Open Access Article
Open Access ArticleUnveiling cellular vitality: peptide fluorescent probes illuminate mitochondrial dynamics and ROS activity†
Ixsoyen
Vázquez-Sandoval
,
Jasmine
Bernal-Escalante
,
Adriana
Romo-Pérez
and
Arturo
Jiménez-Sánchez
 *
*
Instituto de Química – Universidad Nacional Autónoma de México, Ciudad Universitaria, Circuito Exterior s/n., Ciudad de México, Coyoacán 04510, Mexico. E-mail: arturo.jimenez@iquimica.unam.mx
First published on 23rd November 2023
Abstract
This study explored cell-penetrating peptide probes for dynamic cellular vitality tracking. In particular, two mitochondrial-penetrating peptide probes effectively monitored changes in mitochondrial morphology and ROS activity, serving as reliable vitality indicators. Dual-confocal channel monitoring and spectrally-resolved confocal microscopy were employed to trace local structural changes during probe photooxygenation induced by mitochondrial ROS.
Mitochondria are essential organelles for the regulation of cellular bioenergetics and relay the switch to the glycolytic phenotype of malignant cells.1 They modulate Ca2+ ion signaling and reactive oxygen species (ROS) levels.2 It is widely known that mitochondrial function depends on its physical structure which in turn depends on the cellular function.3 All these factors are crucial in the cellular health and apoptotic events. In spite of that, the physical/morphological monitoring of organelles in healthy and disease status is uncommon.4 These changes in mitochondrial morphology, which affect its network organization in response to environmental cues, are known as mitochondrial dynamics.5 Importantly, prior to directly affecting cell viability (defined as the percentage of live cells in a population), these cellular changes may impair the cell ability to divide without necessarily causing cell death, a phenomenon referred to as cell vitality.6 Cell vitality is a rather uncommon process used in tissue culture, microbiology and toxicity research, although represents a powerful tool which involves the physiological capabilities of cells.7 However, while mitochondrial dynamics have been correlated with cell viability,8 the real-time monitoring of cell vitality during mitochondrial dynamics alterations that not necessarily led to cell death, remains an unmet need. In fact, the release of active caspases and cytochrome c before or even in the absence of mitochondrial membrane potential (ΔΨm) depolarization (due to the respiration-driven reestablishment of ΔΨm) represents a source of artifacts when monitoring the initiators of apoptotic events because important morphology alterations and outer membrane rupture can occur before ΔΨm collapse.9 For this reason it is crucial to develop fluorescent probes able to track the cell vitality stage. However, no fluorophore chemistry approaches to track dynamic changes in mitochondria to assess cell vitality have been described to date, and the known molecular probes relay on multiple-assay techniques, i.e. using rhodamine 123 to determine the mitochondrial membrane potential,10 fluorescein diacetate to determine enzymes activity,11 and determination of the cellular ATP content based on the luciferin reaction.12 This type of approach requires a complex workflow and separate signal recordings that may cause phenotypic drift and analytical bias.
Here we developed a mitochondrial penetrating peptide-based fluorescent probes presenting a mitochondrial membrane potential and photooxygenation processes that allow fluorescence recordings in the TxR (λexc = 590 nm, λem = 605 nm) and GFP (λexc = 480 nm, λem = 500) channels. This ROS response and the sensibility of the probes to respond to subtle variations of ΔΨm enable the continuous monitoring of cell vitality. The new mitochondria-penetrating peptide fluorescent probes, termed Dead/Live Cell Cyaninen5 (DeLiCy5) and Coumarin-343 (DeLiC-343), are presented in Fig. 1.
