Open Access Article
Open Access ArticleSustainable dissolution of collagen and the formation of polypeptides in deep eutectic solvents for application as antibacterial agents†
Harmandeep
Kaur
a,
Manpreet
Singh
a,
Navdeep
Kaur
b,
Pratap Kumar
Pati
b,
Monika
Rani
c and
Tejwant Singh
Kang
 *a
*a
aDepartment of Chemistry, UGC Centre for Advanced Studies-II, Guru Nanak Dev University, Amritsar-143005, India. E-mail: tejwantsinghkang@gmail.com; Tel: +91-183-2258802-Ext-3291
bDepartment of Biotechnology, Guru Nanak Dev University, Amritsar-143005, India
cDepartment of Food Science and Technology, Guru Nanak Dev University, Amritsar-143005, India
First published on 24th June 2024
Abstract
Collagen is a protein that is hard to dissolve in water and many other solvents, which limits its applications. Herein, deep eutectic solvents (DESs), i.e. choline chloride![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) lactic acid (ChCl
lactic acid (ChCl![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) LA) = 1
LA) = 1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1 and ethylene glycol
1 and ethylene glycol![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) zinc chloride (EG
zinc chloride (EG![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) ZnCl2) = 4
ZnCl2) = 4![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1, are effectively used to dissolve type I collagen under different conditions. Type I collagen is readily soluble at a concentration of 9.5–22.5 w/v% in DESs, and the solubility is governed by the nature of the DES, temperature (45 °C, 70 °C and 90 °C) and the absence or presence of HCl(aq.) (5 × 10−5 M). The dissolved material is regenerated by employing ethanol as an anti-solvent at 4 °C and investigated for alteration in the polymeric structure using Fourier-transform infrared spectroscopy (FTIR), circular dichroism (CD), UV-vis spectroscopy, X-ray diffraction (XRD), thermogravimetric analysis (TGA), and SDS-PAGE techniques. The increase in temperature and the presence of dilute HCl(aq.) result in a relatively greater disruption of the H-bonded structure of collagen, causing the unwinding of its triple-helical structure coupled with reduction in the helical content of polyproline type-II helices, which exposed vital amino acid residues in the regenerated material. Such an unwinding is accompanied by the formation of low molecular weight polypeptides, which are readily soluble in water and show antimicrobial activity comparable to or more than that exhibited by a model antibiotic Kanamycin towards both Gram-negative and Gram-positive bacteria. DESs are reused for at least 3 cycles for collagen solubilization without alteration in their inherent structure and collagen solubilizing ability, whereas the material regenerated from reused DESs shows properties similar to that shown by the material regenerated from virgin DESs. In this manner, a new sustainable strategy for solubilizing collagen and the direct preparation of essential and active low molecular weight collagen peptides directly from collagen in a single step is established. An inventive approach to using collagen is made possible by the observation that lower molecular weight peptides formed from the sustainable dissolution of collagen with exposed aromatic amino acid residues can demonstrate antibacterial activity.
1, are effectively used to dissolve type I collagen under different conditions. Type I collagen is readily soluble at a concentration of 9.5–22.5 w/v% in DESs, and the solubility is governed by the nature of the DES, temperature (45 °C, 70 °C and 90 °C) and the absence or presence of HCl(aq.) (5 × 10−5 M). The dissolved material is regenerated by employing ethanol as an anti-solvent at 4 °C and investigated for alteration in the polymeric structure using Fourier-transform infrared spectroscopy (FTIR), circular dichroism (CD), UV-vis spectroscopy, X-ray diffraction (XRD), thermogravimetric analysis (TGA), and SDS-PAGE techniques. The increase in temperature and the presence of dilute HCl(aq.) result in a relatively greater disruption of the H-bonded structure of collagen, causing the unwinding of its triple-helical structure coupled with reduction in the helical content of polyproline type-II helices, which exposed vital amino acid residues in the regenerated material. Such an unwinding is accompanied by the formation of low molecular weight polypeptides, which are readily soluble in water and show antimicrobial activity comparable to or more than that exhibited by a model antibiotic Kanamycin towards both Gram-negative and Gram-positive bacteria. DESs are reused for at least 3 cycles for collagen solubilization without alteration in their inherent structure and collagen solubilizing ability, whereas the material regenerated from reused DESs shows properties similar to that shown by the material regenerated from virgin DESs. In this manner, a new sustainable strategy for solubilizing collagen and the direct preparation of essential and active low molecular weight collagen peptides directly from collagen in a single step is established. An inventive approach to using collagen is made possible by the observation that lower molecular weight peptides formed from the sustainable dissolution of collagen with exposed aromatic amino acid residues can demonstrate antibacterial activity.
Sustainable spotlightCollagen is one of the most important and abundant proteins in mammals, and has many important applications. The presence of H, ionic, and hydrophobic bonds as well as electrostatic interactions makes the triple-helical structure of collagen stable; therefore, collagen is considered a hard-to-dissolve polymer that limits its applications. Collagen can otherwise be dissolved in organic solvents or highly acidic media, which is not an environment-friendly option. Therefore, new environment-friendly and cheaper solvents should be used to dissolve collagen and transform it into value-added materials. Herein, the solvents used for the dissolution of collagen, i.e. deep eutectic solvents (DESs), are green, cost-effective, biodegradable, easy to prepare, and environment-friendly compared to other conventional solvents. The material regenerated from DESs after the dissolution of collagen at a relatively lower temperature (45 °C) shows properties similar to native collagen. Moreover, the material regenerated from DESs after the dissolution of collagen at 70 °C and 90 °C exhibits properties akin to gelatin and smaller molecular weight polypeptides. The obtained polypeptides exhibited enhanced antimicrobial activity against both Gram-positive and Gram-negative bacteria, which was even higher than Kanamycin (standard antibiotic). With further research, such materials may be used as biological antimicrobial agents. Further, no harmful organic solvent was used during the synthesis of DESs and thus does not pose any risk to the environment. DESs are recycled and reused, adding to the sustainability of the process. Considering the above premises, the present work certainly emphasizes the following UN sustainable development goals: good health and well-being (SDG3); affordable and clean energy (SDG7); industry, innovation, and infrastructure (SDG9); responsible consumption and production (SDG12), and establish and organize climate action (SDG13). |
1 Introduction
Collagen is one of the most vital proteins in mammals, corresponds to three-quarters of the dry weight of the human skin, and is a widespread component of the extracellular matrix.1–3 Among the various types of collagen,2 type I collagen has gained much attention owing to its wide application. Structurally, type I collagen is a cylindrical entity exhibiting a right-handed triple-helical structure comprising three polyproline type-II helices, which wind around each other to form a rod-like right-handed super helix2 with a molecular weight of ∼300 kDa,4,5 length of ∼280 nm and diameter of ∼1.4 nm.5,6 The three helically twisted polypeptide alpha chains5–7 have repeated units of amino acids, such as Gly-X–Y, where X and Y are proline and 4-hydroxy proline, respectively.1,3,5 Glycine is buried inside the core of the triple-helical structure, and X and Y residues are exposed to the solvent.8,9 The regions at the end of the N- and C-terminus that do not form a triple helical structure are called telopeptides.8,9 Telopeptides typically consist of 15–26 amino acid residues, such as lysine and hydroxylysine, along with their aldehyde derivatives, and are crucial for the creation of both intramolecular and intermolecular covalent crosslinks.Type I collagen exhibits low antigenic and high direct cell adhesion properties and has been extensively used as a biomaterial for the development of tissue engineering constructs and wound dressing systems.9,10 Partial hydrolysis of collagen yields gelatin,11–13 a valuable water-soluble protein that has received considerable attention due to its mineral binding capacity, lipid-lowering effect, immunomodulatory13 and antihypertensive response along with antioxidant13,14 and antimicrobial properties,13 and other applications that are associated with the presence of low molecular weight peptides. The various applications of collagen and its hydrolysed product, gelatin, have made collagen an interesting material to be explored further. Type I collagen is soluble in inorganic (sodium hypochlorite)15 and acts as organic solvents/acids16 but at a relatively higher temperature. The toxic nature of such solvents and the high-temperature conditions of solubilization renders the solubilization non-sustainable and limits the applications of collagen. However, it is non-soluble in water owing to its ordered structure supported by inter- and intra-molecular H-bonds,17 hydrophobic interactions, ionic interactions and van der Waal's forces.18,19 Therefore, new sustainable methods of dissolution, modification or preparation/extraction of collagen peptides/collagen hydrosylates, and subsequent regeneration need to be devised to widen the application of collagen. A detailed comparison of the method in this study with previously reported methods for dissolving collagen and preparing collagen peptides is provided in Annexure S1 (ESI),† revealing that the present method considers all the goals of “Green chemistry”.
