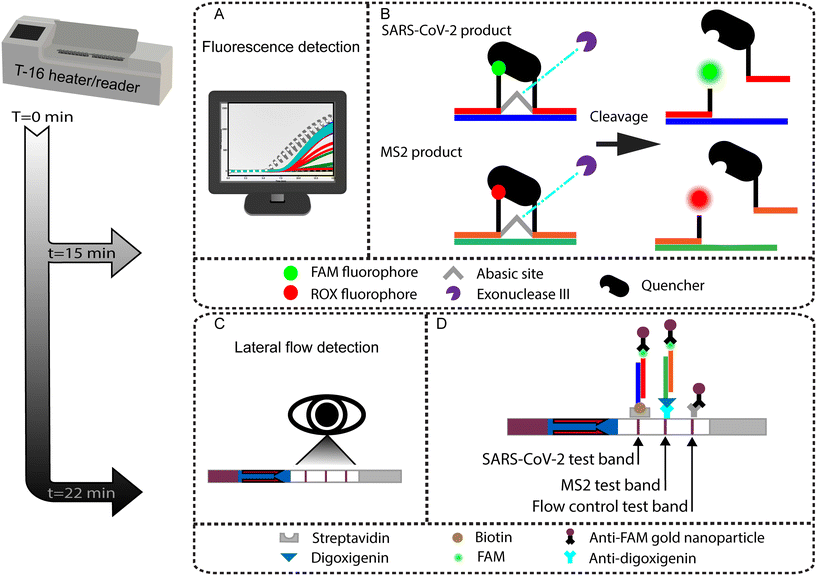
Open Access Article
Open Access ArticleSARS-CoV-2 recombinase polymerase amplification assay with lateral flow readout and duplexed full process internal control†
Coleman D.
Martin
 a,
Andrew T.
Bender
a,
Andrew T.
Bender
 b,
Benjamin P.
Sullivan
b,
Benjamin P.
Sullivan
 b,
Lorraine
Lillis
c,
David S.
Boyle
c and
Jonathan D.
Posner
b,
Lorraine
Lillis
c,
David S.
Boyle
c and
Jonathan D.
Posner
 *abd
*abd
aDepartment of Chemical Engineering, University of Washington, Seattle, Washington, USA
bDepartment of Mechanical Engineering, University of Washington, Seattle, Washington, USA
cPATH, Seattle, Washington, USA
dDepartment of Family Medicine, University of Washington, Seattle, Washington, USA
First published on 12th January 2024
Abstract
Nucleic acid amplification tests for the detection of SARS-CoV-2 have been an important testing mechanism for the COVID-19 pandemic. While these traditional nucleic acid diagnostic methods are highly sensitive and selective, they are not suited to home or clinic-based uses. Comparatively, rapid antigen tests are cost-effective and user friendly but lack in sensitivity and specificity. Here we report on the development of a one-pot, duplexed reverse transcriptase recombinase polymerase amplification SARS-CoV-2 assay with MS2 bacteriophage as a full process control. Detection is carried out with either real-time fluorescence or lateral flow readout with an analytical sensitivity of 50 copies per reaction. Unlike previously published assays, the RNA-based MS2 bacteriophage control reports on successful operation of lysis, reverse transcription, and amplification. This SARS-CoV-2 assay features highly sensitive detection, visual readout through an LFA strip, results in less than 25 minutes, minimal instrumentation, and a useful process internal control to rule out false negative test results.
Introduction
From the onset of the SARS-CoV-2 pandemic, diagnostic testing has played a crucial role in monitoring and diagnosing new infectious cases. SARS-CoV-2 and the associated COVID-19 disease is expected to circulate indefinitely as SARS-CoV-2 continues to mutate, introducing new sub-variants with enhanced immune evasion.1 Diagnostics will continue to play a key role in the timely identification of new cases to stop the spread of disease, inform masking and isolation standards, initiate clinical decisions with patient care plans (e.g., monoclonal antibodies, Paxlovid, etc.), and ensuring safe community gatherings and public openings.2Reverse transcription PCR (RT-PCR) is the gold standard for SARS-CoV-2 molecular testing due to its high sensitivity and specificity, which is crucial for accurate detection during the early phase of infection when viral titers are low.3 Scaled RT-PCR testing is primarily limited to central laboratories due to the need for specialized equipment for nucleic acid extraction, assay preparation, thermocycling, and target detection. These high-resource requirements result in delays from sampling to answer which delays clinical interventions that can prevent disease progression or reduce community transmission.4 Rapid lateral flow assay (LFA) antigen testing is the most widely used SARS-CoV-2 testing because of its low cost, ease of use, speed, and over-the-counter availability for use at home.5 LFAs have poorer clinical accuracy compared to nucleic acid-based tests and have poorer limits of detection, so there are inherent risks that early infection may not be detected when low viral loads are typically present.6–10
Commercial point-of-care (POC) RT-PCR based diagnostics (Abbott ID Now, Cepheid GeneXpert Xpress, Roche cobas Liat, etc.) have been developed and received US FDA emergency use authorization to increase the accessibility of molecular tests with high clinical diagnostic accuracy. These cartridge-based platforms generally have an assay time greater than 30 minutes (ref. 11 and 12) and use desktop readers that automate fluidic handling, amplification, detection, and assay result interpretation. With the goal of increasing accessibility to NAAT testing, government initiatives to accelerate product development (e.g., NIH RADx program) led to several single-use disposable and lower cost NAATs that were specifically targeted for community-based use (physician's office) or home/self-testing (e.g., Lucira CheckIt, Cue COVID-19, Aptitude Metrix, etc.).2,13–15 These commercially available tests use fluorescence or electrochemical detection and require onboard electronics to sense, analyze, and report test results.
Isothermal NAATs with LFA readout can offer the high diagnostic accuracy of RT-PCR and the low-cost and ease of use of antigen-based LFAs. Recombinase polymerase amplification (RPA) is an attractive isothermal amplification method due to its speed (<15 min), accuracy, and low incubation temperature (∼40 °C). Early in the pandemic, multiple SARS-CoV-2 RPA assays were published that targeted either the nucleocapsid (N),16–19 spike (S),18,20 RNA dependent RNA polymerase (RdRp),21 or open reading frame 1 (ORF1ab)20,22 genes with analytical sensitivities as low as 10 RNA copies per reaction while leveraging fluorescent output and/or lateral flow detection. Several RPA assays using LFA detection have incorporated duplexing of multiple SARS targets, RNase P for sampling validation, or simultaneous detection of SARS-CoV-2 and influenza virus targets.23–27 While targeting the human RNase P gene is useful for confirming sampling integrity from a human source, it is not a full process internal control as it is DNA-based and fails to report on potential RNA degradation and reverse transcription.
