Open Access Article
Open Access ArticleDiscovery of penicillic acid as a chemical probe against tau aggregation in Alzheimer's disease†
Jennifer
Shyong‡
 a,
Jinliang
Wang‡
a,
Jinliang
Wang‡
 a,
Quoc-Dung Tran
Huynh
a,
Quoc-Dung Tran
Huynh
 d,
Marina
Fayzullina
d,
Marina
Fayzullina
 a,
Bo
Yuan
a,
Bo
Yuan
 a,
Ching-Kuo
Lee
a,
Ching-Kuo
Lee
 cd,
Thomas
Minehan
cd,
Thomas
Minehan
 e,
Paul M.
Seidler
e,
Paul M.
Seidler
 *a and
Clay C. C.
Wang
*a and
Clay C. C.
Wang
 *ab
*ab
aDepartment of Pharmacology and Pharmaceutical Sciences, Alfred E. Mann School of Pharmacy and Pharmaceutical Sciences, University of Southern California, Los Angeles, California 90089, USA. E-mail: clayw@usc.edu; pseidler@usc.edu
bDepartment of Chemistry, University of Southern California, Dornsife College of Letters, Arts, and Sciences, Los Angeles, California 90089, USA
cSchool of Pharmacy, College of Pharmacy, Taipei Medical University, Taipei 11031, Taiwan
dPhD Program in Clinical Drug Development of Herbal Medicine, College of Pharmacy, Taipei Medical University, Taipei 11031, Taiwan
eDepartment of Chemistry and Biochemistry, California State University, Northridge, Northridge, California 91330, USA
First published on 12th November 2024
Abstract
Alzheimer's Disease (AD) is a neurodegenerative disorder proven to be caused by the aggregation of protein tau into fibrils, resulting in neuronal death. The irreparable neuronal damage leads to irreversible symptoms with no cure; therefore, disaggregation of these tau fibrils could be targeted as a therapeutic approach to AD. Here we have developed a fungal natural product library to screen for secondary metabolites that have bioactive potential towards AD tau. Our initial screenings indicate that penicillic acid demonstrates anti-aggregation activity towards tau, while further in vitro experiments reveal that penicillic acid directly inhibits tau by disaggregating fibrils. Although penicillic acid possesses blood–brain barrier penetrability properties that are computationally predicted to be favorable, it is presumed to contain some mutagenic effects as well. To address this, we used the backbone of penicillic acid as a chemical probe to discover similar compounds that can inhibit AD tau aggregation with limited mutagenicity. This work suggests the potential of discovering chemical probes through natural product screening for small-molecule drug discovery of tauopathies.
Introduction
Alzheimer's Disease (AD) is a progressive, neurodegenerative disease that affects millions of people worldwide.1 It is the most common type of dementia, causing accumulation of beta-amyloid proteins outside neurons and internal neuronal damage by the twisted strands of tau proteins.2 This leads to irreversible symptoms of Alzheimer's, beginning with memory loss, developing into changes of personality and behavior, and eventually progressing to the loss of basic human functions.3 Although there are therapies and medications available to slow development and temporarily improve cognitive symptoms, these do not alter the course of the disease.4 An Alzheimer's diagnosis is fatal, so it is crucial to further investigate new potential therapies.Several natural products, otherwise known as secondary metabolites (SMs), have previously been shown to be a source of medically valuable compounds, including for neuroprotective properties.5,6 One major source of SMs is filamentous fungi, in which the exhibition of these bioactivities originates from its ability to naturally form biofilms.7,8 The development of these biofilms allows for penetration into a host for nutrient acquisition and defense against biological components, such as protein aggregates, that may impact the survival of fungi. This key feature makes the use of filamentous fungi an attractive option for AD drug discovery since biofilms can potentially target tau proteins, a neuropathological characteristic responsible for Alzheimer's progression. For instance, a class of fungal natural products, azaphilones, has previously been demonstrated to be capable of inhibiting tau aggregation.9,10 Azaphilones were also shown to possess disaggregation properties of tau aggregates, suggesting other natural products may have similar characteristics as well.10 Despite the vast amount of fungal secondary metabolites that have been discovered, many have yet to be tested for various bioactivities. In addition, sequence analysis of the fungal genome suggests that the biosynthetic gene clusters (BGCs) that encode proteins that activate the production of SMs are mostly silent.11,12 Studies have demonstrated that the number of predicted BGCs from fungal genomes far outnumber the SMs produced, suggesting that most BGCs are inactive under normal laboratory growth conditions and numerous unknown compounds have yet to be discovered.13–15 Due to the vast potential of using natural products for the treatment of AD, we need new methods to express these cryptic BGCs.
Previous studies have demonstrated that one way to activate cryptic BGCs is through genetic manipulation of the global regulators of fungal SM production.16–18 In particular, knocking out mcrA, a negative global regulator responsible for the inhibition of multiple BGC expression, has allowed for the upregulation of some compounds and the emergence of new products.19 The compounds produced by genetically manipulating our strain of focus, Aspergillus melleus IMV01140, through deletion of mcrA in combination with a large number of predicted BGCs, allowed for the creation of a diverse natural product library. The generated known and novel compounds were tested for bioactivity on tau aggregation through a seeding assay using biosensor cells. Using this method, we discovered penicillic acid as a potential bioactive hit capable of inhibiting tau aggregation. We further analyzed the effectiveness of our target compound using in vitro studies that tested for inhibition of tau monomer aggregation with various assays and quantified the effectiveness of tau fibril disaggregation when treated with penicillic acid using quantitative electron microscopy.
Using penicillic acid as a chemical probe, we generated analogs to further understand the chemical structures that are essential for its bioactive properties. Furthermore, we searched for analogs with the assistance of the ADMET predictor to generate compounds with similar blood–brain barrier permeability and decreased mutagenic effects. Our study demonstrates a promising and expandable method to enable the discovery of new bioactive fungal SMs that are chemical probes for CNS drug discovery.