To advance in this area, ionic liquids (ILs), which comprise only ions20–22 and are accepted as relatively greener solvents, have been tested as media for solubilizing collagen23,24 considering their ability to dissolve many hard-to-dissolve materials.25,26 The solutions of imidazolium-based ILs in acetate buffer have been found to exert stabilizing or destabilizing effects on the helical structure of type I collagen governed by the nature of ions comprising ILs.24 The lyophilized collagen (type I from calf skin) has also been reported to be soluble in concentrated aqueous solutions of imidazolium-based ILs, [C2mim][BF4] and [C2mim][Ac]; however, the maximum yield of dissolution was quite low (3.57 mg ml−1).27 There are few examples of ILs that can dissolve collagen except for 1-butyl-3-methylimidazolium chloride, [C4mim][Cl], which was found to dissolve collagen in good yield by Meng et al.19 The high chloride concentration is effectual in breaking the H-bonds and the ionic bonds in the collagen and thus leads to its dissolution similar to that observed in the case of dissolution of wood,28 cork,29 cellulose,25 wool keratin,30 and silk fibre31 in ILs. Therefore, few advancements in the dissolution of collagen utilizing ILs have been reported. More importantly, the triple helical structure of native collagen and the effectiveness of essential amino acids are noticeably destroyed during the dissolution processes.
The quest to develop new benign solvents has led to another class of widely accepted green solvents called Deep Eutectic Solvents (DESs). DESs are the eutectic mixtures of two or three molecular components (ionic or uncharged), H-bond donor (HBD), and H-bond acceptor (HBA), and they have melting points far below that of the individual components.32–34 The lowering of melting point is ascribed to the charge delocalisation via H-bond interactions of complexing agent (typically H-bond donor) with the halide anion, which in turn reduces the anion's interaction with its parent cation results in the formation of complex anionic species.35 DESs exhibit properties akin to ILs such as low volatility, recyclability, non-flammability, and excellent thermal stability32,36 but are biocompatible, easy to prepare without the use of any organic solvent in a single-step reaction, and cost-effective. This, along with the possibility of offering multiple H-bonding interactions by DESs to polymers/biopolymers,37,38 renders DESs as green solvents of choice for the dissolution and simultaneous preparation of collagen peptides from collagen under optimal conditions. A range of processing methods have been applied to produce collagen peptides or hydrolyzed collagen. However, the collagen structure is severely degraded to the molecular level during the dissolution/extraction processes, leading to a weakened performance of the obtained collagen-based materials with a low yield. What basically hinders the direct extraction of active collagen peptides is the complicated hierarchical structure, non-collagenous proteins and nucleic acid present along with the raw material. This gap drives us to directly obtain lower molecular weight collagen peptides with exposed active amino acids from natural collagen to reserve nanostructures. Although the extraction of collagen peptides from collagen has been reported in DESs,39,40 to the best of our knowledge, there is no report concerned with thorough investigations on the use of DESs for the dissolution and regeneration of collagen along with the formation of gelatin/collagen peptide to be used as antimicrobial agents.
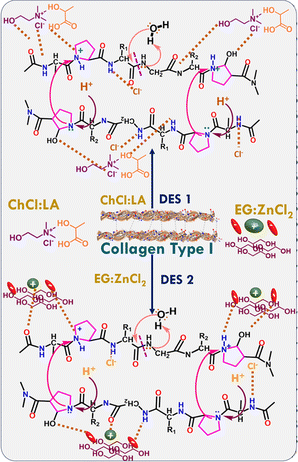
Herein, DESs comprising (i) choline chloride (ChCl) and LA (ChCl![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) LA = 1
LA = 1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1) and (ii) ethylene glycol (EG) and ZnCl2 (EG
1) and (ii) ethylene glycol (EG) and ZnCl2 (EG![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) ZnCl2 = 4
ZnCl2 = 4![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1) have been successfully established as dissolution media for type I collagen. The choice of the components of DESs is because the presence of Cl− and OH-along with the Lewis acidity of ZnCl2 has been found to aid in the dissolution of hard-to-dissolve polymers, such as cellulose,25 keratin,37,41 PET,42,43 polythene44 and wheat straw,45 when present as one of the components in ILs25 or DESs.37,41–45 LA-based DESs have also been used as a solvent for the exfoliation of biopolymers.46,47 ZnCl2 as Lewis acid is expected to activate the –C
1) have been successfully established as dissolution media for type I collagen. The choice of the components of DESs is because the presence of Cl− and OH-along with the Lewis acidity of ZnCl2 has been found to aid in the dissolution of hard-to-dissolve polymers, such as cellulose,25 keratin,37,41 PET,42,43 polythene44 and wheat straw,45 when present as one of the components in ILs25 or DESs.37,41–45 LA-based DESs have also been used as a solvent for the exfoliation of biopolymers.46,47 ZnCl2 as Lewis acid is expected to activate the –C![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) O group of amino acids and –OH groups of EG in the presence of light. Further, Cl− of ZnCl2 could form H-bonds with collagen and is thus supposed to break the amide bonds in collagen. In the case of ChCl
O group of amino acids and –OH groups of EG in the presence of light. Further, Cl− of ZnCl2 could form H-bonds with collagen and is thus supposed to break the amide bonds in collagen. In the case of ChCl![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) LA, the –OH and –C
LA, the –OH and –C![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) O groups of LA and Cl− of ChCl undergo the H-bonding interactions required to dissolve and degrade collagen.
O groups of LA and Cl− of ChCl undergo the H-bonding interactions required to dissolve and degrade collagen.
Various conditions of temperature (45 °C, 70 °C and 90 °C) and the presence of 5 × 10−5 M of dilute HCl(aq.) have been tested for the dissolution and processing of collagen in DESs. A temperature increase is expected to enhance the solubilization of collagen in DESs. Besides, the presence of 5 × 10−5 M concentration of HCl(aq.) in DESs could result in the protonation of the –NH2 group of amino acid residues of collagen, which favour the dissolution of collagen by offering electrostatic repulsion between similarly charged groups. Collagen dissolved in DESs is regenerated using ethanol as an antisolvent at 4 °C. The structure and properties of the regenerated material are explored and compared with those of native collagen using various state-of-the-art techniques. Based on the conditions, the regenerated material is found to be (i) collagen and (ii) low molecular weight collagen peptides. Regenerated collagen and collagen peptides are known to exhibit remarkable antimicrobial activity against Gram-positive and Gram-negative bacteria, which is almost or even better in some cases than that shown by the antibiotic Kanamycin. Compared to ILs and other processing techniques, DESs can disassemble the original triple helical structure and preserve the nanofibrous structure with active vital aromatic amino acids. In a way, a new sustainable method for the dissolution of hard-to-dissolve biopolymer, collagen, in DESs and its transformation to low molecular weight peptides in a single step, which exhibits remarkable antimicrobial activity, is established. The current work is anticipated to offer a new platform not only for the sustainable dissolution and preparation of collagen peptides from collagen but also for the dissolution and processing of many other biologically important polymers for diverse applications.
2 Experimental section
2.1. Materials
Collagen from bovine achilles tendon (type I) was purchased from Sigma-Aldrich. Ethylene glycol (EG) (>98%) and lactic acid (LA) (>88%) were purchased from Loba Chemie, India. Zinc chloride (ZnCl2) (>98%) was bought from Spectrochem Pvt Ltd, Mumbai, India, and choline chloride (ChCl) (>98%) was purchased from Sigma-Aldrich. Ethanol (AR grade) was procured from SD Fine Chemicals Ltd, Mumbai, India. AR grade sodium acetate (>99%) and acetic acid (>99.7%) were used to prepare acetate buffer and were bought from SRL, India. A protein molecular weight marker, a broad range (25 lanes), and 0.25 ml of SKU PMWB1 were purchased from GeNei.2.2. Dissolution and regeneration of collagen at different temperatures
Different Deep Eutectic Solvents (DESs), i.e. (i) ethylene glycol (EG)![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) zinc chloride (ZnCl2) in molar ratio 4
zinc chloride (ZnCl2) in molar ratio 4![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1 and (ii) choline chloride (ChCl)
1 and (ii) choline chloride (ChCl)![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) lactic acid (LA) in molar ratio 1
lactic acid (LA) in molar ratio 1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1, were synthesized as reported earlier.48,49 Thus, the obtained clear and homogenous DESs were stored in a desiccator to avoid the absorption of moisture. The solubility limit of collagen in DESs was first ascertained by adding collagen fibrils in lots (5 mg per lot) to 1 ml of respective DES preheated at 45 °C with continuous stirring at 100 rpm. The solubilization of collagen was monitored at different temperatures (45 °C, 70 °C and 90 °C) in the absence and presence of 5 × 10−5 M concentration of diluted HCl(aq.) using an optical microscope. 5 μL of 0.01 M HCl(aq.) (5 × 10−5 M) was added to 1 ml of the respective DES, which did not alter the pH noticeably of otherwise acidic DESs. The presence of collagen fibrils, even after stirring for a long time under specific conditions, marks the solubility limit of collagen in DESs. Later, for a regeneration process, ethanol (2 ml) was used as an antisolvent to 1 ml of the DES-collagen system at 4 °C. This resulted in the formation of white flocculent precipitations separated by centrifugation, followed by washing with ethanol 5 times. To ensure the complete removal of DESs, the FTIR of regenerated material is compared with native DESs (Fig. S1, ESI†).
