Open Access Article
Open Access ArticleImplications of intracrystalline OC17 on the protection of lattice incorporated proteins†
Huseyin Burak
Caliskan‡
 *ab and
Fatma Isik
Ustok
c
*ab and
Fatma Isik
Ustok
c
aUniversity of Cambridge, Department of Engineering, Trumpington Street, CB2 1PZ Cambridge, UK. E-mail: huseyinburakcaliskan@etu.edu.tr
bUniversity of Cambridge, The Nanoscience Centre, 11 JJ Thomson Avenue, CB3 0FF Cambridge, UK
cUniversity of Cambridge, Cambridge Institute for Medical Research, Department of Haematology, The Keith Peters Building, Hills Road, CB2 0XY Cambridge, UK
First published on 29th May 2024
Abstract
Biogenic CaCO3 formation is regulated by crystallization proteins during crystal growth. Interactions of proteins with nascent mineral surfaces trigger proteins to be incorporated into the crystal lattice. As a result of incorporation, these intracrystalline proteins are protected in the lattice, an example of which is ancient eggshell proteins that have persisted in CaCO3 for thousands of years even under harsh environmental conditions. OC17 is an eggshell protein known to interact with CaCO3 during eggshell formation during which OC17 becomes incorporated into the lattice. Understanding protein incorporation into CaCO3 could offer insights into protein stability inside crystals. Here, we study the protection of OC17 in the CaCO3 lattice. Using thermogravimetric analysis we show that the effect of temperature on intracrystalline proteins of eggshells is negligible below 250 °C. Next, we show that lattice incorporation protects the OC17 structure despite a heat-treatment step that is shown to denature the protein. Because incorporated proteins need to be released from crystals, we verify metal chelation as a safe crystal dissolution method to avoid protein denaturation during reconstitution. Finally, we optimize the recombinant expression of OC17 which could allow engineering OC17 for engineered intracrystalline entrapment studies.
Introduction
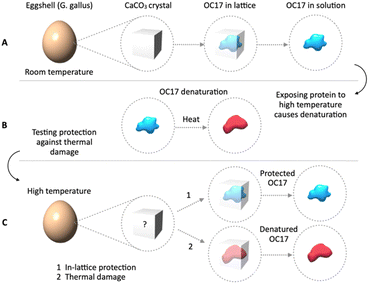
Ovocleidin-17 (OC17) is an eggshell protein whose role is thought to regulate CaCO3 crystal growth during shell formation.1 Simulations suggest that during the formation of an eggshell, OC17 acts like an enzyme to catalyse the amorphous-to-calcite transition.1–3 The existence of an initial amorphous phase in eggshells has previously been verified.4–6 The interaction of OC17 with nascent CaCO3 crystals is in line with the fact that it is a member of calcium-dependent type (C-type) lectin-like protein family.7–11 However, unlike other C-type lectins, which interact with a variety of carbohydrates,12,13 OC17 interacts with inorganic CaCO3, and as a result of interaction, OC17 becomes incorporated into the growing CaCO3 lattice.14,15 Incorporation into the lattice isolates the protein from the environment. A potential result of incorporation is the protection of the incorporated protein against thermal damage at temperatures that would degrade the protein if it were in solution (Fig. 1A and B). Because CaCO3 is a low-cost, non-hazardous material which is synthesized at room temperature (RT), in-lattice protection of proteins in CaCO3 crystals could address the temperature-sensitivity problem of protein-based therapeutics. The stability of proteins at high temperatures is particularly problematic in resource-limited settings where keeping a reliable cold-chain is often challenging. Incorporation of biomolecules in biogenic CaCO3 has been known for decades in the context of wild-type proteins in fossil samples.16–19 Peptide fragments from incorporated proteins are protected for eons in the CaCO3 lattice for which remarkable examples have been documented lately.20,21 Incorporated proteins store valuable information which is utilized for amino acid racemization geochronology, a method for dating archaeological samples.22,23 In recent years, there is growing interest in incorporation of molecules into CaCO3 for other practical purposes, for instance, for organic–inorganic materials24 or for targeted release of drugs on demand.25 Incorporation of a variety of molecules in CaCO3 for bioinspired materials has also been reported.26–28 Importantly, the effects of incorporation on lattice parameters have been thoroughly studied.29–37 There is interest in loading small molecule drugs in crystals for protection under harsh conditions.38 Nanoparticles29,30,38–40 and polymers41–45 have been incorporated, while amino acids have been used to assist in the incorporation of targets46 or to investigate the effect of incorporation on lattice strain.29,32–37 GFP-perlucin fusion47 and cowpea mosaic virus48 have been incorporated in CaCO3. A recent citizen science project also discussed the influence of amino acids and related additives on calcite and vaterite.49 Nonetheless, research on protein incorporation in CaCO3 specifically for high-temperature protection has not attracted enough attention. There is also a gap in our knowledge about whether incorporated proteins in CaCO3 are structurally intact after exposure to high temperatures or after reconstitution from lattice using dissolution agents. It could be insightful to probe intracrystalline proteins for temperature stability. Studying natural intracrystalline proteins may provide a method to address temperature-sensitivity of therapeutics.Here, we probe the effect of heat on intracrystalline proteins of eggshells, particularly focusing on OC17 to investigate whether lattice incorporation protects the protein against thermal damage (Fig. 1C). In order to avoid using harsh reconstitution solutions, we compare acid dissolution and metal chelation to optimize protein release