Open Access Article
Open Access ArticleUniversal cell membrane camouflaged nano-prodrugs with right-side-out orientation adapting for positive pathological vascular remodeling in atherosclerosis†
Xian
Qin‡
abc,
Li
Zhu‡
a,
Yuan
Zhong
a,
Yi
Wang
e,
Xiaoshan
Luo
f,
Jiawei
Li
 bc,
Fei
Yan
b,
Guicheng
Wu
b,
Juhui
Qiu
a,
Guixue
Wang
bc,
Fei
Yan
b,
Guicheng
Wu
b,
Juhui
Qiu
a,
Guixue
Wang
 ad,
Kai
Qu
*abc,
Kun
Zhang
*abc and
Wei
Wu
ad,
Kai
Qu
*abc,
Kun
Zhang
*abc and
Wei
Wu
 *ad
*ad
aKey Laboratory for Biorheological Science and Technology of Ministry of Education, State and Local Joint Engineering Laboratory for Vascular Implants, Bioengineering College of Chongqing University, Chongqing, 400030, China. E-mail: qukaigood@cqu.edu.cn; kunzh01@cqu.edu.cn; david2015@cqu.edu.cn
bChongqing University Three Gorges Hospital, Chongqing Municipality Clinical Research Center for Endocrine and Metabolic Diseases, Chongqing, 404000, China
cSchool of Medicine, Chongqing University, Chongqing, 404010, China
dJinFeng Laboratory, Chongqing, 401329, China
eCollege of Basic Medical Sciences, Chongqing Medical University, Chongqing, 400016, China
fGuizhou Information Engineering University, Bijie, 551700, China
First published on 5th April 2024
Abstract
A right-side-out orientated self-assembly of cell membrane-camouflaged nanotherapeutics is crucial for ensuring their biological functionality inherited from the source cells. In this study, a universal and spontaneous right-side-out coupling-driven ROS-responsive nanotherapeutic approach, based on the intrinsic affinity between phosphatidylserine (PS) on the inner leaflet and PS-targeted peptide modified nanoparticles, has been developed to target foam cells in atherosclerotic plaques. Considering the increased osteopontin (OPN) secretion from foam cells in plaques, a bioengineered cell membrane (OEM) with an overexpression of integrin α9β1 is integrated with ROS-cleavable prodrugs, OEM-coated ETBNPs (OEM-ETBNPs), to enhance targeted drug delivery and on-demand drug release in the local lesion of atherosclerosis. Both in vitro and in vivo experimental results confirm that OEM-ETBNPs are able to inhibit cellular lipid uptake and simultaneously promote intracellular lipid efflux, regulating the positive cellular phenotypic conversion. This finding offers a versatile platform for the biomedical applications of universal cell membrane camouflaging biomimetic nanotechnology.
