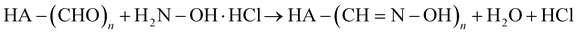Open Access Article
Open Access ArticleStatistical optimization of hydrazone-crosslinked hyaluronic acid hydrogels for protein delivery†
Esther A.
Mozipo‡
ab,
Alycia N.
Galindo‡
 a,
Jenna D.
Khachatourian
ac,
Conor G.
Harris
a,
Jenna D.
Khachatourian
ac,
Conor G.
Harris
 d,
Jonathan
Dorogin
a,
Veronica R.
Spaulding
d,
Jonathan
Dorogin
a,
Veronica R.
Spaulding
 b,
Madeleine R.
Ford
b,
Madeleine R.
Ford
 ac,
Malvika
Singhal
abe,
Kaitlin C.
Fogg
ac,
Malvika
Singhal
abe,
Kaitlin C.
Fogg
 d and
Marian H.
Hettiaratchi
d and
Marian H.
Hettiaratchi
 *abe
*abe
aPhil and Penny Knight Campus for Accelerating Scientific Impact, University of Oregon, Eugene, OR, USA
bDepartment of Chemistry and Biochemistry, University of Oregon, Eugene, OR, USA
cDepartment of Human Physiology, University of Oregon, Eugene, OR, USA
dSchool of Chemical, Biological, and Environmental Engineering, Oregon State University, Corvallis, OR, USA
eInstitute of Molecular Biology, University of Oregon, Eugene, OR, USA. E-mail: mhettiar@uoregon.edu
First published on 5th February 2024
Abstract
Hydrazone-crosslinked hydrogels are attractive protein delivery vehicles for regenerative medicine. However, each regenerative medicine application requires unique hydrogel properties to achieve an ideal outcome. The properties of a hydrogel can be impacted by numerous factors involved in its fabrication. We used design of experiments (DoE) statistical modeling to efficiently optimize the physicochemical properties of a hyaluronic acid (HA) hydrazone-crosslinked hydrogel for protein delivery for bone regeneration. We modified HA with either adipic acid dihydrazide (HA-ADH) or aldehyde (HA-Ox) functional groups and used DoE to evaluate the interactions of three input variables, the molecular weight of HA (40 or 100 kDa), the concentration of HA-ADH (1–3% w/v), and the concentration of HA-Ox (1–3% w/v), on three output responses, gelation time, compressive modulus, and hydrogel stability over time. We identified 100 kDa HA-ADH3.00HA-Ox2.33 as an optimal hydrogel that met all of our design criteria, including displaying a gelation time of 3.7 minutes, compressive modulus of 62.1 Pa, and minimal mass change over 28 days. For protein delivery, we conjugated affinity proteins called affibodies that were specific to the osteogenic protein bone morphogenetic protein-2 (BMP-2) to HA hydrogels and demonstrated that our platform could control the release of BMP-2 over 28 days. Ultimately, our approach demonstrates the utility of DoE for optimizing hydrazone-crosslinked HA hydrogels for protein delivery.
Introduction
Recombinant protein delivery is a promising strategy for the repair of injured or damaged tissues including bone, muscle, nerves, vasculature, and skin.1 Specifically in the case of bone repair, the delivery of bone morphogenetic protein-2 (BMP-2) has demonstrated success as a bone regeneration strategy in both animal models and human patients.2 A wide variety of biomaterials have been investigated for protein delivery to the body, and their properties have been tuned to enable control over protein release rate, material degradation rate, material compressive modulus, and injectability. However, tuning all of these properties simultaneously to develop the ideal biomaterial for protein delivery is challenging. Furthermore, biomaterial delivery vehicles must be individually optimized for different clinical applications, as unique properties may be necessary to control the release of different proteins to different tissues. Hydrogels are chemically or physically crosslinked water-swollen polymer networks that can be effective vehicles for the delivery of a variety of cells, proteins, and drugs due to their ability to support cell viability, provide sustained release of cargo, and mimic the mechanical and biochemical properties of tissues.1,3–5 The relative ease in which a variety of natural and synthetic polymers can be chemically modified with different functional groups has facilitated the fabrication of diverse hydrogels with a wide array of properties suitable for numerous applications. Varying individual input parameters to create a hydrogel, such as polymer concentration, crosslinking ratio, and molecular weight of the polymer, one at a time to determine the combination of parameters that yields a hydrogel with the optimal properties can be time-intensive and expensive. While extensive prior work has been performed to determine the ideal physicochemical properties of hydrogels for a variety of biomedical applications, there has been limited investigation of the collective, combinatorial effects of varying multiple input parameters on the properties of hydrogels in the context of protein delivery.To tackle this challenge, we used a statistical optimization technique known as design of experiments (DoE) to efficiently screen the effects of multiple input parameters on several hydrogel properties and create a mathematical model to predict the optimal and pessimal hydrogel formulations given a specific set of design criteria. Design of experiments (DoE) is a method of experimental design in which a combination of factors are varied simultaneously instead of the typical “one factor at a time” approach. The responses to each set of input parameters is mapped to generate a model for how the different input parameters collectively influence each hydrogel property (i.e., the output responses), known as the response surface methodology.6,7 Mathematical models can then be used to define the response surface and predict expected outcomes. DoE was originally developed in the 1920s for agricultural purposes, but has since been used in a broad array of applications, including the development and optimization of biomaterials.7–10 It lends itself particularly well to applications such as hydrogel design, in which it is highly likely that multiple input parameters will have interactive effects on hydrogel properties, which makes individually varying input parameters less informative. We and others have used DoE to predict responses of hydrogels and cells within hydrogels in systems with multiple input variables.8,11–14 This approach can be widely used in regenerative medicine applications such as scaffold design, cell culture optimization, and the development of in vitro model systems, accelerating the timeframe in which new platforms can be optimized.15–17
In this study, we were specifically interested in tuning the properties of hyaluronic acid hydrogels formed via hydrazone crosslinking. Hyaluronic acid (HA) is an abundant biopolymer found throughout the body. It is a critical extracellular matrix component of connective tissue, skin, and the brain that functions by creating a hydrated polymer matrix for cells to migrate and proliferate.18–20 HA also exists at a range of molecular weights, which can activate different cell signaling pathways.21,22 Although HA itself is soluble in aqueous media, modifications can be made to the carboxylic acid, adjacent alcohol, and secondary alcohol groups on the HA backbone to form crosslinks throughout the polymer, resulting in a stable three-dimensional, water-swollen network. HA hydrogels have been used successfully in several biomedical applications, including protein and cell delivery and tissue regeneration.23–25
While there are numerous criteria that may be considered important for the design of a hydrogel for protein delivery, we narrowed down our desired hydrogel properties to three essential criteria for optimization: compressive modulus, gelation time, and mass change over four weeks. Compressive modulus can have a profound effect on cell differentiation,26,27 while the bulk stiffness of an implanted material must be matched to the target tissue to avoid eliciting mechanical irritation and subsequent inflammation. Hydrogel gelation time should allow for sufficient time to mix components (different polymers, proteins, cells) before gelation. Typically, hydrogel gelation times of less than 10 minutes are preferred.28,29 Finally, the mass change of the hydrogel due to swelling or degradation of the crosslinked network can impact cellular responses over time, such cell spreading, cell-cycle progression, and differentiation.30–32
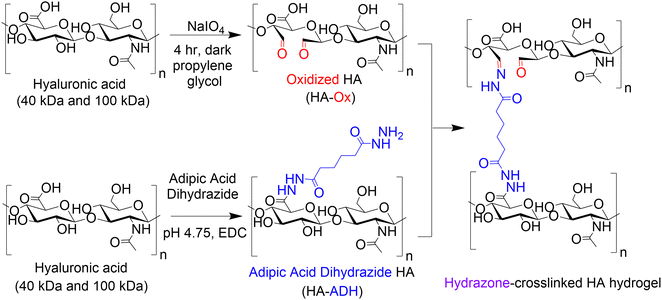
We formed HA hydrogels using dynamic covalent hydrazone crosslinks as this approach can yield a wide range of physicochemical properties, making them an attractive hydrogel platform for numerous regenerative medicine applications.33–40 The properties of hydrazone-crosslinked HA hydrogels can be easily tuned by adjusting the ratios of the two dynamic covalent crosslinking polymers: oxidized HA (HA-Ox) and HA adipic acid dihydrazide (HA-ADH). Thus, we narrowed our input parameters to HA molecular weight, HA-Ox concentration, and HA-ADH concentration. We then used DoE to identify the contributions and combinatorial effects of the concentration of HA-ADH, the concentration of HA-Ox, and the molecular weight of HA on hydrogel compressive modulus, gelation time, and mass change over time. We then created a statistical model that predicted physicochemical properties as a function of input parameters. This model was validated by evaluating the hydrogels predicted to be the optimal and pessimal delivery vehicles. Finally, as a proof-of-concept for protein delivery, we conjugated a protein-binding peptide known as an affibody to the HA hydrogels using thiol–ene click chemistry to enable affinity-controlled delivery of bone morphogenetic protein-2 (BMP-2), which is a potent osteogenic protein that is clinically used as a bone graft substitute. Overall, we developed a hydrogel platform and used DoE analysis to optimize the vast tunability of hydrazone-crosslinked HA hydrogels for protein delivery for tissue regeneration.
