Open Access Article
Open Access ArticleSelective inhibition of cancer cell migration using a pH-responsive nucleobase-modified DNA aptamer†
Yuyuan
Chen‡
,
Kunihiko
Morihiro‡
 *,
Yui
Nemoto
,
Akito
Ichimura
,
Ryosuke
Ueki
,
Shinsuke
Sando
*,
Yui
Nemoto
,
Akito
Ichimura
,
Ryosuke
Ueki
,
Shinsuke
Sando
 and
Akimitsu
Okamoto
and
Akimitsu
Okamoto
 *
*
Department of Chemistry and Biotechnology, Graduate School of Engineering, The University of Tokyo, 7-3-1 Hongo, Bunkyo-ku, Tokyo 113-8656, Japan. E-mail: morihiro@chembio.t.u-tokyo.ac.jp; okamoto@chembio.t.u-tokyo.ac.jp
First published on 24th September 2024
Abstract
Because of the extracellular acidic microenvironment of cancer cells, many pH-responsive molecules have become indispensable materials for bioanalysis and targeted therapy development. pH-Responsive DNA aptamers, which selectively bind to target proteins in cancer cells, have become a key research target in the therapeutic field. However, conventional pH-responsive aptamers have fatal drawbacks, such as complex structures, sequence limitation, and difficulties in mass production, as they require special nucleic acid structures, including the i-motif and DNA triplex. To address these issues, we utilized AnC, which is an unnatural nucleobase with a pKaH of 5.9, to construct a simple pH-responsive DNA aptamer (CSL1-II) for selective binding to the c-Met protein expressed in cancer cells. CSL1-II in a weakly acidic environment had a stronger inhibitory effect on the HGF/c-Met pathway and exerted a strong controlling effect on the spreading and migration of cancer cells. Our strategy provides a simple and versatile method to develop pH-responsive DNA aptamers and represents the first example of a cancer-selective c-Met antagonist that inhibits cell migration.
Introduction
Cancer is a major health threat that often leads to death through metastasis. Cancer cell migration is a crucial step in this process, as it enables invasion from primary tumors and the formation of secondary metastases.1 Therefore, synthetic molecules that inhibit cancer cell migration are promising candidates for antitumor therapy. c-Met signaling is believed to be pivotal among the intricate molecular processes of metastasis.2 c-Met serves as the cell-surface receptor for the hepatocyte growth factor (HGF), and its activation occurs through HGF binding, leading to transphosphorylation of the intracellular domain of c-Met and subsequent downstream signaling events. Many research findings indicate that the overexpression of c-Met promotes the proliferation, migration, and invasion of cancer cells. Therefore, the c-Met pathway is a critical target for cancer therapy.3 To hinder the interaction between c-Met and HGF, as well as the resulting cellular signal transduction, various c-Met inhibitors have been developed, including antibodies and peptide drugs aimed at inhibiting cancer cell migration.4 However, the lower cancer selectivity of these c-Met inhibitors hinders the migration of both cancer and normal cells, leading to adverse effects.The in situ construction of a c-Met inhibitor that targets the tumor microenvironment exclusively is the ideal approach to overcome the challenge of undesired activity in normal cells. We have developed stimuli-responsive materials that react to cues in the form of changes in several environmental parameters, including the redox environment,5 light,6 and the concentration of reactive oxygen species7 and oxygen.8 Among them, pH-sensitive materials have shown promise because of their relevance in biology, given the pH differences that exist between the many tissues and cellular compartments in the human body.9,10 For instance, numerous studies have indicated that the pH of tumor cells is 0.5–1 pH units lower than the pH in surrounding normal cells, because of metabolic glycolysis and lactic acid production.11,12 Based on the weakly acidic nature of tumor tissues, pH-responsive materials have been utilized in cancer detection and drug delivery to develop selective cancer diagnostic and therapeutic strategies.13,14
