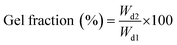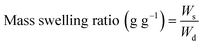Open Access Article
Open Access ArticleEvaluation of gelatin-based hydrogels for colon and pancreas studies using 3D in vitro cell culture†
Regina
Pamplona
 a,
Sandra
González-Lana
a,
Sandra
González-Lana
 bc,
Ignacio
Ochoa
bc,
Ignacio
Ochoa
 cde,
Rafael
Martín-Rapún
cde,
Rafael
Martín-Rapún
 *adf and
Carlos
Sánchez-Somolinos
*adf and
Carlos
Sánchez-Somolinos
 *dg
*dg
aAragón Institute of Nanoscience and Materials (INMA), CSIC-University of Zaragoza, Department of Organic Chemistry, C/Pedro Cerbuna 12, 50009 Zaragoza, Spain. E-mail: rmartin@unizar.es
bBEONCHIP S.L., CEMINEM, Campus Río Ebro. C/Mariano Esquillor Gómez s/n, 50018 Zaragoza, Spain
cTissue Microenvironment (TME) Lab. Aragón Institute of Engineering Research (I3A), University of Zaragoza, C/Mariano Esquillor s/n, 500018 Zaragoza, Spain
dCIBER in Bioengineering, Biomaterials and Nanomedicine (CIBER-BBN), Madrid, Spain
eInstitute for Health Research Aragón (IIS Aragón), Paseo de Isabel La Católica 1-3, 50009 Zaragoza, Spain
fUniversidad de Zaragoza, Facultad de Ciencias, Departamento de Química Orgánica, C/Pedro Cerbuna 12, 50009 Zaragoza, Spain
gAragón Institute of Nanoscience and Materials (INMA), CSIC-University of Zaragoza, Department of Condensed Matter Physics (Faculty of Science), C/Pedro Cerbuna 12, 50009 Zaragoza, Spain. E-mail: carlos.s@csic.es
First published on 4th March 2024
Abstract
Biomimetic 3D models emerged some decades ago to address 2D cell culture limitations in the field of replicating biological phenomena, structures or functions found in nature. The fabrication of hydrogels for cancer disease research enables the study of cell processes including growth, proliferation and migration and their 3D design is based on the encapsulation of tumoral cells within a tunable matrix. In this work, a platform of gelatin methacrylamide (GelMA)-based photocrosslinked scaffolds with embedded colorectal (HCT-116) or pancreatic (MIA PaCa-2) cancer cells is presented. Prior to cell culture, the mechanical characterization of hydrogels was assessed in terms of stiffness and swelling behavior. Modifications of the UV curing time enabled a fine tuning of the mechanical properties, which at the same time, showed susceptibility to the chemical composition and crosslinking mechanism. All scaffolds displayed excellent cytocompatibility with both tumoral cells while eliciting various cell responses depending on the microenvironment features. Individual and collective cell migration were observed for HCT-116 and MIA PaCa-2 cell lines, highlighting the ability of the colorectal cancer cells to cluster into aggregates of different sizes governed by the surrounding matrix. Additionally, metabolic activity results pointed out to the development of a more proliferative phenotype within stiffer networks. These findings confirm the suitability of the presented platform of GelMA-based hydrogels to conduct 3D cell culture experiments and explore biological processes associated with colorectal and pancreatic cancer.
Introduction
Since the first biomimetic strategies in cell culture, the goal of recreating basic aspects of the tumor microenvironment (TME) has been significantly targeted. This surrounding microenvironment comprises different cell types, soluble factors and a complex and dynamic extracellular matrix (ECM). In addition, the composition of the ECM within which cells reside is continuously changing, and some factors, such as increased stiffness, have been related to the development of the cancer disease.1 In this context, colorectal cancer (CRC) in vitro studies, such as the one performed by Tang et al.,2 demonstrated that the alteration of microenvironment stiffness is among the signaling pathways implicated in the initiation of metastasis. Likewise, pancreatic ductal adenocarcinoma (PDAC) is characterized by a particularly pronounced deposition of a dense ECM. Rice and co-workers3 reported how the increasing stiffness of polyacrylamide gels promoted different elements of the epithelial–mesenchymal transition (EMT) in pancreatic cancer cell lines.When considering different biomimetic models, two-dimensional (2D) cell culture studies provide a straightforward, low-cost and high-performance approach to explore cancer biology. Unfortunately, cell–cell interactions established in 2D in vitro experiments are different from how cells would experience in a three-dimensional (3D) environment. This is partly because 2D substrates lack the ability to mimic geometrical confinement or shear stress.4,5 In fact, a two-way communication exists between cells and their surrounding microenvironment, and investigating these cell–matrix interactions in a 3D context constitutes a more physiologically relevant approach.
Amidst the broad domain of 3D scaffolds, gelatin methacrylamide (GelMA) hydrogels have gained increasing attention ever since the initial report from Van Den Bulcke et al.6 These hydrogels originate from a natural polymer, incorporate the cell-adhesive arginine–glycine–aspartic acid (RGD) sequence, and offer extensive photocrosslinking possibilities to achieve tailor-made scaffolds.7 While this material has been particularly useful in breast cancer tumor models,8,9 there are limited studies applied to colon and pancreas and those works are primarily focused on drug screening.10,11
Regarding photo-induced crosslinking reactions which allow for finely-tuned control of biomechanics in hydrogels, the free radical photopolymerization approach stands out as one of the simplest and most rapid method to generate 3D scaffolds.12,13 Nevertheless, GelMA also has the potential to form hydrogels by combining with other photocrosslinkable functional groups, such as allyl14 and diacrylates5 as well as thiol motifs, through a process known as photoclick chemistry.15–17 These thiol-based reactions are orthogonal, highly selective and efficient, yielding the final molecule without any by-products.18 However, when it comes to studies focused on colorectal and pancreatic cancer, there are only a few examples in the literature that demonstrate the use of gelatin-containing hydrogels crosslinked via thiol–ene reactions.12,19,20 In our previous work, we reported the effectiveness of the methacrylate-thiol system (GelMA + 4-arm PEG thiol) as a robust platform for easily tuning hydrogel mechanics. Furthermore, we also confirmed excellent cell viability using semi-3D models (colorectal epithelial cells grown on top of 3D substrates).17
The biomaterials community has shown considerable interest in using 3D cell culture hydrogels to replicate the TME as a means of obtaining a more comprehensive understanding of cancer progression. During this culture process, cells grow and proliferate within a mechanical landscape which is constantly being remodeled. Reproducing the dynamic state of the ECM is a challenging task. However, a number of works focused on the structural architecture, mechanical and biochemical functions can be found in literature, offering valuable insights into cancer research. Referring to colorectal cancer biomimetic studies, Cadamuro and co-workers prepared hyaluronic acid-based hydrogels through 3D bioprinting and they described how the glycosylation of ECM proteins induced several alterations to the proteome of the encapsulated HT-29 cells.21 Williams et al. reported that HCT-116 cells displayed a different phenotype when embedded into fiber-reinforced agarose hydrogels as a result of morphological guidance.22 As well, Jabbari and co-workers found that HCT-116 cells underwent phenotypic changes towards a more aggressive and invasive behavior due to an increase in matrix stiffness.23 Similarly, in the context of pancreatic cancer research, Betriu et al. elucidated how the stiffness of self-assembling peptide scaffolds influenced cellular responses. These authors reported that increased matrix stiffness led to downregulation of focal adhesion kinase (FAK) in PDAC cells.24 In addition, Puls and co-workers encapsulated MIA PaCa-2 cells within collagen hydrogels of varying fibril density and matrix stiffness. They subsequently observed significant changes in cell morphology and proteins expression.25
Despite the efforts of the scientific community, a significant demand remains for the development of biomimetic models tailored to the specific needs of PDAC and colorectal cancer research, in order to further advance our understanding of these diseases. In this study, we exploit our developed methacrylate-thiol system (GelMA + 4-arm PEG thiol) platform to fabricate cell-embedded 3D scaffolds for colorectal and pancreatic cancer research. We aim to investigate how the composition and the hydrogel stiffness influence the cell viability, migration and proliferation of HCT-116 and MIA PaCa-2 cells lines. Cell-free GelMA-based hydrogels were first characterized in terms of swelling behavior and stiffness using atomic force spectroscopy (AFM) measurements. Subsequently, after encapsulating cells, the cell-laden hydrogels were assessed at both day 1 and day 3 for cell viability, mitochondrial activity and DNA content. Furthermore, to study cell migration, the cell-laden hydrogels were placed in the IncuCyte® live-imaging system and continuously monitored in real-time over the course of the 3 days of cell culture assay.