Materials and methods
Materials
Sodium hyaluronate (HA, 40 kDa and 100 kDa) was purchased from Lifecore Biomedical LLC (Chaska, MN). Adipic acid dihydrazide (ADH) was purchased from Spectrum Chemical (Gardena, CA). Hydroxybenzotriazole (HOBt) was purchased from Chem Impex (Wood Dale, IL). 1-Ethyl-3-(3-dimethylaminopropyl)carbodiimide (EDC) was purchased from G Biosciences (St. Louis, MO). Sodium Periodate, AmberLite® MB Ion Exchange Resin, tetrabutylammonium hydroxide (TBA-OH), DMSO, 4-dimethylaminopyridine (DMAP), di-tert-butyl decarbonate (BoC2O) and 5-norbornene-2-carboxylic acid were purchased from Sigma Aldrich (St. Louis, MO). Irgacure 2959 was purchased from Advanced Biomatrix (Carlsbad, CA). Recombinant human bone morphogenetic protein-2 (BMP-2) and the human BMP-2 DuoSet enzyme-linked immunosorbent assay (ELISA) were purchased from R&D Systems (Minneapolis, MN).Synthesis of adipic acid dihydrazide HA (HA-ADH)
HA (200 mg, 0.527 mmol) was dissolved in 20 mL of diH2O to form a 1% w/v solution. Adipic acid dihydrazide (183.75 mg, 1.05 mmol) and hydroxy-benzotriazole (142.53 mg, 1.05 mmol) were added to the HA solution and the pH was adjusted to 4.75. EDC was added (91.00 mg, 0.47 mmol) to the solution, and the pH was monitored and maintained at 4.75 for 4 hours using 1 M HCl and 1 M NaOH. The solution was stirred for 24 hours at room temperature and dialyzed against 0.1 M NaCl in diH2O for 2 days followed by diH2O for 2 days. The solution of HA functionalized with ADH (HA-ADH) was sterile filtered through a 0.22 μm filter, frozen at −80 °C, and lyophilized using a VirTis BenchTop Pro lyophilizer (SP Scientific, Warminster, PA).Synthesis of oxidized HA (HA-Ox)
HA (200 mg, 0.561 mmol) was dissolved in 10 mL of diH2O to form a 2% w/v solution. Sodium periodate (322.00 mg, 0.281 mmol) was added to the solution and stirred for 4 hours at room temperature protected from light. The reaction was quenched with 1 mL of propylene glycol and dialyzed against diH2O for 3 days. The oxidized HA solution (HA-Ox) was sterile filtered through a 0.22 μm filter, frozen at −80 °C, and lyophilized.Synthesis of HA norbornene (HA-Nor)
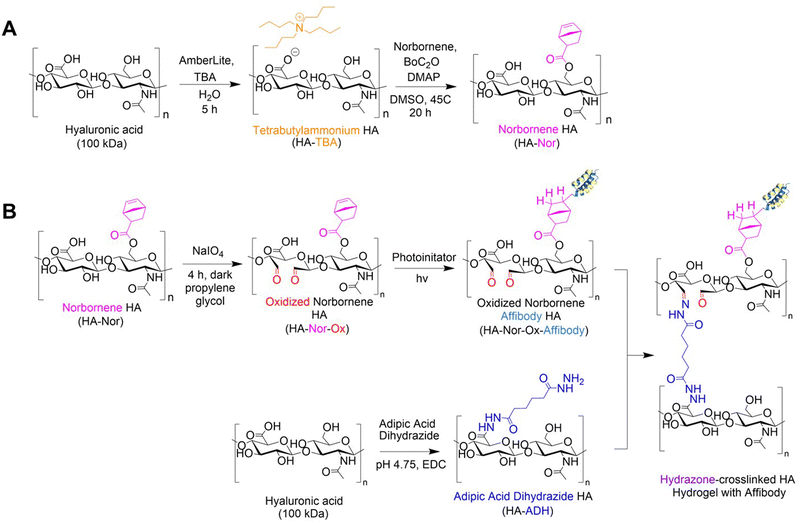
HA (1.01 g, 2.506 mmol) was dissolved in 50 mL diH2O to form a 2% w/v solution. AmberLite™ MB Ion Exchange Resin (3.03 g) was added to the reaction and stirred at room temperature for 5 hours. The resin was vacuum-filtered, and the filtrate was titrated to pH of 7 with TBA-OH and dialyzed against diH2O for 3 days. The HA-TBA solution was sterile filtered through a 0.22 μm filter, frozen at −80 °C, and lyophilized.HA-TBA (153 mg, 0.35 mmol) was dissolved in 0.75 mL DMSO to form a 2% w/v solution. The flask was purged with N2 for 5 minutes, then 5-norbornene-2-carboxylic acid (145 mg, 1.05 mmol) and DMAP (145 mg, 1.049 mmol) was added to the reaction flask. Boc2O was added via syringe (32 μL, 0.14 mmol). The solution was stirred at 45 °C for 20 hours, then quenched with cold diH2O (10 mL) before being dialyzed against diH2O for 3 days. Norbornene-functionalized HA (HA-Nor) was precipitated from the solution by adding NaCl and cold acetone (30 mL), then filtered and dialyzed against diH2O for 3 days. The solution was sterile filtered through a 0.22 μm filter, frozen at −80 °C, and lyophilized.
Oxidization of HA norbornene (HA-Nor-Ox)
HA-Nor (91.75 mg, 0.18 mmol) was dissolved in diH2O to form a 1% w/v solution. Sodium periodate (10.57 mg, 0.049 mmol) was dissolved in diH2O to create a 0.5 M solution and added to the HA-Nor solution. The solution was stirred overnight at room temperature protected from light. The reaction was quenched with 1 mL of propylene glycol and dialyzed against diH2O for 3 days. The oxidized HA-Nor solution (HA-Nor-Ox) was sterile filtered through a 0.22 μm filter, frozen at −80 °C, and lyophilized.Molecular weight analysis of modified HA
Gel permeation chromatography (GPC) was used to determine the molecular weight of unmodified and modified HA at both 40 kDa and 100 kDa. Lyophilized HA samples were sent to Polyanalytik, Inc. (London, ON, Canada) for analysis using a Viscotek TDA 305 and GPCmax (Malvern Panalytical Ltd, Malvern, United Kingdom) fitted with PAA-202 and PAA-206M aqueous columns. Samples were diluted to 1.00–2.50 mg mL−1 in a mobile phase of 8.5 g L−1 NaCl in ultrapure water, allowed to dissolve at room temperature for several hours, and then filtered through a 0.22 μm Nylon syringe filter prior to injection. Samples were run in triplicate at a flow rate of 0.7 mL min−1 and temperature of 30 °C.Affibody bioconjugation
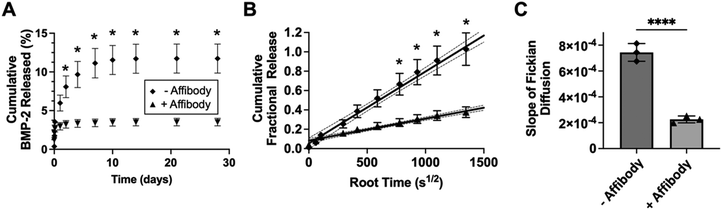
BMP-2-specific affibodies were expressed in E. coli and purified as previously described.41 HA-Nor-Ox (14.5 mg) was dissolved in 1.5 mL of 0.71 mg mL−1 affibody in PBS (1.45 × 10−4 mmol) to create a solution with 1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1 ratio of affibody to HA-Nor-Ox. 15 μL of 10% w/v Irgacure 2595 in methanol was added to the reaction vial, stirred, and illuminated with 365 nm light for 10 minutes. The solution was dialyzed against HEPES buffer pH 7.0 for 1 day and diH2O for 2 days. The affibody-conjugated solution of HA (HA-Nor-Ox-Affibody) was sterile filtered through a 0.22 μm filter, frozen at −80 °C, and lyophilized. The amount of affibody conjugated to the polymer was quantified by dissolving 2–3 mg of HA-Nor-Ox-Affibody in PBS at 2% w/v and determining the protein concentration of the solution using a Pierce 660 Protein Assay Kit (Thermo Fisher Scientific).
