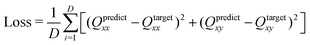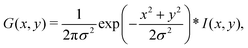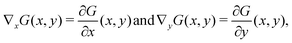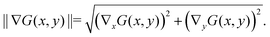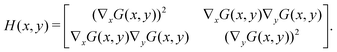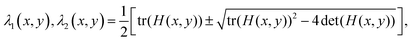Open Access Article
Open Access ArticleA machine learning approach to robustly determine director fields and analyze defects in active nematics†
Yunrui
Li
 a,
Zahra
Zarei
b,
Phu N.
Tran
a,
Zahra
Zarei
b,
Phu N.
Tran
 b,
Yifei
Wang
a,
Aparna
Baskaran
b,
Yifei
Wang
a,
Aparna
Baskaran
 b,
Seth
Fraden
b,
Seth
Fraden
 b,
Michael F.
Hagan
b,
Michael F.
Hagan
 b and
Pengyu
Hong
b and
Pengyu
Hong
 *a
*a
aComputer Science Department, Brandeis University, USA. E-mail: hongpeng@brandeis.edu
bPhysics Department, Brandeis University, USA
First published on 31st January 2024
Abstract
Active nematics are dense systems of rodlike particles that consume energy to drive motion at the level of the individual particles. They exist in natural systems like biological tissues and artificial materials such as suspensions of self-propelled colloidal particles or synthetic microswimmers. Active nematics have attracted significant attention in recent years due to their spectacular nonequilibrium collective spatiotemporal dynamics, which may enable applications in fields such as robotics, drug delivery, and materials science. The director field, which measures the direction and degree of alignment of the local nematic orientation, is a crucial characteristic of active nematics and is essential for studying topological defects. However, determining the director field is a significant challenge in many experimental systems. Although director fields can be derived from images of active nematics using traditional imaging processing methods, the accuracy of such methods is highly sensitive to the settings of the algorithms. These settings must be tuned from image to image due to experimental noise, intrinsic noise of the imaging technology, and perturbations caused by changes in experimental conditions. This sensitivity currently limits automatic analysis of active nematics. To address this, we developed a machine learning model for extracting reliable director fields from raw experimental images, which enables accurate analysis of topological defects. Application of the algorithm to experimental data demonstrates that the approach is robust and highly generalizable to experimental settings that are different from those in the training data. It could be a promising tool for investigating active nematics and may be generalized to other active matter systems.
1 Introduction
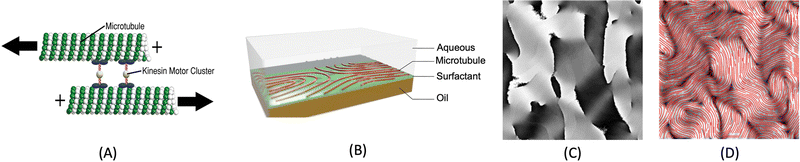
Active materials are ubiquitous non-equilibrium systems that can be found in many natural processes and biological systems, such as cellular cytoskeletons,1–3 bacterial suspensions,4–6 flocks of birds,7 and schools of fish.8 The components of such systems consume energy to drive forces or motion at the scale of individual particles, which leads to emergent macroscale collective dynamics.9,10 In this work, we study a class of active matter known as an active nematic, which describes a system of rodlike particles that are placed at sufficient density to form a nematic liquid crystal with orientational order, but are continually driven out of equilibrium by propulsion of the particles. We focus on a widely studied experimental model active nematic system, consisting of elongated bundles of microtubules and molecular motors (kinesins), in which the kinesins consume adenosine triphosphate (ATP) fuel to slide microtubules relative to each other (Fig. 1).11–13 These motions produce topological defects and chaotic flows that compete with the equilibrium orientational order, producing spectacular spatiotemporal behaviors. Describing and understanding the rich behaviors that arise from this interplay is a challenge at the forefront of modern physics.A remarkable characteristic of active nematics is the continuous proliferation and annihilation of topological defects, which are discontinuities in the orientational order.14,15 Defects arise when the activity of the particles in the nematic is sufficiently high to give rise to a flow that deforms the nematic structure. Gradients in the magnitude and direction of the surrounding director field can be used to identify defects, determine their associated topological charges, and track their motions. Defects in active nematics are important for several reasons. Firstly, they can serve as a measure of the activity of the nematic, with a higher density of defects indicating a higher level of activity. Secondly, defects strongly influence the overall flow patterns and dynamics of the system; in particular, defects with +1/2 topological charge move as if self-propelled. Finally, defects can also affect the transport properties of the nematic, such as its viscosity or diffusivity, which can have important implications for the behavior of the nematic as a whole. Overall, the study of topological defects in active nematics is a rich and active area of research, with many interesting questions still remaining to be answered.3,16–19
1.1 Existing methods to obtain director fields
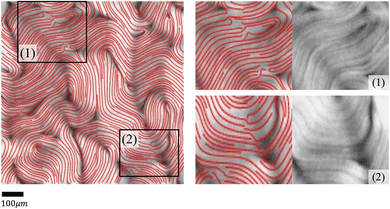
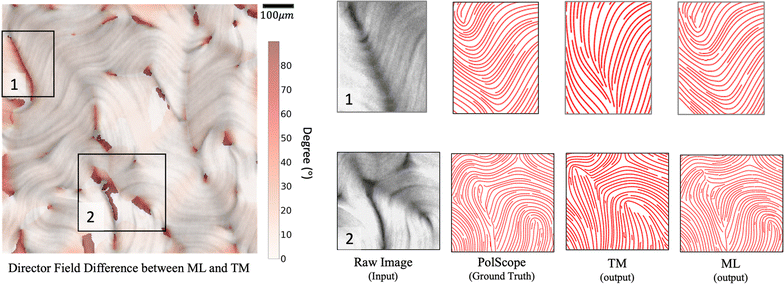
The nematic tensor, or the Q-tensor,10,20–24 describes the director field; that is, the degree of alignment and the direction of the local nematic orientation. A reliable director field is essential to accurately detect, measure, and track defects in active nematic systems. One way to obtain the director field from experiments is to use the PolScope,25–27 which is a powerful tool for quantifying the dynamics of nematic structures. It operates by shining polarized light through the specimen, which interacts with the aligned molecules or particles in the nematic phase. This interaction alters the polarization of the light, allowing the PolScope to measure birefringence quantitatively at every single pixel in the image. The resulting retardation of light allows the PolScope to accurately determine the degree and orientation of microtubule alignment, pixel by pixel, providing important insights into the structural characteristics and behavior of liquid crystal systems. PolScope generates a pair of images – the retardance image and the corresponding director field image, at any given time point. The calculated director field enables a direct comparison between experimental observations and theoretical results. This data also can be used to train dynamics prediction models, such as proposed by Zhou et al.,28 Colen et al.29 Nevertheless, certain experimental setups and controls are not feasible using PolScope. For example, some experiments use light to control the dynamics of an active nematic.30–33 These experiments illuminate the sample from above, which makes the traditional PolScope setup impractical. In addition, high numerical aperture condenser and objective lenses typically introduce distortions that limit the purity of polarization in a light microscope. These distortions present challenges and limitations when using a PolScope.25 Lastly, the nematic material's small retardance values are often overshadowed by stronger background birefringence from microscope objective strain and Fresnel reflectivity. Although background correction is a standard practice, it does not fully account for the microscope slide's birefringence and is sensitive to variations across the image, optical alignment, and temperature. This can result in inaccuracies in local orientation measurements in some regions. For example, discontinuous director fields were observed in the middle of microtubule bundles, see Fig. 2. This may result in false detection and inaccurate localization of defects.Alternatively, other instruments can offer more flexible control and design of the experiments, but cannot generate the corresponding director fields directly. For example, a fluorescence microscope enables direct light control31–33 or ATP control of the system's activities, but does not offer the capability to extract director fields. In some experiments, specialized fluorescent dyes or proteins are added at low concentrations to enable the visualization and tracking of individual microtubule movements. However, such methods only provide sparse director field information that is randomly distributed across the whole view. In these situations, one can use image processing methods to extract orientations from raw images directly from experiments, such as fluorescence images. Usually, an in-house method is developed using softwares such as MATLAB for extracting director fields from raw images.15,34–37 The methods follow a similar approach by first finding the gradient change of a raw image, then coarse-graining the result, and finding the smallest eigenvalue of a matrix formed by the outer product of the gradient. More details of the method we use in this work are presented in Appendix A.1 and Ellis et al.38 We refer to this method as the “TM” in the rest of this article.
