Open Access Article
Open Access ArticleOxygen-bridged W-Pd atomic pairs enable H2O2 activation for sensitive immunoassays†
Chengjie
Chen
a,
Dongbo
Yan
a,
Xiangkun
Jia
a,
Ruimin
Li
a,
Lijun
Hu
a,
Xiaotong
Li
a,
Lei
Jiao
 *a,
Chengzhou
Zhu
*a,
Chengzhou
Zhu
 b,
Yanling
Zhai
*a and
Xiaoquan
Lu
b,
Yanling
Zhai
*a and
Xiaoquan
Lu
 a
a
aInstitute of Molecular Metrology, College of Chemistry and Chemical Engineering, Qingdao University, Qingdao 266071, P. R. China. E-mail: zhaiyanling@qdu.edu.cn; jiaolei@qdu.edu.cn
bNational Key Laboratory of Green Pesticide, International Joint Research Center for Intelligent Biosensing Technology and Health, College of Chemistry, Central China Normal University, Wuhan 430079, P. R. China
First published on 26th August 2024
Abstract
Regulating the performance of peroxidase (POD)-like nanozymes is a prerequisite for achieving highly sensitive and accurate immunoassays. Inspired by natural enzyme catalysis, we design a highly active and selective nanozyme by loading atomically dispersed tungsten (W) sites on Pd metallene (W-O-Pdene) to construct an artificial three-dimensional (3D) catalytic center. The 3D asymmetric W-O-Pd atomic pairs can effectively stretch the O–O bonds in H2O2 and further promote the desorption of H2O to enhance POD-like activity. Moreover, the W-O-Pd sites with unique spatial structures demonstrate satisfactory specificity for H2O2 activation, effectively preventing the interference of dissolved oxygen. Accordingly, the highly active and specific W-O-Pdene nanozymes are utilized for sensitive and accurate prostate-specific antigen (PSA) immunoassay with a low detection limit of 1.92 pg mL−1, superior to commercial enzyme-linked immunosorbent assay.
Introduction
Nanozymes with POD-like activity have become effective substitutes for natural POD and have been extensively investigated for colorimetric immunoassay.1–6 Understanding and regulating the H2O2 activation of POD-like nanozymes plays a key role in improving the sensitivity and accuracy of immunoassay.7–10 Although POD-like nanozymes have witnessed enormous progress in enhancing catalytic activity, limited studies have demonstrated improvement in catalytic specificity.11,12 Although several efforts have been devoted to addressing this issue,13–15 the rational design and controllable preparation of specific POD-like nanozymes are still impeded by an incomplete understanding of the reaction mechanism.16,17 In particular, due to the structural complexity of nanozymes, the selective cleavage of the O–O bond in H2O2 activation is still ambiguous.18–21Horseradish peroxidases (HRP) are state-of-the-art biocatalysts and their catalytic pockets with unique electronic and geometrical structures significantly facilitate H2O2 activation.22,23 In their three-dimensional catalytic pockets, the synergistic effects between catalytic sites and binding sites can accelerate the proton-coupled electron transfer of selective H2O2 activation.24–26 The strong affinity between the H2O2 molecule and enzyme gives HRP excellent catalytic selectivity.27 Hence, to significantly improve the catalytic activity and selectivity of nanozymes for H2O2 activation, rational design of the active sites of nanozymes requires vivid mimicking of the catalytic pocket of HRP. Not only the binding and catalysis sites but also the spatial configuration of the active sites should be designed.
Herein, W-O-Pdene nanozymes with artificial 3D catalytic centers (asymmetric W-O-Pd sites) are synthesized successfully as highly efficient POD mimics. Asymmetric W-O-Pd sites can provide the catalytic and binding sites for H2O2 activation, which not only improve the H2O2 adsorption but also reduce the energy barrier of H2O desorption, thus efficiently achieving the improvement of H2O2 activation efficiency. Besides, owing to the unique spatial configuration, W-O-Pd sites can stretch the O–O bond of H2O2 and compress the O–O bond of O2, resulting in selectively enhanced POD-like activity, effectively preventing the interference of dissolved oxygen. Accordingly, W-O-Pdene nanozymes display more than 6 times higher POD-like activity and better affinity for H2O2 than pure Pd metallene (Pdene) nanozymes. Finally, W-O-Pdene nanozymes are used in the immunoassay of the PSA, demonstrating an ultra-low limit (1.92 pg mL−1) and practical feasibility in clinical diagnosis.