1 ratio of affibody to HA-Nor-Ox. 15 μL of 10% w/v Irgacure 2595 in methanol was added to the reaction vial, stirred, and illuminated with 365 nm light for 10 minutes. The solution was dialyzed against HEPES buffer pH 7.0 for 1 day and diH2O for 2 days. The affibody-conjugated solution of HA (HA-Nor-Ox-Affibody) was sterile filtered through a 0.22 μm filter, frozen at −80 °C, and lyophilized. The amount of affibody conjugated to the polymer was quantified by dissolving 2–3 mg of HA-Nor-Ox-Affibody in PBS at 2% w/v and determining the protein concentration of the solution using a Pierce 660 Protein Assay Kit (Thermo Fisher Scientific).
Degree of modification (DOM) determination
The degree of chemical modification of HA-ADH and HA-Nor was quantified using proton nuclear magnetic resonance spectroscopy (1H NMR, 500 Hz, Bruker, USA). 2–5 mg of modified polymer was dissolved in 600 μL of deuterium oxide (D2O) for 1H NMR (256 scans). To calculate the degree of modification for HA-ADH, the peaks from the aliphatic chain were integrated from 2.5–2.1 ppm and 1.7–1.44 ppm (8H) and normalized to the n-acetyl methyl group peaks from 1.8–2 ppm (3H) (Fig. S1, ESI†).42 The degree of modification for HA-Nor was calculated by integrating the vinyl peaks from 5.7–6.3 ppm and normalizing to the n-acetyl methyl group from 1.8–2 ppm (3H) (Fig. S2, ESI†).42The degree of oxidation for HA-Ox was determined by hydroxylamine hydrochloride titration.43,44 Briefly, 100 mg of oxidized polymer was dissolved in 0.25 N hydroxylamine hydrochloride containing 0.05% w/v methyl orange reagent for 2 hours. The solution was then titrated with 0.1 M NaOH while monitoring the pH until the pH indicator changed from red to yellow at a pH of 4. The reaction of hydroxylamine hydrochloride with the aldehydes on oxidized HA is shown below.
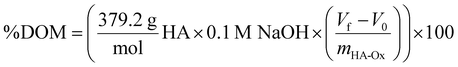 | (1) |
Preparation of HA hydrogels
HA-ADH and oxidized HA (HA-Ox, HA-Nor-Ox, or HA-Nor-Ox-Affibody) were each reconstituted at 1–3% w/v in PBS. Hydrogels were then prepared by mixing 50 μL of each polymer in 2 mL microcentrifuge tube or 8 mm diameter cylindrical silicone mold to create 100 μL hydrogels.Physicochemical characterization of hydrogels
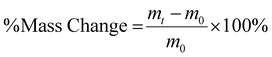 | (2) |
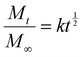 | (3) |
| HA-ADH (% w/v) | HA-Ox (% w/v) | Molecular weight (kDa) |
|---|---|---|
| 1.00 | 1.00 | 40 |
| 1.00 | 3.00 | 40 |
| 1.67 | 1.00 | 40 |
| 2.00 | 2.00 | 40 |
| 3.00 | 1.00 | 40 |
| 3.00 | 3.00 | 40 |
| 1.00 | 1.00 | 100 |
| 1.00 | 3.00 | 100 |
| 1.00 | 1.67 | 100 |
| 1.67 | 3.00 | 100 |
| 2.00 | 2.00 | 100 |
| 2.00 | 2.00 | 100 |
| 2.00 | 2.00 | 100 |
| 2.33 | 1.00 | 100 |
| 3.00 | 1.00 | 100 |
| 3.00 | 3.00 | 100 |
| 3.00 | 2.33 | 100 |
MODDE-Pro 13.0 software (Sartorius, Fremont, CA) was used to generate the D-optimal experimental matrix of 15 hydrogel formulations and analyze the experimental data obtained. Experimental data were collected from each of the hydrogel formulations then used to fit a multiple linear regression model for each response variable as a function of the statistically significant input variables. An input variable was considered statistically significant if it had a p value of < 0.05 as determined by ANOVA. The model performance was further assessed using R2, adjusted R2, and Q2. R2 describes the variation of the response explained by the model for every variable. Adjusted R2 describes the variation of the response explained by the model only for the variables that affect the response. Q2 describes the variation of the response predicted by the model for new data. R2 and adjusted R2 values close to 1.0 indicate a high correlation between observed and predicted values. Q2 above 0.50 and a difference between R2 and Q2 values below 0.30 validate that the model works independently of the specific data used to train the model.51,52 Residual plots, Box–Cox plots, and ANOVA tables were used to diagnose outliers, trends, or lack of fit. The model was then refined by removing any outliers and input variables that were not statistically significant to increase the validity of our model.
Our overall engineering objective was to maximize the compressive modulus, set the gelation time to a value within 1–10 min, and set the mass change at all time points to 0%. Thus, to validate the predictive models, we generated hydrogels predicted to either fulfill these optimization parameters (optimal hydrogels) or fulfill reciprocal objectives (pessimal hydrogels). Specifically, the pessimal hydrogels were predicted to minimize the compressive modulus, maximize the gelation time, and either maximize or minimize the hydrogel mass change at all time points (i.e., induce hydrogel swelling or degradation).
Results and discussion
Functionalization of HA polymers
HA was modified with either aldehyde (HA-Ox) or ADH functional groups (HA-ADH) for hydrazone crosslinking (Fig. 1). Because polymer molecular weight can influence crosslinking density and subsequent physicochemical properties of the hydrogel,28 we functionalized both 40 kDa and 100 kDa HA for hydrogel fabrication and evaluation. To expose aldehyde groups on HA, we oxidized the polymer in the presence of sodium periodate (NaIO4) for 4 h in the dark before quenching the reaction with propylene glycol. Using the same starting material, we added ADH to HA in the presence of EDC at a pH of 4.75 for 24 h to create HA-ADH. The degree of oxidization was characterized using titration with hydroxylamine hydrochloride and was found to be 27.5 ± 5.9% and 41.4 ± 12.8% of HA repeating disaccharide units for 40 kDa and 100 kDa HA-Ox, respectively. The degree of modification of the ADH functional groups on the polymer was 67.6 ± 1.2% for 40 kDa HA-ADH and 50.6 ± 10.1% for 100 kDa HA-ADH as characterized by 1H NMR (Fig. S1, ESI†). The degrees of modification and yields of all modified polymers are summarized in Table S1 (ESI†). Degrees of oxidation between 27–82%33,53 and degrees of modification with ADH between 30–49%54,55 have been previously reported, which is consistent with our results. Gel permeation chromatography was used to determine how the modifications to the HA polymer changed its molecular weight and polydispersity (Table S2, ESI†). Our results revealed a decrease in molecular weight during the oxidation reaction due to the ring break on HA; however, the size of the polymer backbone was not greatly affected by hydrolysis as observed in other HA oxidation reactions.56 Furthermore, the polydisperity indices of the two oxidized HA polymers were similar to unmodified HA. Interestingly, the average molecular weights of both the 40 kDa and 100 kDa HA-ADH were much larger than expected at 366 ± 16.8 kDa and 332 ± 9.58 kDa, respectively, suggesting that ADH reacted with multiple HA polymer chains during synthesis, resulting in larger polymer chains. Similar results have been documented in a previous HA-ADH synthesis in the presence of EDC and a low molar excess of ADH due to the bifunctionality of ADH.57 This finding was further supported by the higher polydispersity indices for both 40 kDa and 100 kDa HA-ADH at 2.02 ± 0.03 and 2.18 ± 0.10, respectively, compared to unmodified HA, which indicate a larger distribution of polymer chain molecular weights.We then used these modified HA polymers to fabricate the 15 hydrogels in Table 1 and evaluated their gelation time, compressive moduli, and mass change over time to generate a predictive model for DoE analysis.
