecules, 2020, 21, 5173–5188 CrossRef CAS PubMed.
- A. I. Carbajo-Gordillo, J. Rodríguez-Lavado, J. L. Jiménez Blanco, J. M. Benito, C. Di Giorgio, I. Vélaz, C. Tros de Ilarduya, C. Ortiz Mellet and J. M. García Fernández, Chem. Commun., 2019, 55, 8227–8230 RSC.
- A. I. Carbajo-Gordillo, M. González-Cuesta, J. L. Jiménez Blanco, J. M. Benito, M. L. Santana-Armas, T. Carmona, C. Di Giorgio, C. Przybylski, C. Ortiz Mellet and C. Tros de Ilarduya, Chem. – Eur. J., 2021, 27, 9429–9438 CrossRef CAS PubMed.
- E. Blanco, H. Shen and M. Ferrari, Nat. Biotechnol., 2015, 33, 941–951 CrossRef CAS PubMed.
- X. Zhou, L. Xu, J. Xu, J. Wu, T. B. Kirk, D. Ma and W. Xue, ACS Appl. Mater. Interfaces, 2018, 10, 35812–35829 CrossRef CAS PubMed.
- C. Xu, B. Yu, Y. Qi, N. Zhao and F. Xu, Adv. Healthcare Mater., 2021, 10, 2001183 CrossRef CAS PubMed.
- Y. J. Ooi, Y. Wen, J. Zhu, X. Song and J. Li, Biomacromolecules, 2020, 21, 1136–1148 CrossRef CAS PubMed.
- T. Fujiwara, S. Hasegawa, N. Hirashima, M. Nakanishi and T. Ohwada, Biochim. Biophys. Acta, Biomembr., 2000, 1468, 396–402 CrossRef CAS PubMed.
- R.-M. Zhao, Y. Guo, H.-Z. Yang, J. Zhang and X.-Q. Yu, New J. Chem., 2021, 45, 13549–13557 RSC.
- Y. Zhang, C.-Y. Li, J. Zhang, W.-J. Yi and X.-Q. Yu, Chem. Biodiversity, 2014, 11, 233–244 CrossRef CAS PubMed.
- Q. Liu, Q.-Q. Jiang, W.-J. Yi, J. Zhang, X.-C. Zhang, M.-B. Wu, Y.-M. Zhang, W. Zhu and X.-Q. Yu, Bioorg. Med. Chem., 2013, 21, 3105–3113 CrossRef CAS PubMed.
- J. Li, Y. Zhu, S. T. Hazeldine, S. M. Firestine and D. Oupický, Biomacromolecules, 2012, 13, 3220–3227 CrossRef CAS PubMed.
- Q.-D. Huang, J. Ren, H. Chen, W.-J. Ou, J. Zhang, Y. Fu, W. Zhu and X.-Q. Yu, ChemPlusChem, 2012, 77, 584–591 CrossRef CAS.
- Z. Huang, Y.-P. Xiao, Y. Guo, H.-Z. Yang, R.-M. Zhao, J. Zhang and X.-Q. Yu, Eur. Polym. J., 2022, 170, 111153 CrossRef CAS.
- Q.-D. Huang, J. Ren, W.-J. Ou, Y. Fu, M.-Q. Cai, J. Zhang, W. Zhu and X.-Q. Yu, Chem. Biol. Drug Des., 2012, 79, 879–887 CrossRef CAS PubMed.
- Q.-D. Huang, W.-J. Ou, H. Chen, Z.-H. Feng, J.-Y. Wang, J. Zhang, W. Zhu and X.-Q. Yu, Eur. J. Pharm. Biopharm., 2011, 78, 326–335 CrossRef CAS PubMed.
- Y. Guo, J.-J. Chen, H.-Z. Yang, J. Zhang, R.-M. Zhao, Z. Huang and X.-Q. Yu, ACS Appl. Bio Mater., 2020, 4, 844–852 CrossRef.
- Z. Chen, S. Liu, W. Liu, P. Jiang, L. Wang, J. Li, H. Zhou and T. Guo, ACS Biomater. Sci. Eng., 2021, 7, 5678–5689 CrossRef CAS PubMed.
- A. Megia-Fernandez, M. Ortega-Muñoz, J. Lopez-Jaramillo, F. Hernandez-Mateo and F. Santoyo-Gonzalez, Adv. Synth. Catal., 2010, 352, 3306–3320 CrossRef CAS.
- S. A. Smith, L. I. Selby, A. P. R. Johnston and G. K. Such, Bioconjugate Chem., 2019, 30, 263–272 CrossRef CAS PubMed.
- C. Ollier, A. Méndez-Ardoy, F. Ortega-Caballero, J. L. Jiménez Blanco, N. Le Bris and R. Tripier, ChemRxiv., 2023, preprint, DOI:10.26434/chemrxiv-2023-85pk4.
Footnote |
| † Electronic supplementary information (ESI) available. See DOI: https://doi.org/10.1039/d3tb02889e |
| This journal is © The Royal Society of Chemistry 2024 |