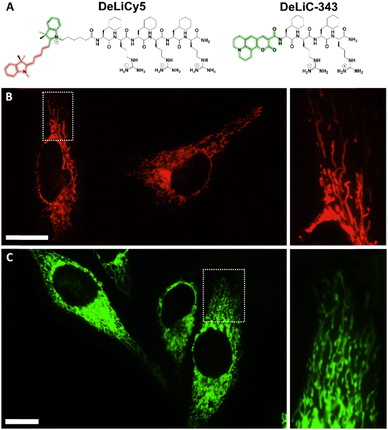 | ||
| Fig. 1 A) Structure of the mitochondrial fluorescent peptide probes, DeLiCy5 and DeLiC-343. Intracellular localization in live HeLa cells for B) DeLiCy5 under the Cy5 confocal channel and C) DeLiC-343 in the GFP channel. The high filamentous-like localization indicates that DeLiC probes are mitochondria-specific. Pearson's coefficient = 0.95 and 0.91 for DeLiCy5 and DeLiC-343, respectively (see ESI† for details). Scale bars = 20 μm. | ||
DeLiC probes are dual Dead/Live cell probes with a mitochondrial ΔΨm dynamic profile enabling a continuous mitochondrial morphology monitoring during apoptotic/necrotic processes as the mitochondrial mass and volume changes are correlated with ΔΨm variations, a feature that is not affordable with cationic fluorophores since their localization is controlled by the same chemical moieties that serve as reporters.13 As these fluorescent probes contain mitochondrial penetrating peptide vectors, they do not require a second probe reporter. Thus, common localization interference and spatial distribution crosstalk problems present in previously reported Dead/Live systems can be overcome.
Notably, a dual-emission channel monitoring was only performed in the case of DeLiCy5 probe, using the photooxygenation features of Cy5 moiety to detect the presence of singlet oxygen (1O2) ROS as previously described,14–16 thus providing an unequivocal cell vitality status tracking.
The DeLiC conjugates were obtained by solid-phase synthesis and characterized as described in the ESI file. We found different localization patterns between these probes. DeLiCy5 is a unique mitochondrial localizer enabling the monitoring of healthy mitochondria in the Cy5 confocal channel with excitation at 647 nm through ΔΨm dynamic monitoring, while a complementary response for compromised proapoptotic or morphologically altered mitochondria upon subtle ROS stimuli monitored in the green GFP confocal channel when exciting at 488 nm. The analogue DeLiC-343 on the other hand, exhibited mitochondrial localization only in the GFP channel with ΔΨm dependence. Colocalizations with MitoLite™ blue localizer are shown in Fig. S7, ESI.† These results highlight the high specificity of the arginine-cyclohexylalanine (Fxr) peptide sequence of the DeLiC probes as previously studied.13,17,18
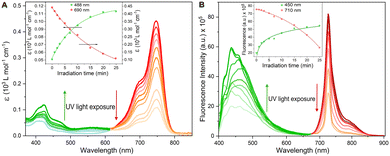
To assess the efficiency of the DeLiCy5 probe undergoing photooxidation, the absorption and emission properties during UV irradiation (300 W Xe lamp) for 25 min translate into a strong deactivation in both, the red-absorption band and the red-fluorescence emission with the simultaneous green-emission increase, highlighting the pi-conjugation breaking, Fig. 2. According to previous literature reports,13,17–19 and the corresponding mass spectrometry (HRMS) studies, this is assigned to the formation of dioxetane and aldehyde species promoted by singlet oxygen photooxygenation (see Fig. S5, ESI† file), thus activating the green-emission response channel.
We then proceeded to analyze the effect of singlet oxygen photoxygenation in vitro, to this end, we utilized 5-aminolevulinic acid (ALA), which is one of the most widely used photosensitizer precursors to produce sufficient singlet oxygen inside the cell in type II photochemical scheme.20
The identification of singlet oxygen in the solutions was established by detecting its emission peak at 1270 nm, which had a natural bandwidth of 18 nm.21 A control experiment was conducted using an air-saturated solution of erythrosin B in anhydrous ethanol, which had an optical density of 0.5. The experiment involved recording single emission peaks at 1270 nm, and this was achieved by employing a 450 W Xe lamp in conjunction with a NIR photomultiplier, (see Fig. S6†). Also, the observed photooxidation and the corresponding selectivity were analysed in terms of in situ produced 1O2 by H2O2 and HClO reaction in the presence and absence of NaN3, a selective 1O2 scavenger, the analysis revealed a strong affinity to 1O2 (Fig. S8†). Then, after NaClO and H2O2 were added, we recorded 10 minutes before starting light irradiation. Notably, no significant oxidation features were observed in the fluorescence profiles, indicating the probe chemical stability (blue spectra, Fig. S8†).