Hydrogel gelation time
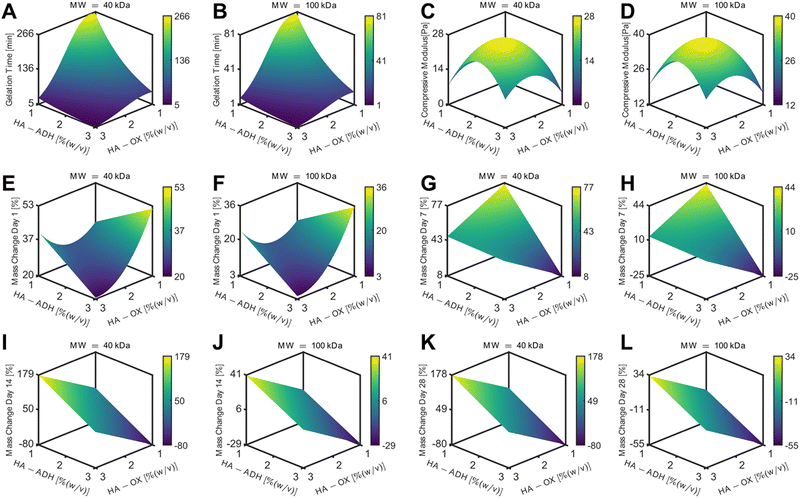
HA hydrogels were fabricated by mixing 50 μL of HA-ADH and 50 μL of HA-Ox at different polymer concentrations and molecular weights. Inversion tests were used to quantify the gelation time of each of the HA hydrogel formulations. As expected, increasing the polymer content of the hydrogels increased the crosslinking density of the polymer network and subsequently decreased the gelation time (Fig. 2A). Changing the polymer concentrations of the 40 kDa hydrogels had a greater effect on gelation time compared to the 100 kDa hydrogels, such that the 40 kDa hydrogels had a larger range of average gelation times ranging from 4.5 to 292.7 min. Additionally, increasing the HA-ADH content in the 40 kDa hydrogels decreased the gelation time of the hydrogels, except for the HA-ADH1.00HA-Ox3.00 hydrogel. The gelation times of the 100 kDa hydrogels all fell between 1.4 and 71.5 min. The 100 kDa HA hydrogels similarly exhibited decreasing gelation time in response to increasing HA-ADH content, except for the HA-ADH1.67HA-Ox3.00 and HA-ADH1.00HA-Ox3.00 hydrogels, which demonstrated faster gelation times than the HA-ADH2.00HA-Ox2.00 hydrogel. Excess HA-Ox content compared to HA-ADH content also resulted in shorter gelation times in both the 40 kDa and 100 kDa hydrogels. Overall, the 100 kDa hydrogels gelled faster than 40 kDa hydrogels, potentially due to increased entanglement of the longer polymer chains.58 Gelation time of the centrally repeated hydrogel condition (100 kDa HA-ADH2.00HA-Ox2.00) was evaluated using 50 μL, 100 μL, and 200 μL hydrogels to determine if the hydrogel size had an effect on the gelation time (Fig. S3, ESI†). As the hydrogel volume increased, gelation time also increased. This was likely due to the increased time required to establish more crosslinks to effect complete gelation of a larger hydrogel.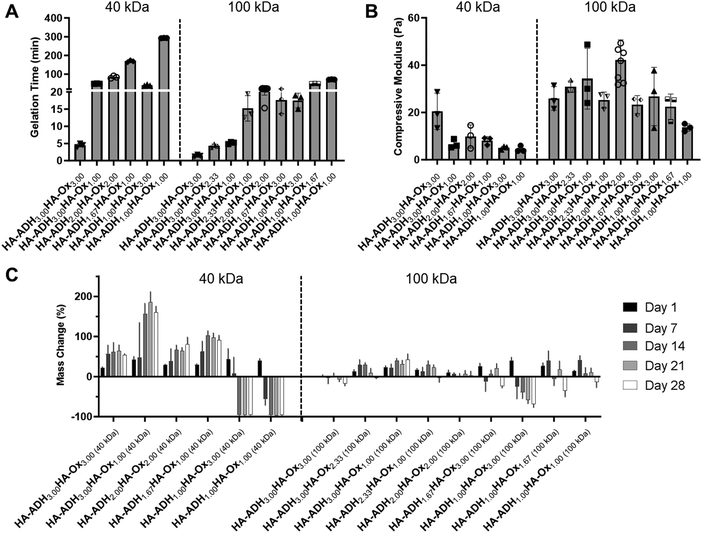 | ||
| Fig. 2 Physicochemical properties for each of the 15 hydrogel formulations for the D-optimal design. HA hydrogels were fabricated using 40 kDa or 100 kDa HA-Ox and HA-ADH at different polymer weight percentages. (A) Gelation time of HA hydrogels. (B) Compressive moduli of HA hydrogels. (C) Mass change of HA hydrogels incubated in phosphate-buffered saline at room temperature for 28 days. Data presented as mean ± SD with 3 replicates for all hydrogel formulations, except for the centrally repeated condition, HA-ADH2.00HA-Ox2.00 (100 kDa), which had 9 replicates. A summary of statistical significance for gelation time, compressive modulus, and mass change can be found in Tables S3–S5 (ESI†), respectively. | ||
Hydrogel compressive modulus
The compressive moduli of the hydrogels ranged from 4.5 to 42.2 Pa (Fig. 2B). The compressive moduli of all the 100 kDa HA hydrogels fell within a narrow range of 20–40 Pa except for the HA-ADH1.00HA-Ox1.00 hydrogel, which exhibited an average compressive modulus of 13.6 Pa, which was significantly lower than HA-ADH3.00HA-Ox2.33 and HA-ADH2.00HA-Ox2.00 hydrogels and likely due to the lower polymer concentration. In contrast, the compressive moduli of all of the 40 kDa HA hydrogels fell within 4.5 to 20 Pa. Increasing the overall polymer content and HA-ADH content of the 40 kDa HA hydrogels resulted in higher compressive moduli, which was similar to the trends of increasing polymer concentration and HA-ADH content decreasing gelation time.Hydrogel mass change over time
The hydrogels were incubated in PBS at room temperature for 28 days, and their masses were periodically measured to observe swelling and/or degradation over time. For the 40 kDa HA hydrogels, the HA-ADH3.00HA-Ox1.00 hydrogel exhibited the maximum swelling of 185% of its original mass at 21 days (Fig. 2C). The HA-ADH1.00HA-Ox3.00 and HA-ADH1.00HA-Ox1.00 hydrogels demonstrated complete degradation (i.e., minimum mass change of −100%) at 14 days. For the 100 kDa HA hydrogels, the HA-ADH1.00HA-Ox3.00 hydrogel demonstrated the greatest degradation of −68% at 28 days, while the HA-ADH2.33HA-Ox1.00 hydrogel demonstrated the maximum swelling of 62% at 4 days (Fig. 2D). Overall, the 40 kDa HA hydrogels demonstrated larger changes in mass (swelling or degradation) than the 100 kDa HA hydrogels. This may be due to the lower degree of modification achieved for 40 kDa HA-Ox compared to 100 kDa HA-Ox, which may have resulted in lower crosslinking densities of the polymer network and less stable hydrogels. Hydrogels that contained either excess ADH or aldehyde content also demonstrated increased degradation or swelling, likely due to lower crosslinking density and a less stable crosslinked polymer network.Our results corroborate the findings of other groups who have fabricated hydrazone-crosslinked HA hydrogels. Similar to the results of our study, others have demonstrated a large range of gelation times, an inverse relationship between gelation time of their HA hydrogels and concentration of ADH crosslinker, and less stable hydrogels with excess aldehydes.34 In another study exploring hydrazone-crosslinked HA hydrogels for a vitreous substitute, the use of higher amounts of free ADH crosslinker stabilized the hydrogels;33 however, we found that excess HA-ADH increased hydrogel swelling at early time points. A wide range of Young's moduli have been observed with hydrazone-crosslinked HA hydrogels with values ranging between 1–2000 Pa reported in the literature.36 In comparison, our HA hydrogels exhibited relatively low compressive moduli within a narrow range of 4.5–42.2 Pa. Others have found that a higher degree of modification for HA-ADH led to stiffer hydrogels, which was similar to what we observed for the 40 kDa hydrogels.35
Ultimately, our method of fabricating hydrazone-crosslinked HA hydrogels yielded hydrogels with a wide range of physicochemical properties, including large ranges in hydrogel gelation time and mass change, with the capacity to swell, degrade, or maintain a relatively constant mass over 28 days depending on the molecular weight and concentrations of HA-ADH and HA-Ox used (Table 2).