The structure and function of biomolecules, especially nucleic acids, are often affected by pH. The i-motif structure, which is a unique quadruplex configuration, is formed in mildly acidic conditions and undergoes disassembly in neutral conditions.15–17 In turn, triplex DNA configurations involve the specific binding of an auxiliary single-stranded DNA to the major groove of duplex DNA via Hoogsteen or reverse Hoogsteen interactions, resulting in the generation of supramolecular DNA assemblies. The C(H+)·G-C triad in DNA triplexes is formed under a mildly acidic pH and dissociates at a neutral pH.18,19 The i-motif and triplex structures are often utilized in designing nucleic acid molecules for pH-responsive applications, such as drug-delivery systems,20 medical materials,21,22 and intracellular pH imaging techniques.23–25
An aptamer is a synthetic single-stranded RNA or DNA molecule with a length typically ranging from approximately 15 to 100 nucleotides that specifically binds to target molecules, such as proteins, nucleic acids, and small molecules.26 The DNA aptamer SL1, a 50-nucleotide-long molecule that acts as a c-Met antagonist, has been developed to compete with HGF and inhibit c-Met phosphorylation and cancer cell migration.27 Tan and co-workers developed pH-responsive DNA aptamers targeting tyrosine kinase-7 by incorporating an i-motif-forming sequence into the stem region. The aptamer exhibited strong binding to target cancer cells under acidic conditions (pH 6.5) compared with neutral-pH conditions (pH 7.3).28 Moreover, Soh and co-workers demonstrated that the insertion of a triplex-forming oligonucleotide (TFO) into an ATP aptamer achieved pH-induced selectivity in affinity of up to 1000-fold.29 However, there would be several challenges, such as ensuring in vivo stability and the technical and cost-related difficulties of synthesizing large quantities of nucleic acids with specific sequences. Furthermore, the design of conventional pH-responsive DNA molecules is limited because i-motifs require a cytosine-rich sequence, and DNA triplexes require homopyrimidine and homopurine sequences. In addition, if pH responsiveness is to be imparted to functional DNA, these methods require the addition of sequences, thus elongating the pH-responsive DNA. Here, we developed a pH-responsive aptamer by regulating DNA duplex formation. This aptamer inhibited the migration of cancer cells without affecting normal cells. Our approach represents a simple method for the development of pH-responsive nucleic acid therapeutics, materials, and nanotechnologies.
Results and discussion
We introduced the pH-responsive artificial nucleobase, AnC,30 into a DNA aptamer targeting c-Met (CSL1) (Fig. 1A). As depicted in Fig. 1B, AnC-modified CSL1 remained unstructured at the physiological pH of normal tissues, thereby impeding binding to c-Met. Subsequently, HGF-dependent c-Met dimerization occurred, leading to phosphorylation at its intracellular domain and consequent activation of signaling pathways that promote cell migration. However, in the weakly acidic microenvironment of tumor tissues, AnC-modified CSL1 formed active structures and bound to c-Met, leading to the inhibition of c-Met dimerization and signal activation, as well as cancer cell migration. The AnC-modified CSL1 aptamer did not require additional sequences, such as the i-motif and TFO; rather, it was obtained by replacing the natural nucleobases in a DNA aptamer with AnC. AnC-modified CSL1 holds potential for application as a selective nucleic acid drug targeting the cancer microenvironment to minimize cytotoxicity to normal tissues by avoiding undesired binding to c-Met. This approach is the first example demonstrating the pH-based control of DNA aptamer activity without relying on the formation of unusual DNA structures, such as the i-motif and DNA triplexes.The unnatural nucleobase AnC, which is capable of responding