In this work, we report on the development of a one-pot, duplexed RT-RPA SARS-CoV-2 assay with an MS2 bacteriophage as a full process control. The duplexed assay can detect amplicons with real-time fluorescence or lateral flow readout using commercially available RPA reagent kits that include an exonuclease for molecular probe cleavage (TwistAmp exo). The MS2 bacteriophage process control reports on successful operation of lysis, reverse transcription, and amplification. The assay has a SARS-CoV-2 RNA sensitivity of 25 copies per reaction when using fluorescence readout and 50 copies per reaction with lateral flow detection. We also demonstrate the ability of the current assay to detect the SARS-CoV-2 Delta and Omicron variants. To our knowledge, this is the first report of a duplexed SARS-CoV-2 lateral flow detection assay to incorporate a full process internal control that reports on lysis, reverse transcription, and amplification. This is also the first reported usage of TwistAmp exo RPA kits for both fluorescence and lateral flow readout, opening the potential for multiple assay readout options with a single reagent format given the resources at the point of testing.
Materials and methods
SARS CoV-2 virus RNA and virion stocks
Purified RNA for influenza A (NR-2773), influenza B (NR-10047), HCoV-NL63 (NR-44105), HCoV-OC43 (NR-52727), HCoV-229E (NR-52728), SARS-CoV-2 Delta Lineage B.1.617.2 (NR-56127), and SARS-CoV-2 Omicron Lineage B.1.1.529 (NR-56494) were used for the specificity analysis and variant detection (BEI resources, USA). Intact bacteriophage MS2 was grown and isolated using an established protocol.28 Quantified SARS-CoV-2 virus inactivated via gamma irradiation (NR-52287) was obtained from BEI Resources. All purified RNA and virus stocks were stored at −80 °C until use.SARS-CoV-2 RT-RPA assay development and fluorescence readout
Monoplex SARS-CoV-2 RPA reactions contained 29.5 μL TwistAmp rehydration buffer (TwistDx Limited, UK), 2.6 μL of 10 μM forward primer, 2.6 μL of 10 μM reverse primer (Integrated DNA Technologies, USA), 0.75 μL of 10 μM FAM-labeled probe (Eurogentec, Belgium), 0.5 μL of 50 U μL−1 reverse transcriptase (Agilent AffinityScript, USA), 5 μL of quantified synthetic SARS-CoV-2 RNA (Twist Bioscience, USA), and 6.55 μL of nuclease-free water added to a TwistAmp exo pellet (TwistDx Limited, UK). 2.5 μL of 280 mM of magnesium acetate (TwistDx Limited, UK) was added to the RPA tube cap, bringing the total reaction volume to 50 μL. We agitated the tubes by repeatedly inverting them for 20 seconds to activate the DNA polymerase with the magnesium cofactor before placing them in a benchtop fluorometer designed for use with isothermal nucleic acid amplification assays (Axxin T16, Axxin, AUS). The RT-RPA protocol consists of an initial five-minute incubation step at 39 °C, after which the reaction tubes are removed from the fluorometer, agitated for 30 seconds, placed back into the fluorometer, and incubated for 10 additional minutes. The LED intensity setting of the T16 fluorometer was set to 7% for the FAM channel.The RT-RPA assay was designed to target the nucleocapsid (N) gene based on the SARS-CoV-2 reference genome (GenBank NC_045512). The assay uses an internally quenched fluorophore probe that was designed using computational and manual methods. Candidate probe sequences were generated using RPA assay design software PrimedRPA.29 The final probe sequence for the assay was selected based on the location of available thymine residues for an internally quenched fluorophore, genome conservation through alignment of other available sequences, and low degree of homology through BLAST alignment of other respiratory viruses. Twelve primer pairs were then designed around this probe sequence using PrimedRPA29 and screened using RT-RPA with fluorescence readout. The best primer candidates were chosen based on time to threshold and slope of the exponential amplification curve. The primers were redesigned with single base-pair shifts and length changes for second round screening, following the TwistDx assay design manual. The primers were again screened for optimal time to threshold. In total, 21 different primer pairs were evaluated with the top performing pair being selected for subsequent experiments (Fig. S2†). The resulting primer pair (Table 1) displayed the shortest time to threshold and the steepest exponential amplification slope. MS2 primers and probe are provided in the ESI† (Table S1).30
| Oligo | Sequence |
|---|---|
| SARS-CoV-2 forward | AAGCCTCTTCTCGTTCCTCATCACGTAG |
| SARS-CoV-2 reverse | GTTGGCCTTTACCAGACATTTTGCTCTCA |
| SARS-CoV-2 reverse lateral flow | GTTGGCCTTTACCAGACATTTTGCTCTCA-[biotin] |
| SARS-CoV-2 probe | GGCGGTGATGCTGCTCTTGCTTTGCTGC-[T(FAM)]-G-dSpacer-[T(BHQ1)]-TGACAGATTGAACCAGC-Spacer C3 |
Sequence-specific amplicon detection of SARS-CoV-2 and MS2 via RT-RPA employs enzymatically-cleaved homologous probes. These probes have 48 base pairs, use a FAM or ROX fluorophore, a tetrahydrofuran (THF) residue, an internal quencher (only in fluorescence readout probe design), and a 3′ block to inhibit probe extension. The fluorescence detection mechanism for SARS-CoV-2 uses a thymine modified with a FAM fluorophore that is 5′ of a proximal THF site and a subsequent thymine modified with an internal fluorophore quencher, as detailed elsewhere.31 Fluorescence detection of MS2 follows the same methodology with the difference of a ROX fluorophore in place of the FAM fluorophore. Given sufficient sequence homology, the DNA exonuclease III (Exo) acts on the THF site, freeing the fluorophore and quencher and allowing for fluorescence. While this assay utilizes a sequence-specific probe, perfect homology across the primer and probe targeting region is not required as RPA has been shown to tolerate up to 15 mismatches, allowing for a high degree of cross variant detection.32In silico alignments displayed two total mismatches for the delta variant with a G to T mutation on the probe's first 5′ base, the other mutation occurs outside the primer or probe regions. Alignments also show 3 consecutive mismatches (GGG to AAC) in the omicron variant. These occur outside the primer and probe hybridization regions, and therefore do not impact the assay's performance.