| Property | Range |
|---|---|
| Gelation time | 1.4–292.7 min |
| Compressive modulus | 4.5–42.2 Pa |
| Mass change day 1 | 1.0–43.1% |
| Mass change day 7 | −56.3 to 62.2% |
| Mass change day 14 | −100 to 156.9% |
| Mass change day 21 | −100 to 185.3% |
| Mass change day 28 | −100 to 160.7% |
Design of experiments analysis
After obtaining response data for our 15 hydrogel formulations, we used this data to generate a multivariate model for DoE analysis to better understand how HA-Ox concentration, HA-ADH concentration, and HA molecular weight interacted to produce a wide range of physicochemical properties demonstrated by our platform and to predict the hydrogel formulations that would yield responses that met our desired design criteria. We fit second order multiple linear regression models for each response (gelation time, compressive modulus, mass change at 1, 7, 14, 21, and 28 days) and eliminated the terms that were not statistically significant unless they contributed to square terms or interaction terms that were statistically significant (p < 0.05) (Table 3). There was a significant effect of HA-ADH concentration on all responses except for mass change at day 7 and a significant effect of HA-Ox concentration on all responses except for mass change at day 1. The molecular weight of the HA had a significant effect on all responses. Notably, several squared and interaction terms also had significant effects on individual responses. We found that all of our models fit the success criteria of R2 and adjusted R2 values close to 1.0, Q2 above 0.50, and the difference between R2 and Q2 values below 0.30. The models with the lowest R2 and Q2 values, indicating the poorest fit, were for mass change at days 1 and 7. This may be due to large variations in the initial swelling of the hydrogel network when incubated in PBS resulting in unpredictable short-term changes in mass compared to hydrogels that have swollen to a stable, more predictable equilibrium at later time points (days 14, 21, and 28).59| Response | Gelation time | Compressive modulus | Mass change day 1 | Mass change day 7 | Mass change day 14 | Mass change day 21 | Mass change day 28 |
|---|---|---|---|---|---|---|---|
| p-Values | |||||||
| [HA-ADH] | <0.0001 | 0.0258 | 0.0136 | 0.612 | <0.0001 | <0.0001 | <0.0001 |
| [HA-Ox] | <0.0001 | 0.173 | 0.725 | 0.000539 | <0.0001 | <0.0001 | <0.0001 |
| Molecular weight (MW) | <0.0001 | 0.00122 | 0.000702 | 0.000222 | 0.000494 | 0.00107 | 0.000234 |
| [HA-ADH]2 | 0.000985 | 0.00375 | 0.0399 | — | — | — | — |
| [HA-Ox]2 | — | 0.000782 | — | — | — | — | — |
| MW2 | — | — | — | — | — | — | — |
| [HA-ADH]* [HA-Ox] | — | — | 0.00213 | 0.00237 | — | — | — |
| [HA-ADH]* MW | — | — | — | — | 0.00166 | 0.000823 | 0.0125 |
| [HA-Ox]* MW | — | — | — | — | 0.000964 | 0.00304 | 0.00763 |
| Lack of fit | 0.177 | 0.363 | 0.948 | 0.213 | 0.410 | 0.472 | 0.548 |
| Total sample size (N) | 17 | 13 | 17 | 16 | 16 | 16 | 16 |
| Degrees of freedom (DF) | 12 | 7 | 11 | 11 | 10 | 10 | 10 |
| R 2 | 0.964 | 0.970 | 0.828 | 0.852 | 0.950 | 0.937 | 0.936 |
| R 2-Adjusted | 0.952 | 0.903 | 0.750 | 0.799 | 0.926 | 0.905 | 0.904 |
| Q 2 | 0.918 | 0.949 | 0.601 | 0.683 | 0.710 | 0.840 | 0.792 |
Three-dimensional response surface plots of gelation time for the 40 kDa HA hydrogels demonstrated that higher polymer content resulted in faster gelation with the shortest gelation time occurring at HA-ADH3.00HA-Ox3.00 and longest gelation time occurring at HA-ADH1.00HA-Ox1.00 (Fig. 3A). This response surface was identical but smaller in magnitude for the 100 kDa HA hydrogels (Fig. 3B). These plots indicate that gelation time was dependent on the concentrations of both HA-ADH and HA-Ox. The response surfaces of compressive modulus for 40 kDa and 100 kDa HA hydrogels both depicted a negative quadratic dependence on the HA-Ox and HA-ADH concentrations with a maximum compressive modulus predicted to occur at HA-ADH2.21HA-Ox2.08. For the 40 kDa HA hydrogels, this maximum compressive modulus was predicted to be 39.2 Pa (Fig. 3C), while, for the 100 kDa HA hydrogels, the maximum compressive modulus was predicted to be 27.3 Pa (Fig. 3D). The relationship between mass change and input parameters was different at different time points. Mass change at day 1 was dependent on interactions between HA-ADH and HA-Ox concentrations and demonstrated a positive quadratic dependence on HA-ADH. The minimal mass change for both 40 kDa and 100 kDa hydrogels occurred at HA-ADH3.00HA-Ox3.00 and was found to be at 4.65% and 21.1%, respectively (Fig. 3E and F). Mass change at day 7 demonstrated a linear dependence on HA-ADH and HA-Ox concentrations, suggesting there were no interactions between HA-ADH and HA-Ox concentrations for this response. For both 40 kDa and 100 kDa hydrogels, the minimum mass change occurred at HA-ADH1.00HA-Ox3.00; however, the predicted minimum was 9.49% for 40 kDa hydrogels (Fig. 3G), and −22.61% for 100 kDa hydrogels (Fig. 3H). At day 14, the model predicted a minimum mass change of −74.87% at HA-ADH1.00HA-Ox3.00 and a maximum mass change of 174.16% at HA-ADH3.00HA-Ox1.00 for 40 kDa hydrogels (Fig. 3I). Minimum and maximum mass changes occurred at the same polymer compositions for 100 kDa hydrogels on day 14, with a minimum at −26.63% and a maximum of 40.23% (Fig. 3J). The response surfaces for 40 kDa and 100 kDa HA hydrogels at day 28 demonstrated linear dependences on HA-ADH and HA-Ox concentrations similar to response surfaces at day 14 (Fig. 3K and L). Overall, the response surfaces and ANOVA table demonstrate that gelation time, compressive modulus, and mass change at day 1 were dependent on interactions between HA-ADH concentration and HA-Ox concentration, while no interactions of the input parameters were observed with mass changes at the other time points.
Optimal and pessimal hydrogel predictions
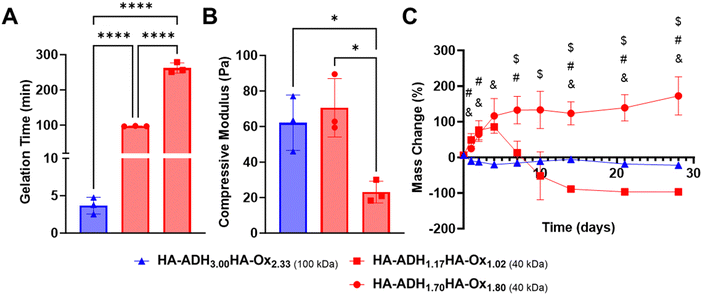
Our overall objective was to use DoE to optimize HA hydrogels for protein delivery for bone regeneration. We aimed to formulate a hydrogel that exhibited a gelation time between 1–10 minutes, minimal mass change (i.e., 0%) over 28 days, and the maximum possible compressive modulus. D-Optimal design generated predictive models describing how HA-ADH polymer concentration, HA-Ox polymer concentration, and HA molecular weight influenced compressive modulus, gelation time, and mass change.To validate this model, we generated a hydrogel formulation that was predicted to optimize our target parameters (Optimal) as well as two hydrogels that were optimized with opposite objectives (Pessimal 1 and 2) (Table 4). More specifically, Pessimal 1 was set to degrade over 28 days, while Pessimal 2 was set to swell over 28 days. Our optimal formulation had a molecular weight of 100 kDa while both pessimal formulations had a molecular weight of 40 kDa. This may be due to the longer gelation times, softer compressive moduli, and larger mass changes over time that were observed in our 40 kDa hydrogels. The optimal formulation, HA-ADH3.00HA-Ox2.33, met our targeted responses with a gelation time of 3.7 min (Fig. 4A), compressive modulus of 62.1 Pa (Fig. 4B), and minimal mass change ranging between −22.2 to 5.5% over 28 days (Fig. 4C). Conversely, our Pessimal 1 formulation, HA-ADH1.17HA-Ox1.02, displayed a gelation time of 262.2 min, compressive modulus of 23.1 Pa, and swelling up to 85.3% of its initial mass on day 4, followed by full degradation by day 21. The Pessimal 2 formulation, HA-ADH1.70HA-Ox1.80 displayed a gelation time of 100 min, a compressive modulus of 70.6 Pa, and swelling up to 172.3% by day 28 with no degradation.