Results
Targeting the mcrA negative global regulator gene for library generation
Using anti-SMASH, a bioinformatics tool that conducts genome-wide annotation, identification, and analysis of BGCs in fungal genomes,20,21 we determined that IMV01140, a strain from the IMV (Institute of Microbiology and Virology, Kyiv, Ukraine) collection has 83 predicted BGCs (Fig. 1A). Because our LC-MS analysis of the wild-type strain of IMV01140 shows fewer peaks than the predicted number of BGCs, this suggests that the strain has multiple BGCs that are currently silenced under normal laboratory conditions. Therefore, by activating cryptic BGCs there is a potential to generate a large natural product library and expand opportunities for the discovery of compounds with undiscovered bioactivities.To activate these silent BGCs, we found a strain that manipulated a global regulator of fungal SM production to induce the expression of previously silent gene clusters. In particular, we selected a strain targeting multicluster regulator A (mcrA), a putative transcription factor responsible for the inhibition of transcription of hundreds of genes of which many are involved in the activation of BGCs (Fig. S1, and Table S1†).19 Previous studies have demonstrated that the deletion of mcrA resulted in the upregulation of the production of BGCs activated under normal laboratory conditions as well as the production of new secondary metabolites.22,23 For this particular strain, mcrA was knocked out to halt the inhibitory property of this gene, improving the production of certain secondary metabolites.22,24 Upon comparison of the metabolomic profile of wild type versus mcrA, we saw the generation of new compounds and the upregulation of others, suggesting that multiple SM pathways have been activated (Fig. 1C).
We proceeded to develop our natural product library through the one-strain-many-compounds (OSMAC) approach in which different culture conditions are used to activate various metabolic pathways of secondary metabolite production.14,25,26 We cultivated both wild-type and mcrAΔ strains in a small-scale screen of twelve different agar plate conditions, including minimal and rich media (Fig. S2†). SMs were extracted for metabolomic analysis, and the profiles were compared. Although all media conditions produced additional metabolites in the mutant strain in comparison to the wild type, the mcrAΔ strain cultivated in malt agar (MB) condition presented with the most potential for a natural product library with several more SMs that appear to be upregulated, and the presence of new compounds not seen in the wild type strain when grown in the same condition (Fig. 1C). IMV01140 was then scaled up using the MB agar condition and fractionated. The library compounds were then tested for AD drug discovery using the workflow shown in Fig. 1D. Select library compounds were characterized (Fig. S3–S17, and Table S2†).
Penicillic acid demonstrates anti-aggregation activity toward tau
We previously demonstrated that knocking out mcrA generates a library of compounds that can be tested for AD bioactivity. Here, we hypothesized that we could discover SMs against tau aggregation using bioactivity-guided fractionation of crude product and a biosensor cell seeding assay as a readout. As a sensor of tau aggregation, initial experiments utilized tau biosensor cells derived from HEK293, a human embryonic kidney cell line modified which expresses an aggregation-prone tau mutant fused with GFP.27 Human AD brain homogenate is used as a seed by transfection in biosensor cells. Compounds that inactivate the seeding power of tau in AD brain homogenate are considered possible inhibitor candidates. We assayed library crudely fractionated SMs from three fractions obtained from a normal phase column (Fig. 2A) by pre-incubating with AD crude brain homogenate overnight prior to transfection in tau biosensor cells. Tau aggregation was ascertained by quantifying fluorescent puncta. Effective inhibitor compounds are recognized as eliciting reductions in the number of puncta compared to control transfections, which contain AD brain homogenate without SM.The biosensor seeding assay demonstrated that our fractions C1 and C2 showed a significant decrease of normalized puncta (p < 0.0001), suggesting a possible hit compound with bioactivity for tau (Fig. 2B). However, upon analysis of cell confluence, which demonstrates cell viability, C2 appeared to be cytotoxic, suggesting possible toxic compounds in the fraction (p < 0.0001) (Fig. 2C). Therefore, we focused our efforts on fractionating the bioactive chemicals from C1. Because F1 contained the potential for a hit, we further fractionated C1 into fractions 1–5 (F1–5) (Fig. 2D). We tested the bioactivity of these higher-resolution fractions by pre-incubating them with AD tau seeds prior to transfecting in biosensor cells. As shown in Fig. 2E and F, F1–F4 showed significant activity (p < 0.01).
Cell-based assays can reveal SMs with direct or indirect effects. Therefore, we evaluated inhibitor activity for F1–5 by in vitro tau aggregation assays using thioflavin T (ThT), a fluorescent dye that emits by binding to beta sheet-rich structures in tau fibrils.28 Only F1 significantly inhibited aggregation (p < 0.05) (Fig. 2H). F1 was tested again using the biosensor seeding assay and was identified by mass spectrometry and nuclear magnetic resonance (NMR) to be penicillic acid (PA) (Fig. S4, and S5†). Since PA is commercially available, as an additional confirmation, we purchased PA from a commercial vendor and confirmed that its tau inhibitor activity was similar to F1 (p < 0.001) (Fig. 3A).
Additionally, to rule out the possibility that PA might block the interaction of ThT with the β-sheet structured tau aggregate, we confirmed that PA inhibited tau aggregation by measuring light scattering as a secondary readout.29 Small Angle X-ray Scattering (SAXS) data was obtained for samples taken directly from the ThT assay plate, shown in Fig. 3E. When we conducted SAXS, the tau sample containing heparin and PA was reduced to buffer levels, whereas tau with heparin produced scattering well above the background of the buffer sample (Fig. 3F), consistent with ThT results. To rule out possible interactions between PA and ThT, we performed an additional ThT assay containing only PA and ThT, demonstrating there is no interaction between the two (Fig. S18†). These results confirm ThT measures indicating PA inhibits aggregation of tau monomers.