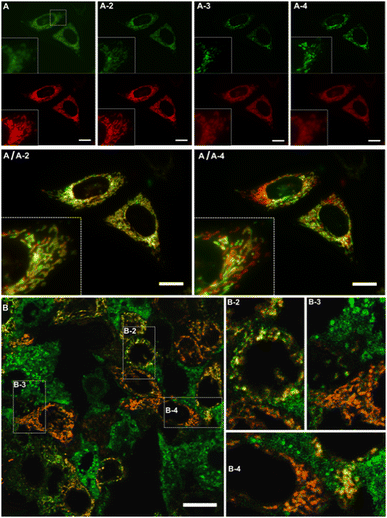
Thus, photosensitized 1O2 production in HeLa cells influences the spectroscopic features of DeLiCy5 to activate the green-emission channel around 500 nm with a concomitant red-emission decrease, Fig. 3. This result corresponds to the observed trend in Fig. 2, indicating that both, dioxetane and the aldehyde derivatives (m/z: 361.2389 and 317.4698, Fig. S5, ESI† file) are formed through the photooxygenation reaction.
The mitochondrial morphology changes induced by ALA-endogenously generated 1O2 were then studied in more detail during time-course experiments, Fig. 3A and B. Specifically, DeLiCy5 probe enabled to evaluate mitochondrial morphology variations under subtle apoptotic conditions using low-dose ALA photosensitization where interconnected filamentous-like frameworks exhibited red fluorescence (λexc = 647, λem = 700 nm) while swelled-spherical morphologies were observed in the GFP green confocal channel, Fig. 3. The robust mitochondrial dynamics indicated a gradual shift from filamentous-like to swollen, rod-shaped, and spherical mitochondria. Some fluorescence stabilization was also noted due to a pronounced signal overlap (depicted in yellow), thus confirming the transitions between these different forms. Notably, as observed in Fig. 3, various cells exhibited a heterogeneous morphology variation when exposed to ALA-induced 1O2 stimuli. However, such cell-to-cell variations gradually became uniform over time.
In fact, 3D (z-scan) monitoring after 36 hours indicated a uniform differentiation between live and dead cells in both channels, Fig. S9, ESI† file.
Importantly, the effect of subtle ALA-1O2 stimuli vs. probes mitochondrial localization was analyzed. Interestingly, CCCP depolarization stimuli evidenced clear mitochondrial release for DeLiC-343, while for DeLiCy5 a mitochondrial retention (no dye release) was observed, suggesting that upon photooxidation the Cy5-aldehyde derivative acts as a mitochondrial immobilization group, as previously described,22 where only mitochondrial permeability transition pore (mPTP) opening resulted in the probe release, Fig. S10.† Additionally, following ALA-induced 1O2 stimuli, 4 hours exposure to 10 μM etoposide, a well-established apoptotic agent for HeLa cells, resulted in the release of the DeLiCy5 probe from mitochondria, a process not observed when the probe undergoes photodegradation alone, Fig. S11 panel A vs. B, ESI† file.
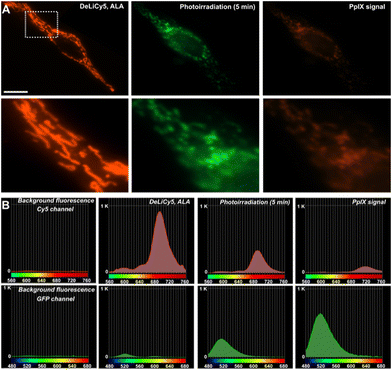
However, to effectively track subtle cellular damage by observing changes in mitochondrial morphology using DeLiCy5 in live cells over time, it is crucial to mitigate issues related to crosstalk, interference, and signal artifacts. One of the primary challenges in achieving this objective is the loss of structural details and local probe spectral resolution during measurements. In this context, spectrally-resolved confocal microscopy offers distinctive opportunities for analytical bioimaging, providing structural insights of probe responses.