Results and discussion
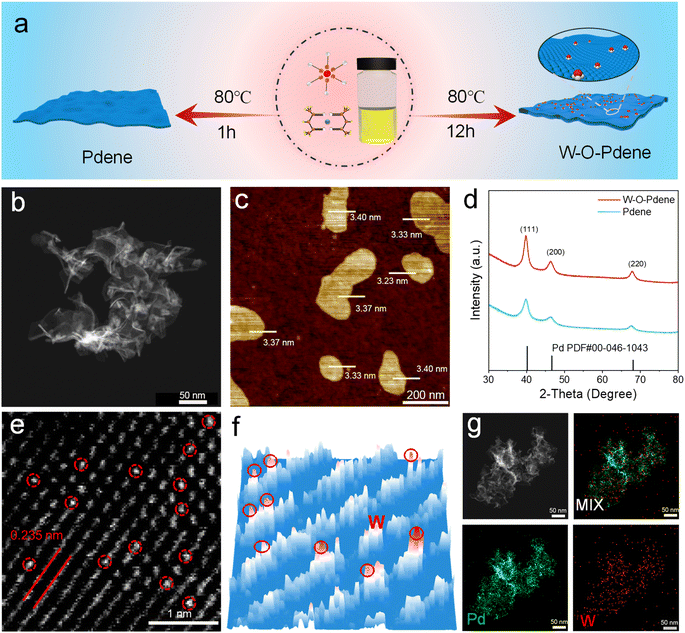
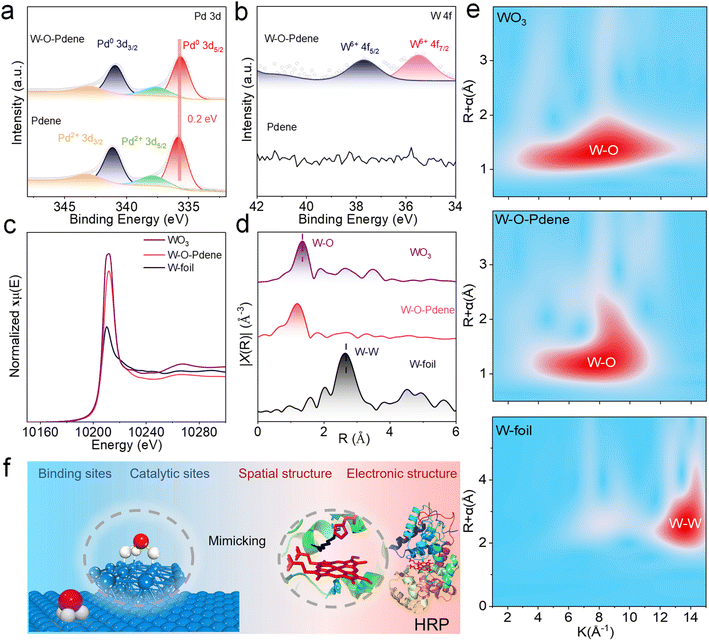
Fig. 1a exhibits a schematic diagram of the fabrication procedure of Pdene and W-O-Pdene nanozymes. The homogeneous reaction mixture, consisting of W(CO)6, ascorbic acid, and Pd(acac)2, was heated at 80 °C for one hour to obtain Pdene. In the process, the temperature-induced decomposition of W(CO)6 led to the emission of carbon monoxide (CO), which acted as a ligand directing the atomic arrangement. The CO ligand exhibits a preference for binding to the low-index (111) facet of Pd, thereby facilitating anisotropic growth. W-O-Pdene with oxygen-bridged W-Pd atomic pairs is obtained after heating for 12 h. Transmission electron microscopy (TEM) and high-angle annular dark-field scanning TEM (HAADF-STEM) images provide evidence that the Pdene and W-O-Pdene exhibit a crosslinked curved 2D nanostructure (Fig. 1b and S1a, b,†). Moreover, the atomic force microscopy (AFM) images of Pdene and W-O-Pdene indicate an average thickness of only 1.2 nm and 3.4 nm, respectively (Fig. 1c and S2†). As described, W-O-Pdene possesses ultra-thin characteristics that can provide more active sites. The X-ray diffraction (XRD) pattern of W-O-Pdene indicates good crystallinity, which is consistent with the selected area electron diffraction (SAED) result (Fig. 1d and S3†). The diffraction peaks at 39.6, 45.8, and 67.3 are indexed to the 111, 200, and 220 planes of the fcc structure of W-O-Pdene. Aberration-corrected HAADF-STEM (AC-HAADF-STEM) characterization was performed. As illustrated in Fig. 1e and f, the heavy element W appears as dense bright contrast dots within the red circles. W-O-Pdene exhibits a lattice spacing of 0.235 nm, consistent with the high-resolution TEM images of Pdene and W-O-Pdene (Fig. S4†). Besides, the energy-dispersive X-ray spectroscopy elemental mapping demonstrates the homogenous distribution of Pd and W elements within the W-O-Pdene (Fig. 1g).The chemical state of W-O-Pdene and Pdene was investigated by X-ray photoelectron spectroscopy (XPS). The location of the Pd0 peaks of W-O-Pdene in the Pd 3d spectra at 335.7 eV and 341.0 eV suggests a negative shift in comparison to Pdene (335.9 eV and 341.2 eV), indicating a clear electronic interaction (Fig. 2a). As displayed in Fig. 2b, two weak peaks at 37.7 eV and 35.5 eV can be attributed to W 4f5/2 and W 4f7/2 in W-O-Pdene. Notably, most of the W exists in the W6+ state, and no discernible peaks attributed to W0 are observed. X-ray absorption near-edge structure (XANES) and extended X-ray absorption fine structure (EXAFS) analyses were utilized to investigate the electronic structure and coordination environment in W-O-Pdene. The white line intensity of W-O-Pdene is between that of W-foil and WO3, indicating that the valence state of W is between 0 and 6 (Fig. 2c). Meanwhile, the Fourier transform W L-edge EXAFS (FT-EXAFS) spectra of W-O-Pdene reveal a single peak centered at 1.2 Å, corresponding to the W–O scattering path (Fig. 2d). In particular, the absence of the W–W peak in the FT-EXAFS spectra confirms the atomic distribution of W. Fig. S5† presents the EXAFS fitting curves, and Table S1† provides the fitting results of W-O-Pdene. The coordination