Testing other natural products for similar biological activity
Given our observation that the common SM PA inhibited tau aggregation, we wondered if other common fungal natural products exhibited similar anti-tau aggregation activity. Perhaps one of the most well-known SM produced by filamentous fungi is penicillin, so we tested the Alzheimer's tau bioactivity of two different forms: penicillin G and penicillin V.30,31 Our seeding assay results indicated that both penicillin G and V did not have significant activity (p > 0.05) when tested with concentrations of 10 μM and 100 μM in comparison to PA (Fig. 3A). Therefore, we investigated the potency and mechanism of PA against tau aggregation. Nonlinear curve fitting using dose-response data shown in Fig. 3B shows PA inhibits tau seeding with an IC50 of 213 nM (Fig. S19†).Testing other tauopathies for similar biological activity
Besides AD, various other neurodegenerative tauopathies cause different pathologies with a direct link established between the conformation of tau fibrils and disease presentation.32–34 Because of the differences in tau fibrils in different tauopathies, we were interested in whether PA would exhibit similar inhibition of tau aggregation. We tested two different tauopathies, human progressive supranuclear palsy (PSP) and corticobasal degeneration (CBD), using the biosensor cell seeding assay, which differs only by the transfected seed. Biosensor seeding assays indicate PA was able to elicit similar inhibition of tau aggregation in PSP compared to AD (Fig. 3C). However, CBD was not inhibited by PA (Fig. 3D). The combination of these results illustrates some degree of specificity of PA for certain tau folds, although the structure–activity relationship requires deeper investigation.PA inhibits tau by fibril disaggregation (qEM)
Since PA inhibited seeding based on results from biosensor assays, and monomer aggregation based on ThT assays, we tested the activity of PA to disaggregate tau fibrils. We investigated disaggregation by incubating PA with AD brain-purified tau fibrils observing fibril counts by quantifying using electron microscopy (EM). When incubated without PA, we can see that numerous AD tau fibrils are visible using negative stain EM (Fig. 3G); however, with the addition of a high assayed concentration of 1 mM of PA nearly eliminates any visible fibrils (Fig. 3H). The EM images were collected from random points on the EM grid and quantified, showing a large reduction in visible fibrils (Fig. 3I). This observation led us to conclude that PA directly inhibits tau by disaggregating fibrils.Using PA as a chemical probe for AD drug discovery
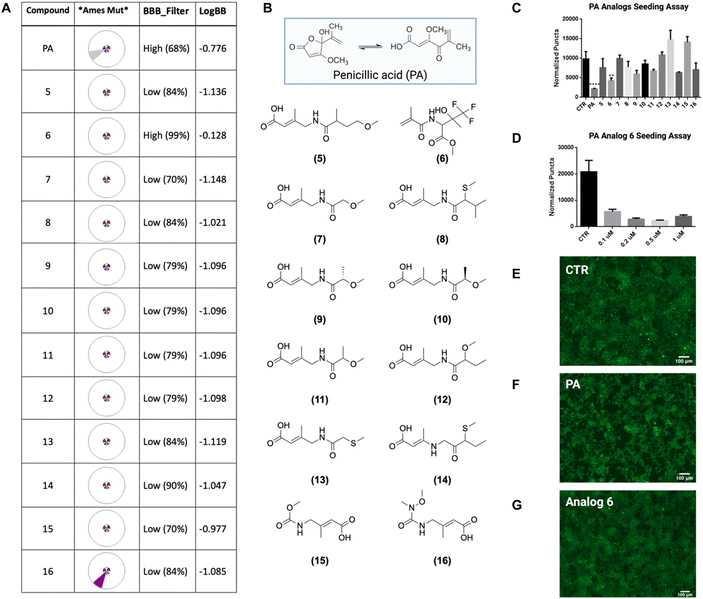
To investigate how PA contributes to tau disaggregation, we explored the structure–activity relationship. PA exists in equilibrium between the lactone and keto acid tautomeric forms, with the lactone predominating at physiological pH.35,36 To determine which tautomer is critical for tau disaggregation, we synthesized stable analogs of both forms. However, when we tested these analogs using a biosensor cell seeding assay, both the lactone and keto acid analogs showed a loss of activity (Fig. S20†). We conclude that the primary activity towards tau stems from the modified moieties, particularly the aliphatic hydroxyl group in the lactone form and/or the carboxylic acid in the keto form.Although we could not definitively determine which tautomer predominates in the active form, we proceeded to use the keto form as a template for drug discovery with ADMET Predictor. Several carboxylic acids from the enamine library similar to the keto form of PA were selected due to the greater electron-withdrawing properties of the carboxyl compared to the aliphatic hydroxyl, potentially making it a stronger inhibitor. In addition, we identified library compounds containing aliphatic hydroxyls, which were represented in the lactone form. ADMET Predictor was employed to assess assets and hazards for the series.37–40 This analysis suggested that PA is blood–brain barrier permeable (Fig. 4A), but also flagged mutagenicity as a potential hazard, indicating that PA may increase mutation frequency (Fig. 4A). Given the predicted mutagenicity, we selected twelve enamine REAL database compounds to test as potential tau inhibitors using the biosensor cell seeding assay (Fig. 4B). Results from our seeding assay indicate that analog 6 exhibits inhibition of tau aggregation activity (p < 0.01) at a high concentration of 50 μM (Fig. 4C, and E–G). To further assess this activity, we tested the dose dependency of analog 3 against tau aggregation by adding 0.1 μM, 0.2 μM, 0.5 μM, and 1 μM of the compound. Interestingly, we observed a dose-independent relationship, where activity was present at both low and high concentrations but was inexplicably lost at intermediate concentrations (Fig. 4D). These findings suggest that bioactive small molecules can serve as chemical probes for discovering new analogs that inhibit AD tau aggregation while minimizing predicted toxicity.
Discussion
Natural product drug discovery is not a novel concept; however, our study proves that the combination of genetics and the one-strain-many-compounds (OSMAC) method is a reliable technique to generate a natural product library that can test compounds that are not commonly found in commercially available natural product libraries. This method demonstrates that although genetically modifying these fungal strains may not activate pathways that result in the production of new compounds, the bioactivities of many previously discovered secondary metabolites have yet to be tested. Furthermore, due to the bioactive nature of fungal secondary metabolites, this technique is not limited to AD bioactivity shown in this study, but many other types of activity as well, including anti-cancer, anti-fungal, anti-bacterial, etc. In other words, the same compounds in this library can be further tested to discover additional bioactivities.With our discovery of PA as a tau aggregation inhibitor, we were able to further predict the toxicity of this compound on humans using ADMET predictor. Whereas ADMET predictor can be seen as a reliable method to determine whether a compound will be successful in making it through the discovery to pipeline market and tremendously speed up the process by giving us information to eliminate compounds with excessive toxicity, the limitation is that it is still a prediction based on past data. In order to firmly conclude that these tested compounds truly have blood–brain barrier permeability and limited toxicities, we still have to conduct physical assays. However, with these predictions, we can still try to limit these red flags before further testing by finding or synthesizing analogs that have all the qualities we want in a drug while keeping similar bioactivity at effective concentrations.
