Open Access Article
Open Access ArticleEnrichment of histone tail methylated lysine residues via cavitand-decorated magnetic nanoparticles for ultra-sensitive proteomics†
Martina
Orlandini‡
 a,
Alex
Bonacini‡
a,
Alessia
Favero
a,
Andrea
Secchi
a,
Alex
Bonacini‡
a,
Alessia
Favero
a,
Andrea
Secchi
 a,
Laura
Lazzarini
b,
Roberto
Verucchi
c,
Enrico
Dalcanale
a,
Laura
Lazzarini
b,
Roberto
Verucchi
c,
Enrico
Dalcanale
 a,
Alessandro
Pedrini
a,
Alessandro
Pedrini
 a,
Simone
Sidoli
*d and
Roberta
Pinalli
a,
Simone
Sidoli
*d and
Roberta
Pinalli
 *a
*a
aDepartment of Chemistry, Life Sciences and Environmental Sustainability, University of Parma, INSTM, UdR Parma, Parco Area delle Scienze 17/A, 43124, Parma, Italy. E-mail: roberta.pinalli@unipr.it
bIMEM-CNR, Institute of Materials for Electronics and Magnetism, National Research Council, Parco Area delle Scienze 37/A, 43124, Parma, Italy
cIMEM-CNR, Institute of Materials for Electronics and Magnetism, National Research Council, Trento Unit, via alla Cascata 56/C, 38123, Trento, Italy
dDepartment of Biochemistry, Albert Einstein College of Medicine, Bronx, NY 10461, USA. E-mail: simone.sidoli@einsteinmed.edu
First published on 15th July 2024
Abstract
Nearly every protein in the human body is modified with post-translational modifications (PTMs). PTMs affect proteins on many levels, including their function, interaction, half-life, and localization. Specifically, for histone proteins, PTMs such as lysine methylation and acetylation play essential roles in chromatin dynamic regulations. For this reason, methods to accurately detect and quantify PTMs are of paramount importance in cell biology, biochemistry, and disease biology. Most protein modifications are sub-stoichiometric, so, to be analyzed, they need methods of enrichment, which are mostly based on antibodies. Antibodies are produced using animals, resulting in high costs, ecological concerns, significant batch variations, and ethical implications. We propose using ferromagnetic nanoparticles functionalized with synthetic receptors, namely tetraphosphonate cavitands, as a tool for selective enrichment of methylated lysines present on histone tails. Before the enrichment step, histone proteins from calf thymus were digested to facilitate the recognition process and to obtain small peptides suitable for mass analyses. Cavitands were anchored on ferromagnetic nanoparticles to easily separate the PTM-peptides of interest from the rest of the proteolytic peptides. Our approach detects more modified peptides with higher signal intensity, rivaling commercial antibodies. This chemical strategy offers a cost-effective and efficient alternative for PTM detection, potentially advancing proteomic research.
Introduction
Post-translational modifications (PTMs) of histone proteins, such as acetylation, methylation, phosphorylation, and ubiquitination, play essential roles in regulating chromatin dynamics. Many PTMs occur on histones and non-histone proteins, regulating the protein–protein interactions, localization, stability, and enzymatic activities of proteins involved in different cellular processes.1,2 Methylation is a conserved post-translational modification of proteins, characterized by the enzymatic transfer of a methyl group from S-adenosyl methionine (SAM) to a lysine or arginine side chain. The ε nitrogen of lysine can be modified with up to three methyl groups, producing a mono-methylated (Kme1), di-methylated (Kme2), or tri-methylated state (Kme3). The presence of methyl groups increases the size and hydrophobicity of lysine and arginine residues, limiting their potential to participate in hydrogen bonding networks. Lysine and arginine methylation has been studied extensively in the context of histones and chromatin biology, and methylation of non-histone proteins has recently emerged as an important factor in many processes of biological regulation.1,3,4 Depending on the methylation site, histone methylation can represent a repressive or activating mark. For example, tri-methylation of lysine 9 on histone H3 (H3K9me3) is associated with silenced chromatin, whereas tri-methylation of lysine 4 on histone H3 (H3K4me3) is related to active chromatin.5,6 Lysine mono-methylation is not nearly as annotated in terms of biological functions. Depending on the position and the histone tail as well, mono-methylation of lysine has been associated with the transcriptional repression in lethal 3 malignant brain tumor 1 (L3MBTL1)7 or with widespread paired helical filament (PHF)-tau modification found in the brain of patients suffering from Alzheimer's disease.8Given their biological importance and relevance in the study of these Non-Communicable Diseases (NCDs)9–12 numerous methods have been developed to identify modified proteins and their specific modification sites. The most straightforward approach uses in vitro methylation assays based on radioactively labelled SAM, which enables the detection of methylated substrates as peptides and full-length proteins. Although this method has been mostly used to confirm a methylation event, i.e. targeted, rather than identifying new methylations, the main drawback of this approach is the poor specificity of the binding of SAM,2 and the lack of single amino acid resolution limiting the possibility of identifying which amino acid undergoes methylation.
Mass spectrometry is the most widespread methodology to identify the type, site and, in case of methylation, state (mono, di or tri) of protein modifications.13–15 The analysis of PTMs represents a challenge since most of them are present in very low stoichiometry; few residues in the target protein contain the modification, and some PTMs produce signal suppression or low-quality MS/MS spectra. Thus, large amounts of starting materials are necessary, and this represents a problem when the samples to be analysed are patients' tissues or biopsies. For these reasons, an enrichment step is always necessary. The use of commercially available pan-methyl antibodies for immunoprecipitation (IP) experiments followed by mass spectrometry analyses has been proposed.16–20 IP is fast and relatively easy compared to affinity chromatography which is time-consuming and involves cycles of binding and washing. Unfortunately, IP is limited by the availability of antibodies that recognize the target protein. Moreover, these antibodies suffer from poor selectivity, low sensitivity and low batch-to-batch reproducibility.21 Furthermore, the use of animals to obtain antibodies, particularly through methods such as immunization and subsequent harvesting of serum, is raising significant ethical and sustainability issues.22,23
As an alternative, the use of the malignant brain tumor (MBT) domain of the protein L3MBTL1, which recognizes mono- and di-methylated proteins, has also been proposed for the enrichment of methylated targets in a stable isotope labelling with amino acids in the cell culture (SILAC) approach.24–26 However, labelling by SILAC depends on the efficient production of labelled SAM and active turnover of methylation, which may vary for different methylation sites and experimental systems.