to a weakly acidic environment, was synthesized previously by our group.30 The pKaH of the AnC nucleoside is 5.9, and it can form an AnC(H+):C base pair through two hydrogen bonds in DNA duplexes exclusively under weakly acidic conditions. In turn, under neutral or alkaline conditions, the introduction of AnC in place of guanine in DNA duplexes leads to the destabilization of the double-stranded structures. The substitution of guanine with AnC enables pH-responsive control over the double-stranded and single-stranded states of DNA without the need for additional sequences. Any double-stranded DNA can achieve pH responsiveness as long as guanines are replaced with AnC nucleobases. SL1 comprises a guanine-rich loop region (G-loop) and a stem region consisting of eight base pairs, including a G:T mismatch. To improve the thermal stability of SL1, we substituted the T in the G:T mismatch with C, resulting in the sequence that was termed CSL1. The anticipated stability of the stem region, as assessed using NUPACK, which is an online software for predicting the thermal stability of nucleic acid structures, suggested that CSL1 is more conducive to aptamer activity compared with the original SL1 (Fig. 2A and B). In addition, by replacing one, two, or three guanines in the stem sequence with AnC, CSL1 variants with three different stem sequences, i.e., CSL1-I, CSL1-II, and CSL1-III, respectively, were designed (Fig. 2C). The synthesis of AnC-modified DNA was carried out using the classical phosphoramidite synthesis method, and the products were identified by HPLC and MALDI-TOF MS (Fig. S1 and S2†). We confirmed that CSL1 showed similar affinity to human lung cancer A549 cells (Kd = 43.1 nM), which express c-Met,31 as SL1 (Kd = 41.2 nM) by flow cytometry. However, CSL1-I, CSL1-II, and CSL1-III exhibited lower affinity (Kd = 58.1, 52.6, and 59.7 nM, respectively) under neutral conditions (Fig. S3†).
According to a previous report, the binding affinity of the c-Met aptamer SL1 to cells expressing c-Met decreases as the stem base is truncated.27 Based on this observation, we hypothesized that the binding capability of CSL1 to c-Met can be predicted by assessing the stability of the stem region. To evaluate the thermal stability of the stem region of CSL1 under neutral and slightly acidic conditions, the melting temperature (Tm) was measured (Table 1). The pH of the solution was adjusted to achieve pH values of 7.4 and 6.4, to mimic the normal and tumor tissue environments, respectively. The melting curves showed that the calculated Tm values reflected the stability of both the stem region and the G-loop (Fig. S4†). Sensitivity to an acidic environment was assessed using the ΔTm value, which was calculated as the difference between the Tm value of the CSL1 series at pH 7.4 and pH 6.4. CSL1 exhibited a ΔTm value of 1 °C, indicating that the stem region of unmodified CSL1 did not have pH sensitivity. In contrast, the ΔTm value of CSL1-I, which carries one AnC, was 5 °C, and that of CSL1-II, which carries two AnC, was 8 °C. As the number of AnC modifications increased, the ΔTm values also increased, indicating that the pH responsiveness of AnC is reflected in the CSL1 aptamer. CSL1-III (with three AnC) did not display a detectable Tm value, even at pH 6.4, probably because of the instability triggered by replacing stable G:C base pairs with AnC(H+):C base pairs, which form only two hydrogen bonds. The results of the CD measurements indicate that the Tm values primarily reflect the stability of the stem region of the DNA aptamers (Fig. S5†). In a 10 mM phosphate buffer (pH 7.4) with 10 mM potassium ions, the CD spectra of all the aptamers showed a negative Cotton effect at 245 nm and a positive Cotton effect at 265 nm, which are characteristic spectral features of a parallel-type G-quadruplex structure. However, the signal intensities were very low without potassium ions, suggesting that almost no G-quadruplexes were formed under the Tm measurement conditions.