Previous studies have shown that another small molecule, epigallocatechin-3 gallate (EGCG), is capable of binding to AD tau; however, it is demonstrated that EGCG is capable of inhibiting not only AD tau aggregation, but other tauopathies as well.41,42 The results that we saw with PA demonstrate a degree of specificity in its activity in that it only inhibits AD and PSP tau aggregation but not CBD tau, which raises a major question on how PA works to inhibit tau proteins. In order to test this, cryo-EM could be used to give further understanding as to how tau is disaggregated and give insight for specifically which parts of PA are vital in this bioactivity. Furthermore, this would unlock a new realm of potential analogs that could be developed to exhibit similar activity while limiting toxicity.
Materials and methods
Genome analysis43
The fungal genome of IMV01140 was downloaded from the NCBI database (GenBank, GCA_030674015.1) and analyzed using antiSMASH v7.1.0 to predict secondary metabolism genes.Molecular genetic procedures for strain development22,23
To confirm the mcrA knockout, diagnostic PCR was performed by designing nesting primers to amplify the resistance gene and extracting DNA from fungal biomass for use as the template.Fermentation and LC-MS analysis
IMV01140 WT and mcrAΔ strains were incubated at 28 °C on different conditions. The conditions are as follows: LMM (15 g L−1 lactose, 50 mL salt solution, 800 μL Hutner's trace element, 1 mL 5.5 M KOH, 15 g L−1 agar), SDA (40 g L−1 dextrose, 100 g L−1 peptone, 15 g L−1 agar), PDA (36 g L−1 criterion cat. no. C6622), TYG (3 g L−1 tryptone, 3 g L−1 yeast extract, 3 g L−1 glucose, 1 g L−1 K2HPO4, 15 g L−1 agar), YAG (yeast extract 5 g L−1, 20 g L−1 glucose, 800 μL Hutner's trace element, 15 g L−1 agar), YES (30 g L−1 sucrose, 5 g L−1 yeast extract, 1 mL Hutner's trace element, 15 g L−1 agar), YEPD (20 g L−1 dextrose, 10 g L−1 yeast extract, 20 g L−1 peptone, 15 g L−1 agar), YM (3 g L−1 yeast extract, 3 g L−1 malt extract, 10 g L−1 dextrose, 5 g L−1 peptone 15 g L−1 agar), MEA (malt extract 20 g L−1, peptone 1 g L−1, dextrose 20 g L−1, 15 g L−1 agar), LCMM (20 g L−1 lactose, 10 g L−1 dextrose, 50 mL salt solution, 800 μL Hutner's trace element, 1 mL 5.5 M KOH, 15 g L−1 agar), CZA (100 mL concentrated Czapek, 1 g K2HPO4, 30 g sucrose, 15 g L−1 agar), MB (130 g L−1 malt extract, 15 g L−1 agar). 1.0 × 107 conidia were inoculated for each strain.After 6 days of incubation, 10 plugs of agar (7 mm diameter) were taken for compound extraction. 5 mL of methanol was used for initial extraction followed by 1 h of sonication. Extract was collected and the agar was further extracted with 5 mL of DCM/MeOH (1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1) followed by 1 h of sonication. The combined extract was evaporated through TurboVap LV. The dried residues were dissolved in equal parts of ethyl acetate and water. The ethyl acetate layer was collected and evaporated through TurboVap. The dried extract was redissolved in dimethyl sulfoxide
1) followed by 1 h of sonication. The combined extract was evaporated through TurboVap LV. The dried residues were dissolved in equal parts of ethyl acetate and water. The ethyl acetate layer was collected and evaporated through TurboVap. The dried extract was redissolved in dimethyl sulfoxide![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) methanol (1
methanol (1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 4) at 1 mg mL−1.
4) at 1 mg mL−1.
HPLC-MS analysis was performed using a RP C18 column on a ThermoFinnigan LCQ Advantage ion trap mass spectrometer. 10 μL of sample was injected. The flow rate used was 125 μL min−1. The solvent gradient consisted of 95% MeCN/H2O (solvent B) and 5% MeCN/H2O (solvent A), both containing 0.05% formic acid. The gradient was run under the following conditions: 0% solvent B from 0 to 5 min, 0–100% solvent B from 5 min to 35 min, 100–0% solvent B from 40 to 45 min, and re-equilibration with 0% solvent B from 45 to 50 min.
Fractionation, isolation, and characterization of metabolites
To isolate metabolites, ethyl acetate was used to extract compounds from 3.5 L of malt agar plates. Agar was extracted with 1 L of methanol followed by 1 h of sonication. Another round of extraction was performed using 1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1 DCM/MeOH followed by 1 h of sonication. The resulting extract was concentrated and re-extracted with EtOAc. After concentrating, crude was subjected to silica-gel column chromatography for fractions tested with biosensor seeding assay. Fractions were further separated through semi-preparative HPLC (Phenomenex Luna 5 μm C18 (2), 250 × 10 mm), with a flow rate of 4.0 mL min−1 and monitored using a UV detector set at 254 nm. NMR spectra were recorded using a Varian VNMRS-400 spectrometer.
1 DCM/MeOH followed by 1 h of sonication. The resulting extract was concentrated and re-extracted with EtOAc. After concentrating, crude was subjected to silica-gel column chromatography for fractions tested with biosensor seeding assay. Fractions were further separated through semi-preparative HPLC (Phenomenex Luna 5 μm C18 (2), 250 × 10 mm), with a flow rate of 4.0 mL min−1 and monitored using a UV detector set at 254 nm. NMR spectra were recorded using a Varian VNMRS-400 spectrometer.