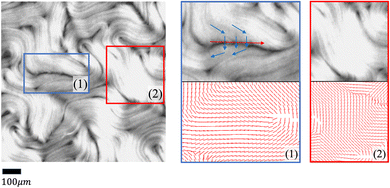
However, the effectiveness of the TM can be limited because raw images contain complex signals arising from variations in experimental settings, noise, limitations of microscopy, etc. Examples of such limitations include the following. First, the analysis of retardance images is intrinsically difficult at regions that have low contrast, are out-of-focus, or have distortions due to a variety of effects. For example, although active nematics are assumed to be two-dimensional by imaging systems, they actually move in a shallow three-dimensional container. This three-dimensionality can influence both the illumination and the focus of the microscope, thereby altering the pixel intensity in the resultant images. Additionally, active nematic filaments, when compressed by adjacent ones, can buckle into a U shape, causing a fissure that exposes the base of the container. Such fissures often run counter to the direction of movement, which can result in a miscalculation of the local orientation. We show some examples of orientation miscalculation by the TM in noisy regions in Fig. 3. Finally, the inherent variability in active nematics systems and the varying levels of activity observed across different experiments contribute to a diverse distribution of noisy regions across successive frames.
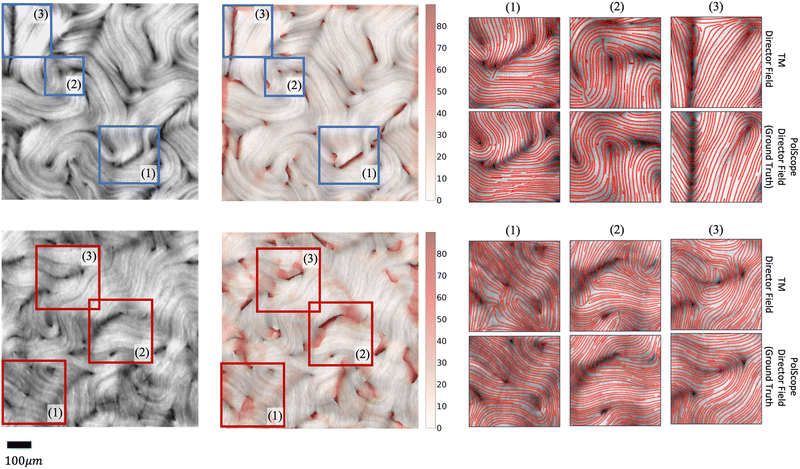
Second, the TM may require extensive manual adjustments of its parameter settings to make it work satisfactorily for different images. In other words, raw images from different experiments or conditions cannot use the same parameter setting. Sometimes, manual adjustment is needed for different images in the same video (see Fig. 4 for an example). This greatly hinders large-scale reliable studies of active nematics. Many factors contribute to this challenge, such as: (1) active nematic materials can exhibit significant variations even under identical experimental conditions; (2) different imaging instruments or settings can lead to different noise distributions; that is, different characteristics or statistical patterns (in space and/or time) of errors in the acquired images; (3) the behavior of nematic materials may not be consistent across the sample (e.g. due to spatially varying activity levels or boundary conditions, which may be according to design or arise from experimentally uncontrollable variations), resulting in defects with varying sizes and mean separation distances that require setting different values for the window size parameter used by the defect detection algorithm.
1.2 Innovation and contribution
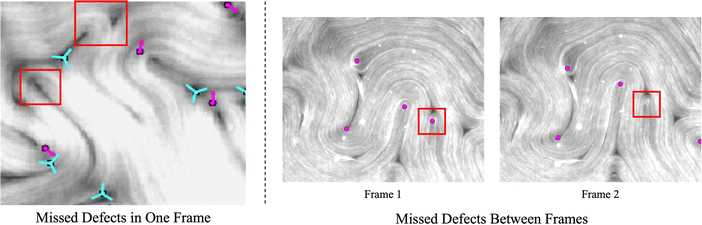
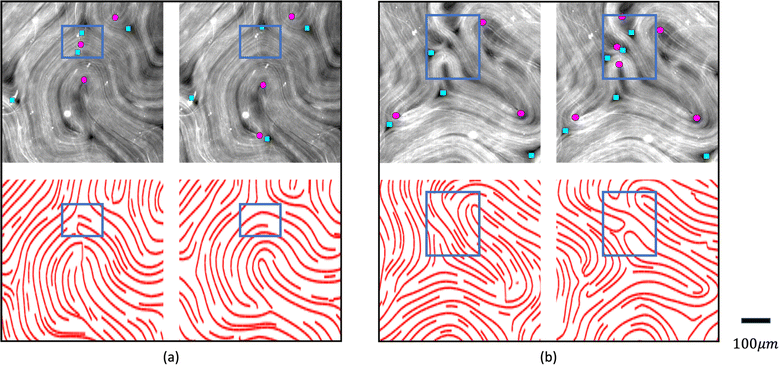
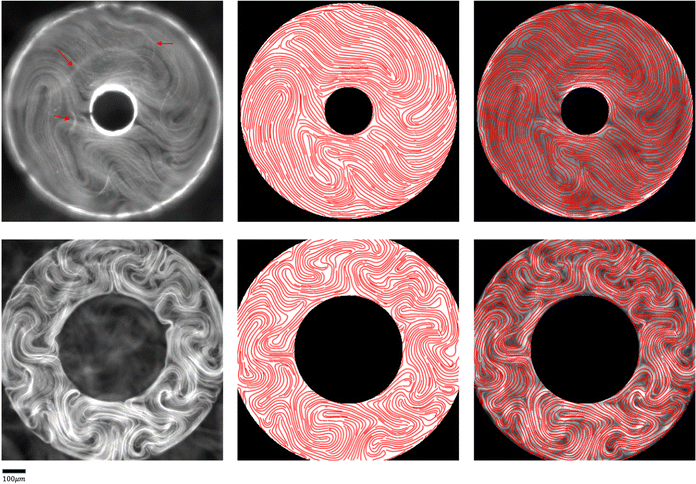
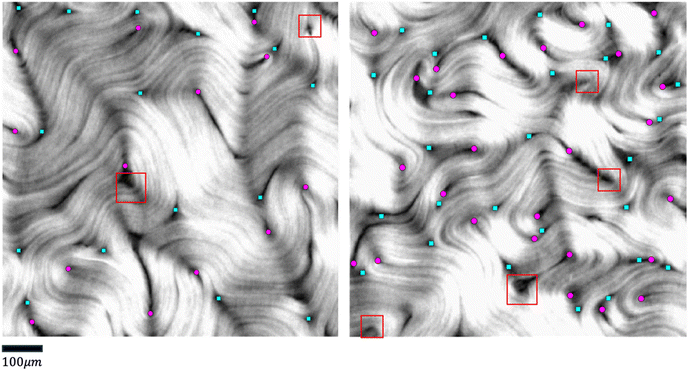
To tackle the above challenges, we have developed a machine learning based technique to robustly and automatically extract director fields from raw images. Machine learning has already made significant impacts on active nematics,39–42 and more applications are being explored. For example, we previously developed a machine learning technique for learning and predicting dynamics of active nematics.28 However, that technique is limited by the availability of of quality director fields. Using our proposed machine learning approach, not only can we reliably and robustly calculate the director fields from raw experimental data directly, but we also enable many downstream tasks such as defect detection, defect tracking, and dynamic prediction. These tasks are significantly simplified, compared to previous approaches, for raw data that does not have the director fields readily available. We benchmark our machine learning model against our in-house implementation of an image processing method (TM) that follows a similar approach to other existing methods.15,34–38 This method takes a gray-scaled image as the input. After denoising the raw image by Gaussian Blur with standard deviation σ, it assumes that image intensities change most dramatically in the direction perpendicular to the alignment of the microtubule bundles at each point. Thus, for each pixel, it calculates the intensity change matrix using convolution, which is then decomposed to obtain its eigenvalues. The local orientation centered at a specific pixel is indicated by the eigenvector associated with the smallest eigenvalue. Once the Q-tensor is obtained, topological defects can be identified by calculating the winding number43 of each point. This is typically done by applying convolutional filters (e.g., Gaussian Kernels) of a user-defined window size on each pixel on the director field obtained. More details of this method can be found in Appendix A.1. Compared with the TM, the director field information obtained by our method is more accurate, which leads to better defect detection results. Fig. 5 shows an example of defects that are missed by the TM. These defects can be recovered using our machine learning models. We note that the TM defect detection results differ slightly when different parameter settings are chosen. Fig. 20 (Appendix A.6) shows examples of changing the parameter settings for individual frames. However, the TM results are consistently less accurate than those obtained by our machine learning approach, and we have tuned the TM parameters for each dataset to achieve reliable results for all frames in that dataset. In addition to increased accuracy, a key advantage of the machine learning approach is that it requires less parameter tuning.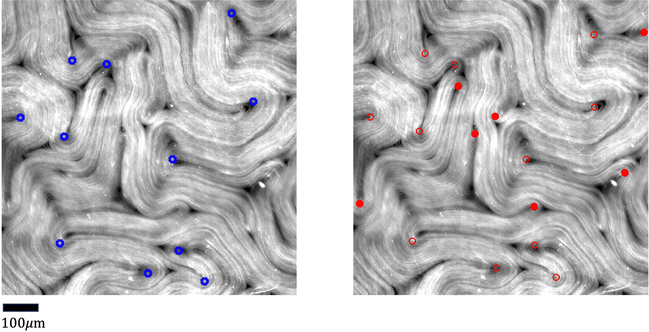 | ||
| Fig. 5 Comparison of defect detection by the TM and our machine learning method from fluorescence microscopy images. Left: Positive defects detected in the director field calculated by the TM. Right: Positive defects calculated by our machine learning method. Defects missed by the TM are highlighted in solid red dots. Empty red dots indicate the defects detected in both settings. Only detected positive defects are shown for clarity. The images are 600 by 600 pixels. The defect detection window in the TM is set to 11 and the defect threshold is set to 0.2. We note that the traditional method defect detection results may be slightly different if these parameters are tuned differently (Fig. 20 in Appendix A.6), but the parameters are tuned for each set of experiments so that all frames in a given experiment achieve acceptable results. Fig. 20 (Appendix A.6) shows how these results depend on parameter settings. | ||
In summary, the main contributions of our work include the following:
