Open Access Article
Open Access ArticleGraphene exfoliation using multidomain peptides†
Ruitao
Jin
 a,
Nermina
Brljak
a,
Nermina
Brljak
 bc,
Joseph M.
Slocik
d,
Rahul
Rao
bc,
Joseph M.
Slocik
d,
Rahul
Rao
 d,
Marc R.
Knecht
d,
Marc R.
Knecht
 *bc and
Tiffany R.
Walsh
*bc and
Tiffany R.
Walsh
 *a
*a
aInstitute for Frontier Materials, Deakin University, Waurn Ponds, VIC 3216, Australia. E-mail: tiffany.walsh@deakin.edu.au
bDepartment of Chemistry, University of Miami, Coral Gables, Florida 33146, USA. E-mail: knecht@miami.edu
cDr. J.T. Macdonald Foundation BioNIUM, University of Miami, Miami, FL 33136, USA
dAir Force Research Laboratory, Wright-Patterson Air Force Base, Ohio 45433, USA
First published on 27th February 2024
Abstract
Liquid-phase exfoliation using biomolecules in aqueous solution is a promising approach to obtain high quality 2D nanosheets. For example, the well-studied graphene-binding peptide, P1 (sequence HSSYWYAFNNKT), has been previously investigated and shown to have a good ability to exfoliate graphene sheets in aqueous conditions under sonication, maintaining colloidal stability. Building on this, the biomolecular exfoliant and assembly motif (BEAM) peptide, that features a graphene-binding domain at one end and a hexagonal boron nitride (h-BN) binding domain at the other, separated by a 10-carbon fatty acid chain in the centre, is shown to exfoliate graphene sheets from bulk graphite in aqueous media. An in-depth examination of the ability of the BEAM to both facilitate sheet exfoliation under sonication conditions and also maintain colliodal stability is provided through molecular dynamics simulations. These findings open new possibilities for designing multi-functional molecules that can both exfoliate and organise 2D materials into heterostructures under ambient conditions in aqueous media.
Introduction
The ability to isolate significant amounts of individual sheets of two dimensional (2D) nanomaterials such as graphene remains a pressing challenge.1 These structures are required for a variety of applications ranging from energy storage to catalysis due to their unique electronic and optical properties.2 While two main paths are known to access these structures for graphene (e.g. synthesis or exfoliation), they either tend to have low yields or generate nanosheets with incorporated defects that can lead to diminished properties. For instance, chemical vapor deposition (CVD) methods can be used to synthesize individual graphene sheets on surfaces;3 however, this low throughput method generates low amounts of the materials. Alternatively, methods have been developed to exfoliate graphene sheets from bulk graphite samples using a variety of ligands such as polymers and non-aqueous solvents.4,5 While these approaches can generate significant quantities of the materials, they tend to lead to the production of defects in the individual sheets. Taken together, new methods must be identified that can be exploited to generate high yields of graphene sheets with minimal defects, preferably under sustainable conditions.As an alternative to conventional methods, bio-inspired approaches have been exploited to isolate peptides with affinity for 2D nanomaterials, including graphene, hexagonal boron nitride (h-BN), and MoS2.6–8 These peptides adsorb to the surface through a complex binding pattern of the residues (typically 12) in the sequence, which, in the case of graphene, can be used to drive nanosheet exfoliation.9–12 For instance, exfoliation of graphene has been demonstrated using the graphene-binding P1 peptide (HSSYWYAFNNKT)7,13 under mild sonication conditions.9,10 In this case, sonication provides sufficient energy to separate the individual sheets in the bulk graphite where peptide adsorption to the exposed surfaces leads to exfoliation. Interestingly, using this bio-based approach results in the isolation of graphene sheets with lower degrees of defect incorporation.10
While the P1 peptide can drive graphene exfoliation, it also provides an opportunity to incorporate additional functionality at the graphene surface. To this end, chemical modification of the peptide could be used to incorporate new functional handles into the sequence that could be docked at the nanosheet surface upon P1 adsorption.14 That said, once these new functionalities are incorporated, they can change the binding of the P1 domain for the target graphene surface, which could lead to modifications in both the peptide adsorbed structure and the ability to drive exfoliation. For instance, incorporation of fatty acid domains to the P1 at either the N- or C-terminus of the sequence demonstrated changes to both the surface adsorbed structure of the P1 as well as the binding affinity of the peptide.14,15