Duplexed RT-RPA assays with fluorescence readout used the same reaction protocol as outlined above, with adjustments to include MS2 bacteriophage RNA, primers, and probe. Duplexed RT-RPA reactions for SARS-CoV-2 and MS2 included 1.08 μL of 10 μM MS2 forward primer, 1.08 μL of 10 μM MS2 reverse primer (Integrated DNA Technologies, USA), 0.19 μL of 10 μM MS2 probe labeled with a ROX fluorophore (LGC Biosearch Technologies, UK), 2 μL of 4.2 × 105 copies per μL (cps/μL) MS2 RNA (Sigma Aldrich, USA), and nuclease-free water as needed to achieve a total reaction volume of 50 μL. As outlined above, tubes were agitated immediately before placing in an Axxin T16 for incubation at 39 °C and fluorescence readout. The LED intensity setting in the FAM channel was set to 7% while the ROX LED intensity was set to 52%.
Lateral flow detection of SARS-CoV-2 RT-RPA assay
For LFA detection assays, both the reverse SARS-CoV-2 primer and MS2 reverse primer had a biotin and digoxigenin moiety added respectively while the MS2 probe utilized a 5′ FAM fluorophore in place of the internal ROX. Reactions were run as described earlier except the final concentration of SARS-CoV-2 forward and reverse primers were 420 nM each with 120 nM of probe and MS2 forward and reverse primers were 210 nM with 60 nM probe. After incubation at 39 °C, we diluted the RPA product 1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 25 in running buffer (HybriDetect 2T, Milenia Biotec, Germany), and pipetted 10 μL of diluted product directly onto a lateral flow test strip (HybriDetect 2T, Milenia Biotec, Germany) before placing the LFA vertically in a tube with 80 μL of running buffer (HybriDetect 2T, Milenia Biotec, Germany). The immediate dilution negated the need to stop the reaction with EDTA.
25 in running buffer (HybriDetect 2T, Milenia Biotec, Germany), and pipetted 10 μL of diluted product directly onto a lateral flow test strip (HybriDetect 2T, Milenia Biotec, Germany) before placing the LFA vertically in a tube with 80 μL of running buffer (HybriDetect 2T, Milenia Biotec, Germany). The immediate dilution negated the need to stop the reaction with EDTA.
After eight minutes of run time, the LFA test strips were removed, pat dried with a Kem wipe (Kimberly-Clark, USA) and scanned with an Epson V370 photo flatbed scanner for use in code-based image analysis. The LFA images were processed by a code (Python 3.10) that calculates the line average intensity of the test line region. A positive result was determined if the line intensity of the test line exceeded the intensity threshold set by the average of all NTCs plus three standard deviations. For qualitative presentation here, the LFA were dried and digitally imaged (EOS Rebel T3, Canon) with a 60 mm macro lens.
Duplexed RT-RPA with lateral flow detection leveraged the chemistry above with the addition of MS2 RNA, primers, and probe. Duplexed reactions included 1.05 μL of 10 μM forward primer, 1.05 μL of 10 μM MS2 reverse primer labeled with digoxigenin, 0.3 μL of 10 μM MS2 probe labeled with a FAM fluorophore (Integrated DNA Technologies), 2 μL of 4.2 × 105 cps/μL MS2 RNA (Sigma Aldrich), and nuclease-free water as needed for a 50 μL total reaction volume. The duplexed assay with LFA readout was conducted in the same manner as outlined for single target detection.
We use two different RPA probe designs in the LFA assay, which are detailed in Table S1.† The first is an internally quenched probe targeting SARS-CoV-2 that is also used for fluorescence readout. The second is a probe with an unquenched 5′ terminal fluorophore label for MS2 amplicon detection as described in the TwistDx design manual and as published in past RPA assays with LFA readout.31,33,34 LFA “sandwich” immunochemistry detection on a nitrocellulose strip uses these RPA probes combined with modified primers: 5′ biotinylated SARS-CoV-2 reverse primer and 5′ digoxigenin-labeled MS2 reverse primer. LFA banding visualization is based on the binding of a dual labeled RPA product being “sandwiched” between the immobilized receptor and an anti-FAM antibody conjugated to a gold nanoparticle. The first binding domain detects the SARS-CoV-2 amplification product which is labeled via the biotinylated reverse primer and cleaved FAM probe, as illustrated in Fig. 1. The second binding domain detects the MS2 amplification product which is similarly labeled with digoxigenin and FAM. The third binding domain serves as the LFA flow control by capturing conjugated gold nanoparticles that did not bind to RPA products.