• We developed a robust machine learning approach to accurately extract director fields from raw microscopy images of active nematics. Our approach is able to handle raw images captured under various experimental settings (e.g., lighting, activity levels of active nematics, microscope lens, etc.).
• We used a masking strategy to correct some regions of director fields calculated by the PolScope that are inaccurate due to sub-optimal retardance images, such as over-/under-exposure, out-of-focus regions, etc.
• We demonstrate promising downstream applications using the director fields generated by our model, including defect detection, defect orientation, defect tracking, etc.
• To facilitate defect tracking as a downstream task, we utilized a matching algorithm across several consecutive frames and evaluated outcomes across frames. This approach enabled us to enhance tracking accuracy by filling in gaps for absent defects in a particular frame, while not in others.
2 Methods
We used the PolScope data (each sample is a pair of a retardance image and its corresponding director field) to train a deep learning model for extracting local orientation information from retardance images. Using the director fields extracted by our approach, we were able to improve defect detection and tracking.2.1 Improving training data
In most cases, PolScope works well in capturing the retardance images of active nematics and correctly calculating the corresponding director fields. However, PolScope occasionally produces errors in the local orientation (examples in Fig. 2), which can hamper the efforts to build a machine learning model to extract director fields. Fortunately, such errors can be automatically corrected using machine learning. Specifically, we adopted the masked image modeling (MIM)44 method (see Fig. 6 Top) that randomly masks out patches in an image decided by a mask size and a masking ratio, and trained a machine learning model to reproduce the masked-out patches using the rest of the image. We chose the mask size as 10 × 10 pixels and the masking ratio as 30% of the whole image. To make this selection, we tried several different mask sizes (6 × 6, 10 × 10 and 20 × 20 pixels) and different masking ratios (20%, 30% and 50%), and found the chosen setting produced the lowest mean absolute error (MAE) of 0.0241. Once trained, the MIM model can be applied to improve director fields not used in training. Two examples are shown in Fig. 6 Bottom to visualize the effects of this process.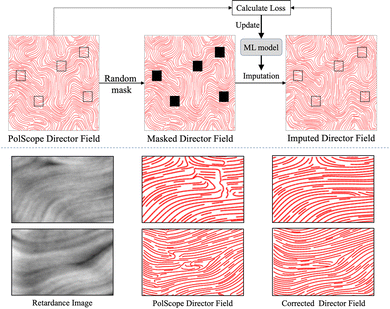 | ||
| Fig. 6 Correcting the PolScope director fields via Masked Image Modeling (MIM), which trains a machine learning model (the Swin Transformer45 in our case) to improve local orientation calculation. Top: In each iteration of the training process, a director field is sampled and several patches in it are randomly masked out (e.g., black patches in the middle image). The MIM machine learning model imputes the masked patches. The loss is calculated as the mean square error between the imputation results and the ground-truth (i.e., the corresponding patches in the input), and is used to update the parameters of the MIM machine learning model. Bottom: Two examples of local orientation correction are shown. The left, middle, and right columns are the retardance images, the PolScope director fields, and the corrected director fields, respectively. | ||
2.2 Director field extraction using machine learning
Using the corrected director field as the target output, we can train our machine learning model to learn the proper mapping between the retardance image to its corresponding director field. Fig. 7 illustrates the architecture of our machine learning model, which consists of a Gabor filter46,47 layer followed by a ResNet structure.48 Gabor filters are a set of filters widely used for texture analysis. Each filter can be tuned to detect spatially local signals with certain frequency components in a certain direction at a certain scale. One can use an array of Gabor filters to help perform multi-scale analysis of spatial patterns in active nematics, such as defects, vortices, or other patterns. More technical details about Gabor filters are provided in Appendix A.2. In our model, we used two sets of Gabor Filters, the first set has a smaller wavelength that aims to detect higher spatial frequencies and finer details, while the second set has a larger wavelength, aiming to capture global features. For each set of filters, 64 filters are used, each with a different combination of 8 orientation values and 8 scale values. For the ResNet, 16 blocks are used, as shown in Fig. 7. Among the 16 ResNet blocks, 4 different channel dimensions are used: 64, 128, 256, and 512. More detailed configurations can be found in Appendix A.3.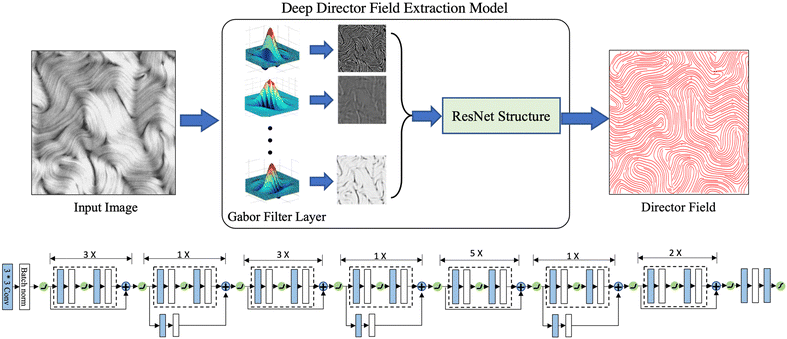 | ||
| Fig. 7 The deep learning model for extracting director fields from PolScope retardance images. Top: Complete model pipeline. Bottom: Details of the ResNet Structure in Top. Blue and white boxes indicate convolution layers and batch normalization operations. A ReLU activation function49 is used except for the output layer, which uses a sigmoid activation function. In total, 16 ResNet blocks are used. | ||
2.3 Defect analysis
One can detect defects by computing the winding numbers of individual points in the director fields43 by counting the local orientation changes centralized at each point within a predefined window size. Nevertheless, the director fields are discretized at the pixel level, which can lead to inaccurate winding number calculations. Some defects may not be detected even from more accurate director fields such as those calculated by PolScope (see Fig. 18 in Appendix A.6) and our machine learning approach (see Fig. 8, Left), due to various factors. These include over-exposed regions in the raw image, leading to an over-smoothed director field; or regions with high local defect densities, where the distance between the defects is smaller than the window size when calculating the winding number. The director fields calculated by our method significantly improve defect detection results (see Fig. 11 in the Experiments section). However, in a non-equilibrium system of active nematics where speed and density change, it is possible that a defect is detected in one frame but missed in the next (see Fig. 8 Right). This can cause the defect tracking algorithm to lose track of defects in consecutive frames and result in inaccurate measurement of defect annihilation and proliferation rates.To maintain the consistency and coherence of defect tracking over time, we match defects detected in three consecutive frames (at time points t − 1, t, and t + 1) using a graduated assignment algorithm,50 and use the matching results to recover missing defects in the t-th frame. The coordinates of the rescued defects are interpolated using the coordinates of their matched defects at time t − 1 and t + 1. By implementing interpolation, our algorithm is better equipped to maintain an accurate representation of defect trajectories, thereby improving the reliability of the tracking process. See Fig. 12–14 for defect detection results over time. Please refer to Sections 3.3 and 3.4 for more results and discussions.