In addition to driving exfoliation, the P1 peptide has also been incorporated into a larger chimeric construct, which displayed highly specific surface binding properties. In this regard, the P1 was conjugated to an h-BN binding peptide (BP7: VDAQSKSYTLHD)16via a spacing fatty acid domain. This new construct is termed a BEAM – biomolecular exfoliating and assembly motif (Fig. 1). In this structure, the BEAM is designed to drive assembly of a graphene/h-BN heterostructure through graphene binding via the P1 domain and h-BN binding through the BP7 region. While prior studies have demonstrated that this sequence can discriminate between the two disparate surfaces and target/peptide specific binding can be achieved,17 including fundamental studies quantifying the binding of the individual peptides P1 and BP7 on both graphene and h-BN surfaces,18 its ability to drive graphene exfoliation remains unknown. This is the first step in achieving heterostructure formation where incorporation of such a large secondary group (e.g. fatty acid and BP7) to the P1 peptide could prevent the exfoliation process. In addition, this additional mass could also lead to significant variations in both exfoliation efficiency and the quality of the exfoliated graphene materials. As such, it is critically important to identify how the global biomolecular structure affects graphene exfoliation for eventual use in material assembly. The design of the BEAM molecule in its application to heterostack formation capability was previously explored with simulations demonstrating the stability of a graphene/h-BN heterostack arrangement,17 along with the proposal of a sequential approach to drive nanosheet assembly based on the P1-only and BP7-only binding data.17
Here we explore the use of the BEAM for driving graphene exfoliation under low temperature, aqueous conditions. Based on earlier work,9,10,19 all-atom molecular dynamics (MD) simulations can provide key insights into this exfoliation process. Here, MD simulations were used to explore the graphene exfoliation and colloidal suspension capabilities of the BEAMs, indicating that these chimeric molecules can successfully produce exfoliated graphene in aqueous media. To experimentally confirm this capability, a probe sonication-based method was exploited to generate graphene sheets in significantly shorter times (1 h) as compared to bath sonication methods (12 h).9,10 Once the materials were obtained, they were fully characterized via spectroscopic and electronic methods to confirm the structure of the graphene nanosheets.
Results and discussion
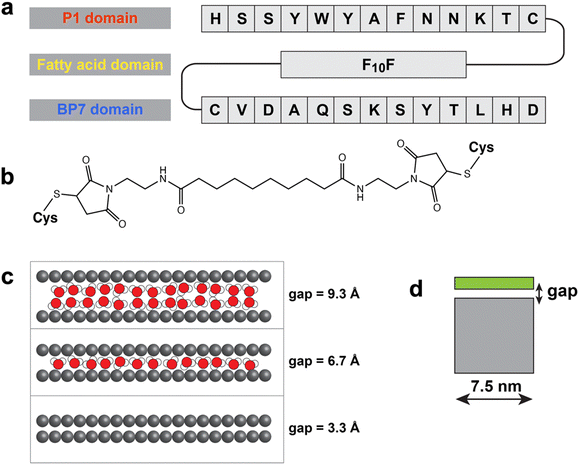
The BEAM molecule (Fig. 1a) was prepared with a P1 peptide domain at the N-terminus, a BP7 peptide domain at the C-terminus, and a 10-carbon-length fatty acid spacer (termed F10F) positioned in between (such that the BEAM is denoted P1C-F10F-CBP7). To connect each peptide domain to the centralized fatty-acid, a cysteine residue was introduced into the C-terminus of P1 and the N-terminus of BP7. This allowed for site specific coupling with two maleimide groups integrated at both ends of the F10F fatty acid (Fig. 1b) using thiol maleimide coupling sequentially as previously described.17The success of the exfoliation process hinges on two stages: first, the sheets must be detached from the bulk material during sonication,9 and second, these detached sheets must be prevented from reunifying upon conclusion of the sonication process.19 Although experimental approaches can ultimately evaluate the success of exfoliation via observation of the resultant colloidal suspension, it is challenging for experiment to directly verify the details of these two phenomena. In contrast, MD simulations can provide atomic-scale insights into each stage of the process. For the simulations, the parameters for describing the F10F domain were taken from previous work of modelling F10CP1 and P1CF10 (essentially P1 with the fatty acid at either termini).10 Comparing with the two parent peptides, this BEAM molecule exhibits stronger binding on the h-BN surface and similar binding free energy on the graphene surface.17 Moreover, replica exchange with solute tempering molecular dynamics (REST-MD) simulations indicated that the P1 domain of the BEAM interacted with the graphene surface strongly, as well as the F10F motif, and that the BP7 domain also maintained sufficient interactions with the surface using several anchor residues such as valine, alanine, glutamine, and tyrosine.17