from crystals. Then, we probe secondary and tertiary structures of a model protein (BSA) after incubation in a metal chelating agent which we show is convenient for reconstitution. Although wild-type OC17 could be extracted from eggshells, it is also desirable to recombinantly express it which would enable engineering OC17 not only for expressing fusion proteins (e.g. for assistive incorporation, such as GFP-OC17) but also for probing the function of OC17, which is yet to be demonstrated. For these reasons, we optimize conditions for recombinant expression of OC17. We also show an alternative method of wild-type OC17 extraction which is beset with the existence of intracrystalline lysozymes.50 Studying natural intracrystalline proteins such as OC17, may help address the temperature-sensitivity problem of protein-based therapeutics. Structural studies probing intracrystalline proteins that are exposed to high ambient temperatures might be beneficial for developing nature-inspired methods to protect, store and deliver proteins outside the cold chain.
Results and discussion
Thermogravimetric analysis (TGA) of eggshell intracrystalline fraction
Thermal degradation of intracrystalline fraction can be probed using TGA which allows quantifying the amount of incorporated material. Because CaCO3 melts at significantly higher temperatures compared to the degradation of biogenic intracrystalline fraction, the percentage of total incorporated content in CaCO3 could be determined. In addition, the temperature at which a substantial weight change occurs is thought to be correlated with the effect of heat on intracrystalline proteins. We studied eggshells to investigate if protein degradation could be inferred during a heat-treatment step in a typical TGA experiment. As a negative control, we used commercial CaCO3 powder. The results are shown in Fig. 2. The weight of commercial CaCO3 did not change from 100 °C up to 500 °C which shows that the difference between pure CaCO3 and eggshell arises from incorporated organic fraction in eggshells. From 100 °C to 300 °C, there is a slight decrease in the weight of the eggshell samples which is thought to be either loosely bound H2O in crystals51 or amorphous CaCO352 which may be present in varying amounts in eggshells. The TGA data suggest that up to 250 °C, intracrystalline proteins are largely unaffected by heat or protein degradation is very low to be detected through weight change. Starting roughly from 250 °C, the observed decrease in weight is most likely related to protein integrity. This change is thought to arise from CO2 release from the organic content.53 It is important to note that changes in the host lattice arising from protein degradation should be taken into account on which there is a significant body of work.29–37 After 400 °C, the rate of weight loss slightly decreases, though we did not continue analysis above 500 °C after which unrelated changes such as CaCO3 transition into CaO,54 would interfere the protein-focused analysis. The fact that purified proteins in solution degrade at much lower temperatures than 250 °C suggests that intracrystalline proteins may be protected against thermal damage as long as they are incorporated into the CaCO3 lattice. Because the focus of this study is to see whether lattice incorporation might be a viable method for protein protection as described above, it might be interesting to compare the protection of intracrystalline lysozyme in eggshells with in vitro incorporated lysozyme. In fact, in vitro incorporated lysozyme has recently been shown to resist high temperature damage in crystals.38 To avoid diverging from the main topic, we focused solely on OC17 to see if a natural intracrystalline protein is protected in the CaCO3 lattice rather than comparing in vivo versus in vitro incorporation.Purification and identification of OC17 for subsequent analyses
Protocols for purification of wild-type OC17 from eggshells have been reported along with the crystal structure of the protein,9 however, the protocols10 require organic solvents (e.g. acetonitrile and trifluoroacetic acid in the HPLC step) which might denature the protein. Instead of using harsh purification media, we used a batch extraction method to purify intact, properly folded OC17 for subsequent heat-treatment tests. Eggshell membranes and surface-bound chemicals (e.g. cuticle, natural dye molecules, etc.) were removed in an EDTA washing step and insoluble materials were filtered. Then, eggshells were ground into powder and dissolved in 5% acetic acid.55,56 After an initial buffer exchange to remove acetic acid in a CM Sepharose column, step-wise pH gradient was used to elute OC17, OC2357 (glycosylated form of OC17) and lysozyme (Fig. 3A). We did not seek further purification of contaminant proteins as their concentrations are negligible as seen in the gel image (Fig. 3B). We verified the proteins using peptide fingerprinting by LC-MS on gel-excised samples corresponding to OC17 and lysozyme. The high coverage of amino acid sequences for OC1758 and lysozyme clearly identifies each protein (Fig. 3C). It is worth noting that removing lysozyme from OC17 is non-trivial, however, it could be removed in a number of ways. Previous protocols utilized size-exclusion steps in tandem to remove lysozyme and OC17.10 We developed an alternative method using a protein–protein interaction between lysozyme and ivy protein. Ivy had likely evolved in certain bacteria as a defence mechanism against lysozyme.59 Ivy binds lysozyme with high affinity which disarms lysozyme's enzymatic function and protects the integrity of the bacterial cell wall.60 We recombinantly tagged ivy with histidine to bind lysozyme to the column through his-tagged ivy which allows eluting OC17 in a single step.Probing lattice protection of OC17 at high temperatures