Finally, we trained a model with four ResNet blocks (32, 64, 128, and 256 channels, respectively) to estimate the orientation of a defect. The input to the model is a 64 × 64 patch centered at a defect, and the output is the angle of the defect. The model error is also measured by the mean absolute error (MAE) between the estimated angles and the manually labeled ground-truth.
3 Application of the model to fluorescence images from experimental data
In this section, we will illustrate our training process and model performance. To create an active nematic system, we combined microtubules, kinesin motors, ATP, and bundling agents, and sedimented the mixture on an oil–water interface. We acquired PolScope and fluorescence data from these samples.In total, 48![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 000 retardance and director field images are used for training and testing the machine learning model. Specifically, we randomly selected 40
000 retardance and director field images are used for training and testing the machine learning model. Specifically, we randomly selected 40![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 000 image pairs to train the neural network, and the remaining 8000 images for performance testing. We present results obtained by applying the model to the test set of data. We chose the PolScope dataset to train the model because it is the only instrument that can produce retardance and director field images at the same time. The director field images are first corrected using the masking strategy, then used as the training targets for the machine learning model. The retardance images are inputs to our machine learning model. The predicted director field is compared with the corrected director field and the learning goal is to achieve maximum similarity. Since the orientations of microtubule bundles have head–tail symmetry, we translate the orientation to the Q-tensor and use the Mean Squared Error (MSE) to measure the learning error as follows.
000 image pairs to train the neural network, and the remaining 8000 images for performance testing. We present results obtained by applying the model to the test set of data. We chose the PolScope dataset to train the model because it is the only instrument that can produce retardance and director field images at the same time. The director field images are first corrected using the masking strategy, then used as the training targets for the machine learning model. The retardance images are inputs to our machine learning model. The predicted director field is compared with the corrected director field and the learning goal is to achieve maximum similarity. Since the orientations of microtubule bundles have head–tail symmetry, we translate the orientation to the Q-tensor and use the Mean Squared Error (MSE) to measure the learning error as follows.
| Qxx = cos2(qi), Qxy = cos(qi)sin(qi), |
where q is the director field orientation angle at each pixel in the input images.
3.1 Extract director fields from retardance images
Fig. 9 illustrates the advantage of our machine learning model over the TM on the PolScope retardance images. Using the director fields calculated by PolScope (and improved by the method described in Section 2.1) as the ground-truth, our trained machine learning model extracts more accurate director fields compared to the TM. We tested our model on 120 randomly selected retardance images excluded from the training set. Using the corresponding director fields calculated by PolScope as the ground-truth, we calculated the error of our model and the TM method. Our approach achieves a mean absolute error (MAE) of 8.8 degrees per location with a standard deviation of 1.46. As a comparison, the MAE of the TM approach is 18.1 per location with a standard deviation of 2.6.3.2 Generalize to fluorescence images
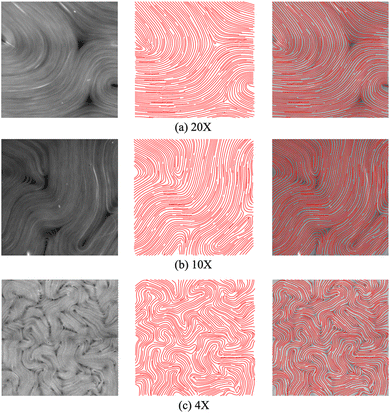
The proposed machine learning approach is trained using PolScope retardance images and calculated director fields. However, once trained, it can be applied to raw images produced at other magnifications, by other microscope instruments, or under different experimental settings (e.g., activity levels or boundary conditions). This is useful because, while the PolScope enables calculating the director field without labeling proteins and is usually highly accurate, it is not suitable for some classes of experiments, such as light-activated active nematics. There are various methods to control the behavior of an active nematic, such as changing the concentration of ATP and motor proteins, applying hard boundaries, etc.23,51–55 However, these methods control activity with only limited precision, and do not enable spatially varying activity profiles. To enable greater control over activity and the ability to change activity in space and time, researchers have developed light-sensitive motor proteins,31–33 with which the active nematic's activity and dynamics can be regulated by varying the spatiotemporal patterns of applied light intensity. In that case, the sample is illuminated by a digital light processor (DLP) projector that projects light (e.g. 460 nm wavelength) from above onto the sample. However, the PolScope setup, which involves a universal compensator and an analyzer for circularly polarized light, limits access to the stage and does not allow us to use the DLP mounted to the stage for light-activated experiments.‡Our proposed machine learning model performs well on fluorescence images of active nematics with different microscope magnifications (see Fig. 10) in an unconfined 2-dimensional experimental setting. Moreover, our model also generalizes well to confined active nematics with different boundary conditions (see Fig. 17 in Appendix A.6, and Videos in the ESI†). Importantly, this figure shows that the model achieves good results even in the presence of unbound microtubules that float above the 2D active nematic layer, and thus have orientations which differ from those of the layer. The machine learning algorithm filters out these extraneous elements, ensuring reliable calculation of the director field.