In this study, MD simulations were first used to examine the possibility and the mechanisms of dynamic insertion of the BEAM molecules into the expanded gap region between two graphene sheets, and the ability of the BEAM to maintain this gap, all done in the medium of liquid water. This process is representative of the first stage of graphene exfoliation from bulk graphite in aqueous media. As in our previous work,9,10 a model of a nine-layer graphite stack was prepared and the gap between the top two graphene sheets in the stack was expanded to 10 Å to mimic the expanded gap under sonication conditions, in the presence or absence of the BEAM in the NVT ensemble (Fig. 1c). Only the very bottom graphene sheet was frozen in space and the rest of the sheets in the stack, including the top-most partially detached sheet, were completely free to move. Given that the BEAM molecule is longer than the P1 peptide or the P1CF10 molecule examined in previous work, the size of the graphene sheet in the stack was also increased compared with previous studies, such that the graphene sheet diameter was ∼7.5 nm (Fig. 1d; compared to a diameter of ∼4.7 nm used previously).9,10
The recombination timescale of the expanded gap, namely the possible reunification of the detached top graphene sheet with the graphite stack in the absence of the BEAM, was first estimated to obtain an average gap closure time. This establishes a baseline timescale against which the closure time in the presence of the BEAMs can be compared, enabling determination of the effectiveness of the BEAMs as exfoliating agents. Upon solvation of the BEAM-absent system, two layers of water molecules were initially observed in the expanded gap region. Four independent simulations in the absence of the BEAM molecules were run, and the average time for gap closure was 73.5 ns with the quickest gap closure time of 20.3 ns. For comparison, the corresponding gap-closure study using a smaller graphene sheet diameter (∼4.7 nm diameter) yielded an averaged closure time of 91 ns in liquid water in the absence of peptide.9 The faster closure time reported here reflects the relatively greater surface area of the basal plane and the concomitant increase in overall van der Waals attraction between the two sheets relative to the smaller sheet dimensions used in previous work. It is noted here that the absolute values of the closure times are not the key point of relevance, but rather provide a relative baseline against which the performance of the BEAMs can be evaluated. In the current study, the two water layers in the expanded gap first reduced to one layer, then diminished completely (Fig. 1c), indicating closure.
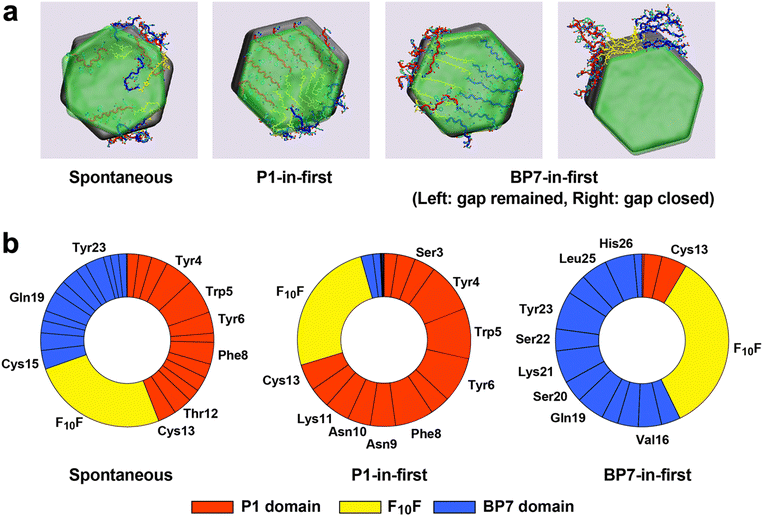
Another two scenarios were then explored using MD simulations involving the BEAM molecules. This was done to explore two key stages of graphene exfoliation: the initial stage to probe the ability of the BEAM to intercalate into the expanded gap and maintain an open gap (i.e., to prevent the top-most layer of graphite from re-uniting with the remainder of the stack), and the final stage probing the ability of the BEAM molecules to decorate free (exfoliated) graphene sheets in solution and inhibit these exfoliated sheets from reuniting in water. In the first scenario, and following from previous studies,9,10 two different initial conditions were investigated, herein termed ‘spontaneous insertion’ and ‘pre-insertion’. In the former case, simulations were first conducted to test if those BEAM molecules could spontaneously insert into the gap. In each scenario, six BEAM molecules were placed in the periodic simulation cell in different arrangements, along with the graphite stack (in the expanded gap configuration) and liquid water. The peptide positions were initially frozen to ensure the ingress of water molecules into the expanded gap, prior to running production simulations in which the peptides were free to move. Ten independent simulations were run for 150 ns for each such arrangement. All ten simulations resulted in spontaneous insertion of the BEAM molecules, as discussed in more detail herein. Next, for the pre-insertion case, BEAM molecules were initially placed inside the expanded gap, and simulations were conducted to test the stability of the expanded gap in this ‘pre-inserted’ state (that is, the stability of the expanded gap was examined following on from the fact that the BEAM molecules could spontaneously insert themselves into the expanded gap). The pre-insertion of the BEAMs was accomplished in two ways: P1-domain in first and BP7-domain in first. For the spontaneous insertion case, BEAM molecules were initially placed near the expanded gap, both parallel to the stack edge and/or situated above the top-most sheet.
