Open Access Article
Open Access ArticleCreative Commons Attribution 3.0 Unported Licence
Adhesion-based high-throughput label-free cell sorting using ridged microfluidic channels
Fatima Ezahra
Chrit
,
Peiru
Li
 ,
Todd
Sulchek
,
Todd
Sulchek
 and
Alexander
Alexeev
and
Alexander
Alexeev
 *
*
George W. Woodruff School of Mechanical Engineering, Georgia Institute of Technology, Atlanta, Georgia 30332, USA. E-mail: alexander.alexeev@me.gatech.edu
First published on 5th February 2024
Abstract
Numerous applications in medical diagnostics, cell engineering therapy, and biotechnology require the identification and sorting of cells that express desired molecular surface markers. We developed a microfluidic method for high-throughput and label-free sorting of biological cells by their affinity of molecular surface markers to target ligands. Our approach consists of a microfluidic channel decorated with periodic skewed ridges and coated with adhesive molecules. The periodic ridges form gaps with the opposing channel wall that are smaller than the cell diameter, thereby ensuring cell contact with the adhesive surfaces. Using three-dimensional computer simulations, we examine trajectories of adhesive cells in the ridged microchannels. The simulations reveal that cell trajectories are sensitive to the cell adhesion strength. Thus, the differential cell trajectories can be leveraged for adhesion-based cell separation. We probe the effect of cell elasticity on the adhesion-based sorting and show that cell elasticity can be utilized to enhance the resolution of the sorting. Furthermore, we investigate how the microchannel ridge angle can be tuned to achieve an efficient adhesion-based sorting of cells with different compliance.
1 Introduction
Identification and isolation of cells that express desired molecular surface markers are required in a variety of applications in the biological sciences, cell therapy, and medical diagnostics.1–4 For example, adhesion-based cell sorting is needed in the manufacturing workflow of chimeric antigen receptor (CAR) T cell therapies. A major challenge to improving these therapies is a lack of efficient methods to evaluate CAR expression efficiency in conjunction with CD4 and CD8 expression, required as part of the release criteria. In addition, there is a lack of separation methods that are sensitive to cell surface expression, while not inducing changes in cell morphology, viability, molecular content, activation state, and phenotype.Labeling methods (for example: FACS and MACS) can be used for adhesion-based cell sorting and offer high purity with high enrichment. However, these techniques provide a binary output for the receptor expression and are not yet able to fractionate into multiple outputs for finer sensitivity to the molecule of interest. Another major drawback of these methods is the changes they induce by labeling tags or tag removal, such as the risk of tag-induced activation of sorted cells, which restrains their downstream use.5,6 To solve this issue, negative selection of target cells can be used. Yet, even this can contaminate therapy products with magnetic beads coated with foreign antibodies.
To overcome these difficulties, label-free approaches can be employed leveraging differences in cells adhesive properties as the mechanism of separation.7–9 As an example of cell adhesion sorting using micro/nano structures, Kwon et al.10 used patterned microchannels to separate and enrich human breast cancer cells from epithelial cells. Detachment assays were used to measure cells adhesion. Specific adhesion of cells using proteins, peptides, and other molecular mediators can be employed to capture desired cells. Antibody coated surfaces in microfluidic channels were demonstrated to be effective for capturing circulating endothelial progenitor cells (EPCs).11 Hashimoto, Kaji, and Nishizawa12 combined dielectrophoresis with adhesion-based microfluidics to selectively capture neutrophils from a mixture of leukocytes. Zhang et al.13 used a microchannel coated with basement membrane extract to separate heterogeneous breast cancer cell lines based on their adhesive capacities. Yang et al.14 achieved circulating tumor cell (CTCs) capture from blood cells using a TA-functionalized film. Negative selection is also an option for enriching rare target cells.15,16 A tandem microfluidic systems was developed to isolate in a high-throughput, label free manner CTCs that exhibit high compliance and low adhesion.17 Singh et al.18 were able to isolate fully reprogrammed induced human pluripotent stem cells (hPSCs) from heterogeneous reprogramming cultures by capturing target cells on a substrate in a microchannel under shear flow. Microfluidic assay was developed to separate genetically-related yeast strains based on adhesion strength enabling the rapid screening and fractionation of yeast based on adhesive properties.19 However, these methods are not continuous since they capture and arrest cells from the flow. Chang, Lee, and Liepmann20 used microstructured fluidic channels with array of pillars coated with E-selectin to capture and arrest cells from the free flow. Bose et al.21 demonstrated affinity flow fractionation where interactions with asymmetric molecular patterns laterally displace cells in a continuous manner. Similarly, Choi, Karp, and Karnik22 described a separation process called “deterministic cell rolling”, which separates cells based on transient molecular interactions with slanted ridges on the channel floor in a continuous process. However, these approaches are limited to low flow rates to maintain rolling interactions with the adhesive surface and to prevent hydrodynamic forces from detaching the cells without any separation.