Introduction
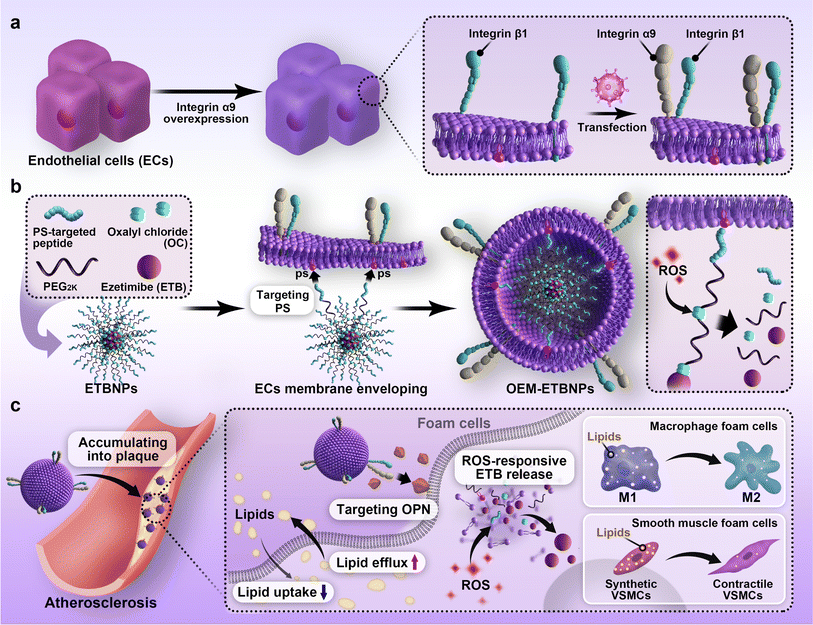
The implementation of a right-side-out-orientated coating holds significant importance in advancing the research and applications of cell membrane camouflaging nanotherapeutics.1,2 By preserving the intrinsic structure and functionality of the cell membrane, including pivotal elements like surface receptors and signal transduction pathways, this approach bestows amplified effectiveness upon the camouflage, enabling precise recognition and targeting capabilities.3 Moreover, it serves to mitigate immune rejection responses, thereby improving the overall biocompatibility and stability of the nanoparticles encapsulated within the right-side-out-orientated cell membrane. However, the achievement of a desired right-side-out orientation during the coating process remains a formidable challenge due to the inherent asymmetry and complex biological properties of the cell membrane. Unfortunately, the process of spontaneous assembly during cell membrane camouflaging frequently encounters obstacles such as a low success rate and significant loss, primarily attributable to the heightened complexity and numerous steps inherent in the procedure. Therefore, it is an ideal case to improve the development of a novel and viable strategy to achieve efficient cell membrane coating, particularly when confronted with situations lacking suitable affinity between certain cell membrane and core pairs. To address this critical issue, our previous research has revealed that the specific binding between the intracellular domain of the transmembrane protein band 3 in red blood cells (RBCs) and the P4.2 peptide ligand can effectively guide the encapsulation of cell membranes with the desired right-side-out orientation,1 addressing the deficiencies of the traditional preparation techniques such as co-extrusion4,5 or ultrasound.6,7 However, it is important to note that the highly specific nature of the transmembrane protein band 3 restricts its applicability to the right-side-out orientation encapsulation of RBC membranes alone, resulting in it being unadaptable for a diverse range of biomimetic cell membrane carriers. To break this limitation, phosphatidylserine (PS), a phospholipid primarily located on the inner leaflet of the cell membrane,8 is utilized. The PS-targeted peptide (CLIKKPF) is designed to bind to PS with remarkable selectivity and affinity.9 It is able to effectively and directionally camouflage the core carrier by harnessing its strong binding capacity to PS. This strategy proves to be highly effective in achieving the inner leaflet-orientated assembly of diverse cell membrane-camouflaged nanoparticles, thereby opening up new possibilities for their application and utilization.In addition to the challenges associated with the assembly of cell membrane camouflaged nanoparticles, achieving controllable cargo release after their local lesion delivery is another critical aspect that must be addressed to