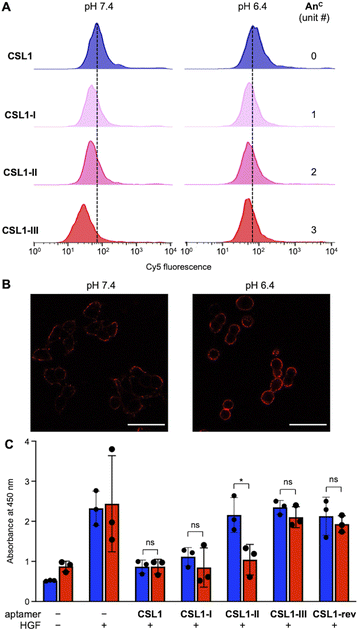
Flow cytometry was performed to investigate the changes in binding affinity between the CSL1 aptamers and c-Met-expressing cells (Fig. 3A). CSL1 aptamers labeled with fluorescent Cy5 were incubated with A549 cells. At pH 7.4, the affinity of CSL1 aptamers to A549 cells decreased as the number of AnC modifications increased. In particular, CSL1-III (containing three AnC) exhibited a significant decrease in binding affinity. The tendency toward decreased fluorescence suggests that the increase in AnC nucleobases in the stem region of CSL1 destabilizes the active structure, leading to a reduced binding affinity to A549 cells under neutral conditions. Conversely, the decrease in binding affinity was less pronounced at pH 6.4. This is believed to be attributed to the stabilization of the stem structure of CSL1 through the formation of AnC(H+):C base pairs, thereby maintaining its binding affinity toward c-Met in A549 cells. We examined the pH-responsive behavior of AnC-modified DNA aptamers in relation to c-Met expression using SNU-1 cells, which express low levels of c-Met (Fig. S6†). No significant change in binding affinity was observed for any of the DNA aptamers under either neutral or acidic conditions, indicating that the observed effects of the pH-responsive aptamers are specific to c-Met-positive cells. We confirmed the presence of the CSL1 aptamers at the cell membrane, rather than within cells, using confocal laser scanning microscopy. CSL1-II was introduced into A549 cells, and the fluorescence emitted by the cells was imaged (Fig. 3B). Cy5 fluorescence was observed surrounding the cells, indicating that CSL1-II bound to c-Met at the cell membrane. Furthermore, the fluorescence intensity was higher at pH 6.4 compared with pH 7.4, further confirming the pH responsiveness of CSL1-II.
To test the inhibitory effect of the CSL1-II aptamers on the c-Met/HGF signaling pathway, the HGF-induced c-Met phosphorylation levels were evaluated in A549 cells using an enzyme-linked immunosorbent assay (ELISA). We initially determined the optimal concentration of HGF for ELISA, i.e., 250 pM (Fig. S7†). Subsequently, different concentrations of CSL1 and the CSL1 G-loop were tested (Fig. S8†). The CSL1 G-loop, which is a stemless sequence rich in guanine that is present in the loop region of CSL1, exhibited a concentration-dependent inhibitory activity on c-Met phosphorylation, suggesting that the guanine tetrad portion of CSL1 is an essential binding site for c-Met (Fig. S9†). To accurately examine the relationship between CSL1 stem stability and HGF activity inhibition, the concentration at which CSL1 exerted HGF activity inhibition, and the CSL1 G-loop did not, was considered ideal. Therefore, 10 nM was determined as the optimal concentration of the CSL1 aptamers for subsequent experiments.
Under the optimized conditions, the pH switching of the antagonist activity of the CSL1 aptamers toward c-Met was evaluated by ELISA (Fig. 3C). CSL1-rev, which is the reverse sequence of CSL1, was used as a negative control that did not bind to c-Met. Compared with cells without the addition of any substance, cells with addition of HGF exhibited higher levels of phosphorylated c-Met. CSL1 composed of natural nucleobases greatly inhibited phosphorylation at both pH 7.4 and 6.4. CSL1-I also inhibited phosphorylation, regardless of the pH value, probably because the stem region is stable, and the aptamer activity was maintained at both pH values. Moreover, CSL1-II yielded a weaker inhibition of c-Met phosphorylation at pH 7.4, whereas it provided a stronger inhibition at pH 6.4. The fine-tuning activity toward a pH change agreed with the result of the Tm measurement and ELISA experiments. CSL1-III afforded a weak inhibition activity at both pH 7.4 and 6.4, indicating that CSL1-III is too unstable to form an active structure. The results of ELISA suggest that CSL1-II, which is modified with two AnC, was the most promising aptamer for the selective inhibition of c-Met phosphorylation in a weakly acidic environment.