We investigated the effect of heat on intracrystalline OC17 by comparing the protein extracted from room temperature-stored eggshells to the protein extracted from the heat-treated eggshell. In other words, intracrystalline protein was first extracted from eggshells and then heat-treated in solution for analysis. For comparison, the eggshell was first heat-treated and then extraction of the protein was carried out from this heat-treated sample. In this way, the protective effect of lattice incorporation could be studied. For this reason, we first extracted OC17 from room temperature-stored eggshells and used circular dichroism (CD) for structural analysis (red line in Fig. 4). Then, the OC17 solution was heat-treated at 50 °C for 60 h. After treatment, the same sample was analysed again to see the effect of heat (black line in Fig. 4). Heat-treated OC17 undergoes a structural change because of heat denaturation. To probe the effect of heat on intracrystalline OC17, we first heat-treated eggshells at 50 °C for 60 h and after heat-treatment, we extracted OC17 and analysed its structure. The CD result shows that OC17 from heat-treated eggshells (blue line in Fig. 4) have almost the same secondary structure compared to OC17 extracted from room temperature-stored eggshells (red line in Fig. 4). The result indicates that the incorporation of OC17 into CaCO3 lattice protects the protein against thermal damage at least at 50 °C for as long as 60 h. Because we focus on protecting proteins using an in vitro incorporation method, we chose experimental conditions in a way that the temperature and the time frame (50 °C, 60 h) is relevant to real-life situations, and we did not test higher temperatures for longer periods. It should be noted that the effect of heat on the lattice incorporated OC17 may be extended to other target proteins. In other words a given target protein could be probed for protection efficiency in the lattice. Because the overarching aim here is to investigate the in-lattice protection of a natural intracrystalline protein, we did not extend the analysis further with different target proteins.Probing the effect of dissolution agents on releasing intracrystalline proteins
Lattice incorporated proteins need to be released from CaCO3 crystals for practical applications. If the incorporated compound is not a protein (e.g. a small molecule drug), then it could be taken out of the crystal using one of the CaCO3 dissolution methods. It is worth noting that incorporated small molecule drugs could be taken orally because CaCO3 would be dissolved in the acidic environment of the stomach. If proteins are incorporated, then releasing these targets needs to be done in a way that does not denature proteins during the reconstitution step.There are two main ways to dissolve CaCO3: acid dissolution and metal chelation. The advantage of acid dissolution is to release intracrystalline content rapidly, however, the caveat is the risk of denaturing incorporated protein at low pH. The advantage of metal chelation is that it is possible to dissolve CaCO3 at neutral pH, thereby avoiding the risk of low pH denaturation. The disadvantage of metal chelation is the slow dissolution rate. We emulated intracrystalline protein releasing step by incubating a model protein, BSA, in either diluted acetic acid or in diluted EDTA.55,56Fig. 5A shows the CD spectra of BSA incubated in 5% acetic acid along with a positive control (BSA in H2O) at the same concentration. The difference in the acetic acid-incubated sample from positive control particularly at small wavelengths indicates that acidic solution causes rearrangements in the protein structure at least to some extent, although the pH of the solution is adjusted to 5. In addition, a decrease in the CD signal compared to BSA in EDTA could be attributed to partial denaturation. In contrast, BSA incubated in 200 mM EDTA solution shows only negligible difference from the positive control (BSA in H2O). Because metal chelation seems to be superior to acetic acid, we further investigated the effect of EDTA on the model protein. Fig. 6A shows extended CD spectra of EDTA-incubated BSA (i.e. extended data from Fig. 5B). The CD signal of the sample superposes with positive control below 210 nm and above 225 nm. The slight difference between roughly 210 and 225 nm does not indicate structural changes of the target, however, to ensure that EDTA incubation does not affect the protein, we probed the 3D structure of the sample using intrinsic tryptophan fluorescence spectroscopy. Tryptophan residues are highly sensitive to structural changes. As a complementary analysis to CD, which provides information on secondary structure, tryptophan fluorescence could reveal changes in the tertiary structure. As is seen in Fig. 6B, EDTA-incubated BSA shows no difference from positive control which suggests that dissolving CaCO3 in EDTA solution is suitable to release incorporated proteins at neutral pH without denaturing the protein structure. In addition, EDTA dissolution also offers a way to study the effect of incorporation itself on the protein structure (i.e. the structure of the same protein before and after incorporation) because any difference after releasing the target from the crystal lattice could be attributed to incorporation-related effects. It is reasonable to use EDTA dissolution despite its slower dissolution rate compared to acidic solution. An exception could be a target that is resistant to unfolding under low pH conditions.
