Open Access Article
Open Access ArticleTumor diagnosis based on nucleolus labeling
Caiwei
Jia†
 a,
Jiani
Gao†
a,
Jiani
Gao†
 b,
Dong
Xie
*b and
Jin-Ye
Wang
b,
Dong
Xie
*b and
Jin-Ye
Wang
 *a
*a
aSchool of Biomedical Engineering, Shanghai Jiao Tong University, 800 Dongchuan Road, Shanghai 200240, China. E-mail: jinyewang@sjtu.edu.cn
bDepartment of Thoracic Surgery, Shanghai Pulmonary Hospital, Tongji University School of Medicine, Shanghai 200433, China. E-mail: kongduxd@163.com
First published on 16th September 2024
Abstract
The nucleolus is crucial for ribonucleoprotein particle assembly. Vital molecular regulators such as RB (retinoblastoma protein) and p53 (tumor suppressor protein) influence nucleolar function and tumorigenesis. The absence or inactivation of these proteins often leads to nucleolar dysfunction and alteration, which is a key indicator among the primary histopathological features of malignancy. These changes are closely related to the proliferation, differentiation, and survival of tumor cells, such as abnormalities in the number, size, and shape of nucleoli. In recent years, as the relationship between nucleoli and tumorigenesis has been further explored, various nucleolar labeling techniques have been developed for pathological analysis and tumor diagnosis, such as immunohistochemistry (IHC)/immunofluorescence (IF), and fluorescence labeling. These methods complement the traditional use of transmission electron microscopy (TEM) for observing nucleoli. In this review, we explore the relationship between the nucleolus and tumorigenesis and evaluate current methods for diagnosing tumors by examining nucleolar characteristics. We discuss the advantages, disadvantages, and applications of diagnostic techniques such as TEM, IHC/IF, and fluorescence labeling for analyzing the nucleolus.
1 Introduction
The nucleolus is an essential structure within the cell nucleus, primarily responsible for synthesizing ribosomal RNA (rRNA) and assembling ribosomal subunits. This process is crucial for cell growth and proliferation.1–3 In tumor cells, the morphology and function of nucleoli often undergo significant changes.4 These changes are mainly reflected in abnormalities in the number, size, and shape of the nucleoli.5 Such alterations are closely related to the proliferation, differentiation, and survival of tumor cells,6 and also with their metabolic activity and stress response.7 Consequently, tumor diagnosis methods based on nucleolar markers have become a significant focus in oncology research in recent years.8,9Abnormal changes in the nucleolus primarily result from genetic and molecular alterations in tumor cells, abnormal cell cycle regulation, and cellular stress responses.7 These changes are closely related to the upregulation of ribosome biosynthesis within the cells.10 Studies have found that the enlargement of the nucleolus in most tumor cells is associated with active transcription of rRNA genes.11 This is because tumor cells require substantial protein synthesis to support their rapid proliferation, with ribosomes serving as the protein synthesis factories.12 Therefore, the enhanced function of the nucleolus is a manifestation of tumor cells adapting to their high proliferation state.11
RB (retinoblastoma protein) and p53 (tumor suppressor protein) play key roles in the molecular mechanisms regulating nucleolar function and tumorigenesis. RB protein is a cell cycle regulator that influences nucleolar function by regulating the expression of proteins related to nucleolar formation. p53 is widely involved in various cellular processes, such as cell cycle progression, DNA repair, and apoptosis. p53 regulates nucleolar function by controlling the nucleolar stress response and rRNA transcription. When cells experience stress or DNA damage, p53 inhibits rRNA synthesis, thereby reducing ribosome biosynthesis and preventing further cell proliferation.
Clinically, nucleoli have been used for tumor diagnosis.13–16 For example, AgNOR polymorphism increased gradually according to the grade of histological lesions and could be used as a prognostic factor for squamous cell carcinoma progression.13 The presence and number of large nucleoli in uveal melanoma are positively correlated with the maximum basal diameter of the tumor. Increased nucleolar counts in tumor cells were positively correlated with the primary tumor stage. The presence of prominent nucleoli and multiple nucleoli is associated with a significant decrease in overall survival and disease-free survival.14
TEM is the traditional method for observing nucleoli.17,18 In recent years, as the link between nucleoli and tumorigenesis has been studied further, scientists have developed nucleolar labeling techniques for pathology and tumor diagnosis,17 including immunohistochemistry (IHC)/immunofluorescence (IF), and fluorescence labeling.
TEM is a high-resolution microscopy technique that can observe ultra-fine details of the internal structure of cells.19 Using TEM, the morphological changes of nucleoli, such as their size, number, and internal structure, can be observed.20 While observing the nucleolus, TEM can also reveal changes in other ultrastructures within the cell, providing additional pathological information.17 However, the operation of TEM is complex and costly, which limits its widespread application in clinical practice.17,18
Immunohistochemistry (IHC) and immunofluorescence (IF) are labeling techniques based on antigen–antibody reactions, used to detect specific proteins in tissue sections.21 By employing specific antibodies against nucleolar-related proteins, IHC and IF can accurately locate and quantitatively analyze changes in nucleoli within tumor tissues.22,23 Commonly used nucleolar marker proteins include nucleolar phosphorylation protein (NPM),24 fibrillarin (FBL),25 nucleolar organizing region protein (nucleolin, NCL),26 and so on. IHC is easy to perform, highly sensitive, and can be observed under an optical microscope, making it suitable for clinical pathological diagnosis.22 IF can simultaneously detect multiple nucleolar-related proteins, distinguishing them by the different colors of fluorescence signals.27 However, the results of IHC and IF depend on the specificity and sensitivity of the antibodies and are also affected by the tissue sample processing procedures.28
Fluorescence labeling techniques use fluorescent probes to label specific nucleolar proteins or nucleic acids, enabling visualization and quantitative analysis of nucleoli.29 Fluorescence in situ hybridization (FISH) is a commonly used fluorescence labeling method that detects rRNA genes in the nucleolus formation region through specific probes to observe the distribution and quantitative changes of nucleoli.30 Additionally, fluorescence labeling techniques have the advantages of high sensitivity and high specificity.31 They can monitor the dynamic changes of nucleoli in real-time in living cells, providing an important tool for tumor diagnosis.32 However, fluorescence labeling technology requires professional operating techniques, and labeling and optimizing labeled probes are also challenging.
The nucleolus has 6 nucleolar sub-regions, including fibrillar centers (FC), dense fibrillar components (DFC), granular components (GC), nucleolar rim (NR), and peri-nucleolar compartment (PNC), and the periphery of the dense fibrillar component (PDFC)33 (Fig. 1). The morphological and functional changes of nucleoli are of great significance in the occurrence and development of tumors, providing important tools for the diagnosis and pathological analysis of tumors.34 In this review, we present the role of the nucleolus in tumor diagnosis, examine the relationship between the nucleolus and tumorigenesis, and introduce commonly used nucleolar labeling methods. We believe that this comprehensive review will be a valuable resource for researchers, physicians, and students interested in this field.
2 The relationship between nucleoli and tumorigenesis
2.1 Nucleolar changes in tumor cells: unraveling the complex causes
The nucleolus has long been a focal point of research in cancer biology due to its critical role in ribosome biogenesis and cellular stress response. Nucleolar changes in tumor cells indicate altered cellular processes, and understanding the causes behind these changes is crucial for unraveling the complexities of cancer development and progression. We explored the multifaceted factors contributing to nucleolar alterations in tumor cells, drawing on recent literature to provide a comprehensive overview.Ribosomal proteins (RPs), fundamental components of the ribosome machinery, are encoded by a large family of genes. Mutations or dysregulation of these genes can lead to nucleolar stress, impaired ribosome assembly, and subsequent disruption of cellular processes.38–41 This stress response can activate various signaling pathways, including p53, which usually protects against genomic instability and tumorigenesis. However, when overwhelmed or mutated, p53 dysfunction can facilitate cancer development.42,43
c-Myc is a prominent player in driving cellular transformation among the many oncogenes implicated in cancer.44,45 Its aberrant activation, often through gene amplification or mutation, sets off a cascade of events that fuel tumor growth.45,46 One of c-Myc's pivotal roles is in upregulating ribosomal RNA (rRNA) synthesis,47,48 a fundamental step in ribosome biogenesis and protein production. Elevated rRNA synthesis supports the heightened biosynthetic demands of rapidly dividing cancer cells and contributes to nucleolar hypertrophy, a phenomenon observed in numerous cancer types.49,50 This hypertrophy signifies a shift towards a more proliferative state, indicative of the cell's commitment to uncontrolled growth and division.12
Furthermore, c-Myc's impact extends beyond ribosome biogenesis, influencing multiple cellular processes such as metabolism, cell cycle progression, and angiogenesis, converging to create a tumor-permissive environment. For instance, the oncogene's capacity to reprogram cellular metabolism redirects resources toward biomass production and energy supply necessary for rapid cell multiplication.51,52
One significant consequence of p53 dysfunction is the hyperactivation of ribosomal biogenesis, which is closely associated with nucleolar function.39,58 Normally, p53 ensures that ribosome production aligns with cellular needs, but in its absence, there is an excessive increase in rRNA synthesis and ribosome assembly, supporting the high metabolic demands of proliferating cancer cells.65,66 This results in morphological changes within the nucleolus, indicating the substantial impact of p53 on nucleolar dynamics.62,63,67,68
Moreover, p53 dysfunction impairs the nucleolar stress response,64 a protective mechanism that senses disturbances in ribosome biogenesis and activates compensatory pathways, including p53. Under normal conditions, when nucleolar function is disturbed, p53 is activated to halt the cell cycle, allowing time for repair or, if necessary, triggering cell death.69 However, when p53 is non-functional, this safety net fails, allowing cells to continue proliferating despite accumulating damage, further potentiating tumor development.68
One such disruption arises from disturbances in nucleolar proteostasis, the delicate balance of protein folding, trafficking, and degradation within the nucleolus. Perturbations to this balance, whether through mutations, misexpression of ribosomal proteins, or imbalances in chaperone proteins that assist in protein folding, can lead to significant nucleolar changes observable in tumor cells.71,72 These alterations reflect an adaptation to the altered cellular environment and actively contribute to the transformation and maintenance of the cancerous phenotype.