Viral lysis and MS2 bacteriophage internal control
For viral lysis studies, inactivated SARS-CoV-2 and MS2 viral stocks were diluted in PBS pH 8.0 buffer to 600 genome copies per μL. To ensure no extracellular RNA was present in the samples prior to lysis, 5 μL of the viral samples were treated with 5 μL of 10 mg mL−1 RNase A (ThermoFisher) followed by an inactivation step of 5 μL of RNasin™ Plus Ribonuclease Inhibitor (Promega, USA). These steps were carried out sequentially at 37 °C for 5 minutes each to degrade free RNA prior to lysis. Heat lysis was completed with incubation at 95 °C for 5 minutes. 5 μL of the lysate was then spiked directly into RT-RPA reactions. Non-lysed controls without the 95 °C heat lysis step were run to verify the absence of free RNA in viral stocks.Results and discussion
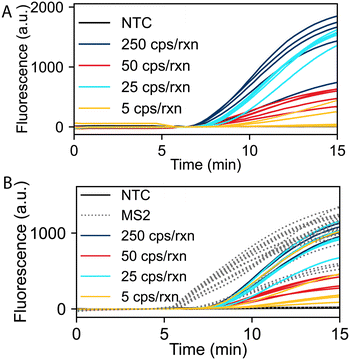
We evaluated the limit of detection (LOD) of the RT-RPA assay for SARS-CoV-2 and duplexed detection of SARS-CoV-2 and the MS2 internal control using fluorescence readout. Fig. 2A shows real-time RT-RPA fluorescence intensities over a 15-minute incubation for reactions ranging from 5 to 250 SARS-CoV-2 RNA copies per reaction (cps/rxn) with five replicates at each concentration. Fig. 2B shows SARS-CoV-2 and MS2 duplexed RT-RPA reactions with varying SARS-CoV-2 RNA concentrations and 8 × 104 cps/rxn of MS2 RNA for all experiments. For all duplexed reactions at varying SARS-CoV-2 input copies, the corresponding MS2 reactions successfully amplified, except the no template controls (NTCs) which contained no SARS-CoV-2 or MS2 RNA. For both the mono and duplex RT-RPA reactions, we observe strong amplification in less than 10 minutes down to 25 cps/rxn of each replicate. We also evaluated the RT-RPA assay at copy loads greater than 250 cps/rxn, up to 5 × 105 cps/rxn, which all amplified consistently (Fig. S1 in the ESI†). We observed more variability in exponential amplification slope and time to threshold in the low copy replicates. A similar effect has been observed in other RPA studies.23,35Fig. 2B also demonstrates the effect of multiplexing on the reaction as the time to threshold and total overall fluorescence output of the SARS-CoV-2 assay is inhibited when compared to the monoplexed reaction. This inhibition is due to the competition of the SARS-CoV-2 and MS2 primers and probes for the limited RPA reagents needed for amplification.Within the duplex RT-RPA assay, the MS2 input copies and concentrations of primers and probe were optimized to retain a low LOD for SARS-CoV-2 detection, while also consistently amplifying the MS2 internal control. The SARS-CoV-2 primer and probe concentrations were conserved throughout both the mono and duplexed assays. MS2 primer and probe concentrations were determined through a systematic screening of lower MS2 oligo concentrations while maintaining SARS-CoV-2 assay performance. Concurrent with the oligo concentration, we evaluated MS2 input copies on the amplification behavior. The final concentration of MS2 RNA in the assay was determined through optimization such that the time to threshold of the MS2 amplification was less than 7 minutes and all MS2 reactions consistently amplified regardless of input copy load of SARS-CoV-2 RNA. We observed that the optimal total oligonucleotide concentration in the reaction was 1540 μM. Further increases in the oligo concentration demonstrated increased time to threshold and reduced exponential amplification slope, resulting in worse duplexed assay performance. We hypothesize that the decrease in performance is due to oligo saturation of the single stranded binding proteins and/or recombinase proteins, which inhibits the strand invasion and primer hybridization steps of RPA.
In order to implement LFA detection of RT-RPA products, we explored different probe designs and TwistAmp RPA enzyme kits for LFA detection. Many previously published RPA assays with LFA readout use the TwistAmp nfo kit and a 5′ unquenched fluorophore probe as in the TwistAmp RPA design manual.31,33,34 At the time of experimentation, the TwistAmp nfo kit was discontinued and no longer available for purchase from TwistDx Limited (now a subsidiary of Abbott Laboratories). Attempts were made to spike purchased endonuclease IV (nfo) enzyme to a TwistAmp basic kit with mild success (data not shown); however, through experimentation we found that sensitivity of the LFA assay was improved using the TwistAmp exo kits, which are typically reserved for real-time fluorescence readout. We evaluated the performance of the internally quenched fluorescence detection probe design compared to a 5′ unquenched fluorophore LFA probe design and found the detection limit to be equivalent. To demonstrate the utility of these probe designs, we carried out all LFA readout experiments using the 5′ unquenched FAM fluorophore probe design for MS2 detection and the internally quenched FAM fluorophore probe design for SARS-CoV-2 detection. We observed that employing TwistAmp exo kits and internally quenched probes enables dual readout, either through real-time fluorescence or endpoint LFA analysis. This is notable because RPA primer and probe screening is most efficiently performed using real-time fluorescence data to indicate optimal primer and probe combinations, which can then be leveraged directly for LFA readout if desired.
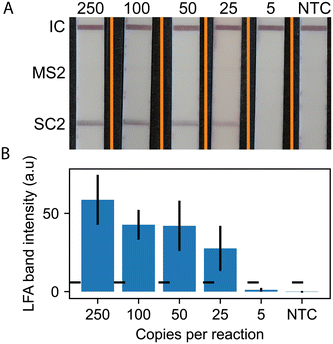
Using the RPA probe design, we conducted experiments to detect SARS-CoV-2 using RT-RPA paired with lateral flow readout. Fig. 3 shows representative LFA strip images and quantitative measurement of LFA line intensities (5 strip average) for SARS-CoV-2. The images show positive lines for the flow controls and SARS-CoV-2 for all concentrations down to 25 copies per reaction. The MS2 LFA line is not present, as expected, because the MS2 RNA, primers, and probe are not included in the monoplex version of the assay. The quantitative results in Fig. 3B show that the line intensity increases with increasing input copy numbers.
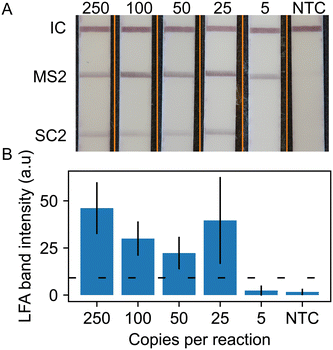
We repeated these lateral flow readout experiments with the duplex RT-RPA assay for SARS-CoV-2 with MS2 internal control. Fig. 4 shows representative strip images of the duplexed LFA and quantitative measurement of LFA line intensities averaged over 5 replicates. The images show a positive line for the flow control, MS2 internal control, and the SARS-CoV-2 strip regions for all concentrations down to 25 copies per reaction. We observe that the MS2 line intensity levels are greater than the SARS-CoV-2 test lines. This is likely due to the excess MS2 RNA in the reaction. The quantitative analysis shows that the SARS-CoV-2 line intensity decreases with copy number, except at 25 cps/rxn where the average line intensity and standard deviation is much greater than expected. This is due to two replicates, one where the SARS-CoV-2 test stripe intensity was much greater than three replicates and one replicate that failed to develop a line at an intensity reaching the determination cut off both visually and by the objective code method. This low line intensity replicate was retested with the same result. As observed in the fluorescence readout data in Fig. 2, high variation in RT-RPA amplification is common near the limit of detection of the assay.