Materials and methods
Materials
Gelatin from porcine skin (type A, 300 Bloom), methacrylic anhydride (MAA) (94%), 3-(trimethylsilyl)propionic-2,2,3,3-d4 acid sodium salt (TMSP), dialysis tubing cellulose membranes (MWCO: 12–14 kDa), 2,4,6-trinitrobenzenesulfonic acid 5% w/v solution (TNBS), sodium n-dodecyl sulfate 20% w/v solution (SDS), glycine (ReagentPlus, ≥99%), photoinitiator 2-hydroxy-4′-(2-hydroxyethoxy)-2-methylpropiophenone, also known as Irgacure 2959 (I2959), NaOH 1 N and DMEM 5× were obtained from Sigma Aldrich. Deuterium oxide was purchased from Eurisotop. 4-Arm poly(ethylene glycol) thiol (4PEGSH) was supplied by JenKem, USA. Poly(dimethylsiloxane) (PDMS) elastomer was prepared from Sylgard-184 (Dow Corning). Type I rat tail collagen (10.9 mg mL−1) was purchased from Corning, USA. Phosphate buffered saline (PBS) pH 7.4, high glucose Dulbecco's modified eagle's medium (DMEM) without Phenol red, Advanced DMEM, Glutamax, penicillin/streptomycin (10![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 000 U mL−1) and non-essential amino acids (10×) were purchased from Gibco, Life Technologies. Fetal bovine serum (FBS), trypsin, Calcein AM (CAM), propidium iodide (PI) and Triton X-000 were obtained from Sigma-Aldrich. 3-(4,5-Dimethylthiazol-2-yl)-5-(3-carboxymethoxyphenyl)-2-(4-sulfophenyl)-2H-tetrazolium (MTS) reagent was supplied by Abcam. Paraformaldehyde and peqGold Blood & Tissue DNA Mini Kit protocol were purchased from VWR. Fluorescent dyes Cell Tracker Orange CMTMR, 4′,6-diamidino-2-phenylindole (DAPI) and Alexa Fluor 488 Phalloidin were supplied by Invitrogen. All purchased materials were used without further purification.
000 U mL−1) and non-essential amino acids (10×) were purchased from Gibco, Life Technologies. Fetal bovine serum (FBS), trypsin, Calcein AM (CAM), propidium iodide (PI) and Triton X-000 were obtained from Sigma-Aldrich. 3-(4,5-Dimethylthiazol-2-yl)-5-(3-carboxymethoxyphenyl)-2-(4-sulfophenyl)-2H-tetrazolium (MTS) reagent was supplied by Abcam. Paraformaldehyde and peqGold Blood & Tissue DNA Mini Kit protocol were purchased from VWR. Fluorescent dyes Cell Tracker Orange CMTMR, 4′,6-diamidino-2-phenylindole (DAPI) and Alexa Fluor 488 Phalloidin were supplied by Invitrogen. All purchased materials were used without further purification.
GelMA synthesis
Gelatin methacrylamide was synthesized on a 3-gram scale according to Shirahama et al.26 In brief, a 10% w/v solution of type A gelatin (300 Bloom) in carbonate–bicarbonate (CB) buffer (0.25 M, pH 9) was prepared. The solution contained in a round-bottom flask was heated in an oil bath at 50 °C for 20 min without stirring, and then vigorously stirred for 1 h until complete dissolution. Next, methacrylic anhydride (MAA) was added dropwise to the gelatin solution with an amine![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) MAA feed molar ratio of 1
MAA feed molar ratio of 1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1.5, where the free amino groups of gelatin were determined following the Habeeb method27 also used by Van Den Bulcke et al.6 The reaction was allowed to continue under stirring at 50 °C for 3 h. Afterward, the pH was adjusted to 7.4 to halt the methacrylation reaction. The resulting solution was dialyzed against distilled water using dialysis membranes. Dialysis took place inside an incubator at 37 °C with gentle orbital shaking and was performed four times within 24 h to remove salts, methacrylic acid, and anhydride. Following purification, the product was subjected to an overnight freezing at −80 °C. Finally, it was lyophilized for 1 week and stored at −20 °C, shielded from light until further use.
1.5, where the free amino groups of gelatin were determined following the Habeeb method27 also used by Van Den Bulcke et al.6 The reaction was allowed to continue under stirring at 50 °C for 3 h. Afterward, the pH was adjusted to 7.4 to halt the methacrylation reaction. The resulting solution was dialyzed against distilled water using dialysis membranes. Dialysis took place inside an incubator at 37 °C with gentle orbital shaking and was performed four times within 24 h to remove salts, methacrylic acid, and anhydride. Following purification, the product was subjected to an overnight freezing at −80 °C. Finally, it was lyophilized for 1 week and stored at −20 °C, shielded from light until further use.
NMR spectroscopy
NMR spectra were acquired at room temperature (RT) using a Bruker AV-400 spectrometer and operating at a proton Larmor frequency of 400.16 MHz. Both gelatin and GelMA macromer samples were prepared at a final concentration of 25 mg mL−1 in deuterium oxide. To serve as an internal standard, TMSP was employed at a concentration of 1 mg mL−1. Data processing was conducted using MestReNova software.Degree of functionalization: TNBS assay
The degree of functionalization (DF) was determined as the percentage of amino groups (originating from lysine and hydroxylysine) derivatized in GelMA, as described in literature.28 The quantification involved measuring the remaining free amino groups using TNBS based on Habeeb method27 and Lee et al. modifications.29 Each sample was prepared in duplicate following the outlined protocol. In brief, separate solutions of GelMA and gelatin were dissolved in 0.1 M CB buffer pH 8.5, with concentrations of 5 mg mL−1 for GelMA and 1 mg mL−1 for gelatin. Next, 50 μL of each gelatin solution were pipetted into 96-well plates and 25 μL of 0.1% w/v TNBS were added. Microwell plate was then incubated for 2.5 h at 37 °C in the dark with gentle shaking to allow the nucleophilic substitution and formation of the yellow-colored amine derivative.30 After incubation, 25 μL of 10% w/v SDS solution and 12.5 μL of 1 M HCl were added to each sample to stop the reaction. The absorbance of samples was measured at 330 nm using a microwell plate reader (Multiskan Go, Thermo Scientific). A glycine standard curve was used to calculate the amino group content with standard sample solutions prepared at 0, 0.16, 0.32, 0.48, 0.64, 0.72 mM. All glycine solutions were subjected to the same TNBS procedure as GelMA samples. DF was calculated as shown in the following formula:Hydrogels preparation
GelMA hydrogels were formed through free radical photopolymerization of the GelMA macromer in high glucose DMEM supplemented with 1% penicillin/streptomycin and 1% v/v of non-essential amino acids (named as DG0 throughout the manuscript), along with a photoinitiator. During photopolymerization, undesirable effects such as UV light absorption and attenuation may be generated by the presence of phenol red in cell culture media.31 For this reason, gelatin-based solutions were all prepared in DMEM without phenol red, with a final concentration of 6% w/v GelMA macromer and 0.1% w/v I2959. The hydrogel precursor mixture in DG0 was incubated at 37 °C for 1 h while protected from light to ensure complete dissolution. In the case of GelMA–SH hydrogels, 5% w/v 4-arm PEG thiol solution in DG0 was then added to the mixture at a final functional group ratio methacrylamide![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) SH 1
SH 1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 0.5. As presented in our previous work,17 PDMS molds with two different dimensions enabled the fabrication of hydrogels for mechanical testing. AFM measurements were performed with disc-shaped scaffolds hydrogels (D = 10 mm, thickness = 1 mm) prepared by pouring 120 μL of mixture into PDMS molds mounted on top of glass slides. On the other hand, 130 μL of GelMA hydrogel precursor were used to fabricate cylindrical hydrogels (D = 6 mm, thickness = 3 mm) for swelling experiments. Subsequently, hydrogels were incubated at RT for 20 min to allow for physical gelation. Finally, hydrogels were exposed to UV light (320–390 nm, 10 mW cm−2) for 10 s, 30 s or 150 s using and OmniCure S2000 UV Lamp, resulting in the formation of the final photopolymerized hydrogels. The UV time applied to each hydrogel is indicated in the name given to each material, i.e. for GelMA-30 the mixture was exposed to UV light for 30 s.
0.5. As presented in our previous work,17 PDMS molds with two different dimensions enabled the fabrication of hydrogels for mechanical testing. AFM measurements were performed with disc-shaped scaffolds hydrogels (D = 10 mm, thickness = 1 mm) prepared by pouring 120 μL of mixture into PDMS molds mounted on top of glass slides. On the other hand, 130 μL of GelMA hydrogel precursor were used to fabricate cylindrical hydrogels (D = 6 mm, thickness = 3 mm) for swelling experiments. Subsequently, hydrogels were incubated at RT for 20 min to allow for physical gelation. Finally, hydrogels were exposed to UV light (320–390 nm, 10 mW cm−2) for 10 s, 30 s or 150 s using and OmniCure S2000 UV Lamp, resulting in the formation of the final photopolymerized hydrogels. The UV time applied to each hydrogel is indicated in the name given to each material, i.e. for GelMA-30 the mixture was exposed to UV light for 30 s.
To create the control collagen hydrogels, a mixture was made by combining 10.9 mg mL−1 of type I rat tail collagen with 1 N NaOH and DMEM 5× in a proportion of 1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 40 and 1
40 and 1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 8 (v/v of collagen volume), respectively. After pouring the warm mixture into the PDMS molds, collagen hydrogels were physically crosslinked at 37 °C for 20 min inside an incubator. Furthermore, to include the UV condition in the control samples, some collagen hydrogels were irradiated with the maximum UV time employed within this work, that is, 150 s.
8 (v/v of collagen volume), respectively. After pouring the warm mixture into the PDMS molds, collagen hydrogels were physically crosslinked at 37 °C for 20 min inside an incubator. Furthermore, to include the UV condition in the control samples, some collagen hydrogels were irradiated with the maximum UV time employed within this work, that is, 150 s.