 | ||
| Fig. 5 CD spectra showing the effect of reconstitution solution on the model protein structure. The common method of dissolving synthetic CaCO3 crystals with an incorporated target is to use an acidic solution to release as much intracrystalline content as possible in a short period of time. The caveat of using acid solution is the risk of target denaturation at low pH. The comparison of (A) 5% acetic acid (pH 5) and (B) 200 mM EDTA solutions on protein structure indicates that EDTA avoids protein denaturation. The increasing difference towards 220 nm compared to positive control (BSA in H2O) suggests that acetic acid triggers partial unfolding. A decrease in ellipticity in acetic acid compared to EDTA may also reflect changes in protein structure. Note that acetic acid interferes strongly with CD signal below 220 nm. For extended spectra of BSA in EDTA see Fig. 6. | ||
Cloning and optimization of recombinant OC17 expression
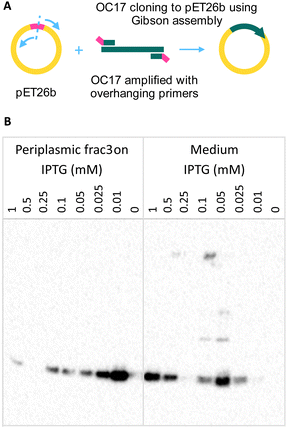
Although OC17 could be extracted from eggshells, it would be desirable to express it recombinantly for a number of reasons. Because the function of OC17 has been proposed to regulate amorphous-to-crystal transition during shell formation, recombinant expression could allow designing experiments with engineered protein to verify molecular simulations.1 For instance, site-directed mutagenesis could help revealing residues that were proposed to be key for interaction with the growing crystal.2,3 Recombinant OC17 not only offers an experimental system to investigate the function of the protein but it could also advance our understanding of the amorphous-to-crystal transition in biogenic minerals. In addition, recombinant expression could allow further engineering of the protein such as insertion of small molecules into OC17 through non-canonical amino acids. Labelling OC17 might be beneficial to conclusively show that the protein binds to amorphous CaCO3 and detaches once the structure transforms into calcite. Furthermore, from a practical perspective, genetic fusion of a target protein to OC17 could incorporate both proteins at once if the target cannot incorporate itself. Although an eggshell is a low-cost material, recombinant expression could yield high concentrations of protein unaffected by incorporation-related (if any) structural changes. For these reasons, we developed and optimized the expression of OC17 in E. coli. We designed primers to amplify the OC17 sequence with overhanging regions that target the insert site for Gibson assembly (Fig. 7A). After verifying the plasmid sequence, we tested protein expression in two different media: LB and TY supplemented with 1% glucose because of the effect of glucose on expression using pET plasmids.§ At two temperatures and different IPTG concentrations, we found that there is almost no expression in the TY medium at 25 °C (see the dot blot image in the ESI†). The pelB sequence was included in the construct for secretion of the target protein into the periplasmic space, however, low concentrations of OC17 were detected in the medium after overnight incubation at 30 °C, probably because of the small size of OC17. In subsequent analyses, both periplasmic fraction and medium were tested for the target protein. Expression in LB medium was found to be more efficient, however, at 30 °C, there is no expression above 0.1 mM IPTG. Expression markedly increases at 25 °C. We further tested lower IPTG concentrations (0.01, 0.025, 0.05, 0.1, 0.25 and 0.5 mM) to see if expression could be enhanced (Fig. 7B). His-tagged OC17 samples were analysed with western blot using anti-Histag antibodies (Fig. 7B). The highest expression was observed with 0.01 mM IPTG in the periplasmic space, however, because the protein also leaks into the medium at 0.05 mM, collection of both medium and periplasmic space fractions at this concentration would be reasonable.As described in the previous sections, it would be illuminating to show OC17 in action, although, to our knowledge, there is no such report. The absence of experimental verification of simulations probably arises from the difficulty of designing a setup with which OC17–CaCO3 interaction could be investigated directly in real-time. With recombinant OC17 at hand, it can be labelled in a way that allows monitoring the OC17–CaCO3 interaction, for instance using FLIM-FRET, with which we hope to show OC17 in action.
