Open Access Article
Open Access ArticleNanopore-based glycan sequencing: state of the art and future prospects
Guangda
Yao†
abc,
Wenjun
Ke†
ad,
Bingqing
Xia
*ad and
Zhaobing
Gao
 *ade
*ade
aState Key Laboratory of Drug Research, Shanghai Institute of Materia Medica, Chinese Academy of Sciences, 201203, Shanghai, China. E-mail: zbgao@simm.ac.cn; xiabingqing@simm.ac.cn
bSchool of Life Science and Technology, Shanghai Tech University, 201210, Shanghai, China
cLingang Laboratory, 200031, Shanghai, China
dUniversity of Chinese Academy of Sciences, 100049, Beijing, China
eZhongshan Institute for Drug Discovery, Shanghai Institute of Materia Medica, Chinese Academy of Sciences, 528400, Zhongshan, China
First published on 3rd April 2024
Abstract
Sequencing of biomacromolecules is a crucial cornerstone in life sciences. Glycans, one of the fundamental biomolecules, derive their physiological and pathological functions from their structures. Glycan sequencing faces challenges due to its structural complexity and current detection technology limitations. As a highly sensitive sensor, nanopores can directly convert nucleic acid sequence information into electrical signals, spearheading the revolution of third-generation nucleic acid sequencing technologies. However, their potential for deciphering complex glycans remains untapped. Initial attempts demonstrated the significant sensitivity of nanopores in glycan sensing, which provided the theoretical basis and insights for the realization of nanopore-based glycan sequencing. Here, we present three potential technical routes to employ nanopore technology in glycan sequencing for the first time. The three novel technical routes include: strand sequencing, capturing glycan chains as they translocate through nanopores; sequential hydrolysis sequencing, capturing released monosaccharides one by one; splicing sequencing, mapping signals from hydrolyzed glycan fragments to an oligosaccharide database/library. Designing suitable nanopores, enzymes, and motors, and extracting characteristic signals pose major challenges, potentially aided by artificial intelligence. It would be highly desirable to design an all-in-one high-throughput glycan sequencer instrument by integrating a sample processing unit, nanopore array, and signal acquisition system into a microfluidic device. The nanopore sequencer invention calls for intensive multidisciplinary cooperation including electrochemistry, glycochemistry, engineering, materials, enzymology, etc. Advancing glycan sequencing will promote the development of basic research and facilitate the discovery of glycan-based drugs and disease markers, fostering progress in glycoscience and even life sciences.
1. Introduction
Glycans represent the most abundant constituent in all life and are composed of multiple monosaccharides linked through glycosidic bonds.1,2 Glycans are present either in the form of free glycans or glycan-attached glycoconjugates such as glycoproteins,3 glycolipids,4 and glycol-RNA,5 which play essential roles in biological processes involving energy storage, shape regulation, molecular recognition, etc.6 The complex structure of glycans underpins their diverse functions.7 Therefore, elucidating the structure of glycans is crucial for understanding their physiological and pathological roles, which can accelerate the discovery of glycan-based disease biomarkers8,9 and new drugs.10,11 However, glycan sequencing is still challenging due to the intricate heterogeneity of the glycan structure and the limitations of current detection technologies.12–14The primary structure of glycan, comprising the monosaccharide sequence, chain length, glycosidic linkages, anomeric configurations, substituents, and branch,14 is much more complex compared to that of linear nucleic acids and peptides. Nowadays, nuclear magnetic resonance (NMR) spectroscopy, mass spectrometry (MS) and related hyphenated methods are still the mainstream tools for glycan structure analysis.15,16 NMR can be used to conclude α/β anomers, linkage position, and even the monosaccharide order, but the analysis will become more challenging for longer glycans.16–18 MS and MS-based technologies can also elucidate glycan sequences via glycosidic dissociation and cross-ring dissociation, but the resolution requires further improvement, especially for glycans consisting of repeating units.19–22 These limitations urgently require us to develop innovative technologies for efficient glycan sequencing.12
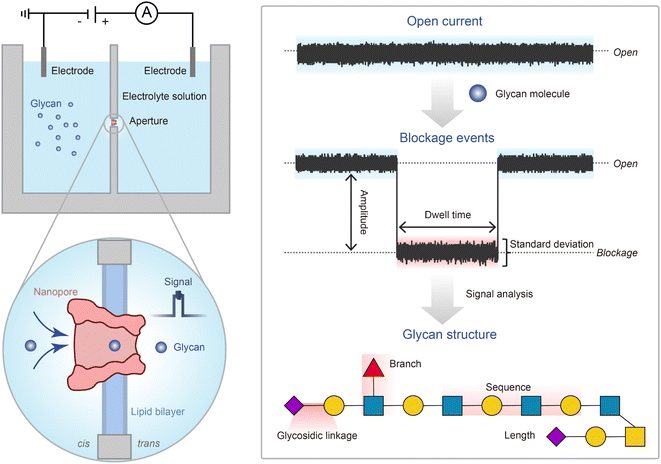
Some emerging technologies just like nanopore technology,23 recognition tunnelling (RT),24 and glycan microarrays25 have been employed for glycan structure characterization.13 Among them, the nanopore technology is the most time-saving, portable, and low-cost method with high sensitivity and high spatial and temporal resolution, providing full composition and structural information of analytes at the single-molecule level.23,26 During the translocation of different analytes through the same nanopore lumen, the different analyte molecules in nanopore confined space induce characteristic ionic current modulation due to volume exclusion and interaction (Fig. 1).27,28 The specific features of current signals including the blockage amplitude, dwell time, and stand deviation (Std) are extracted to elucidate the analytes' structural information of composition, charge distribution, sequence, etc.29,30 The nanopore technology offers comprehensive single-molecule structural insights, which makes it particularly advantageous for the sequencing of biomolecules (e.g., glycans and proteins) which cannot be amplified in vitro like nucleic acids. Over the past few decades, nanopore technology emerged as a successful platform for long-read-length nucleic acid sequencing31–33 and a promising approach for protein sequencing.34,35 Given the compatibility of the size and chemical properties of glycans with nanopores, the nanopore technology holds considerable promise for glycan sequencing as well. The increasingly more attempts at nanopore-based glycan sensing confirmed this speculation to a certain extent,36–45 which provided the theoretical basis and insights for the realization of nanopore-based glycan sequencing.38,39 However, the applicability of nanopores towards glycan sequencing has not been formally proposed yet.
Here, we give a comprehensive overview of the application of nanopore technology in glycan sensing from the 1990s to the present and present the perspective of nanopore-based glycan sequencing for the first time. Three potential technical routes of nanopore-based glycan sequencing are proposed including strand sequencing, sequential hydrolysis sequencing, and splicing sequencing. Additionally, we outline the anticipated process of nanopore glycan sequencing. Although there will be some challenges on the way to glycan sequencing, we believe that with interdisciplinary collaboration, nanopore-based glycan sequencing will become a reality someday. The advancements in glycan sequencing technology will promote the development of glycoscience and its applications in medicine and beyond.46
2. Nanopore-based glycan sensing: from detection to sequencing
The reports about nanopore-based glycan sensors were summarized and classified into three stages. The first stage is nanopore discovery since the 1990s when the nanopore was presented as the sensor. In the second stage, from 2010 to day, glycans started to receive attention from the nanopore or channel researchers. Especially starting in 2020, more and more researchers delved into employing nanopores to discriminate glycans with minor structural differences. Lastly, we classify several recent studies proposing the concept of nanopore glycan sequencing and the future development process into the third stage. At the end of the third stage, the goal of nanopore-based glycan sequencing will come true in the future (Fig. 2).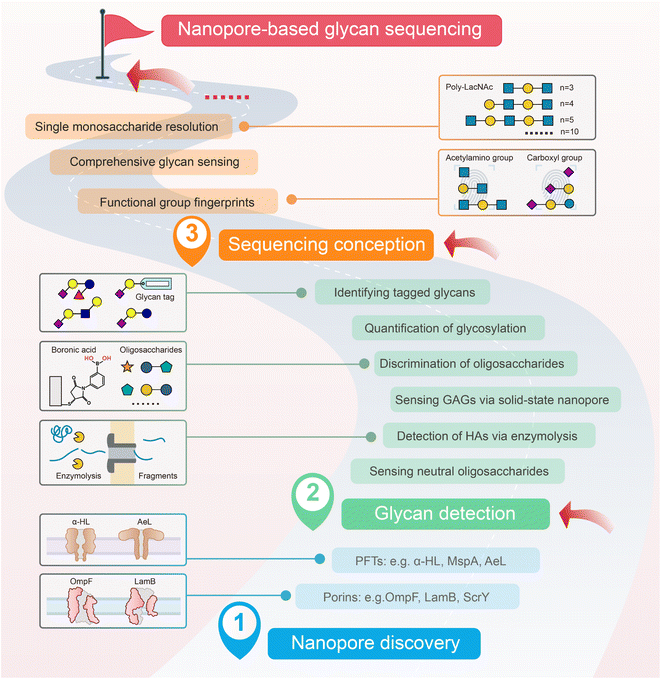 | ||
| Fig. 2 Development stages of the research on nanopore-based glycan sequencing. This development of nanopore application in glycan analysis was classified into three stages: nanopore discovery, glycan detection, and sequencing conception. The final goal is nanopore-based glycan sequencing. Some representative research studies at each stage were briefly described. The representative reports in seven boxes from the bottom to the top were from ref. 51, ref. 63, ref. 89, ref. 40 and 41, ref. 37, ref. 38, and ref. 39. PFTs, pore-forming toxin proteins. α-HL, α-hemolysin. MspA, Mycobacterium smegmatis porin A. AeL, aerolysin. OmpF, outer membrane protein F. LamB, maltoporin. ScrY, sucrose-specific porin. HAs, hyaluronic acids. GAGs, glycosaminoglycans. LacNAc, N-acetyl-D-lactosamine. | ||
2.1 Nanopore discovery
In general, the natural sources of biological nanopores involve bacterial porins and pore-forming toxin proteins (PFTs).47–49 The porins refer to the transmembrane proteins located at the outer membrane of Gram-negative bacteria.48,50 By structurally forming a hollow pore, the porins allow passive diffusion of small biomolecules including oligosaccharides, amino acids, nucleosides, etc.51 According to the specificity, the porins can be classified into general (substrate-nonspecific) porins such as outer membrane protein F (OmpF) and PhoE52,53 and substrate-specific porins just like the sucrose-specific porin (ScrY).54 The general porins (<10 Å) non-specifically allow diffusion of solutes with a size lower than 600 Da, while the specific porins have stronger substrate selectivity and the exclusion limit is 200 Da or lower.48,51 Early in the 1990s, some single-channel studies explored the transport mechanism of glycans via porins such as LamB55 and chitooligosaccharide-uptake porins ChiP (chitoporin).56 Although porins were proposed to be used as the earliest glycan sensors,57 their low conductance, unavoidable gating signals, strong translocation specificity, and high blockage rate hindered their further application as glycan sensors.57 By contrast, the general porins OmpF and OmpG with higher signal stability were developed as biosensors58,59 and possibly applicable for glycan sensing. Nevertheless, the natural transport mechanism of glycan-specific porins seems very enlightening for nanopore design.56,60 For example, the asymmetric charge distribution within most glycan porins could enhance their sensing resolution.60 The aromatic residues lining the pore work to assist the glycan translocation with their hydrophobic core.61 With the help of genetic and protein engineering, the glycan porins may be transformed into satisfactory novel nanopore sensors for glycans.62Compared with the bacterial porins, the nanoscale bacterial pore-forming toxin proteins (PFTs) were developed as widely used nanopores owing to their better perforation activity, stable open state, and strong tolerance of various conditions. Each PFT nanopore consists of several identical subunits that can self-assemble into β-barrel-shaped pores on the phospholipid bilayer,49,63 with the narrowest region (usually called the constriction site) as the primary sensing region. The adequate diameter with lower substrate selectivity enabled them to be developed as the universal sequencing tools for different biopolymers.64–69 Since the first heptameric nanopore α-hemolysin (a-HL) from Staphylococcus aureus was developed as a single-molecule sensor,64 more PFTs including Mycobacterium smegmatis porin A (MspA),65,70 aerolysin (AeL),71,72 fragaceatoxin C (FraC),73,74Escherichia coli curli transport channel (CsgG),75,76etc. have been confirmed to allow the detection or/and sequencing of various analytes including peptides, nucleic acids, etc.30,77 For instance, MspA and CsgG have been employed in DNA sequencing because of their narrower sensing region (less than 1 nm) and pore radius (1 nm approximately).75,78 Although the wild-type nanopores appeared less sensitive to glycans than glycan-specific porins,48,79 their structure and properties have been elucidated more sufficiently, which will guide us to optimize their performance in glycan sensing.80