Preparation of crude Alzheimer's brain-derived tau seeds
Human Alzheimer's brain autopsy samples were obtained from the UCLA Pathology Department following HHS regulations, with prior consent from the patients. Approximately 0.2 grams of hippocampal tissue were excised and homogenized using a Kinematica PT 10–35 POLYTRON at level 4–5. The homogenization process was performed in 10% sucrose buffer containing 1 mM ethylene glycol tetraacetic acid (EGTA) and 5 mM ethylenediaminetetraacetic acid (EDTA) in 15 mL Falcon tubes.K18CY cell culture
HEK293T cell lines stably expressing tau-K18CY labeled with green fluorescent protein (GFP), obtained from Marc Diamond's laboratory at the University of Texas Southwestern Medical Center, were used in this study.44 The cells were cultured in a T25 flask containing Dulbecco's Modified Eagle Medium (DMEM) (Life Technologies, cat. 11965092) supplemented with 10% fetal bovine serum (FBS) (Life Technologies, cat. A3160401), 1% penicillin/streptomycin (Life Technologies, cat. 15140122), and 1% GlutaMAX (Life Technologies, cat. 35050061). They were maintained at 37 °C and 5% CO2 in a humidified incubator. For inhibitor testing on the biosensor cells, 100 μl of the cell suspension was plated in 96-well plates and incubated at 37 °C and 5% CO2 for 16 to 24 hours prior to transfection.Biosensor seeding assay
Penicillic acid (Santa Cruz, CAS 90-65-3) was diluted in dimethyl sulfoxide (DMSO) to create 1.4 mM stock solutions. Homogenized Alzheimer's disease brain tissue was diluted in Opti-MEM (Thermo Fisher Scientific, cat. 31985062) at a 1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 20 ratio. The diluted brain homogenate was incubated with the specified inhibitors overnight at 4 °C. The inhibitor-treated samples were then sonicated in a Cup Horn (Qsonica, MPH) water bath for 3 minutes at 40% power and subsequently mixed with a 1
20 ratio. The diluted brain homogenate was incubated with the specified inhibitors overnight at 4 °C. The inhibitor-treated samples were then sonicated in a Cup Horn (Qsonica, MPH) water bath for 3 minutes at 40% power and subsequently mixed with a 1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 20 solution of Lipofectamine 2000 (Thermo Fisher Scientific, cat. 11668019) and Opti-MEM. After 20 minutes, 10 μl of the inhibitor-treated fibrils were added to the previously plated 100 μl of cells in triplicate, avoiding the perimeter wells, to achieve a final ligand concentration of 10 mM on the cells.
20 solution of Lipofectamine 2000 (Thermo Fisher Scientific, cat. 11668019) and Opti-MEM. After 20 minutes, 10 μl of the inhibitor-treated fibrils were added to the previously plated 100 μl of cells in triplicate, avoiding the perimeter wells, to achieve a final ligand concentration of 10 mM on the cells.
Quantification of tau aggregation
The number of seeded aggregates was determined using the BioTek Cytation 5 Imaging Multimode Reader in the GFP channel to image the entire 96-well plate. Exposure and contrast were adjusted to clearly distinguish the seeded aggregates, which appear as bright green puncta, from the cells. The aggregates in each image were quantified using a script in ImageJ 2.3.0, a Java-based image processing program.45 This script subtracts the background fluorescence from unseeded cells and then employs a built-in particle analyzer to count the number of puncta, representing seeded aggregates, as peaks contrasting against the background fluorescence. The puncta count was normalized across all images according to cell confluence using a separate ImageJ 2.3.0 script for confluence measurement. The average number of seeded aggregates per well, along with standard deviations from triplicate measurements normalized to confluence, was plotted to compare the inhibitory activity.Preparation of purified Alzheimer's brain-derived tau fibrils
2–6 grams of human Alzheimer's brain autopsy samples was homogenized in 10 mL of sucrose buffer supplemented with 1 mM EGTA, using 2 × 50 mL Falcon tubes sealed with parafilm. The homogenate was transferred to 2 mL Eppendorf tubes and centrifuged at 20![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 000×g for 20 minutes. The supernatant was saved in a separate tube. Sarkosyl was added to a final concentration of 1%, and the mixture was incubated with orbital shaking at 190 rpm for 1 hour at room temperature in a 50 mL Falcon tube containing five glass beads to enhance mixing. The mixture was ultracentrifuged at 95
000×g for 20 minutes. The supernatant was saved in a separate tube. Sarkosyl was added to a final concentration of 1%, and the mixture was incubated with orbital shaking at 190 rpm for 1 hour at room temperature in a 50 mL Falcon tube containing five glass beads to enhance mixing. The mixture was ultracentrifuged at 95![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 000×g for 60 minutes. The ultra pellet was resuspended in approximately 1/4 volume of sucrose buffer supplemented with 5 mM EDTA and 1 mM EGTA, then centrifuged at 20
000×g for 60 minutes. The ultra pellet was resuspended in approximately 1/4 volume of sucrose buffer supplemented with 5 mM EDTA and 1 mM EGTA, then centrifuged at 20![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 100×g for 30 minutes at 4 °C. The sample was ultracentrifuged again at 95
100×g for 30 minutes at 4 °C. The sample was ultracentrifuged again at 95![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 000×g for 60 minutes. Finally, the pellet was resuspended in 250 μl of 20 mM Tris–HCl (pH 7.4) and 100 mM NaCl.
000×g for 60 minutes. Finally, the pellet was resuspended in 250 μl of 20 mM Tris–HCl (pH 7.4) and 100 mM NaCl.
Negative stain grid preparation
Purified Alzheimer's brain-derived tau fibrils were diluted 1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 10 in PBS and incubated with penicillic acid for 24 hours at 4 °C. For electron microscopy, negatively stained grids were prepared by depositing 6 μl of the fibril samples onto formvar/carbon-coated copper grids (400 mesh) for 3 minutes, following the 24 hours inhibitor incubation. The sample was then rapidly blotted using filter paper without drying the grid, stained with 4% uranyl acetate for 2 minutes, and finally wicked dry with filter paper.
10 in PBS and incubated with penicillic acid for 24 hours at 4 °C. For electron microscopy, negatively stained grids were prepared by depositing 6 μl of the fibril samples onto formvar/carbon-coated copper grids (400 mesh) for 3 minutes, following the 24 hours inhibitor incubation. The sample was then rapidly blotted using filter paper without drying the grid, stained with 4% uranyl acetate for 2 minutes, and finally wicked dry with filter paper.
Quantitative EM (qEM) imaging
For quantitative electron microscopy (qEM), negatively stained EM grids of each sample were screened using the JEOL 2100 TEM at a magnification of ×12![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 000.
000.