Conclusions
Protein incorporation into CaCO3 lattice might address the protection of temperature-sensitive proteins for long periods outside the cold-chain. The structural characterization of natural intracrystalline proteins for their heat-resistance might be beneficial. These proteins could be attractive and accessible models. As a step towards this goal, we focused on eggshells which are not only a low-cost, ubiquitous biomineral but also contain one of the intriguing intracrystalline proteins, OC17, reported to date. We investigated whether or not lattice incorporation provides heat-resistance to eggshell protein OC17, the study of which could inspire efficient synthetic incorporation pathways. Apart from loading target proteins in CaCO3, an equally important step is to release the target protein into solution without affecting the function of the protein. For this reason, we studied the effect of CaCO3 dissolution agents on a model protein. Compared to acid dissolution, we found that metal chelation using EDTA is superior because it allows dissolving CaCO3 at neutral pH, though it takes longer than acid dissolution. Having recombinant OC17 in hand, it would be possible to design experiments to show the protein in action which could be interesting based on the proposed function of OC17. Looking from a practical perspective, if extended to other molecules such as nucleic acids, lattice incorporation might be applied to different biological compounds. The temperature-sensitive nature of therapeutics is a global health problem, unveiled again with challenges in vaccine delivery during the COVID-19 pandemic. Incorporation of protein-based vaccines in CaCO3 may offer an alternative way to keep vaccines safe at high ambient temperatures as long as needed. Intracrystalline proteins such as OC17 may offer insights for developing biomimetic, low-cost and effective vaccine stabilization technology.Experimental
Materials
The E. coli K-12 MG 1655 cells were donated by Dr Gillian Fraser from the Department of Pathology, University of Cambridge, UK. The primers and plasmid pET26b were purchased from Sigma Aldrich (Darmstadt, Germany). The XL1-Blue Competent cells were purchased from Stratagene, USA. The Q5 High-Fidelity 2X Master Mix was purchased from New England Biolabs, UK. The 2X ReddyMix PCR Master Mix and FastDigest DpnI were purchased from Thermo Scientific, UK. PCR purification, gel extraction, plasmid purification kits were purchased from the QIAGEN, UK. The OC17 gene was synthesized by Invitrogen Thermo Scientific, UK. LB growth medium, agar plates, and buffer solutions were obtained from the Department of Haematology, University of Cambridge, UK. All purification columns were purchased from GE Healthcare, UK, unless otherwise stated. The SDS-PAGE gels were purchased from Life Technologies., UK. The western blot membranes and ultrafiltration centrifugal filters were purchased from Merck Millipore, UK. All chemicals for buffer preparations, cell culture, protein purification, and proteins (chicken egg white lysozyme and lyophilized powder) and bovine serum albumin (lyophilized powder) were purchased from Sigma Aldrich (Darmstadt, Germany), unless otherwise stated. All solutions were prepared with deionized water and pH adjustment was done with concentrated HCl or NaOH if needed. The solutions were filtered either through vacuum filtration with 0.45 or 0.2 μm filter papers or using syringe-driven filters. Buffer exchange of the protein solutions was done using Amicon Ultra Centrifugal Filters (Merck, Darmstadt, Germany) along with a Sorvall ST 8 bench-top centrifuge (Thermo Scientific, Massachusetts, USA).Methods
Author contributions
H. B. C. and F. I. U. conceptualized the project, developed the methodology, designed and performed the experiments and wrote the manuscript.Conflicts of interest
There are no conflicts to declare.Acknowledgements
H. B. C. would like to thank the Cambridge Commonwealth, European & International Trust, the Department of Engineering, the Nanoscience Centre, and Prof Jim Huntington from the Cambridge Institute for Medical Research, Department of Haematology, Cambridge, UK.Notes and references
- C. L. Freeman, J. H. Harding, D. Quigley and P. M. Rodger, Angew. Chem., Int. Ed., 2010, 49, 5135–5137 CrossRef CAS PubMed.
- C. L. Freeman, J. H. Harding, D. Quigley and P. M. Rodger, J. Phys. Chem. C, 2011, 115, 8175–8183 CrossRef CAS.
- C. L. Freeman, J. H. Harding, D. Quigley and P. M. Rodger, Phys. Chem. Chem. Phys., 2015, 17, 17494–17500 RSC.