Methylated proteins can also be enriched using chemical methods that exploit biorthogonal reactive chemical moieties, such as azides or alkynes, which enable the attachment of affinity tags in place of the methyl group.27–30 Yet, these approaches require complex chemical modifications of peptides or SAM and are time-consuming since they require analytical reverse phase HPLC purification.
Accordingly, the development of an efficient, fast, recyclable and cost-effective tool for the enrichment of methylated proteins is still a challenge. Even more demanding is finding an efficient method for the enrichment of a single modification among mono-, di- and tri-methylation on lysine residues. In the last few years, the use of synthetic receptors has emerged as a viable alternative route to detect histone PTMs.31 The major target so far has been lysine methylation. Most of the attention focused on the recognition of tri-methylated lysine, using p-sulfonatocalix[4]arenes32–34 and cavitands35–37 as complexing agents. Cucurbit[7]uril is effective in binding di-methylated lysines in proteins,38,39 while the most elusive mono-methylated lysines are selectively targeted by tetraphosphonate cavitands.
Tetraphosphonate cavitands are a family of resorcinarene-based receptors able to selectively complex biologically relevant mono-methylated ammonium derivatives via synergistic host–guest interactions namely the combination of cation–dipole, cation–π and H-bonding interactions between the aromatic cavity and the phosphonate moieties of the cavitand and the N-methylammonium residue.31 These complexes can be dissociated on demand making the system fully reversible.40 The synthetic versatility of resorcinarene-based cavitands allows them to be functionalized both at the upper and at the lower rim with suitable groups to modulate their recognition properties and to be anchored to a specific support.41,42 In the past few years, the specificity of tetraphosphonate cavitands in complexing mono-methylated lysine in water (Ka = 1.49 × 103 M−1 at 298 K) has been demonstrated.43 This ability was successfully applied to the detection of the mono-methylation state of lysines present in human histone H3 tails.44 To this end, one of the four cavitand feet was equipped with a carboxylic acid moiety to enable its grafting onto TiO2 nanoparticles, and the obtained hybrid system was used to isolate the peptide–cavitand complex from the solution for its subsequent detection via non-plasmonic surface enhanced Raman scattering (SERS). This system demonstrated the potential of tetraphosphonate cavitand-functionalized nanoparticles in detecting mono-methylated lysine residues present in histone tails.
In this paper, we present a versatile methodology for the selective enrichment of mono-methylated lysine residues of histone proteins based on the use of tetraphosphonate cavitands supported on ferromagnetic nanoparticles (FeNPs). Iron oxide nanoparticles were selected for their magnetic properties, which facilitate the isolation of the host–guest complexes, as well as for their biocompatibility, chemical stability, and their accessible surface functionalization. To validate the effectiveness of the proposed strategy, the functionalized nanoparticles were incubated with a digested solution of calf thymus histones. The peptides extracted from the NPs were then analysed via High-Resolution Mass Spectrometry (HR-MS) to quantify modified and unmodified histone peptides for estimating the enrichment of methylation. The results were compared with the ones obtained by immunoprecipitation using a commercially available antibody-based enrichment kit.
Results and discussion
Cavitand-grafted ferromagnetic nanoparticle preparation and characterization
Two different cavitands were synthesized (Scheme 1), namely an “active” tetraphosphonate cavitand 1, bearing four P![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) O groups at its upper rim pointing into the cavity, and a reference cavitand 2, bearing four methylene bridges at its upper rim. Cavitand 2 is depleted by the aforementioned interacting sites (e.g. cation–dipole, and H-bonding interactions), and therefore it is inefficient in recognizing mono-methylated ammonium salts.45 For their grafting onto FeNPs, both the cavitands were functionalized at the lower rim with four carboxyl groups. Compound 1 was prepared in six steps with a 54% overall yield, while cavitand 2 was obtained in two steps with a 62% overall yield. A detailed synthesis of both cavitands is reported in the ESI (Schemes S1 and S2,† respectively).
O groups at its upper rim pointing into the cavity, and a reference cavitand 2, bearing four methylene bridges at its upper rim. Cavitand 2 is depleted by the aforementioned interacting sites (e.g. cation–dipole, and H-bonding interactions), and therefore it is inefficient in recognizing mono-methylated ammonium salts.45 For their grafting onto FeNPs, both the cavitands were functionalized at the lower rim with four carboxyl groups. Compound 1 was prepared in six steps with a 54% overall yield, while cavitand 2 was obtained in two steps with a 62% overall yield. A detailed synthesis of both cavitands is reported in the ESI (Schemes S1 and S2,† respectively).