1, were synthesized as reported earlier.48,49 Thus, the obtained clear and homogenous DESs were stored in a desiccator to avoid the absorption of moisture. The solubility limit of collagen in DESs was first ascertained by adding collagen fibrils in lots (5 mg per lot) to 1 ml of respective DES preheated at 45 °C with continuous stirring at 100 rpm. The solubilization of collagen was monitored at different temperatures (45 °C, 70 °C and 90 °C) in the absence and presence of 5 × 10−5 M concentration of diluted HCl(aq.) using an optical microscope. 5 μL of 0.01 M HCl(aq.) (5 × 10−5 M) was added to 1 ml of the respective DES, which did not alter the pH noticeably of otherwise acidic DESs. The presence of collagen fibrils, even after stirring for a long time under specific conditions, marks the solubility limit of collagen in DESs. Later, for a regeneration process, ethanol (2 ml) was used as an antisolvent to 1 ml of the DES-collagen system at 4 °C. This resulted in the formation of white flocculent precipitations separated by centrifugation, followed by washing with ethanol 5 times. To ensure the complete removal of DESs, the FTIR of regenerated material is compared with native DESs (Fig. S1, ESI†).
2.3. Characterisation of native collagen and regenerated material
Fourier Transform-Infrared (FTIR) spectroscopy was used to study the chemical composition of native collagen and regenerated material using an Agilent Cary 630 spectrometer in the range of 400–4000 cm−1. Circular dichroism (CD) spectroscopic measurements were performed by employing a BioLogic MOS-500 spectrometer in the wavelength range of 190–250 nm using a cuvette of path length 1 mm at a scan rate of 50 nm min−1. A UV-vis spectrophotometer (Cary 5000 UV-vis-NIR) was used for UV-vis absorption measurements in the wavelength range of 400–800 nm using a quartz cuvette of unit path length. UV-vis and CD spectroscopic measurements were made by dissolving the respective material in acetate buffer (pH ∼5.6). The crystallinity of native collagen and regenerated material was probed using X-ray diffraction (XRD) measurements employing a SHIMADZU MAXIMA ∼70![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 000 instruments in the 2θ range of 5–80°. The thermal stability of the material was monitored using a HITACHI STA7200 thermal analyzer under an N2 atmosphere at temperatures ranging from 25 to 1000 °C at a heating rate of 10 °C min−1. The molecular weight of the native collagen and regenerated material (collagen and collagen peptides) was determined using SDS-Page (Mini-Protean Tetra Cell, Bio-Rad Laboratories, Hercules, USA) at a constant current of 25 mA in 8% resolving gel. Optical density was measured at 600 nm using a UV-visible true double-beam spectrometer (Motras Scientific).
000 instruments in the 2θ range of 5–80°. The thermal stability of the material was monitored using a HITACHI STA7200 thermal analyzer under an N2 atmosphere at temperatures ranging from 25 to 1000 °C at a heating rate of 10 °C min−1. The molecular weight of the native collagen and regenerated material (collagen and collagen peptides) was determined using SDS-Page (Mini-Protean Tetra Cell, Bio-Rad Laboratories, Hercules, USA) at a constant current of 25 mA in 8% resolving gel. Optical density was measured at 600 nm using a UV-visible true double-beam spectrometer (Motras Scientific).
2.4. Antibacterial activity assay
Antibacterial activity was evaluated against two Gram negative (Escherichia coli and Pseudomonas syringae) and two Gram positive (Bacillus subtilis and Staphylococcus aureus) bacterial strains. Antimicrobial activity assays were performed as per the method described by Ennaas et al. in 2016 with minor modifications.50 All the samples with 1 mg ml−1 concentration of the material were serially diluted 2-fold in Luria Bertani (LB) broth. 100 μL of bacterial suspension (∼1 × 106 CFU per ml) of the given bacterial strain was seeded into all the samples. The samples were incubated at 37 °C in the case of E. coli and S. aureus, 28 °C in the case of P. syringae, and 30 °C for B. subtilis. Kanamycin (1 mg ml−1) was used as a reference. All the samples were incubated for 24 h with a given bacterial strain, and absorbance was measured at 600 nm. The data were statistically analysed using a one-way analysis of variance (ANOVA) (the Fischer LSD) (Sigma Stat version 3.5).3 Results and discussion
3.1. Solubility of collagen in DESs
The synthesized DESs, i.e. (i) EG![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) ZnCl2 (4
ZnCl2 (4![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1) and ChCl
1) and ChCl![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) LA (1
LA (1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1), are tested for dissolving collagen (type I) under different conditions. The extent of dissolution of collagen and the yield of the regenerated material in the investigated DESs under different conditions are illustrated in Table 1. Collagen dissolves in both the investigated DESs in appreciable amounts, where the solubility of collagen is found to be in the range of 9.5–22.5 w/v%. At any temperature, in the absence and presence of HCl(aq.), EG
1), are tested for dissolving collagen (type I) under different conditions. The extent of dissolution of collagen and the yield of the regenerated material in the investigated DESs under different conditions are illustrated in Table 1. Collagen dissolves in both the investigated DESs in appreciable amounts, where the solubility of collagen is found to be in the range of 9.5–22.5 w/v%. At any temperature, in the absence and presence of HCl(aq.), EG![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) ZnCl2 shows a higher solubilizing ability towards collagen compared to ChCl
ZnCl2 shows a higher solubilizing ability towards collagen compared to ChCl![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) LA. An increase in temperature results in an increase in the solubility of collagen in both DESs. However, the increase is more pronounced in the case of EG
LA. An increase in temperature results in an increase in the solubility of collagen in both DESs. However, the increase is more pronounced in the case of EG![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) ZnCl2 (∼2.2 mg ml−1 °C−1) compared to that observed in ChCl
ZnCl2 (∼2.2 mg ml−1 °C−1) compared to that observed in ChCl![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) LA (∼1.2 mg ml−1 °C−1) (Fig. S2, ESI†). The presence of HCl(aq.) in the investigated DESs at any of the investigated temperatures enhances the solubility of collagen (Table 1). To have a broader idea about the change in the molecular structure of collagen upon dissolution, followed by regeneration, the solubility of the regenerated material was checked in water at room temperature. Interestingly, some parts of the regenerated material obtained from the dissolution of collagen at 70 °C in the presence of HCl(aq.) in both of the DESs are found to be soluble in water, whereas the entire regenerated material obtained after the dissolution of collagen at 90 °C in the presence and absence of HCl(aq.) is dissolved in water. The details about the role of DESs, temperature and acidic conditions in the dissolution of collagen and modification in the structural arrangement of collagen along with the formation of water-soluble material under certain conditions are thoroughly discussed in the later sections of this paper.