As was previously reported for the parent P1 peptide,9 all spontaneous insertion tests ended with the expanded gap remaining in place at the end of simulations, in which at least two BEAM molecules were found to remain inserted into the gap. Next, based on the simulations from the second case, the spontaneous insertion mechanism for the BEAM was found to be reminiscent of the P1-based mechanism,9 namely via a sideswiping movement (Fig. 2a), indicating the capability of the BEAM to find the solvent-accessible surface of the graphene sheet and bind onto it by moving into the gap. In addition, during the insertion progress of the biomolecules, the detached graphene sheet was observed to have a greater mobility of rotation and/or large tilting angle caused by the local assembling of BEAMs within the gap, whereas the top sheet in BEAM-free tests lacked such flexibility, which may also contribute to the exfoliation progress under sonication.
For the pre-insertion runs, in most instances the expanded gap remained open. For the P1-in-first or BP7-in-first cases, the majority of the pre-inserted peptide domain remained within the gap, together with nearly all of F10F and part of the other peptide domain adsorbed into the gap as well (Fig. 2b). The packing of BEAMs within the gap showed some evidence of partial ordering for the pre-insertion cases. However, the spontaneous insertion tests resulted in a quite diverse arrangement of those conformations within the gap. Further analysis of which parts of the BEAM remained inside the gap using the last frame of the spontaneous insertion test trajectories for each production run revealed that a greater number of P1 domain atoms were found in the gap compared with the BP7 domain, consistent with the slightly higher binding affinity for P1 on graphene over BP7. Together with the 60% success rate of pre-insertion tests performed with BP7 domains placed in the gap first, this indicates that the BP7 domain was certainly able to be inserted into the gap as long as this included the presence of the exfoliant domain (P1) or linker (F10F). More specifically, residues including the YWY motif and the phenylalanine residue in the P1 domain, and the tyrosine residues in the BP7 domain, clearly acted as the main binder of the BEAM as well as the F10F hydrocarbon spacer. All of these results suggested that the BEAM is able to exfoliate the graphite under sonication conditions, similar to the behaviour of P1 or P1CF10 in experiment.
With the computational analysis indicating that the BEAM can support graphene exfoliation, its use in liquid phase graphene exfoliation was analysed experimentally. Previous binding studies confirmed that the BEAM can bind the graphene surface with a ΔG value of −35.3 ± 1.9 kJ mol−1.17 For graphene exfoliation, 1 mg of the BEAM was dissolved in 10 mL of water to which 50 mg of bulk graphite was added. This mixture was then subjected to probe ultra-sonication for 1 h. After sonication, the sample was centrifuged to separate bulk graphite where the supernatant containing the exfoliated graphene sheets was collected for further characterization.
UV-vis spectroscopy was initially used to quantify the degree of exfoliation achieved as shown in Fig. 3a and b. From the supernatant of the exfoliated sample, a dark solution was achieved, which demonstrated substantial scattering in the sample (Fig. 3a), consistent with significant exfoliation. When the same sonication procedure was performed in the absence of the BEAM, a clear and colourless solution was achieved, which demonstrated negligible absorbance. To further evaluate the samples, the degree of scattering at 660 nm was compared (Fig. 3b), giving rise to a value of 1.31 ± 0.098 and 0.065 ± 0.024 cm−1 for the reactions with and without the BEAM present, respectively, confirming significant graphene exfoliation using the biomolecules.
In addition to UV-vis analysis, fluorescence spectroscopy of the sample before and after exfoliation was studied (Fig. 3c). The BEAM is generally fluorescent with an emission at 378 nm due to the aromatic residues within the peptides. As the BEAM is bound to the surface of graphene, quenching of the fluorescence intensity was observed, consistent with previous studies.9,20 This suggests that the BEAM molecules are adsorbed to the graphene surface to maintain material colloidal stability. Based upon visual inspection of the samples, the BEAM stabilized materials remained colloidally suspended for at least two months without notable precipitation.