- A. B. Rodriguez-Navarro, P. Marie, Y. Nys, M. T. Hincke and J. Gautron, J. Struct. Biol., 2015, 190, 291–303 CrossRef CAS PubMed.
- L. Stapane, N. Le Roy, J. Ezagal, A. B. Rodriguez-Navarro, V. Labas, L. Combes-Soia, M. T. Hincke and J. Gautron, J. Biol. Chem., 2020, 295, 15853–15869 CrossRef CAS PubMed.
- J. Gautron, L. Stapane, N. Le Roy, Y. Nys, A. B. Rodriguez-Navarro and M. T. Hincke, BMC Mol. Cell Biol., 2021, 22, 11 CrossRef CAS PubMed.
- M. Hincke, C. Tsang, M. Courtney, V. Hill and R. Narbaitz, Calcif. Tissue Int., 1995, 56, 578–583 CrossRef CAS PubMed.
- J. Reyes-Grajeda, D. Jauregui-Zuniga, A. Rodriguez-Romero, A. Hernandez-Santoyo, V. Bolanos-Garcia and A. Moreno, Protein Pept. Lett., 2002, 9, 253–257 CrossRef CAS PubMed.
- J. Reyes-Grajeda, A. Moreno and A. Romero, J. Biol. Chem., 2004, 279, 40876–40881 CrossRef CAS PubMed.
- K. Mann and F. Siedler, Biochem. Mol. Biol. Int., 1999, 47, 997–1007 CAS.
- Q. Zhang, L. Liu, F. Zhu, Z. Ning, M. T. Hincke, N. Yang and Z. Hou, PLoS One, 2014, 9(3), e93452 CrossRef PubMed.
- K. Drickamer, Curr. Opin. Struct. Biol., 1999, 9, 585–590 CrossRef CAS PubMed.
- A. Zelensky and J. Gready, FEBS J., 2005, 272, 6179–6217 CrossRef CAS PubMed.
- Y. Nys, J. Gautron, J. Garcia-Ruiz and M. Hincke, C. R. Palevol, 2004, 3, 549–562 CrossRef.
- M. T. Hincke, Y. Nys, J. Gautron, K. Mann, A. B. Rodriguez-Navarro and M. D. McKee, Front. Biosci.-Landmark, 2012, 17, 1266–1280 CrossRef CAS PubMed.
- G. Sykes, M. Collins and D. Walton, Org. Geochem., 1995, 23, 1059–1065 CrossRef CAS.
- S. Weiner, H. A. Lowenstam and L. Hood, Proc. Natl. Acad. Sci. U. S. A., 1976, 73, 2541–2545 CrossRef CAS PubMed.
- M. Salamon, N. Tuross, B. Arensburg and S. Weiner, Proc. Natl. Acad. Sci. U. S. A., 2005, 102, 13783–13788 CrossRef CAS PubMed.
- C. L. Oskam, J. Haile, E. Mclay, P. Rigby, M. E. Allentoft, M. E. Olsen, C. Bengtsson, G. H. Miller, J.-L. Schwenninger, C. Jacomb, R. Walter, A. Baynes, J. Dortch, M. Parker-Pearson, M. T. P. Gilbert, R. N. Holdaway, E. Willerslev and M. Bunce, Proc. R. Soc. B, 2010, 277, 1991–2000 CrossRef CAS PubMed.
- B. Demarchi, S. Hall, T. Roncal-Herrero, C. L. Freeman, J. Woolley, M. K. Crisp, J. Wilson, A. Fotakis, R. Fischer, B. M. Kessler, R. R. Jersie-Christensens, J. V. Olsen, J. Haile, J. Thomas, C. W. Marean, J. Parkington, S. Presslee, J. Lee-Thorp, P. Ditchfield, J. F. Hamilton, M. W. Ward, C. M. Wang, M. D. Shaw, T. Harrison, M. Dominguez-Rodrigo, R. D. E. MacPheel, A. Kwekason, M. Ecker, L. K. Horwitz, M. Chazan, R. Kroger, J. Thomas-Oates, J. H. Harding, E. Cappellini, K. Penkman and M. J. Collins, eLife, 2016, 5, e17092 CrossRef PubMed.
- B. Demarchi, M. Mackie, Z. Li, T. Deng, M. J. Collins and J. Clarke, eLife, 2022, 11, e82849 CrossRef CAS PubMed.
- M. Crisp, B. Demarchi, M. Collins, M. Morgan-Williams, E. Pilgrim and K. Penkman, Quat. Geochronol., 2013, 16, 110–128 CrossRef.
- G. Miller, P. Beaumont, H. Deacon, A. Brooks, P. Hare and A. Jull, Quat. Sci. Rev., 1999, 18, 1537–1548 CrossRef.