LA (∼1.2 mg ml−1 °C−1) (Fig. S2, ESI†). The presence of HCl(aq.) in the investigated DESs at any of the investigated temperatures enhances the solubility of collagen (Table 1). To have a broader idea about the change in the molecular structure of collagen upon dissolution, followed by regeneration, the solubility of the regenerated material was checked in water at room temperature. Interestingly, some parts of the regenerated material obtained from the dissolution of collagen at 70 °C in the presence of HCl(aq.) in both of the DESs are found to be soluble in water, whereas the entire regenerated material obtained after the dissolution of collagen at 90 °C in the presence and absence of HCl(aq.) is dissolved in water. The details about the role of DESs, temperature and acidic conditions in the dissolution of collagen and modification in the structural arrangement of collagen along with the formation of water-soluble material under certain conditions are thoroughly discussed in the later sections of this paper.
| DESs | 45 °C | 70 °C | 90 °C | ||||||
|---|---|---|---|---|---|---|---|---|---|
| Time (h) | Solubility (mg ml−1) | Yield (mg) | Time (h) | Solubility (mg ml−1) | Yield (mg) | Time (h) | Solubility (mg ml−1) | Yield (mg) | |
ChCl![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) : :![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) LA LA |
4.0 | 95 | 55 | 3.0 | 120 | 90 | 2.0 | 150 | 100 |
EG![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) : :![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) ZnCl2 ZnCl2 |
2.5 | 110 | 70 | 2.0 | 185 | 160 | 1.0 | 210 | 170 |
ChCl![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) : :![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) LA + HCl(aq.) LA + HCl(aq.) |
3.0 | 120 | 90 | 2.0 | 130 | 95 | 1.0 | 195 | 157 |
| EG : ZnCl2 + HCl(aq.) | 2.0 | 166 | 145 | 1.0 | 170 | 155 | 0.5 | 225 | 195 |
3.2. Structural characterization of regenerated material
The collagen is dissolved in different DESs under different conditions of temperature in the presence and absence of HCl(aq.). Therefore, for better understanding, the regenerated material is abbreviated in accordance with the condition as R1-(T), R2-(T) and R1-(T)-A and R2-(T)-A, where R represents regenerated material; 1 and 2 denote ChCl![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) LA (1
LA (1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1) and EG
1) and EG![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) ZnCl2 (4
ZnCl2 (4![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1), respectively; T is the temperature of dissolution; and A corresponds to acidic conditions (in the presence of HCl(aq.)).
1), respectively; T is the temperature of dissolution; and A corresponds to acidic conditions (in the presence of HCl(aq.)).
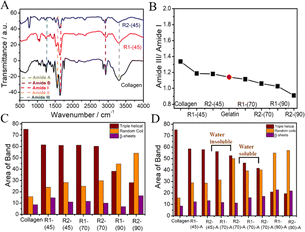
It is deduced that although collagen mainly preserves its structure, some disorders occur in the triple-helical structure of collagen. With an increase in dissolution temperature, the amide-A band (3287 cm−1) is broadened, where the broadening follows the order R1-(45) < R1-(70) < R1-(90) and is more in the case of R2 compared to R1 at all of the investigated temperatures (Fig. S3, ESI†). This suggests an increasing extent of disruption of the H-bonding network of collagen at higher temperatures, which is more in the case of R2 compared to R1. The ratio of the amide-III to amide-I band decreases as temperature increases (Fig. 1B) following the order R1-(45) < R1-(70) < R1-(90), which falls even below that shown by gelatin, a linear polypeptide and hydrolysis product of collagen in the case of R1-(90) and R2-(90).
This indicates the complete loss of 2°-structure and unfolding of collagen at higher temperatures, leading to the formation of random coils and other structures. The heterogeneity in the carbonyl groups and their coupling in stretching modes results in complexity in the amide-I region; thus, it is important to focus on this area to understand the different structural components of collagen. Therefore, deconvolution of the amide-I region is performed to compare the 2°-structure of native collagen and regenerated materials.55 The different structural components in native collagen, such as β-sheets, random coils and triple-helical structures, are centred around 1612 cm−1, 1630 cm−1 and 1664 cm−1, respectively, in the amide-I band.55 The change in band area and position in regenerated material represents the changes in the relative content of 2° structural components under different conditions (Fig. 1C and Table S1, ESI†). In comparison with collagen, R1-(45) shows a ∼15% increase in the content of random coils at the cost of a decrease in the content of β-sheets to a similar extent, which is relatively more in the case of R2-(45), where the content of triple-helical structures remains approximately the same. Relatively more loss in the content of β-sheets is observed in the case of R1-(70) and R2-(70). However, a drastic decrease in the content of the triple-helical structure of collagen from 75% to 40% and 27%, and an increase in the content of random coils from 15% in collagen to 45% and 55% in the case of R1-(90) and R2-(90), respectively, suggests complete disruption of the triple-helical structure of collagen at elevated temperatures. The observance of low molecular weight peptides along with the partially unfolded collagen in the case of R1-(70) and R2-(70) and the peptides only at R1-(90) and R2-(90) supports the above inference (discussed later). At different temperatures, the impact of HCl(aq.) in DESs on the properties of regenerated material is also investigated. No significant change in the characteristic properties of R1-(45)-A and R2-(45)-A is observed, as inferred from FTIR investigations (Fig. S4A, ESI†). Moreover, R1-(70)-A and R2-(70)-A are found to be partially soluble in water, whereas R1-(90)-A and R2-(90)-A are completely soluble in water similar to that observed in the case of R1-(90) and R2-(90).
FTIR spectra of water-insoluble material (from R1-(70)-A and R2-(70)-A) (Fig. S4B, ESI†) resemble those of collagen (Fig. S3A, ESI†). Furthermore, water-soluble material from R1-(70)-A and R2-(70)-A (Fig. S4B, ESI†) and (R1-(90)-A and R2-(90)-A) (Fig. S4C, ESI†), expected to be low molecular weight peptides, exhibits a relatively broadened amide-A band (3290 cm−1), red-shifted amide-III (1650 cm−1) and amide II (1550 cm−1) bands mimicking the bands shown by gelatin (Fig. S3B, ESI†). This suggests the formation of water-soluble polypeptides structurally similar to that of gelatin devoid of any 2°-structure, where the complete loss of triple-helical content is supported by the absence of three bands in the amide-III region (1200–1237 cm−1). No significant difference in the 2°-structural components between R1-(45) and R1-(45)-A is observed from the deconvolution of the amide-I band (Fig. 1D). R1-(70)-A exhibits marginal loss in the triple-helical content at the cost of a similar increase in the content of random coils compared to R1-(70), whereas a significant increase in the content of random coils and loss in the triple-helical structure is observed in the case of R2-(70)-A compared to R2-(70) (Fig. 1D). The presence of HCl(aq.) results in the unwinding of the triple-helical structure of collagen, the effect of which is more at higher temperatures, as suggested by relative values of amide III/amide I ratio (Fig. S4D, ESI†), which is in line with the observations made from deconvolution of the amide-I band (Fig. 1D).
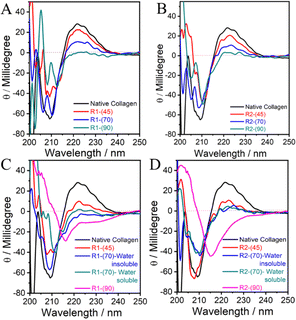 | ||
| Fig. 2 (A–D) CD spectra of native collagen and the material regenerated from collagen dissolved in DESs under different conditions. | ||
The ellipticity of the positive band decreases with an increase in the temperature of dissolution, which almost disappears in the case of R1-(90) and R2-(90) without any discrimination towards DES used (Fig. 2A and B). This indicates the complete disruption of the “polyproline-II” helical structure via the breakage of H-bonds between proline/hydroxy-proline amino acid residues.
A single negative band peculiar to the triple-helical structure in collagen transforms into two narrowly separated bands located at ∼205 and ∼210 nm in the case of R1-(70) and R2-(70), respectively, which further shifts red accompanied by a decrease in ellipticity in the case of R1-(90) and R2-(90) (Fig. 2A and B). The band at ∼205 nm closely resembles that shown by polyproline-II56 with a helical structure, the content of which decreases with an increase in temperature as also suggested by the disappearance of the positive band ∼222 nm at higher temperatures. The appearance of the band around 210 nm, which shifts red at higher temperatures, is assigned to the formation of the β-sheet structure, which may be present in the random coils.57 Further, a red shift suggests the breaking of peptide bonds, which could result in the formation of low molecular weight polypeptides with a structure similar to random coils or unordered peptides. Many unordered polypeptides with different compositions stabilized via hydration show such features in the CD spectra.57,58 The presence of HCl(aq.) does not affect the line shape of both CD bands in the case of R1-(45)-A and R2-(45)-A to an appreciable extent compared to that of collagen (Fig. 2C and D) in line with the observations made from FTIR-spectroscopy. Water insoluble R1-(70)-A retains the shape of a negative CD band, and it shifts towards red, whereas a change is observed in the shape of the CD band in the case of water-insoluble R2-(70)-A, water-soluble R1-(70)-A and R2-(70)-A. This along with the complete loss of a positive CD band in the case of water-soluble and water-insoluble R1-(70)-A and R2-(70)-A (Fig. 2C and D) suggests that the triple-helical structure of collagen is appreciably unfolded at higher dissolution temperatures in the presence of HCl(aq.), resulting in its partial transformation to water soluble low molecular weight polypeptides displaying no characteristic band of “polyproline-II” type helices. The appearance of a symmetric negative band at 215 nm and the complete loss of a positive band at 222 nm specific to the “polyproline-II” helical structure in the case of R1-(90)-A and R2-(90)-A (Fig. 2C and D) reveals the complete disruption of the triple-helical structure of collagen and the formation of β-sheets and random coils. On comparing, it is observed that although there is disruption of the H-bonded network of amino-acid residues in both the “polyproline-II” helices and triple-helical structure in the absence of HCl(aq.). However, the presence of HCl(aq.) completely transforms the collagen into polypeptides with random coils and β-sheet structure.