achieve efficient therapeutic outcomes. The persistent strong interaction between the cargo and carrier may not fulfill the diverse requirements at different stages of delivery. For instance, while the strong coupling interaction between the cell membrane and nanoparticles is beneficial for enhancing cargo delivery and reducing premature cargo release during circulation in the bloodstream, it will significantly limit therapeutic efficacy at the target site or lesion. It is imperative to optimize the core nanoparticles to respond to both endogenous and exogenous stimuli, enabling “on-demand” cargo release in the pathological lesion.10 Specifically, the core nanoparticles can be designed to selectively and responsively release the cargo upon encountering specific pathological stimuli. This approach allows for self-adaptive regulation and localized cargo release, thereby improving therapeutic efficacy and minimizing toxic side effects.11 In the context of pathological stimuli, the development of a prodrug strategy holds promise for enhancing the pharmacokinetic efficacy of drugs, particularly in target drug delivery.12 Reactive oxygen species (ROS), which play a significant role in the pathogenesis of atherosclerotic plaques, are involved in processes such as oxidative stress, inflammation, and apoptosis, thereby accelerating the formation and progression of atherosclerotic plaques.13,14 Therefore, ROS-responsive prodrugs have immense potential for precise treatment of atherosclerosis (AS), allowing for targeted and controlled cargo release specifically in response to ROS levels.15,16
Moreover, within the pathology of AS, osteopontin (OPN) emerges as a pivotal cell adhesion and chemotactic factor, exhibiting heightened expression within plaque foam cells (FCs).17,18 OPN plays a crucial role in recruiting monocyte-derived macrophages and facilitating their retention within chronically inflamed sites, thereby accelerating AS progression and vulnerable plaque formation.19,20 Recent investigations have shed light on the critical involvement of integrin α9β1, serving as a receptor for OPN, and its associated signaling pathways in the development of various autoimmune disorders.21,22 Consequently, within the realm of biomimetic nanotherapeutic systems for AS target delivery, a design strategy involves the utilization of nanocarriers engineered with overexpressed α9β1 on the cell membrane surface (OEM). These specialized nanocarriers possess the capacity to proficiently discern and engage with OPN within FCs present in atherosclerotic plaques. Leveraging the α9β1/OPN axis and its inherent homing effect holds immense potential in enabling highly efficient drug delivery for AS.