Besides the above easily accessible and widely used PFT nanopores, it is also possible to obtain novel glycan-matched nanopores through de novo design,81 porin screening or engineering,82 and shape-tunable nanopores.83 Beyond biological nanopores, artificial nanopores including solid-state nanopores,2 nanopipettes,84 and chemosynthetic membrane channels,85 are also optional glycan sensing tools. However, site-directed modification on solid-state nanopores is not as convenient as that on biological nanopores. The surface coatings could improve their sensitivity and control the pore diameter.86 Compared with biological nanopores, most artificial nanopores are quite stable for long-term sequencing, but the fabrication of artificial nanopores with high-precision size is still challenging.2 A hybrid nanopore, obtained by inserting a single protein nanopore into a solid-state nanopore, shows better robustness while retaining high sensitivity.87,88 The hybrid nanopore array could be created for high-throughput sequencing.28
2.2 Sensing strategies for glycan detection
In the 2010s, some researchers began to explore the capability of nanopores to detect glycans. The feasibility was preliminarily confirmed in 2011 by the attempt to discriminate maltose and dextran oligosaccharides with different glycosidic linkages and polymerization degrees according to the differentiated dwell time.43 In the subsequent years, more and more researchers further explored the feasibility of glycan detection using nanopores. Some ingenious nanopore-based glycan sensing strategies have been devised such as site-directed mutation of nanopores,38 combination of enzymatic hydrolysis and nanopore sensing,89,90 capture of glycan molecules by boronic acid covalently bound to nanopores,40–42 assistance of glycan binding proteins (GBPs),91,92 surface modification of solid-state nanopores,45,93–95 and chemical tags of glycans.37 These approaches aimed to enhance sensitivity and resolution in glycan detection and discrimination. Various types of glycan structural differences were successfully identified using engineered or unmodified nanopores, including diverse building blocks,38,41,42 distinct glycosidic linkages,37,39,40 varying lengths,37–39 and branches.37 It should be noticed that different strategies could be challenging when applied to different model glycans. For example, boronic acid-bound nanopores expressed high sensitivity in discriminating both monosaccharides and disaccharides.40,41,96 But they may face challenges in identifying large-sized glycans due to blurred event clusters. Comparatively, wild-type (WT) aerolysin nanopores were utilized to characterize glycosaminoglycan oligosaccharides with various sulfate patterns, glycosidic bonds, and epimers of uronic acid residues, which marked the success of nanopores in comprehensive elucidation of longer charged glycans,36 although the sensitivity seems not as high as some engineered nanopores. Besides, the analysis ability of solid-state nanopores is also examined on plant polysaccharides45 and glycosaminoglycans (GAGs),94,95 but the resolution needs further improvement. Anyway, these meaningful efforts advanced the application of nanopores in glycan detection and provided vitally important insights for future research on nanopore glycan sensing. Beyond these reports related to nanopore glycan sensing, since nanopores are currently most widely used in nucleic acids and peptides, many sensing strategies and experiences such as analyte carriers97 and host–guest interaction69 can be learned. Meanwhile, developing glycan-specific sensing strategies remains a crucial task.2.3 Step towards glycan sequencing
Following glycan detection, our ultimate goal is to achieve nanopore-based glycan sequencing. Glycan sequencing required comprehensive and accurate elucidation of the monosaccharide sequence, anomeric carbons' configuration, glycosidic linkages, branches (position, sequence, and modification pattern), and substituents, demanding the further development of a powerful nanopore platform. Numerous challenges remain to be addressed before achieving glycan sequencing: recognition of the functional group substituent, achieving single monosaccharide resolution on the glycan chain, discrimination of successive addition of single monosaccharides, etc. Previous research demonstrated that the fingerprints of glycan functional groups can be established by nanopores.38 The glycan fingerprints were utilized for the identification of glycans with different functional groups and varying lengths. In this work, we presented the conception of nanopore-based glycan sequencing for the first time.38 Subsequently, we screened out a more versatile engineered nanopore for comprehensive identification of the glycan sequence, isomers, and length with a more direct and rapid procedure. The highly sensitive nanopore achieved single monosaccharide resolution on the glycan chain. The resolution of the single building block was considered a necessity for nanopore-based biopolymer sequencing.98–101 The nanopore also allowed the accurate discrimination of the chain length from pentasaccharides to decasaccharides, which has never been reported and implied the ability to sense the glycan chain. We also discriminated glycan isomers at the tetrasaccharide level.39 These published studies not only confirmed the comprehensive sensing ability of nanopores for larger-sized glycans but also marked the first step on the way toward future nanopore-based glycan sequencing.All of the above significant efforts provided the theoretical basis and insights for the realization of nanopore-based glycan sequencing and boosted the progress toward the promising final goal, although it must be admitted that there is still a long way to go. The next task is to design highly general and applicable nanopore sequencing routes that can achieve the determination of unknown glycan sequences, laying the foundation for the ultimate nanopore glycan sequencing machine (glycan sequencer).
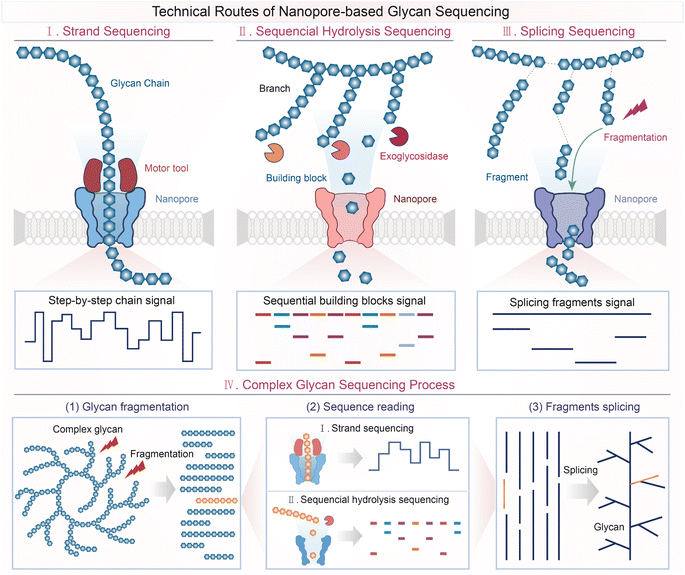
3. Technical routes of nanopore-based glycan sequencing
To achieve nanopore-based glycan sequencing, three technique routes were proposed: strand sequencing, sequential hydrolysis sequencing, and splicing sequencing (Fig. 3). We have presented the basic sequencing principles, the key points, and the application scope of each sequencing scheme, respectively.3.1 Strand sequencing
Nowadays, based on the strand sequencing strategy, Oxford Nanopore Technology (ONT) has achieved long-read DNA sequencing.28,102,103 Moreover, a few researchers have confirmed the possibility of peptide strand sequencing using nanopores by conjugating the peptide with an oligonucleotide recently.34,104,105 Similarly, nanopore technology may be also applicable in glycan strand sequencing. The basic principle of glycan strand sequencing is that when the glycan chain translocates through the nanopore, the monosaccharides on the glycan interact with the sensing region in order, which will be reflected in the ‘step-by-step’ ionic current alteration, each alteration corresponding to an individual monosaccharide if the resolution is high enough, so the sequence information of the glycan can be elucidated manually or automatically with the assistance of machine learning. Theoretically, this strategy can be utilized for sequencing any chained glycan especially when the glycan chain itself is highly charged to make itself fully stretched, such as glycosaminoglycans (GAGs) which are the major linear glycans and participate in various biochemical processes.106 For neutral glycans with flexible structures, it could be helpful to introduce charged groups to stretch them out. The branched glycans may not fit strand sequencing route as their larger size makes the entrance into the nanopore and translocation difficult.Several issues in glycan strand sequencing need to be taken into consideration to improve the resolution, many of which are based on the experience in DNA/peptide strand sequencing. First, the interactions between many natural glycans and the nanopore lumen are not strong enough to induce significant signals. The solutions include site-directed mutation based on the amino acid properties inside the nanopore, and chemical modification of the nanopore or the analytes (covalent or non-covalent, including aptamers, chemical tags, etc.).37,99,107–109 Additionally, the dwell time of the analytes in the pore needs to be controlled to make sure that the analytes can interact with the nanopore sufficiently so that the resolution can be improved. Narrowing the translocation path by mutation/modification of the nanopore or the glycan might also work in this issue. The radius of solid-state nanopores can be controlled to the sub-nanometer level, which may be comparable to the size of glycan molecules, but enhancing the interaction needs to be considered carefully.2 Besides, using a motor protein to control the movement of the glycan molecule is another idea.110 The development of gene and protein engineering may provide insights into the design of a glycan motor protein. Finally, unlike nucleic acids, many glycans are neutral, without high charge density to induce electrostatic repulsion so they tend to form secondary structures while translocating through the nanopore, which would bring great challenges to nanopore-based glycan sequencing. Modifying the glycans with charged groups might be necessary to keep the glycan chain as straight as possible. In theory, the strand sequencing strategy could be applicable to any chained glycan if the problems above are solved.