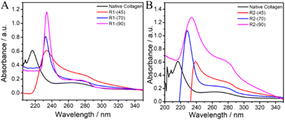 | ||
Fig. 3 UV-vis spectra of native collagen and the material regenerated after dissolution of collagen in (A) ChCl![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) : :![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) LA and (B) EG LA and (B) EG![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) : :![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) ZnCl2 at different temperatures. ZnCl2 at different temperatures. | ||
Generally, the hydrolysis of collagen occurs via the breakdown of amide bonds in polypeptide chains, which results in the appearance of free –NH2 groups on the side chains of peptides, leading to a red shift in the absorption spectra. Similarly, the retention of the absorption bands in the range 240–300 nm suggests no alteration in the molecular structure of aromatic amino acid residues, whereas a red shift indicates the change in the molecular environment of these amino acid residues in regenerated material caused by disruption of the H-bonded network of collagen.
An increase in temperature results in red shift from ∼215 nm to 230 and 240 nm in the material regenerated from ChCl![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) LA and EG
LA and EG![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) ZnCl2, respectively. Besides, an increase in hyperchromicity of the bands (∼215 nm and ∼280 nm) with an increase in temperature for both of the investigated DESs is observed. This depicts the formation of random coil structure and exposure of free –NH2 groups along with unmasking of aromatic amino acid residues (phenylalanine, tyrosine and tryptophan) upon hydrolysis, the extent of which is more in the case of R2 compared to R1.62 This observation is in line with the results obtained from FTIR studies. Further, the absorption band observed at ∼230 nm in the case of material regenerated in the presence of HCl(aq.) after dissolution at 70 °C and 90 °C resembles well the absorbance spectra of gelatin and peptides owing to prominent π–π* transitions (Fig. S5, ESI†). From the observations made from FTIR spectroscopy, CD spectroscopy and UV-vis absorption spectroscopy, it is inferred that there is a negligible effect of HCl(aq.) on the inherent structure and properties of material regenerated after dissolving collagen at 45 °C, whereas the presence of HCl(aq.) and high-temperature synergistically enhances the dissolution of collagen accompanied by the relatively greater unwinding of the triple-helical structure and “polyproline-II” helices, resulting in the formation of water-soluble polypeptides.
ZnCl2, respectively. Besides, an increase in hyperchromicity of the bands (∼215 nm and ∼280 nm) with an increase in temperature for both of the investigated DESs is observed. This depicts the formation of random coil structure and exposure of free –NH2 groups along with unmasking of aromatic amino acid residues (phenylalanine, tyrosine and tryptophan) upon hydrolysis, the extent of which is more in the case of R2 compared to R1.62 This observation is in line with the results obtained from FTIR studies. Further, the absorption band observed at ∼230 nm in the case of material regenerated in the presence of HCl(aq.) after dissolution at 70 °C and 90 °C resembles well the absorbance spectra of gelatin and peptides owing to prominent π–π* transitions (Fig. S5, ESI†). From the observations made from FTIR spectroscopy, CD spectroscopy and UV-vis absorption spectroscopy, it is inferred that there is a negligible effect of HCl(aq.) on the inherent structure and properties of material regenerated after dissolving collagen at 45 °C, whereas the presence of HCl(aq.) and high-temperature synergistically enhances the dissolution of collagen accompanied by the relatively greater unwinding of the triple-helical structure and “polyproline-II” helices, resulting in the formation of water-soluble polypeptides.
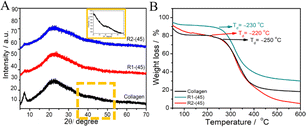 | ||
Fig. 4 (A) X-ray diffraction pattern and (B) TGA profiles of native collagen and the material regenerated from ChCl![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) : :![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) LA and EG LA and EG![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) : :![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) ZnCl2. ZnCl2. | ||
Thermogravimetric analysis (TGA) profiles of the native collagen and R1-(45) and R2-(45) (Fig. 4B) show the relatively lower thermal stability of regenerated material with degradation temperature, Td ∼230 °C, compared to native collagen with Td ∼250 °C. The presence of a relatively strong and greater number of H-bonds and ionic bonds in native collagen compared to those present in regenerated material offers higher thermal stability. The weight loss of native and regenerated collagen as a function of temperature is also examined. A small weight loss of ∼100 °C represents the evaporation of physiosorbed and bound water in native collagen and regenerated material (Fig. 4B). Another weight loss in the temperature ranging from 215 to 420 °C is ascribed to the degradation of side-chain groups of amino acids and thermal decomposition of polypeptide chains or higher molecular weight fractions.63 At a temperature above 420 °C, another slight weight loss occurs in the case of native collagen and R1-(45). This loss ensues from the breakdown of the residual organic components such as derivatives of carboxy-terminal crosslinked telopeptides, helical peptides, or some residues with carbonaceous or nitrogenous content, which are the degradation products of collagen and are non-volatile.64–66 A weight loss of ∼20% in the temperature ranging from 420 °C to 600 °C in the case of R2-(45) is assigned to the decomposition of highly unordered polypeptide chains originated by the partial unfolding of triple-helical structure as suggested by FTIR results. Few changes in the thermal stability of the material regenerated at higher temperatures in the presence and absence of HCl(aq.) (Fig. S8 and S9, ESI†) are observed.
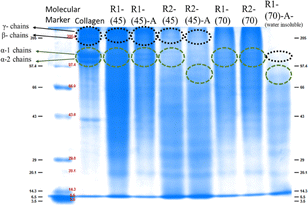 | ||
Fig. 5 SDS-PAGE displaying the molecular weight of the marker used, native collagen and the material regenerated after dissolution of collagen at 45 °C and 70 °C in ChCl![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) : :![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) LA and EG LA and EG![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) : :![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) ZnCl2. ZnCl2. | ||
However, in the case of the regenerated material obtained from collagen dissolved at 90 °C, band positions at lower molecular weight with weak intensity represent the intense fragmentation of collagen molecules with the complete disruption of the triple-helical structure both in the absence and presence of HCl(aq.), leading to the formation of polypeptides in both the employed DESs.
![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) O and –NH2 groups of different amino acid residues of collagen interact with Cl− of ChCl, –OH and –C
O and –NH2 groups of different amino acid residues of collagen interact with Cl− of ChCl, –OH and –C![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) O groups of LA via H-bonding interactions in the case of ChCl
O groups of LA via H-bonding interactions in the case of ChCl![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) LA. Similarly, the –OH groups of EG and Cl− of ZnCl2 form H-bonds with collagen and thus break the amide bonds (Scheme 1) in the case of EG
LA. Similarly, the –OH groups of EG and Cl− of ZnCl2 form H-bonds with collagen and thus break the amide bonds (Scheme 1) in the case of EG![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) ZnCl2. Owing to its Lewis acidity, ZnCl2 activates the –C
ZnCl2. Owing to its Lewis acidity, ZnCl2 activates the –C![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) O group of amino acids, lowering the bond energy and thus making the bond easily breakable. Therefore, a relatively large number of –OH groups in EG and the presence of ZnCl2 in EG
O group of amino acids, lowering the bond energy and thus making the bond easily breakable. Therefore, a relatively large number of –OH groups in EG and the presence of ZnCl2 in EG![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) ZnCl2 DES help in greater solubilization of collagen44 at a faster rate compared to that observed in the case of ChCl
ZnCl2 DES help in greater solubilization of collagen44 at a faster rate compared to that observed in the case of ChCl![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) LA. The solubility of collagen increases as temperature increases, which is more pronounced at higher temperatures in both DESs (Table 1).
LA. The solubility of collagen increases as temperature increases, which is more pronounced at higher temperatures in both DESs (Table 1).