Table 2 summarizes the observed LODs of the RT-RPA assay with fluorescence or LFA readout for both monoplex detection of SARS-CoV-2 and duplexed detection of SARS-CoV-2 and the MS2 internal control. We list the detection fraction of SARS-CoV-2 RNA from 250 cps/rxn to 5 cps/rxn stratified by readout method and monoplexed versus duplexed target detection. RT-RPA with fluorescence readout demonstrated lower LOD with mixed positive results at 5 cps/rxn and a limit of detection of 25 cps/rxn in both single target and duplexed assays. Monoplexed RT-RPA with LFA readout exhibited a 100% SARS-CoV-2 detection rate down to 25 cps/rxn. The duplexed LFA of variable SARS-CoV-2 target and fixed MS2 target demonstrated 100% detection rate down to 50 cps/rxn and 80% detection rate at 25 cps/rxn. Neither monoplexed nor duplexed LFA could detect any LFA banding through either visual or analytical detection at 5 cps/rxn.
| Detection method | 250 cps/rxn | 100 cps/rxn | 50 cps/rxn | 25 cps/rxn | 5 cps/rxn | NTC |
|---|---|---|---|---|---|---|
| Singleplex RT-RPA with fluorescence | 5/5 | 5/5 | 5/5 | 5/5 | 3/5 | 0/5 |
| Duplex RT-RPA with fluorescence | 5/5 | 5/5 | 5/5 | 5/5 | 2/5 | 0/5 |
| Singleplex RT-RPA with LFA | 5/5 | 5/5 | 5/5 | 5/5 | 0/5 | 0/5 |
| Duplex RT-RPA with LFA | 5/5 | 5/5 | 5/5 | 4/5 | 0/5 | 0/5 |
The duplex RT-RPA assay with LFA readout was tested with other respiratory viruses in order to assess its analytical specificity. Table 3 shows detection of SARS-CoV-2 variants Delta B.1.617.2 and Omicron B.1.1.529 and cross-reactivity screening against genomic RNA from other common respiratory viruses. The data shows that the assay has no cross-reactivity with influenza A, influenza B, human coronavirus 229E (HCoV-229E), HCoV-NL63, or HCoV-OC43. The SARS-CoV-2 Delta and Omicron variants RNA were detected in all three replicates at input copy loads of 103 cps/rxn (Fig. S6 and S7†). We did not thoroughly evaluate the LOD for these variants of concern, but we expect similar results to Table 2 which used RNA from the ancestral SARS-CoV-2 strain. In alignments of these SARS-CoV-2 variant sequences with our RPA primers and probe, we discovered no mismatches in the hybridization regions; therefore, we anticipate no significant effect on limit of detection. There are some sublineages that introduce minimal mismatches with our RPA sequences, but we do not expect significant impacts to our assay's performance due to the mismatch tolerance of RPA that has been reported previously by our research group and others.32,36
| Virus | Concentration (cps/rxn) | Positive replicates |
|---|---|---|
| Flu A | 103 | 0/3 |
| Flu B | 103 | 0/3 |
| HCoV-229E | 103 | 0/3 |
| HCoV-NL63 | 103 | 0/3 |
| HCoV-OC43 | 103 | 0/3 |
| Delta | 103 | 3/3 |
| Omicron | 103 | 3/3 |
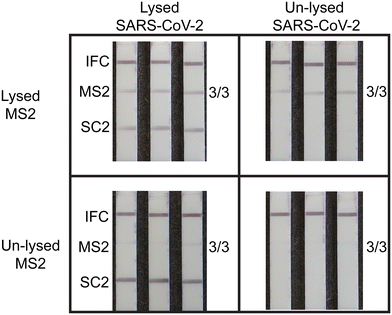
We validated the internal MS2 viral control using heat for viral lysis and sample preparation. Heat lysis has been demonstrated as an effective viral lysis technique compatible with nucleic acid amplification assays.18,37,38 We conducted a series of duplexed RT-RPA experiments with LFA readout at 103 copies per reaction (N = 3) to demonstrate efficacy of heat-based lysis and the MS2 full process internal control. Prior to RT-RPA analysis, we pre-treated intact SARS-CoV-2 virus and MS2 bacteriophage to generate four different samples: MS2 and SARS-CoV-2 both heat-lysed, only SARS-CoV-2 lysed, only MS2 lysed, and no lysing. Fig. 5 shows representative images of LFA strips for RT-RPA of pretreated samples directly added to reactions. All four conditions yielded the expected results as using lysed materials permitted successful RT-RPA and LFA detection, while unlysed materials did not amplify and therefore gave negative test results. In instances where MS2 detection fails, this indicates an invalid test result due to a failure of one or more integral steps, and retesting is required. This result could be due to RNA degradation or unsuccessful viral lysis, reverse transcription, or amplification. Failure to use a full process internal control may increase risk of false negatives and misdiagnosis, which prevents immediate clinical interventions that mitigate community spread.