Aberrations in protein synthesis, a direct consequence of disrupted ribosomal biogenesis, are particularly detrimental. They can induce a state of nucleolar stress.73,74 Central to this response is the activation of tumor suppressor pathways, most notably the p53 pathway, which can halt cell cycle progression, induce DNA repair mechanisms, or initiate programmed cell death if the damage is irreparable.77,78
However, these protective mechanisms are themselves compromised in many cancer scenarios. Mutations or dysfunction of p53 and related pathways can convert the nucleolar stress response from a protective measure into a facilitator of cancer progression. Instead of inducing cell cycle arrest or apoptosis, the altered signaling promotes survival mechanisms that allow cancer cells to persist despite the ongoing stress.79,80 This includes upregulation of pro-survival genes, enhancement of DNA repair capabilities, and modulation of metabolic pathways to sustain the high-energy demands of uncontrolled proliferation.81,82
Furthermore, nucleolar stress can contribute to chromatin remodeling, epigenetic modifications, and microRNA dysregulation, further skewing the cellular landscape in favor of cancer cell survival and proliferation.83,84 It fosters an environment where cells can adapt and thrive under otherwise lethal conditions to normal cells, enabling them to evade apoptosis and continue their uncontrolled growth.
In essence, the disturbances in nucleolar proteostasis and the resulting nucleolar stress represent a critical node in the complex network of cancer pathogenesis.85 Understanding the intricate interplay between ribosomal biogenesis, nucleolar function, and cancer cell survival mechanisms holds the promise of uncovering novel therapeutic targets and strategies for intervention.86,87 By manipulating these pathways, researchers and clinicians aim to tip the balance back in favor of normal cellular homeostasis, halting or reversing cancer progression.
The nucleolus has increasingly been recognized as a sophisticated sensor and responder to cellular stress, adeptly integrating signals from genetic, epigenetic, and proteostasis networks.27 Within this intricate cellular framework, the maintenance of protein homeostasis plays a pivotal role. Disruptions in ribosomal protein balance or dysfunctions in molecular chaperones can precipitate nucleolar abnormalities, thereby contributing to the complex and multifaceted interactions that promote a pro-tumorigenic environment.97,98
Technological innovations, such as next-generation sequencing and high-resolution imaging, empower researchers to explore the nucleolus with unprecedented depth, uncovering its intricate complexities and hidden vulnerabilities. These breakthroughs are progressively illuminating the nucleolus's multifaceted role in cancer, from its involvement in cell cycle regulation to its emerging potential as a biomarker for early-stage tumor detection and disease monitoring. As these discoveries unfold, they are poised to redefine our understanding of the nucleolus in the oncological landscape.99,100
In conclusion, the nucleolar alterations characteristic of tumor cells serve as a vivid testament to the complex interplay of genetic, epigenetic, and proteomic influences. Deciphering this intricate web is vital for developing therapeutic and diagnostic strategies targeting disease causation's foundation. As scientific inquiry continues to shed light on the previously obscure realms of nucleolar biology, it paves the way for groundbreaking advancements in cancer treatment and diagnosis, heralding a future where therapies are more effective and finely tuned to each patient's unique molecular profile. The ongoing exploration of nucleolar dynamics offers hope in the relentless fight against cancer, underscoring the potential for transformative breakthroughs in personalized medicine.101–103
2.2 Rb and p53 influence the nucleolar function
The intricate dance of life within each cell is masterfully orchestrated by a cadre of molecular regulators, with the retinoblastoma protein (pRb) and p53 as central figures in this complex choreography. Central to the cell cycle's progression, pRb acts as a vigilant sentinel at the G1 phase's juncture. With meticulous precision, it evaluates whether the cell is sufficiently prepared for the rigorous process of DNA replication, ensuring that no premature steps disrupt the orderly progression of the cell cycle.104,105 This crucial function of pRb is paramount in preserving the accuracy and integrity of cell division, much like a disciplined conductor leading a symphony of cellular events with unwavering precision.Conversely, p53, revered as the guardian of the genome, vigilantly monitors the cellular environment for any signs of distress or damage.79 Upon detecting DNA lesions or other stress signals, p53 rapidly activates a protective response. This can involve inducing programmed cell death (apoptosis) to eliminate cells bearing irreparable damage or arresting the cell cycle at the G2 checkpoint, thereby preventing the transmission of defective genetic material to daughter cells and safeguarding the fidelity of gene inheritance.57,106–109 In essence, p53 functions as a crucial firewall, averting the propagation of genetic errors that could otherwise lead to oncogenesis. The lack of functional PTEN, or inappropriate activation of PI3K–AKT will ring from downstream target MDM2 of PTEN, which will decrease p53 activity and disable cancer cells to make a proper response to DNA damage110 (Fig. 2).
 | ||
| Fig. 2 Rb-driven entry of MDM2 into the nucleus prevents transcriptional activation of p53 and promotes p53 degradation. | ||
Beyond their quintessential roles in orchestrating cell cycle checkpoints, pRb and p53 go into the nucleolus, where the ribosomes, the intricate machinery of protein synthesis, are meticulously assembled.111–114 Here, they take on a new role as transcriptional repressors, modulating the expression of ribosomal genes. pRb accomplishes this by engaging in a molecular tango with the upstream binding factor UBF,115 while p53 interacts with the selectivity factor SL1,116 both of which are integral components of the UBF–SL1 complex that recruits RNA polymerase I to the ribosomal RNA (rRNA) gene promoter.117–119 By deftly hindering the formation of this vital complex, pRb and p53 can dampen rRNA transcription, exerting a powerful regulatory influence over ribosome biogenesis and protein synthesis—fundamental processes that are the bedrock of cell growth and proliferation.
However, pRb and p53 are frequently compromised in human malignancies, and their dysfunction dismantles the safeguards against uncontrolled cell division. It accelerates ribosomal RNA transcription, fueling the insatiable biosynthetic appetite of cancer cells and fostering their malignant transformation.120–125 This impairment releases the brakes on cellular proliferation and leads to hyperactive rRNA transcription, fueling the heightened biosynthetic requirements of cancer cells and contributing to their malignant transformation.120,123–125 This dysfunction manifests in an enlarged nucleolus, a telltale sign of the tumor suppressors' compromised status and a diagnostic clue to the presence of cancer.126,127 The nucleolus's size thus becomes a diagnostic biomarker, revealing a disturbance in the cellular balance that often precedes a more aggressive cancer phenotype.
Unraveling the intricate interplay between nucleolar function, tumor suppressor proteins, and cancer progression is paramount. It enriches our understanding of the disease's molecular underpinnings and paves the way for the development of innovative diagnostic tools and therapeutic strategies. By manipulating the activities of pRb, p53, or their downstream effectors within the nucleolus, scientists aim to recalibrate the disrupted balance between cell cycle control and ribosome biogenesis, inhibiting cancer's relentless progression.128,129 This targeted approach represents a promising frontier in cancer therapy, offering the potential to intervene at the heart of the disease's machinery.
Furthermore, exploring the nucleolar dynamics associated with the dysfunction of pRb and p53 offers profound insights into broader biological themes such as cellular stress responses, senescence, and aging. This research frames the nucleolus as an active contributor rather than a passive observer within the complex network of signaling pathways that maintain cellular homeostasis.57,130 This perspective accentuates the pivotal role of the nucleolus in preserving the delicate balance within cells. It underscores its significance as a central hub for elucidating and addressing the multifaceted nature of cancer.
As research in this field progresses, each discovery adds a piece to the puzzle, gradually elucidating the complex interplay between nucleolar function, tumor suppressors, and cancer biology. These advancements bring us closer to realizing the ambitious goal of transforming cancer from an ominous foe into a manageable and potentially curable condition through the application of molecular biology's vast arsenal. The ongoing exploration of the nucleolus and its regulators is not merely a scientific pursuit; it is a quest driven by the hope of alleviating human suffering and reclaiming lives from the clutches of this relentless disease. In this journey, every new insight illuminates a path toward a future where cancer is no longer a death sentence but a manageable chapter in the story of human health and resilience.