A high throughput adhesion based sorting method was recently reported23 that consists of a microchannel decorated with diagonal ridges and coated with adhesion molecules. Flow cytometry data for cells collected at the microchannel outlets showed different cell distributions correlated with the differences in cell adhesiveness indicating sorting dependence on adhesion levels. A unique aspect of this approach is the ability to operate at high throughput. The experiments demonstrated flow of cells at rates up to 0.2 m s−1, which is over two orders of magnitude higher than previous methods. Results indicated that sorting did not activate the cells due to the brief interaction time of cells with adhesion ligands beneficial for downstream cell analysis. We note that ridged microfluidic channels have been previously used to sort cells and microparticles by size and elasticity.24–27
Computational modeling has been used to probe adhesive behavior of cells, especially in the context of cell rolling.28–30 These models are usually based on theoretical frameworks which describe adhesive interactions using chemical reaction kinetics and relate the kinetic rates to the applied force. Bell31 and Dembo et al.32 adapted this kinetic theory and proposed an exponential constitutive law between the dissociation rate and the force. Evans, Berk, and Leung33 and Evans and Ritchie34 employed a power law instead of the exponential law used in the Bell model.31 Jadhav, Eggleton, and Konstantopoulos35 developed a three-dimensional computational model to predict receptor-mediated rolling of deformable cells in shear flow. Hammer and Apte36 simulated the rolling and adhesion of selectin-mediated neutrophil using a kinetics framework. Khismatullin and Truskey37 used a modified Dembo model and evaluated the effect of cell deformability and viscoelasticity on receptor-mediated leukocyte adhesion to ligand coated surface. Interactions of compliant adhesive capsules and cells in patterned microfluidic channels have been studied using computer simulations with applications to the sorting and separation.38–41
In this work, we first examine the rolling of deformable cells represented by a fluid-filled elastic particle on a flat substrate patterned with diagonal adhesive stripes. We show that the cells follow different trajectories based on the adhesiveness and explain the sorting mechanism. We further show that this separation method is limited to low flow rates due to the hydrodynamic lift detaching cells from the substrate. We then demonstrate high-throughput sorting of adhesive cells in ridged microfluidic channels. The periodic diagonal ridges compress cells propelled by the flow, thereby ensuring cell contact with adhesive surfaces even at high flow rates. We systematically investigate the effects of cell adhesion and elasticity on cell trajectories in the ridged microchannel and demonstrate that cells with different adhesiveness follow different trajectories. The simulations reveal that cell elasticity can be used to enhance sorting of adhesive cells. Furthermore, we show that the resolution of the adhesion-based cell sorting can be increased by adjusting the angle of the microchannel ridges. Together these studies suggest ways to improve the sensitivity of separation to adhesion in the limit of transient interactions characterized by fast timescales.
2 Methods
We employ a three-dimensional computational approach42–45 to model the motion of compliant neutrally-buoyant adhesive cells in a microfluidic channel filled with a viscous fluid. The simulations are performed using an inhouse three-dimensional fluid-structure interaction solver based on the lattice Boltzmann method (LBM) for fluid flow coupled in a two-way manner with a lattice spring model (LSM) for solid mechanics.LBM uses time integration of the discretized Boltzmann equation fi(r + ciΔt, t + Δt) = fi(r, t) + Δi[f(r, Δt)] for a distribution function fi(r, t) describing the mass density of fluid particles at a lattice node r and time t propagating along a lattice direction i with a constant velocity ci.46,47 Here, Δt is the time step. The collision operator Δi describes the change in fi due to collisions at lattice nodes. In three dimensions, we use a cubic lattice D3Q19 with 19 discrete velocities.
The compliant cell is modelled as a fluid-filled elastic membrane. LSM uses a triangulated network of harmonic springs that connect regularly spaced mass nodes on the cell surface to represent an elastic membrane.48,49 The stretching elastic energy associated with a node at a position ri is 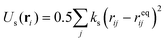 where rij = |ri − rj| and reqij are respectively the length and the equilibrium length of a spring between two nodes with positions ri and ri, and ks is the spring constant. Bending rigidity of cell membrane is included by considering the change of dihedral angle of adjacent pairs of triangles. The bending energy is given by
where rij = |ri − rj| and reqij are respectively the length and the equilibrium length of a spring between two nodes with positions ri and ri, and ks is the spring constant. Bending rigidity of cell membrane is included by considering the change of dihedral angle of adjacent pairs of triangles. The bending energy is given by 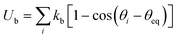 , where kb is the bending spring constant, θi is the instantaneous angle between two adjacent triangles with a common edge i, and θeq is the equilibrium angle.
, where kb is the bending spring constant, θi is the instantaneous angle between two adjacent triangles with a common edge i, and θeq is the equilibrium angle.
To model adhesion between the cell and microchannel surfaces, we use the Morse potential that has a repulsive and attractive components, depending on the separation distance. The Morse potential associated with two nodes separated by a distance r is given by 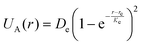 , where De is the depth of the potential well, Ke is a parameter controlling the width of the potential, and re is the equilibrium distance. The force derived from the Morse potential is given by
, where De is the depth of the potential well, Ke is a parameter controlling the width of the potential, and re is the equilibrium distance. The force derived from the Morse potential is given by 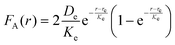 . Each LSM node on the particle surface can adhesively interact with nodes on the microchannel surfaces located within a cut-off distance rcut = 1.5re.