3.2 Sequential hydrolysis sequencing
Nanopore-based sequential hydrolysis sequencing was initially proposed for nucleic acid sequencing and used to achieve continuous identification of nucleotides cleaved from ssDNA using an exonuclease.109 Recently, Zhang et al. presented a proof-of-concept demonstration of peptide hydrolysis sequencing using a nanopore with the help of carboxypeptidases.69 This breakthrough confirmed the feasibility of sequential hydrolysis sequencing in biopolymers. There were also various exoglycosidases (EXGases) functioning to cleave external glycosidic bonds and remove terminal monosaccharides from the glycan non-reducing end, which made glycan sequential hydrolysis sequencing also feasible theoretically. To date, some attempts have been made to identify glycans using EXGases combined with classic detection methods such as MS.111,112 Presently, we proposed the technical route of nanopore-based glycan sequential hydrolysis sequencing. Sequential hydrolysis of glycans using EXGases causes monosaccharides on glycans to be released and captured by the nanopore and induces real-time translocation signals sequentially. The order of distinct translocation signals corresponds to the sequence of monosaccharides while the EXGases specificity corresponds to the stereochemistry information (e.g., conformation and configuration of the glycosidic bond). Compared with strand sequencing, the advantages of sequential hydrolysis sequencing are mainly reflected in smaller-sized and branched oligosaccharides containing limited types of glycosidic linkages. For example, N-glycan and O-glycan which play essential roles in numerous biological processes and participate in immune regulation, signal transduction, pathogenic infections, etc.,113,114 could be sequenced through sequential hydrolysis sequencing.Likewise, there are a few key points in this technical route. To begin with, the temperature and substrate concentration should be adequate so that the hydrolysis rate can match the nanopore capture rate to ensure that the order of monosaccharide signals corresponds to the glycan sequence exactly. By manipulating the active site and activity of the exonuclease through enzyme engineering and fusion expression or chemical attachment of the exonuclease to the entrance of the nanopore, it is expected that the released monosaccharides will enter the pore at a suitable trajectory and speed. If the capture rate is lower than the hydrolysis rate, it is better to perform the enzymatic hydrolysis first and then enrich the monosaccharide products for nanopore detection. In this way, the abundance of monosaccharide-specific translocation signals could be analyzed to reflect the glycan sequence information. In addition, as mentioned above, the stereochemistry information of the glycan is concluded based on the specificity of the EXGases, so EXGases with high specificity are needed to ensure accurate sequencing. Nevertheless, the species and specificity of EXGases are quite limited, which could be solved by the collaboration of enzyme engineering and computer-aided design. Lastly, sequential hydrolysis of one glycan molecule should synchronize with the others to make sure that the hydrolysis product is relatively homogeneous so that the translocation signal can fit the monosaccharide specificity well. Adjusting the hydrolysis time according to signal quality might be helpful.101 Considering the difficulty of discrimination of all the monosaccharides,14 the sequential hydrolysis sequencing routes could be initially applied to a series of glycans such as a mammalian glycan which consists of only nine monosaccharides.115
3.3 Splicing sequencing
Splicing sequencing is a classic sequencing strategy in DNA sequencing, which has been used as early as the 1990s when Human Genome Project (HGP) was ongoing and was defined as short-gun sequencing.116,117 At present, this strategy is utilized in almost all whole genome sequencing.118,119 The basic principle of splicing sequencing is to obtain the complete sequence of biopolymers by splicing their fragment sequence. The molecular masses of some polysaccharide polymers can reach up to 1000 kDa.115 For glycans with multiple branches or/and extremely large size, it is challenging to sequence them directly with strand sequencing or sequential hydrolysis sequencing due to some limitations (e.g., limited read length in strand sequencing and low specificity of glycosidases in hydrolysis sequencing). Splicing sequencing methods offer a solution to sequencing these large-sized glycan molecules.In the direct glycan splicing sequencing we defined here, first, the glycan molecules were broken into shorter fragments. The glycan fragmentation methods involve acid hydrolysis, enzymatic hydrolysis (endoglycosidases (ENGases)), ultrasonic treatment, and microwave radiation, and their mechanism and characteristics were reviewed by Chen et al.120 Unlike EXGases in sequential hydrolysis sequencing, the fragmentation induced by these methods occurs inside the glycan molecules rather than at the ends. Next, the fragments will be directly sensed by the nanopore and identified via mapping the characteristic signals to the pre-created library of glycan signals. Finally, we could obtain the complete glycan sequence by splicing these glycan fragments. This splicing sequencing method will be relatively easier to implement when the fragments are small enough because the library of smaller oligosaccharides is relatively more accessible. However, building a database of larger fragments or fragments consisting of diverse building blocks would be a tricky business. Therefore, the direct splicing sequencing method is unpractical for glycans which cannot be completely fragmented using existing hydrolysis methods or containing diverse monosaccharides. Considering the unimaginable workload of building the database due to the diverse glycan building blocks, the splicing sequencing methods can be firstly applied to some specific important glycans through signal profiling.121,122
Inspired by DNA and protein sequencing,123,124 the future nanopore-based glycan sequence process would combine several sequencing methods or strategies to cope with the complexity and diversity of glycan structures. The strand sequencing method or sequential sequencing method can be combined with the splicing strategy to achieve sequencing of complex glycans which cannot be elucidated by the direct splicing sequencing method as mentioned above. Specifically speaking, after the glycan molecules are broken incompletely, the fragments could be sequenced by the strand sequencing or sequential sequencing method. Notably, as described above, strand sequencing and sequential sequencing methods are suitable for sequencing distinct types of glycan. At an early stage, combining other technologies such as mass spectrometry (MS) could be helpful for the evaluation and control of glycan fragment size, to determine the applicability of strand sequencing and sequential hydrolysis sequencing. In the future, it is necessary to develop hydrolysis methods and protocols for sample preparation, which could generate glycan fragments suitable for different nanopore sequencing routes. In this way, the full glycan sequence can be obtained by splicing the fragment sequence assisted by bioinformatics tools. This sequencing process will reduce the difficulty of sequencing complex glycans by avoiding the difficulty in chain sequencing caused by branches, the trouble in hydrolysis sequencing caused by long chains, and the adversity in fragment library construction caused by high complexity. Up to this point, we presented three possible methods and proposed a possible sequencing process of glycan sequencing.
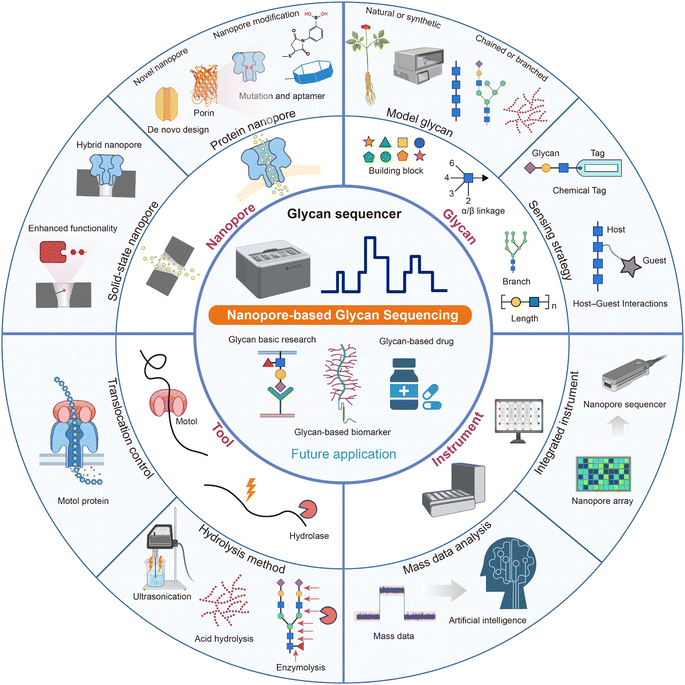
Each method has its most suitable type of glycan structure, which depends on their respective characteristics. But they share some common challenges as well. First, the resolution and sensitivity of nanopore sensing to glycans especially neutral glycans need further improvement. It will be solved by nanopore engineering methods, which include site-directed mutation,38 genetic code expansion (GCE),125 glycan recognition module grafting,126,127 integrated adaptors,128 glycan derivatization including tags with different sizes or properties,37 reversible host–guest interactions,129etc.130 The future novel sensors derived from the de novo protein design81 and glycan porins/transporters will be more glycan-matched.62 Modification with glycan binding proteins (GBPs) such as lectins and antibodies would enable solid-state nanopores to detect glycans with higher sensitivity.91,92,131,132 Hybrid nanopores can be designed and fabricated as nanopore arrays for rapid, high-throughput glycan sequencing.87 Besides, optimization of detection conditions including temperature, applied potentials, and pH values is also helpful to provide additional discrimination capability. Second, the presence of diverse modifications (e.g., sulfation, acetylation, etc.), which could have different impacts on the interactions between the analytes and the pore, needs particular attention.38,133,134 In the future, we will attempt to design a suitable nanopore or even a nanopores platform for different modifications to guarantee a higher accuracy. Third, no matter which sequencing method we use, it is an almost inescapable task to build a huge glycan structure-translocation signal database. The small-sized branched glycan signals are necessary to be concluded in the signal library for attribution of glycan fragment signals to the side or main chain, unless other analysis tools (MS or scanning tunnelling microscopy (STM)) are combined to obtain branch information. In view of the complexity of the sample and the hugeness of the database, processing the sequencing data needs the assistance of artificial intelligence and multi-team collaboration.135 Unification of data standards is necessary for convenient data sharing. Last but not least, the low availability of high-purity glycans impedes the sequencing method training, so developing highly efficient synthesis methods such as automatic glycan synthesis136 and purification methods of natural or synthesized glycans137,138 is urgently needed. With developments in related methods, we believe there will be some excellent proof-of-concept studies of nanopore glycan sequencing based on these technical routes or possibly other ingenious designs in the near future.