Imaging of the exfoliated graphene sheets was achieved using both transmission electron microscopy (TEM) and atomic force microscopy (AFM). Fig. 3d shows a TEM image of a single graphene sheet exfoliated using the BEAMs. Significantly large samples were obtained with lateral dimensions of greater than 100 nm. An additional, high-resolution TEM image of the materials is presented in the ESI,† Fig. S1. AFM analysis was also conducted to complement TEM imaging to measure and assess the thickness of the graphene sheets. Fig. 3e shows the AFM image of the graphene sample exfoliated by the BEAM and the statistical analysis of the height profiles. Fig. S2 in the ESI† provides a larger AFM image of the exfoliated materials. The average height of the BEAM-exfoliated material was 1.8 ± 0.4 nm, which is similar to the thickness of the samples exfoliated with the parent P1 peptide previously.9
In addition, Raman spectroscopy analysis was conducted to provide insights into the defect density. This was done by depositing the exfoliated graphene sheets onto a gold-coated glass slide and exciting at 514.5 nm. Fig. 3f displays a Raman spectrum averaged by collecting spectra from at least six spots from the BEAM-exfoliated graphene sheets. Three peaks can be observed in the spectrum at 1350, 1585, and 2720 cm−1, corresponding to the D, G, and 2D peaks, respectively. The G peak represents in-plane stretching vibrations involving the sp2 hybridized carbon atoms. The D peak corresponds to lattice defects and the 2D peak is the overtone of the D peak resulting from two phonon vibrations within the lattice. The intensity and shape of the 2D peak is known to correlate to the number of graphene layers.21 A single sharp high intensity 2D peak corresponding to monolayer graphene was not observed; however, the lineshape and intensity of the 2D peak in Fig. 3f suggests the presence of few layers of graphene, which was confirmed via a Lorentzian linesphape analysis (ESI,† Fig. S2).22,23 To determine the quality of the graphene sheets, the (integrated) intensity ratio between the D peak and the G peak (ID/IG) can be used. The ratio calculated for the graphene sheets exfoliated by BEAM is 0.57 ± 0.12 which is higher than the ratio previously measured for P1 ∼0.4,9 suggesting more defects on the surface.
The post-sonication stability of the exfoliated graphene is achieved via BEAM adsorption onto the nanosheets; this sterically prevents recombination of the materials to regenerate the bulk system. To explore this effect, the free energy profile of the reunification of two BEAM-decorated graphene sheets in liquid water was examined computationally. The two sheets were arranged in a face-to-face configuration. These simulations were conducted to mimic post-sonication events at 300 K, using the umbrella sampling MD simulation method. Following a procedure similar to that previously published to investigate the free energy profile for the face-to-face approach of two P1-decorated sheets,19 extensive tests were first conducted to determine an appropriate level of loading (i.e., how many BEAM molecules could be loaded onto the ∼7.5 nm diameter graphene sheet). The BEAM-loading test simulations determined a maximum stable loading of 12 BEAMs distributed over both sides (six per side) of the graphene sheet, resulting in a pseudo-film thickness of ∼1.5 nm. The two BEAM-decorated sheet complexes were then arranged face-to-face and separated vertically with the closest distance between two complexes being ∼2.5 nm (between a BEAM atom on sheet one/BEAM atom on sheet two) while the initial centre-to-centre distance between the two graphene sheets was ∼5.5 nm (Fig. 4a).
The potential of mean force (PMF) profile for the decorated-sheet reunification is shown in Fig. 4b with representative snapshots of the system at different values of the reaction coordinate (RC, the vertical centre-to-centre distance between the two graphene sheets). Similar to what was previously reported for the PMF profile of two P1-decorated graphene sheets, two apparent regimes were present: the low-repulsion region and the high-repulsion region with the transition point for a value of the RC of ∼2.5 nm. The free energy penalty for the two complexes to approach each other at a distance greater than 2.5 nm inter-sheet separation was minimal. However, an abrupt onset indicating a repulsion effect was observed when the two complexes approached more closely than this, which was attributed to the close contact between the BEAM molecules absorbed on the opposing graphene surfaces. This made the reunification of the two graphene sheets extremely unlikely. Between the RC values of 2.5 to 1.0 nm, no entire BEAM chain was observed to be squeezed out the inter-sheet gap, whereas parts of the BEAMs were forced from the gap with different expulsion rates as indicated in Fig. 4c. Compared with the P1 domain, the BP7 domains were more readily expelled from the gap with a higher expulsion rate, as indicated by the number of atoms belonging to the BP7 domains decreasing more rapidly. The F10F domain had the slowest expulsion rate, indicating its strong binding affinity. Water molecules were always observed to be found in the gap, even at the very close inter-sheet separations with higher repulsion. However, the total number of water molecules also decreased as the inter-sheet gap decreased; therefore, one source of the high-repulsion regime could be attributed to effective dehydration in the gap leaving only a highly-dehydrated BEAM layer in the centre. As such, similar to its parent P1 peptide, the BEAM molecule exhibited similar behaviour in the post-sonication scenario in that the BEAM-decorated graphene sheets were able to inhibit reunification in the aqueous environment to maintain the long-lived dispersions of exfoliated graphene sheets.