Hence, in this study, an OEM camouflaged biomimetic ROS-sensitive nanotherapeutic approach was employed to develop AS management. As a model drug, Ezetimibe (ETB) was chosen because of its pharmacological activity in reducing cholesterol absorption associated with AS. To achieve a target delivery approach, the prodrug was functionalized with a right-side-out orientated coupling. Specifically, the ETB and the PS-targeted peptide were conjugated using oxalyl chloride (OC) modified HO-PEG2K-Mal, leading to the formation of ETBNPs. The resulting OEM-coated ETBNPs (OEM-ETBNPs) spontaneously self-assembled in a right-side-out orientation on the inner leaflet of the OEM, facilitated by the specific affinity between the intracellular domain of PS on the cell membrane and the paired PS-targeted peptide-modified nanoparticles. Notably, this assembly process was achieved without the need for additional co-extrusion steps. The constructed OEM-ETBNPs were demonstrated to significantly improve accumulation within AS plaques, and the local ROS stimulus triggered an “on-demand” release of the prodrug, effectively inhibiting lipid uptake and promoting lipid efflux by FCs through phenotypic conversion. These mechanisms synergistically enhanced the efficacy of AS therapy (Fig. 1).
Results
Synthesis and characterization of OEM-ETBNPs
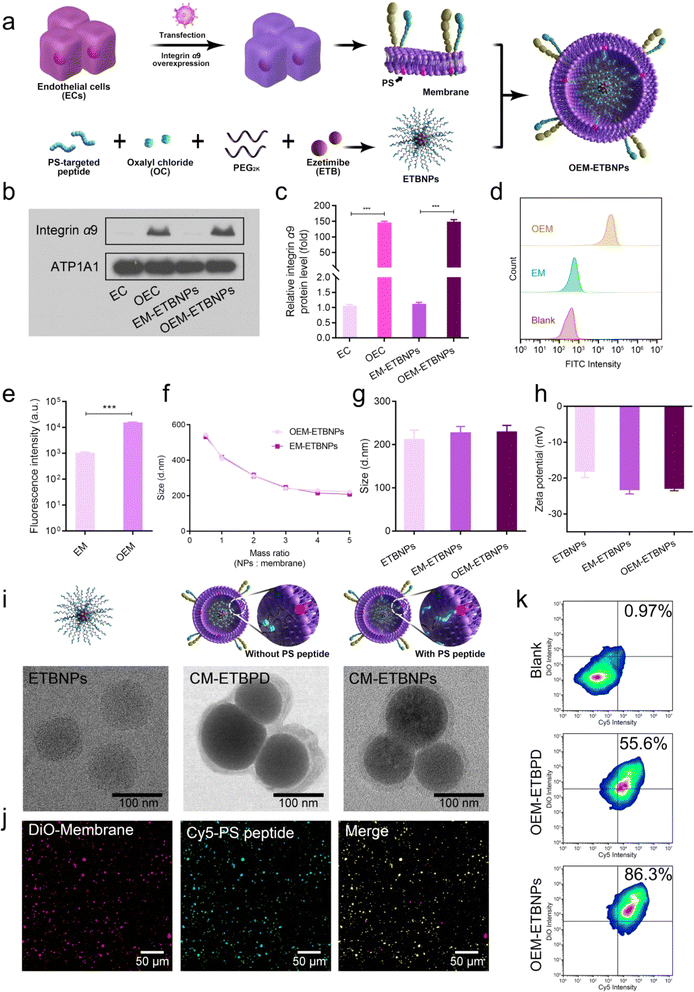
For the synthesis of the ROS-sensitive polymeric prodrug (ESI Fig. 1†), HO-PEG2K-OH was conjugated with ETB, followed by acylation with oxalyl chloride (OC) and HO-PEG2K-Mal to obtain Mal-PEG2K-ETB-PEG2K-Mal (ETBPD). The successful synthesis was confirmed through 1H NMR analysis (ESI Fig. 2a†). To further functionalize the polymeric prodrug for right-side-out orientated coupling on the inner leaflet of the cell membrane, the PS-targeted peptide sequence CLIKKPF was reacted with Mal-PEG2K-ETB-PEG2K-Mal via the Michael reaction between the maleimide and sulfhydryl groups, resulting in a functional polymeric prodrug (PS-Mal-PEG2K-ETB-PEG2K-Mal-PS). The results were successfully confirmed based on 1H NMR and 13C NMR analysis (ESI Fig. 2b, c, ESI Tables 1 and 2†) and the gel permeation chromatography traces (GPC) (ESI Fig. 3†).To achieve stable overexpression of integrin alpha-9/beta-1 (α9β1) in endothelial cells (ECs), α9 lentivirus was transfected into ECs that already exhibit high expression of β1.23,24 The α9 in a normal EC and overexpressed EC (OEC) was detected using western blotting (WB). As shown in Fig. 2a–c, the expression level of α9 on the OEC was significantly higher compared to the normal EC, with an approximately 150-fold increase in protein-level expression. Correspondingly, the expression level in OEM-ETBNPs was also approximately 150-fold higher than that in normal EC membrane coated ETBNPs (EM-ETBNPs). Furthermore, cell membranes were extracted from both the EC and the OEC respectively, and flow cytometry analysis was performed to determine the expression of integrin α9 on the cell membranes. As illustrated in Fig. 2d and e, the fluorescence intensity of anti-integrin α9 on the OEM was significantly higher compared to the EM. In addition, SDS-PAGE protein analysis showed that OEC, OEM, and OEM-ETBNPs had high consistency in protein bands (ESI Fig. 4†).