number of the W–O shell is about 3. Furthermore, wavelet transform (WT) analysis was employed (Fig. 2e). The maximum intensity of the W-foil is about 13.6 Å−1, which corresponds to the W–W bond. The maximum intensity of W-O-Pdene is about 8.1 Å−1, which is like that of WO3 and corresponds to the W–O bond. Based on these results, the geometric configuration is optimized through first-principles calculations (Fig. S6†). In the most stable configuration of W-O-Pdene, W atoms are anchored on Pdene with three oxygen bridges. As displayed in Fig. 2f, the W-O-Pdene nanozyme possesses a spatially stable 3D structure. W-O-Pd asymmetric sites are successfully constructed as artificial 3D catalytic centers, and the W atom provides binding sites and catalytic sites for H2O2 activation. Like the HRP catalytic pocket, the artificial 3D catalytic centers possess unique spatial and electronic structures and can potentially enhance nanozyme activity and specificity.28–31
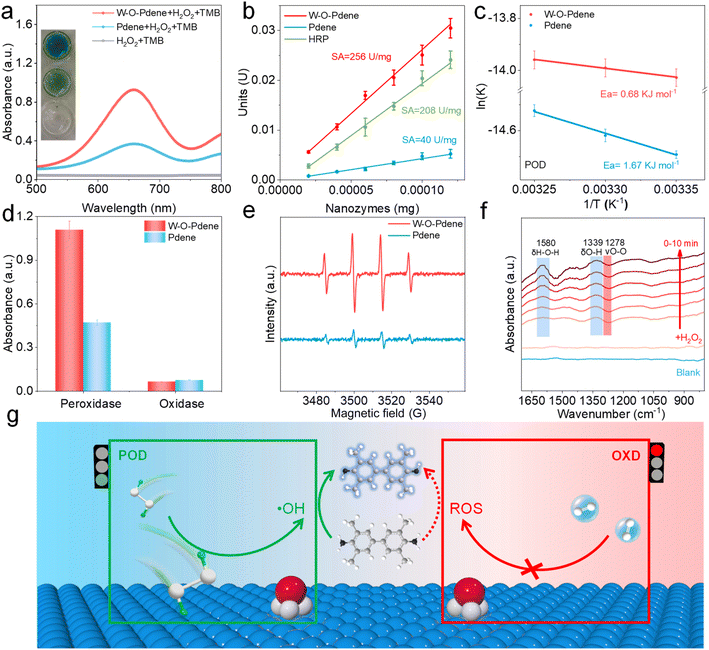
The typically chromogenic reaction (TMB/H2O2) was used to verify the POD-like activity of W-O-Pdene and Pdene. As displayed in Fig. 3a, the W-O-Pdene can rapidly oxidize TMB and produce an obvious absorbance change at 652 nm with the assistance of H2O2, and the absorbance of the W-O-Pdene system is higher than that of the Pdene system. Without nanozymes, the TMB/H2O2 system cannot trigger the absorbance changes. As shown in Fig. S7,† the Michaelis–Menten equation verified the kinetic analysis of the POD-like activity of different nanozymes at different concentrations of H2O2 and TMB, respectively. Table S2† displays the kinetic constants (Vmax and Km). On the one hand, W-O-Pdene nanozymes possess higher Vmax for both H2O2 and TMB than Pdene nanozymes, showing their higher catalytic activity; on the other hand, W-O-Pdene nanozymes have lower Km value than Pdene, which indicates that W-O-Pdene has a better affinity to H2O2. The specific activity (SA) is defined as activity units per milligram of nanozymes which can quantitatively evaluate the POD-like activity.32 As shown in Fig. 3b, the SA of W-O-Pdene (256 U mg−1) increases more than 6 times compared with Pdene (40 U mg−1) and is higher than that of HRP (208 U mg−1), which means that W-O-Pdene has better H2O2 catalytic activity than Pdene and HRP. As shown in Table S3,† compared to noble metal nanoparticle nanozymes, W-O-Pdene nanozymes have the best POD-like activity. The activation energy (Ea) of POD-like activity was calculated based on the Arrhenius equation to reveal the catalytic mechanism.33–35 As displayed in Fig. 3c, W-O-Pdene nanozymes (0.68 KJ mol−1) have a lower Ea value for POD-like activity than that of Pdene nanozymes (1.67 KJ mol−1), indicating that W-O-Pdene nanozymes can better express POD-like activity. Like natural HRP, W-O-Pdene nanozymes display pH-dependent characteristics (Fig. S8a†). Compared with natural HRP, W-O-Pdene nanozymes possess high-temperature tolerance (Fig. S8b†). Meanwhile, W-O-Pdene has better recyclability than HRP; after five cycles the POD-like activity remained unchanged (Fig. S8c†). XRD (Fig. S9a†) and TEM (Fig. S9b†) of W-O-Pdene after the reaction have not changed, indicating that W-O-Pdene has excellent stability.