. Each LSM node on the particle surface can adhesively interact with nodes on the microchannel surfaces located within a cut-off distance rcut = 1.5re.
The fluid and solid models are coupled via the boundary conditions at the fluid–solid interface that utilize the interpolated bounce back method50,51 and the momentum exchange method.52 The computational methodology for fluid structure interactions has been previously extensively validated by simulating flows with different rigid and deformable particles including mesh independence study, and found good agreement with relevant theories and experiments.24,26,53–56
3 Computational setup
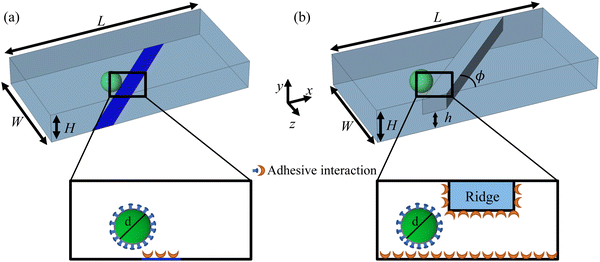
The computational setup for simulating cells propelled by a fluid flow in a microchannel where the substrate is patterned with adhesive patches is illustrated in Fig. 1a, whereas the setup for simulating cell motion in a microfluidic channel with diagonal ridges is presented in Fig. 1b. The following dimensions are used for the computational setups: height H = 1.5d, width W = 5d, and length L = 10d. The adhesive stripe and ridge have a width of Wr = 1.5d and are inclined at an angle ϕ with respect to the channel axis. The ridge forms a gap h with the bottom wall of the channel. The magnitude of the gap λ = h/d is normalized by the particle diameter d = 20. Note that we express the dimensional quantities in LB units unless indicated otherwise.To characterize the flow within the channel, we define a channel Reynolds number 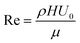 , which represents the ratio between the fluid inertia and viscous forces. The Reynolds number is based on the channel height H, the fluid density ρ and viscosity μ, and the characteristic flow velocity in the channel U0 due to a pressure gradient ΔP/L, defined as
, which represents the ratio between the fluid inertia and viscous forces. The Reynolds number is based on the channel height H, the fluid density ρ and viscosity μ, and the characteristic flow velocity in the channel U0 due to a pressure gradient ΔP/L, defined as 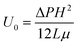 . The compliant particle is characterized by a capillary number
. The compliant particle is characterized by a capillary number 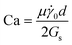 . Here,
. Here, 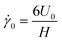 is the characteristic shear rate, and
is the characteristic shear rate, and 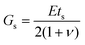 is the two-dimensional shear modulus of the particle elastic shell, where E is the particle Young's modulus, ts is the shell thickness, and ν is the Poisson's ratio. Thus, the capillary number represents the ratio between viscous stresses on the particle and elastic stiffness of the particle shell. The adhesion level is controlled by varying the dimensionless adhesion strength
is the two-dimensional shear modulus of the particle elastic shell, where E is the particle Young's modulus, ts is the shell thickness, and ν is the Poisson's ratio. Thus, the capillary number represents the ratio between viscous stresses on the particle and elastic stiffness of the particle shell. The adhesion level is controlled by varying the dimensionless adhesion strength 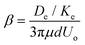 .
.
In our simulations the particles are initially positioned at the channel centerline. The particle trajectory is represented using dimensionless particle's center of mass coordinates Cx = cx/d and Cz = cz/d. The particle motion is quantified using the dimensionless particle displacement per ridge 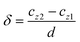 where cz1 and cz2 are the particle's center of mass z-coordinate at x = L/3 and x = L, respectively.
where cz1 and cz2 are the particle's center of mass z-coordinate at x = L/3 and x = L, respectively.
4 Results and discussion
4.1 Cell motion over adhesive stripes
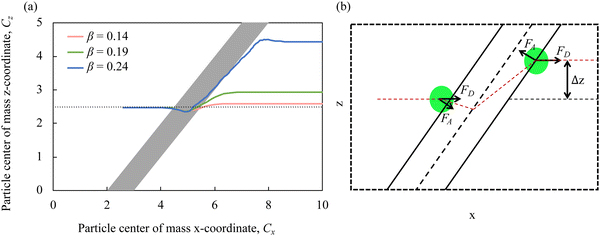
We first consider the case of compliant cells rolling on a substrate patterned with diagonal adhesive stripes, as shown in Fig. 1a. Here, only the diagonal patch shown in blue is adhesive, whereas the remaining channel surfaces are non-sticky. The motion of adhesive cells over microfluidic surfaces patterned with stripes was previously examined computationally and experimentally.39,57,58Fig. 2a presents the trajectories of the particle's center of mass in the x–z plane for particles with different adhesion strengths β and Ca = 3.5 × 10−3. As the adhesion strength β increases, the deflection δ increases. In Fig. 2b, we show a schematic illustration of the particle trajectory across an adhesive diagonal stripe and the main forces acting on the particle. Before encountering the stripe, the particle moves along the channel centerline due to a drag force FD. When the particle approaches the adhesive stripe, it is pulled towards the centerline of the stripe by an adhesive force FA. The adhesive force deflects the particle trajectory in the negative x direction. When the particle crosses the middle of the stripe, the adhesive force flips its direction and acts to keep the particle on the stripe to minimize the energy.39 The combined action of FA and FD propels the particle along the stripe until it reaches the back edge of the stripe where the action of the adhesive force diminishes. After that the particle moves along the flow direction due to the drag force FD. As a result, the particle experiences a net positive lateral displacement Δz with increasing adhesion.