3.3 Defect detection
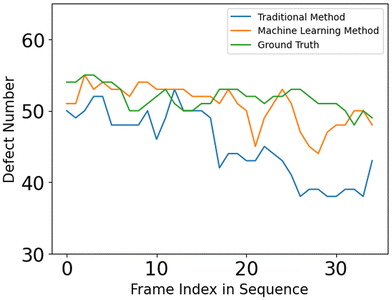
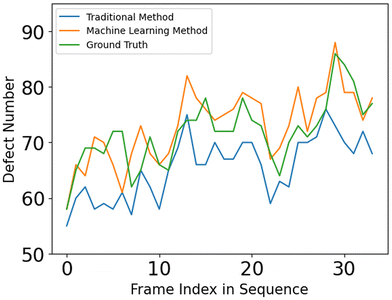
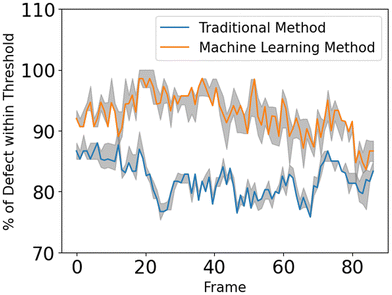
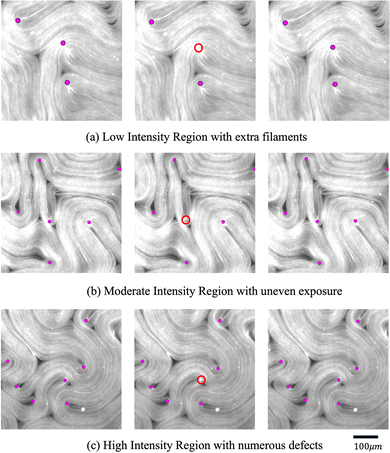
The defect detection results using the director fields extracted by our approach are substantially better than those obtained using the director fields extracted by the TM (details in Appendix A.1). Fig. 11 visualizes the defect detection results from raw images in one experiment using fluorescence microscopy, which was not used to train our director field extraction model.To evaluate the defect detection results, we manually labeled defects in 40 fluorescence images (not used in model training) from one experiment as the ground-truth. Fluorescence microscopy enables a light-responsive system that allows researchers to combine a considerable quantity of ATP with microtubules and regulate its behavior using light intensity. When the light intensity is increased, a larger number of motor proteins and microtubules become attached, leading to increased system activity and ultimately resulting in more defects. Fig. 12 and 13 show the performance of our approach in both low and high light intensity experiments. Note that the high-intensity sample (Fig. 13) has not reached steady-state, and thus the number of defects is increasing over time. Using the director fields extracted by our machine learning model, the numbers of defects closely track the hand-labeled ground-truth. Moreover, we closely observed the defect location generated by both methods. Compared with the manually annotated location, if the detected location is within a 40-pixel radius, which is roughly half of the averaged defect size (100 um, 77 pixels), it is considered a precise detection. The size of the raw image is 1296 × 1280. Fig. 14 compares the percentage of precise detections using both methods. The director field extracted by our machine learning model enables better defect detection results (both defect detection rate and location).
3.4 Defect tracking
Once defects are detected, we can track them over time. This can be achieved by matching the locations of defects across different frames using the graduated assignment algorithm.50 This article proposed a fast and accurate graph matching algorithm using nonlinear optimization and graduated non-convexity to avoid poor local minima. This algorithm allows matching between graphs of different node numbers. This fits well with our system because it enables handling the spontaneous birth and annihilation of defects. However, it is possible that one defect is detected in one frame, but disappears in the next frame. This could be due to defect annihilation or a mistake in the defect detection process. To deal with the latter, we apply the matching algorithm to three consecutive frames. Using the matching results, we can interpolate the locations of the defects that are missed in the middle frame but are detected in the preceding and succeeding frames. Fig. 15 illustrates the tracking results on three image sequences captured under different lighting conditions.3.5 Defect angle estimation
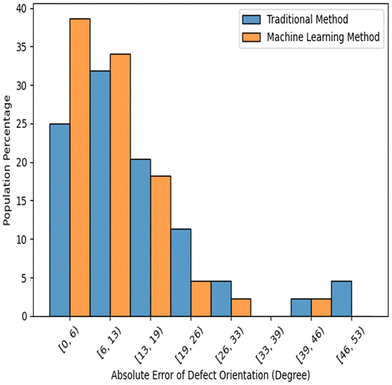
One natural extension from the foundation of an accurate director field is to identify the microtubule angles at the defect locations, which may provide insights into the dynamics of the active nematic system. This can be conveniently achieved using a similar CNN network introduced in Section 2.2 and 2.3. The image processing method (TM) can also derive approximate defect orientations (details are in Appendix A.1). To obtain accurate angles at defect locations, we manually labeled 800 defects on fluorescence images using the Python package “PyQt5”, out of which, 390 were negative and 410 were positive defects. We ran our model and the TM on the same images and compared the angle differences with the ground-truth. Among these defects, our model and the TM resulted in average errors (defined as the difference in detected angle from the ground-truth angle) of 9.86° and 14.33° respectively. The comparison results are summarized in Fig. 16.4 Conclusions and discussions
We present a machine learning based approach for automatically extracting director fields from images of active nematics, which enables downstream analyses, such as topological defect detection and tracking, defect orientation estimation, and related characteristics. Our approach is less sensitive to noise than the traditional method (TM) and can generalize to images captured by settings (e.g., microscope instruments, lighting conditions, and camera focus) not encountered in producing the training images. Comparison of results from applying our method and (TM) to experimental data demonstrates the superior performance of the machine learning approach. Despite the advances relative to traditional image processing methods, we note that our method does not entirely compensate for all kinds of noise, particularly in suboptimal regions with over-/under-exposure or containing additional microtubule filaments above the plane of the active nematic. Furthermore, as our model is trained on PolScope images, its performance may vary slightly when applied to raw images from other microscopy techniques due to differences in noise distributions. Nevertheless, machine learning models have consistently demonstrated their potential in the realm of soft matter research, providing precise and automated solutions for diverse tasks, from dynamic predictions28 to defect analyses.56,57 Our algorithm can be further improved by training on other types of microscopy images.By employing the suggested machine learning method, we can obtain director fields with greater accuracy than the TM. This enhances the reliability of detecting and pinpointing defects. Such improved data is vital for addressing scientific questions regarding the time taken for a system to reach a steady state, the spatial distribution of defects, and their rates of annihilation and proliferation. In the future, we plan to use the calculated director field to do dynamics forecasting. In other words, given a short time sequence of images from an active nematic experiment, we aim to predict the subsequent time evolution of the sample. The more accurate calculation of director fields and detection of defects should lead to improvements over current algorithms.28,29 Forecasting is the first step toward being able to control the behaviors of an active nematic system, by actuating with light or other external control inputs. Similarly, we hope to use this approach to derive statistics of the system's characteristics and link them with the dynamics of active nematic systems. More broadly, our approach could be generalized to other active matter and soft matter systems.