Summary
We developed a rapid, duplexed, single-pot RT-RPA nucleic acid amplification assay with lateral flow readout for SARS-CoV-2 with a MS2 bacteriophage as a full process internal control. We demonstrated the duplexed amplification and detection of SARS-CoV-2 and MS2 in real time using both fluorescence readout with an LOD of 25 copies per reaction sensitivity with a reaction time of 15 minutes and end point visual readout using lateral flow strips with 50 copies per reaction sensitivity with test results in under 25 minutes. The assay detects both the delta and omicron SARS-CoV-2 variants and does not exhibit any cross-reactivity with influenza A, influenza B, or other common human coronaviruses (HCoV-NL63, HCoV-OC43 and HCoV-229E). We demonstrated that MS2 serves as a process control for lysis, reverse transcription, amplification, and readout. While we used an Axxin T16 to incubate the RPA reactions, other low-cost heat blocks, ambient temperature, self-regulating chemical reactions, and body heat39–41 have been effective and demonstrate that the heating energy required for the RPA reaction does not require mains electricity. Additional work on lysis, fluid handling, reagent storage, heating, and amplicon containment is required to realize an integrated POC device suitable for commercialization.This SARS-CoV-2 assay meets multiple characteristics for assay performance as set forth by the World Health Organization's COVID-19 Target Product Profile (TPP) for priority diagnostics to support response to the COVID-19 pandemic.42 The TPP lists the desirable analytical test sensitivity to be equivalent to 104 genome copies per mL. We have demonstrated a duplexed assay with an analytical sensitivity of 50 copies per reaction. The TPP further lists desirable analytical specificity as the ability to detect all SARS-CoV-2 viral strains, not reacting with interferants, and not cross reacting with other common viral diseases that present with common signs and symptoms of COVID-19 like influenza A/B. Here, we have demonstrated that the assay detects both the delta and omicron variants while not cross reacting with other human coronaviruses nor influenza A or B. For interpretation of test results, the TPP lists visual manual readout in both the acceptable and desired categories which is achieved with our lateral flow readout mechanism. We have demonstrated this assay to be rapid with lateral flow results in 28 minutes from raw sample lysis to answer. The use of RPA also has inherent benefits as the lyophilized reaction pellets have been proven to be stable outside of cold chain storage for up to 12 weeks.43
To our knowledge, this assay is the first reported duplexed SARS-CoV-2 RPA assay for lateral flow strip detection format that incorporates an internal full process internal control that reports on the successful lysis, reverse transcription, and amplification of each RNA test reaction. It is also the first reported use of the RPA TwistAmp exo kits for both fluorescence and lateral flow detection formats opening the possibility for multiple assay readout options with a single kit, albeit with slight differences in assay performance. This utility of exo-based kits for LFA also enables convenient RPA assay design and optimization as primer and probe combinations can be tested with real-time readout instead of endpoint analysis.
Conflicts of interest
There are no conflicts to declare.Acknowledgements
The work reported in this publication was supported by the National Institute of Biomedical Imaging and Bioengineering of the National Institutes of Health under Award Number R01EB022630 and by the National Center for Advancing Translational Sciences of the National Institutes of Health under Award Number TL1TR002318. Part of this work was conducting using equipment in the Biochemical Diagnostics Foundry for Translational Research supported by the M. J. Murdock Charitable Trust. The content is solely the responsibility of the authors and does not necessarily represent the official views of the National Institutes of Health.References
- N. Phillips, The Coronavirus Is Here to Stay — Here's What That Means, Nature, 2021, 590(7846), 382–384, DOI:10.1038/d41586-021-00396-2.
- R. W. Peeling, D. L. Heymann, Y.-Y. Teo and P. J. Garcia, Diagnostics for COVID-19: Moving from Pandemic Response to Control, Lancet, 2022, 399(10326), 757–768, DOI:10.1016/S0140-6736(21)02346-1.
- O. Puhach, B. Meyer and I. Eckerle, SARS-CoV-2 Viral Load and Shedding Kinetics, Nat. Rev. Microbiol., 2022, 21, 147–161, DOI:10.1038/s41579-022-00822-w.
- A. Brihn, J. Chang, K. OYong, S. Balter, D. Terashita, Z. Rubin and N. Yeganeh, Diagnostic Performance of an Antigen Test with RT-PCR for the Detection of SARS-CoV-2 in a Hospital Setting — Los Angeles County, California, June–August 2020, MMWR Morb. Mortal. Wkly. Rep., 2021, 70(19), 702–706, DOI:10.15585/mmwr.mm7019a3.
- P. K. Drain, Rapid Diagnostic Testing for SARS-CoV-2, N. Engl. J. Med., 2022, 386(3), 264–272, DOI:10.1056/NEJMcp2117115.
- G. A. Perchetti, M.-L. Huang, M. G. Mills, K. R. Jerome and A. L. Greninger, Analytical Sensitivity of the Abbott BinaxNOW COVID-19 Ag Card, J. Clin. Microbiol., 2021, 59(3), e02880-20, DOI:10.1128/JCM.02880-20.
- V. T. Chu, N. G. Schwartz, M. A. P. Donnelly, M. R. Chuey, R. Soto, A. R. Yousaf, E. N. Schmitt-Matzen, S. Sleweon, J. Ruffin, N. Thornburg, J. L. Harcourt, A. Tamin, G. Kim, J. M. Folster, L. J. Hughes, S. Tong, G. Stringer, B. A. Albanese, S. E. Totten, M. M. Hudziec, S. R. Matzinger, E. A. Dietrich, S. W. Sheldon, S. Stous, E. C. McDonald, B. Austin, M. E. Beatty, J. E. Staples, M. E. Killerby, C. H. Hsu, J. E. Tate, H. L. Kirking and A. Matanock, COVID-19 Household Transmission Team. Comparison of Home Antigen Testing With RT-PCR and Viral Culture During the Course of SARS-CoV-2 Infection, JAMA Intern. Med., 2022, 182(7), 701–709, DOI:10.1001/jamainternmed.2022.1827.
- M. Kahn, L. Schuierer, C. Bartenschlager, S. Zellmer, R. Frey, M. Freitag, C. Dhillon, M. Heier, A. Ebigbo, C. Denzel, S. Temizel, H. Messmann, M. Wehler, R. Hoffmann, E. Kling and C. Römmele, Performance of Antigen Testing for Diagnosis of COVID-19: A Direct Comparison of a Lateral Flow Device to Nucleic Acid Amplification Based Tests, BMC Infect. Dis., 2021, 21(1), 798, DOI:10.1186/s12879-021-06524-7.
- L. E. Brümmer, S. Katzenschlager, S. McGrath, S. Schmitz, M. Gaeddert, C. Erdmann, M. Bota, M. Grilli, J. Larmann, M. A. Weigand, N. R. Pollock, A. Macé, B. Erkosar, S. Carmona, J. A. Sacks, S. Ongarello and C. M. Denkinger, Accuracy of Rapid Point-of-Care Antigen-Based Diagnostics for SARS-CoV-2: An Updated Systematic Review and Meta-Analysis with Meta-Regression Analyzing Influencing Factors, PLoS Med., 2022, 19(5), e1004011, DOI:10.1371/journal.pmed.1004011.
- W. Ashagre, A. Atnafu, L. Wassie, R. Tschopp, D. Fentahun, G. Assefa, T. Wegayehu, B. Wondale, A. Mulu, A. Miheret and K. Bobosha, Evaluation of the Diagnostic Performance of PanbioTM Abbott SARS-CoV-2 Rapid Antigen Test for the Detection of COVID-19 from Suspects Attending ALERT Center, PLoS One, 2022, 17(11), e0277779, DOI:10.1371/journal.pone.0277779.