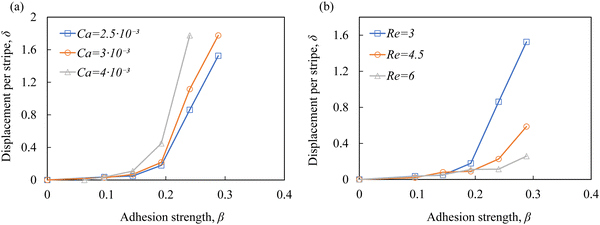
Fig. 3a shows how the dimensionless particle displacement δ = Δz/d depends on the adhesion strength β for particles with different elasticity Ca. We find that δ increases with increasing β. Furthermore, δ increases with increasing Ca. Thus, softer particles experience larger lateral displacement as they cross the adhesive stripe, which can be explained by a larger contact area between softer particles and the adhesive stripe increasing the free energy change and, therefore, FA.39
Fig. 3b shows δ as function of β for different channel flow rates expressed in terms of the Reynolds number Re. We find that the deflection decreases as Re increases due to a greater contribution of the hydrodynamic drag force FD. Furthermore, the increase of Re above the values reported in Fig. 1b results in the particle failure to interact with the adhesive stripe due to an increased lift force translating the particle away from the substrate. Thus, the method while can be efficient in separating cells is limited to relatively low flow rates57,58 and, therefore, low cell throughput.
4.2 Cell motion in ridged microchannel
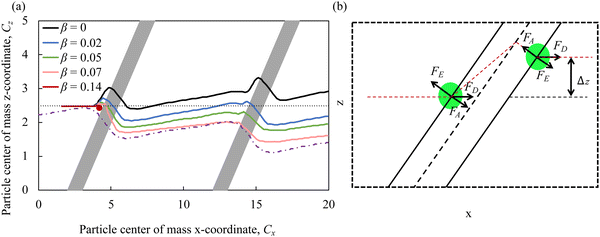
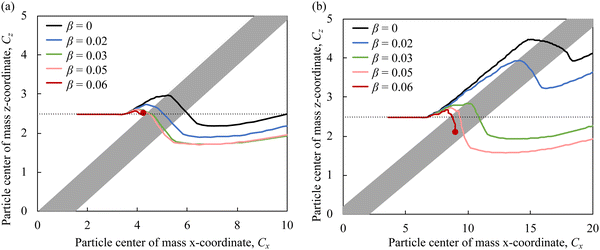
To ensure the contact of cells with adhesive microchannel surfaces independently of the channel flow rate, we use diagonal ridges that form a gap h between the ridge surface and the channel wall that is smaller than the cell diameter d, as shown in Fig. 1b. Furthermore, we consider that all internal surfaces of the ridged microchannel are coated with adhesion molecules, thereby eliminating the need for adhesive patterning.Fig. 4a shows the trajectories of the particle's center of mass in the x–z plane for particles with different adhesion strength β. The figure also includes a typical streamline in the fluid flow passing through the microchannel middle plane. We find that the particles follow different trajectories depending on the magnitude of the adhesion β. The less adhesive particles deflect more along the ridge, whereas more adhesive particles more closely follows the flow streamline. Thus, the adhesion facilitates the particle crossing the ridge and, as a result, the negative transverse z-displacement increases with increasing β. Furthermore, we find that when the adhesion strength β exceeds a critical value βcr, the particles are unable to cross the ridge as they get immobilized by the adhesive interaction with the ridge.
The motion of compliant adhesive particles in a ridged microchannel is governed by three main forces, as illustrated in Fig. 4b. The fluid drag force FD acts along the flow direction due to the relative velocity between the fluid and the particle. The elastic force FE arises due to the particle elastic deformation by the ridge and is directed away from the ridge. The adhesive force FA is proportional to the gradient of the adhesive energy and is directed towards the ridge. Thus, the elastic force FE and the adhesive force FA have opposing effect on the particle trajectory and effectively counteract each other. Indeed, when FA is weak, FE causes the particle to move along the ridge leading to particle cross-channel displacement in the positive z direction. On the other hand, when FA is comparable or exceeds FE, it cancels out the effect of FE on the particle trajectory and the particle follows the flow streamline. When FA is too strong, FD is unable to translate an adhered particle and the particle becomes immobilized by the ridge. We note that the idealized particle trajectory shown in Fig. 4b somewhat deviates from the trajectories obtained in the simulations reported in Fig. 4a. This can be related to the non-linearity associated with particle deformation by the ridge. The particle deformation gradually grows until approximately a half of the particle enters in the gap formed by the ridge. After that the elastic resistance due to particle deformation rapidly weakens and the particle can readily traverse the ridge under the action of the hydrodynamic and adhesion forces. Furthermore, diagonal ridges in the microchannel give rise to a circulatory secondary flow that transport fluid in the cross stream direction, thereby deflection particle trajectory in the positive z direction.24–26
In Fig. 5, we plot the particle displacement per ridge δ as function of the adhesion strength β for particles with different elasticity Ca. The figure shows that at higher adhesion levels with β > 0.05, δ only slightly changes with Ca indicating that the motion under the ridge is dominated by the adhesive interactions. However, for weaker adhesion the particle displacement increases as adhesion decreases. The difference in the displacement between particles with weak and high adhesiveness is the most significant for stiffer particles with low Ca. Thus, the sorting resolution increases as particle stiffness increases.