4. Outlook and future prospects
4.1 Glycan sequencer instrument
Compared with classic analysis technology NMR and MS, nanopore sensors are more easily miniaturized and integrated into portable devices.139,140 The incorporation of complementary metal-oxide-semiconductor (CMOS)-integrated electronics with high-integration microfluidic systems offers an opportunity for parallel and high-throughput nanopore glycan sequencing.140 It is possible to integrate the sample processing unit and nanopore array (including the necessary molecules, e.g., enzymes) with the current signal detecting and processing unit to assemble a portable glycan sequencing machine. As this field is still in its early stages, massive obstacles are waiting to be overcome. The methods surrounding the nanopore-based glycan sequencer are presented in Fig. 4. The essential nanopore sensors, motor protein or highly specific EXGases, model glycans, sequencing algorithm, and instruments need to be either redesigned or improved. Despite the advantages of nanopore-based glycan sequencing, it must be admitted that MS and NMR have their unique advantages, e.g., MS can be used for the assessment and control of glycan size and help identify branches' positions with higher efficiency. Therefore, the combination of nanopores with other classic analysis methods (MS and NMR) or even optical tools141–143 will help to fully resolve the full structure of complex glycans. To sum up, the nanopore sequencer invention calls for interdisciplinary collaborations including chemistry, glycochemistry, engineering, materials, enzymology, etc.4.2 Future applications
4.2.2.1 Disease diagnosis kit. The interactions between glycans and proteins are implicated in the development of diverse diseases, such as pathogen infection,145 inflammation,146 and cancer.9 Therefore, glycans hold considerable promise as a disease marker.147,148 On the one hand, through nanopore glycan sequencing, various undefined physiological and pathological processes related to glycans will be elucidated more comprehensively, which will promote the discovery of some reliable disease markers. On the other hand, the glycan-related disease biomarkers provide an application scenario for nanopore-based glycan sequencing. Nanopore-based glycan sequencing methods might be developed as a diagnosis tool to analyze specific glycan biomarkers in body fluid samples directly.149 With the development of methods of sample handling, nanopore sequencing, and signal processing, a fast, disease diagnosis kit might be developed, which will improve the convenience and compliance of the diagnosis for both the doctors and the patients.
4.2.2.2 Glycan-based drugs. Glycans have great promise in drug discovery.10 The glycan-based drugs and therapeutics represent a great market.150 The approved drugs include polysaccharides/oligosaccharides, glycomimetics, small molecule glycosides, glycoproteins, glycopeptides, and glycan-based vaccines.11,151,152 On the one hand, nanopore-based glycan sequencing could provide access to well-defined glycans (endogenous or exogenous), which will make the design of glycan-based drugs more rational and less risky. On the other hand, even subtle differences in glycan molecular weight, monosaccharide compositions, and glycosidic linkages could affect their biological activities.152 Heparin, one of the most successful anticoagulant drugs, has drawn global attention due to the heparin contamination crisis in 2007, which suggested the importance of precise glycan structure elucidation for the quality and safety of glycan-based drugs.153 Actually, quality analysis of heparin and other drugs using nanopores has been reported in recent years,36,154 which indicates its potential in drug quality control. The nanopore-based glycan sequencer will be widely applied to the quality control of glycan-based drugs.
5. Conclusion
Glycoscience, a pivotal and burgeoning field, intricately intertwines with all branches of biology.46 However, challenges in glycan sequencing impede the development progress of glycoscience. Recently, significant progress has been made in nanopore-based glycan sensing, shedding light on the feasibility of nanopore-based glycan sequencing. We presented the conception of nanopore-based glycan sequencing in our recent work for the first time.38 In this perspective, we summarized the major advances in nanopore-based glycan analysis. These meaningful reports were manually classified into three stages: nanopore discovery, glycan detection, and sequencing conception. These efforts paved the way to the ultimate goal of nanopore-based glycan sequencing, although there is still a long way to go. Based on the characteristics of the glycan structure and the nanopore sequencing technology, we presented three potential glycan sequencing technical routes and a complex glycan sequencing process. To simplify the sequencing process, we proposed the idea of integrating all the necessary units into a glycan sequencing machine (glycan sequencer). Undoubtedly, the invention of a glycan sequencer will meet different hurdles and require multidisciplinary collaborations. We provided some strategies to improve the sensitivity and resolution of nanopores for glycan sensing, which could provide some insights for overcoming the technical barriers in nanopore-based glycan sequencing. Finally, an outlook was given on the future application of nanopore-based glycan sequencing. We believe that the advancement of glycan sequencing would facilitate the development of glycoscience and even the entire life sciences.Author contributions
Z. Gao and B. Xia provided the key advice and supervised the preparation of the manuscript. G. Yao and W. Ke conducted literature search and analysis of published results. All the authors wrote the manuscript.Conflicts of interest
There are no conflicts to declare.Acknowledgements
We are grateful to the Shanghai Municipal Science and Technology Major Project, the National Science Fund of Distinguished Young Scholars (Grant 81825021), the Fund of Youth Innovation Promotion Association (Grants 2019285, 2021333, and 2022077), the National Natural Science Foundation of China (Grants 92169202 and 82341058), the Fund of Shanghai Science and Technology Innovation Action Plan (Grant 20ZR1474200), the Shanghai Rising-Star Program (Grant 22QA1411000), supported by grants from the Science and Technology Commission of Shanghai Municipality (23JC1404301), the Natural Science Foundation of Shanghai (Grant 22ZR1474000), the National Key Laboratory Program of China (Grants LG202101-01-04 and LG202102-01-01), the National Key Research and Development Program of China (Grant 2020YFC0842000), and the Strategic Leading Science and Technology Projects of Chinese Academy of Sciences (Grant XDA12050308) for financial support.References
- R. F. Service, Cell biology. Looking for a sugar rush, Science, 2012, 338, 321–323 CrossRef CAS.
- L. Xue, H. Yamazaki, R. Ren, M. Wanunu, A. P. Ivanov and J. B. Edel, Solid-state nanopore sensors, Nat. Rev. Mater., 2020, 5, 931–951 CrossRef CAS.
- I. Bagdonaite, S. A. Malaker, D. A. Polasky, N. M. Riley, K. Schjoldager, S. Y. Vakhrushev, A. Halim, K. F. Aoki-Kinoshita, A. I. Nesvizhskii, C. R. Bertozzi, H. H. Wandall, B. L. Parker, M. Thaysen-Andersen and N. E. Scott, Glycoproteomics, Nat. Rev. Methods Primers, 2022, 2, 48 CrossRef CAS.
- C. Kirschbaum, K. Greis, E. Mucha, L. Kain, S. Deng, A. Zappe, S. Gewinner, W. Schöllkopf, G. von Helden, G. Meijer, P. B. Savage, M. Marianski, L. Teyton and K. Pagel, Unravelling the structural complexity of glycolipids with cryogenic infrared spectroscopy, Nat. Commun., 2021, 12, 1201 CrossRef CAS.
- R. A. Flynn, K. Pedram, S. A. Malaker, P. J. Batista, B. A. H. Smith, A. G. Johnson, B. M. George, K. Majzoub, P. W. Villalta, J. E. Carette and C. R. Bertozzi, Small RNAs are modified with N-glycans and displayed on the surface of living cells, Cell, 2021, 184, 3109–3124.e3122 CrossRef CAS PubMed.
- A. Varki, Biological roles of glycans, Glycobiology, 2017, 27, 3–49 CrossRef CAS PubMed.
- C. R. Bertozzi and D. Rabuka, in Essentials of Glycobiology, ed. A. Varki, R. D. Cummings, J. D. Esko, H. H. Freeze, P. Stanley, C. R. Bertozzi, G. W. Hart and M. E. Etzler, Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY, 2009 Search PubMed.
- M. Hu, Y. Lan, A. Lu, X. Ma and L. Zhang, Glycan-based biomarkers for diagnosis of cancers and other diseases: Past, present, and future, Prog. Mol. Biol. Transl. Sci., 2019, 162, 1–24 CAS.
- N. Taniguchi and Y. Kizuka, Glycans and cancer: role of N-glycans in cancer biomarker, progression and metastasis, and therapeutics, Adv. Cancer Res., 2015, 126, 11–51 CrossRef CAS PubMed.
- P. Valverde, A. Ardá, N. C. Reichardt, J. Jiménez-Barbero and A. Gimeno, Glycans in drug discovery, MedChemComm, 2019, 10, 1678–1691 RSC.
- L. Pan, C. Cai, C. Liu, D. Liu, G. Li, R. J. Linhardt and G. Yu, Recent progress and advanced technology in carbohydrate-based drug development, Curr. Opin. Biotechnol., 2021, 69, 191–198 CrossRef CAS PubMed.
- C. J. Gray, L. G. Migas, P. E. Barran, K. Pagel, P. H. Seeberger, C. E. Eyers, G. J. Boons, N. L. B. Pohl, I. Compagnon, G. Widmalm and S. L. Flitsch, Advancing Solutions to the Carbohydrate Sequencing Challenge, J. Am. Chem. Soc., 2019, 141, 14463–14479 CrossRef CAS PubMed.
- A. Lakshminarayanan, M. Richard and B. G. Davis, Studying glycobiology at the single-molecule level, Nat. Rev. Chem, 2018, 2, 148–159 CrossRef CAS.