All the protocols with collagen, GelMA and GelMA–SH mixtures were performed under sterile conditions.
Hydrogels characterization
On the other hand, the hydrogel swelling ratio refers to its capacity to absorb an aqueous medium. The experimental protocol to assess the mass swelling ratio (MSR) began with the incubation of photocrosslinked hydrogels in DG0 at 37 °C for 24 h under sterile conditions. Then, the excess of DG0 was gently removed with a Kimwipe paper and samples were weighed (Ws). Finally, the dry mass of hydrogels (Wd) was recorded after freezing in liquid nitrogen and lyophilizing overnight. MSR (g g−1) was defined as:
Three replicate samples of each condition were tested for all calculations regarding gel fraction and MSR.
Cell culture
For 3D in vitro cell culture experiments, hydrogels were directly prepared in 96 well-plates. HCT-116 and MIA PaCa-2 cells were cultured in flasks in Advanced DMEM supplemented with 10% v/v FBS and 1% penicillin/streptomycin (named as DG10 throughout the manuscript). Cells were maintained at 37 °C in a humidified incubator with 5% CO2 (standard conditions), exchanging the medium every 2–3 days until they reached 90% confluence. For the fabrication of cell-laden hydrogels, HCT-116 and MIA PaCa-2 cells were first trypsinized, counted and re-suspended in DG10. Following centrifugation, the pellet cells were resuspended in warm mixtures of collagen, GelMA, or GelMA–SH to achieve a final density of 4 × 106 cells per mL. For GelMA and GelMA–SH hydrogels, each well was filled with 50 μL of the cell mixture, and the well plates were incubated at RT for 10–15 min, shielded from light. After this incubation period, the well plates were turned upside down to ensure proper 3D distribution of cells within the hydrogel, and further incubated at RT for an additional 5–10 min. Subsequently, each well was irradiated the corresponding UV time (from 10 s to 150 s) and washing was performed twice with DG0 to remove the unreacted photoinitiator. Then, DG10 was added to each well. For collagen hydrogels, the same protocol as previously described for GelMA and GelMA–SH was followed unless for the incubation process which occurred inside an incubator at 37 °C instead of RT.Finally, 96-well plates were cultured for 3 days under standard conditions, with DG10 being exchanged every other day. Three to four cell-laden hydrogels were prepared for each experimental condition.
Cell viability
Cell viability assessment was conducted on days 1 and 3 of cell culture using a live/dead staining protocol. The cell-laden hydrogels were incubated with 2 μg mL−1 of calcein-AM (CAM) and 6 μg mL−1 of propidium iodide (PI) in DG10 for 30 min under standard culture conditions. An inverted fluorescence microscope (Leica DMi8) was used to monitor cell viability. The obtained fluorescence images were processed with ImageJ software, and manual thresholding was applied to quantify cell viability. Analysis of HCT-116 cell clusters was performed by randomly selecting 150 clusters of each condition and manually fitting their areas to spheroidal/elliptical shapes.DNA extraction
On days 1 and 3, well plates containing the cell-laden hydrogels were frozen at −80 °C until further use. To prepare for DNA isolation, each hydrogel was finely powdered in liquid nitrogen using a cold mortar and pestle. The collected samples were then processed following the peqGold Blood & Tissue DNA Mini Kit protocol from VWR. Total DNA was isolated and quantified using a Nanodrop spectrophotometer (Synergy HT, BioTek). For DNA quantification, calibration was carried out using solutions with increasing cell concentrations. Cell pellets were treated similarly to the hydrogel samples. For each experimental condition, three samples were tested.![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 400) for 90 min at RT while protected from light. After rinsing three times with PBS, nuclei staining was performed with DAPI (1
400) for 90 min at RT while protected from light. After rinsing three times with PBS, nuclei staining was performed with DAPI (1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1000) for 60 min at RT protected from light. Confocal images were obtained with a 63× oil-immersion objective (confocal Zeiss LSM 880 with a Plan-Apochromat 63X/1.4 oil (DT 0.19), DIC objective).
1000) for 60 min at RT protected from light. Confocal images were obtained with a 63× oil-immersion objective (confocal Zeiss LSM 880 with a Plan-Apochromat 63X/1.4 oil (DT 0.19), DIC objective).
Data analysis and statistics
All results are presented as mean ± standard deviation (SD). Graph plotting and statistical analysis were performed using OriginPro 2020 software (OriginLab). Prior to significance testing, normality was assessed using the Shapiro-Wilk test, and the equality of variances between datasets was examined. For determining significant differences (p < 0.05), Student's t-tests and one-way ANOVAs with Tukey's post hoc tests were employed.Results and discussion
Functionalization of gelatin
The methacrylation of gelatin is a commonly employed technique to produce photocrosslinkable derivatives, facilitating the creation of a chemically bonded network upon exposure to UV light. In the present work, the method described by Shirahama et al.26 was utilized to adjust the experimental parameters and optimize the incorporation of methacryloyl groups into gelatin as previously described.17 Successful methacrylation of primary amines present in lysine and hydroxylysine residues was confirmed using 1H-NMR spectroscopy30 (Fig. S1, ESI†). The signals at 5.4 and 5.7 ppm, corresponding to the vinyl protons, and the peak at 1.9 ppm, attributed to the methyl groups, provided evidence of the presence of methacrylamide groups. Additionally, the characteristic lysine signal at 3.0 ppm almost vanished in the GelMA spectra, indicating the reaction of the ε-amino groups with MAA. The TNBS assay results indicated an 85% yield in the transformation of free amines to methacrylamide groups. Notably, this reaction does not involve arginine residues, ensuring that the RGD motifs remained intact, and GelMA retained its favorable cell adhesive properties.32Preparation and mechanical characterization of cell-free hydrogels
As described in our previous work,17 GelMA and GelMA–SH hydrogels constitute an excellent bioscaffolding platform for in vitro cellular studies with epithelial cells. Given the good results obtained, the present study was conducted using the same hydrogels for 3D in vitro cell culture studies. We decided to keep most of the experimental parameters, such as the photoinitiator system I2959 and the GelMA concentration (6% w/v), since previous studies in the literature had already demonstrated good cell viability33 and biocompatibility.34Taking into account the ultimate 3D in vitro cell culture experiments and the fact that the aqueous media used in photopolymerization strongly influences the mechanical properties of hydrogels,35 all gelatin derivative and collagen solutions were prepared in DG0. In the case of gelatin-based precursor mixtures, they were poured into PDMS molds and kept at RT, shielded from light, for 20 min before UV irradiation, leading to the formation of a physical gel. As time progresses, the random coil GelMA chains come together and form triple helices through hydrogen bonds, leading to the process of physical gelation (Fig. 1).36 This physical gel formation taking place before UV irradiation is known to yield stiffer networks with enhanced mechanical properties.37
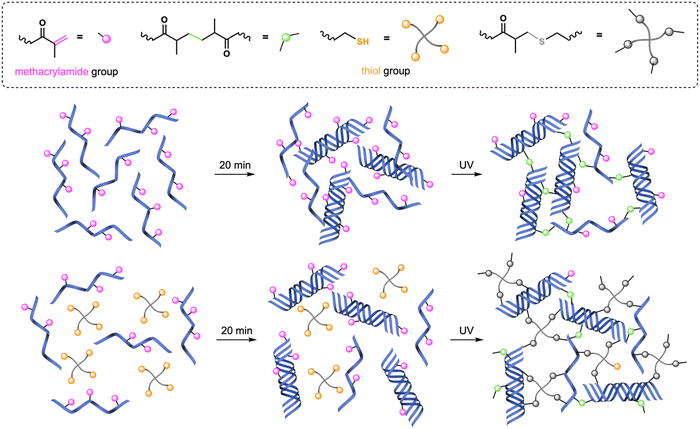 | ||
| Fig. 1 Crosslinking procedures for GelMA and GelMA–SH hydrogels. Physical gelation at RT takes place during the 20 min incubation period. | ||
Consequently, after the 20-min period incubation at RT, GelMA hydrogels were fabricated following a chain-growth mechanism38 by exposing the precursor mixture to actinic light for 30 s or 150 s (named as GelMA-30 and GelMA-150, respectively). Regarding GelMA–SH hydrogels, the introduction of thiol-methacrylamide chemistry enabled a mixed-mode crosslinking upon UV irradiation (chain-growth and step-growth mechanisms) which is related to higher conversion of functional groups.39 This higher conversion has been previously demonstrated to lead to stiffer hydrogels by using GelMA in combination with the 4-arm thiol also employed in this work. That is, keeping the same UV dose for curing, GelMA–SH hydrogels are stiffer than their GelMA analogues.17 We have applied this alternative mixed-mode crosslinking strategy to fabricate two different GelMA–SH hydrogels. Following a similar preparation method that that used in GelMA hydrogels, after the incubation period, we have employed two different UV light doses: 10 s and 30 s for hydrogels named SH-10 and SH-30, respectively. It is worth to mention that GelMA–SH hydrogels were prepared at a methacrylamide![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) thiol ratio of 1
thiol ratio of 1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 0.5 (6% w/v GelMA and 1% w/v 4-arm thiol) to achieve a good balance between the double bonds and sulfhydryl nucleophiles, preventing dangling structures caused by an excessive thiol proportion.40
0.5 (6% w/v GelMA and 1% w/v 4-arm thiol) to achieve a good balance between the double bonds and sulfhydryl nucleophiles, preventing dangling structures caused by an excessive thiol proportion.40
We decided not to dissolve the photoinitiator I2959 in methanol since cells embedded within the network would be affected by the presence of the organic solvent.41 This choice was grounded in a preliminary assay carried out with GelMA-150 hydrogels encapsulating HCT-116 cells prepared by keeping or removing the methanol from the precursor mixture preparation protocol. As shown in Fig. S2 (ESI†), cell toxicity was considerably reduced (55-fold) in hydrogels without methanol. Consequently, all gelatin derivative solutions were prepared by just thoroughly dissolving I2959 together with GelMA macromer in warm DG0.