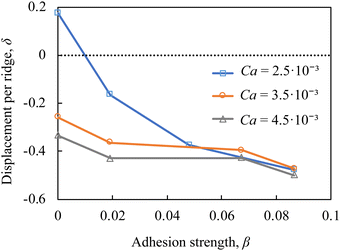 | ||
| Fig. 5 Particle lateral displacement per ridge δ = Δz/d as a function of the adhesion strength β in a ridged microchannel with Re = 3, λ = 0.75, and ϕ = 45°. | ||
Fig. 6a and b compare the trajectories of particles with Ca = 3.5 × 10−3 and different adhesiveness in microfluidic channels with ridge angles ϕ = 30° and ϕ = 15°, respectively. Note that the two figures have different scales of the x-axes, so the ridge angles are not shown to scale. We find that the separation between particles with different β is greater using ridges with ϕ = 15° compared to ϕ = 30°. Furthermore, the trajectories of particles with β = 0.03 and β = 0.05 practically coincide when the ridge angle is ϕ = 30°, whereas for ϕ = 15°, the trajectories of such particles are well separated. Thus, the use of lower ridge angle not only increases the separation resolution, but also enables separation of particles with greater adhesiveness.
To explain the effect of ridge angle ϕ on the trajectories of adhesive particles, we consider how relative direction of forces acting on the particle change with ϕ (Fig. 4b). The elastic force FE and the adhesive force FA are oriented normal to the ridge. Thus the direction of FE and FA changes with ϕ, whereas their magnitudes are independent of ϕ. The drag force FD acts along the flow direction independently of ϕ. Since the component of FD acting normal to the ridge balances FE and FA, the magnitude of this component of FD does not change with ϕ. This requires that the magnitude of FD increases with decreasing ϕ, and therefore, the component of FD directed along the ridge increases for smaller ϕ. This, in turn, results in a greater translation of the particles along the ridge enhancing the trajectory separation for particles with different adhesiveness.
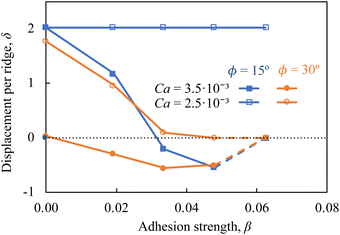
Fig. 7 presents the particle displacement per ridge δ in microchannels with ϕ = 15° and ϕ = 30° as a function of the adhesion strength β for particles with Ca = 2.5 × 10−3 and Ca = 3.5 × 10−3. Note that the displacement δ = 2 corresponds to the condition where particles deflect along the ridge all the way until they reach the channel sidewall. Furthermore, the dashed lines in Fig. 7 indicate the conditions where particle motion is arrested at the ridge due to the large adhesion force. For stiffer particles with Ca = 2.5 × 10−3, the microchannel with ϕ = 30° provides significant separation among nonadhesive particles and particles with the adhesiveness up to β = 0.05. The same particles do not separate in the microchannel with ϕ = 15°, where they are deflected by the ridge to the channel side wall. On the one hand, softer particles with Ca = 3.5 × 10−3 and different β display minor differences in δ in channels with ϕ = 30° and, therefore, cannot be separated in such channels. The use of ridges with ϕ = 15° significantly enhances the separation of softer particles. In fact, the channel with ϕ = 15° yields separation of softer particles with Ca = 3.5 × 10−3 comparable to the separation of stiffer particles with Ca = 2.5 × 10−3 in microchannels with ϕ = 30°. This result suggests that adjustment of the ridge angle can be used to achieve an efficient adhesion-based sorting of particles with specific stiffness.
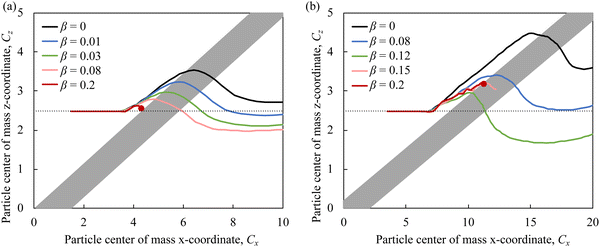
Finally, we assess the impact of higher Re on adhesion-based particle separation in the ridged microchannels. We modify Re by changing the fluid viscosity, in which case Ca does not change with Re. Fig. 8a and b show the trajectories of particles with Ca = 2.5 × 10−3 and different adhesiveness in microfluidic channels with Re = 60 and ridge angles ϕ = 30° and ϕ = 15°, respectively. Note that the two figures are not shown to scale. We find that the separation between particles with different β occurs even at high Re that significantly exceeds the maximum Re that can be used in microchannels with patterned stripes (Fig. 1a). Similarly to lower Re (Fig. 6), ridges with smaller angle ϕ result in greater separation of adhesive particles. Thus, our simulations show that microfluidic channels with diagonal ridges can be effectively used for high-throughput sorting of adhesive particles.