ThT plate-reader assay
The concentrated tau protein was diluted into PBS buffer (pH 7.4) to achieve the final concentration at 100 μm. The proteins were agitated in solutions containing 75 μM ThT, 0.5 mg mL−1 heparin (Sigma cat. no. H3393), 1 mM DTT, and a two-fold molar excess of penicillic acid in 96-well plates with a plastic bead to enhance mixing. ThT fluorescence was measured using excitation and emission wavelengths of 440 nm and 480 nm, respectively. Averaged curves were generated from triplicate measurements, as described in the figure legend. Error bars represent the standard deviation of replicate measurements.Tau K18+ monomer purification
Prior to purification of tau monomer via ion exchange and size exclusion chromatography, K18+ tau expressed in pNG2 plasmid was transformed into BL21DE3 E. coli competent cells (New England Biolabs, cat. C2527I). Single colonies were inoculated into 80 mL LB broth, supplemented with 100 μg mL−1 ampicillin antibiotic, and grown overnight at 30 °C overnight with orbital shaking at 250 rpm. The following morning, the starter was inoculated into 1 L LB broth + ampicillin and grown at 37 °C with 250 rpm orbital shaking. The cells were grown until reaching OD 600 = 0.8–1.00. Protein expression was induced with 0.5 mM IPTG for 3 h. The cells were centrifuged at 4500 rpm and lysed in: 100 mL 20 mM MES|pH6.8| (Grainger, cat. M22040-1000.0), 1 mM EDTA (Sigma, cat. E9884), 1 mM magnesium chloride, 5 mM mercaptoethanol, and 100 μl HALT protease inhibitor cocktail (Life Technologies, cat. 78438). Then, the cells were sonicated on ice using Misonix S-4000 Sonicator, following boiling for 20 min with the addition of 500 mM NaCl. The samples were centrifuged at 15![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 000 rpm for 15 minutes at 4 °C. The supernatant was dialyzed against 20 mM MES|pH6.8|, 50 mM NaCl, 5 mM BME buffer. The dialyzed tau was purified using Biologic Duoflow Chromatography system with HiTrap SP FF column. The presence of K18+ tau monomer was confirmed by SDS-PAGE. The suitable fractions were combined, and the tau protein was concentrated using Pierce™ Protein Concentrator PES, 3 K MWCO (Life Technologies, cat. 88525).
000 rpm for 15 minutes at 4 °C. The supernatant was dialyzed against 20 mM MES|pH6.8|, 50 mM NaCl, 5 mM BME buffer. The dialyzed tau was purified using Biologic Duoflow Chromatography system with HiTrap SP FF column. The presence of K18+ tau monomer was confirmed by SDS-PAGE. The suitable fractions were combined, and the tau protein was concentrated using Pierce™ Protein Concentrator PES, 3 K MWCO (Life Technologies, cat. 88525).
Small Angle X-ray scattering (SAXS)
Each sample (25 μl) was loaded in a Xenocs thermalized BioCube flow-cell with camera and pump. All measurements were made in an air-evacuated space at a pressure below 1.0 mbar and at room temperature and were performed using a Xeuss 3.0 (Xenocs) with Microfocus GeniX 3D X-ray microsource and Eiger2 R 1M Dectris hybrid pixel silicon sensor detector. The scattering experiments were performed with the q range of 0.055–0.25 Å−1, d-space of 25–1150 Å, and incidence beam of 9.3 Mph s−1 for a total of 20 minutes. The data was then plotted using Xenocs XSACT software on a log/log graph.Computational studies
The pharmacokinetics and toxicity profiles for penicillic acid, penicillins, and synthetic analogs were evaluated in silico using ADMET Predictor™ 10.4 (Simulations Plus, Inc., Lancaster, CA, USA). ADMET properties including mutagenicity and blood–brain barrier (BBB) permeability were evaluated by comparison for synthetic analogs with penicillic acid. Reported in silico mutagenicity was assessed using an Ames model. BBB permeability results are reported as LogBB and BBB Filter.Conclusions
Due to the lack of available treatments for AD, we are constantly investigating various sources of small molecules that can be tested for bioactivity for the development of better therapeutic interventions for AD patients. As tau is a proven neuropathological characteristic responsible for Alzheimer's progression, we opted to focus on discovering compounds with bioactivity targeting tau. Historically, secondary metabolites have demonstrated bioactive potential; however, many have not been tested. Here, we present experimental data to reveal that genetically manipulating Aspergillus melleus, a strain of filamentous fungi, can allow for the discovery of chemical probes for Alzheimer's disease therapeutics. Specifically, we found that penicillic acid can inhibit tau seeding, which indicates bioactivity towards AD tau. Following this observation, we systemically investigated the in vitro effects of penicillic acid on AD tau aggregation through various techniques. Doing so uncovered that penicillic acid directly inhibits tau by disaggregating fibrils.Using ADMET predictor resulted in the prediction of mutagenic effects of penicillic acid on humans, initiating the utilization of the penicillic acid backbone as a chemical probe. Our screening data reveals that the discovery of compounds with a similar backbone is an effective method for finding similar compounds with inhibition in AD tau aggregation. Together, our usage of cell-based screening assays in combination with our computational studies resulted in the discovery of an additional small-molecule compound that inhibits AD tau aggregation while keeping blood–brain barrier permeability and limiting predicted toxicity.
Data availability
The data supporting this article have been included as part of the ESI.†Author contributions
The project was conceived and designed by J. S., J. L., C. W., and P. M. S. Genetic knockouts were performed by J. S. and B. Y. Generation of compound library and isolation of compounds were completed by J. S., Q. H., C. L. Biosensor seeding assays were carried out by J. L. and J. S. In vitro screening assays were performed by J. L. and M. F. Computational analysis was performed by P. M. S. The manuscript was prepared by J. S., J. L., P. M. S., and C. W. with contributions from all other authors.Conflicts of interest
There are no conflicts to declare.Acknowledgements
We thank the donors and their families; without whom this work would not have been possible. We are grateful to Simulations Plus for providing a license for ADMET Predictor® through the University + program. We also thank BrightFocus Foundation (A2023016S). Figures were created with https://www.biorender.com/.References
- K. Blennow, M. J. de Leon and H. Zetterberg, Alzheimer's disease, Lancet, 2006, 368, 387–403 CrossRef CAS PubMed
.
- R. Medeiros, D. Baglietto-Vargas and F. M. LaFerla, The role of tau in Alzheimer's disease and related disorders, CNS Neurosci. Ther., 2011, 17, 514–524 CrossRef CAS PubMed
.