- J. Y. Zhang, A. T. Bender, D. S. Boyle, P. K. Drain and J. D. Posner, Current State of Commercial Point-of-Care Nucleic Acid Tests for Infectious Diseases, Analyst, 2021, 146(8), 2449–2462, 10.1039/D0AN01988G.
- O. I. Wilner, D. Yesodi and Y. Weizmann, Point-of-Care Nucleic Acid Tests: Assays and Devices, Nanoscale, 2023, 15(3), 942–952, 10.1039/D2NR05385C.
- M. M. Islam and D. Koirala, Toward a Next-Generation Diagnostic Tool: A Review on Emerging Isothermal Nucleic Acid Amplification Techniques for the Detection of SARS-CoV-2 and Other Infectious Viruses, Anal. Chim. Acta, 2022, 1209, 339338, DOI:10.1016/j.aca.2021.339338.
- T. Kang, J. Lu, T. Yu, Y. Long and G. Liu, Advances in Nucleic Acid Amplification Techniques (NAATs): COVID-19 Point-of-Care Diagnostics as an Example, Biosens. Bioelectron., 2022, 206, 114109, DOI:10.1016/j.bios.2022.114109.
- Y. Mardian, H. Kosasih, M. Karyana, A. Neal and C.-Y. Lau, Review of Current COVID-19 Diagnostics and Opportunities for Further Development, Front. Med., 2021, 8, 615099, DOI:10.3389/fmed.2021.615099.
- O. Behrmann, I. Bachmann, M. Spiegel, M. Schramm, A. Abd El Wahed, G. Dobler, G. Dame and F. T. Hufert, Rapid Detection of SARS-CoV-2 by Low Volume Real-Time Single Tube Reverse Transcription Recombinase Polymerase Amplification Using an Exo Probe with an Internally Linked Quencher (Exo-IQ), Clin. Chem., 2020, 66(8), 1047–1054, DOI:10.1093/clinchem/hvaa116.
- Y. L. Lau, I. binti Ismail, N. I. binti Mustapa, M. Y. Lai, T. S. T. Soh, A. H. Hassan, K. M. Peariasamy, Y. L. Lee, M. K. B. A. Kahar, J. Chong and P. P. Goh, Development of a Reverse Transcription Recombinase Polymerase Amplification Assay for Rapid and Direct Visual Detection of Severe Acute Respiratory Syndrome Coronavirus 2 (SARS-CoV-2), PLoS One, 2021, 16(1), e0245164, DOI:10.1371/journal.pone.0245164.
- J. Qian, S. A. Boswell, C. Chidley, Z. Lu, M. E. Pettit, B. L. Gaudio, J. M. Fajnzylber, R. T. Ingram, R. H. Ward, J. Z. Li and M. Springer, An Enhanced Isothermal Amplification Assay for Viral Detection, Nat. Commun., 2020, 11, 5920, DOI:10.1038/s41467-020-19258-y.
- D. Liu, H. Shen, Y. Zhang, D. Shen, M. Zhu, Y. Song, Z. Zhu and C. Yang, A Microfluidic-Integrated Lateral Flow Recombinase Polymerase Amplification (MI-IF-RPA) Assay for Rapid COVID-19 Detection, Lab Chip, 2021, 21(10), 2019–2026, 10.1039/D0LC01222J.
- G. Xue, S. Li, W. Zhang, B. Du, J. Cui, C. Yan, L. Huang, L. Chen, L. Zhao, Y. Sun, N. Li, H. Zhao, Y. Feng, Z. Wang, S. Liu, Q. Zhang, X. Xie, D. Liu, H. Yao and J. Yuan, Reverse-Transcription Recombinase-Aided Amplification Assay for Rapid Detection of the 2019 Novel Coronavirus (SARS-CoV-2), Anal. Chem., 2020, 92(14), 9699–9705, DOI:10.1021/acs.analchem.0c01032.
- L. Farrera-Soler, A. Gonse, K. T. Kim, S. Barluenga and N. Winssinger, Combining Recombinase Polymerase Amplification and DNA-Templated Reaction for SARS-CoV-2 Sensing with Dual Fluorescence and Lateral Flow Assay Output, Biopolymers, 2022, 113(4), e23485, DOI:10.1002/bip.23485.
- X. Meng, S. Zou, D. Li, J. He, L. Fang, H. Wang, X. Yan, D. Duan and L. Gao, Nanozyme-Strip for Rapid and Ultrasensitive Nucleic Acid Detection of SARS-CoV-2, Biosens. Bioelectron., 2022, 217, 114739, DOI:10.1016/j.bios.2022.114739.
- D. Cherkaoui, D. Huang, B. S. Miller, V. Turbé and R. A. McKendry, Harnessing Recombinase Polymerase Amplification for Rapid Multi-Gene Detection of SARS-CoV-2 in Resource-Limited Settings, Biosens. Bioelectron., 2021, 189, 113328, DOI:10.1016/j.bios.2021.113328.
- T. R. Shelite, A. C. Uscanga-Palomeque, A. Castellanos-Gonzalez, P. C. Melby and B. L. Travi, Isothermal Recombinase Polymerase Amplification-Lateral Flow Detection of SARS-CoV-2, the Etiological Agent of COVID-19, J. Virol. Methods, 2021, 296, 114227, DOI:10.1016/j.jviromet.2021.114227.
- E. Xiong, L. Jiang, T. Tian, M. Hu, H. Yue, M. Huang, W. Lin, Y. Jiang, D. Zhu and X. Zhou, Simultaneous Dual-Gene Diagnosis of SARS-CoV-2 Based on CRISPR/Cas9-Mediated Lateral Flow Assay, Angew. Chem., Int. Ed., 2021, 60(10), 5307–5315, DOI:10.1002/anie.202014506.
- G. Su, M. Zhu, D. Li, M. Xu, Y. Zhu, Y. Zhang, H. Zhu, F. Li and Y. Yu, Multiplexed Lateral Flow Assay Integrated with Orthogonal CRISPR-Cas System for SARS-CoV-2 Detection, Sens. Actuators, B, 2022, 371, 132537, DOI:10.1016/j.snb.2022.132537.