| HA-ADH (%w/v) | HA-Ox (%w/v) | Molecular weight (kDa) | |
|---|---|---|---|
| Optimal | 3.00 | 2.33 | 100 |
| Pessimal 1 | 1.17 | 1.02 | 40 |
| Pessimal 2 | 1.70 | 1.80 | 40 |
The gelation time of the optimal formulation was significantly lower than that of the pessimal formulations. Mass change for the optimal formulation was closer to the 0% target mass change at all timepoints, while both pessimal formulations exhibited initial swelling before either degrading completely or continuing to swell excessively. However, there were no significant differences between the compressive moduli of the optimal formulation and the two pessimal formulations. Thus, trying to independently target each of our desired physicochemical properties with our hydrogel formulation would not have been successful, highlighting the importance of using DoE to simultaneously optimize multiple output responses. Scanning electron microscopy of the optimal hydrogel formulation revealed interconnected pores of different sizes (Fig. S5, ESI†). This porous morphology resembles the morphology observed in other HA hydrogels.60
BMP-2 release from affibody-containing hydrogels
As proof-of-concept of affinity-controlled protein delivery from our HA hydrogels, we modified HA-Ox to contain a norbornene functional group for bioconjugation of a previously discovered BMP-2-specific affibody with high affinity for BMP-2 (dissociation constant, KD = 10.7 nM).41 Affibodies are small proteins with 58 amino acids folded into a bundle of three α-helices.61,62 Affibodies can be engineered to bind to different proteins with a range of affinities and can be used to release proteins at various rates by tuning the strength of the affinity interactions.41,63,64 HA was functionalized with norbornene through BoC2O activated coupling (Fig. 5A). The resulting product was then modified a second time by the previously described oxidation reaction, resulting in oxidized, norbornene-functionalized HA (HA-Nor-Ox; Fig. 5B). Two times molar excess affibody to HA was then added to HA-Nor-Ox in the presence of a photoinitiator and exposed to light at 365 nm to create affibody-conjugated HA-Nor-Ox (HA-Nor-Ox-Affibody). The average concentration of affibody conjugated to HA-Nor-Ox was 2.80 ± 1.95 nmol mg−1 HA polymer (Fig. S6, ESI†). This modified polymer was crosslinked with HA-ADH to form a hydrazone-crosslinked, affibody-containing HA hydrogel.Release of BMP-2 was evaluated over 28 days from unmodified HA hydrogels or hydrogels conjugated with affibodies with high affinity and specificity for BMP-2 (dissociation constant, KD = 10.7 nM).41 Hydrogels were formed with BMP-2 and incubated in 1% w/v BSA in PBS. Aliquots were taken from the supernatant over 28 days to evaluate a release profile of BMP-2 from the HA hydrogel. The HA hydrogel with BMP-2 affibody reduced the release of BMP-2 compared to a HA hydrogel without affibody (Fig. 6A). After 28 days, 11.7% of BMP-2 was released from the hydrogel without the affibodies, whereas 3.5% of BMP-2 was released from the hydrogel with the BMP-2-specific affibodies. Fig. 6B depicts the cumulative fraction release at each time point (Mt/M∞) as a function of the square root of time of the linear region of the BMP-2 release profile. The diffusivity (i.e., release rate) or the Fickian diffusion slope, k, was calculated using the slope of the cumulative fractional release (eqn (3)), demonstrating that the release rate of BMP-2 was significantly slower in the affibody-containing hydrogel compared to the hydrogel without affibodies (Fig. 6C). Our goal was to use BMP-2-specific affibodies to increase BMP-2 retention within the HA hydrogel for future applications in bone regeneration, since increased BMP-2 retention has been shown to stimulate localized bone formation within an injury site.65 In other work, we have demonstrated that BMP-2 release from polyethylene glycol (PEG) hydrogels can be tuned over a range of release rates and amounts by changing the affinity between the affibodies and BMP-2.41 In the future, we plan to further explore how changes to the HA hydrogel formulation, affibody amount, and affibody affinity strength can influence BMP-2 release to enhance bone regeneration.
Conclusion
Although hydrazone chemistry is similar in many applications, each hydrogel requires distinct properties for a specific use. Designing an appropriate hydrogel for a biomedical application requires the consideration of many design criteria, such as gelation time, compressive modulus, and stability over time, that can be affected by many factors, including polymer molecular weight and concentration. In this work, we optimized hydrazone-crosslinked HA hydrogels for protein delivery for bone regeneration using DoE to better understand how various input parameters interacted to affect physicochemical properties, while reducing the number of experiments required to obtain this information. We generated models to describe how HA-ADH concentration, HA-Ox concentration, and HA molecular weight affected hydrogel compressive modulus, gelation time, and mass change over time. We identified an optimized hydrogel formulation that simultaneously maximized compressive modulus (62.1 Pa), achieved a target gelation time between 1–10 minutes (3.7 minutes), and was stable over 28 days with minimal mass change. We confirmed the validity of our model by evaluating two pessimal hydrogel formulations that were predicted to provide responses opposite to our optimal formulation. We also demonstrated that protein release from hydrazone-crosslinked HA hydrogels could be sustained over 28 days by conjugated BMP-2-specific affibodies to the hydrogels to control the release of BMP-2. Ultimately, hydrazone-crosslinked HA hydrogels are a promising platform for protein delivery and DoE is a valuable tool to optimize hydrogels for this application.Conflicts of interest
A. N. G., J. D., V. R. S., and M. H. H. are authors on a pending patent application encompassing some of this work.Acknowledgements
We are grateful for funding from the Collins Medical Trust, National Institutes of Health (R21-EB032112), and Wu Tsai Human Performance Alliance. JD is supported by a doctoral-level post-graduate scholarship (PGS-D) from the Natural Sciences and Engineering Research Council of Canada (NSERC). EAM and JK were supported by the Knight Campus Undergraduate Scholars Program. MF was supported by the University of Oregon's Summer Program in Undergraduate Research (SPUR) and Vice President for Research and Innovation (VPRI) Fellowship. We would like to thank Dr Casey Check and Professor Willie (Skip) Rochefort for their guidance on rheology, Patrick Hall for his guidance on scanning electron microscopy, and Professor Danielle Benoit for her guidance on gel permeation chromatography. We would also like to thank Chandler Asnes and Payton Jefferis for their thoughtful feedback on this work.References
- J. Dorogin, J. M. Townsend and M. H. Hettiaratchi, Biomaterials for protein delivery for complex tissue healing responses, Biomater. Sci., 2021, 9(7), 2339–2361 RSC.
- A. W. James, G. LaChaud, J. Shen, G. Asatrian, V. Nguyen and X. Zhang, et al., A Review of the Clinical Side Effects of Bone Morphogenetic Protein-2, Tissue Eng., Part B, 2016, 22(4), 284–297 CrossRef CAS PubMed.
- M. Bartnikowski, R. M. Wellard, M. Woodruff and T. Klein, Tailoring Hydrogel Viscoelasticity with Physical and Chemical Crosslinking, Polymers, 2015, 7(12), 2650–2669 CrossRef CAS.
- T. Vermonden, R. Censi and W. E. Hennink, Hydrogels for Protein Delivery, Chem. Rev., 2012, 112(5), 2853–2888 CrossRef CAS PubMed.
- S. Naahidi, M. Jafari, M. Logan, Y. Wang, Y. Yuan and H. Bae, et al., Biocompatibility of hydrogel-based scaffolds for tissue engineering applications, Biotechnol. Adv., 2017, 35(5), 530–544 CrossRef CAS PubMed.
- M. Darnell and D. J. Mooney, Leveraging advances in biology to design biomaterials, Nat. Mater., 2017, 16(12), 1178–1185 CrossRef CAS PubMed.
- J. Telford. A Brief Introduction to Design of Experiments [Internet]. 2007 [cited 2023 May 7]. Available from: https://secwww.jhuapl.edu/techdigest/Home/Detail?Journal=J&VolumeID=27&IssueID=3.
- J. Lam, S. T. Carmichael, W. E. Lowry and T. Segura, Hydrogel Design of Experiments Methodology to Optimize Hydrogel for iPSC-NPC Culture, Adv. Healthcare Mater., 2015, 4(4), 534–539 CrossRef CAS PubMed.
- A. S. H. Ahmed, U. K. Mandal, M. Taher, D. Susanti and J. M. D. Jaffri, PVA-PEG physically cross-linked hydrogel film as a wound dressing: experimental design and optimization, Pharm. Dev. Technol., 2018, 23(8), 751–760 CrossRef CAS PubMed.
- L. R. Nih, P. Moshayedi, I. L. Llorente, A. R. Berg, J. Cinkornpumin and W. E. Lowry, et al., Engineered HA hydrogel for stem cell transplantation in the brain: biocompatibility data using a design of experiment approach, Data Brief., 2017, 10, 202–209 CrossRef PubMed.