Author contributions
Yunrui Li: methodology, investigation, visualization, writing – original draft. Zahra Zarei: data curation, writing – review & editing. Phu N. Tran: validation. Yifei Wang: validation. Aparna Baskaran: writing – review & editing. Seth Fraden: writing – review & editing. Michael F. Hagan: writing – review & editing. Pengyu Hong: conceptualization, supervision, writing – review & editing.Conflicts of interest
There are no conflicts of interest to declare.Appendices
A.1 Traditional image processing method used
The traditional image processing method used as a benchmark in this paper is from Michael M. Norton. The steps are below:• Calculate the intensity change of a given image.
– Apply a Gaussian filter:
– Compute the gradient:
| ∇G(x,y) = (∇xG,∇yG), |
– Compute the gradient magnitude:
Optionally, compute the gradient direction:
θ(x,y) = atan![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 2(∇yG(x,y),∇xG(x,y)), 2(∇yG(x,y),∇xG(x,y)), |
![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 2 is the four-quadrant inverse tangent that returns values in the closed interval [−π, π].
2 is the four-quadrant inverse tangent that returns values in the closed interval [−π, π].
• Derive orientation.
– Since the microtubule bundles have head–tail symmetry, we can use the rank-2 gradient tensor:
– Compute the eigenvalues of the gradient tensor:
– The eigenvalues λ1 and λ2 represent the strength of the gradient at each pixel in the direction of the two corresponding eigenvectors. The director field should be the smaller of the two eigenvalues.35
After the director field is calculated, topological defects can be identified from regions of rapid rotation of the director field.15,34 The winding number is used to measure the change. The winding number is a mathematical concept that describes the number of times a curve, typically a closed curve, wraps around a given point in a plane. In the context of liquid crystals and active nematics, the winding number is a topological concept that captures the number of times the director rotates around a closed loop in the director field. More formally, it quantifies how many times the director completes a full rotation (360 degrees) when one traverses a closed curve in the plane of the field. It is a useful tool in topology and geometry for characterizing the behavior of curves and shapes. To calculate the winding number using the director field, we can choose a point in the space and numerically count the local orientation change around this point. Applying a threshold, the topological defect can be detected.
To deduce the intrinsic orientation of a defect, a circular window is defined around every detected defect location. For every pixel along this window, the local orientation of the director field is noted. Then, the angle originating from this pixel to the defect's center is computed. Next, we compute the scalar product of these two angles. Then, the pixel yielding the maximum scalar product value indicates the direction in the director field most closely aligned with a line radiating from the defect's core. We use the orientation of the director field in this specific pixel as a proxy to infer the intrinsic orientation of the defect.
A.2 Details of the Gabor filter
A Gabor filter46 is a type of linear filter used in image processing that is designed to respond to specific frequencies or orientations in an image. In the context of Polscope images of active nematics, Gabor filters can be used to enhance the visibility of the nematic texture and reduce the effects of noise. Eqn (1) gives the equation of the Gabor filter. It is composed of two components: a complex sinusoidal wave and a Gaussian kernel. The Gaussian kernel allows the filter to respond to features of varying sizes, while the sinusoidal wave allows the filter to respond to features of varying orientations. By computing the Gabor response at multiple scales and orientations, features such as texture, edges, and corners can be identified and used to segment and classify the image.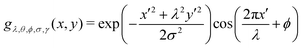 | (1) |
x′ = x![[thin space (1/6-em)]](https://www.rsc.org/images/entities/i_char_2009.gif) cos cos![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) θ + y θ + y![[thin space (1/6-em)]](https://www.rsc.org/images/entities/i_char_2009.gif) sin sin![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) θ, y′ = x θ, y′ = x![[thin space (1/6-em)]](https://www.rsc.org/images/entities/i_char_2009.gif) sin sin![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) θ + y θ + y![[thin space (1/6-em)]](https://www.rsc.org/images/entities/i_char_2009.gif) cos cos![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) θ. θ. |
Here, x and y are the coordinates of image pixels, σ is the width of the Gaussian kernel, θ is the angle of the wave function, ϕ is the phase, and λ is the wavelength.
A.3 Model parameters of the Gabor filter and ResNet blocks
For each wavelength, we use 64 Gabor filters with different orientations and scale values. The orientations take values with an additive interval of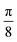 from
from 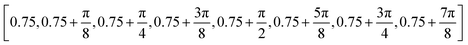 . The scales take values with a multiplicative interval of
. The scales take values with a multiplicative interval of 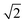 from 0.75 to
from 0.75 to 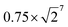 .
.
In the ResNet structure shown in Fig. 7, each block comprises two sets of convolutional and batch normalization layers linked by ReLU activation.49 The input is reintroduced to the output after the second batch normalization layer to guarantee reliable and resilient gradient computation. To begin the ResNet architecture, a single convolutional and batch normalization layer is employed to project the grayscale input image onto 64 channels. The subsequent three blocks continue to extract image features on 64 channels. In the subsequent four blocks, the results are projected to 128 channels. Following this, six blocks are used to capture higher-level features on 256 channels. The last three blocks extract global features on 512 channels. Following the completion of the ResNet blocks, the 512-channel image is projected back to grayscale using two convolutional layers. The purpose of increasing the number of dimensions is to allow the network to capture more complex and abstract features from the input image. With a higher number of channels, the convolutional layers can learn to detect more diverse patterns and structures within the image, leading to better performance.
A.4 Data augmentation
Data augmentation is a technique used to increase the size and diversity of a training dataset. In our model, we have applied a set of random transformations to the original data, including cropping, rotating, or adding noise. In active nematics experiments, free or extra microtubule filaments may float in the container, making the local orientation calculation difficult or inaccurate. To alleviate this, we proactively add random white lines in the input image to simulate the extra filaments. The transformed data is then added to the original dataset, increasing its size and variety, which helps to improve the performance of the machine learning model. Data augmentation can also help to reduce overfitting and make the model more robust to unseen data. By augmenting the training data with a variety of random transformations, we can help the model learn more generalizable features, which can improve its performance on new data.A.5 Tracking visualization over longer sequences
With the matching algorithm discussed in Section 3.4, we can perform defect tracking over a long sequence of images. Please refer to ESI† for a video of tracking results.A.6 More supporting figures
Fig. 17 illustrates the machine learning model's capability in processing fluorescence images of a 2D active nematic system with varying boundary conditions. The authors highlight that even in the presence of unbound microtubules floating along the z-axis of the 3D container, which is not aligned with the local orientation within the 2D nematic layer, the machine learning model adeptly filters out these elements, ensuring an accurate calculation of the director field.Fig. 18 showcases some examples in which some defects cannot be detected from the PolScope-calculated director field. We note that the local orientation by PolScope calculation is mostly accurate and the defect detection accuracy is generally good. The main contribution of this work is to develop a robust model that is less sensitive to noises in raw images and can be applied to experimental images from a variety of instruments.