5 Conclusions
Using three-dimensional computer simulations, we examine a microfluidic method for label-free separation and sorting of compliant cells by their adhesiveness. The approach consists of flowing cells in a microchannel decorated with skewed periodic ridges coated with adhesive molecules. The ridges form a gap smaller than the cell diameter thereby ensuring cell contact with adhesive surfaces independently of the channel flow rate. Thus the method opens up separation opportunities at high flowrate and high cell throughput.Our simulations identified three modes of adhesive particle transport through the ridged microchannels. In the case of small ridge angle, cell rolling can occur along the diagonal ridges in which a threshold of adhesion eliminates cell sliding. In the case of high adhesion, cells can occlude the ridge leading to extended contact times and irreversible binding. Finally, cell trajectories can be modified by adhesive ridges in a manner that greater displacement differences were observed for stiffer particles with low Ca number.
Our results demonstrate that cell trajectories within ridged microchannels are sensitive to the adhesion strength, which can be used to continuously sort cells based on their adhesiveness by collecting cells at different microchannel outlets. The simulations reveal that particle elasticity is essential for the adhesion-based sorting in ridged microchannels and can be used to enhance the sorting resolution by counteracting the adhesion force on the cells. We also show that the ridge angle can be tuned to achieve efficient adhesion-based sorting for cells with different mechanical properties. Furthermore, our simulations for Re = 60 show that ridged microfluidic channels can be effectively used for high-throughput adhesion-based particle sorting.
It is further interesting to examine the role of adhesion type on cell-ridge interactions. In these studies, we considered interactions represented by the Morse potential in which bond strength develops instantaneously as in the case of electrostatic interactions. In the case of specific bond adhesion, which is contact time dependent such as described by the Bell model, additional parameter space emerges affecting the cell transport that can be explored.
Data availability
All the data for the current study are available from the corresponding authors on reasonable request.Conflicts of interest
There are no conflicts to declare.Acknowledgements
Financial support from the National Science Foundation (Grant No. CBET-1928262) is gratefully acknowledged.References
- K. Watanabe, S. Terakura, A. C. Martens, T. Van Meerten, S. Uchiyama, M. Imai, R. Sakemura, T. Goto, R. Hanajiri and N. Imahashi, et al., Target antigen density governs the efficacy of anti–CD20-CD28-CD3 ζ chimeric antigen receptor–modified effector CD8+ T cells, J. Immunol., 2015, 194, 911–920 CrossRef CAS PubMed.
- R. Tinoco, F. Carrette, M. L. Barraza, D. C. Otero, J. Magaña, M. W. Bosenberg, S. L. Swain and L. M. Bradley, PSGL-1 is an immune checkpoint regulator that promotes T cell exhaustion, Immunity, 2016, 44, 1190–1203 CrossRef CAS PubMed.
- R. Tinoco, D. C. Otero, A. A. Takahashi and L. M. Bradley, PSGL-1: a new player in the immune checkpoint landscape, Trends Immunol., 2017, 38, 323–335 CrossRef CAS PubMed.
- J. M. Karp and G. S. L. Teo, Mesenchymal stem cell homing: the devil is in the details, Cell Stem Cell, 2009, 4, 206–216 CrossRef CAS PubMed.
- W. Lee, P. Tseng and D. Di Carlo, Microtechnology for cell manipulation and sorting, Springer, 2017 Search PubMed.
- N. E. Stone, A. P. Voigt, R. F. Mullins, T. Sulchek and B. A. Tucker, Microfluidic processing of stem cells for autologous cell replacement, Stem Cells Transl. Med., 2021, 10, 1384–1393 CrossRef PubMed.
- S. Zhu, F. Jiang, Y. Han, N. Xiang and Z. Ni, Microfluidics for label-free sorting of rare circulating tumor cells, Analyst, 2020, 145, 7103–7124 RSC.
- L. Sun, W. Yang, S. Cai, Y. Chen, H. Chu, H. Yu, Y. Wang and L. Liu, Recent advances in microfluidic technologies for separation of biological cells, Biomed. Microdevices, 2020, 22, 1–16 CrossRef PubMed.
- G. V. Grigorev, A. V. Lebedev, X. Wang, X. Qian, G. V. Maksimov and L. Lin, Advances in microfluidics for single red blood cell analysis, Biosensors, 2023, 13, 117 CrossRef CAS PubMed.
- K. W. Kwon, S. S. Choi, S. H. Lee, B. Kim, S. N. Lee, M. C. Park, P. Kim, S. Y. Hwang and K. Y. Suh, Label-free, microfluidic separation and enrichment of human breast cancer cells by adhesion difference, Lab Chip, 2007, 7, 1461–1468 RSC.
- B. D. Plouffe, T. Kniazeva, J. E. Mayer Jr, S. K. Murthy and V. L. Sales, Development of microfluidics as endothelial progenitor cell capture technology for cardiovascular tissue engineering and diagnostic medicine, FASEB J., 2009, 23, 3309 CrossRef CAS PubMed.