3 Nucleolus methods for tumor diagnosis
3.1 Transmission electron microscopy (TEM)
As a high-resolution imaging technique, TEM can observe the ultrastructural changes of the nucleolus.131 The morphological abnormalities of tumor nucleoli can be directly observed through TEM, making it a valuable tool for tumor diagnosis (Fig. 3).69Clinically, acute monocytic leukemia (AML-M5) can be classified into four subtypes: typical monoblast (TMB), atypical monoblast (AMB), atypical promonocyte (APM), and typical promonocyte (TPM).136 The TMB subtype is poorly differentiated, while the TPM subtype is well differentiated. AMB and APM exhibit an intermediate level of differentiation. These differences in differentiation are characterized by features such as the larger size of AMB compared to TMB; numerous vacuoles and granules in TPM and APM compared to TMB and AMB; and larger nuclei and nucleoli in AMB and APM compared to TMB and TPM. These characteristics illustrate consecutive differentiation stages, aiding in the precise definition of monoblasts and promonocytes and refining the light microscopy criteria in M5.136
A study compared the ultrastructure of chronic gastritis, gastric cancer, and gastric precancer using TEM. Clinically, only a few chronic gastritis patients' gastric mucosal epithelial cells are likely to develop into cancer cells. Significant differences were found between CG and GC in terms of ultrastructure and molecular biology. Nuclear chromatins were either scattered, associated with nucleoli, or aligned along nuclear perimeters. The light-bright zones between heterochromatins in the nucleoli are euchromatins. The nucleoli exhibited high electron density without a capsule, and features such as nucleolar margination, multinucleoli, and nucleolar division were observed.137
TEM requires ultrathin sectioning of samples, and the thickness of the samples is usually less than 100 nm.138 This process is complicated and time-consuming, and it is easy to introduce human errors.139 In addition, the preparation process of ultrathin sections may cause sample deformation or structural damage, thus affecting the accuracy of the results.140 Since TEM uses high-energy electron beams to image samples, high-energy electron beams may cause damage to biological samples, especially to structures such as nucleoli that are rich in RNA and proteins.141 Radiation damage from electron beams may change or destroy the structure of nucleoli and affect the observation results.142 TEM provides only two-dimensional projection images of samples and cannot directly obtain three-dimensional structural information of nucleoli.143 Although three-dimensional reconstructed images can be obtained by electron tomography (ET) technology, this process requires more time and complex computational processing.144,145 The nucleolus is an important structure in the cell nucleus, rich in RNA and proteins, and has a low natural contrast.146 In order to improve the contrast, heavy metal stains (such as uranium, lead, etc.) are often used to stain the sample.147 However, the staining process may introduce artifacts or nonspecific staining, thus affecting the observation results.147 Although the resolution of TEM is very high, reaching the sub-nanometer level, the resolution may still not be enough to reveal the details of certain tiny structures when observing large-area samples.148 In addition, TEM can only image fixed samples and cannot observe the dynamic changes of the nucleolus in real time.149 The dynamic changes of the nucleolus during the cell cycle are very important for understanding its function and mechanism, which is difficult to achieve in TEM.
3.2 Immunohistochemistry (IHC)/immunofluorescence (IF)
IHC/IF have become core tools for tumor diagnosis.150–152 Both immunohistochemistry and immunofluorescence are techniques that locate and quantify specific proteins in tissue sections through color development reactions based on antigen–antibody reactions.153–158 IHC plays an important role in pathological diagnosis, especially in the classification and grading of tumors.159 IF uses fluorescent dyes to label antibodies and observe the location and expression levels of markers through fluorescence microscopy.160 IF provides high-resolution protein distribution maps at the cellular and tissue levels and is widely used in basic research and clinical diagnosis (Fig. 3).When combined with other diagnostic methods such as gene sequencing and liquid biopsy, they could form a comprehensive diagnostic system to improve the accuracy and efficiency of diagnosis. Labeling nucleoli by IHC and IF can not only help diagnose tumors but also provide detailed molecular information about tumor cells, serving as a basis for the formulation of personalized treatment plans. For example, the expression levels of different nucleolar markers can indicate a patient's response to certain chemotherapy drugs or targeted therapies.161
In prostate cancer, enlargement of the nucleolus is the key diagnostic feature of high-grade prostatic intraepithelial neoplasia (PIN), an early stage that appears to be the precursor to the majority of invasive prostate cancers. Several cancer genes implicated in PIN are known to augment ribosome production, including c-Myc, p27, retinoblastoma, p53, and growth factors that impact ERK signaling.170 p120 nucleolar protein has indeed been used in immunohistochemical analysis, showing significantly elevated expression levels in prostate cancer. Moreover, p120 nucleolar protein is closely associated with cell proliferation, and its expression typically reflects the proliferative activity of cells.171
In lung cancer research, IHC/IF labeling of NPM and NCL can clarify nucleolar structural changes and functional abnormalities.172–174 NPM1 overexpression correlates with 18F-FDG PET/CT metabolic parameters and improves diagnostic accuracy in lung adenocarcinoma.175 IHC on TMAs from 92 NSCLC samples and 42 non-cancerous lung tissues showed high nucleolin and phosphorylation of nucleolin (P-nucleolin) expression in the cytoplasm and nucleus of NSCLC tissues, while nucleolin was primarily nuclear and P-nucleolin was low in non-cancerous tissues.176
A proliferating cell nuclear antigen (PCNA) was detected in the nucleoli of human cell lines, including HeLa, Hep-2, and Namalwa, as well as in solid tumors from human renal and prostate carcinomas. Both strong and weak nucleolar fluorescence signals were observed in the renal and prostate carcinomas, indicating varying levels of proliferation among the tumor cells. Two human colon carcinoma cell lines with different growth rates were compared: Ω, an aggressive, fast-growing clone of the HCT 116 cell line, and CBS, a slower-growing cell line (group 3). The fast-growing Ω cells showed a higher percentage of nucleolar fluorescence (28.5%) compared to the slower-growing CBS cells (13.6%).177
3.3 Fluorescence labeling techniques
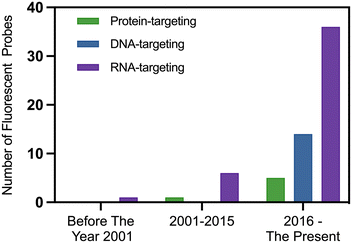
Fluorescence labeling techniques have shown significant potential in tumor diagnosis, particularly in the study of nucleoli.179 By utilizing these techniques, changes in nucleoli can be detected efficiently and accurately, providing crucial information for the diagnosis and treatment of tumors. Fluorescence labeling employs fluorescent dyes or fluorescent proteins to label nucleoli, allowing them to be clearly observed and analyzed under a microscope.179 This offers an effective means for tumor diagnosis. Immunofluorescence has already been introduced in the previous section, so it will not be covered here. Instead, this section focuses on fluorescent probe labeling. Some fluorescent probe technologies can monitor the dynamic changes of nucleoli in real-time within living cells by labeling nucleoli, offering a novel approach for the early-stage diagnosis and treatment of tumors (Fig. 3). The markers used in fluorescence labeling methods can be DNA, RNA, or proteins. Since 2016, there has been a significant increase in the number of small-molecule fluorescent probes targeting nucleoli in published papers, with those targeting RNA being the most prevalent (Fig. 4).Naphthalimide derivatives (NI-1 to NI-5) penetrate both the cell membrane and nuclear membrane, achieving clear nucleolar staining in live cells (Fig. 5). The presence of amino groups on the side chains of the naphthalimide backbone enhances their targeting specificity to the nucleolus. Molecular docking results indicate that NI-1 to NI-5 form hydrogen bonds and hydrophobic interactions with RNA, resulting in fluorescence enhancement upon RNA binding. These findings provide valuable support for the future diagnosis and treatment of nucleolar-related diseases.186
 | ||
| Fig. 5 Nucleolus imaging based on naphthalimide derivatives. Ref. 186 (Copyright is used with permission). | ||
A novel chaperone@DNA molecular tool, the phenylboronic acid-modified avidin conjugated with an abasic site-containing DNA probe (PB-ACP), has been developed for the real-time observation of APE1 in the nucleus and nucleolus of living cells (Fig. 6). The phenylboronic acid-modified avidin not only serves as a chaperone to protect the AP-DNA from nonspecific degradation but also facilitates the targeted delivery of the probe to the nucleus. PB-ACP shows high specificity and sensitivity to APE1 due to the strong binding affinity of APE1 to both avidin and the AP site in DNA. The probe efficiently migrates from the cytoplasm to the nucleus, specifically displaying the distribution and in vivo activity of endogenous nuclear APE1. This tool offers a powerful method to investigate the cellular behavior of APE1 in living cells and improve cancer therapies targeting APE1.188 These fluorescent probes targeting nucleolar proteins provide valuable support for the diagnosis of tumors.