Additionally, to compare cell behavior against a 3D control network, collagen hydrogels were fabricated. The vast majority of articles on collagen hydrogels work at concentrations lower than 4 mg mL−142,43 which constitute a lower collagen content than that of native tissue. Cancerous tissue presents a variable collagen concentration ranging from 9 to 45 mg mL−1,44 however, the high viscosity of the precursor mixtures complicates the experimental process. For this reason, we decided to increase the collagen concentration compared to most of the works published in the literature to approach a more physiological content. Thus, the collagen hydrogels in this study were prepared at 6 mg mL−1. As described in the previous section, the collagen precursor mixture was prepared in DG0, poured into PDMS molds and incubated at 37 °C the same 20-min period time than GelMA-based hydrogels. During this time, self-assembly of collagen molecules takes place rapidly to generate a consistent scaffold.42 Apart from these widely prepared collagen hydrogels (named as “Col” throughout the manuscript) and to also include the UV light parameter into the control samples, we decided to irradiate some of the collagen hydrogels with the highest UV time among all conditions, that is, 150 s (named as Col-150). Thus, the following mechanical and biological discussion is focused on six different bioscaffolds varying the network crosslinking procedure and the UV-exposure time: Col-0, Col-150, GelMA-30, GelMA-150, SH-10 and SH-30 (photocrosslinked cell-free hydrogel images are shown in Fig. S3, ESI†).
To create the closest physiologically relevant environment possible, crosslinked hydrogels were allowed to reach equilibrium swelling at standard culture conditions before nanoindentation. After 24 h of swelling, AFM measurements were conducted in DG0 at 37 °C (Fig. 2).
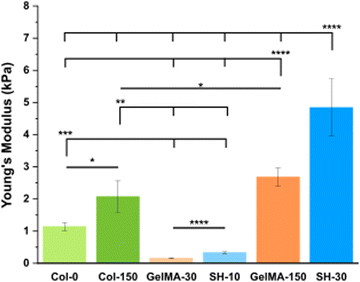 | ||
| Fig. 2 Stiffness results for collagen, GelMA and GelMA–SH hydrogels prepared in DG0 after different UV exposure times. Error bars SD. Note: ****p < 0.0001, ***p < 0.001, **p < 0.01, *p < 0.05. | ||
Physically crosslinked collagen control hydrogels (Col-0) exhibited a Young's Modulus of 1.1 kPa. This result is in good agreement with several studies in the literature which have reported stiffness in the range 0.5–2 kPa, for collagen hydrogel scaffolds formed under similar collagen concentration and polymerization temperature.43,49 The irradiation with 150 s of UV light yielded to stiffer hydrogels with a Young's Modulus of 2.1 kPa. This UV-derived hardening mechanism has been previously observed for collagen hydrogels. Ishibashi et al. recently reported that the storage modulus for pre-gel collagen solutions increased with the UV dose.50 Maeda analyzed the effect of different UV wavelengths in the mechanical properties of collagen gels in vitro.51 This study confirmed that tyrosine crosslinks (dityrosines) increased with radiation at wavelengths between 300–340 nm, having the radiation at 330 nm the greatest effect. Despite the fact that the maximum emission peak from our UV lamp is 365 nm, the equipment covers a wavelength range from 320 to 390 nm. The shortest wavelengths emitted by our lamp could cause dityrosine formation contributing to the observed stiffening of collagen hydrogels.
Free radical crosslinked GelMA scaffolds were prepared by irradiating with UV light for 30 s or 150 s (referred to as GelMA-30 and GelMA-150, respectively). The Young's Modulus obtained for GelMA-150 was 2.7 kPa, almost 17 times higher than GelMA-30, that was 0.16 kPa. Comparison between GelMA–SH hydrogels yielded a similar rise of approximately 15 times from SH-10 (0.33 kPa) to SH-30 (4.8 kPa). As previously reported for homologous pre-hydrogels generated using methanol to facilitate the photoinitiator solubilization, both chemistries allow to tune the stiffness of the hydrogel over a large range of values with biological interest, however the SH hydrogels lead to similar values with much lower UV doses, when compared with GelMA scaffolds.
Interestingly, comparing these results with the ones obtained for hydrogels formulated with methanol (Fig. S4A, ESI†), an almost identical stiffness trend was observed, increasing in the following order: GelMA-30, SH-10, GelMA-150 and SH-30.17 Nevertheless, a notable aspect was that all gelatin-based formulations without methanol yielded softer hydrogels. Besides, the shorter the UV exposure time applied, the greater the decrease in stiffness with respect the methanol-containing formulations. SH-10, GelMA-30, SH-30 and GelMA-150 underwent a 9-fold, 7-fold, 4.5-fold and 2-fold reduction in Young's Moduli. To elucidate a feasible explanation, it seems obvious the key role of methanol in properly dissolving the photoinitiator I2959. In general, one of the main requirements of photonitiators is a good solubility in the precursor solution.52 The efficiency of the derived photochemical processes is strongly influenced by the efficiency of the photoinitiator generating radicals. We assume that removing methanol from the fabrication protocol led to a poor solubility of I2959 in the cell culture medium DG0 and consequently to a lower crosslinking degree in all cases. Additionally, methanol could also have a role in the physical gelation taking place while incubation at RT. As described above, at the initial stage of gelation, random coil GelMA chains aggregate through hydrogen bonding. The presence of methanol in the precursor mixture could promote structural stability owing to the enhanced intramolecular bonding.53
Diseased tissue typically exhibits higher stiffness than healthy tissue. Several studies have confirmed that normal tissues present Young's Moduli around 1 kPa for both healthy colon54,55 and pancreas.3,43 In contrast, colorectal cancer tissue exhibits variations in stiffness based on the degree of disease progression54 and pancreatic cancer tissue is characterized by a heterogeneous map of stiffness as a result of fibrosis.3 Nebuloni et al. performed AFM measurements with CRC samples derived from three donors and found that the vast majority of results ranged from ∼3 to ∼30 kPa.55 The median elastic modulus obtained by Kawano and coworkers for CRC samples from 106 tissue donors was 7.51 kPa but the whole range covered from 1.1 to 68.0 kPa.54 As for human pancreatic tumors, Rubiano et al. employed a custom indentation equipment to test the resected samples, reporting a mean value of 5.46 ± 3.18 kPa43 while Rice and coworkers showed an upper quartile mean stiffness of 4 kPa for PDAC.3 Considering our platform of hydrogels and the Young's Moduli obtained, GelMA-30 (0.16 kPa) and SH-10 (0.33 kPa) scaffolds would be even softer than healthy tissue. On the other hand, Col-0 (1.1 kPa), Col-150 (2.1 kPa), GelMA-150 (2.7 kPa) and SH-30 (4.8 kPa) hydrogels showed stiffness matching with the initial stages of CRC but also with the metastatic tumors and the highly fibrotic PDAC stroma.
As a conclusion for this section, a precise control over stiffness with remarkable reproducibility led to the preparation of custom-tailored hydrogels upon well-defined and easily-adjustable irradiation UV times. The mechanical properties of the prepared hydrogels closely resemble those of both healthy and tumoral pancreas or colon tissues, making them a promising platform for cancer research.
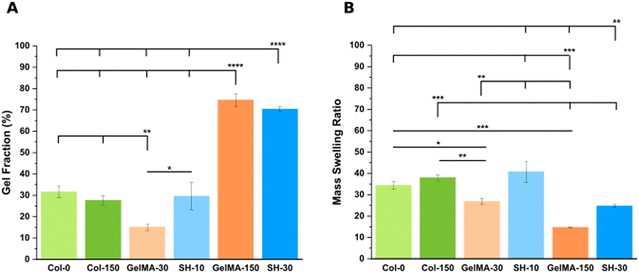
Considering the definition of gel fraction, this property indicates the percentage of macromer molecules forming the 3D network. It is hence expected that the softest hydrogels with low degrees of crosslinking (GelMA-30 and SH-10) exhibit also low values of gel fraction (15.1 ± 1.5% and 29.6 ± 6.3%, respectively) as a result of a higher proportion of non-bounded polymer chains leaching out from the network during swelling. In contrast, the stiffest scaffolds (GelMA-150 and SH-30) displayed higher gel fractions (74.6 ± 2.9% and 70.4 ± 1.1%, respectively) according to also higher final polymer concentrations. These results agree with the works of several authors for related materials.34,56 Notably, even though the gel fraction outcomes are almost identical, SH-30 scaffolds exhibited a significantly higher stiffness (4.8 kPa) than GelMA-150 (2.7 kPa) as described in the section above. Again, this result is originated by the thiol click chemistry: a mixed-mode crosslinking strategy comprised a larger amount of reactive groups, yielding a higher crosslinking density and stiffness.