CSL1-II, which was the most promising pH-responsive c-Met DNA aptamer, afforded selective inhibition of cancer cell migration. A wound healing assay was performed using human pancreatic cancer SUIT-2 cells and normal embryonic lung fibroblast MRC-5 cells. Both cell types express c-Met on their surface.32,33 Under conditions lacking the addition of HGF and pH-responsive CSL1-II, there was no significant closure of the gap between cells (Fig. 4A). In contrast, the addition of HGF alone and both HGF and the negative control sequence CSL1-rev led to the observation of numerous small cells filling the gap between the cell spaces. However, in the presence of pH-responsive CSL1-II, this cell migration was inhibited, and a significant amount of space remained in the artificial gap. The difference in cell-migration-inhibiting ability observed between pH-responsive CSL1-II and CSL1-rev clearly demonstrates that pH-responsive CSL1-II effectively inhibited the cancer cell migration induced by c-Met signaling. In contrast, the inhibitory effect of pH-responsive CSL1-II on the migration of MRC-5 cells was much weaker compared with that observed for SUIT-2 cells (Fig. 4B). The migration rate was quantified by comparing the area of the blank region before and after the artificial gap between cells. The addition of pH-responsive CSL1-II effectively suppressed the HGF-induced migration of SUIT-2 cells, clearly indicating that pH-responsive CSL1-II inhibited the motility of cancer cells activated by c-Met (Fig. 4C). However, we did not observe a significant inhibitory effect of pH-responsive CSL1-II in MRC-5 cells (Fig. 4D). The comparison of the cell recovery rates between SUIT-2 and MRC-5 cells indicated the occurrence of cancer-selective cell migration inhibition by pH-responsive CSL1-II (Fig. 4E). A time-course analysis revealed that the migration was almost complete within 16 h in the absence of pH-responsive CSL1-II, thus demonstrating the powerful migration-inhibition ability of our system (Fig. 4F and Movie S1†).
It is well known that the culture medium of cancer cells becomes gradually more acidic than that of normal cells because of the increase in the levels of lactate.34,35 We measured the pH values of the culture media of SUIT-2 and MRC-5 cells, and observed that the SUIT-2 culture medium alone exhibited a gradual decrease in pH over 7 days (Fig. S10†). This reduction in pH indicated that the microenvironment of SUIT-2 cells is sufficiently acidic for the protonation of AnC. The disparity in migration rates observed between SUIT-2 and MRC-5 cells suggests that pH-responsive CSL1-II responds to the weakly acidic environment, thereby exerting its anticancer function. Cell wound assays further confirmed that the pH-responsive CSL1-II aptamer designed here formed an active structure, bound to c-Met, inhibited the c-Met/HGF signaling pathway, and consequently suppressed cancer cell migration and dissemination specifically within the cancer microenvironment.