- K. F. Lopes, V. S. Bahia, J. C. Natividade, R. V. S. Bastos, W. A. Shiguti, K. E. R. da Silva and W. C. de Souza, Changes in personality traits in patients with Alzheimer's Disease, Dement. Neuropsychol., 2022, 16, 187–193 CrossRef PubMed
.
- D. P. Veitch, M. W. Weiner, P. S. Aisen, L. A. Beckett, N. J. Cairns, R. C. Green, D. Harvey, C. R. Jack, W. Jagust, J. C. Morris, R. C. Petersen, A. J. Saykin, L. M. Shaw, A. W. Toga, J. Q. Trojanowski and A. s. D. N. Initiative, Understanding disease progression and improving Alzheimer's disease clinical trials: Recent highlights from the Alzheimer's Disease Neuroimaging Initiative, Alzheimers Dement., 2019, 15, 106–152 CrossRef PubMed
.
- N. P. Keller, G. Turner and J. W. Bennett, Fungal secondary metabolism - from biochemistry to genomics, Nat. Rev. Microbiol., 2005, 3, 937–947 CrossRef CAS PubMed
.
- X. Chen, J. Drew, W. Berney and W. Lei, Neuroprotective Natural Products for Alzheimer’s Disease, Cells, 2021, 10, 1309 CrossRef CAS PubMed
.
- L. R. Martinez and B. C. Fries, Fungal Biofilms: Relevance in the Setting of Human Disease, Curr. Fungal Infect. Rep., 2010, 4, 266–275 CrossRef PubMed
.
- S. Fanning and A. P. Mitchell, Fungal biofilms, PLoS Pathog., 2012, 8, e1002585 CrossRef CAS PubMed
.
- K. Williams, C. Greco, A. M. Bailey and C. L. Willis, Core Steps to the Azaphilone Family of Fungal Natural Products, Chembiochem, 2021, 22, 3027–3036 CrossRef CAS PubMed
.
- S. R. Paranjape, A. P. Riley, A. D. Somoza, C. E. Oakley, C. C. Wang, T. E. Prisinzano, B. R. Oakley and T. C. Gamblin, Azaphilones inhibit tau aggregation and dissolve tau aggregates in vitro, ACS Chem. Neurosci., 2015, 6, 751–760 CrossRef CAS PubMed
.
- B. C. Covington, F. Xu and M. R. Seyedsayamdost, A Natural Product Chemist's Guide to Unlocking Silent Biosynthetic Gene Clusters, Annu. Rev. Biochem., 2021, 90, 763–788 CrossRef CAS PubMed
.
- A. Rokas, M. E. Mead, J. L. Steenwyk, H. A. Raja and N. H. Oberlies, Biosynthetic gene clusters and the evolution of fungal chemodiversity, Nat. Prod. Rep., 2020, 37, 868–878 RSC
.
-
Y.-M. Chiang, C. C. C. Wang and B. R. Oakley, in Natural Products, 2014, pp. 171–193 Search PubMed
.
- H. Wei, Z. Lin, D. Li, Q. Gu and T. Zhu, [OSMAC (one strain many compounds) approach in the research of microbial metabolites–a review], Weishengwu Xuebao, 2010, 50, 701–709 CAS
.
- I. Kjærbølling, U. H. Mortensen, T. Vesth and M. R. Andersen, Strategies to establish the link between biosynthetic gene clusters and secondary metabolites, Fungal Genet. Biol., 2019, 130, 107–121 CrossRef PubMed
.
- J. W. Bok and N. P. Keller, LaeA, a regulator of secondary metabolism in Aspergillus spp, Eukaryotic Cell, 2004, 3, 527–535 CrossRef CAS PubMed
.
- M. F. Grau, R. Entwistle, C. E. Oakley, C. C. C. Wang and B. R. Oakley, Overexpression of an LaeA-like Methyltransferase Upregulates Secondary Metabolite Production in, ACS Chem. Biol., 2019, 14, 1643–1651 CrossRef CAS PubMed
.
- W. Yin and N. P. Keller, Transcriptional regulatory elements in fungal secondary metabolism, J. Microbiol., 2011, 49, 329–339 CrossRef CAS PubMed
.
- C. E. Oakley, M. Ahuja, W. W. Sun, R. Entwistle, T. Akashi, J. Yaegashi, C. J. Guo, G. C. Cerqueira, J. Russo Wortman, C. C. Wang, Y. M. Chiang and B. R. Oakley, Discovery of mcrA, a master regulator of Aspergillus secondary metabolism, Mol. Microbiol., 2017, 103, 347–365 CrossRef CAS PubMed
.
- M. H. Medema, K. Blin, P. Cimermancic, V. de Jager, P. Zakrzewski, M. A. Fischbach, T. Weber, E. Takano and R. Breitling, antiSMASH: rapid identification, annotation and analysis of secondary metabolite biosynthesis gene clusters in bacterial and fungal genome sequences, Nucleic Acids Res., 2011, 39, W339–W346 CrossRef CAS PubMed
.
- K. Blin, S. Shaw, K. Steinke, R. Villebro, N. Ziemert, S. Y. Lee, M. H. Medema and T. Weber, antiSMASH 5.0: updates to the secondary metabolite genome mining pipeline, Nucleic Acids Res., 2019, 47, W81–W87 CrossRef CAS PubMed
.
- B. Yuan, M. F. Grau, R. M. Murata, T. Torok, K. Venkateswaran, J. E. Stajich and C. C. C. Wang, Identification of the Neoaspergillic Acid Biosynthesis Gene Cluster by Establishing an, ACS Omega, 2023, 8, 16713–16721 CrossRef CAS PubMed
.
- B. Yuan, N. P. Keller, B. R. Oakley, J. E. Stajich and C. C. C. Wang, Manipulation of the Global Regulator mcrA Upregulates Secondary Metabolite Production in Aspergillus wentii Using CRISPR-Cas9 with In Vitro Assembled Ribonucleoproteins, ACS Chem. Biol., 2022, 17, 2828–2835 CrossRef CAS PubMed
.
- Q. Al Abdallah, W. Ge and J. R. Fortwendel, A Simple and Universal System for Gene Manipulation in Aspergillus fumigatus: In Vitro-Assembled Cas9-Guide RNA Ribonucleoproteins Coupled with Microhomology Repair Templates, mSphere, 2017, 2, e00446 CrossRef CAS PubMed
.