- M. Hashimoto, H. Kaji and M. Nishizawa, Selective capture of a specific cell type from mixed leucocytes in an electrode-integrated microfluidic device, Biosens. Bioelectron., 2009, 24, 2892–2897 CrossRef CAS PubMed.
- Y. Zhang, M. Wu, X. Han, P. Wang and L. Qin, High-Throughput, Label-Free Isolation of Cancer Stem Cells on the Basis of Cell Adhesion Capacity, Angew. Chem., Int. Ed., 2015, 54, 10838–10842 CrossRef CAS PubMed.
- L. Yang, H. Sun, W. Jiang, T. Xu, B. Song, R. Peng, L. Han and L. Jia, A chemical method for specific capture of circulating tumor cells using label-free polyphenol-functionalized films, Chem. Mater., 2018, 30, 4372–4382 CrossRef CAS.
- J. V. Green, T. Kniazeva, M. Abedi, D. S. Sokhey, M. E. Taslim and S. K. Murthy, Effect of channel geometry on cell adhesion in microfluidic devices, Lab Chip, 2009, 9, 677–685 RSC.
- J. V. Green and S. K. Murthy, Microfluidic enrichment of a target cell type from a heterogenous suspension by adhesion-based negative selection, Lab Chip, 2009, 9, 2245–2248 RSC.
- Y. Jia, P. Shen, T. Yan, W. Zhou, J. Sun and X. Han, Microfluidic tandem mechanical sorting system for enhanced cancer stem cell isolation and ingredient screening, Adv. Healthcare Mater., 2021, 10, 2100985 CrossRef CAS PubMed.
- A. Singh, S. Suri, T. Lee, J. M. Chilton, M. T. Cooke, W. Chen, J. Fu, S. L. Stice, H. Lu and T. C. McDevitt, et al., Adhesion strength–based, label-free isolation of human pluripotent stem cells, Nat. Methods, 2013, 10, 438–444 CrossRef CAS PubMed.
- K. Reinmets, A. Dehkharghani, J. S. Guasto and S. M. Fuchs, Microfluidic quantification and separation of yeast based on surface adhesion, Lab Chip, 2019, 19, 3481–3489 RSC.
- W. C. Chang, L. P. Lee and D. Liepmann, Biomimetic technique for adhesion-based collection and separation of cells in a microfluidic channel, Lab Chip, 2005, 5, 64–73 RSC.
- S. Bose, R. Singh, M. Hanewich-Hollatz, C. Shen, C. H. Lee, D. M. Dorfman, J. M. Karp and R. Karnik, Affinity flow fractionation of cells via transient interactions with asymmetric molecular patterns, Sci. Rep., 2013, 3, 1–8 Search PubMed.
- S. Choi, J. M. Karp and R. Karnik, Cell sorting by deterministic cell rolling, Lab Chip, 2012, 12, 1427–1430 RSC.
- B. Tasadduq, B. McFarland, M. Islam, A. Alexeev, A. F. Sarioglu and T. Sulchek, Continuous sorting of cells based on differential P selectin glycoprotein ligand expression using molecular adhesion, Anal. Chem., 2017, 89, 11545–11551 CrossRef CAS PubMed.
- J. P. Arata and A. Alexeev, Designing microfluidic channel that separates elastic particles upon stiffness, Soft Matter, 2009, 5, 2721–2724 RSC.
- G. Wang, W. Mao, R. Byler, K. Patel, C. Henegar, A. Alexeev and T. Sulchek, Stiffness dependent separation of cells in a microfluidic device, PLoS One, 2013, 8, e75901 CrossRef CAS PubMed.
- W. Mao and A. Alexeev, Hydrodynamic sorting of microparticles by size in ridged microchannels, Phys. Fluids, 2011, 23, 051704 CrossRef.
- B. Tasadduq, W. Lam, A. Alexeev, A. F. Sarioglu and T. Sulchek, Enhancing size based size separation through vertical focus microfluidics using secondary flow in a ridged microchannel, Sci. Rep., 2017, 7, 17375 CrossRef PubMed.
- C. Pozrikidis Computational hydrodynamics of capsules and biological cells. CRC press, 2010 Search PubMed.
- D. A. Hammer and M. Tirrell, Biological adhesion at interfaces, Annu. Rev. Mater. Sci., 1996, 26, 651–691 CrossRef CAS.
- R. P. McEver and C. Zhu, Rolling cell adhesion, Annu. Rev. Cell Dev. Biol., 2010, 26, 363–396 CrossRef CAS PubMed.
- G. I. Bell, Models for the specific adhesion of cells to cells, Science, 1978, 200, 618–627 CrossRef CAS PubMed.
- M. Dembo, D. Torney, K. Saxman and D. Hammer, The reaction-limited kinetics of membraneto-surface adhesion and detachment, Proc. R. Soc. London, Ser. B, 1988, 234, 55–83 CAS.
- E. Evans, D. Berk and A. Leung, Detachment of agglutinin-bonded red blood cells. I. Forces to rupture molecular-point attachments, Biophys. J., 1991, 59, 838–848 CrossRef CAS PubMed.