 | ||
| Fig. 6 Visualization of APE1 in the nucleus and nucleolus using the PB-ACP-based chaperone@DNA probe. Ref. 188 (Copyright is used with permission). | ||
Green-emitting carbon dots (m-CDs) have a significantly higher affinity for nucleolar RNA. Visualizing nucleoli in cells using m-CDs can accurately determine their number and morphology, distinguishing cancerous cells from normal cells (Fig. 7). Additionally, m-CDs can be used to monitor nucleolar dynamics during the apoptosis of malignant cells induced by DOX. m-CDs show great potential as nucleolar probes, with promising applications in cancer cell screening and therapeutic efficacy evaluation.179
 | ||
| Fig. 7 Schematic illustration of the synthesis of m-CDs and the selective staining of nucleolus. Ref. 179 (Copyright is used with permission). | ||
Protein-bound fluorescent probes (such as fibrillarin-GFP fusion protein) usually require genetic engineering or antibody labeling, which are complex and costly to operate and are not suitable for large-scale clinical testing. These complex procedures and high costs limit the widespread application of these probes in clinical practice.201
4 Perspectives and outlook
In the future, the efficiency and accuracy of TEM observations can be improved by enhancing sample preparation methods, allowing for a better revelation of fine structural changes in nucleoli. Developing automated image analysis tools using artificial intelligence and machine learning can increase the efficiency of identifying and analyzing changes in nucleolar ultrastructure. Integrating TEM with other imaging techniques, such as confocal microscopy and scanning electron microscopy, for multimodal imaging can provide more comprehensive cellular ultrastructural information, aiding in the comprehensive diagnosis of tumors. With the development of nanotechnology and molecular biology, new labeling methods such as nanoparticle labeling and the CRISPR/Cas9 system are expected to improve the specificity and sensitivity of labeling, reducing false positive and false negative results for IHC and IF. Low-toxicity fluorescently labeled nucleolar probes have been studied for dynamic observation of nucleoli. Combining fluorescent probes with other imaging techniques (such as super-resolution microscopy) can achieve multimodal and high-resolution imaging of nucleoli, and yield detailed tumor cell information at the single-cell level.202 Multimodal imaging technology can integrate different types of signals, improving the resolution and information content of imaging to support accurate diagnosis. Combining big data analysis and artificial intelligence can lead to the development of automated analysis tools for fluorescence imaging data, improving diagnostic accuracy and efficiency. By training deep learning models, nucleolar features in fluorescence imaging can be automatically identified and analyzed, providing quantitative analysis results to assist doctors in making diagnostic decisions.Data availability
All data created or analyzed in this study are available on request from the corresponding author.Conflicts of interest
The authors declare that they don't have any competing financial interests or personal relationships that could have appeared to influence the work reported in this review.Acknowledgements
This study was supported by the National Key R&D Program of China (SQ2024YFE0104607; 2019YFE0101200), the Science and Technology Commission Shanghai Municipality (13JC1403400, 18490740200), and the Plan of Jiaxing Innovation and Elites Leading.Notes and references
- D. L. Lafontaine and D. Tollervey, Nat. Rev. Mol. Cell Biol., 2001, 2, 514–520 CrossRef.
- Y. Cheng, C. Li, R. Mu, Y. Li, T. Xing, B. Chen and C. Huang, Anal. Chem., 2018, 90, 11358–11365 CrossRef.
- J. T. Sczepanski and G. F. Joyce, Nature, 2014, 515, 440–442 CrossRef.
- D. Zink, A. H. Fischer and J. A. Nickerson, Nat. Rev. Cancer, 2004, 4, 677–687 CrossRef.
- L. Montanaro, D. Trere and M. Derenzini, Am. J. Pathol., 2008, 173, 301–310 CrossRef.
- J. van Riggelen, A. Yetil and D. W. Felsher, Nat. Rev. Cancer, 2010, 10, 301–309 CrossRef PubMed.
- M. S. Shim, A. Nettesheim, J. Hirt and P. B. Liton, Autophagy, 2020, 16, 1248–1261 CrossRef.
- Z. Zhang, H. Yu, W. Yao, N. Zhu, R. Miao, Z. Liu, X. Song, C. Xue, C. Cai, M. Cheng, K. Lin and D. Qi, Cell Commun. Signaling, 2022, 20, 188 CrossRef.
- D. L. J. Lafontaine, J. A. Riback, R. Bascetin and C. P. Brangwynne, Nat. Rev. Mol. Cell Biol., 2021, 22, 165–182 CrossRef PubMed.
- Z. Wang, Z. Li, K. Zhou, C. Wang, L. Jiang, L. Zhang, Y. Yang, W. Luo, W. Qiao, G. Wang, Y. Ni, S. Dai, T. Guo, G. Ji, M. Xu, Y. Liu, Z. Su, G. Che and W. Li, Nat. Commun., 2021, 12, 6500 CrossRef PubMed.
- X. T. Nguyen le, A. Raval, J. S. Garcia and B. S. Mitchell, J. Cell. Physiol., 2015, 230, 1181–1188 CrossRef PubMed.
- S. Kofuji, A. Hirayama, A. O. Eberhardt, R. Kawaguchi, Y. Sugiura, O. Sampetrean, Y. Ikeda, M. Warren, N. Sakamoto, S. Kitahara, H. Yoshino, D. Yamashita, K. Sumita, K. Wolfe, L. Lange, S. Ikeda, H. Shimada, N. Minami, A. Malhotra, S. Morioka, Y. Ban, M. Asano, V. L. Flanary, A. Ramkissoon, L. M. L. Chow, J. Kiyokawa, T. Mashimo, G. Lucey, S. Mareninov, T. Ozawa, N. Onishi, K. Okumura, J. Terakawa, T. Daikoku, T. Wise-Draper, N. Majd, K. Kofuji, M. Sasaki, M. Mori, Y. Kanemura, E. P. Smith, D. Anastasiou, H. Wakimoto, E. C. Holland, W. H. Yong, C. Horbinski, I. Nakano, R. J. DeBerardinis, R. M. Bachoo, P. S. Mischel, W. Yasui, M. Suematsu, H. Saya, T. Soga, I. Grummt, H. Bierhoff and A. T. Sasaki, Nat. Cell Biol., 2019, 21, 1003–1014 CrossRef PubMed.
- C. Alarcon-Romero Ldel, B. Illades-Aguiar, E. Flores-Alfaro, M. A. Teran-Porcayo, V. Antonio-Vejar and E. Reyes-Maldonado, Rev. Invest. Salud Publica, 2009, 51, 134–140 Search PubMed.
- T. Berus, A. Markiewicz, P. Biecek, J. Orlowska-Heitzman, A. Halon, B. Romanowska-Dixon and P. Donizy, Anticancer Res., 2020, 40, 3505–3512 CrossRef.
- K. A. Elsharawy, M. S. Toss, S. Raafat, G. Ball, A. R. Green, M. A. Aleskandarany, L. W. Dalton and E. A. Rakha, Histopathology, 2020, 76, 671–684 CrossRef PubMed.
- P. Donizy, P. Biecek, A. Halon, A. Maciejczyk and R. Matkowski, Diagn. Pathol., 2017, 12, 88 CrossRef.
- R. D. Jiang, H. Shen and Y. J. Piao, Rom. J. Morphol. Embryol., 2010, 51, 663–667 Search PubMed.
- H. Leek and M. Albertsson, Scanning, 2000, 22, 326–331 CrossRef PubMed.
- S. Besztejan, S. Keskin, S. Manz, G. Kassier, R. Bucker, D. Venegas-Rojas, H. K. Trieu, A. Rentmeister and R. J. Miller, Microsc. Microanal., 2017, 23, 46–55 CrossRef PubMed.
- D. S. Zhang, L. Liu, L. Q. Jin, M. L. Wan and Q. H. Li, World J. Gastroenterol., 2004, 10, 1551–1554 CrossRef.
- H. M. Hussaini, B. Seo and A. M. Rich, Methods Mol. Biol., 2023, 2588, 439–450 CrossRef PubMed.
- N. Yang, Y. Huang, P. Yang, W. Yan, S. Zhang, N. Li and Z. Feng, Diagn. Pathol., 2023, 18, 25 CrossRef PubMed.
- N. Tulchin, M. Chambon, G. Juan, S. Dikman, J. Strauchen, L. Ornstein, B. Billack, N. T. Woods and A. N. Monteiro, Am. J. Pathol., 2010, 176, 1203–1214 CrossRef.
- C. Mascaux, F. Bex, B. Martin, A. Burny, A. Haller, M. Paesmans, K. Willard-Gallo, V. Ninane and J. P. Sculier, Eur. Respir. J., 2008, 32, 678–686 CrossRef PubMed.
- R. W. Yao, G. Xu, Y. Wang, L. Shan, P. F. Luan, Y. Wang, M. Wu, L. Z. Yang, Y. H. Xing, L. Yang and L. L. Chen, Mol. Cell, 2019, 76, 767–783 CrossRef PubMed , e711.
- K. Fujita, C. P. Pavlovich, G. J. Netto, Y. Konishi, W. B. Isaacs, S. Ali, A. De Marzo and A. K. Meeker, Hum. Pathol., 2009, 40, 924–933 CrossRef PubMed.
- I. Orsolic, D. Jurada, N. Pullen, M. Oren, A. G. Eliopoulos and S. Volarevic, Semin. Cancer Biol., 2016, 37–38, 36–50 CrossRef PubMed.
- M. Bzorek, Sr., B. L. Petersen and L. Hansen, Appl. Immunohistochem. Mol. Morphol., 2008, 16, 279–286 CrossRef PubMed.
- B. Unnikrishnan, R. S. Wu, S. C. Wei, C. C. Huang and H. T. Chang, ACS Omega, 2020, 5, 11248–11261 CrossRef.
- C. Grandori, N. Gomez-Roman, Z. A. Felton-Edkins, C. Ngouenet, D. A. Galloway, R. N. Eisenman and R. J. White, Nat. Cell Biol., 2005, 7, 311–318 CrossRef.
- F. Ma, Y. Li, B. Tang and C. Y. Zhang, Acc. Chem. Res., 2016, 49, 1722–1730 CrossRef PubMed.
- B. Zhou, W. Liu, H. Zhang, J. Wu, S. Liu, H. Xu and P. Wang, Biosens. Bioelectron., 2015, 68, 189–196 CrossRef PubMed.