In addition to POD-like activity, nanozymes usually have oxidase (OXD)-like activity which can catalyze O2 and oxidize chromogenic substrates, seriously affecting the accuracy of immunoassay. Herein, the OXD-like and POD-like activities of W-O-Pdene nanozymes and Pdene nanozymes are investigated (Fig. 3d). Impressively, compared with Pdene, W-O-Pdene nanozymes possess lower OXD-like and higher POD-like activity, demonstrating that W-O-Pdene nanozymes can enhance POD-like activity and decrease OXD-like activity. As shown in Fig. S10,† the Michaelis–Menten equation verifies the kinetic analysis of the OXD-like activity of the W-O-Pdene nanozymes and Pdene nanozymes at different concentrations of TMB. W-O-Pdene nanozymes have lower Vmax and higher Km than that of Pdene nanozymes, indicating that Pdene nanozymes have better O2 catalytic activity (Table S4†). Electrochemical tests were performed to further verify the specificity of the nanozymes, using nanozyme-modified glassy carbon electrodes to evaluate the catalytic activity of H2O2 and O2. As displayed in Fig. S11a,† W-O-Pdene nanozymes demonstrate a larger H2O2 reduction current than that of Pdene nanozymes, indicating that W-O-Pdene nanozymes have outstanding H2O2 reduction activity. In contrast, Fig. S11b† indicates that W-O-Pdene nanozymes have excellent specificity for H2O2 activation. These results convincingly demonstrate that W-O-Pdene nanozymes possess excellent specificity for H2O2 activation. Next, the active intermediates of the POD-like reaction system were confirmed by using isopropanol (IPA) as the ·OH scavenger. As shown in Fig. S12a,† when IPA is added to the POD-like catalytic system, a significant decrease in the absorbance value emerges, indicating that the produced ·OH is one of the active intermediates. Moreover, terephthalic acid (TA) can be used as a typical fluorescent probe to further detect the resulting ·OH. As shown in Fig. S12b and c,† the Fenton reaction system can react with TA and generate an obvious fluorescence signal, indicating that TA can be used as an ·OH fluorescent probe. Similarly, both Pdene and W-O-Pdene reaction systems can also react with TA to generate significant fluorescence signals, which can determine the existence of ·OH intermediates. To obtain deeper insights into the catalytic process, electron paramagnetic resonance (EPR) tests were conducted. 5,5-dimethyl-1-pyrroline N-oxide (DMPO) is used as the spin trap of ·OH. As displayed in Fig. 3e, the higher EPR signal of the W-O-Pdene catalytic system is observed, indicating that more ·OH is produced than that of the Pdene. To examine the other reactive oxygen species, nitro blue tetrazolium chloride (NBT) and Dimethyl sulfoxide (DMSO) were used to detect ·O2−and absorbed oxygen (*O) respectively. NBT can react with ·O2− to produce blue absorption at about 560 nm. Fig. S13a† indicates that there is no ·O2− in the catalytic system. DMSO can scavenge *O and ·OH, and tert-butanol (TBA) can only scavenge ·OH. As displayed in Fig. S13b,† ·OH is the main reactive intermediate in the Pdene nanozyme and W-O-Pdene nanozyme catalytic systems. Then, in situ ATR-FTIR spectroscopy was employed to monitor the process of H2O2 activation. After the addition of H2O2, two signals at 1580 cm−1 and 1339 cm−1 can be detected respectively (Fig. 3f), which can be assigned to the H–O–H bending mode and O–H of absorbed H2O2.36,37 Besides, peaks at 1278 cm−1 can be assigned to the stretching vibration of O–O in H2O2, which gradually decreased because of the partial dissociation of H2O2.38 These results indicate that the O–O bond in H2O2 can be stretched in the catalytic process (Fig. 3g).