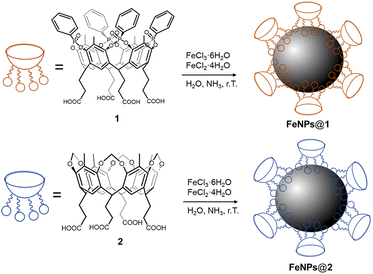 | ||
| Scheme 1 Synthesis of cavitand-grafted ferromagnetic nanoparticles with tetraphosphonate cavitand 1 (above) and reference cavitand 2 (below). | ||
The functionalization of FeNPs with cavitands 1 and 2 was performed directly via in situ co-precipitation, reacting FeCl3 and FeCl2 in the presence of the receptor, which acts as a surfactant to stabilize the FeNP surface. Non-functionalized ferromagnetic nanoparticles were also synthesized to account for non-specific interactions in complexation experiments following an analogous co-precipitation method.46
Both bare and functionalized NPs were fully characterized. FT-IR analysis confirmed the presence of the receptor on the NP surface. In the FT-IR spectra of FeNPs@1 (Fig. S5,† green line) and FeNPs@2 (Fig. S5,† red line), the stretching bands of C![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) O bonds, belonging to the carboxyl groups decorating the lower rim of the cavitands, are present at 1644 cm−1 and 1624 cm−1, respectively. The diagnostic band associated with the stretching of Fe–O is visible around 600 cm−1, in agreement with the FT-IR spectrum of FeNPs reported in the literature.47 Moreover, for FeNPs@1, the FT-IR spectrum showed diagnostic bands at 1070 cm−1 and 898 cm−1, corresponding to P
O bonds, belonging to the carboxyl groups decorating the lower rim of the cavitands, are present at 1644 cm−1 and 1624 cm−1, respectively. The diagnostic band associated with the stretching of Fe–O is visible around 600 cm−1, in agreement with the FT-IR spectrum of FeNPs reported in the literature.47 Moreover, for FeNPs@1, the FT-IR spectrum showed diagnostic bands at 1070 cm−1 and 898 cm−1, corresponding to P![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) O and P–O–P bond stretching. Thermogravimetric analysis (TGA) was used to determine the NP functionalization degree (Fig. S6–S10 and Table S2†). From the thermogram of FeNPs@1 a mass percentage of 17% of cavitand onto the NPs was estimated, while for FeNPs@2 the percentage is around 14% (Table S2†). The surface charge of the NPs was characterized via ζ-potential measurements (Fig. S11†), highlighting the effect of the coating.
O and P–O–P bond stretching. Thermogravimetric analysis (TGA) was used to determine the NP functionalization degree (Fig. S6–S10 and Table S2†). From the thermogram of FeNPs@1 a mass percentage of 17% of cavitand onto the NPs was estimated, while for FeNPs@2 the percentage is around 14% (Table S2†). The surface charge of the NPs was characterized via ζ-potential measurements (Fig. S11†), highlighting the effect of the coating.
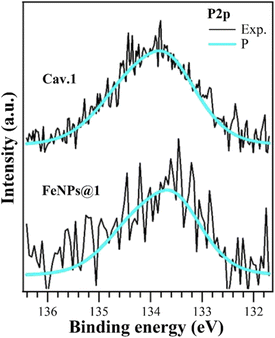
To unambiguously confirm the grafting of the cavitands onto the FeNPs, X-ray photoelectron spectroscopy (XPS) analyses were performed. The XPS analysis of FeNPs@1 shows a complex scenario. Bare FeNPs present a significant amount of adventitious carbon and oxygen (Table S3 and Fig. S12†); thus, the successful functionalization with cavitand 1 would lead to the superposition of very similar photoemission peaks for C 1s and O 1s core levels. We found a binding energy (BE) shift of about +0.6 eV due to charging phenomena (all data and peak BEs for FeNPs@1 are reported in Table S5†). The Fe 2p signal has a low S/N ratio, probably due to the presence of the coating functionalizing molecules (Fig. S14†). The presence of phosphorus on the surface is evidenced by the P 2p emission peak (Fig. 1) at the same BE of the isolated molecule, which is diagnostic of the presence of cavitand 1 on the FeNP surface. This emission peak is absent both in bare FeNPs and in FeNPs@2. The C 1s and O 1s (Fig. S14†) core levels can be reproduced as the superposition of components from the iron oxide and adventitious species on FeNPs with the organic features; however, a more detailed comprehension is not fully reliable. O 1s core level (Fig. S14†) analysis revealed a significant increase of the C![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) O peak (531.92 eV) intensity and can be related to the formation of COO– groups, and thus deprotonated carboxyl. This is in agreement also with the COH peak (533.97 eV) intensity reduction,48,49 as well as with the higher BE of the COOH group (+0.2 eV) in the C 1s core level.50–52 All of this evidence confirms the chemical bonds between the carboxylate groups of cavitand 1 with FeNPs. We can conclude that functionalization occurs via deprotonation of a carboxyl group, with the formation of different oxygen-based species on the FeNP surface. Analogously, XPS analyses performed on FeNPs@2 confirmed the presence of cavitand 2 on the NP surface (see the ESI†).
O peak (531.92 eV) intensity and can be related to the formation of COO– groups, and thus deprotonated carboxyl. This is in agreement also with the COH peak (533.97 eV) intensity reduction,48,49 as well as with the higher BE of the COOH group (+0.2 eV) in the C 1s core level.50–52 All of this evidence confirms the chemical bonds between the carboxylate groups of cavitand 1 with FeNPs. We can conclude that functionalization occurs via deprotonation of a carboxyl group, with the formation of different oxygen-based species on the FeNP surface. Analogously, XPS analyses performed on FeNPs@2 confirmed the presence of cavitand 2 on the NP surface (see the ESI†).
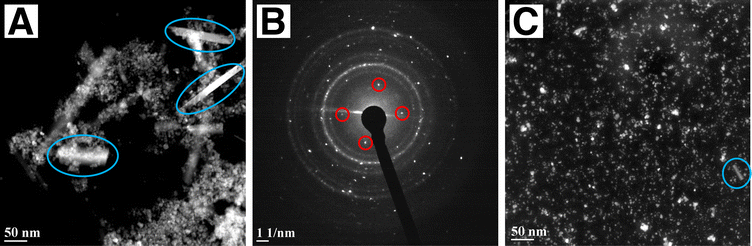
The crystalline phase of bare and functionalized FeNPs was also investigated via X-ray diffraction (XRD) analyses. The XRD diffraction pattern of the non-functionalized FeNPs perfectly reflected the pattern of standard Fe3O4, present as the predominant crystalline phase (Fig. S17†). Both the XRD diffractograms of the functionalized nanoparticles FeNPs@1 and FeNPs@2 revealed the simultaneous presence of two crystalline phases, magnetite and goethite (Fig. S18†),53 suggesting that the co-precipitation in the presence of the receptors promotes the formation of an additional phase. To determine the structure, size, shape and surface morphology, the synthetized magnetic nanoparticles were characterized using transmission electron microscopy (TEM) imaging in the high-resolution (HR-TEM) and the High-Angle Annular Dark-Field Scanning TEM (HAADF-STEM) mode. The TEM images of the bare FeNPs collected in conventional TEM and HAADF-STEM showed a strong tendency to aggregate into large agglomerates, making difficult an accurate study of the single nanoparticle (Fig. S19A and B†). However, from the study of the agglomerate's periphery, an irregular and often faceted particle shape can be deduced (Fig. S19C†), in agreement with the literature.54,55 The estimated size is in the range of 10–20 nm. Also, no individual particles were observed outside of aggregates.