- I. A. Cadena, M. R. Buchanan, C. G. Harris, M. A. Jenne, W. E. Rochefort and D. Nelson, et al., Engineering high throughput screening platforms of cervical cancer, J. Biomed. Mater. Res., Part A, 2023, 111(6), 747–764 CrossRef CAS PubMed.
- M. G. Arafa and B. M. Ayoub, DOE Optimization of Nano-based Carrier of Pregabalin as Hydrogel: New Therapeutic & Chemometric Approaches for Controlled Drug Delivery Systems, Sci. Rep., 2017, 7(1), 41503 CrossRef CAS PubMed.
- A. Nastruzzi, G. Pitingolo, G. Luca and C. Nastruzzi, DoE Analysis of Approaches for Hydrogel Microbeads’ Preparation by Millifluidic Methods, Micromachines, 2020, 11(11), 1007 CrossRef PubMed.
- M. M. Pakulska, K. Vulic, R. Y. Tam and M. S. Shoichet, Hybrid Crosslinked Methylcellulose Hydrogel: A Predictable and Tunable Platform for Local Drug Delivery, Adv. Mater., 2015, 27(34), 5002–5008 CrossRef CAS PubMed.
- A. M. Yousefi, J. Liu, R. Sheppard, S. Koo, J. Silverstein and J. Zhang, et al., I-Optimal Design of Hierarchical 3D Scaffolds Produced by Combining Additive Manufacturing and Thermally Induced Phase Separation, ACS Appl. Bio Mater., 2019, 2(2), 685–696 CrossRef CAS PubMed.
- M. Asadi-Eydivand, M. Solati-Hashjin, A. Fathi, M. Padashi and N. A. Abu Osman, Optimal design of a 3D-printed scaffold using intelligent evolutionary algorithms, Appl. Soft Comput., 2016, 39, 36–47 CrossRef.
- M. Kuterbekov, P. Machillot, F. Baillet, A. M. Jonas, K. Glinel and C. Picart, Design of experiments to assess the effect of culture parameters on the osteogenic differentiation of human adipose stromal cells, Stem Cell Res. Ther., 2019, 10(1), 256 CrossRef PubMed.
- M. F. P. Graça, S. P. Miguel, C. S. D. Cabral and I. J. Correia, Hyaluronic acid-Based wound dressings: a review, Carbohydr. Polym., 2020, 241, 116364 CrossRef PubMed.
- J. Necas, L. Bartosikova, P. Brauner and J. Kolar, Hyaluronic acid (hyaluronan): a review, Veterinární Med., 2008, 53(8), 397–411 CrossRef CAS.
- M. Dovedytis, Z. J. Liu and S. Bartlett, Hyaluronic acid and its biomedical applications: a review, Eng. Regener., 2020, 1, 102–113 Search PubMed.
- M. K. Cowman, H. G. Lee, K. L. Schwertfeger, J. B. McCarthy and E. A. Turley, The Content and Size of Hyaluronan in Biological Fluids and Tissues, Front. Immunol., 2015, 6 DOI:10.3389/fimmu.2015.00261.
- A. G. Tavianatou, I. Caon, M. Franchi, Z. Piperigkou, D. Galesso and N. K. Karamanos, Hyaluronan: molecular size-dependent signaling and biological functions in inflammation and cancer, FEBS J., 2019, 286(15), 2883–2908 CrossRef CAS PubMed.
- J. Yang, C. C. Hsu, T. T. Cao, H. Ye, J. Chen and Y. Q. Li, A hyaluronic acid granular hydrogel nerve guidance conduit promotes regeneration and functional recovery of injured sciatic nerves in rats, Neural Regener. Res., 2023, 18(3), 657 CrossRef CAS PubMed.
- M. Halperin-Sternfeld, A. Pokhojaev, M. Ghosh, D. Rachmiel, R. Kannan and I. Grinberg, et al., Immunomodulatory fibrous hyaluronic acid-Fmoc-diphenylalanine-based hydrogel induces bone regeneration, J. Clin. Periodontol., 2023, 50(2), 200–219 CrossRef CAS PubMed.
- S. J. Yoon, Y. H. Fang, C. H. Lim, B. S. Kim, H. S. Son and Y. Park, et al., Regeneration of ischemic heart using hyaluronic acid-based injectable hydrogel, J. Biomed. Mater. Res., Part B, 2009, 91B(1), 163–171 CrossRef CAS PubMed.
- A. J. Engler, S. Sen, H. L. Sweeney and D. E. Discher, Matrix Elasticity Directs Stem Cell Lineage Specification, Cell, 2006, 126(4), 677–689 CrossRef CAS PubMed.
- D. A. Young, Y. S. Choi, A. J. Engler and K. L. Christman, Stimulation of adipogenesis of adult adipose-derived stem cells using substrates that mimic the stiffness of adipose tissue, Biomaterials, 2013, 34(34), 8581–8588 CrossRef CAS PubMed.
- X. Xue, K. Liang, W. Huang, H. Yang, L. Jiang and Q. Jiang, et al., Molecular Engineering of Injectable, Fast Self-Repairing Hydrogels with Tunable Gelation Time: Characterization by Diffusing Wave Spectroscopy, Macromolecules, 2022, 55(15), 6474–6486 CrossRef CAS.
- V. Delplace, P. E. B. Nickerson, A. Ortin-Martinez, A. E. G. Baker, V. A. Wallace and M. S. Shoichet, Nonswelling, Ultralow Content Inverse Electron-Demand Diels–Alder Hyaluronan Hydrogels with Tunable Gelation Time: Synthesis and In Vitro Evaluation, Adv. Funct. Mater., 2020, 30(14), 1903978 CrossRef CAS.
- H. Park, X. Guo, J. S. Temenoff, Y. Tabata, A. I. Caplan and F. K. Kasper, et al., Effect of Swelling Ratio of Injectable Hydrogel Composites on Chondrogenic Differentiation of Encapsulated Rabbit Marrow Mesenchymal Stem Cells In Vitro, Biomacromolecules, 2009, 10(3), 541–546 CrossRef CAS PubMed.
- W. Mubarok, K. C. M. L. Elvitigala, M. Nakahata, M. Kojima and S. Sakai, Modulation of Cell-Cycle Progression by Hydrogen Peroxide-Mediated Cross-Linking and Degradation of Cell-Adhesive Hydrogels, Cells, 2022, 11(5), 881 CrossRef CAS PubMed.
- C. Wang, X. Tong, X. Jiang and F. Yang, Effect of Matrix Metalloproteinase-Mediated Matrix Degradation on Glioblastoma Cell Behavior in 3D PEG-based Hydrogels, J. Biomed. Mater. Res., Part A, 2017, 105(3), 770–778 CrossRef CAS PubMed.
- W. Y. Su, K. H. Chen, Y. C. Chen, Y. H. Lee, C. L. Tseng and F. H. Lin, An Injectable Oxidated Hyaluronic Acid/Adipic Acid Dihydrazide Hydrogel as a Vitreous Substitute, J. Biomater. Sci., Polym. Ed., 2011, 22(13), 1777–1797 CrossRef CAS PubMed.
- C. G. França, D. P. Sacomani, D. G. Villalva, V. F. Nascimento, J. L. Dávila and M. H. A. Santana, Structural changes and crosslinking modulated functional properties of oxi-HA/ADH hydrogels useful for regenerative purposes, Eur. Polym. J., 2019, 121, 109288 CrossRef.
- L. Vítková, L. Musilová, E. Achbergerová, R. Kolařík, M. Mrlík and K. Korpasová, et al., Formulation of Magneto-Responsive Hydrogels from Dually Cross-Linked Polysaccharides: Synthesis, Tuning and Evaluation of Rheological Properties, Int. J. Mol. Sci., 2022, 23(17), 9633 CrossRef PubMed.
- C. G. França, D. G. Villalva and M. H. A. Santana, Oxi-HA/ADH Hydrogels: A Novel Approach in Tissue Engineering and Regenerative Medicine, Polysaccharides, 2021, 2(2), 477–496 CrossRef.
- Department of Orthopaedics, National Taiwan University College of Medicine and National Taiwan University Hospital, No. 1, Sec. 1, JenAi Road, Taipei 100, Taiwan, M. Hu, K. Yang, Y. Sun, Y. Chen and S. Yang, et al., In situ forming oxidised hyaluronic acid/adipic acid dihydrazide hydrogel for prevention of epidural fibrosis after laminectomy, eCM., 2017, 307–320 CrossRef CAS PubMed.