To reveal the internal mechanism of artificial 3D catalytic centers to enhance the activity and specificity of nanozymes, density functional theory (DFT) calculations were performed. At first, the impacts of active sites are evaluated by calculating the O–O bond distance of H2O2 and O2 adsorbed on Pdene and W-O-Pdene (Fig. 4a). The dual active sites (W-O-Pd) of W-O-Pdene nanozymes can alter the adsorption conformation of H2O2 and thus achieve the extension of the O–O bond, which facilitates the activation of H2O2. Impressively, when O2 is adsorbed on the W-O-Pd site, the O–O bond of O2 is compressed and thus inhibits the O2 activation. Moreover, the analysis of charge density difference (Fig. 4b) reveals the significant electronic interaction in W-O-Pdene. Then, the projected density of states (PDOS) of the Pd d-band is employed further to indicate the electronic interaction effects (Fig. 4c). The d-band center of W-O-Pdene (−1.768 eV) exhibits a downshift relative to that of Pdene (−1.465 eV), indicating that the electronic interaction of W-O-Pd can optimize the electronic structure of W-O-Pdene nanozymes and accelerate the desorption process of intermediates. Furthermore, Fig. 4d displays the schematic representation of the fundamental reaction step and energy change diagram of H2O2 activation on nanozymes. H2O2 is first absorbed on the W-O-Pd active sites. Then H2O2 is cleaved into two absorbed hydroxyl groups (OH*). Next, the two absorbed OH* are protonated to constitute H2O* and desorb subsequently. W-O-Pdene nanozymes have a better capacity for capturing H2O2 because the adsorption energy of W-O-Pdene (−1.53 eV) is lower than that of Pdene (−1.03 eV). The step of desorption of the H2O can be regarded as the rate-determining step (RDS). The RDS energy barrier of W-O-Pdene (0.82 eV) is lower than that of Pdene (1.19 eV), explaining the superiority of the POD-like activity of W-O-Pdene nanozymes. This result is consistent with the PDOS analysis. Besides, the weakened H2O* binding on W-O-Pdene can be further evidenced by crystal orbital Hamilton population (COHP) analysis (Fig. 4e); pCOHP analysis reveals that the -ICOHP value at the Fermi level is lower for W-O-Pdene (2.434 eV) than that of Pdene (2.916 eV), which means that the H2O* is easier to desorb on W-O-Pdene. Accordingly, W-O-Pdene nanozymes have high activity and specificity for the catalysis of H2O2, which is due to the synergistic effect of Pd sites and axial W atoms in the artificial 3D catalytic centers. On the one hand, the axial W atoms play an electronic role in the regulation of the Pd sites, resulting in a better desorption to enhance POD. On the other hand, the axial W atoms play an important role in binding for catalysis of H2O2, superior spatial configuration improving the specificity (Fig. S14†).
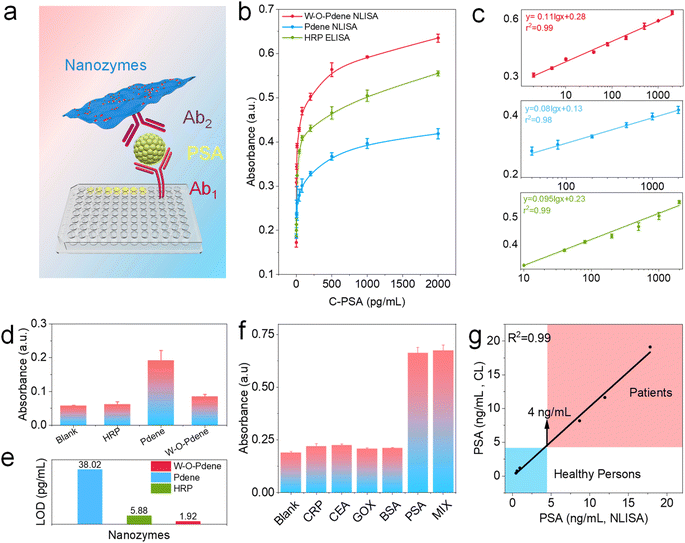
As the specific marker for prostate cancer, PSA has been widely used for the early diagnosis of prostate cancer during routine check-ups.39–42 To investigate the potential application of W-O-Pdene for immunoassays, a colorimetric nanozyme-linked immunosorbent assay (NLISA) of PSA was performed.43–46 As displayed in Fig. 5a, NLISA is implemented through the typical sandwich immunoassay; anti-PSA coating monoclonal antibody (Ab1) can capture the target PSA, subsequently, PSA is captured by anti-PSA labeling monoclonal antibody (Ab2) to form the sandwich immunocomplexes. As displayed in Fig. 5b and c, with the increase of PSA concentration, the associated absorbance signals are consequently increased, demonstrating the feasibility of NILISA for the detection of PSA. The interference of dissolved oxygen with the background signal was investigated. As displayed in Fig. 5d, HRP has low background signals because of the superior specificity. Compared with