The Diffraction Patterns (DPs) collected on the sample confirmed that the particles present the magnetite phase (Fig. S19D†). As for FeNPs@2, the particles are smaller in size with respect to the non-functionalized ones and present two different shapes (Fig. 2A). In particular, along with the quasi-spherical particles attributable to the magnetite phase, there are also crystalline rods of highly variable length and diameter (in the blue ovals). The analysis of the DPs (Fig. 2B) showed that these rods mainly present the goethite phase, in agreement with XRD analysis.54 The presence of the goethite phase does not affect the magnetic properties of the NPs. TEM images of FeNPs@1 showed that they prefer to form small agglomerates of a few units, as depicted in Fig. 2C. Crystalline rods related to the goethite phase are still occasionally visible, but are smaller in size with respect to FeNPs@2. In this sample, the predominance of the magnetite phase is also confirmed, in agreement with the XRD results.
Enrichment experiments
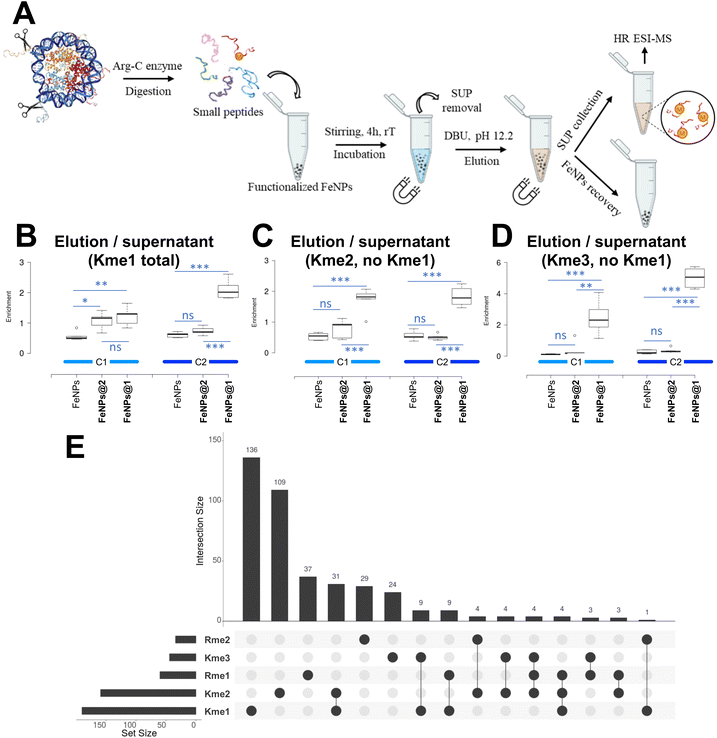
Bare and functionalized NPs were then used in the subsequent enrichment experiments. Commercial histones from calf thymus were chosen as a model source of methylated proteins. In our experimental design, FeNPs@1 should complex the methylated peptides, while FeNPs@2 was used as a control experiment to highlight the role of molecular recognition in this binding process. Additionally, bare FeNPs were employed as a blank to consider physisorption phenomena associated with the nanoparticle surface. The proposed methodology is depicted in Fig. 3A. In brief, the histone proteins were subjected to proteolytic digestion in the presence of Arg-C to obtain small peptides suitable for the subsequent mass-spectrometry analysis. Additionally, cutting the protein into small fragments allows the exposure of all the possible modifications, some of them otherwise hidden by the protein folding. After the digestion step, the peptides were incubated with functionalized and bare FeNPs. Magnetic separation of the NPs was then performed, leading to the removal of the supernatant solution.The next step involved the extraction of the retained peptides from the nanoparticle surface to allow their characterization by LC-MS/MS spectrometry. This can be obtained by straightforwardly exploiting the reversibility of ammonium complexation by tetraphosphonate cavitands. By increasing the pH upon base addition, the deprotonation of the guest severely reduced its interaction with the cavity leading to decomplexation.40 In particular, we used 1,8-diazobicyclo[5.4.0]undec-7-ene (DBU) as a base. Both the extracted samples and the supernatant were analysed via nano-chromatography coupled online with tandem mass spectrometry to quantify the relative enrichment of methylated peptides (see the ESI† for protocol details). For each NP system, two enrichment cycles (C1 = cycle 1; C2 = cycle 2) following this protocol were performed on the same batches and five replicates were carried out for each cycle.