Conclusions
In summary, this work has demonstrated that the newly proposed hybrid molecule, BEAM, was able to bind the solvent-exposed graphene surface in a similar manner to the P1 peptide or the fatty acid conjugated molecule P1CF10 to both enable the exfoliation of the graphene sheets under sonication conditions and stabilize the resultant colloidal suspension of the graphene sheets in water. This suggests that incorporation of additional functionality (e.g., the BP7 peptide) into the biomolecular construct does not interfere with graphene exfoliation. The results further suggest a stronger interaction between the P1 domain of the BEAM with the graphene surface, as anticipated from the original target affinity of the peptide. This indicates that the BP7 domain may remain available for h-BN binding for eventual heterostructure formation. Significant more experimental analysis is required to achieve and confirm such capabilities.Experimental
Materials
The peptides used for the synthesis of the bifunctional BEAM molecules were obtained commercially from Genscript with a purity of >99% and used without purification. Sebacoyl chloride was purchased from Sigma. Triethylamine (TEA), N,N-dimethylformamide (DMF), and ethyl ether were sourced from EMD Millipore. Ammonium persulfate was acquired from BDH Chemicals. Dichloromethane (DCM) and N-(2-amino-ethyl)-maleimide hydrochloride were obtained from Macron Fine Chemicals and TCI, respectively. Silicon wafers were acquired from University Wafer. Milli-Q water (18.2 MΩ cm) was used for all analysis and synthesis methods.Synthesis
The synthesis of the BEAM was based on previous methods.17 The bifunctional molecule was comprised of a maleimide-modified fatty acid spacer that was sandwiched between two peptides: P1 and BP7. The fatty acid spacer, termed F10F, is a 10-carbon fatty acid synthesized with a maleimide on each end using previous methods.17 The peptides, P1 and BP7 were attached to each maleimide sequentially through thiol/maleimide coupling. These peptides were purchased containing a cysteine on either the C- or N-terminus of the peptide sequence labelled as P1C and CBP7. For the synthesis of the BEAM, the peptide with the cysteine on the N-terminus, CBP7, was first coupled to the fatty acid in a 5.21![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1 molar ratio (F10F
1 molar ratio (F10F![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) CBP7). This was done by forming two solutions, one with 39.7 mg of F10F in 6 mL of DMF and another containing 25 mg of CBP7 in 6 mL of DMF. The two solutions were combined together dropwise and allowed to stir for 3 days at room temperature. After completion, the sample underwent a diethyl ether wash to remove any unreacted F10F. The new conjugate (labelled F10FCBP7) was then dissolved in water, lyophilized, purified with reverse phase HPLC, and then confirmed with MADLI-TOF mass spectrometry. The next step was to attach the other peptide with a cysteine on the C-terminus, P1C, to the other end of the modified fatty acid. For this, one solution containing 13.4 mg of F10FCBP7 in 3 mL of water was made. A separate solution containing 11.4 mg of the P1C in 3 mL of water was also prepared. The two solutions were combined together in a dropwise manner resulting in a 1
CBP7). This was done by forming two solutions, one with 39.7 mg of F10F in 6 mL of DMF and another containing 25 mg of CBP7 in 6 mL of DMF. The two solutions were combined together dropwise and allowed to stir for 3 days at room temperature. After completion, the sample underwent a diethyl ether wash to remove any unreacted F10F. The new conjugate (labelled F10FCBP7) was then dissolved in water, lyophilized, purified with reverse phase HPLC, and then confirmed with MADLI-TOF mass spectrometry. The next step was to attach the other peptide with a cysteine on the C-terminus, P1C, to the other end of the modified fatty acid. For this, one solution containing 13.4 mg of F10FCBP7 in 3 mL of water was made. A separate solution containing 11.4 mg of the P1C in 3 mL of water was also prepared. The two solutions were combined together in a dropwise manner resulting in a 1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1 molar ratio. After the solution was stirred for 3 days at room temperature, the bioconjugate was collected, purified via reverse phase HPLC, and confirmed by MALDI-TOF mass spectrometry.
1 molar ratio. After the solution was stirred for 3 days at room temperature, the bioconjugate was collected, purified via reverse phase HPLC, and confirmed by MALDI-TOF mass spectrometry.