Pdene nanozymes, W-O-Pdene nanozymes can decrease the background signal of immunoassay. Fig. 5e displays the limit of detection (LOD) of NLISA and ELISA. Interestingly, the LOD of W-O-Pdene-involved NLISA is determined to be 1.92 pg mL−1, and the LOD of HRP-involved ELISA and Pdene-involved NLISA is calculated to be 5.88 and 38.02 pg mL−1, respectively. The enhancement in the detection sensitivity for W-O-Pdene NLISA can be ascribed to the high catalytic efficiency and specificity of the W-O-Pdene. Furthermore, the specificity of PSA detection is demonstrated by the negligible responses observed for other biomarkers such as C-reactive protein (CRP), carcinoembryonic antigen (CEA), glucose oxidase (GOX), and bovine serum albumin (BSA) at a concentration of 2 ng mL−1 (Fig. 5f). Due to the superior sensing performance of NLISA, it is hopeful to be used in clinical diagnosis. Therefore, to verify the practicability in clinical diagnosis, NLISA was employed to detect human serum samples. As displayed in Fig. 5g and Table S5,† NLISA gets a linear fit greater than 99% with the chemiluminescence (CL) method in the hospital, verifying the practicability of NLISA in practical applications.
Conclusions
Inspired by the natural HRP catalytic pocket, W-O-Pdene nanozymes with artificial 3D catalytic centers are prepared successfully and demonstrate specific H2O2 activation efficiency. The catalytic activity and selectivity of W-O-Pdene nanozymes are systematically investigated based on experimental and theoretical analysis. 3D asymmetric W-O-Pd atomic pairs can selectively regulate the O–O bonding in H2O2 and O2, and thus achieve the selective H2O2 activation. Besides, the asymmetric W-O-Pd atomic pairs can regulate the d-band center to accelerate the desorption of H2O, which improves the catalytic activity of W-O-Pdene nanozymes. As a typical application, a colorimetric immunoassay for PSA is established, demonstrating better sensitivity than an enzyme-linked immunosorbent assay. This work offers new insight into fabricating unique POD-like nanozymes with high activity and selectivity.Data availability
The data supporting this article have been included as part of the ESI.†Author contributions
Conceptualization: L. J.; formal analysis: C. C., D. Y., X. J., R. L., L. H., and X. L.; funding acquisition: X. L., L. J., and Y. Z.; methodology: C. C. and L. J.; writing – original draft: C. C.; writing – review & editing: L. J., Y. Z., C. Z., and X. L. All authors discussed the results and assisted during manuscript preparation.Conflicts of interest
There are no conflicts to declare.Acknowledgements
The authors gratefully acknowledge the financial support of the National Natural Science Foundation of China (22174110, 22127803, and 22304096), the project ZR2023QB006 supported by Shandong Provincial Natural Science Foundation, supported by the China Postdoctoral Science Foundation under Grant Number 2023M741853, the Qingdao Postdoctoral Science Foundation (QDBSH20230202031), and the Plan for Youth Innovation Team of Colleges in Shandong Province.Notes and references
- H. Wei and E. Wang, Chem. Soc. Rev., 2013, 42, 6060–6093 RSC.
- H. Fan, J. Zheng, J. Xie, J. Liu, X. Gao, X. Yan, K. Fan and L. Gao, Adv. Mater., 2024, 36, e2300387 CrossRef PubMed.
- L. Zhang, H. Wang and X. Qu, Adv. Mater., 2024, 36, e2211147 CrossRef PubMed.
- C. Peng, R. Pang, J. Li and E. Wang, Adv. Mater., 2024, 36, e2211724 CrossRef.
- A. Zhang, A. Gao, C. Zhou, C. Xue, Q. Zhang, J. M. Fuente and D. Cui, Adv. Mater., 2023, 35, e2303722 CrossRef PubMed.
- S. Zhang, Y. Li, S. Sun, L. Liu, X. Mu, S. Liu, M. Jiao, X. Chen, K. Chen, H. Ma, T. Li, X. Liu, H. Wang, J. Zhang, J. Yang and X. D. Zhang, Nat. Commun., 2022, 13, 4744 CrossRef CAS.
- Y. Wang and Y. Xianyu, Small Methods, 2022, 6, e2101576 CrossRef PubMed.