Fig. 19 visualizes the 40-pixel threshold used to evaluate defect detection accuracy.
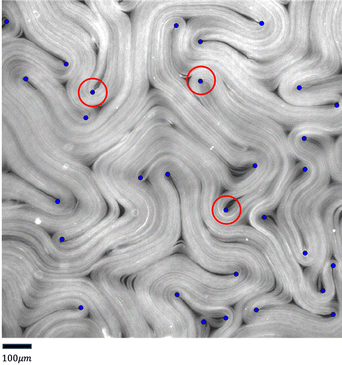 | ||
| Fig. 19 Visualization of the 40-pixel threshold (red circle) on fluorescence images, used in the evaluation of defect detection accuracy discussed in Section 3.3 and Fig. 14. | ||
Fig. 20 demonstrates the defect detection results using the TM generated director field. It is an extended analysis to Fig. 5.
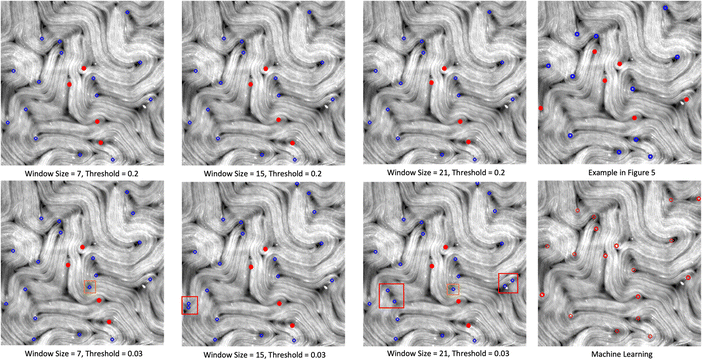 | ||
| Fig. 20 An extension to Fig. 5 to show how the results of the TM depend on the key parameter values of window size and defect detection threshold. We find that changes in the window size parameter may alter the defect detection results, but 4–5 defects remain undetected (solid red dots for each parameter setting) even for optimal values. Setting the defect detection threshold lower can lead to finding more defects, but also leads to multiple defects in one location (red box), or detection of non-existing defects (orange box). | ||
Acknowledgements
This work was primarily supported by the Department of Energy (DOE) DE-SC0022291 (YL, PNT, YW, SF, MFH, PH). The experimental data acquisition and curation were supported by the Brandeis NSF MRSEC, Bioinspired Soft Materials, DMR-2011846 (ZZ). We also acknowledge computational support from the Brandeis HPCC which is partially supported by NSF OAC-1920147.Notes and references
- T. B. Saw, A. Doostmohammadi, V. Nier, L. Kocgozlu, S. Thampi, Y. Toyama, P. Marcq, C. T. Lim, J. M. Yeomans and B. Ladoux, Nature, 2017, 544, 212–216 CrossRef CAS PubMed.
- K. Kawaguchi, R. Kageyama and M. Sano, Nature, 2017, 545, 327–331 CrossRef CAS PubMed.
- J. Prost, F. Jülicher and J.-F. Joanny, Nat. Phys., 2015, 11, 111–117 Search PubMed.
- A. Sokolov, I. S. Aranson, J. O. Kessler and R. E. Goldstein, Phys. Rev. Lett., 2007, 98, 158102 CrossRef PubMed.
- H. H. Wensink, J. Dunkel, S. Heidenreich, K. Drescher, R. E. Goldstein, H. Löwen and J. M. Yeomans, Proc. Natl. Acad. Sci. U. S. A., 2012, 109, 14308–14313 CrossRef CAS PubMed.
- J. Dunkel, S. Heidenreich, K. Drescher, H. H. Wensink, M. Bär and R. E. Goldstein, Phys. Rev. Lett., 2013, 110, 228102 CrossRef PubMed.
- J. Toner and Y. Tu, Phys. Rev. Lett., 1995, 75, 4326 CrossRef CAS PubMed.
- Y. Katz, K. Tunstrøm, C. C. Ioannou, C. Huepe and I. D. Couzin, Proc. Natl. Acad. Sci. U. S. A., 2011, 108, 18720–18725 CrossRef CAS PubMed.
- M. C. Marchetti, J. F. Joanny, S. Ramaswamy, T. B. Liverpool, J. Prost, M. Rao and R. A. Simha, Rev. Mod. Phys., 2013, 85, 1143–1189 CrossRef CAS.
- S. Ramaswamy, Annu. Rev. Condens. Matter Phys., 2010, 1, 323–345 CrossRef.
- S. J. DeCamp, G. S. Redner, A. Baskaran, M. F. Hagan and Z. Dogic, Nat. Mater., 2015, 14, 1110–1115 CrossRef CAS PubMed.
- T. Sanchez, D. T. Chen, S. J. DeCamp, M. Heymann and Z. Dogic, Nature, 2012, 491, 431–434 CrossRef CAS PubMed.
- G. Henkin, S. J. DeCamp, D. T. Chen, T. Sanchez and Z. Dogic, Philos. Trans. R. Soc., A, 2014, 372, 20140142 CrossRef PubMed.
- E. Putzig, G. S. Redner, A. Baskaran and A. Baskaran, Soft Matter, 2016, 12, 3854–3859 RSC.
- A. J. Tan, E. Roberts, S. A. Smith, U. A. Olvera, J. Arteaga, S. Fortini, K. A. Mitchell and L. S. Hirst, Nat. Phys., 2019, 15, 1033–1039 Search PubMed.
- M. Serra, L. Lemma, L. Giomi, Z. Dogic and L. Mahadevan, Nat. Phys., 2023, 1–7 Search PubMed.
- J. Hardoüin, C. Doré, J. Laurent, T. Lopez-Leon, J. Ignés-Mullol and F. Sagués, Nat. Commun., 2022, 13.1, 6675 Search PubMed.
- M. F. Hagan and A. Baskaran, Curr. Opin. Cell Biol., 2016, 38, 74–80 CrossRef CAS PubMed.
- M. Bär, R. Großmann, S. Heidenreich and F. Peruani, Annu. Rev. Condens. Matter Phys., 2020, 11, 441–466 CrossRef.
- O. Bantysh, J. Nambisan, B. Martnez-Prat, A. Fernández-Nieves, F. Sagués and J. Ignés-Mullol, arXiv, 2023, preprint, arXiv:2303.05589 DOI:10.48550/arXiv.2303.05589.
- M. Golden, R. O. Grigoriev, J. Nambisan and A. Fernandez-Nieves, Sci. Adv., 2023, 9, eabq6120 CrossRef CAS PubMed.
- A. Doostmohammadi, J. Ignés-Mullol, J. M. Yeomans and F. Sagués, Nat. Commun., 2018, 9, 3246 CrossRef PubMed.
- F. C. Keber, E. Loiseau, T. Sanchez, S. J. DeCamp, L. Giomi, M. J. Bowick, M. C. Marchetti, Z. Dogic and A. R. Bausch, Science, 2014, 345, 1135–1139 CrossRef CAS PubMed.