Conclusions
We designed a series of weakly acidic environment-responsive c-Met aptamers (CSL1 aptamers) via the incorporation of the artificial nucleobase AnC. As the number of AnC nucleobases in the stem region increased, we observed an increasing difference in melting temperatures between pH 7.4 and pH 6.4 (i.e., weak acid responsiveness). The assessment of the binding affinity of the CSL1 aptamers to c-Met-expressing cancer cells revealed that it decreased as the number of AnC increased at pH 7.4, whereas the affinity was maintained at pH 6.4, indicating that the formation of AnC(H+):C base pairs is important for the binding to c-Met. CSL1-II, which contains two AnC nucleobases, selectively inhibited c-Met phosphorylation at pH 6.4, which corresponded to a weak acid responsiveness. Furthermore, pH-responsive CSL1-II effectively suppressed cancer cell migration while being ineffective against normal cells, which confirmed its specificity toward cancer cells. Because of this unique property, pH-responsive CSL1-II is likely to emerge as a promising molecular platform for preventing tumor development and metastasis without adverse effects on normal tissues. The application of the pH-responsive nucleobase AnC to other DNA aptamers and pH-responsive nanodevices holds promise in fields such as cancer therapy and diagnosis.Data availability
All the data supporting this study are included in the main text and the ESI.†Author contributions
K. M. and A. O. conceived and directed the study. Y. C., Y. N, and A. I. performed the experiments and analyzed the data with the aid of S. S. All the authors prepared the manuscript.Conflicts of interest
There are no conflicts to declare.Acknowledgements
This work was supported by JSPS KAKENHI grants (23H00317 to K. M. and A. O., 23K17969 and 24H02214 to A. O.), AMED grant (23ak0101194h0001 to K. M., 24ak0101231h0001 to A. O.), the Shorai Foundation for Science and Technology (to K. M.), and the Noguchi Institute (to K. M.). The table of contents graphic and Fig. 1B were created using the tools provided by https://www.biorender.com/.Notes and references
- F. van Zijl, G. Krupitza and W. Mikulits, Mutat. Res., 2011, 728, 23–34 CAS.
- L. Trusolino and P. M. Comoglio, Nat. Rev. Cancer, 2002, 2, 289–300 CrossRef PubMed.
- Y. Zhang, M. Xia, K. Jin, S. Wang, H. Wei, C. Fan, Y. Wu, X. Li, G. Li, Z. Zeng and W. Xiong, Mol. Cancer, 2018, 17, 45 CrossRef PubMed.
- A. Puccini, N. I. Marín-Ramos, F. Bergamo, M. Schirripa, S. Lonardi, H. J. Lenz, F. Loupakis and F. Battaglin, Drug Saf., 2019, 42, 211–233 CrossRef CAS PubMed.
- K. Morihiro, T. Kodama, Kentefu, Y. Moai, R. N. Veedu and S. Obika, Angew. Chem., Int. Ed., 2013, 52, 5074–5078 CrossRef CAS PubMed.
- K. Morihiro, T. Kodama, R. Waki and S. Obika, Chem. Sci., 2014, 5, 744–750 RSC.
- S. Mori, K. Morihiro, T. Okuda, Y. Kasahara and S. Obika, Chem. Sci., 2018, 9, 1112–1118 RSC.
- K. Morihiro, T. Ishinabe, M. Takatsu, H. Osumi, T. Osawa and A. Okamoto, J. Am. Chem. Soc., 2021, 143, 3340–3347 CrossRef CAS PubMed.
- W. Aoi and Y. Marunaka, BioMed Res. Int., 2014, 2014, 598986 Search PubMed.
- S. H. Kuo, C. J. Shen, C. F. Shen and C. M. Cheng, Diagnostics, 2020, 10, 107 CrossRef CAS PubMed.
- M. Stubbs, P. M. McSheehy, J. R. Griffiths and C. L. Bashford, Mol. Med. Today, 2000, 6, 15–19 CrossRef CAS PubMed.
- N. M. Anderson and M. C. Simon, Curr. Biol., 2020, 30, R921–R925 CrossRef CAS PubMed.
- M. Damaghi, J. W. Wojtkowiak and R. J. Gillies, Front. Physiol., 2013, 4, 370 Search PubMed.
- N. M. AlSawaftah, N. S. Awad, W. G. Pitt and G. A. Husseini, Polymers, 2022, 14, 936 CrossRef CAS PubMed.
- K. Gehring, J. L. Leroy and M. Guéron, Nature, 1993, 363, 561–565 CrossRef CAS PubMed.
- Y. Dong, Z. Yang and D. Liu, Acc. Chem. Res., 2014, 47, 1853–1860 CrossRef CAS PubMed.