- D. Jiang, D. Ni, Z. T. Rosenkrans, P. Huang, X. Yan and W. Cai, Chem. Soc. Rev., 2019, 48, 3683–3704 RSC.
- M. Liang and X. Yan, Acc. Chem. Res., 2019, 52, 2190–2200 CrossRef CAS PubMed.
- M. Zandieh and J. Liu, Adv. Mater., 2024, 36, e2211041 CrossRef PubMed.
- C. Shang, Q. Wang, H. Tan, S. Lu, S. Wang, Q. Zhang, L. Gu, J. Li, E. Wang and S. Guo, JACS Au, 2022, 2, 2453–2459 CrossRef CAS.
- L. Jiao, H. Yan, Y. Wu, W. Gu, C. Zhu, D. Du and Y. Lin, Angew. Chem., Int. Ed., 2020, 59, 2565–2576 CrossRef CAS PubMed.
- Y. Xu, Z. Zhou, N. Deng, K. Fu, C. Zhu, Q. Hong, Y. Shen, S. Liu and Y. Zhang, Sci. China: Chem., 2023, 66, 1318–1335 CrossRef CAS.
- Z. Zhang, X. Zhang, B. Liu and J. Liu, J. Am. Chem. Soc., 2017, 139, 5412–5419 CrossRef CAS PubMed.
- X. Chen, L. Zhao, K. Wu, H. Yang, Q. Zhou, Y. Xu, Y. Zheng, Y. Shen, S. Liu and Y. Zhang, Chem. Sci., 2021, 12, 8865–8871 RSC.
- Z. Wang, R. Zhang, X. Yan and K. Fan, Mater. Today, 2020, 41, 81–119 CrossRef CAS.
- Y. Chen, L. Jiao, H. Yan, W. Xu, Y. Wu, L. Zheng, W. Gu and C. Zhu, Anal. Chem., 2021, 93, 12353–12359 CrossRef CAS PubMed.
- Y. Wang, G. Jia, X. Cui, X. Zhao, Q. Zhang, L. Gu, L. Zheng, L. H. Li, Q. Wu, D. J. Singh, D. Matsumura, T. Tsuji, Y.-T. Cui, J. Zhao and W. Zheng, Chem, 2021, 7, 436–449 CAS.
- J. Han, H. Gong, X. Ren and X. Yan, Nano Today, 2021, 41, 101295 CrossRef CAS.
- Y. Xu, A. Huang, W. Yi, G. Chen, S. Huang and G. Ouyang, Coord. Chem. Rev., 2024, 500, 215517 CrossRef CAS.
- Y. Xu, Z. Mao, J. Zhang, J. Ji, Y. Zou, M. Dong, B. Fu, M. Hu, K. Zhang, Z. Chen, S. Chen, H. Yin, P. Liu and H. Zhao, Angew. Chem., Int. Ed., 2024, 63, e202316029 CrossRef CAS.
- A. P. Green, T. Hayashi, P. R. E. Mittl and D. Hilvert, J. Am. Chem. Soc., 2016, 138, 11344–11352 CrossRef CAS.
- R. Geng, R. Chang, Q. Zou, G. Shen, T. Jiao and X. Yan, Small, 2021, 17, e2008114 CrossRef.
- L. Xie, X.-P. Zhang, B. Zhao, P. Li, J. Qi, X. Guo, B. Wang, H. Lei, W. Zhang, U.-P. Apfel and R. Cao, Angew. Chem., Int. Ed., 2021, 60, 7576–7581 CrossRef CAS.
- G. Li, H. Liu, T. Hu, F. Pu, J. Ren and X. Qu, J. Am. Chem. Soc., 2023, 145, 16835–16842 CrossRef CAS PubMed.
- P. Campomanes, U. Rothlisberger, M. Alfonso-Prieto and C. Rovira, J. Am. Chem. Soc., 2015, 137, 11170–11178 CrossRef CAS PubMed.
- W. Zhang, J. J. Dynes, Y. Hu, P. Jiang and S. Ma, Nat. Commun., 2019, 10, 1913 CrossRef PubMed.
- Z. Lian, Y. Lu, S. Zhao, Z. Li and Q. Liu, Adv. Sci., 2023, 10, 2205975 CrossRef CAS.
- Z. Li, S. Ji, C. Xu, L. Leng, H. Liu, J. H. Horton, L. Du, J. Gao, C. He, X. Qi, Q. Xu and J. Zhu, Adv. Mater., 2023, 35, 2209644 CrossRef CAS.
- X. Li, G. Hai, J. Liu, F. Zhao, Z. Peng, H. Liu, M. K. H. Leung and H. Wang, Appl. Catal., B, 2022, 314, 121531 CrossRef CAS.