- A. Doostmohammadi, M. F. Adamer, S. P. Thampi and J. M. Yeomans, Nat. Commun., 2016, 7, 10557 CrossRef CAS PubMed.
- R. Oldenbourg, et al., Live cell imaging: A laboratory manual, 2005, pp. 205–237 Search PubMed.
- S. Zhou, A. Sokolov, O. D. Lavrentovich and I. S. Aranson, Proc. Natl. Acad. Sci. U. S. A., 2014, 111, 1265–1270 CrossRef CAS PubMed.
- M. Shribak and R. Oldenbourg, Appl. Opt., 2003, 42, 3009–3017 CrossRef PubMed.
- Z. Zhou, C. Joshi, R. Liu, M. M. Norton, L. Lemma, Z. Dogic, M. F. Hagan, S. Fraden and P. Hong, Soft Matter, 2021, 17, 738–747 RSC.
- J. Colen, M. Han, R. Zhang, S. A. Redford, L. M. Lemma, L. Morgan, P. V. Ruijgrok, R. Adkins, Z. Bryant and Z. Dogic, et al. , Proc. Natl. Acad. Sci. U. S. A., 2021, 118, e2016708118 CrossRef CAS PubMed.
- Z. Zarei, J. Berezney, A. Hensley, L. Lemma, N. Senbil, Z. Dogic and S. Fraden, Soft Matter, 2023, 19, 6691–6699 RSC.
- R. Zhang, S. A. Redford, P. V. Ruijgrok, N. Kumar, A. Mozaffari, S. Zemsky, A. R. Dinner, V. Vitelli, Z. Bryant and M. L. Gardel, et al. , Nat. Mater., 2021, 20, 875–882 CrossRef CAS PubMed.
- L. M. Lemma, M. Varghese, T. D. Ross, M. Thomson, A. Baskaran and Z. Dogic, PNAS Nexus, 2023, 2(5), gad130 CrossRef PubMed.
- T. D. Ross, H. J. Lee, Z. Qu, R. A. Banks, R. Phillips and M. Thomson, Nature, 2019, 572, 224–229 CrossRef CAS PubMed.
- X.-q Shi and Y.-q Ma, Nat. Commun., 2013, 4, 3013 CrossRef PubMed.
- R. Blackwell, O. Sweezy-Schindler, C. Baldwin, L. E. Hough, M. A. Glaser and M. Betterton, Soft Matter, 2016, 12, 2676–2687 RSC.
- G. Duclos, R. Adkins, D. Banerjee, M. S. Peterson, M. Varghese, I. Kolvin, A. Baskaran, R. A. Pelcovits, T. R. Powers and A. Baskaran, et al. , Science, 2020, 367, 1120–1124 CrossRef CAS PubMed.
- R. Rezakhaniha, A. Agianniotis, J. T. C. Schrauwen, A. Griffa, D. Sage, C. V. Bouten, F. Van De Vosse, M. Unser and N. Stergiopulos, Biomech. Model. Mechanobiol., 2012, 11, 461–473 CrossRef CAS PubMed.
- P. W. Ellis, D. J. Pearce, Y.-W. Chang, G. Goldsztein, L. Giomi and A. Fernandez-Nieves, Nat. Phys., 2018, 14, 85–90 Search PubMed.
- F. Cichos, K. Gustavsson, B. Mehlig and G. Volpe, Nat. Mach. Intell., 2020, 2, 94–103 Search PubMed.
- J. Rabault, J. Kolaas and A. Jensen, Meas. Sci. Technol., 2017, 28, 125301 CrossRef.
- M. D. Hannel, A. Abdulali, M. OBrien and D. G. Grier, Opt. Express, 2018, 26, 15221–15231 CrossRef CAS PubMed.
- G. Muñoz-Gil, M. A. Garcia-March, C. Manzo, J. D. Martn-Guerrero and M. Lewenstein, New J. Phys., 2020, 22, 013010 CrossRef.
- N. D. Mermin, Rev. Mod. Phys., 1979, 51, 591 CrossRef CAS.
- Z. Xie, Z. Zhang, Y. Cao, Y. Lin, J. Bao, Z. Yao, Q. Dai and H. Hu, Proceedings of the IEEE/CVF Conference on Computer Vision and Pattern Recognition, 2022, pp. 9653–9663.
- Z. Liu, J. Ning, Y. Cao, Y. Wei, Z. Zhang, S. Lin and H. Hu, Proceedings of the IEEE/CVF conference on computer vision and pattern recognition, 2022, pp. 3202–3211.
- I. Fogel and D. Sagi, Biol. Cybern., 1989, 61, 103–113 CrossRef.
- F. Bianconi and A. Fernández, Pattern Recogn., 2007, 40, 3325–3335 CrossRef.
- K. He, X. Zhang, S. Ren and J. Sun, Proceedings of the IEEE Conference on Computer Vision and Pattern Recognition (CVPR), 2016.
- A. L. Maas, A. Y. Hannun and A. Y. Ng, et al. , Proc. ICML, 2013, 3 Search PubMed.
- S. Gold and A. Rangarajan, IEEE Trans. Pattern Anal. Mach. Intell., 1996, 18, 377–388 CrossRef.
- A. Opathalage, M. M. Norton, M. P. Juniper, B. Langeslay, S. A. Aghvami, S. Fraden and Z. Dogic, Proc. Natl. Acad. Sci. U. S. A., 2019, 116, 4788–4797 CrossRef CAS PubMed.
- K. Thijssen, D. A. Khaladj, S. A. Aghvami, M. A. Gharbi, S. Fraden, J. M. Yeomans, L. S. Hirst and T. N. Shendruk, Proc. Natl. Acad. Sci. U. S. A., 2021, 118, e2106038118 CrossRef CAS PubMed.
- L. Zhao, L. Yao, D. Golovaty, J. Ignés-Mullol, F. Sagués and M. Carme Calderer, Chaos, 2020, 30, 113105 CrossRef PubMed.
- J. Hardoüin, R. Hughes, A. Doostmohammadi, J. Laurent, T. Lopez-Leon, J. M. Yeomans, J. Ignés-Mullol and F. Sagués, Commun. Phys., 2019, 2, 121 CrossRef.
- L. M. Lemma, S. J. DeCamp, Z. You, L. Giomi and Z. Dogic, Soft Matter, 2019, 15, 3264–3272 RSC.
- E. N. Minor, S. D. Howard, A. A. Green, M. A. Glaser, C. S. Park and N. A. Clark, Soft Matter, 2020, 16, 1751–1759 RSC.
- R. A. Chowdhury, A. A. Green, C. S. Park, J. E. Maclennan and N. A. Clark, Phys. Rev. E, 2023, 107, 044701 CrossRef CAS PubMed.
Footnotes |
| † Electronic supplementary information (ESI) available. See DOI: https://doi.org/10.1039/d3sm01253k |
| ‡ Additionally, data acquisition with PolScope requires taking 5 images in different polarization settings to calculate the retardance image and its director field,27 which limits the frame rate for taking data. For example, with a camera exposure time of 500 ms, it takes 670 ms to capture a single image and roughly 3.4 sec for five images. The impact of these exposure times on the observations becomes noticeable when the nematic speed is high, as the motion between frames is difficult to follow, and consequently, tracking defects becomes less precise. |
| This journal is © The Royal Society of Chemistry 2024 |