The thermal energy offered by high temperatures naturally lowers the activation energy of dissolution via the greater breakage of H-bonds between collagen fibrils. This results in the unfolding of a triple-helical structure as suggested by FTIR, CD and UV-vis absorption measurements and consequently offers more sites for components of DESs to engage in H-bonding with collagen in a synergistic manner and thus enhances the solubility. The solubility of collagen is also observed to increase under mild acidic conditions (Table 1). HCl(aq.) is expected to aid in the breaking of amide bonds in the polypeptides comprising collagen, leading to the appearance of low molecular weight polypeptides. Excess H+ interacts with the imino of proline and hydroxyproline of collagen, resulting in the formation of ammonium salt, which further attracts more of Cl− of DESs as well as of HCl, thus weakening inter- and intra-molecular H-bonding of collagen. An increase in temperature results in greater ionization of HCl in DESs; consequently, relatively more H+ and Cl− interacts with amino acid residues of collagen, leading to the breaking of H-bonds.16 The protonation of the –NH2 group of amino acid residues of collagen favours dissolution by offering electrostatic repulsion between similarly charged groups and facilitating the breakage of the H-bonding network in collagen at the cost of establishing H-bonding with Cl− present in DESs. Comparing the effects of HCl(aq.) and temperature, it is observed that HCl(aq.) does not affect the extent of solubilization of collagen and properties of R1-(45)-A, R2-(45)-A and R1-(70)-A, whereas the effect is more noticeable in the case of R2-(70)-A. Moreover, the structural alterations caused by collagen during dissolution are found to be comparable in the case of material regenerated at 90 °C in both DESs in the presence of HCl(aq.). To ascertain the formation of low molecular weight peptides, especially at higher temperatures and in the presence of HCl(aq.) as suggested by SDS-PAGE measurements, the solubility of the recovered material was examined. It is observed that the R1-(45), R2-(45), R1-(45)-A and R2-(45)-A are not water-soluble, whereas a part of the R1-(70)-A and R2-(70)-A is found to be water soluble. However, the material regenerated after dissolution at 90 °C is spontaneously dissolved in water in both the presence and absence of HCl(aq.). This led to the conclusion that the presence of HCl(aq.) as well as a high dissolution temperature enhances the solubilization of collagen and breaks the collagen into low molecular weight peptides. This happens via the enhanced ionization of HCl(aq.), which in turn hydrolyse the collagen into water-soluble peptides with ionized amino acid residues available to be hydrated by water. During the regeneration process, DESs are washed away, and the H-bonding can be partially restored between the amino acids of collagen chains. Nevertheless, the regenerated material does not have the same amount and location of H-bonds as the native collagen. This results in variations in the structure and properties of regenerated material in comparison to native collagen.
![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) LA (ChCl: –OH(str.) ∼3400–3500 cm−1, C–H(str.) ∼2800 cm−1 of the tertiary amine group, C–N(str.) and C–H(bending) in the range of 1300–1000 cm−1, and LA: C
LA (ChCl: –OH(str.) ∼3400–3500 cm−1, C–H(str.) ∼2800 cm−1 of the tertiary amine group, C–N(str.) and C–H(bending) in the range of 1300–1000 cm−1, and LA: C![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) O(str.) in the range of 1700–1725 cm−1, and the C–O(str.) in the range of 1000–1300 cm−1). Most of the significant bands ∼3000–3500 cm−1 related to –OH(str.) and bands ∼2800–2950 cm−1 corresponding to C–H(sym.) and C–H(asym.) methylene group stretching are also retained upon recycling EG
O(str.) in the range of 1700–1725 cm−1, and the C–O(str.) in the range of 1000–1300 cm−1). Most of the significant bands ∼3000–3500 cm−1 related to –OH(str.) and bands ∼2800–2950 cm−1 corresponding to C–H(sym.) and C–H(asym.) methylene group stretching are also retained upon recycling EG![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) ZnCl2 DES.
ZnCl2 DES.
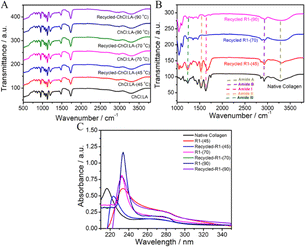
Further, the material regenerated after dissolution of collagen in recycled DESs displays bands similar to those observed in the case of materials regenerated from virgin DESs (Fig. 6B and S13, ESI†). A similar shift in amide bands in comparison to native collagen is observed in samples obtained from recycled DESs. Furthermore, the UV-vis absorbance spectra of regenerated material from recycled DESs (Fig. 6C) are observed to have similar bands at ∼210–230 nm and ∼270–300 nm as those observed for the material regenerated from the original DESs. A similar trend is observed in recycled DESs in the presence of HCl(aq.) (Fig. S14, ESI†). A comparable bathochromic and hyperchromic shift is also observed. This demonstrates the recyclable nature of DESs, which contributes to the sustainability of the dissolution process.
| Sample | Escherichia coli | Pseudomonas syringae | Bacillus subtilis | Staphylococcus aureus |
|---|---|---|---|---|
| a Data represent the mean ± SE of three independent biological replicates. Different letters (a–n) within the column represent values that were significantly different among different samples (Fisher LSD; p ≤ 0.05). | ||||
| Buffer | 2.69 ± 0.13g | 10.41 ± 0.54i | 6.16 ± 0.28j | 4.88 ± 0.62n |
| Kanamycin | 86.02 ± 1.30a | 90.58 ± 1.73a | 83.21 ± 0.56b | 91.95 ± 0.26a |
| Native | 40.30 ± 0.39f | 12.54 ± 0.42h | 27.95 ± 1.03i | 13.53 ± 0.92m |
| R1-(45) | 73.41 ± 0.79d | 81.80 ± 0.36c | 71.27 ± 0.58f | 85.39 ± 0.83c |
| R2-(45) | 77.33 ± 1.12b | 81.12 ± 0.28c | 62.61 ± 0.21h | 87.35 ± 0.37b |
| R1-(45)-A | 75.44 ± 1.13c | 86.46 ± 0.32b | 77.17 ± 0.53d | 54.33 ± 1.31l |
| R2-(45)-A | 85.86 ± 1.41a | 59.23 ± 0.28f | 80.03 ± 0.59c | 76.70 ± 1.60g |
| R1-(70) | 73.52 ± 1.29d | 70.09 ± 0.34e | 74.88 ± 0.39e | 66.42 ± 0.94i |
| R2-(70) | 66.98 ± 1.37e | 80.09 ± 0.37c | 62.57 ± 0.20h | 56.14 ± 0.52k |
| R1-(70)-A-water insoluble | 86.66 ± 1.30a | 82.89 ± 0.46c | 65.50 ± 0.50g | 77.22 ± 0.67f |
| R2-(70)-A-water insoluble | 86.75 ± 0.92a | 87.51 ± 0.46b | 87.76 ± 0.47a | 91.86 ± 0.58a |
| R1-(70)-A-water soluble | 85.94 ± 1.39a | 86.18 ± 0.45b | 82.60 ± 0.38b | 80.84 ± 0.65e |
| R2-(70)-A-water soluble | 86.82 ± 1.45a | 86.85 ± 0.61b | 65.66 ± 0.27g | 78.15 ± 0.44f |
| R1-(90) | 75.26 ± 1.83c | 71.67 ± 0.73e | 75.32 ± 0.39e | 69.27 ± 0.33h |
| R1-(90) | 76.81 ± 1.82c | 50.43 ± 0.74g | 78.60 ± 0.39d | 59.58 ± 0.94j |
| R1-(90)-A | 85.79 ± 1.38a | 76.97 ± 1.03d | 76.23 ± 0.51e | 82.00 ± 0.13d |
| R2-(90)-A | 78.42 ± 1.07b | 86.67 ± 0.84b | 85.31 ± 0.90a | 86.88 ± 0.42b |
Such positively charged peptides undergo electrostatic interactions with the negatively charged bacterial membranes.13,50 Following this, the aromatic amino acid residues of collagen and collagen peptides are exposed towards solvent upon unfolding during dissolution, especially at high temperatures, and the presence of HCl(aq.) facilitates the anchoring of such hydrophobic amino acid residues to the hydrophobic lipid core of the bacterial membrane.71 According to the barrel-stave model, small cationic peptides adsorb on the surface of bacteria and their hydrophobic amino acid groups are embedded into the membrane, resulting in the formation of pores.72
These pores block bacterial functioning by interacting with their DNAs and RNAs. Further, the efficient antimicrobial activity shown by water soluble R1-(70)-A and R2-(70)-A compared to that of R1-(90), R2-(90), R1-(90)-A and R2-(90)-A can correlate with the decreasing number of peptides in the given fraction with an increase in temperature as evidenced by SDS PAGE as a higher number of collagen peptides are considered to exhibit better antimicrobial properties.13
4 Conclusions
A simple, sustainable, and robust method for the dissolution and regeneration of type I collagen in DESs, along with its transformation to water soluble collagen peptides, is established. DESs, i.e. ChCl![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) LA (1
LA (1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1) and EG
1) and EG![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) ZnCl2 (4
ZnCl2 (4![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1), have shown remarkable dissolving ability towards collagen, which increases with an increase in the temperature of dissolution. EG
1), have shown remarkable dissolving ability towards collagen, which increases with an increase in the temperature of dissolution. EG![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) ZnCl2 is more effective in dissolving collagen than ChCl
ZnCl2 is more effective in dissolving collagen than ChCl![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) LA at any investigated temperature. The presence of [HCl(aq.)] = 5 × 10−5 M increases the solubility of collagen by 2–5 w/v% depending on the nature of DES and temperature although no change in pH occurs. Collagen is dissolved in DESs following alterations in the 2°-structure, where a higher dissolution temperature and the presence of H+ result in greater unwinding of the triple-helical structure of collagen. Alterations in the 2°-structure accompanied by cleavage of peptide bonds lead to the formation of low molecular weight collagen peptides that are readily soluble in water. The regenerated material shows good antimicrobial activity towards both Gram-positive and Gram-negative bacteria, which is even more than that shown by the standard antibiotic Kanamycin, especially in the case of collagen peptides. After regeneration of the dissolved collagen, the recovered DESs show no structural alterations compared to native DESs and are reused for the dissolution of collagen. Along with the previous studies on the extraction of collagen using DESs,39,40 the present work would pave a platform for the dissolution and stabilization of other biologically important polymers in DESs with new implications for human health and food safety.