LC-MS/MS spectrometry analysis results
By LC-MS/MS spectrometry, we detected 410 peptides, including methylated (407) and unmodified (3) ones (see the dataset supplemental table†). By using the area underneath the curve of the extracted ion chromatogram for each peptide, we calculated the sum of all the methylated forms and compared them with the same signal in the unbound fraction, i.e. supernatant. Interestingly, among the identified peptides we detected peptides with non-specific cleavage, i.e. cleaved after lysine residues by ArgC, and peptides with methylated residues at the C-terminus. Unfortunately, ArgC specificity is not as high as that of trypsin.56 Furthermore, it is possible that the sample preparation introduced a percentage of chemically methylated lysine residues, as discussed in other publications.57 We took these aspects into consideration but proceeded to compare our enrichments regardless due to the technical nature of this work. By comparing bare beads, FeNPs@1 and FeNPs@2 we observed a remarkable enrichment of mono-methylated peptides by the FeNPs@1 system (Fig. 3B), confirming that the cavitand has the capability of binding and partitioning methylated histone peptides. Interestingly, we obtained a similar enrichment for di-methylated peptides (Fig. 3C) and an apparently even higher enrichment for tri-methylated peptides (Fig. 3D). As for di-methylated lysines, it is known that the tetraphosphonate cavitand is able to bind also di-methylated ammonium salts but with reduced affinity with respect to the mono-methylated ones.45 It is worth noting that at the solid/liquid interface, other factors such as surface work can affect the association constants.58 Moreover, on a surface functionalized with a large number of hosts, the simultaneous occurrence of multiple interactions between the binding partners, which characterizes multivalency, can occur.59 Regarding the tri-methylated lysine results, it is worth noting that tri-methylated peptides are far less abundant than the other two considered PTMs, so the enrichment might be biased by the overall lower signal. Notably, we performed the enrichment using the same beads twice to assess whether saturation would have a beneficial effect on their usage, i.e. utilizing them more than once leads to an improved enrichment compared to single use. Reusability is common in other proteomics experiments, such as phosphoproteomics.60,61 It is important to note that the performance of the FeNPs@1 system in enriching peptides bearing mono-methylated lysines is 2.2 fold higher with respect to the bare FeNPs after the first cycle, and up to 3.5 fold higher after the second cycle (Fig. 3B). Notably, part of the enrichment is biased because not all peptides are modified with a single modification. Histone peptides in particular are commonly decorated with multiple PTMs.62 As described in Fig. 3E, the Venn diagram highlights that the majority of detected histone peptides are modified with a single PTM, but there are also a considerable number of co-modified ones. In the diagram, we considered all the peptides bearing a modification, including arginine (R) methylation. It is also important to note that this analysis is likely over-representing the relative abundance of modified histone peptides compared to the unmodified ones, mostly because no derivatization was utilized in contrast to common practices63–66 leading to unmodified peptides being digested into shorter (sometimes undetectable) sequences. It is therefore important to clarify that the abundance of detected methylations might be overestimated in terms of stoichiometry, as discussed elsewhere.67407 peptides were identified, of which 136 bear exclusively Kme1 as modification, while 54 bear other modifications besides Kme1. Also, the developed system is able to enrich Kme2, detecting 109 peptides bearing exclusively this modification and more than 50 peptides Kme2 bearing other modifications. Importantly, the results highlight that lysine mono-methylation is by far the preferred peptide to be enriched by our functionalized beads. Notably, a few notorious histone peptides were not detected in this study. Arguably, the most known methylations absent in our results are H3K4me and H4K20me. These two peptides are very hydrophilic as they have basic residues and short sequences, i.e. TKQTAR and KVLR, respectively. It is therefore unlikely that they will bind to a hydrophobic C18 chromatography column in the absence of prior derivatisation.
Comparison of the performance between FeNPs@1 and commercial antibodies
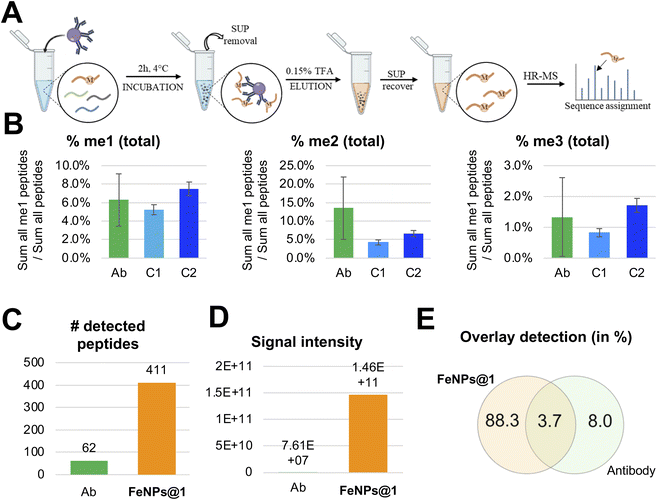
Next, we assessed the specificity of cavitands compared to antibodies by repeating the experiment using a well-known kit, the Pan-Methyl Lysine kit, for methylation enrichment (see the ESI† for the protocol and Fig. 4A). For this experiment, we used the same batch of digested histone proteins employed for the enrichment experiments with functionalized and bare NPs. Also in this case, five replicates were performed. By calculating the relative abundance of the methylated peptides vs. all the detectable peptides, we estimated a comparable enrichment between FeNPs@1 and antibodies for each of the three methylated states (Fig. 4B). However, we noticed two important differences: (i) antibodies have a preference for di-methylated peptides, while the FeNPs@1 system at the second cycle showed a preference for the mono-methylated ones; (ii) the overall experiment with the antibody produced a lower yield in our hands, leading to fewer identified peptides (Fig. 4C) and an overall lower signal intensity (Fig. 4D). The lower intensity of the signal in the antibody analysis justifies the larger error bars of these replicates (Fig. 4B). Finally, it should be noted that the peptide sequences identified by the two experiments tend to be different (Fig. 4E), with a longer list for the cavitand experiment due to the higher sensitivity. Notably, these results show comparable enrichment compared to antibodies and detection of more modified peptides with higher signal intensity.Conclusions