- X. Jia, G. Colombo, R. Padera, R. Langer and D. S. Kohane, Prolongation of sciatic nerve blockade by in situ cross-linked hyaluronic acid, Biomaterials, 2004, 25(19), 4797–4804 CrossRef CAS PubMed.
- Y. Yeo, C. B. Highley, E. Bellas, T. Ito, R. Marini and R. Langer, et al., In situ cross-linkable hyaluronic acid hydrogels prevent post-operative abdominal adhesions in a rabbit model, Biomaterials, 2006, 27(27), 4698–4705 CrossRef CAS PubMed.
- N. de Paiva Narciso, R. S. Navarro, A. E. Gilchrist, M. L. M. Trigo, G. Aviles Rodriguez and S. C. Heilshorn, Design Parameters for Injectable Biopolymeric Hydrogels with Dynamic Covalent Chemistry Crosslinks, Adv. Healthcare Mater., 2023, 2301265 CrossRef CAS PubMed.
- J. Dorogin, H. B. Hochstatter, S. O. Shepherd, J. E. Svendsen, M. A. Benz and A. C. Powers, et al., Moderate-Affinity Affibodies Modulate the Delivery and Bioactivity of Bone Morphogenetic Protein-2, Adv. Healthcare Mater., 2023, 2300793 CrossRef CAS PubMed.
- M. D. Davidson, E. Ban, A. C. M. Schoonen, M. H. Lee, M. D’Este and V. B. Shenoy, et al., Mechanochemical Adhesion and Plasticity in Multifiber Hydrogel Networks, Adv. Mater., 2020, 32(8), 1905719 CrossRef CAS PubMed.
- H. Zhao and N. D. Heindel, Determination of Degree of Substitution of Formyl Groups in Polyaldehyde Dextran by the Hydroxylamine Hydrochloride Method, Pharm. Res., 1991, 8(3), 400–402 CrossRef CAS PubMed.
- Q. Wang, Z. Wang, D. Zhang, J. Gu, Y. Ma and Y. Zhang, et al., Circular Patterns of Dynamic Covalent Hydrogels with Gradient Stiffness for Screening of the Stem Cell Microenvironment, ACS Appl. Mater. Interfaces, 2022, 14(42), 47461–47471 CrossRef CAS PubMed.
- J. M. Alonso, J. Andrade Del Olmo, R. Perez Gonzalez and V. Saez-Martinez, Injectable Hydrogels: From Laboratory to Industrialization, Polymers (Basel), 2021, 13(4), 650 CrossRef CAS PubMed.
- N. R. Raia, B. P. Partlow, M. McGill, E. P. Kimmerling, C. E. Ghezzi and D. L. Kaplan, Enzymatically Crosslinked Silk-Hyaluronic Acid Hydrogels, Biomaterials, 2017, 131, 58–67 CrossRef CAS PubMed.
- P. Rosaming, J. Jirayupapong, S. Thamnium, Y. Y. Win, V. Limprasutr and R. Rodsiri, et al., Interpenetrating Low-Molecular Weight Hyaluronic Acid in Hyaluronic Acid-Based In Situ Hydrogel Scaffold for Periodontal and Oral Wound Applications, Polymers, 2022, 14(22), 4986 CrossRef CAS PubMed.
- V. G. Muir, S. Weintraub, A. P. Dhand, H. Fallahi, L. Han and J. A. Burdick, Influence of Microgel and Interstitial Matrix Compositions on Granular Hydrogel Composite Properties, Adv. Sci., 2023, 10(10), 2206117 CrossRef CAS PubMed.
- P. L. Ritger and N. A. Peppas, A simple equation for description of solute release I. Fickian and non-fickian release from non-swellable devices in the form of slabs, spheres, cylinders or discs, J. Controlled Release, 1987, 5(1), 23–36 CrossRef CAS.
- M. M. Pakulska, K. Vulic and M. S. Shoichet, Affinity-based release of chondroitinase ABC from a modified methylcellulose hydrogel, J. Controlled Release, 2013, 171(1), 11–16 CrossRef CAS PubMed.
- R. E. Bruns, I. S. Scarminio and B. B. de Neto, Statistical Design – Chemometrics, Elsevier Science, Amsterdam; Boston, 2006. 1st edn, p. 422 Search PubMed.
- MODDE® 12 User Guide. June 21, 2017. Sartorius Stedim Data Analytics AB.
- C. G. França, D. P. Sacomani, D. G. Villalva, V. F. Nascimento, J. L. Dávila and M. H. A. Santana, Structural changes and crosslinking modulated functional properties of oxi-HA/ADH hydrogels useful for regenerative purposes, Eur. Polym. J., 2019, 121, 109288 CrossRef.
- S. Uman, L. L. Wang, S. L. Thorn, Z. Liu, J. S. Duncan and A. J. Sinusas, et al., Imaging of injectable hydrogels delivered into myocardium with SPECT/CT, Adv. Healthcare Mater., 2020, 9(14), e2000294 CrossRef PubMed.
- Y. Wang, Y. Chen, J. Zheng, L. Liu and Q. Zhang, Three-Dimensional Printing Self-Healing Dynamic/Photocrosslinking Gelatin-Hyaluronic Acid Double-Network Hydrogel for Tissue Engineering, ACS Omega, 2022, 7(14), 12076–12088 CrossRef CAS PubMed.
- L. L. Wang, C. B. Highley, Y. C. Yeh, J. H. Galarraga, S. Uman and J. A. Burdick, 3D extrusion bioprinting of single- and double-network hydrogels containing dynamic covalent crosslinks, J. Biomed. Mater. Res., Part A, 2018, 106(4), 865–875 CrossRef CAS PubMed.
- S. Kadi, D. Cui, E. Bayma, T. Boudou, C. Nicolas and K. Glinel, et al., Alkylamino Hydrazide Derivatives of Hyaluronic Acid: Synthesis, Characterization in Semidilute Aqueous Solutions, and Assembly into Thin Multilayer Films, Biomacromolecules, 2009, 10(10), 2875–2884 CrossRef CAS PubMed.
- N. Lagneau, P. Tournier, B. Halgand, F. Loll, Y. Maugars and J. Guicheux, et al., Click and bioorthogonal hyaluronic acid hydrogels as an ultra-tunable platform for the investigation of cell-material interactions, Bioact. Mater., 2023, 24, 438–449 CAS.
- N. R. Richbourg, M. Wancura, A. E. Gilchrist, S. Toubbeh, B. A. C. Harley and E. Cosgriff-Hernandez, et al., Precise control of synthetic hydrogel network structure via linear, independent synthesis-swelling relationships, Sci. Adv., 2021, 7(7), eabe3245 CrossRef CAS PubMed.
- J. M. Alonso, J. Andrade Del Olmo, R. Perez Gonzalez and V. Saez-Martinez, Injectable Hydrogels: From Laboratory to Industrialization, Polymers, 2021, 13(4), 650 CrossRef CAS PubMed.
- J. Löfblom, J. Feldwisch, V. Tolmachev, J. Carlsson, S. Ståhl and F. Y. Frejd, Affibody molecules: engineered proteins for therapeutic, diagnostic and biotechnological applications, FEBS Lett., 2010, 584(12), 2670–2680 CrossRef PubMed.
- S. Ståhl, T. Gräslund, A. Eriksson Karlström, F. Y. Frejd, P. Å. Nygren and J. Löfblom, Affibody Molecules in Biotechnological and Medical Applications, Trends Biotechnol., 2017, 35(8), 691–712 CrossRef PubMed.
- C. J. Teal, M. H. Hettiaratchi, M. T. Ho, A. Ortin-Martinez, A. N. Ganesh and A. J. Pickering, et al., Directed Evolution Enables Simultaneous Controlled Release of Multiple Therapeutic Proteins from Biopolymer-Based Hydrogels, Adv. Mater., 2022, 34(34), 2202612 CrossRef CAS PubMed.
- C. Bostock, C. J. Teal, M. Dang, A. W. Golinski, B. J. Hackel and M. S. Shoichet, Affibody-mediated controlled release of fibroblast growth factor 2, J. Controlled Release, 2022, 350, 815–828 CrossRef CAS PubMed.
- M. H. Hettiaratchi, L. Krishnan, T. Rouse, C. Chou, T. C. McDevitt and R. E. Guldberg, Heparin-mediated delivery of bone morphogenetic protein-2 improves spatial localization of bone regeneration, Sci. Adv., 2020, 6(1), eaay1240 CrossRef CAS PubMed.
Footnotes |
| † Electronic supplementary information (ESI) available. See DOI: https://doi.org/10.1039/d3tb01588b |
| ‡ These authors contributed equally to this work. |
| This journal is © The Royal Society of Chemistry 2024 |