As for the swelling properties (Fig. 3(B)), Col-0 and Col-150 hydrogels exhibited no significant differences in their elevated mass swelling ratios (34.4 ± 1.9 and 38.0 ± 1.3, respectively). Indeed, it is known that collagen stands out for its exceptional degree of hydration which plays a crucial role in the stabilization of the triple helical conformation.57 Interestingly, in a very similar manner as the comparison described in the previous section, MSR results displayed the same trend as the one obtained for methanol-containing GelMA-derived hydrogels (Fig. S4B, ESI†), although absolute values were higher in all cases (matching with the lower stiffness). Less crosslinked hydrogels, which are softer, exhibit higher capacity to absorb aqueous medium. This was confirmed by comparing pairs of scaffolds created through the same synthesis strategy: GelMA-30 (MSR: 26.9 ± 1.5) demonstrated a 1.8-fold increase compared to GelMA-150 (MSR: 14.7 ± 0.1). Likewise, SH-10 scaffolds (MSR: 40.8 ± 4.9) were characterized by a 1.6-fold higher MSR than SH-30 (24.8 ± 0.6). However, the comparison between the two softest hydrogels showed an inverse trend: SH-10 condition exhibited an unexpectedly high MSR compared to GelMA-30 (1.5-fold increase) while it was also twice as stiff. Similarly, SH-30 displayed a 1.7-fold rise in MSR than GelMA-150 whereas stiffness was 1.8 times higher. This increased swelling capacity is attributed to the presence of hydrophilic PEG-thiol chains in the GelMA–SH networks. Additionally, Bertlein et al.34 explained the different swelling behavior depending on the network crosslinking mechanism. Free radical photopolymerization is associated to heterogeneous networks58,59 with reduced swelling capacity. In contrast, hydrogels fabricated through thiol click chemistry are defined by a more uniform distribution of crosslinking density with enhanced swelling properties.
Summing up, the comparison between GelMA-based scaffolds fabricated through the same chemical crosslinking mechanism (either chain-growth or mixed-mode photopolymerization) confirmed that softer hydrogels displayed lower values of gel fraction and hence, greater MSR values. In addition, the introduction of the PEG chains within the hydrogel network led to a remarkable rise in the swelling capacity.
Cell culture
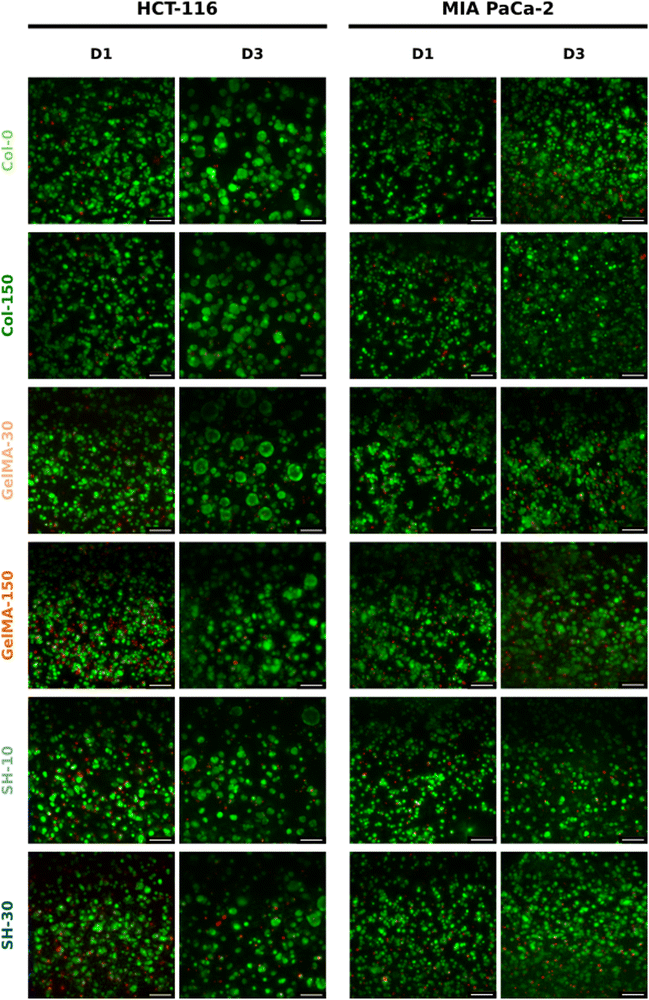
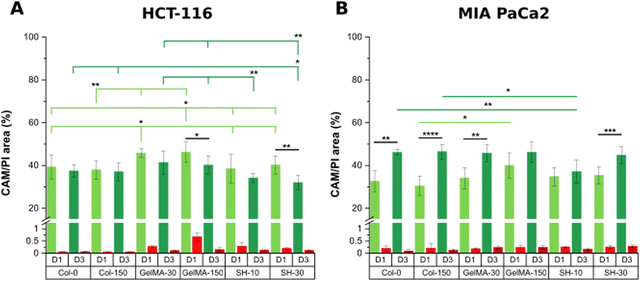
On the other hand, considering the area occupied by living cells (green area in live/dead micrographs), it seems that both GelMA scaffolds presented the highest composition affinity towards HCT-116 cells with CAM areas around 50% while the living cell occupancy for the other four scaffolds was near 40% (Fig. 5(A)).
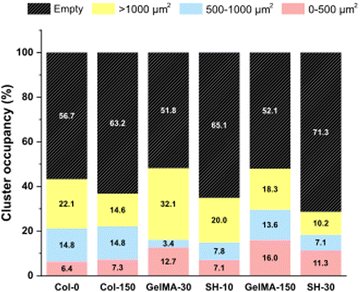
Interestingly, the initial single cell suspension of HCT-116 cells underwent proliferation, resulting in all cases, in the formation of compact cell clusters throughout the 3-day experimental period with particularly distinct features depending on the hydrogel (Fig. 6). In order to gain insight in the cell cluster formation, we sorted the cluster in three groups attending to their size: clusters with areas below 500 μm2, in the range of 500–1000 μm2 or larger than 1000 μm2 were defined as “small”, “medium” and “large”, respectively (Fig. S5, ESI†). On one hand, Col-0 and Col-150 hydrogels exhibited the most homogeneous distributions in terms of cluster sizes, although a higher amount of medium (blue) and large (yellow) structures were observed rather than small ones (pink). In GelMA-30, the softest biomaterial in this study, there was a clear predominance of large clusters, being the cell area occupied by this type of aggregates (32.1%) the highest among all scaffolds. Notably, medium cell aggregates had a very low occupancy (3.4%), almost 4 times less than the smallest clusters (12.7%). On the other hand, the cell area occupied in GelMA-150 hydrogels, which showed the nearest stiffness to Col-0 and Col-150, was equally distributed among the three sizes of clusters, as well. Similar to GelMA-30, the also soft SH-10 condition promoted an area occupancy dominated by the larger clusters (20.0%) while medium and small structures occupied a similar area (7.8% and 7.1%, respectively). Finally, the stiffest hydrogel SH-30 area was also evenly occupied by large, medium and small clusters although it is worth mentioning that this condition displayed the lowest occupancy by large clusters (10.2%) among the six hydrogels. Looking at each pair of hydrogels with the same chemical composition (Col, GelMA and SH), it is clear the effect of stiffness on the aggregates size: the higher area occupied by large clusters was observed when HCT-116 cells were cultured in the softer hydrogels. Our results agree with other works found in the literature on epithelial breast cancer and ovarian cancer where larger cell aggregates presented a progressively lower area occupancy with increasing stiffness.61–63 This could be explained by the degree of restriction established by the matrix: low-stiffness hydrogels offer a permissive environment for cell migration and growth. In contrast, high-stiffness hydrogels correspond to matrices that may restrict cell cluster growth. Above all, it should be highlighted that these 3D cellular structures emerged without the need for any biochemical stimuli, showcasing the adaptability of collagen and GelMA in creating a tunable biomimetic tumor microenvironment. Histograms and visual scheme on cluster size quantification are shown in Fig. S5 (ESI†). Finally, and as a consequence of cellular clustering, CAM areas at day 3 decreased in all cases, being especially significant in the stiffest conditions: GelMA-150 and SH-30 (Fig. 5(A)).
Regarding MIA PaCa-2 cell line, a non-aggregating pattern was observed and all scaffolds showed excellent biocompatibility at days 1 and 3 (Fig. 4). In addition, cells quickly proliferated and CAM areas increased from 35–40% (day 1) up to near 50% (day 3) in all cases, with the exception of SH-10 (Fig. 5(B)). Conversely to HCT-116, matrix stiffness and composition did not apparently affect biological behavior in terms of cell growth and morphology.
To sum up, the platform of bioscaffolds generated to encapsulate HCT-116 and MIA PaCa-2 cells yielded physiologically relevant matrices where cells remained viable, grew and successfully proliferated (3D renders of the three-dimensional distribution of cells within the hydrogels are shown in Fig. S6, ESI†).