In this study, we proposed an alternative tool to immunoprecipitation of target proteins. To this end, we synthesized, via in situ precipitation, ferromagnetic nanoparticles functionalized with molecular receptors selective for the binding of methylated lysines. These FeNPs were obtained from tetraphosphonate (1) and tetramethylene (2) bridged cavitands equipped with four carboxylic acids at the lower rim. Both the obtained systems, namely FeNPs@1 and FeNPs@2, were fully characterized to assess the success and the amount of functionalization. FeNPs@1 and FeNPs@2 were used in enrichment experiments to extract methylated peptides from a digested solution of calf thymus histones. In particular, the FeNPs@2 system was used as a control experiment together with bare FeNPs to account for any physisorption effects. Two cycles were performed: the first one for conditioning the system, and the second cycle to assess the elution efficiency. The LC-MS/MS analyses revealed that FeNPs@1 is an efficient tool for the enrichment of mono- and di-methylated lysines, while bare FeNPs and FeNPs@2 are considerably less effective. Finally, the performances of FeNPs@1 were compared to those of commercial antibodies. The developed system showed comparable enrichment results to the commonly used antibodies, with FeNPs@1 able to detect a larger number of modified peptides with higher overall signal intensity. In addition, FeNPs@1 displayed a preference for the enrichment of mono-methylated lysines with respect to the di-methylated ones. In conclusion, the functionalized FeNPs developed in this work are cheaper with respect to antibodies, easy to be produced, scalable, and amenable to long-term storage. In our opinion, this work will encourage the development of synthetic receptors to replace immunoprecipitation as the enrichment step in the most critical histone epigenetic modifications. Significantly, opting out of antibody usage will yield greater long-term benefits by promoting animal-free alternatives. Additional studies are ongoing in our lab to prove the recyclability of the functionalized nanoparticles and to improve the extraction method. Furthermore, our enrichment strategy should be optimized with the entire cell methylome to exploit its full potential. Our laboratory will work on this project in the near future.Data availability
Experimental details; experimental synthetic procedures; characterization data; protocol for proteolytic digestion; protocol for enrichment experiments; protocol for immunoprecipitation; sample preparation for LC-MS/MS analysis and theoretical calculation results are available in the ESI.† Additional supporting material available: Supplementary table containing HR-HPLC-MS analysis data.Author contributions
M. O., A. B. and A. F. synthesized the cavitands and the functionalized ferromagnetic nanoparticles. M. O. performed the enrichment experiments; A. S. supervised the ferromagnetic nanoparticle synthesis and functionalization; L. L. performed TEM analyses; R. V. performed XPS analyses; S. S. performed HR-MS analyses; E. D., A. P., S. S. and R. P. designed the research and analyzed the data; R. P. oversaw the project; E. D., A. P., S. S. and R. P. wrote the paper.Conflicts of interest
There are no conflicts to declare.Acknowledgements
This work has benefited from the equipment and framework of the COMP-HUB and COMP-R Initiatives, funded by the ‘Departments of Excellence’ program of the Italian Ministry for University and Research (MIUR, 2018–2022 and MUR, 2023–2027). M. O. thanks the University of Parma intramural funding “Bando di Ateneo per la Ricerca 2021”. A. P., E. D. and R. P. thank the project ENRICH, funded through the European Union Horizon Europe Program (Horizon-MSCA-2022-SE under grant agreement no. 101131120). R. P. declares that this study was carried out within the “Innovative amplification strategies for ultrasensitive detection of molecular targets (P2022ANCEK, E53D23015640001)” project – funded by European Union – Next Generation EU within the PRIN 2022 PNRR program (D.D.1409 del 14/09/2022 Ministero dell’Università e della Ricerca). This manuscript reflects only the authors' views and opinions and neither the European Union nor the granting authority nor the Ministry can be held responsible for them. We thank the Centro Interfacoltà di Misure “G. Casnati” of the University of Parma for the use of NMR facilities and Prof. Francesco Mezzadri for the XRD analyses.Notes and references
- X. Zhang, H. Wen and X. Shi, Acta Biochim. Biophys. Sin., 2012, 44, 14–27 CrossRef CAS PubMed.
- D. Levy, Cell. Mol. Life Sci., 2019, 76, 2873–2883 CrossRef CAS PubMed.
- A. Di Lorenzo and M. T. Bedford, FEBS Lett., 2011, 585, 2024–2031 CrossRef CAS.
- Y. Yang and M. T. Bedford, Nat. Rev. Cancer, 2013, 13, 37–50 CrossRef CAS PubMed.
- E. L. Greer and Y. Shi, Nat. Rev. Genet., 2012, 13, 343–357 CrossRef CAS.
- T. Kouzarides, Cell, 2007, 128, 693–705 CrossRef CAS.
- N. Kalakonda, W. Fischle, P. Boccuni, N. Gurvich, R. Hoya-Arias, X. Zhao, Y. Miyata, D. Macgrogan, J. Zhang, J. K. Sims, J. C. Rice and S. D. Nimer, Oncogene, 2008, 27, 4293–4304 CrossRef CAS PubMed.
- S. N. Thomas, K. E. Funk, Y. Wan, Z. Liao, P. Davies, J. Kuret and A. J. Yang, Acta Neuropathol., 2012, 123, 105–117 CrossRef CAS PubMed.
- K. K. Biggar and S.-C. L. Shawn, Nat. Rev. Mol. Cell Biol., 2014, 16, 5–17 CrossRef PubMed.
- E. L. Greer and Y. Shi, Nat. Rev. Genet., 2012, 13, 343–357 CrossRef CAS.
- P. K. Mazur, N. Reynoird, P. Khatri, W. T. T. P. Jansen, W. A. Wilkinson, S. Liu, O. Barbash, S. G. Van Aller, M. Huddleston, D. Dhanak, J. P. Tummino, G. R. Kruger, A. B. Garcia, J. A. Butte, M. Vermeulwn, J. Sage and O. Gozani, Nature, 2014, 510, 283–287 CrossRef CAS PubMed.
- K. E. Moore and O. Gozani, Biochim. Biophys. Acta, 2014, 1839, 1395–1403 CrossRef CAS.
- M. I. Maron, M. S. Lehman, S. Gayatri, J. D. DeAngelo, S. Hegde, B. M. Lorton, Y. Sun, D. L. Bai, S. Sidoli, V. Gupta, M. R. Marunde, J. R. Bone, Z. Sun, M. T. Bedford, J. Shabanowitz, H. Chen, D. F. Funt and D. Shechter, iScience, 2021, 24, 102971 CrossRef CAS PubMed.