- Essentials of Glycobiology, ed. A. Varki, R. D. Cummings, J. D. Esko, P. Stanley, G. W. Hart, M. Aebi, D. Mohnen, T. Kinoshita, N. H. Packer, J. H. Prestegard, R. L. Schnaar and P. H. Seeberger, Cold Spring Harbor Laboratory Press, Cold Spring Harbor, New York, 2022, DOI:10.1101/9781621824213.
- M. Grabarics, M. Lettow, C. Kirschbaum, K. Greis, C. Manz and K. Pagel, Mass Spectrometry-Based Techniques to Elucidate the Sugar Code, Chem. Rev., 2022, 122, 7840–7908 CrossRef CAS PubMed.
- C. Fontana and G. Widmalm, Primary Structure of Glycans by NMR Spectroscopy, Chem. Rev., 2023, 123, 1040–1102 CrossRef CAS PubMed.
- H. Y. Yao, J. Q. Wang, J. Y. Yin, S. P. Nie and M. Y. Xie, A review of NMR analysis in polysaccharide structure and conformation: Progress, challenge and perspective, Food Res. Int., 2021, 143, 110290 CrossRef CAS PubMed.
- I. Speciale, A. Notaro, P. Garcia-Vello, F. Di Lorenzo, S. Armiento, A. Molinaro, R. Marchetti, A. Silipo and C. De Castro, Liquid-state NMR spectroscopy for complex carbohydrate structural analysis: A hitchhiker's guide, Carbohydr. Polym., 2022, 277, 118885 CrossRef CAS PubMed.
- J. Zaia, Mass spectrometry of oligosaccharides, Mass Spectrom. Rev., 2004, 23, 161–227 CrossRef CAS PubMed.
- M. J. Kailemia, L. R. Ruhaak, C. B. Lebrilla and I. J. Amster, Oligosaccharide analysis by mass spectrometry: a review of recent developments, Anal. Chem., 2014, 86, 196–212 CrossRef CAS PubMed.
- L. Veillon, Y. Huang, W. Peng, X. Dong, B. G. Cho and Y. Mechref, Characterization of isomeric glycan structures by LC-MS/MS, Electrophoresis, 2017, 38, 2100–2114 CrossRef CAS PubMed.
- X. Dong, Y. Huang, B. G. Cho, J. Zhong, S. Gautam, W. Peng, S. D. Williamson, A. Banazadeh, K. Y. Torres-Ulloa and Y. Mechref, Advances in mass spectrometry-based glycomics, Electrophoresis, 2018, 39, 3063–3081 CrossRef CAS.
- M. MacKenzie and C. Argyropoulos, An Introduction to Nanopore Sequencing: Past, Present, and Future Considerations, Micromachines, 2023, 14, 459 CrossRef.
- J. Im, S. Biswas, H. Liu, Y. Zhao, S. Sen, S. Biswas, B. Ashcroft, C. Borges, X. Wang, S. Lindsay and P. Zhang, Electronic single-molecule identification of carbohydrate isomers by recognition tunnelling, Nat. Commun., 2016, 7, 13868 CrossRef CAS.
- Y. Kim, J. Y. Hyun and I. Shin, Glycan microarrays from construction to applications, Chem. Soc. Rev., 2022, 51, 8276–8299 RSC.
- S. Howorka and Z. Siwy, Nanopore analytics: sensing of single molecules, Chem. Soc. Rev., 2009, 38, 2360–2384 RSC.
- S. M. Lu, Y. Y. Peng, Y. L. Ying and Y. T. Long, Electrochemical Sensing at a Confined Space, Anal. Chem., 2020, 92, 5621–5644 CrossRef CAS PubMed.
- Y. Wang, Y. Zhao, A. Bollas, Y. Wang and K. F. Au, Nanopore sequencing technology, bioinformatics and applications, Nat. Biotechnol., 2021, 39, 1348–1365 CrossRef CAS PubMed.
- W. Liu, Z. L. Yang, C. N. Yang, Y. L. Ying and Y. T. Long, Profiling single-molecule reaction kinetics under nanopore confinement, Chem. Sci., 2022, 13, 4109–4114 RSC.
- Y. L. Ying, Z. L. Hu, S. Zhang, Y. Qing, A. Fragasso, G. Maglia, A. Meller, H. Bayley, C. Dekker and Y. T. Long, Nanopore-based technologies beyond DNA sequencing, Nat. Nanotechnol., 2022, 17, 1136–1146 CrossRef CAS PubMed.
- E. L. van Dijk, Y. Jaszczyszyn, D. Naquin and C. Thermes, The Third Revolution in Sequencing Technology, Trends Genet., 2018, 34, 666–681 CrossRef CAS PubMed.
- D. Deamer, M. Akeson and D. Branton, Three decades of nanopore sequencing, Nat. Biotechnol., 2016, 34, 518–524 CrossRef CAS PubMed.
- D. Branton and D. Deamer, Nanopore Sequencing, World Scientific, 2018 Search PubMed.
- H. Brinkerhoff, A. S. W. Kang, J. Liu, A. Aksimentiev and C. Dekker, Multiple rereads of single proteins at single-amino acid resolution using nanopores, Science, 2021, 374, 1509–1513 CrossRef CAS PubMed.
- N. Varongchayakul, J. Song, A. Meller and M. W. Grinstaff, Single-molecule protein sensing in a nanopore: a tutorial, Chem. Soc. Rev., 2018, 47, 8512–8524 RSC.
- P. Bayat, C. Rambaud, B. Priem, M. Bourderioux, M. Bilong, S. Poyer, M. Pastoriza-Gallego, A. Oukhaled, J. Mathé and R. Daniel, Comprehensive structural assignment of glycosaminoglycan oligo- and polysaccharides by protein nanopore, Nat. Commun., 2022, 13, 5113 CrossRef CAS.
- M. Li, Y. Xiong, Y. Cao, C. Zhang, Y. Li, H. Ning, F. Liu, H. Zhou, X. Li, X. Ye, Y. Pang, J. Zhang, X. Liang and G. Qing, Identification of tagged glycans with a protein nanopore, Nat. Commun., 2023, 14, 1737 CrossRef CAS PubMed.
- B. Xia, J. Fang, S. Ma, M. Ma, G. Yao, T. Li, X. Cheng, L. Wen and Z. Gao, Mapping the Acetylamino and Carboxyl Groups on Glycans by Engineered α-Hemolysin Nanopores, J. Am. Chem. Soc., 2023, 145, 18812–18824 CrossRef CAS PubMed.
- G. Yao, Y. Tian, W. Ke, J. Fang, S. Ma, T. Li, X. Cheng, B. Xia, L. Wen and Z. Gao, Direct Identification of Complex Glycans via a Highly Sensitive Engineered Nanopore, J. Am. Chem. Soc., 2024 DOI:10.1021/jacs.4c02081.
- S. Zhang, Z. Cao, P. Fan, W. Sun, Y. Xiao, P. Zhang, Y. Wang and S. Huang, Discrimination of Disaccharide Isomers of Different Glycosidic Linkages Using a Modified MspA Nanopore, Angew Chem. Int. Ed. Engl., 2024, 63, e202316766 CrossRef CAS PubMed.
- S. Zhang, Z. Cao, P. Fan, Y. Wang, W. Jia, L. Wang, K. Wang, Y. Liu, X. Du, C. Hu, P. Zhang, H. Y. Chen and S. Huang, A Nanopore-Based Saccharide Sensor, Angew Chem. Int. Ed. Engl., 2022, 61, e202203769 CrossRef CAS PubMed.
- W. J. Ramsay and H. Bayley, Single-Molecule Determination of the Isomers of d-Glucose and d-Fructose that Bind to Boronic Acids, Angew Chem. Int. Ed. Engl., 2018, 57, 2841–2845 CrossRef CAS PubMed.
- L. Bacri, A. Oukhaled, E. Hémon, F. B. Bassafoula, L. Auvray and R. Daniel, Discrimination of neutral oligosaccharides through a nanopore, Biochem. Biophys. Res. Commun., 2011, 412, 561–564 CrossRef CAS PubMed.
- A. Fennouri, J. Ramiandrisoa, L. Bacri, J. Mathé and R. Daniel, Comparative biosensing of glycosaminoglycan hyaluronic acid oligo- and polysaccharides using aerolysin and α-hemolysin nanopores, Eur. Phys. J. E: Soft Matter Biol. Phys., 2018, 41, 127 CrossRef CAS PubMed.
- Y. Cai, B. Zhang, L. Liang, S. Wang, L. Zhang, L. Wang, H. L. Cui, Y. Zhou and D. Wang, A solid-state nanopore-based single-molecule approach for label-free characterization of plant polysaccharides, Plant Commun., 2021, 2, 100106 CrossRef CAS.
- S. L. Flitsch, Introduction: Glycosciences, Chem. Rev., 2022, 122, 15501–15502 CrossRef CAS PubMed.
- S. F. Mayer, C. Cao and M. Dal Peraro, Biological nanopores for single-molecule sensing, iScience, 2022, 25, 104145 CrossRef CAS.
- J. Vergalli, I. V. Bodrenko, M. Masi, L. Moynié, S. Acosta-Gutiérrez, J. H. Naismith, A. Davin-Regli, M. Ceccarelli, B. van den Berg, M. Winterhalter and J. M. Pagès, Porins and small-molecule translocation across the outer membrane of Gram-negative bacteria, Nat. Rev. Microbiol., 2020, 18, 164–176 CrossRef CAS.
- F. R. Ulhuq and G. Mariano, Bacterial pore-forming toxins, Microbiology, 2022, 168, 001154 CrossRef CAS PubMed.
- M. Masi, M. Winterhalter and J. M. Pagès, Outer Membrane Porins, Subcell. Biochem., 2019, 92, 79–123 CrossRef CAS.
- J. D. Prajapati, U. Kleinekathöfer and M. Winterhalter, How to Enter a Bacterium: Bacterial Porins and the Permeation of Antibiotics, Chem. Rev., 2021, 121, 5158–5192 CrossRef CAS PubMed.