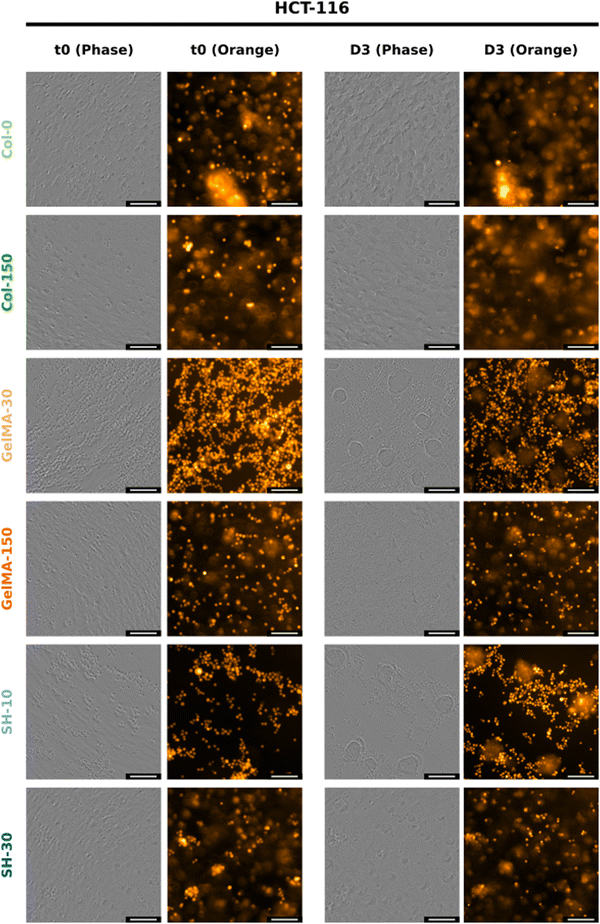 | ||
| Fig. 7 Phase-contrast and orange fluorescence images from IncuCyte® of HCT-116-embedded hydrogels at t = 0 (left) and day-3 (right). The scale bar represents 100 μm for 20× micrographs. (The twin figure for MIA PaCa-2 cells corresponds to Fig. S12, ESI†). | ||
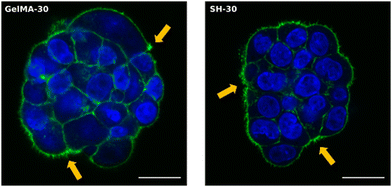
The truly appealing feature of HCT-116 cells to be analyzed with the Incucyte® system was the cluster-like conformation observed in all scaffolds. The progressive formation of these 3D cellular aggregates was easily followed in the softest hydrogels GelMA-30 and SH-10. Time-lapse movies can be found in the ESI.† As confirmed in Movies 1 and 2, (ESI†) cells started to move within the network and group together from the very beginning. Individual migration of single cells was observed according to Fig. S8 (ESI†). Small multicellular clusters were already observed after 17 h of cell culture both in GelMA-30 and SH-10 hydrogels and kept growing over time not only because of cell proliferation but also the assembly of cell aggregates that were initially separated in close proximity and moved toward each other progressively. Upon coalescence, the newly formed cluster undergoes a remodeling and compacting event, matching with the results reported by Palmiero et al. with several cell lines in their study about collective directional migration.64 Furthermore, during the experimental process, cell aggregates had the ability to rotate within the crosslinked network as shown in Fig. S9 (ESI†). This event was also reported by Peela et al. in their developed tumor model for breast cancer with MCF7 and MCF10A clusters formed in GelMA hydrogels.9 Likewise, Tanner and coworkers thoroughly described cellular rotation during the formation of glandular tissue and proposed this phenomenon to be promoted by the temporal loss of cell–ECM interactions and local degradation of ECM.65 As mentioned above, we additionally observed how clusters approximated to each other to finally assemble into a larger aggregate (Fig. S10, ESI†), probably due to the secretion of autocrine and paracrine factors that can modulate and drive the migration of the surrounding cells. In this respect, a wealth of studies has focused on collective cell migration taking place during metastasis. Although several phenomena remain to be investigated, there is evidence that tumor cell communication is supported by the expression of cell adhesion molecules such as E-cadherin (essential for adherent junctions)66 and mediated by upstream signaling. The autocrine secretion of soluble factors such as chemoattractants are partly responsible of cluster-cluster crosstalk at distance and collective directional migration occurs due to chemotaxis (concentration gradients).64,66 Regarding MIA PaCa-2 cell line, we also witnessed cell migration as a function of time, however and conversely to HCT-116 cells, MIA PaCa-2 cells grouped together while keeping intact their membranes as well as the fluorescence, not fusing into 3D clusters (Fig. S11, ESI†).
Thus, real-time cell monitoring disclosed individual and collective cell migration of both HCT-116 and MIA PaCa-2 cells. Notably, it was observed that the progressive approach between HCT-116 cells along with cell proliferation generated well-defined clusters with the ability to rotate and migrate.
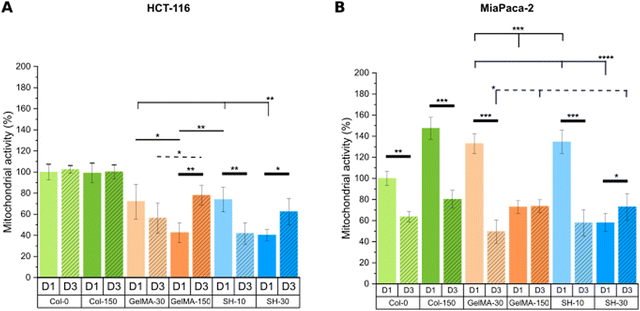 | ||
| Fig. 9 Mitochondrial metabolic activity was assessed through MTS assay on day-1 and day-3 of cell culture for HCT-116-laden (A) and MIA PaCa-2-laden (B) hydrogels. Error bars SD. Note: ****p < 0.0001, ***p < 0.001, **p < 0.01, *p < 0.05. Thin and dashed lines represent statistics among scaffolds on day-1 and day-3, respectively. Thick lines correspond to statistical differences between day-1 and day-3 within the same condition. (Statistical differences towards Col-0 and Col-150 and non-statistical differences are not drawn in the graph – see Fig. S13, ESI†). | ||
On the other hand, MIA PaCa-2 cells exhibited a slightly different behavior compared to HCT-116 cell line (Fig. 9(B)). Looking at the soft and medium stiffness hydrogels, Col-150, GelMA-30 and SH-10 exhibited an increased metabolic activity compared to the control Col-0 hydrogel. Although I2959 photoinitiator system has demonstrated excellent cytocompatibility, the irradiation with UV light may cause weak cell damage and as a result, MIA PaCa-2 cells might have become metabolically more active to overcome this initial harm. Similar findings were reported by Kremslehner and colleagues, who found a glucose 6-phosphate dehydrogenase (G6PD) upregulation in keratinocytes as an immediate UV response. G6PD constitutes the initial stage in the metabolic pathway for processing glucose via the oxidative branch of the pentose phosphate pathway (PPP). Cells that have previously experienced UV-induced oxidative stress and DNA damage generally exhibit a heightened demand for the products generated by the PPP. Thus, the authors verified that G6PD activity was upregulated in keratinocytes exposed to UV and that this enzymatic activity was increased in cells which initiated nucleotide damage response mechanisms.74
Remarkably, all scaffolds with stiffness below 2 kPa (Col-0, Col-150, GelMA-30 and SH-10) displayed a drastic decrease in metabolic activity after 3 days of culture despite the increased live cell area occupancy observed in live/dead micrographs. In contrast, no significant differences were found in GelMA-150 condition at days 1 and 3 while cells encapsulated within the stiffest hydrogel SH-30 showed a slightly higher metabolic activity over time.
In conclusion, the microenvironment created by the softest GelMA-based scaffolds (GelMA-30 and SH-10) promoted a decreased cell metabolic activity for both tumoral cell lines HCT-116 and MIA PaCa-2, according to healthy tissue-like conditions. Instead, stiffer GelMA-150 and SH-30 hydrogels markedly generated a rise in HCT-116 metabolic activity while they had minimal effect on MIA PaCa-2 cells.
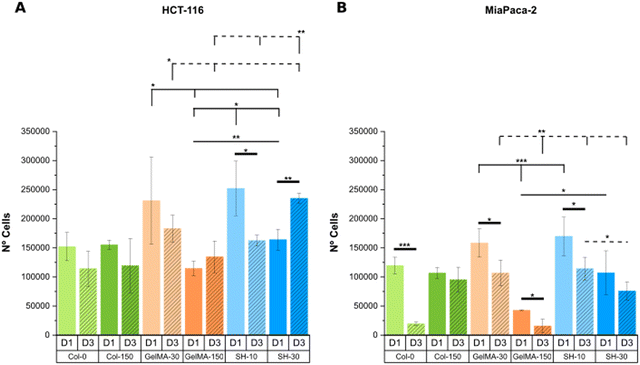 | ||
| Fig. 10 DNA content is expressed as the number of HCT-116 (A) and MIA PaCa-2 cells (B) embedded within collagen and GelMA-based hydrogels on day-1 and day-3 (calibration curves are shown in Fig. S14, ESI†). Error bars SD. Note: ****p < 0.0001, ***p < 0.001, **p < 0.01, *p < 0.05. Thin and dashed lines represent statistics among scaffolds on day-1 and day-3, respectively. Thick lines correspond to statistical differences between day-1 and day-3 within the same condition. (Statistical differences towards Col-0 and Col-150 and non-statistical differences are not drawn in the graph – see Fig. S15, ESI†). | ||
To conclude, a good alignment of results was generally found for the softest conditions (GelMA-30 and SH-10) in both cell lines HCT-116 and MIA PaCa-2, matching the decreased metabolic activity at day 3 with a lower quantified DNA content.