- S. Stephanie, R. Cutler, J. Aguilan, E. Nieves and S. Sidoli, Epigenet. Chromatin, 2022, 15, 35 CrossRef.
- S. Sidoli, J. Simithy, K. R. Karch, K. Kulej and B. A. Garcia, Anal. Chem., 2015, 87, 11448 CrossRef CAS PubMed.
- S. E. Ong, G. Mittler and M. Mann, Nat. Methods, 2004, 1, 119–126 CrossRef CAS PubMed.
- X. J. Cao, A. M. Arnaud and B. A. Garcia, Epigenetics, 2013, 8, 477–485 CrossRef CAS PubMed.
- X. J. Cao and B. A. Garcia, Curr. Protoc. Protein Sci., 2016, 86, 24 Search PubMed.
- A. Guo, H. Gu, J. Zhou, D. Mulhern, Y. Wang, K. A. Lee, V. Yang, M. Aguiar, J. Kornhauser, X. Jia, J. Ren, S. A. Beausoleil, J. C. Silva, V. Vemulapalli, M. T. Bedford and M. J. Comb, Mol. Cell. Proteom, 2014, 13, 372–387 CrossRef CAS PubMed.
- J. Gil, A. Ramírez-Torres and S. Encarnación-Guevara, J. Proteomics, 2017, 150, 297–309 CrossRef CAS PubMed.
- D. Levy, C. L. Liu, Z. Yang, A. M. Newman, A. A. Alizadeh, P. J. Utz and O. Gozani, Epigenet. Chromatin, 2011, 4, 19 CrossRef CAS PubMed.
- A. C. Gray, S. S. Sidhu, P. C. Chandrasekera, C. F. M. Hendrikse and C. A. K. Borrebaeck, Trends Biotechnol., 2016, 34, 960–969 CrossRef CAS.
- A. Gray, A. R. M. Bradbury, A. Knappik, A. Plückthun, C. A. K. Borrebaeck and S. Dübel, Nat. Biotechnol., 2020, 38, 1234–1239 CrossRef CAS PubMed.
- J. Min, A. Allali-Hassani, N. Nady, C. Qi, H. Ouyang, Y. Liu, F. MacKenzie, M. Vedadi and C. H. Arrowsmith, Nat. Struct. Mol. Biol., 2007, 14, 1229–1230 CrossRef CAS.
- N. Nady, L. Krichevsky, N. Zhong, S. Duan, W. Tempel, M. F. Amaya, M. Ravichandran and C. H. Arrowsmith, J. Mol. Biol., 2012, 423, 702–718 CrossRef CAS.
- S. M. Carlson, K. E. Moore, E. M. Green and G. M. Martìn, Nat. Protoc., 2014, 9, 37–50 CrossRef CAS.
- K. Islam, R. B. Sarangi, V. V. Gurchenkov, B. Sinha, R. T. Kießling, L. Fetler, F. Rico, S. Scheuring, C. Lamaze, A. Simon, S. Geraldo, D. Vignjevic, H. Doméjean, L. Rolland, A. Funfak, J. Bibette, N. Bremond and P. Nassoy, Proc. Natl. Acad. Sci. U. S. A., 2013, 110, 16778–16783 CrossRef CAS PubMed.
- G. Blum, R. I. Bothwell, K. Islam and M. Luo, Curr. Protoc. Chem. Biol., 2013, 5, 45–66 CrossRef PubMed.
- K. Islam, I. Bothwell, Y. Chen, C. Sengelaub, R. Wang, H. Deng and M. Luo, J. Am. Chem. Soc., 2012, 134, 5909–5915 CrossRef CAS PubMed.
- O. Binda, M. Boyce, S. J. Rush, K. K. Palaniappan, R. C. Bertozzi and O. Gozani, ChemBioChem., 2011, 12, 330–334 CrossRef CAS PubMed.
- R. Pinalli, A. Pedrini and E. Dalcanale, Chem. Soc. Rev., 2018, 47, 7006–7026 RSC.
- C. S. Beshara, C. E. Jones, K. D. Daze, B. J. Lilgert and F. Hof, ChemBioChem., 2010, 11, 63–66 CrossRef CAS PubMed.
- K. D. Daze, T. Pinter, C. S. Beshara, A. Ibraheem, A. Minaker, M. C. F. Ma, R. J. M. Courtemanche, R. E. Campbell and F. Hof, Chem. Sci., 2012, 3, 2695–2699 RSC.
- Y. Kimura, N. Saito, K. Hanada, J. Liu, T. Okabe, S. A. Kawashima, K. Yamatsugu and M. Kanai, ChemBioChem., 2015, 16, 2599–2604 CrossRef CAS PubMed.
- Liu, L. Perez, M. Mettry, C. J. Easley, R. J. Hooley and W. Zhong, J. Am. Chem. Soc., 2016, 138, 10746–10749 CrossRef CAS PubMed.
- Y. Liu, L. Perez, A. D. Gill, M. Mettry, L. Li, Y. Wang, R. J. Hooley and W. Zhong, J. Am. Chem. Soc., 2017, 139, 10964–10967 CrossRef CAS.
- J. Lee, L. Perez, Y. Liu, H. Wang, R. J. Hooley and W. Zhong, Anal. Chem., 2018, 90, 1881–1888 CrossRef CAS.
- F. Guagnini, P. M. Antonik, M. L. Rennie, P. O'Byrne, A. R. Khan, R. Pinalli, E. Dalcanale and P. B. Crowley, Angew. Chem., Int. Ed., 2018, 57, 7126–7130 CrossRef CAS PubMed.
- L. Li, M. Liu, L. Yue, R. Wang, N. Zhang, Y. Liang, L. Zhang, L. Cheng and J. Wang, Anal. Chem., 2020, 92, 9322–9329 CrossRef CAS.
- E. Biavardi, G. Battistini, M. Montalti, R. M. Yebeutchou, L. Prodi and E. Dalcanale, Chem. Commun., 2008, 14, 1638–1640 RSC.