- S. O. Hagge, H. de Cock, T. Gutsmann, F. Beckers, U. Seydel and A. Wiese, Pore formation and function of phosphoporin PhoE of Escherichia coli are determined by the core sugar moiety of lipopolysaccharide, J. Biol. Chem., 2002, 277, 34247–34253 CrossRef CAS PubMed.
- S. W. Cowan, R. M. Garavito, J. N. Jansonius, J. A. Jenkins, R. Karlsson, N. König, E. F. Pai, R. A. Pauptit, P. J. Rizkallah, J. P. Rosenbusch, G. Rummel and T. Schirmer, The structure of OmpF porin in a tetragonal crystal form, Structure, 1995, 3, 1041–1050 CrossRef CAS PubMed.
- D. Forst, W. Welte, T. Wacker and K. Diederichs, Structure of the sucrose-specific porin ScrY from Salmonella typhimurium and its complex with sucrose, Nat. Struct. Biol., 1998, 5, 37–46 CrossRef CAS PubMed.
- L. Kullman, M. Winterhalter and S. M. Bezrukov, Transport of maltodextrins through maltoporin: a single-channel study, Biophys. J., 2002, 82, 803–812 CrossRef CAS PubMed.
- S. Sanram, A. Aunkham, R. Robinson and W. Suginta, Structural displacement model of chitooligosaccharide transport through chitoporin, J. Biol. Chem., 2023, 299, 105000 CrossRef CAS.
- E. Berkane, F. Orlik, A. Charbit, C. Danelon, D. Fournier, R. Benz and M. Winterhalter, Nanopores: maltoporin channel as a sensor for maltodextrin and lambda-phage, J. Nanobiotechnol., 2005, 3, 3 CrossRef CAS PubMed.
- J. Wang, J. D. Prajapati, F. Gao, Y. L. Ying, U. Kleinekathöfer, M. Winterhalter and Y. T. Long, Identification of Single Amino Acid Chiral and Positional Isomers Using an Electrostatically Asymmetric Nanopore, J. Am. Chem. Soc., 2022, 144, 15072–15078 CrossRef CAS PubMed.
- M. A. Fahie and M. Chen, Electrostatic Interactions between OmpG Nanopore and Analyte Protein Surface Can Distinguish between Glycosylated Isoforms, J. Phys. Chem. B, 2015, 119, 10198–10206 CrossRef CAS PubMed.
- F. Dumas, R. Koebnik, M. Winterhalter and P. Van Gelder, Sugar transport through maltoporin of Escherichia coli. Role of polar tracks, J. Biol. Chem., 2000, 275, 19747–19751 CrossRef CAS PubMed.
- A. Charbit, J. Wang, V. Michel and M. Hofnung, A cluster of charged and aromatic residues in the C-terminal portion of maltoporin participates in sugar binding and uptake, Mol. Gen. Genet., 1998, 260, 185–192 CrossRef CAS.
- D. Vikraman, R. Satheesan, M. Rajendran, N. A. Kumar, J. B. Johnson, S. K. R and K. R. Mahendran, Selective Translocation of Cyclic Sugars through Dynamic Bacterial Transporter, ACS Sens., 2022, 7, 1766–1776 CrossRef CAS.
- M. Zhang, C. Chen, Y. Zhang and J. Geng, Biological nanopores for sensing applications, Proteins, 2022, 90, 1786–1799 CrossRef CAS.
- L. Song, M. R. Hobaugh, C. Shustak, S. Cheley, H. Bayley and J. E. Gouaux, Structure of staphylococcal alpha-hemolysin, a heptameric transmembrane pore, Science, 1996, 274, 1859–1866 CrossRef CAS.
- M. Faller, M. Niederweis and G. E. Schulz, The structure of a mycobacterial outer-membrane channel, Science, 2004, 303, 1189–1192 CrossRef CAS PubMed.
- R. Strack, Aerolysin nanopores, Nat. Methods, 2020, 17, 29 Search PubMed.
- M. Jain, R. Abu-Shumays, H. E. Olsen and M. Akeson, Advances in nanopore direct RNA sequencing, Nat. Methods, 2022, 19, 1160–1164 CrossRef CAS PubMed.
- A. H. Laszlo, I. M. Derrington, B. C. Ross, H. Brinkerhoff, A. Adey, I. C. Nova, J. M. Craig, K. W. Langford, J. M. Samson, R. Daza, K. Doering, J. Shendure and J. H. Gundlach, Decoding long nanopore sequencing reads of natural DNA, Nat. Biotechnol., 2014, 32, 829–833 CrossRef CAS PubMed.
- Y. Zhang, Y. Yi, Z. Li, K. Zhou, L. Liu and H. C. Wu, Peptide sequencing based on host-guest interaction-assisted nanopore sensing, Nat. Methods, 2024, 21, 102–109 CrossRef CAS PubMed.
- S. Punthambaker, Detection of modified RNA with an engineered nanopore, Nat. Nanotechnol., 2022, 17, 1044–1045 CrossRef CAS PubMed.
- I. Iacovache, S. De Carlo, N. Cirauqui, M. Dal Peraro, F. G. van der Goot and B. Zuber, Cryo-EM structure of aerolysin variants reveals a novel protein fold and the pore-formation process, Nat. Commun., 2016, 7, 12062 CrossRef CAS PubMed.
- M. Z. Huo, Z. L. Hu, Y. L. Ying and Y. T. Long, Enhanced identification of Tau acetylation and phosphorylation with an engineered aerolysin nanopore, Proteomics, 2022, 22, e2100041 CrossRef PubMed.
- K. Tanaka, J. M. Caaveiro, K. Morante, J. M. González-Mañas and K. Tsumoto, Structural basis for self-assembly of a cytolytic pore lined by protein and lipid, Nat. Commun., 2015, 6, 6337 CrossRef PubMed.
- R. C. A. Versloot, F. L. R. Lucas, L. Yakovlieva, M. J. Tadema, Y. Zhang, T. M. Wood, N. I. Martin, S. J. Marrink, M. T. C. Walvoort and G. Maglia, Quantification of Protein Glycosylation Using Nanopores, Nano Lett., 2022, 22, 5357–5364 CrossRef CAS PubMed.
- S. E. Van der Verren, N. Van Gerven, W. Jonckheere, R. Hambley, P. Singh, J. Kilgour, M. Jordan, E. J. Wallace, L. Jayasinghe and H. Remaut, A dual-constriction biological nanopore resolves homonucleotide sequences with high fidelity, Nat. Biotechnol., 2020, 38, 1415–1420 CrossRef CAS PubMed.
- P. Goyal, P. V. Krasteva, N. Van Gerven, F. Gubellini, I. Van den Broeck, A. Troupiotis-Tsaïlaki, W. Jonckheere, G. Péhau-Arnaudet, J. S. Pinkner, M. R. Chapman, S. J. Hultgren, S. Howorka, R. Fronzes and H. Remaut, Structural and mechanistic insights into the bacterial amyloid secretion channel CsgG, Nature, 2014, 516, 250–253 CrossRef CAS PubMed.
- X. Wei, T. Penkauskas, J. E. Reiner, C. Kennard, M. J. Uline, Q. Wang, S. Li, A. Aksimentiev, J. W. F. Robertson and C. Liu, Engineering Biological Nanopore Approaches toward Protein Sequencing, ACS Nano, 2023, 17, 16369–16395 CrossRef CAS PubMed.
- I. M. Derrington, T. Z. Butler, M. D. Collins, E. Manrao, M. Pavlenok, M. Niederweis and J. H. Gundlach, Nanopore DNA sequencing with MspA, Proc. Natl. Acad. Sci. U. S. A., 2010, 107, 16060–16065 CrossRef PubMed.
- J. M. Jeckelmann and B. Erni, Transporters of glucose and other carbohydrates in bacteria, Pflugers Arch., 2020, 472, 1129–1153 CrossRef CAS PubMed.
- J. T. Hagan, B. S. Sheetz, Y. Bandara, B. I. Karawdeniya, M. A. Morris, R. B. Chevalier and J. R. Dwyer, Chemically tailoring nanopores for single-molecule sensing and glycomics, Anal. Bioanal. Chem., 2020, 412, 6639–6654 CrossRef CAS PubMed.
- K. Shimizu, B. Mijiddorj, M. Usami, I. Mizoguchi, S. Yoshida, S. Akayama, Y. Hamada, A. Ohyama, K. Usui, I. Kawamura and R. Kawano, De novo design of a nanopore for single-molecule detection that incorporates a β-hairpin peptide, Nat. Nanotechnol., 2022, 17, 67–75 CrossRef CAS PubMed.
- D. Vikraman, R. Satheesan, K. S. Kumar and K. R. Mahendran, Nanopore Passport Control for Substrate-Specific Translocation, ACS Nano, 2020, 14, 2285–2295 CrossRef CAS.
- Y. Xing, A. Dorey, L. Jayasinghe and S. Howorka, Highly shape- and size-tunable membrane nanopores made with DNA, Nat. Nanotechnol., 2022, 17, 708–713 CrossRef CAS.
- Z. R. Zhou, X. Y. Wang, J. Lv, B. B. Chen, Y. B. Tang and R. C. Qian, Nanopipette-assisted single cell metabolic glycan labeling, RSC Adv., 2019, 9, 30666–30670 RSC.
- D. Qiao, H. Joshi, H. Zhu, F. Wang, Y. Xu, J. Gao, F. Huang, A. Aksimentiev and J. Feng, Synthetic Macrocycle Nanopore for Potassium-Selective Transmembrane Transport, J. Am. Chem. Soc., 2021, 143, 15975–15983 CrossRef CAS.
- O. M. Eggenberger, C. Ying and M. Mayer, Surface coatings for solid-state nanopores, Nanoscale, 2019, 11, 19636–19657 RSC.
- A. R. Hall, A. Scott, D. Rotem, K. K. Mehta, H. Bayley and C. Dekker, Hybrid pore formation by directed insertion of α-haemolysin into solid-state nanopores, Nat. Nanotechnol., 2010, 5, 874–877 CrossRef CAS.
- D. F. Cairns-Gibson and S. L. Cockroft, Functionalised nanopores: chemical and biological modifications, Chem. Sci., 2022, 13, 1869–1882 RSC.