LA at any investigated temperature. The presence of [HCl(aq.)] = 5 × 10−5 M increases the solubility of collagen by 2–5 w/v% depending on the nature of DES and temperature although no change in pH occurs. Collagen is dissolved in DESs following alterations in the 2°-structure, where a higher dissolution temperature and the presence of H+ result in greater unwinding of the triple-helical structure of collagen. Alterations in the 2°-structure accompanied by cleavage of peptide bonds lead to the formation of low molecular weight collagen peptides that are readily soluble in water. The regenerated material shows good antimicrobial activity towards both Gram-positive and Gram-negative bacteria, which is even more than that shown by the standard antibiotic Kanamycin, especially in the case of collagen peptides. After regeneration of the dissolved collagen, the recovered DESs show no structural alterations compared to native DESs and are reused for the dissolution of collagen. Along with the previous studies on the extraction of collagen using DESs,39,40 the present work would pave a platform for the dissolution and stabilization of other biologically important polymers in DESs with new implications for human health and food safety.
Data availability
The data will be made available on request.Author contributions
Harmandeep Kaur (experimentation, data analysis and drafting of manuscript); Navdeep Kaur (antimicrobial activity and concerned data analysis); Pratap Kumar Pati (data analysis, supervision); Monika Rani (SDS-PAGE); Tejwant Singh Kang (conceptualization, supervision, data analysis and manuscript writing). All authors have given approval to the final version of the manuscript.Conflicts of interest
There are no conflicts to declare.Acknowledgements
This work was supported by CSIR, Govt of India wide project number 01(3018)/21/ EMR-II and DST-SERB (CRG/2021/005897). The infrastructure provided for this work under the UPE grant and UGC-CAS (Centre for Advanced Studies) program is highly acknowledged. H. K. is thankful to CSIR, Govt of India, M. S. is thankful to UGC, Govt of India, N. K. is thankful to DST-Woman Scientist-A (WOS-A) for fellowship.References
- S. Noorzai, C. J. R. Verbeek, M. C. Lay and J. Swan, Waste Biomass Valorization, 2020, 11, 5687–5698 CrossRef CAS.
- A. León-López, A. Morales-Peñaloza, V. M. Martínez-Juárez, A. Vargas-Torres, D. I. Zeugolis and G. Aguirre-Álvarez, Molecules, 2019, 24, 4031–4047 CrossRef PubMed.
- S. W. Chang, S. J. Shefelbine and M. J. Buehler, Biophys. J., 2012, 102(3), 640–648 CrossRef CAS PubMed.
- J. Lei, B. Zou, R. Zhang, K. Zhang, R. Xie, W. Zhang, J. Wu, S. Li, B. Zheng and F. Huo, J. Leather Sci. Eng., 2019, 1(1), 1–9 CrossRef.
- N. O. Metreveli, K. K. Jariashvili, L. O. Namicheishvili, D. V. Svintradze, E. N. Chikvaidze, A. Sionkowska and J. Skopinska, Ecotoxicol. Environ. Saf., 2010, 73, 448–455 CrossRef CAS PubMed.
- G. C. Na, Top. Catal., 1988, 8, 315–330 CAS.
- T. Miyahara, A. Murai, T. Tanaka, S. Shiozawa and M. Kameyama, J. Gerontol., 1982, 37, 651–655 CrossRef CAS PubMed.
- M. D. Shoulders and R. T. Raines, Annu. Rev. Biochem., 2009, 78, 929–958 CrossRef CAS PubMed.
- N. Rajan, J. Habermehl, M. F. Coté, C. J. Doillon and D. Mantovani, Nat. Protoc., 2007, 1, 2753–2758 CrossRef PubMed.
- P. Mokrejs, F. Langmaier, M. Mladek, D. Janacova, K. Kolomaznik and V. Vasek, Waste Manage. Res., 2009, 27, 31–37 CrossRef CAS PubMed.
- D. Liu, M. Nikoo, G. Boran, P. Zhou and J. M. Regenstein, Annu. Rev. Food Sci. Technol., 2015, 6, 527–557 CrossRef CAS PubMed.
- G. Singh, G. Singh, K. Damarla, P. K. Sharma, A. Kumar and T. S. Kang, ACS Sustain. Chem. Eng., 2017, 5, 6568–6577 CrossRef CAS.
- M. C. Gomez-Guillen, B. Gimenez, M. E. Lopez-Caballero and M. P. Montero, Food Hydrocolloids, 2011, 25, 1813–1827 CrossRef CAS.
- J. Ao and B. Li, Food Sci. Technol. Int., 2012, 18, 425–434 CrossRef CAS PubMed.
- D. Dumitriu and T. Dobre, J. Endod., 2015, 41, 903–906 CrossRef PubMed.
- P. Qi, Y. Zhou, D. Wang, Z. He and Z. Li, RSC Adv., 2015, 5, 87180–87186 RSC.
- K. Jariashvili, B. Madhan, B. Brodsky, A. Kuchava, L. Namicheishvili and N. Metreveli, Biopolymers, 2012, 97, 189–198 CrossRef CAS PubMed.
- Y. Hu, L. Liu, W. Dan, N. Dan and Z. Gu, J. Appl. Polym. Sci., 2013, 130, 2245–2256 CrossRef CAS.
- Z. Meng, X. Zheng, K. Tang, J. Liu, Z. Ma and Q. Zhao, Int. J. Biol. Macromol., 2012, 51, 440–448 CrossRef CAS PubMed.
- Z. Lei, B. Chen, Y. M. Koo and D. R. Macfarlane, Chem. Rev., 2017, 117, 6633–6635 CrossRef PubMed.
- C. Mukesh, D. Mondal, M. Sharma and K. Prasad, Chem. Commun., 2013, 49, 6849–6851 RSC.
- J. S. Wilkes, Green Chem., 2002, 4, 73–80 RSC.
- A. Tarannum, A. Adams, B. Blümich and N. N. Fathima, J. Phys. Chem. B, 2018, 122, 1060–1065 CrossRef CAS PubMed.
- A. Tarannum, R. R. Jonnalagadda and N. F. Nishter, Spectrochim. Acta, Part A, 2019, 212, 343–348 CrossRef CAS PubMed.
- H. Zhang, J. Wu, J. Zhang and J. He, Macromolecules, 2005, 38, 8272–8277 CrossRef CAS.
- K. Singh, S. Mehra and A. Kumar, Green Chem., 2022, 24, 9629–9642 RSC.
- S. Liu, Q. Li and G. Li, J. Leather Sci. Eng., 2019, 1, 1–12 CrossRef.