Graphene exfoliation
A probe sonicator (QSonica 500) was used to drive graphene exfoliation. The sample was prepared by dispersing 50 mg of graphite and 1.0 mg of the BEAM in 10 mL of water. A 0.25 in titanium probe was used for the exfoliation experiments, which was immersed halfway into the solution sample. The sample was sonicated for 1 h at a pulse rate of 3s/3s (on/off) and with an amplitude of 100%. The sample was kept in an ice bath during sonication to prevent the system from overheating. The samples were then subjected to centrifugation for 1 h at 3000 rpm in which the exfoliated graphene sheets were found in the supernatant and removed for analysis.Analysis and characterization
The absorbance of the obtained exfoliated sheets was measured by UV-vis spectroscopy with an Agilent 8453 spectrometer and a quartz cuvette with a 1.0 cm pathlength. Each sample was blanked against water prior to analysis. Fluorescence measurements were obtained using a PerkinElmer LS55 fluorescence spectrometer. The analysis was completed using a disposable plastic cuvette with a 1 cm pathlength. AFM images of the graphene sheets were obtained on a Veeco AFM (Dimension 3100) in tapping mode. For this analysis, the samples were prepped by drop-casting exfoliated sheets (80–100 μL) onto a silicon wafer and left to sit for 3 h. After this time, the wafers were dried with air and stored in a desiccator overnight. TEM images were measured by using an FEI-Cs-corrected Titan TEM microscope and were obtained at 300 kV. The samples for TEM were prepared by depositing a drop of the exfoliated graphene solution (10 μL) onto a lacey carbon-coated 200 mesh copper TEM grid. The samples were allowed to dry overnight prior to measuring. Raman measurements were performed by drop-casting a solution of exfoliated graphene onto a gold-coated glass slide and measured using a Renishaw inVia Raman microscope operating at a power of 1 μW and an excitation at 514.5 nm.Molecular dynamics simulations
All simulations were performed using the Gromacs software package (version 2021.4).24 For the pre-inserted and spontaneous insertion simulations, the simulation system comprised a vertical stack of nine hexagonal graphene sheets with a diameter ∼7.5 nm, six BEAM chains, and a sufficient number of water molecules in the liquid state. The flake at the bottom of the stack was frozen in all three dimensions mimicking the surface rigidity of graphite at sub-nm depth. For the pre-insertion runs, the BEAMs were placed into the expanded gap either P1-in-first or BP7-in-first. For the spontaneous insertion simulations, four of the six BEAM molecules were placed outside the expanded gap but nearby to the stack, parallel to the edge of graphene sheets, whereas the other two chains were placed at the top of upper-most graphene sheet. Ten independent replica simulations were completed for each of the three types of runs described above, each for a maximum duration of 150 ns, post-equilibration.For the BEAM-decorated sheet free energy simulations, the system comprised two identical BEAM-decorated graphene sheets (12 BEAMs per graphene sheet) that were initially arranged face-to-face along the z-axis (perpendicular to the graphene plane), where the distance between two central rings of the graphene sheets was 5.5 nm. The system was solvated with sufficient water molecules. A total of 105 configurations were used in the umbrella sampling simulations at 300 K in the NVT ensemble, for which the two sheets were drawn closer together and the reaction coordinate for the free energy profile was defined by the vertical inter-sheet distance. Each umbrella sampling window was simulated for 100 ns, and the accumulated length of the umbrella sampling simulation was 10.5 μs in total. Full methodological details of all simulations are provided in the ESI.†
Conflicts of interest
There are no conflicts to declare.Acknowledgements
This material is based upon work supported by the Air Force Office of Scientific Research, Grant FA9550-18-1-0329. TRW acknowledges computational resources provided by the National Computing Infrastructure (NCI), Canberra, and the Pawsey Supercomputing Centre, Perth, Australia, under the NCMAS scheme. Part of this work was also undertaken using the LIEF HPC-GPGPU Facility established with the assistance of LIEF Grant LE170100200, hosted at the University of Melbourne.Notes and references
- M. Yusuf, M. Kumar, M. A. Khan and M. Sillanpää, Adv. Colloid Interface Sci., 2019, 273, 102036 CrossRef CAS PubMed.
- V. Georgakilas, J. N. Tiwari, C. Kemp, J. A. Perman, A. B. Bourlinos, K. S. Kim and R. Zboril, Chem. Rev., 2016, 116, 5464–5519 CrossRef CAS PubMed.