Conclusions
The creation and evaluation of 3D scaffolds for cancer disease research has become a crucial step halfway from affordable simple 2D substrates and expensive complex in vivo models. Herein, we explore a platform of photopolymerized GelMA-based hydrogels with tunable stiffness as biomimetic 3D models for colorectal and pancreas diseases. We found that hydrogels prepared without methanol in the initial formulation exhibited lower mechanical properties than those with methanol but same trends and capability to tune stiffness although in a lower range of values. Besides, cell viability displayed an exceptional increase in hydrogels formulated without methanol.Thus, in addition to the outstanding biological performance shown in semi-3D experiments from our earlier work, GelMA-based scaffolds also exhibited excellent cytocompatibility in 3D cell culture assays performed with two tumoral cell lines, HCT-116 and MIA PaCa-2 cells. HCT-116 cells cultured in collagen and GelMA-based hydrogels showed a clustering growth pattern with cell aggregates of different sizes over time which changed in response to the surrounding network. These results evidenced that less densely crosslinked and softer matrices represent permissive microenvironments for cell growth and migration. Linked to this cell motility, real-time monitoring revealed individual and collective migration of HCT-116 and MIA PaCa-2 cells. On the other hand, the formation of HCT-116 cellular clusters as a function of time pointed out the suitability of collagen and GelMA scaffolds to recreate the tumor cell microenvironment. HCT-116 cell line presented abundant filopodia formation which is in good agreement with its migratory behavior. Finally, the measured metabolic activity underwent a significant increase in stiffer hydrogels over the cell culture time while soft scaffolds promoted a drastic reduction. This proliferative and metabolically active phenotype developed within stiff networks matches with the malignant behavior observed in cancerous tissue.
In conclusion, we have demonstrated the ability of our GelMA-based material platform to create three dimensional microenvironments with different chemical composition, swelling behavior and stiffness and how HCT-116 and MIA PaCa-2 cells responded distinctly to the surrounding photocrosslinked matrix. This research establishes an attractive basis for further and more complex tumor models comprising concentration and stiffness gradients or co-culture with cancer associated fibroblasts (CAF), among others.
Author contributions
The manuscript was written through contributions of all authors. All authors have given approval to the final version of the manuscript.Conflicts of interest
There are no conflicts of interest to declare.Acknowledgements
Authors acknowledge the use of instrumentation as well as the technical advice provided by the NMR Service of CEQMA (UZ-CSIC), Microscopy and Imaging Core Facility, (IACS-IISAragon) and “Servicio General de Apoyo a la Investigación (SAI)” This work was funded through projects PID2020-118485RB-I00 and PID2022-141276OB-I00 granted by MICIU/AEI/10.13039/501100011033 and by “ERDF A way of making Europe”; through grant LMP221_21 funded by Gobierno de Aragón and “ERDF A way of making Europe”; and, through the “Fondo Social Europeo” (DGA E15_23R). This research was also supported by CIBER -Consorcio Centro de Investigación Biomédica en Red-(CB06/01/00263), Instituto de Salud Carlos III, Ministerio de Ciencia e Innovación. RPC acknowledges Gobierno de Aragón for a predoctoral fellowship (2017-2021). SGL was funded by Spanish MINECO fellowship (DI-17-09585).References
- L. Chin, Y. Xia, D. E. Discher and P. A. Janmey, Curr. Opin. Chem. Eng., 2016, 11, 77–84 CrossRef PubMed.
- X. Tang, T. B. Kuhlenschmidt, J. Zhou, P. Bell, F. Wang, M. S. Kuhlenschmidt and T. A. Saif, Biophys. J., 2010, 99, 2460–2469 CrossRef CAS PubMed.
- A. J. Rice, E. Cortes, D. Lachowski, B. C. H. Cheung, S. A. Karim, J. P. Morton and A. Del Río Hernández, Oncogenesis, 2017, 6, 1–9 CrossRef PubMed.
- S. Lee and S. Kumar, F1000Research, 2016, 5, 2261 Search PubMed.
- A. Singh, N. Dalal and P. Tayalia, Biomed. Mater., 2023, 18, 25010 CrossRef PubMed.
- A. I. Van Den Bulcke, Biomacromolecules, 2000, 1, 31–38 CrossRef CAS PubMed.
- Y. Zhang, H. Chen and J. Li, Int. J. Biol. Macromol., 2022, 221, 91–107 CrossRef CAS PubMed.
- A. D. Aryaa, P. M. Hallur, A. G. Karkisaval, A. Gudipati, S. Rajendiran, V. Dhavale, B. Ramachandran, A. Jayaprakash, N. Gundiah and A. Chaubey, ACS Appl. Mater., 2016, 8, 22005–22017 CrossRef PubMed.
- N. Peela, F. S. Sam, W. Christenson, D. Truong, A. W. Watson, G. Mouneimne, R. Ros and M. Nikkhah, Biomaterials, 2016, 81, 72–83 CrossRef CAS PubMed.
- R. Mazrouei, V. Velasco and R. Esfandyarpour, Sci. Rep., 2020, 10, 14669 CrossRef CAS PubMed.
- F. Fu, L. Shang, F. Zheng, Z. Chen, H. Wang, J. Wang, Z. Gu and Y. Zhao, ACS Appl. Mater. Interfaces, 2016, 8, 13840–13848 CrossRef CAS PubMed.
- C.-Y. Chang, H. C. Johnson, O. Babb, M. L. Fishel and C.-C. Lin, Acta Biomater., 2021, 130, 161–171 CrossRef CAS PubMed.
- A. Rosenfeld, T. Göckler, M. Kuzina, M. Reischl, U. Schepers and P. A. Levkin, Adv. Healthcare Mater., 2021, 10, 2100632 CrossRef CAS PubMed.
- Y. Li, C. Liu, W. Liu, X. Cheng, A. Zhang, S. Zhang, C. Liu, N. Li and X. Jian, Macromol. Biosci., 2021, 21, 2100262 CrossRef CAS PubMed.
- G. C. J. Brown, K. S. Lim, B. L. Farrugia, G. J. Hooper and T. B. F. Woodfield, Macromol. Biosci., 2017, 17, 1–13 CrossRef PubMed.
- N. Rajabi, M. Kharaziha, R. Emadi, A. Zarrabi, H. Mokhtari and S. Salehi, J. Colloid Interface Sci., 2020, 564, 155–169 CrossRef CAS PubMed.
- R. Pamplona, S. González-Lana, P. Romero, I. Ochoa, R. Martín-Rapún and C. Sánchez-Somolinos, ACS Appl. Polym. Mater., 2023, 5, 1487–1498 CrossRef CAS PubMed.
- C. E. Hoyle and C. N. Bowman, Angew. Chem., Int. Ed., 2010, 49, 1540–1573 CrossRef CAS PubMed.
- H. Y. Liu, M. Korc and C. C. Lin, Biomaterials, 2018, 160, 24–36 CrossRef CAS PubMed.
- M. Devarasetty, E. Wang, S. Soker and A. Skardal, Biofabrication, 2017, 9, 021002 CrossRef PubMed.
- F. Cadamuro, L. Marongiu, M. Marino, N. Tamini, L. Nespoli, N. Zucchini, A. Terzi, D. Altamura, Z. Gao, C. Giannini, G. Bindi, A. Smith, F. Magni, S. Bertini, F. Granucci, F. Nicotra and L. Russo, Carbohydr. Polym., 2023, 302, 120395 CrossRef CAS PubMed.
- A. Williams, J. F. Nowak, R. Dass, J. Samuel and K. L. Mills, Front. Physiol., 2018, 9, 1–10 Search PubMed.
- E. Jabbari, S. K. Sarvestani, L. Daneshian and S. Moeinzadeh, PLoS One, 2015, 10, e0132377 CrossRef PubMed.
- N. Betriu, A. Andreeva, A. Alonso and C. E. Semino, Biomedicines, 2022, 10, 1835 CrossRef CAS PubMed.
- T. J. Puls, X. Tan, C. F. Whittington and S. L. Voytik-Harbin, PLoS One, 2017, 12, e0188870 CrossRef CAS PubMed.
- H. Shirahama, B. H. Lee, L. P. Tan and N. J. Cho, Sci. Rep., 2016, 6, 1–11 CrossRef PubMed.
- A. F. S. A. Habeeb, Anal. Biochem., 1966, 14, 328–336 CrossRef CAS PubMed.
- E. Hoch, C. Schuh, T. Hirth, G. E. M. Tovar and K. Borchers, J. Mater. Sci.: Mater. Med., 2012, 23, 2607–2617 CrossRef CAS PubMed.
- B. H. Lee, H. Shirahama, N. J. Cho and L. P. Tan, RSC Adv., 2015, 5, 106094–106097 RSC.
- C. Claaßen, M. H. Claaßen, V. Truffault, L. Sewald, G. E. M. Tovar, K. Borchers and A. Southan, Biomacromolecules, 2018, 19, 42–52 CrossRef PubMed.
- M. Monfared, M. D. Nothling, D. Mawad and M. H. Stenzel, Biomacromolecules, 2021, 22, 4295–4305 CrossRef CAS PubMed.