- L. Shan, G. Xu, R. W. Yao, P. F. Luan, Y. Huang, P. H. Zhang, Y. H. Pan, L. Zhang, X. Gao, Y. Li, S. M. Cao, S. X. Gao, Z. H. Yang, S. Li, L. Z. Yang, Y. Wang, C. C. L. Wong, L. Yu, J. Li, L. Yang and L. L. Chen, Nature, 2023, 615, 526–534 CrossRef CAS.
- A. Sava, C. F. Costea, R. Vatavu, M. Grigore, M. D. Turliuc, G. F. Dumitrescu, L. Eva, A. G. M. Motoc, C. I. Stan, L. C. Gavril and S. I. Scripcariu, Rom. J. Morphol. Embryol., 2021, 62, 435–444 CrossRef PubMed.
- A. Bowry, R. D. W. Kelly and E. Petermann, Trends Cancer, 2021, 7, 863–877 CrossRef.
- C. Johnson, D. L. Burkhart and K. M. Haigis, Cancer Discovery, 2022, 12, 913–923 CrossRef.
- D. S. Kim, C. V. Camacho, A. Nagari, V. S. Malladi, S. Challa and W. L. Kraus, Mol. Cell, 2019, 75, 1270–1285 CrossRef PubMed , e1214.
- M. Girbig, A. D. Misiaszek and C. W. Müller, Nat. Rev. Mol. Cell Biol., 2022, 23, 603–622 CrossRef PubMed.
- J. Kang, N. Brajanovski, K. T. Chan, J. Xuan, R. B. Pearson and E. Sanij, Signal Transduction Targeted Ther., 2021, 6, 323 CrossRef.
- Y. Luan, N. Tang, J. Yang, S. Liu, C. Cheng, Y. Wang, C. Chen, Y. N. Guo, H. Wang, W. Zhao, Q. Zhao, W. Li, M. Xiang, R. Ju and Z. Xie, Nucleic Acids Res., 2022, 50, 6601–6617 CrossRef CAS.
- A. J. W. Te Velthuis, J. M. Grimes and E. Fodor, Nat. Rev. Microbiol., 2021, 19, 303–318 CrossRef CAS PubMed.
- L. Jiang, N. Kon, T. Li, S. J. Wang, T. Su, H. Hibshoosh, R. Baer and W. Gu, Nature, 2015, 520, 57–62 CrossRef.
- A. J. Levine, Nat. Rev. Cancer, 2020, 20, 471–480 CrossRef.
- C. V. Dang, Cell, 2012, 149, 22–35 CrossRef PubMed.
- R. Dhanasekaran, A. Deutzmann, W. D. Mahauad-Fernandez, A. S. Hansen, A. M. Gouw and D. W. Felsher, Nat. Rev. Clin. Oncol., 2022, 19, 23–36 CrossRef.
- Y. Zhu, B. Zhou, X. Hu, S. Ying, Q. Zhou, W. Xu, L. Feng, T. Hou, X. Wang, L. Zhu and H. Jin, Clin. Transl. Med., 2022, 12, e703 CrossRef.
- A. R. Elhamamsy, B. J. Metge, H. A. Alsheikh, L. A. Shevde and R. S. Samant, Cancer Res., 2022, 82, 2344–2353 CrossRef.
- A. Babaian, K. Rothe, D. Girodat, I. Minia, S. Djondovic, M. Milek, S. E. Spencer Miko, H. J. Wieden, M. Landthaler, G. B. Morin and D. L. Mager, Cell Rep., 2020, 31, 107611 CrossRef CAS PubMed.
- I. Barbieri and T. Kouzarides, Nat. Rev. Cancer, 2020, 20, 303–322 CrossRef.
- V. Matson, J. Fessler, R. Bao, T. Chongsuwat, Y. Zha, M. L. Alegre, J. J. Luke and T. F. Gajewski, Science, 2018, 359, 104–108 CrossRef PubMed.
- A. Soufi, M. F. Garcia, A. Jaroszewicz, N. Osman, M. Pellegrini and K. S. Zaret, Cell, 2015, 161, 555–568 CrossRef PubMed.
- P. Xia, H. Zhang, H. Lu, K. Xu, X. Jiang, Y. Jiang, X. Gongye, Z. Chen, J. Liu, X. Chen, W. Ma, Z. Zhang and Y. Yuan, Cancer Commun., 2023, 43, 338–364 CrossRef.
- C. Whibley, P. D. Pharoah and M. Hollstein, Nat. Rev. Cancer, 2009, 9, 95–107 CrossRef PubMed.
- J. Hu, J. Cao, W. Topatana, S. Juengpanich, S. Li, B. Zhang, J. Shen, L. Cai, X. Cai and M. Chen, J. Hematol. Oncol., 2021, 14, 157 CrossRef.
- H. Arakawa, Cell Death Differ., 2005, 12, 1057–1065 CrossRef CAS PubMed.
- H. Wang, M. Guo, H. Wei and Y. Chen, Signal Transduction Targeted Ther., 2023, 8, 92 CrossRef CAS.
- K. Engeland, Cell Death Differ., 2018, 25, 114–132 CrossRef PubMed.
- M. S. Lindström, J. Bartek and A. Maya-Mendoza, Cell Death Differ., 2022, 29, 972–982 CrossRef.
- N. J. Boon, R. A. Oliveira, P. R. Körner, A. Kochavi, S. Mertens, Y. Malka, R. Voogd, S. E. M. van der Horst, M. A. Huismans, L. P. Smabers, J. M. Draper, L. F. A. Wessels, P. Haahr, J. M. L. Roodhart, T. N. M. Schumacher, H. J. Snippert, R. Agami and T. R. Brummelkamp, Science, 2024, 384, 785–792 CrossRef.
- Q. Peng, X. Shi, D. Li, J. Guo, X. Zhang, X. Zhang and Q. Chen, Cell Death Differ., 2023, 30, 1849–1867 CrossRef PubMed.
- I. Rodriguez-Pastrana, E. Birli and A. S. Coutts, Cell Death Differ., 2023, 30, 1636–1647 CrossRef.
- G. E. Jimenez-Gutierrez, R. Mondragon-Gonzalez, L. A. Soto-Ponce, W. L. Gómez-Monsiváis, I. García-Aguirre, R. A. Pacheco-Rivera, R. Suárez-Sánchez, A. Brancaccio, J. J. Magaña and R. C. R. Perlingeiro, Int. J. Mol. Sci., 2020, 21, 4961 CrossRef PubMed.
- J. Zarka, N. J. Short, R. Kanagal-Shamanna and G. C. Issa, Genes, 2020, 11, 649 CrossRef CAS PubMed.
- I. Ohbayashi and M. Sugiyama, Front. Plant Sci., 2017, 8, 2247 CrossRef.
- K. Boulias and E. L. Greer, Nat. Rev. Genet., 2023, 24, 143–160 CrossRef.
- A. Sivan, L. Corrales, N. Hubert, J. B. Williams, K. Aquino-Michaels, Z. M. Earley, F. W. Benyamin, Y. M. Lei, B. Jabri, M. L. Alegre, E. B. Chang and T. F. Gajewski, Science, 2015, 350, 1084–1089 CrossRef.
- K. M. Hannan, P. Soo, M. S. Wong, J. K. Lee, N. Hein, P. Poh, K. D. Wysoke, T. D. Williams, C. Montellese, L. K. Smith, S. J. Al-Obaidi, L. Núñez-Villacís, M. Pavy, J. S. He, K. M. Parsons, K. E. Loring, T. Morrison, J. Diesch, G. Burgio, R. Ferreira, Z. P. Feng, C. M. Gould, P. B. Madhamshettiwar, J. Flygare, T. J. Gonda, K. J. Simpson, U. Kutay, R. B. Pearson, C. Engel, N. J. Watkins, R. D. Hannan and A. J. George, Cell Rep., 2022, 41, 111571 CrossRef PubMed.
- X. Jia, H. Liu, X. Ren, P. Li, R. Song, X. Li, Y. Guo and X. Li, Oncogene, 2022, 41, 4474–4484 CrossRef.
- M. C. Lafita-Navarro and M. Conacci-Sorrell, Semin. Cell Dev. Biol., 2023, 136, 64–74 CrossRef PubMed.
- P. Carotenuto, A. Pecoraro, G. Palma, G. Russo and A. Russo, Cells, 2019, 8, 1090 CrossRef.
- J. Pelletier, G. Thomas and S. Volarević, Nat. Rev. Cancer, 2018, 18, 51–63 CrossRef.
- R. Y. Ebright, S. Lee, B. S. Wittner, K. L. Niederhoffer, B. T. Nicholson, A. Bardia, S. Truesdell, D. F. Wiley, B. Wesley, S. Li, A. Mai, N. Aceto, N. Vincent-Jordan, A. Szabolcs, B. Chirn, J. Kreuzer, V. Comaills, M. Kalinich, W. Haas, D. T. Ting, M. Toner, S. Vasudevan, D. A. Haber, S. Maheswaran and D. S. Micalizzi, Science, 2020, 367, 1468–1473 CrossRef.
- X. Chen and J. R. Cubillos-Ruiz, Nat. Rev. Cancer, 2021, 21, 71–88 CrossRef PubMed.