In aqueous solution, the EM and OEM were co-incubated with ETBNPs, whereas the EM and OEM were co-extruded with ETBPD. The hydrodynamic diameter (Dh) of OEM-ETBNPs decreased as the mass ratio of ETBNPs to the cell membrane increased to 4![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1, and a stable Dh plateau was maintained at a higher mass ratio. Therefore, OEM-ETBNPs with the mass ratio of ETBNPs to EM/OEM at 4
1, and a stable Dh plateau was maintained at a higher mass ratio. Therefore, OEM-ETBNPs with the mass ratio of ETBNPs to EM/OEM at 4![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1 were utilized for all the subsequent studies (Fig. 2f). The Dh and zeta potential of ETBNPs, EM-ETBNPs, and OEM-ETBNPs were further characterized using dynamic light scattering (DLS). As shown in Fig. 2g, h and ESI Fig. 5,† the mean Dh values of ETBNPs, EM-ETBNPs, and OEM-ETBNPs were 200 ± 2.1 nm, 214 ± 3.4 nm, and 215 ± 1.9 nm, respectively. For the group prepared using the conventional co-extrusion method without the PS-targeted peptide, ETBPD, EM-ETBPD, and OEM-ETBPD were further investigated (ESI Fig. 6†). The mean Dh values of ETBPD, EM-ETBPD, and OEM-ETBPD were 198 ± 3.2 nm, 220 ± 2.5 nm, and 224 ± 2.8 nm, respectively. The low polydispersity index results indicated a favorable dispersity in ETBPD, ETBNPs, EM-ETBPD, OEM-ETBPD, EM-ETBNPs, and OEM-ETBNPs (ESI Fig. 7†).
1 were utilized for all the subsequent studies (Fig. 2f). The Dh and zeta potential of ETBNPs, EM-ETBNPs, and OEM-ETBNPs were further characterized using dynamic light scattering (DLS). As shown in Fig. 2g, h and ESI Fig. 5,† the mean Dh values of ETBNPs, EM-ETBNPs, and OEM-ETBNPs were 200 ± 2.1 nm, 214 ± 3.4 nm, and 215 ± 1.9 nm, respectively. For the group prepared using the conventional co-extrusion method without the PS-targeted peptide, ETBPD, EM-ETBPD, and OEM-ETBPD were further investigated (ESI Fig. 6†). The mean Dh values of ETBPD, EM-ETBPD, and OEM-ETBPD were 198 ± 3.2 nm, 220 ± 2.5 nm, and 224 ± 2.8 nm, respectively. The low polydispersity index results indicated a favorable dispersity in ETBPD, ETBNPs, EM-ETBPD, OEM-ETBPD, EM-ETBNPs, and OEM-ETBNPs (ESI Fig. 7†).
To validate the morphologies, transmission electron microscopy (TEM) was utilized to visually observe ETBNPs, as well as cell membrane coated ETBPD (CM- ETBPD) and ETBNPs (CM-ETBNPs). The results shown in Fig. 2i indicated that both ETBNPs exhibited a consistent spherical morphology and displayed a homogeneous distribution. After being coated with a cell membrane, ETBPD and ETBNPs maintained their uniformity while adopting a nanoscale spherical morphology with a diameter of approximately 210 nm. Interestingly, it was noteworthy that the corona layer of CM-ETBNPs exhibited a significantly reduced thickness compared with that of CM-ETBPD. Furthermore, free ETB, EM-ETBPD, OEM-ETBPD, EM-ETBNPs, and OEM-ETBNPs all exhibited a favorable stability throughout a 60 h investigation (ESI Fig. 8†). This sustained stability underscored their suitability for future studies.
To quantify the amount of cell membrane and PS-targeted peptide present on CM-ETBNPs, double-fluorescent labeling was employed, enabling the co-localization of the DiO-labeled cell membrane and Cy5-labeled PS-targeted peptide. Confocal laser scanning microscopy (CLSM) was used to overlay the cell membrane with the PS-targeted peptide, as depicted in Fig. 2j. Flow cytometry analysis was then conducted, demonstrating that the double positive rates were 55.6% and 86.3% for OEM-ETBPD and OEM-ETBNPs, respectively (Fig. 2k). These findings suggested the successful camouflaging of the cell membrane onto ETBNPs based on the high affinity between PS on the inner leaflet of the cell membrane and the PS-targeted peptide of ETBNPs.