Using a confocal array coupled with a spectral detector unit, accurate spectral unmixing enabled DeLiCy5 to be spectrally resolved in real time, Fig. 4. As evident from the real-time observations, during apoptosis, there was a steady rise in the diffuse green-fluorescent signal from the outset of the experiment, coupled with a noticeable decline in mitochondrial red fluorescence when employing 488 and 647 nm excitation. This phenomenon consistently aligned with the findings from spectrally-resolved images in Fig. 4B, indicating that the green channel is primarily influenced by the photooxidized Cy5 fluorescence following singlet oxygen photogeneration.
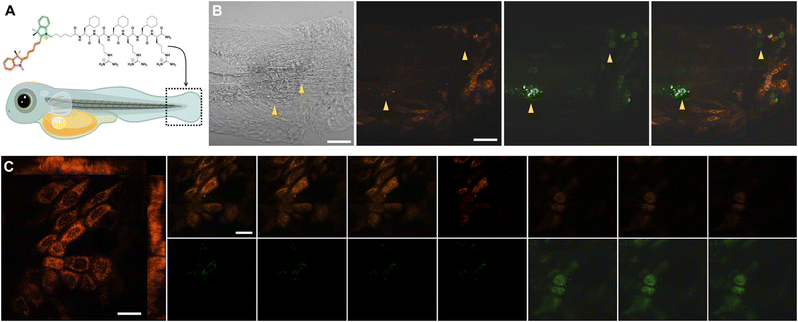
Finally, we investigated the DeLiCy5 probe imaging capabilities through in vivo investigations performed in the zebrafish (Danio rerio) model. Then, to demonstrate the DeLiCy5 ability to sense native 1O2, we examined the injury response in a caudal fin wound, as depicted in Fig. 5. Zebrafish serves as a model system for studying tissue regeneration,19 aiding in the comprehension of tissue maintenance and turnover processes. The wounded area is predominantly characterized by dead cells. Nonetheless, to investigate subtle, in vivo wound healing processes that affect cellular vitality, we employed laser irradiation. Using an erbium:YAG laser (Asclepion) with a frequency of 5 Hz and 7 J cm−2 power density, we could swiftly and consistently induce full-thickness wounds on the flank of adult zebrafish.23 These wounds undergo extremely rapid re-epithelialization, independent of blood clot formation and inflammation.
Through this approach, we successfully tracked the emergence of the initial green-emission signal immediately following erbium:YAG laser tissue injury in real-time and continuously over an extended duration. We then assessed the injury response by employing z-stack time-lapse imaging of the caudal fin, Fig. 5C.
As observed, the region of injured tissue displayed the real-time development of morphologically altered proapoptotic and deceased epithelial cells, marked by a green fluorescence signal. It's recognized that tissue damage leads to the release of various reactive oxygen species (ROS), including superoxide (O2˙−) and hydroxyl (˙OH) radicals, which could potentially trigger lateral reactions of the probe, resulting in fluorescence quenching over extended time periods. However, these outcomes underscore the remarkable sensitivity of DeLiCy5 in detecting native ROS within the proapoptotic and deceased cells in the wounded tissue.
In conclusion, although DeLiC-343 probe is also able to localize mitochondria through a ΔΨm Nernstian interaction, we focused on the DeLiCy5 fluorescent probe given the Cyanine5 photooxygenation features. DeLiCy5 has been effectively employed in the exploration of various cell-penetrating peptide probes for the dynamic tracking of cellular vitality. This mitochondrial-penetrating peptide probe reliably monitor alterations in mitochondrial morphology by detecting ROS activity, thus serving as robust vitality indicator. The application of dual-confocal channel monitoring and spectrally-resolved confocal microscopy allowed us to observe and analyze local structural changes during probe photooxygenation, which was triggered by mitochondrial ROS. These findings highlight the utility of DeLiCy5 in bioimaging applications and provide valuable insights into the assessment of cellular vitality.
Author contributions
Ixsoyen Vazquez-Sandoval: synthesis and HPLC purification, methodology, analysis; Jasmine Bernal-Escalante: synthesis and chemical characterization, Adriana Romo-Pérez: methodology, tissue culture; Arturo Jiménez-Sánchez: supervision, data analysis, writing – original draft, review & editing.Conflicts of interest
There are no conflicts to declare.Notes and references
- Mitochondria: The Anti-cancer Target for the Third Millennium, ed. J. Neuzil, S. Pervaiz and S. Fulda, Springer-Dordrecht, 2014 Search PubMed.