- L. Deng, T. Meng, L. Chen, W. Wei and P. Wang, Signal Transduction Targeted Ther., 2020, 5, 11 CrossRef.
- X. Wang, H. Zhang, R. Sapio, J. Yang, J. Wong, X. Zhang, J. Y. Guo, S. Pine, H. Van Remmen, H. Li, E. White, C. Liu, M. Kiledjian, D. G. Pestov and X. F. Steven Zheng, Nat. Commun., 2021, 12, 2259 CrossRef.
- J. Baßler and E. Hurt, Annu. Rev. Biochem., 2019, 88, 281–306 CrossRef PubMed.
- C. L. Ford, L. Randal-Whitis and S. R. Ellis, Cancer Res., 1999, 59, 704–710 Search PubMed.
- H. Liu, L. Xu, Y. Zhang, Y. Xie, L. Wang, Y. Zhou, Z. Wang, Y. Pan, W. Li, L. Xu, X. Xu, T. Wang, K. Meng, J. He, Y. Qiu, G. Xu, W. Ge, Y. Zhu and L. Wang, Adv. Healthcare Mater., 2023, 12, e2300913 CrossRef PubMed.
- Y. Liu, Z. Su, O. Tavana and W. Gu, Cancer Cell, 2024, 42, 946–967 CrossRef.
- J. Chen, Cold Spring Harbor Perspect. Med., 2016, 6, a026104 CrossRef.
- B. A. Rybicki, C. Neslund-Dudas, C. H. Bock, A. Rundle, A. T. Savera, J. J. Yang, N. L. Nock and D. Tang, Clin. Cancer Res., 2008, 14, 750–757 CrossRef PubMed.
- M. L. Castro, M. J. McConnell and P. M. Herst, Free Radical Biol. Med., 2014, 74, 200–209 CrossRef PubMed.
- J. A. Belk, W. Yao, N. Ly, K. A. Freitas, Y. T. Chen, Q. Shi, A. M. Valencia, E. Shifrut, N. Kale, K. E. Yost, C. V. Duffy, B. Daniel, M. A. Hwee, Z. Miao, A. Ashworth, C. L. Mackall, A. Marson, J. Carnevale, S. A. Vardhana and A. T. Satpathy, Cancer Cell, 2022, 40, 768–786 CrossRef PubMed , e767.
- A. E. Baxter, H. Huang, J. R. Giles, Z. Chen, J. E. Wu, S. Drury, K. Dalton, S. L. Park, L. Torres, B. W. Simone, M. Klapholz, S. F. Ngiow, E. Freilich, S. Manne, V. Alcalde, V. Ekshyyan, S. L. Berger, J. Shi, M. S. Jordan and E. J. Wherry, Immunity, 2023, 56, 1320–1340 CrossRef PubMed , e1310.
- D. R. Bublik, S. Bursać, M. Sheffer, I. Oršolić, T. Shalit, O. Tarcic, E. Kotler, O. Mouhadeb, Y. Hoffman, G. Fuchs, Y. Levin, S. Volarević and M. Oren, Proc. Natl. Acad. Sci. U. S. A., 2017, 114, E496–E505 CrossRef.
- Y. Liu, C. Deisenroth and Y. Zhang, Trends Cancer, 2016, 2, 191–204 CrossRef.
- A. M. Lüchtenborg, P. Metzger, M. Cosenza Contreras, V. Oria, M. L. Biniossek, F. Lindner, K. Fröhlich, A. Malyi, T. Erbes, N. Gensch, J. Maurer, A. Thomsen, M. Boerries, O. Schilling, M. Werner and P. Bronsert, Breast Cancer Res., 2022, 24, 65 CrossRef.
- W. Zhao, N. Ma, S. Wang, Y. Mo, Z. Zhang, G. Huang, K. Midorikawa, Y. Hiraku, S. Oikawa, M. Murata and K. Takeuchi, J. Exp. Clin. Cancer Res., 2017, 36, 88 CrossRef.
- Y. Huang, Y. Liu, X. Guo, C. Fan, C. Yi, Q. Shi, H. Su, C. Liu, J. Yuan, D. Liu, W. Yang and F. Han, Plant J., 2023, 115, 1298–1315 CrossRef.
- M. G. Bacalini, A. Pacilli, C. Giuliani, M. Penzo, D. Treré, C. Pirazzini, S. Salvioli, C. Franceschi, L. Montanaro and P. Garagnani, BMC Cancer, 2014, 14, 361 CrossRef PubMed.
- M. Yue, M. Gautam, Z. Chen, J. Hou, X. Zheng, H. Hou and L. Li, Physiol. Plant., 2021, 172, 2079–2089 CrossRef PubMed.
- D. Sharma, M. Singh and R. Rani, Semin. Cancer Biol., 2022, 87, 184–195 CrossRef PubMed.
- S. He, D. Nakada and S. J. Morrison, Annu. Rev. Cell Dev. Biol., 2009, 25, 377–406 CrossRef CAS PubMed.
- H. Wu, W. Qin, S. Lu, X. Wang, J. Zhang, T. Sun, X. Hu, Y. Li, Q. Chen, Y. Wang, H. Zhao, H. Piao, R. Zhang and M. Wei, Mol. Cancer, 2020, 19, 95 CrossRef PubMed.
- Q. Zhang, C. Thakur, J. Shi, J. Sun, Y. Fu, P. Stemmer and F. Chen, Semin. Cancer Biol., 2019, 57, 27–35 CrossRef.
- Y. Yi, Y. Li, Q. Meng, Q. Li, F. Li, B. Lu, J. Shen, L. Fazli, D. Zhao, C. Li, W. Jiang, R. Wang, Q. Liu, A. Szczepanski, Q. Li, W. Qin, A. B. Weiner, T. L. Lotan, Z. Ji, S. Kalantry, L. Wang, E. M. Schaeffer, H. Niu, X. Dong, W. Zhao, K. Chen and Q. Cao, Nat. Cell Biol., 2021, 23, 341–354 CrossRef.
- K. Qin, S. Yu, Y. Liu, R. Guo, S. Guo, J. Fei, Y. Wang, K. Jia, Z. Xu, H. Chen, F. Li, M. Niu, M. S. Dai, L. Dai, Y. Cao, Y. Zhang, Z. J. Xiao and Y. Yi, Nat. Commun., 2023, 14, 6473 CrossRef PubMed.
- W. Qin, J. S. Cheah, C. Xu, J. Messing, B. D. Freibaum, S. Boeynaems, J. P. Taylor, N. D. Udeshi, S. A. Carr and A. Y. Ting, Cell, 2023, 186, 3307–3324 CrossRef PubMed , e3330.
- J. Li and B. Z. Stanger, Trends Immunol., 2020, 41, 859–863 CrossRef PubMed.
- J. Bai, Y. Li and G. Zhang, Cancer Biol. Med., 2017, 14, 348–362 CrossRef CAS PubMed.
- D. Chakravarty, A. Johnson, J. Sklar, N. I. Lindeman, K. Moore, S. Ganesan, C. M. Lovly, J. Perlmutter, S. W. Gray, J. Hwang, C. Lieu, F. André, N. Azad, M. Borad, L. Tafe, H. Messersmith, M. Robson and F. Meric-Bernstam, J. Clin. Oncol., 2022, 40, 1231–1258 CrossRef.
- N. Nishida and M. Kudo, Liver Cancer, 2014, 3, 417–427 CrossRef.
- N. Chen, C. Peng and D. Li, Front. Immunol., 2022, 13, 869307 CrossRef.
- A. C. Mandigo, S. A. Tomlins, W. K. Kelly and K. E. Knudsen, Clin. Cancer Res., 2022, 28, 255–264 CrossRef.
- K. Simin, H. Wu, L. Lu, D. Pinkel, D. Albertson, R. D. Cardiff and T. Van Dyke, PLoS Biol., 2004, 2, E22 CrossRef.
- R. A. Weinberg, Cell, 1995, 81, 323–330 CrossRef PubMed.
- B. Vogelstein and K. W. Kinzler, Nat. Med., 2004, 10, 789–799 CrossRef CAS PubMed.
- C. Giacinti and A. Giordano, Oncogene, 2006, 25, 5220–5227 CrossRef.
- F. Bunz, A. Dutriaux, C. Lengauer, T. Waldman, S. Zhou, J. P. Brown, J. M. Sedivy, K. W. Kinzler and B. Vogelstein, Science, 1998, 282, 1497–1501 CrossRef.
- L. D. Mayo and D. B. Donner, Trends Biochem. Sci., 2002, 27, 462–467 CrossRef PubMed.
- R. Voit, K. Schäfer and I. Grummt, Mol. Cell. Biol., 1997, 17, 4230–4237 CrossRef.
- A. Budde and I. Grummt, Oncogene, 1999, 18, 1119–1124 CrossRef.
- W. Zhai and L. Comai, Mol. Cell. Biol., 2000, 20, 5930–5938 CrossRef PubMed.
- Z. Andrysik, K. D. Sullivan, J. S. Kieft and J. M. Espinosa, Nat. Commun., 2022, 13, 7400 CrossRef CAS PubMed.
- A. Theophanous, A. Christodoulou, C. Mattheou, D. S. Sibai, T. Moss and N. Santama, J. Biol. Chem., 2023, 299, 105203 CrossRef PubMed.
- K. Srivastava, K. E. Lines, D. Jach and T. Crnogorac-Jurcevic, Oncogene, 2023, 42, 3422–3434 CrossRef PubMed.