- X. Xin, J. Chen, L. Ma, T. Ma, W. Xin, H. Xu, W. Ren and Y. Liu, Small Methods, 2023, 7, 2300156 CrossRef CAS PubMed.
- A. Amiri, M. Naraghi, G. Ahmadi, M. Soleymaniha and M. Shanbedi, FlatChem, 2018, 8, 40–71 CrossRef CAS.
- Y. Wei and Z. Sun, Curr. Opin. Colloid Interface Sci., 2015, 20, 311–321 CrossRef CAS.
- T. R. Walsh and M. R. Knecht, Bioconjugate Chem., 2019, 30, 2727–2750 CrossRef CAS PubMed.
- S. N. Kim, Z. Kuang, J. M. Slocik, S. E. Jones, Y. Cui, B. L. Farmer, M. C. McAlpine and R. R. Naik, J. Am. Chem. Soc., 2011, 133, 14480–14483 CrossRef CAS PubMed.
- C. R. So, Y. Hayamizu, H. Yazici, C. Gresswell, D. Khatayevich, C. Tamerler and M. Sarikaya, ACS Nano, 2012, 6, 1648–1656 CrossRef CAS PubMed.
- A. D. Parab, A. Budi, J. M. Slocik, R. Rao, R. R. Naik, T. R. Walsh and M. R. Knecht, J. Phys. Chem. C, 2020, 124, 2219–2228 CrossRef CAS.
- A. D. Parab, R. Dureja, R. Rao, J. M. Slocik, R. R. Naik, T. R. Walsh and M. R. Knecht, Langmuir, 2021, 37, 1152–1163 CrossRef CAS PubMed.
- L. Zhang, Y. Sheng, A. Z. Yazdi, K. Sarikhani, F. Wang, Y. Jiang, J. Liu, T. Zheng, W. Wang, P. Ouyang and P. Chen, Nanoscale, 2019, 11, 2999–3012 RSC.
- M. Cao, N. Wang, L. Wang, Y. Zhang, Y. Chen, Z. Xie, Z. Li, E. Pambou, R. Li, C. Chen, F. Pan, H. Xu, J. Penny, J. R. P. Webster and J. R. Lu, J. Mater. Chem. B, 2016, 4, 152–161 RSC.
- Y. Cui, S. N. Kim, S. E. Jones, L. L. Wissler, R. R. Naik and M. C. McAlpine, Nano Lett., 2010, 10, 4559–4565 CrossRef CAS PubMed.
- A. D. Parab, A. Budi, N. Brljak, M. R. Knecht and T. R. Walsh, Adv. Mater. Interfaces, 2021, 8, 2001659 CrossRef CAS.
- Y. Perdomo, R. Jin, A. D. Parab, M. R. Knecht and T. R. Walsh, J. Mater. Chem. B, 2022, 10, 6018–6025 RSC.
- H. Zhang, T. Yamazaki, C. Zhi and N. Hanagata, Nanoscale, 2012, 4, 6343–6350 RSC.
- R. Jin, N. Brljak, R. Sangrigoli, T. R. Walsh and M. R. Knecht, Nanoscale, 2022, 14, 14113–14121 RSC.
- N. Brljak, A. D. Parab, R. Rao, J. M. Slocik, R. R. Naik, M. R. Knecht and T. R. Walsh, Chem. Commun., 2020, 56, 8834–8837 RSC.
- R. Jin, F. Vuković and T. R. Walsh, J. Phys. Chem. Lett., 2021, 12, 11945–11950 CrossRef CAS PubMed.
- S. Zhao, S. Xie, Z. Zhao, J. Zhang, L. Li and Z. Xin, ACS Sustainable Chem. Eng., 2018, 6, 7652–7661 CrossRef CAS.
- A. C. Ferrari and D. M. Basko, Nat. Nanotechnol., 2013, 8, 235–246 CrossRef CAS PubMed.
- D. Graf, F. Molitor, K. Ensslin, C. Stampfer, A. Jungen, C. Hierold and L. Wirtz, Nano Lett., 2007, 7, 238–242 CrossRef CAS PubMed.
- R. Podila, R. Rao, R. Tsuchikawa, M. Ishigami and A. M. Rao, ACS Nano, 2012, 6, 5784–5790 CrossRef CAS PubMed.
- D. V. D. Spoel, E. Lindahl, B. Hess, G. Grownhof, A. E. Mark and H. J. C. Berendsen, J. Comput. Chem., 2005, 26, 1701–1718 CrossRef PubMed.
Footnote |
| † Electronic supplementary information (ESI) available: Computational methodological details. See DOI: https://doi.org/10.1039/d3tb02109b |
| This journal is © The Royal Society of Chemistry 2024 |