- A. Görlach, K. Bertram, S. Hudecova and O. Krizanova, Redox Biol., 2015, 6, 260 CrossRef PubMed.
- J. R. Friedman and J. Nunnari, Nature, 2014, 505, 335 CrossRef CAS PubMed.
- D. R. Green and J. C. Reed, Science, 1998, 281, 1309 CrossRef CAS PubMed.
- M. Giacomello, A. Pyakurel, C. Glytsou and L. Scorrano, Nat. Rev. Mol. Cell Biol., 2020, 21, 204 CrossRef CAS PubMed.
- M. Kwolek-Mirek and R. Zadrag-Tecza, FEMS Yeast Res., 2014, 14, 1068 CAS.
- E. Colombo, A. De Angelis, C. Bassani, F. Ruffini, L. Ottoboni, L. Garzetti, A. Finardi, G. Martino, R. Furlan and C. Farina, BMC Neurosci., 2023, 24, 33 CrossRef CAS PubMed.
- L. Simula and S. Campello, Mitochondrial Bioenergetics, Springer Protocols, New York, 2018, ch. 15 Monitoring the Mitochondrial Dynamics in Mammalian Cells Search PubMed.
- F. Ichas, L. S. Jouaville and J.-P. Mazat, Cell, 1997, 89, 1145 CrossRef CAS PubMed.
- E. Marchi and D. Cavalieri, Genes Nutr., 2008, 3, 159 CrossRef CAS PubMed.
- P. Breeuwer, J. L. Drocourt, N. Bunschoten, M. H. Zwietering, F. M. Rombouts and T. Abee, Appl. Environ. Microbiol., 1995, 61, 1614 CrossRef CAS PubMed.
- S. Ansehn and L. Nilsson, Antimicrob. Agents Chemother., 1984, 26, 22 CrossRef CAS PubMed.
- A. Jiménez-Sánchez, E. Lei and S. O. Kelley, Angew. Chem., Int. Ed., 2018, 57, 8891 CrossRef PubMed.
- Y. Cho, H. J. An, T. Kim, C. Lee and N. K. Lee, J. Am. Chem. Soc., 2021, 143, 14125 CrossRef CAS PubMed.
- Q. Zheng and L. D. Lavis, Curr. Opin. Chem. Biol., 2017, 39, 32 CrossRef CAS PubMed.
- J. B. Grimm and L. D. Lavis, Nat. Methods, 2022, 19, 149 CrossRef CAS PubMed.
- S. B. Fonseca, M. P. Pereira and S. O. Kelley, Adv. Drug Delivery Rev., 2009, 61, 953 CrossRef CAS PubMed.
- S. R. Jean, M. Ahmed, E. K. Lei, S. P. Wisnovsky and S. O. Kelley, Acc. Chem. Res., 2016, 49, 1983 CrossRef PubMed.
- J. Bernal-Escalante, T. Molina-Villa, F. López-Casillas and A. Jiménez-Sánchez, ACS Sens., 2022, 7, 2303 CrossRef CAS PubMed.
- J. Chi, Q. Ma, Z. Shen, C. Ma, W. Zhu, S. Han, Y. Liang, J. Cao and Y. Sun, Nanoscale, 2020, 12, 11008 RSC.
- M. Bregnhøj, M. Westberg, B. F. Minaev and P. R. Ogilby, Acc. Chem. Res., 2017, 50, 1920 CrossRef PubMed.
- S. Zhang, H. Chen, L. Wang, C. Liu, L. Liu, Y. Sun and X. Shen, J. Mater. Chem. B, 2021, 9, 1089 RSC.
- R. Richardson, K. Slanchev, C. Kraus, P. Knyphausen, S. Eming and M. Hammerschmidt, J. Invest. Dermatol., 2013, 133, 1655 CrossRef CAS PubMed.
Footnote |
| † Electronic supplementary information (ESI) available. See DOI: https://doi.org/10.1039/d3sd00279a |
| This journal is © The Royal Society of Chemistry 2024 |