- H. Beckmann, J. L. Chen, T. O'Brien and R. Tjian, Science, 1995, 270, 1506–1509 CrossRef PubMed.
- W. M. Hempel, A. H. Cavanaugh, R. D. Hannan, L. Taylor and L. I. Rothblum, Mol. Cell. Biol., 1996, 16, 557–563 CrossRef PubMed.
- J. C. Tuan, W. Zhai and L. Comai, Mol. Cell. Biol., 1999, 19, 2872–2879 CrossRef PubMed.
- P. Hallenborg, S. Feddersen, L. Madsen and K. Kristiansen, Expert Opin. Ther. Targets, 2009, 13, 235–246 CrossRef PubMed.
- C. J. Sherr, Cell, 2004, 116, 235–246 CrossRef CAS PubMed.
- C. J. Sherr and F. McCormick, Cancer Cell, 2002, 2, 103–112 CrossRef CAS.
- C. Cordon-Cardo, Am. J. Pathol., 1995, 147, 545–560 Search PubMed.
- M. Ghosh, S. Saha, J. Bettke, R. Nagar, A. Parrales, T. Iwakuma, A. W. M. van der Velden and L. A. Martinez, Cancer Cell, 2021, 39, 494–508 CrossRef , e495.
- L. Chen, S. Liu and Y. Tao, Signal Transduction Targeted Ther., 2020, 5, 90 CrossRef PubMed.
- C. Caggiano, E. Guida, F. Todaro, P. Bielli, M. Mori, F. Ghirga, D. Quaglio, B. Botta, F. Moretti, P. Grimaldi, P. Rossi, E. A. Jannini, M. Barchi and S. Dolci, Cell Death Discovery, 2020, 6, 111 CrossRef.
- L. Montanaro, D. Treré and M. Derenzini, Am. J. Pathol., 2008, 173, 301–310 CrossRef.
- M. VanInsberghe, J. van den Berg, A. Andersson-Rolf, H. Clevers and A. van Oudenaarden, Nature, 2021, 597, 561–565 CrossRef PubMed.
- D. Simsek, G. C. Tiu, R. A. Flynn, G. W. Byeon, K. Leppek, A. F. Xu, H. Y. Chang and M. Barna, Cell, 2017, 169, 1051–1065 CrossRef CAS PubMed , e1018.
- L. Zhu, Z. Lu and H. Zhao, Oncogene, 2015, 34, 4547–4557 CrossRef.
- K. Y. Wang, C. H. Wu, L. Y. Zhou, X. H. Yan, R. L. Yang, L. M. Liao, X. M. Ge, Y. S. Liao, S. J. Li, H. Z. Li, L. L. Gao, J. S. Lin and S. Y. Huang, Eur. Neurol., 2015, 74, 28–35 CrossRef.
- M. Boussada, I. Hammami, R. Ben Ali, A. B. Ammar, M. Alves, P. F. Oliveira, A. B. Akacha, I. L. Abdelkarim, S. Zekri and M. V. El May, Andrologia, 2022, 54, e14634 CrossRef PubMed.
- A. Stepanovic, N. Terzic Jovanovic, A. Korac, M. Zlatovic, I. Nikolic, I. Opsenica and M. Pesic, Biomed. Pharmacother., 2024, 174, 116496 CrossRef.
- P. F. Islas-Morales, A. Cardenas, M. J. Mosqueira, L. F. Jimenez-Garcia and C. R. Voolstra, Front. Microbiol., 2023, 14, 1075071 CrossRef.
- N. G. Ordonez, Mod. Pathol., 2012, 25, 1011–1022 CrossRef.
- Y. X. Ru, Y. C. Mi, J. H. Liu, H. J. Wang, S. X. Zhao, W. Cui, C. W. Li, Q. H. Li, X. F. Zhu, Z. J. Xiao, J. X. Pang and J. X. Wang, Ultrastruct. Pathol., 2009, 33, 67–75 CrossRef PubMed.
- G. Y. Yin, W. N. Zhang, X. J. Shen, Y. Chen and X. F. He, World J. Gastroenterol., 2003, 9, 851–857 CrossRef PubMed.
- K. E. Ramohlola, E. I. Iwuoha, M. J. Hato and K. D. Modibane, J. Anal. Methods Chem., 2020, 2020, 8896698 Search PubMed.
- A. Valery, A. Pofelski, L. Clement, F. Lorut and E. F. Rauch, Micron, 2017, 92, 43–50 CrossRef.
- P. Tizro, C. Choi and N. Khanlou, Methods Mol. Biol., 2019, 1897, 417–424 CrossRef.
- A. Velazco, A. Beche, D. Jannis and J. Verbeeck, Ultramicroscopy, 2022, 232, 113398 CrossRef.
- S. Kakoti, M. Yamauchi, W. Gu, R. Kato, T. Yasuhara, Y. Hagiwara, S. Laskar, T. Oike, H. Sato, K. D. Held, T. Nakano and A. Shibata, Oncol. Rep., 2019, 42, 2293–2302 Search PubMed.
- S. Radulovic, S. Sunkara, R. Rachel and G. Leitinger, Histochem. Cell Biol., 2022, 158, 203–211 CrossRef PubMed.
- Y. Ju, H. J. Ro, Y. S. Yi, T. Cho, S. I. Kim, C. W. Yoon, S. Jun and J. Kim, Anal. Chem., 2021, 93, 2871–2878 CrossRef.
- P. Tchelidze, H. Kaplan, A. Beorchia, M. F. O'Donohue, H. Bobichon, N. Lalun, L. Wortham and D. Ploton, Methods Mol. Biol., 2008, 463, 137–158 CrossRef.
- T. R. Thyagarajan and H. B. Naylor, J. Bacteriol., 1962, 83, 127–136 CrossRef PubMed.
- M. Bendayan and E. Paransky, Microsc. Res. Tech., 2014, 77, 999–1004 CrossRef.
- C. Pan, Y. Tong, H. Qian, A. V. Krasavin, J. Li, J. Zhu, Y. Zhang, B. Cui, Z. Li, C. Wu, L. Liu, L. Li, X. Guo, A. V. Zayats, L. Tong and P. Wang, Nat. Commun., 2024, 15, 2840 CrossRef PubMed.
- D. Wilburn, E. Fletcher, A. Ismaeel, D. Miserlis, B. Zechmann and P. Koutakis, Ultramicroscopy, 2022, 241, 113600 CrossRef.
- Y. Yatabe, S. Dacic, A. C. Borczuk, A. Warth, P. A. Russell, S. Lantuejoul, M. B. Beasley, E. Thunnissen, G. Pelosi, N. Rekhtman, L. Bubendorf, M. Mino-Kenudson, A. Yoshida, K. R. Geisinger, M. Noguchi, L. R. Chirieac, J. Bolting, J. H. Chung, T. Y. Chou, G. Chen, C. Poleri, F. Lopez-Rios, M. Papotti, L. M. Sholl, A. C. Roden, W. D. Travis, F. R. Hirsch, K. M. Kerr, M. S. Tsao, A. G. Nicholson, I. Wistuba and A. L. Moreira, J. Thorac. Oncol., 2019, 14, 377–407 CrossRef PubMed.
- W. Sheng, C. Zhang, T. M. Mohiuddin, M. Al-Rawe, F. Zeppernick, F. H. Falcone, I. Meinhold-Heerlein and A. F. Hussain, Int. J. Mol. Sci., 2023, 24, 3086 CrossRef.
- Y. Xu, Y. Wang, R. Zhou, H. Li, H. Cheng, Z. Wang and J. Zhang, Chin. J. Cancer Res., 2016, 28, 72–79 Search PubMed.
- C. C. Wang, H. Peng, Z. Wang, J. Yang, R. G. Hu, C. Y. Li and W. J. Geng, Mil. Med. Res., 2022, 9, 35 Search PubMed.
- W. C. C. Tan, S. N. Nerurkar, H. Y. Cai, H. H. M. Ng, D. Wu, Y. T. F. Wee, J. C. T. Lim, J. Yeong and T. K. H. Lim, Cancer Commun., 2020, 40, 135–153 CrossRef.
- C. W. Jia, Y. C. Zhang, Y. Y. Wang, J. N. Gao, A. Raza, T. Ogawa, S. Wada, D. Xie and J. Y. Wang, J. Drug Delivery Sci. Technol., 2023, 80, 104081 CrossRef.
- X. Xu, C. Li, X. Gao, K. Xia, H. Guo, Y. Li, Z. Hao, L. Zhang, D. Gao, C. Xu, H. Xu, Z. Q. Xiong, Z. Qiu, L. Mei, X. Xie, K. Ruan and R. Hu, Cell Res., 2018, 28, 48–68 CrossRef CAS.
- C. Li, T. Han, Q. Li, M. Zhang, R. Guo, Y. Yang, W. Lu, Z. Li, C. Peng, P. Wu, X. Tian, Q. Wang, Y. Wang, V. Zhou, Z. Han, H. Li, F. Wang and R. Hu, Nucleic Acids Res., 2021, 49, 3796–3813 CrossRef CAS PubMed.
- C. Jia, H. Tang, Y. Yang, S. Yuan, T. Han, M. Fang, S. Huang, R. Hu, C. Li and W. Geng, Biochem. Biophys. Res. Commun., 2020, 529, 43–50 CrossRef CAS.