- A. Fennouri, C. Przybylski, M. Pastoriza-Gallego, L. Bacri, L. Auvray and R. Daniel, Single molecule detection of glycosaminoglycan hyaluronic acid oligosaccharides and depolymerization enzyme activity using a protein nanopore, ACS Nano, 2012, 6, 9672–9678 CrossRef CAS PubMed.
- A. Fennouri, R. Daniel, M. Pastoriza-Gallego, L. Auvray, J. Pelta and L. Bacri, Kinetics of enzymatic degradation of high molecular weight polysaccharides through a nanopore: experiments and data-modeling, Anal. Chem., 2013, 85, 8488–8492 CrossRef CAS PubMed.
- N. S. Galenkamp, M. Soskine, J. Hermans, C. Wloka and G. Maglia, Direct electrical quantification of glucose and asparagine from bodily fluids using nanopores, Nat. Commun., 2018, 9, 4085 CrossRef PubMed.
- D. K. Kwak, J. S. Kim, M. K. Lee, K. S. Ryu and S. W. Chi, Probing the Neuraminidase Activity of Influenza Virus Using a Cytolysin A Protein Nanopore, Anal. Chem., 2020, 92, 14303–14308 CrossRef CAS PubMed.
- B. I. Karawdeniya, Y. Bandara, J. W. Nichols, R. B. Chevalier and J. R. Dwyer, Surveying silicon nitride nanopores for glycomics and heparin quality assurance, Nat. Commun., 2018, 9, 3278 CrossRef.
- F. Rivas, O. K. Zahid, H. L. Reesink, B. T. Peal, A. J. Nixon, P. L. DeAngelis, A. Skardal, E. Rahbar and A. R. Hall, Label-free analysis of physiological hyaluronan size distribution with a solid-state nanopore sensor, Nat. Commun., 2018, 9, 1037 CrossRef.
- K. Xia, J. T. Hagan, L. Fu, B. S. Sheetz, S. Bhattacharya, F. Zhang, J. R. Dwyer and R. J. Linhardt, Synthetic heparan sulfate standards and machine learning facilitate the development of solid-state nanopore analysis, Proc. Natl. Acad. Sci. U. S. A., 2021, 118, e2022806118 CrossRef CAS.
- X. Wu, Z. Li, X. X. Chen, J. S. Fossey, T. D. James and Y. B. Jiang, Selective sensing of saccharides using simple boronic acids and their aggregates, Chem. Soc. Rev., 2013, 42, 8032–8048 RSC.
- X. Wang, T. M. Thomas, R. Ren, Y. Zhou, P. Zhang, J. Li, S. Cai, K. Liu, A. P. Ivanov, A. Herrmann and J. B. Edel, Nanopore Detection Using Supercharged Polypeptide Molecular Carriers, J. Am. Chem. Soc., 2023, 145, 6371–6382 CrossRef CAS PubMed.
- D. Stoddart, A. J. Heron, J. Klingelhoefer, E. Mikhailova, G. Maglia and H. Bayley, Nucleobase recognition in ssDNA at the central constriction of the alpha-hemolysin pore, Nano Lett., 2010, 10, 3633–3637 CrossRef CAS PubMed.
- D. Stoddart, A. J. Heron, E. Mikhailova, G. Maglia and H. Bayley, Single-nucleotide discrimination in immobilized DNA oligonucleotides with a biological nanopore, Proc. Natl. Acad. Sci. U. S. A., 2009, 106, 7702–7707 CrossRef CAS PubMed.
- N. Ashkenasy, J. Sánchez-Quesada, H. Bayley and M. R. Ghadiri, Recognizing a single base in an individual DNA strand: a step toward DNA sequencing in nanopores, Angew Chem. Int. Ed. Engl., 2005, 44, 1401–1404 CrossRef CAS PubMed.
- J. Jiang, M. Y. Li, X. Y. Wu, Y. L. Ying, H. X. Han and Y. T. Long, Protein nanopore reveals the renin-angiotensin system crosstalk with single-amino-acid resolution, Nat. Chem., 2023, 15, 578–586 CrossRef CAS PubMed.
- D. Sheka, N. Alabi and P. M. K. Gordon, Oxford nanopore sequencing in clinical microbiology and infection diagnostics, Briefings Bioinf., 2021, 22, bbaa403 CrossRef PubMed.
- K. Dumschott, M. H. Schmidt, H. S. Chawla, R. Snowdon and B. Usadel, Oxford Nanopore sequencing: new opportunities for plant genomics?, J. Exp. Bot., 2020, 71, 5313–5322 CrossRef CAS PubMed.
- Z. Chen, Z. Wang, Y. Xu, X. Zhang, B. Tian and J. Bai, Controlled movement of ssDNA conjugated peptide through Mycobacterium smegmatis porin A (MspA) nanopore by a helicase motor for peptide sequencing application, Chem. Sci., 2021, 12, 15750–15756 RSC.
- S. Yan, J. Zhang, Y. Wang, W. Guo, S. Zhang, Y. Liu, J. Cao, Y. Wang, L. Wang, F. Ma, P. Zhang, H. Y. Chen and S. Huang, Single Molecule Ratcheting Motion of Peptides in a Mycobacterium smegmatis Porin A (MspA) Nanopore, Nano Lett., 2021, 21, 6703–6710 CrossRef CAS PubMed.
- N. Afratis, C. Gialeli, D. Nikitovic, T. Tsegenidis, E. Karousou, A. D. Theocharis, M. S. Pavão, G. N. Tzanakakis and N. K. Karamanos, Glycosaminoglycans: key players in cancer cell biology and treatment, FEBS J., 2012, 279, 1177–1197 CrossRef CAS PubMed.
- Y. Astier, O. Braha and H. Bayley, Toward single molecule DNA sequencing: direct identification of ribonucleoside and deoxyribonucleoside 5'-monophosphates by using an engineered protein nanopore equipped with a molecular adapter, J. Am. Chem. Soc., 2006, 128, 1705–1710 CrossRef CAS PubMed.
- N. Mitchell and S. Howorka, Chemical tags facilitate the sensing of individual DNA strands with nanopores, Angew Chem. Int. Ed. Engl., 2008, 47, 5565–5568 CrossRef CAS PubMed.
- J. Clarke, H. C. Wu, L. Jayasinghe, A. Patel, S. Reid and H. Bayley, Continuous base identification for single-molecule nanopore DNA sequencing, Nat. Nanotechnol., 2009, 4, 265–270 CrossRef CAS PubMed.
- G. M. Cherf, K. R. Lieberman, H. Rashid, C. E. Lam, K. Karplus and M. Akeson, Automated forward and reverse ratcheting of DNA in a nanopore at 5-Å precision, Nat. Biotechnol., 2012, 30, 344–348 CrossRef CAS.
- E. McLeod, P. Magnelli and X. Shi, Use of Exoglycosidases for the Structural Characterization of Glycans, Methods Mol. Biol., 2021, 2271, 273–280 CrossRef CAS PubMed.
- A. Guttman, Multistructure sequencing of N-linked fetuin glycans by capillary gel electrophoresis and enzyme matrix digestion, Electrophoresis, 1997, 18, 1136–1141 CrossRef CAS PubMed.
- W. Ma, Z. Xu, Y. Jiang, J. Liu, D. Xu, W. Huang and T. Li, Divergent Enzymatic Assembly of a Comprehensive 64-Membered IgG N-Glycan Library for Functional Glycomics, Adv. Sci., 2023, 10, e2303832 CrossRef.
- H. Wilkinson and R. Saldova, Current Methods for the Characterization of O-Glycans, J. Proteome Res., 2020, 19, 3890–3905 CrossRef CAS PubMed.
- M. E. Griffin and L. C. Hsieh-Wilson, Tools for mammalian glycoscience research, Cell, 2022, 185, 2657–2677 CrossRef CAS.
- E. R. Zabarovsky, V. I. Kashuba, B. Pettersson, N. Petrov, V. Zakharyev, R. Gizatullin, T. Lebedeva, V. Bannikov, E. S. Pokrovskaya and V. I. Zabarovska, et al., Shot-gun sequencing strategy for long-range genome mapping: a pilot study, Genomics, 1994, 21, 495–500 CrossRef CAS PubMed.
- X. Perret, R. Fellay, A. J. Bjourson, J. E. Cooper, S. Brenner and W. J. Broughton, Subtraction hybridisation and shot-gun sequencing: a new approach to identify symbiotic loci, Nucleic Acids Res., 1994, 22, 1335–1341 CrossRef CAS PubMed.
- M. R. Wilson, E. Brown, C. Keys, E. Strain, Y. Luo, T. Muruvanda, C. Grim, J. Jean-Gilles Beaubrun, K. Jarvis, L. Ewing, G. Gopinath, D. Hanes, M. W. Allard and S. Musser, Whole Genome DNA Sequence Analysis of Salmonella subspecies enterica serotype Tennessee obtained from related peanut butter foodborne outbreaks, PLoS One, 2016, 11, e0146929 CrossRef PubMed.
- M. Kuroda, T. Ohta, I. Uchiyama, T. Baba, H. Yuzawa, I. Kobayashi, L. Cui, A. Oguchi, K. Aoki, Y. Nagai, J. Lian, T. Ito, M. Kanamori, H. Matsumaru, A. Maruyama, H. Murakami, A. Hosoyama, Y. Mizutani-Ui, N. K. Takahashi, T. Sawano, R. Inoue, C. Kaito, K. Sekimizu, H. Hirakawa, S. Kuhara, S. Goto, J. Yabuzaki, M. Kanehisa, A. Yamashita, K. Oshima, K. Furuya, C. Yoshino, T. Shiba, M. Hattori, N. Ogasawara, H. Hayashi and K. Hiramatsu, Whole genome sequencing of meticillin-resistant Staphylococcus aureus, Lancet, 2001, 357, 1225–1240 CrossRef CAS PubMed.
- S. K. Chen, X. Wang, Y. Q. Guo, X. X. Song, J. Y. Yin and S. P. Nie, Exploring the partial degradation of polysaccharides: Structure, mechanism, bioactivities, and perspectives, Compr. Rev. Food Sci. Food Saf., 2023, 22, 4831–4870 CrossRef CAS.
- F. L. R. Lucas, R. C. A. Versloot, L. Yakovlieva, M. T. C. Walvoort and G. Maglia, Protein identification by nanopore peptide profiling, Nat. Commun., 2021, 12, 5795 CrossRef CAS.