- Y.-Y. Kim, J. D. Carloni, B. Demarchi, D. Sparks, D. G. Reid, M. E. Kunitake, C. C. Tang, M. J. Duer, C. L. Freeman, B. Pokroy, K. Penkman, J. H. Harding, L. A. Estroff, S. P. Baker and F. C. Meldrum, Nat. Mater., 2016, 15, 903 CrossRef CAS PubMed.
- G. Magnabosco, M. Di Giosia, I. Polishchuk, E. Weber, S. Fermani, A. Bottoni, F. Zerbetto, P. G. Pelicci, B. Pokroy, S. Rapino, G. Falini and M. Calvaresi, Adv. Healthcare Mater., 2015, 4, 1510–1516 CrossRef CAS PubMed.
- M. Di Giosia, I. Polishchuk, E. Weber, S. Fermani, L. Pasquini, N. M. Pugno, F. Zerbetto, M. Montalti, M. Calvaresi, G. Falini and B. Pokroy, Adv. Funct. Mater., 2016, 26, 5569–5575 CrossRef CAS.
- Y.-Y. Kim, L. Ribeiro, F. Maillot, O. Ward, S. J. Eichhorn and F. C. Meldrum, Adv. Mater., 2010, 22, 2082 CrossRef CAS PubMed.
- Y.-Y. Kim, A. S. Schenk, D. Walsh, A. N. Kulak, O. Cespedes and F. C. Meldrum, Nanoscale, 2014, 6, 852–859 RSC.
- B. Pokroy, A. N. Fitch, F. Marin, M. Kapon, N. Adir and E. Zolotoyabko, J. Struct. Biol., 2006, 155, 96–103 CrossRef CAS PubMed.
- E. Zolotoyabko, Adv. Mater. Interfaces, 2017, 4 Search PubMed.
- B. Pokroy, J. P. Quintana, E. N. Caspi, A. Berner and E. Zolotoyabko, Nat. Mater., 2004, 3, 900–902 CrossRef CAS PubMed.
- S. Borukhin, L. Bloch, T. Radlauer, A. H. Hill, A. N. Fitch and B. Pokroy, Adv. Funct. Mater., 2012, 22, 4216–4224 CrossRef CAS.
- I. Ben Shir, S. Kababya, I. Katz, B. Pokroy and A. Schmidt, Chem. Mater., 2013, 25, 4595–4602 CrossRef CAS.
- E. Seknazi and B. Pokroy, Adv. Mater., 2018, 30, 1707263 CrossRef PubMed.
- E. Seknazi, S. Mijowska, I. Polishchuk and B. Pokroy, Inorg. Chem. Front., 2019, 6, 2696–2703 RSC.
- M. Różycka, I. Coronado, K. Brach, J. Olesiak-Bańska, M. Samoć, M. Zarębski, J. Dobrucki, M. Ptak, E. Weber, I. Polishchuk, B. Pokroy, J. Stolarski and A. Ożyhar, Chem. – Eur. J., 2019, 25, 12740–12750 CrossRef PubMed.
- S. Mijowska, I. Polishchuk, A. Lang, E. Seknazi, C. Dejoie, S. Fermani, G. Falini, N. Demitri, M. Polentarutti, A. Katsman and B. Pokroy, Chem. Mater., 2020, 32, 4205–4212 CrossRef CAS.
- O. Nahi, A. N. Kulak, T. Kress, Y.-Y. Kim, O. G. Grendal, M. J. Duer, O. J. Cayre and F. C. Meldrum, Chem. Sci., 2021, 12, 9839–9850 RSC.
- A. N. Kulak, M. Semsarilar, Y.-Y. Kim, J. Ihli, L. A. Fielding, O. Cespedes, S. P. Armes and F. C. Meldrum, Chem. Sci., 2014, 5, 738–743 RSC.
- Y. Ning, L. Han, M. Douverne, N. J. W. Penfold, M. J. Derry, F. C. Meldrum and S. P. Armes, J. Am. Chem. Soc., 2019, 141, 2481–2489 CrossRef CAS PubMed.
- Y. Ning, L. Han, M. J. Derry, F. C. Meldrum and S. P. Armes, J. Am. Chem. Soc., 2019, 141, 2557–2567 CrossRef CAS PubMed.
- A. N. Kulak, P. Yang, Y.-Y. Kim, S. P. Armes and F. C. Meldrum, Chem. Commun., 2014, 50, 67–69 RSC.
- Y.-Y. Kim, K. Ganesan, P. Yang, A. N. Kulak, S. Borukhin, S. Pechook, L. Ribeiro, R. Kroeger, S. J. Eichhorn, S. P. Armes, B. Pokroy and F. C. Meldrum, Nat. Mater., 2011, 10, 890–896 CrossRef CAS PubMed.