- K. S. Janardhan, H. Jensen, N. P. Clayton and R. A. Herbert, Toxicol. Pathol., 2018, 46, 488–510 CrossRef CAS.
- Y. Yang, Y. Luo, C. Yang, R. Hu, X. Qin and C. Li, Biochim. Biophys. Acta, Gene Regul. Mech., 2023, 1866, 194954 CrossRef CAS PubMed.
- C. Liu, D. Liu, F. Wang, Y. Liu, J. Xie, J. Xie and Y. Xie, Front. Immunol., 2022, 13, 1038927 CrossRef CAS.
- M. Masiuk, M. Lewandowska, L. Teresinski, E. Dobak and E. Urasinska, Folia Histochem. Cytobiol., 2019, 57, 139–145 CAS.
- A. Gandjeva and R. M. Tuder, Methods Mol. Biol., 2018, 1809, 315–329 CrossRef CAS.
- F. Varghese, A. B. Bukhari, R. Malhotra and A. De, PLoS One, 2014, 9, e96801 CrossRef.
- A. Bonnet-Garnier, P. Feuerstein, M. Chebrout, R. Fleurot, H. U. Jan, P. Debey and N. Beaujean, Int. J. Dev. Biol., 2012, 56, 877–887 CrossRef CAS.
- M. T. Spooner, J. E. Alex, J. A. Greer, D. R. Delorey, R. A. Kiser, C. Petersen, T. Polk and K. Gunzelman, Mil. Med., 2019, 184, e141–e146 CrossRef.
- D. Hanahan and R. A. Weinberg, Cell, 2011, 144, 646–674 CrossRef CAS.
- Q. Wu, C. Yuan, N. Liu, J. Shu, J. Wang, J. Qian, L. Zeng, H. Zhang, X. Wang and W. Mei, J. Exp. Clin. Cancer Res., 2022, 41, 201 CrossRef CAS PubMed.
- H. Choi, H. D. Kim, Y. W. Choi, H. Lim, K. W. Kim, K. S. Kim, Y. C. Lee and C. H. Kim, Arch. Biochem. Biophys., 2023, 750, 109810 CrossRef CAS PubMed.
- S. J. Barfeld, L. Fazli, M. Persson, L. Marjavaara, A. Urbanucci, K. M. Kaukoniemi, P. S. Rennie, Y. Ceder, A. Chabes, T. Visakorpi and I. G. Mills, Onco Targets Ther, 2015, 6, 12587–12602 Search PubMed.
- M. Migaldi, G. Barbolini, D. Trere, M. Criscuolo, A. M. Martinelli and E. Zunarelli, Histochem. J., 1997, 29, 661–668 CrossRef CAS.
- X. S. Liu, L. M. Zhou, L. L. Yuan, Y. Gao, X. Y. Kui, X. Y. Liu and Z. J. Pei, Front. Immunol., 2021, 12, 724741 CrossRef CAS PubMed.
- F. Ezzatifar, A. Rafiei and M. Jeddi-Tehrani, Pathol., Res. Pract., 2022, 240, 154160 CrossRef CAS PubMed.
- M. Derenzini, L. Montanaro and D. Trere, Histopathology, 2009, 54, 753–762 CrossRef.
- L. M. Zhou, L. L. Yuan, Y. Gao, X. S. Liu, Q. Dai, J. W. Yang and Z. J. Pei, Eur. J. Nucl. Med. Mol. Imaging, 2021, 48, 904–912 CrossRef CAS PubMed.
- F. Huang, Y. Wu, H. Tan, T. Guo, K. Zhang, D. Li and Z. Tong, Oncol. Rep., 2019, 41, 590–598 CAS.
- P. K. Chan, R. Frakes, E. M. Tan, M. G. Brattain, K. Smetana and H. Busch, Cancer Res., 1983, 43, 3770–3777 CAS.
- A. M. Gown, Arch. Pathol. Lab. Med., 2016, 140, 893–898 CrossRef PubMed.
- B. Y. Zhang, Q. Q. Duan, H. C. Zhao, Y. X. Zhang, X. N. Li, Y. F. Xi, Z. F. Wu, L. Guo, P. C. Li and S. B. Sang, Sens. Actuators, B, 2021, 329, 129156 CrossRef CAS.
- B. Chazotte, Cold Spring Harb. Protoc., 2011, 2011, pdb prot5556 CrossRef PubMed.
- R. Chen, K. Qiu, G. Han, B. K. Kundu, G. Ding, Y. Sun and J. Diao, Chem. Sci., 2023, 14, 10236–10248 RSC.
- P. Santangelo, N. Nitin, L. LaConte, A. Woolums and G. Bao, J. Virol., 2006, 80, 682–688 CrossRef CAS.
- J. A. Kilgore and N. J. Dolman, Curr. Protoc., 2023, 3, e752 CrossRef CAS PubMed.
- R. Lamba, A. Salam, F. Anjum, A. Yadav, R. Garg, K. Kaushik, S. Sharma and C. K. Nandi, Nanoscale, 2024, 16, 11739–11748 RSC.
- J. C. Stockert, A. Blazquez-Castro and R. W. Horobin, Methods Mol. Biol., 2014, 1094, 25–38 CrossRef CAS.
- Y. Yang, D. X. Yan, R. X. Rong, B. Y. Shi, M. Zhang, J. Liu, J. Xin, T. Xu, W. J. Ma, X. L. Li and K. R. Wang, Bioorg. Chem., 2024, 142, 106969 CrossRef CAS.
- D. Dilworth, R. P. Hanley, R. Ferreira de Freitas, A. Allali-Hassani, M. Zhou, N. Mehta, M. R. Marunde, S. Ackloo, R. A. Carvalho Machado, A. Khalili Yazdi, D. D. G. Owens, V. Vu, D. Y. Nie, M. Alqazzaz, E. Marcon, F. Li, I. Chau, A. Bolotokova, S. Qin, M. Lei, Y. Liu, M. M. Szewczyk, A. Dong, S. Kazemzadeh, T. Abramyan, I. K. Popova, N. W. Hall, M. J. Meiners, M. A. Cheek, E. Gibson, D. Kireev, J. F. Greenblatt, M. C. Keogh, J. Min, P. J. Brown, M. Vedadi, C. H. Arrowsmith, D. Barsyte-Lovejoy, L. I. James and M. Schapira, Nat. Chem. Biol., 2022, 18, 56–63 CrossRef CAS PubMed.
- X. Cao, J. Zheng, R. Zhang, Y. Sun and M. Zhao, Nucleic Acids Res., 2024, 52, e41 CrossRef PubMed.
- J. Lippincott-Schwartz and G. H. Patterson, Science, 2003, 300, 87–91 CrossRef CAS.
- H. Xue, J. Lu, H. Yan, J. Huang, H. B. Luo, M. S. Wong, Y. Gao, X. Zhang and L. Guo, Talanta, 2022, 237, 122898 CrossRef CAS.
- J. R. Luo, W. Long, Z. X. Chen, S. M. Wang, Y. X. Zeng, Y. J. Lu, B. X. Zheng, M. T. She and W. L. Wong, ACS Sens., 2024, 9, 1545–1554 CrossRef CAS.
- J. Liu, S. Zhang, J. Zhu, X. Liu, G. Yang and X. Zhang, Anal. Bioanal. Chem., 2019, 411, 5223–5231 CrossRef CAS PubMed.
- I. C. Mondal, M. Galkin, S. Sharma, N. A. Murugan, D. A. Yushchenko, K. Girdhar, A. Karmakar, P. Mondal, P. Gaur and S. Ghosh, Chem. – Asian J., 2022, 17, e202101281 CrossRef CAS PubMed.
- M. J. Kim, Y. Li, J. A. Junge, N. K. Kim, S. E. Fraser and C. Zhang, ACS Chem. Biol., 2023, 18, 1523–1533 CrossRef CAS.
- L. Wang, Q. Xia, M. Hou, C. Yan, Y. Xu, J. Qu and R. Liu, J. Mater. Chem. B, 2017, 5, 9183–9188 RSC.
- A. J. Pickard and U. Bierbach, ChemMedChem, 2013, 8, 1441–1449 CrossRef CAS PubMed.
- S. Song, M. Manook, J. Kwun, A. M. Jackson, S. J. Knechtle and G. Kelsoe, Commun. Biol., 2021, 4, 1338 CrossRef CAS.
- M. Wu, G. Xu, C. Han, P. F. Luan, Y. H. Xing, F. Nan, L. Z. Yang, Y. Huang, Z. H. Yang, L. Shan, L. Yang, J. Liu and L. L. Chen, Science, 2021, 373, 547–555 CrossRef CAS PubMed.
- L. L. Chen, Nat. Methods, 2022, 19, 1152–1155 CrossRef CAS.
- P. Jiang, W. Pang, S. Ding, D. Wang, X. Wei and B. Gu, Anal. Chim. Acta, 2021, 1154, 338309 CrossRef CAS PubMed.
- R. N. Day and M. W. Davidson, Chem. Soc. Rev., 2009, 38, 2887–2921 RSC.
- A. Rangel-Pozzo, S. Booth, P. L. I. Yu, M. Singh, G. Selivanova and S. Mai, J. Clin. Med., 2020, 9, 598 CrossRef CAS PubMed.
Footnote |
| † These authors contributed equally to this work. |
| This journal is © The Royal Society of Chemistry 2024 |