- H. B. Bode, B. Bethe, R. Höfs and A. Zeeck, Big effects from small changes: possible ways to explore nature's chemical diversity, Chembiochem, 2002, 3, 619–627 CrossRef CAS
.
- R. Pan, X. Bai, J. Chen, H. Zhang and H. Wang, Exploring Structural Diversity of Microbe Secondary Metabolites Using OSMAC Strategy: A Literature Review, Front. Microbiol., 2019, 10, 294 CrossRef PubMed
.
- B. D. Hitt, J. Vaquer-Alicea, V. A. Manon, J. D. Beaver, O. M. Kashmer, J. N. Garcia and M. I. Diamond, Ultrasensitive tau biosensor cells detect no seeding in Alzheimer's disease CSF, Acta Neuropathol. Commun., 2021, 9, 99 CrossRef CAS PubMed
.
- D. Sui, M. Liu and M. H. Kuo, In vitro aggregation assays using hyperphosphorylated tau protein, J. Visualized Exp., 2015, e51537 Search PubMed
.
- N. V. Gorantla, A. V. Shkumatov and S. Chinnathambi, Conformational Dynamics of Intracellular Tau Protein Revealed by CD and SAXS, Methods Mol. Biol., 2017, 1523, 3–20 CrossRef CAS PubMed
.
- A. J. Wright, The penicillins, Mayo Clin. Proc., 1999, 74, 290–307 CrossRef CAS PubMed
.
- A. Fleming, On the antibacterial action of cultures of a penicillium, with special reference to their use in the isolation of B. influenzae. 1929, Bull. W. H. O., 2001, 79, 780–790 CAS
.
- Y. Shi, W. Zhang, Y. Yang, A. G. Murzin, B. Falcon, A. Kotecha, M. van Beers, A. Tarutani, F. Kametani, H. J. Garringer, R. Vidal, G. I. Hallinan, T. Lashley, Y. Saito, S. Murayama, M. Yoshida, H. Tanaka, A. Kakita, T. Ikeuchi and S. H. W. Scheres, Structure-based classification of tauopathies, Nature, 2021, 598, 359–363 CrossRef CAS PubMed
.
- C. W. Chang, E. Shao and L. Mucke, Tau: Enabler of diverse brain disorders and target of rapidly evolving therapeutic strategies, Science, 2021, 371, eabb8255 CrossRef CAS PubMed
.
- J. Vaquer-Alicea, M. I. Diamond and L. A. Joachimiak, Tau strains shape disease, Acta Neuropathol., 2021, 142, 57–71 CrossRef CAS PubMed
.
- C. W. MUNDAY, Tautomerism of penicillic acid, Nature, 1949, 163, 443 CrossRef CAS PubMed
.
- E. SHAW, A synthesis of protoanemonin; the tautomerism of acetylacrylic acid and of penicillic acid, J. Am. Chem. Soc., 1946, 68, 2510–2513 CrossRef CAS PubMed
.
- J. Dulsat, B. López-Nieto, R. Estrada-Tejedor and J. I. Borrell, Evaluation of Free Online ADMET Tools for Academic or Small Biotech Environments, Molecules, 2023, 28, 776 CrossRef CAS PubMed
.
- J. Ghosh, M. S. Lawless, M. Waldman, V. Gombar and R. Fraczkiewicz, Modeling ADMET, Methods Mol. Biol., 2016, 1425, 63–83 CrossRef CAS PubMed
.
- F. Cheng, W. Li, G. Liu and Y. Tang, In silico ADMET prediction: recent advances, current challenges and future trends, Curr. Top. Med. Chem., 2013, 13, 1273–1289 CrossRef CAS PubMed
.
- A. K. Sohlenius-Sternbeck and Y. Terelius, Evaluation of ADMET Predictor in Early Discovery Drug Metabolism and Pharmacokinetics Project Work, Drug Metab. Dispos., 2022, 50, 95–104 CrossRef CAS PubMed
.
- P. M. Seidler, K. A. Murray, D. R. Boyer, P. Ge, M. R. Sawaya, C. J. Hu, X. Cheng, R. Abskharon, H. Pan, M. A. DeTure, C. K. Williams, D. W. Dickson, H. V. Vinters and D. S. Eisenberg, Structure-based discovery of small molecules that disaggregate Alzheimer's disease tissue derived tau fibrils in vitro, Nat. Commun., 2022, 13, 5451 CrossRef CAS PubMed
.
- S. K. Sonawane, H. Chidambaram, D. Boral, N. V. Gorantla, A. A. Balmik, A. Dangi, S. Ramasamy, U. K. Marelli and S. Chinnathambi, EGCG impedes human Tau aggregation and interacts with Tau, Sci. Rep., 2020, 10, 12579 CrossRef CAS PubMed
.
- K. Blin, S. Shaw, H. E. Augustijn, Z. L. Reitz, F. Biermann, M. Alanjary, A. Fetter, B. R. Terlouw, W. W. Metcalf, E. J. N. Helfrich, G. P. van Wezel, M. H. Medema and T. Weber, antiSMASH 7.0: new and improved predictions for detection, regulation, chemical structures and visualisation, Nucleic Acids Res., 2023, 51, W46–W50 CrossRef CAS PubMed
.
- D. W. Sanders, S. K. Kaufman, S. L. DeVos, A. M. Sharma, H. Mirbaha, A. Li, S. J. Barker, A. C. Foley, J. R. Thorpe, L. C. Serpell, T. M. Miller, L. T. Grinberg, W. W. Seeley and M. I. Diamond, Distinct tau prion strains propagate in cells and mice and define different tauopathies, Neuron, 2014, 82(6), 1271–1288 CrossRef CAS PubMed
.
- J. Schindelin, I. Arganda-Carreras, E. Frise, V. Kaynig, M. Longair, T. Pietzsch, S. Preibisch, C. Rueden, S. Saalfeld, B. Schmid, J. Y. Tinevez, D. J. White, V. Hartenstein, K. Eliceiri, P. Tomancak and A. Cardona, Fiji: An open-source platform for biological-image analysis, Nat. Methods, 2012, 9(7), 676–682 CrossRef CAS PubMed
.
Footnotes |
| † Electronic supplementary information (ESI) available. See DOI: https://doi.org/10.1039/d4sc05469e |
| ‡ These authors contributed equally. |
| This journal is © The Royal Society of Chemistry 2024 |