- X. Chen, Human Milk Oligosaccharides (HMOS): Structure, Function, and Enzyme-Catalyzed Synthesis, Adv. Carbohydr. Chem. Biochem., 2015, 72, 113–190 CrossRef PubMed.
- J. Shendure, S. Balasubramanian, G. M. Church, W. Gilbert, J. Rogers, J. A. Schloss and R. H. Waterston, DNA sequencing at 40: past, present and future, Nature, 2017, 550, 345–353 CrossRef CAS PubMed.
- O. Gotoh, Direct mapping and alignment of protein sequences onto genomic sequence, Bioinformatics, 2008, 24, 2438–2444 CrossRef CAS PubMed.
- X. Y. Wu, M. Y. Li, S. J. Yang, J. Jiang, Y. L. Ying, P. R. Chen and Y. T. Long, Controlled Genetic Encoding of Unnatural Amino Acids in a Protein Nanopore, Angew Chem. Int. Ed. Engl., 2023, 62, e202300582 CrossRef CAS.
- R. D. Cummings, M. Etzler, M. G. Hahn, A. Darvill, K. Godula, R. J. Woods and L. K. Mahal, in Essentials of Glycobiology, ed. A. Varki, R. D. Cummings, J. D. Esko, P. Stanley, G. W. Hart, M. Aebi, D. Mohnen, T. Kinoshita, N. H. Packer, J. H. Prestegard, R. L. Schnaar and P. H. Seeberger, Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY, 2022, pp. 645–662, DOI:10.1101/glycobiology.4e.48.
- L. Zhao, F. Wei, X. He, A. Dai, D. Yang, H. Jiang, L. Wen and X. Cheng, Identification of a carbohydrate recognition motif of purinergic receptors, eLife, 2023, 12, e85449 CrossRef PubMed.
- L. Q. Gu, O. Braha, S. Conlan, S. Cheley and H. Bayley, Stochastic sensing of organic analytes by a pore-forming protein containing a molecular adapter, Nature, 1999, 398, 686–690 CrossRef CAS PubMed.
- T. Li, L. Liu, Y. Li, J. Xie and H. C. Wu, A universal strategy for aptamer-based nanopore sensing through host-guest interactions inside α-hemolysin, Angew Chem. Int. Ed. Engl., 2015, 54, 7568–7571 CrossRef CAS PubMed.
- L. Wang, H. Wang, X. Chen, S. Zhou, Y. Wang and X. Guan, Chemistry solutions to facilitate nanopore detection and analysis, Biosens. Bioelectron., 2022, 213, 114448 CrossRef CAS PubMed.
- M. E. Taylor, K. Drickamer, R. L. Schnaar, M. E. Etzler and A. Varki, in Essentials of Glycobiology, ed. A. Varki, R. D. Cummings, J. D. Esko, P. Stanley, G. W. Hart, M. Aebi, A. G. Darvill, T. Kinoshita, N. H. Packer, J. H. Prestegard, R. L. Schnaar and P. H. Seeberger, Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY, 2015, pp. 361–372, DOI:10.1101/glycobiology.3e.028.
- S. Tommasone, F. Allabush, Y. K. Tagger, J. Norman, M. Köpf, J. H. R. Tucker and P. M. Mendes, The challenges of glycan recognition with natural and artificial receptors, Chem. Soc. Rev., 2019, 48, 5488–5505 RSC.
- D. P. Hoogerheide, P. A. Gurnev, T. K. Rostovtseva and S. M. Bezrukov, Effect of a post-translational modification mimic on protein translocation through a nanopore, Nanoscale, 2020, 12, 11070–11078 RSC.
- S. M. Muthana, C. T. Campbell and J. C. Gildersleeve, Modifications of glycans: biological significance and therapeutic opportunities, ACS Chem. Biol., 2012, 7, 31–43 CrossRef CAS PubMed.
- D. Bojar and F. Lisacek, Glycoinformatics in the Artificial Intelligence Era, Chem. Rev., 2022, 122, 15971–15988 CrossRef CAS PubMed.
- Z. Wang, W. Zhao, G. F. Hao and B. A. Song, Automated synthesis: current platforms and further needs, Drug Discovery Today, 2020, 25, 2006–2011 CrossRef CAS PubMed.
- M. Tudu and A. Samanta, Natural polysaccharides: Chemical properties and application in pharmaceutical formulations, Eur. Polym. J., 2023, 184, 111801 CrossRef CAS.
- I. Benalaya, G. Alves, J. Lopes and L. R. Silva, A Review of Natural Polysaccharides: Sources, Characteristics, Properties, Food, and Pharmaceutical Applications, Int. J. Mol. Sci., 2024, 25, 1322 CrossRef CAS PubMed.
- J. Wang, Y. L. Ying, C. B. Zhong, L. M. Zhang, F. Yan and Y. T. Long, Instrumentational implementation for parallelized nanopore electrochemical measurements, Analyst, 2021, 146, 4111–4120 RSC.
- J. K. Rosenstein, M. Wanunu, C. A. Merchant, M. Drndic and K. L. Shepard, Integrated nanopore sensing platform with sub-microsecond temporal resolution, Nat. Methods, 2012, 9, 487–492 CrossRef CAS PubMed.
- M. Delbianco and Y. Ogawa, Visualizing the structural diversity of glycoconjugates, Nat. Chem. Biol., 2024, 20, 11–12 CrossRef CAS PubMed.
- H.-W. Wang, S. M. Lu, M. Chen and Y. Long, Optical-Facilitated Single-Entity Electrochemistry, Curr. Opin. Electrochem., 2022, 100999 CrossRef CAS.
- K. Anggara, L. Sršan, T. Jaroentomeechai, X. Wu, S. Rauschenbach, Y. Narimatsu, H. Clausen, T. Ziegler, R. L. Miller and K. Kern, Direct observation of glycans bonded to proteins and lipids at the single-molecule level, Science, 2023, 382, 219–223 CrossRef CAS PubMed.
- M. Thaysen-Andersen, D. Kolarich and N. H. Packer, Glycomics & Glycoproteomics: From Analytics to Function, Mol. Omics, 2021, 17, 8–10 RSC.
- A. E. Dugan, A. L. Peiffer and L. L. Kiessling, Advances in glycoscience to understand viral infection and colonization, Nat. Methods, 2022, 19, 384–387 CrossRef CAS PubMed.
- R. L. Schnaar, Glycobiology simplified: diverse roles of glycan recognition in inflammation, J. Leukocyte Biol., 2016, 99, 825–838 CrossRef CAS PubMed.
- C. Reily, T. J. Stewart, M. B. Renfrow and J. Novak, Glycosylation in health and disease, Nat. Rev. Nephrol., 2019, 15, 346–366 CrossRef PubMed.
- H. J. An, S. R. Kronewitter, M. L. de Leoz and C. B. Lebrilla, Glycomics and disease markers, Curr. Opin. Chem. Biol., 2009, 13, 601–607 CrossRef CAS PubMed.
- X. Chen, S. Zhou, Y. Wang, L. Zheng, S. Guan, D. Wang, L. Wang and X. Guan, Nanopore Single-molecule Analysis of Biomarkers: Providing Possible Clues to Disease Diagnosis, TrAC, Trends Anal. Chem., 2023, 162, 117060 CrossRef CAS PubMed.
- B. A. H. Smith and C. R. Bertozzi, The clinical impact of glycobiology: targeting selectins, Siglecs and mammalian glycans, Nat. Rev. Drug Discovery, 2021, 20, 217–243 CrossRef CAS PubMed.
- X. Cao, X. Du, H. Jiao, Q. An, R. Chen, P. Fang, J. Wang and B. Yu, Carbohydrate-based drugs launched during 2000-2021, Acta Pharm. Sin. B, 2022, 12, 3783–3821 CrossRef CAS PubMed.
- C. Anish, M. Beurret and J. Poolman, Combined effects of glycan chain length and linkage type on the immunogenicity of glycoconjugate vaccines, npj Vaccines, 2021, 6, 150 CrossRef CAS PubMed.
- H. Liu, Z. Zhang and R. J. Linhardt, Lessons learned from the contamination of heparin, Nat. Prod. Rep., 2009, 26, 313–321 RSC.
- P. Fan, S. Zhang, Y. Wang, T. Li, H. Zhang, P. Zhang and S. Huang, Nanopore analysis of salvianolic acids in herbal medicines, Nat. Commun., 2024, 15, 1970 CrossRef CAS PubMed.
- S. Goodwin, J. D. McPherson and W. R. McCombie, Coming of age: ten years of next-generation sequencing technologies, Nat. Rev. Genet., 2016, 17, 333–351 CrossRef CAS PubMed.
- W. Gu, S. Miller and C. Y. Chiu, Clinical Metagenomic Next-Generation Sequencing for Pathogen Detection, Annu. Rev. Pathol.: Mech. Dis., 2019, 14, 319–338 CrossRef CAS PubMed.
- X. Cao, X. Du, H. Jiao, Q. An, R. Chen, P. Fang, J. Wang and B. Yu, Carbohydrate-based drugs launched during 2000–2021, Acta Pharm. Sin. B, 2022, 12, 3783–3821 CrossRef CAS PubMed.
- A. Arima, I. H. Harlisa, T. Yoshida, M. Tsutsui, M. Tanaka, K. Yokota, W. Tonomura, J. Yasuda, M. Taniguchi, T. Washio, M. Okochi and T. Kawai, Identifying Single Viruses Using Biorecognition Solid-State Nanopores, J. Am. Chem. Soc., 2018, 140, 16834–16841 CrossRef CAS PubMed.
- Y. Zhou, M. Ren, P. Zhang, D. Jiang, X. Yao, Y. Luo, Z. Yang and Y. Wang, Application of Nanopore Sequencing in the Detection of Foodborne Microorganisms, Nanomaterials, 2022, 12, 1534 CrossRef CAS PubMed.
- G. M. Roozbahani, X. Chen, Y. Zhang, L. Wang and X. Guan, Nanopore detection of metal ions: Current status and future directions, Small Methods, 2020, 4, 2000266 CrossRef CAS PubMed.
Footnote |
| † Guangda Yao and Wenjun Ke contributed equally to this work. |
| This journal is © The Royal Society of Chemistry 2024 |