- L. Lannes, S. Halder, Y. Krishnan and H. Schwalbe, ChemBioChem, 2015, 16, 1647–1656 CrossRef CAS PubMed.
- P. A. Beal and P. B. Dervan, Science, 1991, 251, 1360–1363 CrossRef CAS PubMed.
- Y. Hu, A. Cecconello, A. Idili, F. Ricci and I. Willner, Angew. Chem., Int. Ed., 2017, 56, 15210–15233 CrossRef CAS PubMed.
- D. Miao, Y. Yu, Y. Chen, Y. Liu and G. Su, Mol. Pharm., 2020, 17, 1127–1138 CrossRef CAS PubMed.
- Y. Li, S. Yue, J. Cao, C. Zhu, Y. Wang, X. Hai, W. Song and S. Bi, Theranostics, 2020, 10, 8250–8263 CrossRef CAS PubMed.
- H. Park, J. Kim, S. Jung and W. Kim, Adv. Funct. Mater., 2018, 1705416 CrossRef.
- S. Modi, C. Nizak, S. Surana, S. Halder and Y. Krishnan, Nat. Nanotechnol., 2013, 8, 459–467 CrossRef CAS PubMed.
- A. S. Boutorine, D. S. Novopashina, O. A. Krasheninina, K. Nozeret and A. G. Venyaminova, Molecules, 2013, 18, 15357–15397 CrossRef CAS PubMed.
- P. Peng, Y. Du, J. Zheng, H. Wang and T. Li, Angew. Chem., Int. Ed., 2019, 58, 1648–1653 CrossRef CAS PubMed.
- K. Morihiro, Y. Kasahara and S. Obika, Mol. Biosyst., 2017, 13, 235–245 RSC.
- R. Ueki and S. Sando, Chem. Commun., 2014, 50, 13131–13134 RSC.
- L. Li, Y. Jiang, C. Cui, Y. Yang, P. Zhang, K. Stewart, X. Pan, X. Li, L. Yang, L. Qiu and W. Tan, J. Am. Chem. Soc., 2018, 140, 13335–13339 CrossRef CAS PubMed.
- A. E. Rangel, A. A. Hariri, M. Eisenstein and H. T. Soh, Adv. Mater., 2020, 32, e2003704 CrossRef PubMed.
- K. Morihiro, Y. Moriyama, Y. Nemoto, H. Osumi and A. Okamoto, J. Am. Chem. Soc., 2021, 143, 14207–14217 CrossRef CAS PubMed.
- P. C. Ma, R. Jagadeeswaran, S. Jagadeesh, M. S. Tretiakova, V. Nallasura, E. A. Fox, M. Hansen, E. Schaefer, K. Naoki, A. Lader, W. Richards, D. Sugarbaker, A. N. Husain, J. G. Christensen and R. Salgia, Cancer Res., 2005, 65, 1479–1488 CrossRef CAS PubMed.
- J. Yu, K. Ohuchida, K. Mizumoto, N. Ishikawa, Y. Ogura, D. Yamada, T. Egami, H. Fujita, S. Ohashi, E. Nagai and M. Tanaka, World J. Gastroenterol., 2006, 12, 3878–3882 CrossRef CAS PubMed.
- M. Tokunou, T. Niki, K. Eguchi, S. Iba, H. Tsuda, T. Yamada, Y. Matsuno, H. Kondo, Y. Saitoh, H. Imamura and S. Hirohashi, Am. J. Pathol., 2001, 158, 1451–1463 CrossRef CAS PubMed.
- S. Lee and A. Shanti, Int. J. Mol. Sci., 2021, 22, 9910 CrossRef CAS PubMed.
- J. Michl, K. C. Park and P. Swietach, Commun. Biol., 2019, 2, 144 CrossRef PubMed.
Footnotes |
| † Electronic supplementary information (ESI) available. See DOI: https://doi.org/10.1039/d4sc04424j |
| ‡ These authors contributed equally to this work. |
| This journal is © The Royal Society of Chemistry 2024 |