- M. Giannetto, A. Pedrini, S. Fortunati, D. Brando, S. Milano, C. Massera, R. Tatti, R. Verucchi, M. Careri, E. Dalcanale and R. Pinalli, Sens. Actuators, B, 2018, 276, 340–348 CrossRef CAS.
- M. Amorini, N. Riboni, L. Pesenti, V. Dini, A. Pedrini, C. Massera, C. Gualandi, F. Bianchi, R. Pinalli and E. Dalcanale, Small, 2022, 18, 2104946 CrossRef CAS PubMed.
- R. Pinalli, G. Brancatelli, A. Pedrini, D. Menozzi, D. Hernàndez, P. Ballester, S. Geremia and E. Dalcanale, J. Am. Chem. Soc., 2016, 8, 8569–8580 CrossRef.
- N. Bontempi, E. Biavardi, D. Bordiga, G. Candiani, I. Alessandri, P. Bergese and E. Dalcanale, Nanoscale, 2017, 9, 8639–8646 RSC.
- M. Dionisio, G. Oliviero, D. Menozzi, S. Federici, R. M. Yebeutchou, F. P. Schmidtchen, E. Dalcanale and P. Bergese, J. Am. Chem. Soc., 2012, 134, 2392–2398 CrossRef CAS PubMed.
- A. Xiao, C. Xu, Y. Lin, H. Ni, Y. Zhu and H. Cai, Electron. J. Biotechnol., 2016, 19, 1–7 CrossRef.
- M. I. Khalil, Arabian J. Chem., 2015, 8, 279–284 CrossRef CAS.
- D. Briggs and G. Beamson, Anal. Chem., 1993, 85, 1517–1523 CrossRef.
- V. M. Nardi, M. Timpel, L. Pasquardini, T. Toccoli, M. Scarpa and R. Verucchi, Materials, 2023, 16, 5390 CrossRef PubMed.
- S. Garcia-Gil, A. Arnau and A. Garcia-Lekue, Surf. Sci., 2013, 613, 102–107 CrossRef CAS.
- Y. Liu, Z. Xu, J. Zhang, J. Cheng, M. Miao and D. Zhang, Pigment, 2019, 170, 107586 CrossRef.
- F. M. Pantano, R. Tatti, L. Aversa, R. Verucchi and M. N. Pugno, Front. Mater., 2020, 7, 197 CrossRef.
- S. Mahadevan, G. Gnanaprakash, J. Philip, B. P. C. Rao and T. Jayakumar, Phys. E, 2007, 39, 20–25 CrossRef CAS.
- M. Salvador, G. Gutiérrez, S. Noriega, A. Moyano, C. M. Blanco-Lopez and M. Matos, Int. J. Mol. Sci., 2021, 22, 427 CrossRef CAS PubMed.
- S. F. Chin, M. Makha, L. C. Raston and M. Saunders, Chem. Commun., 2007, 19, 1948–1950 RSC.
- R. J. Krueger, T. R. Hobbs, K. A. Mihal, J. Tehrani and M. G. Zeece, J. Chromatogr., 1991, 543, 451–461 CrossRef CAS.
- S. Willems, M. Dhaenens, E. Govaert, L. De Clerck, P. Meert, C. Van Neste, F. Van Nieuwerburgh and D. Deforce, J. Proteome Res., 2017, 16, 655–664 CrossRef CAS PubMed.
- K. Manoli, M. Magliulo, M. Y. Mulla, M. Singh, L. Sabbatini, G. Palazzo and L. Torsi, Angew. Chem., Int. Ed., 2015, 54, 12562 CrossRef CAS PubMed.
- R. Haag, Beilstein J. Org. Chem., 2015, 11, 848 CrossRef CAS PubMed.
- T. E. Thingholm, O. N. Jensen, P. J. Robinson and M. R. Larsen, Mol. Cell. Proteomics, 2008, 7, 661–671 CrossRef CAS PubMed.
- T. E. Thingholm and M. R. Larsen, Methods Mol. Biol., 2016, 1355, 147–160 CrossRef CAS PubMed.
- S. Sidoli, C. Lu, M. Coradin, X. Wang, K. R. Karch, C. Ruminowicz and B. A. Garcia, Epigenet. Chromatin, 2017, 10, 34 CrossRef.
- B. A. Garcia, S. Mollah, B. M. Ueberheide, S. A. Busby, T. L. Muratore, J. Shabanowitz and D. F. Hunt, Nat. Protoc., 2007, 2, 933–938 CrossRef CAS PubMed.
- E. Hersman, D. M. Nelson, W. P. Griffith, C. Jelinek and R. J. Cotter, Int. J. Mass Spectrom., 2012, 312, 5–16 CrossRef CAS PubMed.
- R. Liao, H. Wu, H. Deng, Y. Yu, M. Hu, H. Zhai, P. Yang, S. Zhou and W. Yi, Anal. Chem., 2013, 85, 2253–2259 CrossRef CAS PubMed.
- S. Sidoli, Z. F. Yuan, S. Lin, K. Karch, X. Wang, N. Bhanu, A. M. Arnaudo, L. M. Britton, X. J. Cao, M. Gonzales-Cope, Y. Han, S. Liu, R. C. Molden, S. Wein, L. Afjehi-Sadat and B. A. Garcia, Proteomics, 2015, 15, 1459–1469 CrossRef CAS.
- S. Sidoli, S. Lin, K. R. Karch and B. A. Garcia BA, Anal. Chem., 2015, 87, 3129–3133 CrossRef CAS PubMed.
Footnotes |
| † Electronic supplementary information (ESI) available: Synthesis and characterization (NMRs, ATR, TGA, XPS, TEM, XRD, ζ-potential) of cavitands and ferromagnetic nanoparticles, proteolytic digestion, enrichment and immunological protocols. Full MS analysis data are reported in the dataset supplemental table. See DOI: https://doi.org/10.1039/d4sc02076f |
| ‡ These authors contributed equally. |
| This journal is © The Royal Society of Chemistry 2024 |