- Y. Ning, D. J. Whitaker, C. J. Mable, M. J. Derry, N. J. W. Penfold, A. N. Kulak, D. C. Green, F. C. Meldrum and S. P. Armes, Chem. Sci., 2018, 9, 8396–8401 RSC.
- Y. Ning, L. A. Fielding, L. P. D. Ratcliffe, Y.-W. Wang, F. C. Meldrum and S. P. Armes, J. Am. Chem. Soc., 2016, 138, 11734–11742 CrossRef CAS PubMed.
- B. Marzec, D. C. Green, M. A. Holden, A. S. Cote, J. Ihli, S. Khalid, A. Kulak, D. Walker, C. Tang, D. M. Duffy, Y.-Y. Kim and F. C. Meldrum, Angew. Chem., Int. Ed., 2018, 57, 8623–8628 CrossRef CAS PubMed.
- E. Weber, L. Bloch, C. Guth, A. N. Fitch, I. M. Weiss and B. Pokroy, Chem. Mater., 2014, 26, 4925–4932 CrossRef CAS.
- M. B. Al-Handawi, P. Commins, S. Shukla, P. Didier, M. Tanaka, G. Raj, F. A. Veliz, R. Pasricha, N. F. Steinmetz and P. Naumov, Adv. Biosyst., 2018, 2(5), e1700176 CrossRef PubMed.
- C. A. Murray, P. M. Scientists, L. Holland, R. O’Brien, A. Richards, A. R. Baker, M. Basham, D. Bond, L. D. Connor, S. J. Day, J. Filik, S. Fisher, P. Holloway, K. Levik, R. Mercado, J. Potter, C. C. Tang, S. P. Thompson and J. E. Parker, CrystEngComm, 2024, 26, 753–763 RSC.
- M. Hincke, J. Gautron, M. Panheleux, J. Garcia-Ruiz, M. McKee and Y. Nys, Matrix Biol., 2000, 19, 443–453 CrossRef CAS PubMed.
- Z. Zou, X. Yang, M. Alberic, T. Heil, Q. Wang, B. Pokroy, Y. Politi and L. Bertinetti, Adv. Funct. Mater., 2020, 30, 2000003 CrossRef CAS.
- A. V. Radha, T. Z. Forbes, C. E. Killian, P. U. P. A. Gilbert and A. Navrotsky, Proc. Natl. Acad. Sci. U. S. A., 2010, 107, 16438–16443 CrossRef CAS PubMed.
- Y. Levi-Kalisman, S. Raz, S. Weiner, L. Addadi and I. Sagi, J. Chem. Soc., Dalton Trans., 2000, 3977–3982 RSC.
- J. Ihli, W. C. Wong, E. H. Noel, Y.-Y. Kim, A. N. Kulak, H. K. Christenson, M. J. Duer and F. C. Meldrum, Nat. Commun., 2014, 5, 3169 CrossRef PubMed.
- F. Marin and G. Luquet, C. R. Palevol, 2004, 3, 469–492 CrossRef.
- F. Marin, B. Pokroy, G. Luquet, P. Layrolle and K. De Groot, Biomaterials, 2007, 28, 2368–2377 CrossRef CAS PubMed.
- K. Mann, FEBS Lett., 1999, 463, 12–14 CrossRef CAS PubMed.
- K. Mann and F. Siedler, Comp. Biochem. Physiol., Part B: Biochem. Mol. Biol., 2006, 143, 160–170 CrossRef PubMed.
- C. Abergel, V. Monchois, D. Byrne, S. Chenivesse, F. Lembo, J.-C. Lazzaroni and J.-M. Claverie, Proc. Natl. Acad. Sci. U. S. A., 2007, 104, 6394–6399 CrossRef CAS PubMed.
- V. Monchois, C. Abergel, J. Sturgis, S. Jeudy and J. Claverie, J. Biol. Chem., 2001, 276, 18437–18441 CrossRef CAS PubMed.
Footnotes |
| † Electronic supplementary information (ESI) available: Dot blot image of the optimization of OC17 expression. See DOI: https://doi.org/10.1039/d4sm00371c |
| ‡ Present address: TOBB University of Economics and Technology, Department of Biomedical Engineering, Söğütözü Caddesi No. 43, 06560, Ankara, Turkey. |
| § R. Novy and B. Morris, Use of glucose to control basal expression in the pET system, inNovations, 2001, 13, 8–10. https://www.sigmaaldrich.com/deepweb/assets/sigmaaldrich/product/documents/386/947/innovations-013-nvg.pdf. |
| This journal is © The Royal Society of Chemistry 2024 |