- B. Soares, A. M. da Costa Lopes, A. J. Silvestre, P. C. R. Pinto, C. S. Freire and J. A. Coutinho, Ind. Crops Prod., 2021, 160, 113128 CrossRef CAS.
- H. Garcia, R. Ferreira, M. Petkovic, J. L. Ferguson, M. C. Leitão, H. N. Gunaratne, R. Seddonab Kenneth, N. Rebelo Luís Paulo and C. S. Pereira, Green Chem., 2010, 12(3), 367–369 RSC.
- A. Idris, R. Vijayaraghavan, U. A. Rana, A. F. Patti and D. R. Macfarlane, Green Chem., 2014, 16, 2857–2864 RSC.
- D. M. Phillips, L. F. Drummy, D. G. Conrady, D. M. Fox, R. R. Naik, M. O. Stone, P. C. Trulove, H. C. De Long and R. A. Mantz, J. Am. Chem. Soc., 2004, 126, 14350–14351 CrossRef CAS PubMed.
- E. L. Smith, A. P. Abbott and K. S. Ryder, Chem. Rev., 2014, 114, 11060–11082 CrossRef CAS PubMed.
- A. Abo-Hamad, M. Hayyan, M. A. H. AlSaadi and M. A. Hashim, J. Chem. Eng., 2015, 273, 551–567 CrossRef CAS.
- Y. Dai, J. van Spronsen, G. J. Witkamp, R. Verpoorte and Y. H. Choi, Anal. Chim. Acta, 2013, 766, 61–68 CrossRef CAS PubMed.
- D. Carriazo, M. C. Serrano, M. C. Gutiérrez, M. L. Ferrer and F. del Monte, Chem. Soc. Rev., 2012, 41, 4996–5014 RSC.
- A. S. Khan, T. H. Ibrahim, N. A. Jabbar, M. I. Khamis, P. Nancarrow and F. S. Mjalli, RSC Adv., 2021, 11, 12398–12422 RSC.
- E. M. Nuutinen, P. Willberg-Keyriläinen, T. Virtanen, A. Mija, L. Kuutti, R. Lantto and A. S. Jääskeläinen, RSC Adv., 2019, 9, 19720–19728 RSC.
- D. Wang, X. H. Yang, R. C. Tang and F. Yao, Polymers, 2018, 10(9), 993 CrossRef PubMed.
- C. Bai, Q. Wei and X. Ren, ACS Sustain. Chem. Eng., 2017, 5, 7220–7227 CrossRef CAS.
- M. Bisht, M. Martins, A. C. R. V. Dias, S. P. M. Ventura and J. A. P. Coutinho, Green Chem., 2021, 23, 8940–8948 RSC.
- H. Zhang, J. Lang, P. Lan, H. Yang, J. Lu and Z. Wang, Mater. Polym., 2018, 10(9), 993 Search PubMed.
- Q. Wang, X. Yao, Y. Geng, Q. Zhou, X. Lu and S. Zhang, Green Chem., 2015, 17, 2473–2479 RSC.
- L. Zhou, X. Lu, Z. Ju, B. Liu, H. Yao, J. Xu, Q. Zhou, Y. Hu and S. Zhang, Green Chem., 2019, 21, 897–906 RSC.
- H. Kaur, M. Singh, H. Singh, M. Kaur, G. Singh, K. Sekar and T. S. Kang, Green Chem., 2022, 24, 2953–2961 RSC.
- H. Kaur, M. Singh, K. Singh, A. Kumar and T. S. Kang, Green Chem., 2023, 25, 5172–5181 RSC.
- X. Tan, Y. Wang, W. Du and T. Mu, ChemSusChem, 2020, 13, 321–327 CrossRef CAS PubMed.
- X. Tan, W. Zhao and T. Mu, Green Chem., 2018, 20, 3625–3633 RSC.
- P. Kalhor, K. Ghandi, H. Ashraf and Z. Yu, Phys. Chem. Chem. Phys., 2021, 23, 13136–13147 RSC.
- R. Alcalde, A. Gutiérrez, M. Atilhan and S. Aparicio, J. Mol. Liq., 2019, 290, 110916 CrossRef CAS.
- N. Ennaas, R. Hammami, A. Gomaa, F. Bédard, É. Biron, M. Subirade, L. Beaulieu and I. Fliss, Biochem. Biophys. Res. Commun., 2016, 473, 642–647 CrossRef CAS PubMed.
- B. De Campos Vidal and M. L. S. Mello, Micron, 2011, 42, 283–289 CrossRef PubMed.
- M. G. Bridelli, in Fourier Transforms – HigH-Tech Application and Current Trends, InTech, 2017 Search PubMed.
- S. Y. Bak, S. W. Lee, C. H. Choi and H. W. Kim, Materials, 2018, 11(12), 2518 CrossRef PubMed.
- M. Vedhanayagam, S. Anandasadagopan, B. U. Nair and K. J. Sreeram, Mater. Sci. Eng., C, 2020, 108, 110378 CrossRef CAS PubMed.
- K. J. Payne and A. Veis, Biopolymers, 1988, 27(11), 1749–1760 CrossRef CAS PubMed.
- V. Gauba and J. D. Hartgerink, J. Am. Chem. Soc., 2007, 129, 2683–2690 CrossRef CAS PubMed.
- N. J. Greenfield, Nat. Protoc., 2006, 1(6), 2876–2890 CrossRef CAS PubMed.
- R. W. Woody, J. Am. Chem. Soc., 2009, 131, 8234–8245 CrossRef CAS PubMed.
- N. T. Chinh, V. Q. Manh, V. Q. Trung, T. D. Lam, M. D. Huynh, N. Q. Tung, N. D. Trinh and T. Hoang, Nat. Prod. Commun., 2019, 14(7), 7–19 CrossRef.
- F. Zsila, Anal. Biochem., 2022, 639, 114512 CrossRef CAS PubMed.
- Q. Y. Han, T. Koyama, S. Watabe, Y. Nagashima and S. Ishizaki, Molecules, 2023, 28(2), 889 CrossRef CAS PubMed.
- D. Li, C. Mu, S. Cai and W. Lin, Ultrason. Sonochem., 2009, 16, 605–609 CrossRef CAS PubMed.
- F. Zhang, A. Wang, Z. Li, S. He and L. Shao, Food Nutr. Sci., 2011, 02, 818–823 CAS.
- M. Ahmad, N. P. Nirmal and J. Chuprom, RSC Adv., 2016, 6, 33868–33879 RSC.
- A. Sionkowska, J. Skopinska-Wisniewska, M. Gawron, J. Kozlowska and A. Planecka, Int. J. Biol. Macromol., 2010, 47, 570–577 CrossRef CAS PubMed.
- D. Kathyayani, B. Mahesh, N. A. Chamaraja, B. S. Madhukar and D. C. Gowda, Colloids Surf., A, 2022, 649, 129503 CrossRef CAS.
- C. Hermida-Merino, D. Cabaleiro, C. Gracia-Fernández, J. Valcarcel, J. A. Vázquez, N. Sanz, M. Pérez-Rodríguez, M. Arenas-Moreira, D. Banerjee, A. Longo, C. Moya-Lopez, L. Lugo, P. Bourson, A. B. Pereiro, G. Salloum-Abou-Jaoude, I. Bravo, M. M. Piñeiro and D. Hermida-Merino, Gels, 2022, 8(9), 594 CrossRef CAS PubMed.
- O. S. Rabotyagova, P. Cebe and D. L. Kaplan, Mater. Sci. Eng., C, 2008, 28(8), 1420–1429 CrossRef CAS PubMed.
- J. Wu, Z. Li, X. Yuan, P. Wang, Y. Liu and H. Wang, Trans. Tianjin Univ., 2011, 17, 111–117 CrossRef CAS.
- G.-G. Mc, L.-C. Me, A. A. López, L. A. Giménez and B. Montero, Sea By-Products as a Real Material: New Ways of Application, 2010, vol. 37, pp. 89–115 Search PubMed.
- N. Ennaas, R. Hammami, L. Beaulieu and I. Fliss, Biochem. Biophys. Res. Commun., 2015, 462, 195–200 CrossRef CAS PubMed.
- J. Lei, L. Sun, S. Huang, C. Zhu, P. Li, J. He, V. Mackey, D. H. Coy and Q. He, Am. J. Transl. Res., 2019, 11(7), 3919 CAS.
Footnote |
| † Electronic supplementary information (ESI) available. See DOI: https://doi.org/10.1039/d4su00122b |
| This journal is © The Royal Society of Chemistry 2024 |