- Q. Wang, K. Chen, H. Jiang, C. Chen, C. Xiong, M. Chen, J. Xu, X. Gao, S. Xu, H. Zhou and Y. Wu, Nat. Commun., 2023, 14, 5338 CrossRef CAS.
- B. Jiang, D. Duan, L. Gao, M. Zhou, K. Fan, Y. Tang, J. Xi, Y. Bi, Z. Tong, G. F. Gao, N. Xie, A. Tang, G. Nie, M. Liang and X. Yan, Nat. Protoc., 2018, 13, 1506–1520 CrossRef CAS.
- T. Chen, Y. Zhang and W. Xu, J. Am. Chem. Soc., 2016, 138, 12414–12421 CrossRef CAS PubMed.
- Z. D. Hood, H. Wang, A. Samuthira Pandian, J. K. Keum and C. Liang, J. Am. Chem. Soc., 2016, 138, 1768–1771 CrossRef CAS.
- S. E. Henkelis, M. Mazur, C. M. Rice, P. S. Wheatley, S. E. Ashbrook and R. E. Morris, J. Am. Chem. Soc., 2019, 141, 4453–4459 CrossRef CAS PubMed.
- S. Mondal, D. Bagchi, M. Riyaz, S. Sarkar, A. K. Singh, C. P. Vinod and S. C. Peter, J. Am. Chem. Soc., 2022, 144, 11859–11869 CrossRef CAS PubMed.
- Y. Wu, J. Wen, W. Xu, J. Huang, L. Jiao, Y. Tang, Y. Chen, H. Yan, S. Cao, L. Zheng, W. Gu, L. Hu, L. Zhang and C. Zhu, Small, 2021, 17, e2101907 CrossRef PubMed.
- Y. Tang, Y. Chen, Y. Wu, W. Xu, Z. Luo, H. R. Ye, W. Gu, W. Song, S. Guo and C. Zhu, Nano Lett., 2023, 23, 267–275 CrossRef CAS.
- Z. Xi, K. Wei, Q. Wang, M. J. Kim, S. Sun, V. Fung and X. Xia, J. Am. Chem. Soc., 2021, 143, 2660–2664 CrossRef CAS.
- S. Xie, X. Fei, J. Wang, Y.-C. Zhu, J. Liu, X. Du, X. Liu, L. Dong, Y. Zhu, J. Pan, B. Dong, J. Sha, Y. Luo, W. Sun and W. Xue, Adv. Sci., 2023, 10, 2206494 CrossRef CAS.
- L. Kachuri, T. J. Hoffmann, Y. Jiang, S. I. Berndt, J. P. Shelley, K. R. Schaffer, M. J. Machiela, N. D. Freedman, W. Y. Huang, S. A. Li, R. Easterlin, P. J. Goodman, C. Till, I. Thompson, H. Lilja, S. K. Van Den Eeden, S. J. Chanock, C. A. Haiman, D. V. Conti, R. J. Klein, J. D. Mosley, R. E. Graff and J. S. Witte, Nat. Med., 2023, 29, 1412–1423 CrossRef CAS PubMed.
- E. Shenderov, A. M. De Marzo, T. L. Lotan, H. Wang, S. Chan, S. J. Lim, H. Ji, M. E. Allaf, C. Chapman, P. A. Moore, F. Chen, K. Sorg, A. M. White, S. E. Church, B. Hudson, P. A. Fields, S. Hu, S. R. Denmeade, K. J. Pienta, C. P. Pavlovich, A. E. Ross, C. G. Drake, D. M. Pardoll and E. S. Antonarakis, Nat. Med., 2023, 29, 888–897 CrossRef CAS PubMed.
- M. Broto, M. M. Kaminski, C. Adrianus, N. Kim, R. Greensmith, S. Dissanayake-Perera, A. J. Schubert, X. Tan, H. Kim, A. S. Dighe, J. J. Collins and M. M. Stevens, Nat. Nanotechnol., 2022, 17, 1120–1126 CrossRef CAS.
- R. Li, H. Fan, H. Zhou, Y. Chen, Q. Yu, W. Hu, G. L. Liu and L. Huang, Adv. Sci., 2023, 10, 2301658 CrossRef CAS.
- Z. Farka, V. Čunderlová, V. Horáčková, M. Pastucha, Z. Mikušová, A. Hlaváček and P. Skládal, Anal. Chem., 2018, 90, 2348–2354 CrossRef CAS.
- N. Ye, S. Huang, H. Yang, T. Wu, L. Tong, F. Zhu, G. Chen and G. Ouyang, Anal. Chem., 2021, 93, 13981–13989 CrossRef CAS PubMed.
Footnote |
| † Electronic supplementary information (ESI) available. See DOI: https://doi.org/10.1039/d4sc04711g |
| This journal is © The Royal Society of Chemistry 2024 |