- X. Li, S. Chen, J. Li, X. Wang, J. Zhang, N. Kawazoe and G. Chen, Polymers, 2016, 8, 269 CrossRef PubMed.
- L. Li, C. Lu, L. Wang, M. Chen, J. White, X. Hao, K. M. McLean, H. Chen and T. C. Hughes, ACS Appl. Mater. Interfaces, 2018, 10, 13283–13292 CrossRef CAS PubMed.
- S. Bertlein, G. Brown, K. S. Lim, T. Jungst, T. Boeck, T. Blunk, J. Tessmar, G. J. Hooper, T. B. F. Woodfield and J. Groll, Adv. Mater., 2017, 29, 1–6 CrossRef PubMed.
- R. Pamplona, S. González-Lana, P. Romero, I. Ochoa, R. Martín-Rapún and C. Sánchez-Somolinos, Macromol. Biosci., 2023, 23, 2300227 CrossRef CAS PubMed.
- R. Schrieber and H. Gareis, Gelatine Handbook: Theory and Industrial Practice, 2007 Search PubMed.
- J. Van Hoorick, P. Gruber, M. Markovic, M. Tromayer, J. Van Erps, H. Thienpont, R. Liska, A. Ovsianikov, P. Dubruel and S. Van Vlierberghe, Biomacromolecules, 2017, 18, 3260–3272 CrossRef CAS PubMed.
- S. Xiao, T. Zhao, J. Wang, C. Wang, J. Du, L. Ying, J. Lin, C. Zhang, W. Hu, L. Wang and K. Xu, Stem Cell Rev. Rep., 2019, 15, 664–679 CrossRef CAS PubMed.
- J. Van Hoorick, L. Tytgat, A. Dobos, H. Ottevaere, J. Van Erps, H. Thienpont, A. Ovsianikov, P. Dubruel and S. Van Vlierberghe, Acta Biomater., 2019, 97, 46–73 CrossRef CAS PubMed.
- G. Miquelard-Garnier, S. Demeures, C. Creton and D. Hourdet, Macromolecules, 2006, 39, 8128–8139 CrossRef CAS.
- S. T. Nguyen, H. T.-L. Nguyen and K. D. Truong, Biomed. Res. Ther., 2020, 7, 3855–3859 CrossRef.
- E. E. Antoine, P. P. Vlachos and M. N. Rylander, Tissue Eng., Part B, 2014, 20, 683–696 CrossRef CAS PubMed.
- A. Rubiano, D. Delitto, S. Han, M. Gerber, C. Galitz, J. Trevino, R. M. Thomas, S. J. Hughes and C. S. Simmons, Acta Biomater., 2018, 67, 331–340 CrossRef PubMed.
- S. Ramanujan, A. Pluen, T. D. Mckee, E. B. Brown, Y. Boucher and R. K. Jain, Biophys. J., 2002, 83, 1650–1660 CrossRef CAS PubMed.
- B. N. Mason, J. P. Califano and C. A. Reinhart-King, Matrix Stiffness: a regulator of cellular behavior and tissue formation, 2012 Search PubMed.
- H. Yu, J. K. Mouw and V. M. Weaver, Trends Cell Biol., 2011, 21, 47–56 CrossRef PubMed.
- S. Van Helvert and P. Friedl, ACS Appl. Mater. Interfaces, 2016, 8, 21946–21955 CrossRef CAS PubMed.
- N. R. Richbourg, M. K. Rausch and N. A. Peppas, Polymer, 2022, 258, 125316 CrossRef CAS.
- C. B. Raub, A. J. Putnam, B. J. Tromberg and S. C. George, Acta Biomater., 2010, 6, 4657–4665 CrossRef CAS PubMed.
- Y. Ishibashi, R. Haraguchi, S. Aoki, Y. Oishi and T. Narita, Gels, 2023, 9, 458 CrossRef CAS PubMed.
- K. Maeda, Cosmetics, 2018, 5, 14 CrossRef.
- W. Tomal and J. Ortyl, Polymers, 2020, 12, 1073 CrossRef CAS PubMed.
- M. Wang, J. Bai, K. Shao, W. Tang, X. Zhao, D. Lin, S. Huang, C. Chen, Z. Ding and J. Ye, Int. J. Polym. Sci., 2021, 2021, 1–16 CAS.
- S. Kawano, M. Kojima, Y. Higuchi, M. Sugimoto, K. Ikeda, N. Sakuyama, S. Takahashi, R. Hayashi, A. Ochiai and N. Saito, Cancer Sci., 2015, 106, 1232–1239 CrossRef CAS PubMed.
- M. Nebuloni, L. Albarello, A. Andolfo, C. Magagnotti, L. Genovese, I. Locatelli, G. Tonon, E. Longhi, P. Zerbi, R. Allevi, A. Podestà, L. Puricelli, P. Milani, A. Soldarini, A. Salonia and M. Alfano, Sci. Rep., 2016, 6, 22522 CrossRef CAS PubMed.
- T. Billiet, B. Van Gasse, E. Gevaert, M. Cornelissen, J. C. Martins and P. Dubruel, Macromol. Biosci., 2013, 13, 1531–1545 CrossRef CAS PubMed.
- A. Sionkowska, Polym. Degrad. Stab., 2000, 68, 147–151 CrossRef CAS.
- Z. Mũnoz, H. Shih and C. Lin, Biomater. Sci., 2014, 2, 1063 RSC.
- B. D. Polizzotti, B. D. Fairbanks and K. S. Anseth, Biomacromolecules, 2008, 9, 1084–1087 CrossRef CAS PubMed.
- D. J. Lavalley, P. G. Miller and M. L. Shuler, Biotechnol. Prog., 2020, 37, e3105 CrossRef PubMed.
- L. Shah, A. Latif, K. J. Williams and A. Tirella, Acta Biomater., 2022, 152, 273–289 CrossRef CAS PubMed.
- M. Cavo, M. Fato, L. Peñuela, F. Beltrame, R. Raiteri and S. Scaglione, Sci. Rep., 2016, 6, 35367 CrossRef CAS PubMed.
- D. Loessner, K. S. Stok, M. P. Lutolf, D. W. Hutmacher, J. A. Clements and S. C. Rizzi, Biomaterials, 2010, 31, 8494–8506 CrossRef CAS PubMed.
- M. Palmiero, I. Cantarosso, L. di Blasio, V. Monica, B. Peracino, L. Primo and A. Puliafito, Mol. Oncol., 2023, 17, 1699–1725 CrossRef CAS PubMed.
- K. Tanner, H. Mori, R. Mroue, A. Bruni-Cardoso and M. J. Bissell, Proc. Natl. Acad. Sci. U. S. A., 2012, 109, 1973–1978 CrossRef CAS PubMed.
- A. Yamamoto, A. E. Doak and K. J. Cheung, Annu. Rev. Pathol.: Mech. Dis., 2023, 18, 231–256 CrossRef CAS PubMed.
- M. M. Brás, T. B. Cruz, A. F. Maia, M. J. Oliveira, S. R. Sousa, P. L. Granja and M. Radmacher, Cancers, 2022, 14, 5053 CrossRef PubMed.
- Y. F. Zeng, Y. S. Xiao, Y. Liu, X. J. Luo, L. D. Wen, Q. Liu and M. Chen, World J. Gastroenterol., 2018, 24, 3884–3897 CrossRef CAS PubMed.
- P. K. Mattila and P. Lappalainen, Nat. Rev. Mol. Cell Biol., 2008, 9, 446–454 CrossRef CAS PubMed.
- A.-M. Baker, D. Bird, G. Lang, T. R. Cox and J. T. Erler, Oncogene, 2013, 32, 1863–1868 CrossRef CAS PubMed.
- N. Davidenko, C. F. Schuster, D. V. Bax, R. W. Farndale, S. Hamaia, S. M. Best and R. E. Cameron, J. Mater. Sci.: Mater. Med., 2016, 27, 148 CrossRef PubMed.
- K. M. Pawelec, S. M. Best and R. E. Cameron, J. Mater. Chem. B, 2016, 4, 6484–6496 RSC.
- R. Ata and C. Antonescu, Int. J. Mol. Sci., 2017, 18, 189 CrossRef PubMed.
- C. Kremslehner, A. Miller, R. Nica, I. M. Nagelreiter, M. S. Narzt, B. Golabi, V. Vorstandlechner, M. Mildner, J. Lachner, E. Tschachler, F. Ferrara, K. Klavins, M. Schosserer, J. Grillari, A. Haschemi and F. Gruber, Redox Biol., 2020, 37, 101583 CrossRef CAS PubMed.
- B. Deng, Z. Zhao, W. Kong, C. Han, X. Shen and C. Zhou, J. Transl. Med., 2022, 20, 540 CrossRef CAS PubMed.
- K. S. Lim, B. J. Klotz, G. C. J. Lindberg, F. P. W. Melchels, G. J. Hooper, J. Malda, D. Gawlitta and T. B. F. Woodfield, Macromol. Biosci., 2019, 19, 1–14 CrossRef PubMed.
- J. S. Brand, L. Forster, T. Böck, P. Stahlhut, J. Teßmar, J. Groll and K. Albrecht, Macromol. Biosci., 2022, 22, 2100274 CrossRef CAS PubMed.
Footnote |
| † Electronic supplementary information (ESI) available. See DOI: https://doi.org/10.1039/d3tb02640j |
| This journal is © The Royal Society of Chemistry 2024 |