- E. Evans and K. Ritchie, Dynamic strength of molecular adhesion bonds, Biophys. J., 1997, 72, 1541–1555 CrossRef CAS PubMed.
- S. Jadhav, C. D. Eggleton and K. Konstantopoulos, A 3-D computational model predicts that cell deformation affects selectin-mediated leukocyte rolling, Biophys. J., 2005, 88, 96–104 CrossRef CAS PubMed.
- D. A. Hammer and S. M. Apte, Simulation of cell rolling and adhesion on surfaces in shear flow: general results and analysis of selectin-mediated neutrophil adhesion, Biophys. J., 1992, 63, 35–57 CrossRef CAS PubMed.
- D. B. Khismatullin and G. A. Truskey, Three-dimensional numerical simulation of receptor-mediated leukocyte adhesion to surfaces: Effects of cell deformability and viscoelasticity, Phys. Fluids, 2005, 17, 031505 CrossRef.
- A. Alexeev and A. C. Balazs, Designing smart systems to selectively entrap and burst microcapsules, Soft Matter, 2007, 3, 1500–1505 RSC.
- A. Alexeev, R. Verberg and A. C. Balazs, Patterned surfaces segregate compliant microcapsules, Langmuir, 2007, 23, 983–987 CrossRef CAS PubMed.
- G. Zhu, A. Alexeev and A. C. Balazs, Designing constricted microchannels to selectively entrap soft particles, Macromolecules, 2007, 40, 5176–5181 CrossRef CAS.
- G. Zhu, A. Alexeev, E. Kumacheva and A. C. Balazs, Modeling the interactions between compliant microcapsules and pillars in microchannels, J. Chem. Phys., 2007, 127, 034703 CrossRef PubMed.
- A. Alexeev, R. Verberg and A. C. Balazs, Modeling the motion of microcapsules on compliant polymeric surfaces, Macromolecules, 2005, 38, 10244–10260 CrossRef CAS.
- A. Alexeev, R. Verberg and A. C. Balazs, Designing compliant substrates to regulate the motion of vesicles, Phys. Rev. Lett., 2006, 96, 148103 CrossRef PubMed.
- A. Alexeev, R. Verberg and A. C. Balazs, Modeling the interactions between deformable capsules rolling on a compliant surface, Soft Matter, 2006, 2, 499–509 RSC.
- O. B. Usta, A. Alexeev, G. Zhu and A. C. Balazs, Modeling microcapsules that communicate through nanoparticles to undergo self-propelled motion, ACS Nano, 2008, 2, 471–476 CrossRef CAS PubMed.
- A. Ladd and R. Verberg, Lattice-Boltzmann simulations of particle-fluid suspensions, J. Stat. Phys., 2001, 104, 1191–1251 CrossRef CAS.
- C. K. Aidun and J. R. Clausen, Lattice-Boltzmann method for complex flows, Annu. Rev. Fluid Mech., 2010, 42, 439–472 CrossRef.
- G. A. Buxton, C. M. Care and D. J. Cleaver, A lattice spring model of heterogeneous materials with plasticity, Modell. Simul. Mater. Sci. Eng., 2001, 9, 485 CrossRef.
- A. J. Ladd, J. H. Kinney and T. M. Breunig, Deformation and failure in cellular materials, Phys. Rev. E, 1997, 55, 3271 CrossRef CAS.
- M. Bouzidi, M. Firdaouss and P. Lallemand, Momentum transfer of a Boltzmann-lattice fluid with boundaries, Phys. Fluids, 2001, 13, 3452–3459 CrossRef CAS.
- B. Chun and A. Ladd, Interpolated boundary condition for lattice Boltzmann simulations of flows in narrow gaps, Phys. Rev. E, 2007, 75, 066705 CrossRef CAS PubMed.
- A. J. C. Ladd, Numerical simulations of particulate suspensions via a discretized Boltzmann equation. Part 1. Theoretical foundation, J. Fluid Mech., 1994, 271, 285–309 CrossRef CAS.
- W. Mao and A. Alexeev, Motion of spheroid particles in shear flow with inertia, J. Fluid Mech., 2014, 749, 145 CrossRef CAS.
- A. Kilimnik, W. Mao and A. Alexeev, Inertial migration of deformable capsules in channel flow, Phys. Fluids, 2011, 23, 123302 CrossRef.
- J. Branscomb and A. Alexeev, Designing ciliated surfaces that regulate deposition of solid particles, Soft Matter, 2010, 6, 4066–4069 RSC.
- H. Masoud and A. Alexeev, Modeling magnetic microcapsules that crawl in microchannels, Soft Matter, 2010, 6, 794–799 RSC.
- R. Karnik, S. Hong, H. Zhang, Y. Mei, D. G. Anderson, J. M. Karp and R. Langer, Nanomechanical control of cell rolling in two dimensions through surface patterning of receptors, Nano Lett., 2008, 8, 1153–1158 CrossRef CAS PubMed.
- A. K. Dasanna and U. S. Schwarz, Adhesion-based sorting of blood cells: an adhesive dynamics simulation study, Soft Matter, 2018, 14, 9061–9070 RSC.
| This journal is © The Royal Society of Chemistry 2024 |