- Y. Sun, P. Qin, J. He, W. Li, Y. Shi, J. Xu, Q. Wu, Q. Chen, W. Li, X. Wang, G. Liu and W. Chen, Rapid and Simultaneous Visual Screening of SARS-CoV-2 and Influenza Virufses with Customized Isothermal Amplification Integrated Lateral Flow Strip, Biosens. Bioelectron., 2022, 197, 113771, DOI:10.1016/j.bios.2021.113771.
- J. Dreier, M. Störmer and K. Kleesiek, Use of Bacteriophage MS2 as an Internal Control in Viral Reverse Transcription-PCR Assays, J. Clin. Microbiol., 2005, 43(9), 4551–4557, DOI:10.1128/JCM.43.9.4551-4557.2005.
- M. Higgins, M. Ravenhall, D. Ward, J. Phelan, A. Ibrahim, M. S. Forrest, T. G. Clark and S. Campino, PrimedRPA: Primer Design for Recombinase Polymerase Amplification Assays, Bioinformatics, 2019, 35(4), 682–684, DOI:10.1093/bioinformatics/bty701.
- A. T. Bender, B. P. Sullivan, J. Y. Zhang, D. C. Juergens, L. Lillis, D. S. Boyle and J. D. Posner, HIV Detection from Human Serum with Paper-Based Isotachophoretic RNA Extraction and Reverse Transcription Recombinase Polymerase Amplification, Analyst, 2021, 146(9), 2851–2861, 10.1039/D0AN02483J.
- O. Piepenburg, C. H. Williams, D. L. Stemple and N. A. Armes, DNA Detection Using Recombination Proteins, PLoS Biol., 2006, 4(7), e204, DOI:10.1371/journal.pbio.0040204.
- L. Lillis, D. A. Lehman, J. B. Siverson, J. Weis, J. Cantera, M. Parker, O. Piepenburg, J. Overbaugh and D. S. Boyle, Cross-Subtype Detection of HIV-1 Using Reverse Transcription and Recombinase Polymerase Amplification, J. Virol. Methods, 2016, 230, 28–35, DOI:10.1016/j.jviromet.2016.01.010.
- B. A. Rohrman and R. R. Richards-Kortum, A Paper and Plastic Device for Performing Recombinase Polymerase Amplification of HIV DNA, Lab Chip, 2012, 12(17), 3082, 10.1039/c2lc40423k.
- M. S. Cordray and R. R. Richards-Kortum, A Paper and Plastic Device for the Combined Isothermal Amplification and Lateral Flow Detection of Plasmodium DNA, Malar. J., 2015, 14(1), 472, DOI:10.1186/s12936-015-0995-6.
- Z. A. Crannell, B. Rohrman and R. Richards-Kortum, Development of a Quantitative Recombinase Polymerase Amplification Assay with an Internal Positive Control, J. Visualized Exp., 2015,(97), e52620, DOI:10.3791/52620.
- M. Euler, Y. Wang, O. Nentwich, O. Piepenburg, F. T. Hufert and M. Weidmann, Recombinase Polymerase Amplification Assay for Rapid Detection of Rift Valley Fever Virus, J. Clin. Virol., 2012, 54(4), 308–312, DOI:10.1016/j.jcv.2012.05.006.
- C. Myhrvold, C. A. Freije, J. S. Gootenberg, O. O. Abudayyeh, H. C. Metsky, A. F. Durbin, M. J. Kellner, A. L. Tan, L. M. Paul, L. A. Parham, K. F. Garcia, K. G. Barnes, B. Chak, A. Mondini, M. L. Nogueira, S. Isern, S. F. Michael, I. Lorenzana, N. L. Yozwiak, B. L. MacInnis, I. Bosch, L. Gehrke, F. Zhang and P. C. Sabeti, Field-Deployable Viral Diagnostics Using CRISPR-Cas13, Science, 2018, 360(6387), 444–448, DOI:10.1126/science.aas8836.
- G. Ruano, E. M. Pagliaro, T. R. Schwartz, K. Lamy, D. Messina, R. E. Gaensslen and H. C. Lee, Heat-Soaked PCR: An Efficient Method for DNA Amplification with Applications to Forensic Analysis, BioTechniques, 1992, 13(2), 266–274 CAS.
- R. Wang, F. Zhang, L. Wang, W. Qian, C. Qian, J. Wu and Y. Ying, Instant, Visual, and Instrument-Free Method for On-Site Screening of GTS 40-3-2 Soybean Based on Body-Heat Triggered Recombinase Polymerase Amplification, Anal. Chem., 2017, 89(8), 4413–4418, DOI:10.1021/acs.analchem.7b00964.
- Z. A. Crannell, B. Rohrman and R. Richards-Kortum, Equipment-Free Incubation of Recombinase Polymerase Amplification Reactions Using Body Heat, PLoS One, 2014, 9(11), e112146, DOI:10.1371/journal.pone.0112146.
- L. Lillis, D. Lehman, M. C. Singhal, J. Cantera, J. Singleton, P. Labarre, A. Toyama, O. Piepenburg, M. Parker, R. Wood, J. Overbaugh and D. S. Boyle, Non-Instrumented Incubation of a Recombinase Polymerase Amplification Assay for the Rapid and Sensitive Detection of Proviral HIV-1 DNA, PLoS One, 2014, 9(9), e108189, DOI:10.1371/journal.pone.0108189.
- COVID-19 Target product profiles for priority diagnostics to support response to the COVID-19 pandemic v.1.0, https://www.who.int/publications/m/item/covid-19-target-product-profiles-for-priority-diagnostics-to-support-response-to-the-covid-19-pandemic-v.0.1, (accessed 2023-02-28).
- L. Lillis, J. Siverson, A. Lee, J. Cantera, M. Parker, O. Piepenburg, D. A. Lehman and D. S. Boyle, Factors Influencing Recombinase Polymerase Amplification (RPA) Assay Outcomes at Point of Care, Mol. Cell. Probes, 2016, 30(2), 74–78, DOI:10.1016/j.mcp.2016.01.009.
Footnote |
| † Electronic supplementary information (ESI) available. See DOI: https://doi.org/10.1039/d3sd00246b |
| This journal is © The Royal Society of Chemistry 2024 |