Universal cell membrane right-side-out-orientated camouflaged nanoparticles
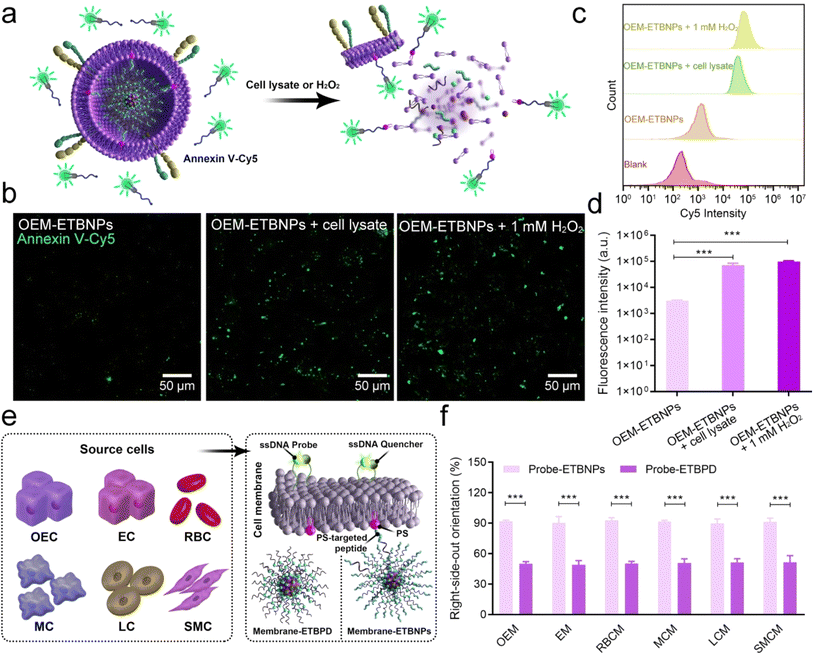
To validate the right-side-out orientation of the cell membrane on the nanoparticles and the distribution of PS on the inner leaflet of the cell membrane in OEM-ETBNPs under normal and stress conditions, an initial experiment was conducted using an Annexin V-Cy5 fluorescent probe. Annexin V, a protein with high affinity for PS, is conventionally situated on the inner leaflet of the cell membrane. As shown in Fig. 3a and b, the fluorescence intensity emitted by the OEM-ETBNP group was rarely observed under normal conditions, confirming the general distribution of PS on the inner leaflet of the cell membrane. However, upon the introduction of a cell lysate and hydrogen peroxide (H2O2), the cellular membrane enveloping OEM-ETBNPs underwent rupture, leading to the translocation of PS molecules from the inner leaflet to the outer leaflet of the cell membrane. This PS translocation was easily detected through a fluorescent probe with a vivid and robust fluorescence signal. Furthermore, the results were confirmed and quantified through flow cytometry analysis (Fig. 3c and d).In addition, a comprehensive investigation was conducted to assess the universal applicability of the PS-targeted peptide based right-side-out orientation in several typical cell membrane-coated nanoparticles, employing a cell membrane outer leaflet-specific DNA fluorescent probe. As illustrated in Fig. 3e, nanoparticles with PS-targeted peptide functionalization were co-incubated with cell membranes derived from various sources (designated as Probe-ETBNPs), including OEC, EC, RBC, macrophage cells (MCs), liver cancer cells (LCs), and smooth muscle cells (SMC). As the negative control, nanoparticles without PS-targeted peptide functionalization and several cell membranes were prepared using the conventional co-extrusion method (designated as Probe-ETBPD). As shown in Fig. 3f and ESI Fig. 9,† the right-side-out orientation rates of the Probe-ETBPD formulations without the PS-targeted peptide ranged from 48.5% to 50.4%. Surprisingly, the remarkable right-side-out orientation of the Probe-ETBNP formulations with the PS-targeted peptide was up to a high level ranging from 90.5% to 92.3%. These results indicated that the right-side-out orientated cell membrane coating based on the PS-specific interaction could be universally applied to multiple cell types.
Characterization of ROS-responsive drug release and target functions in vitro
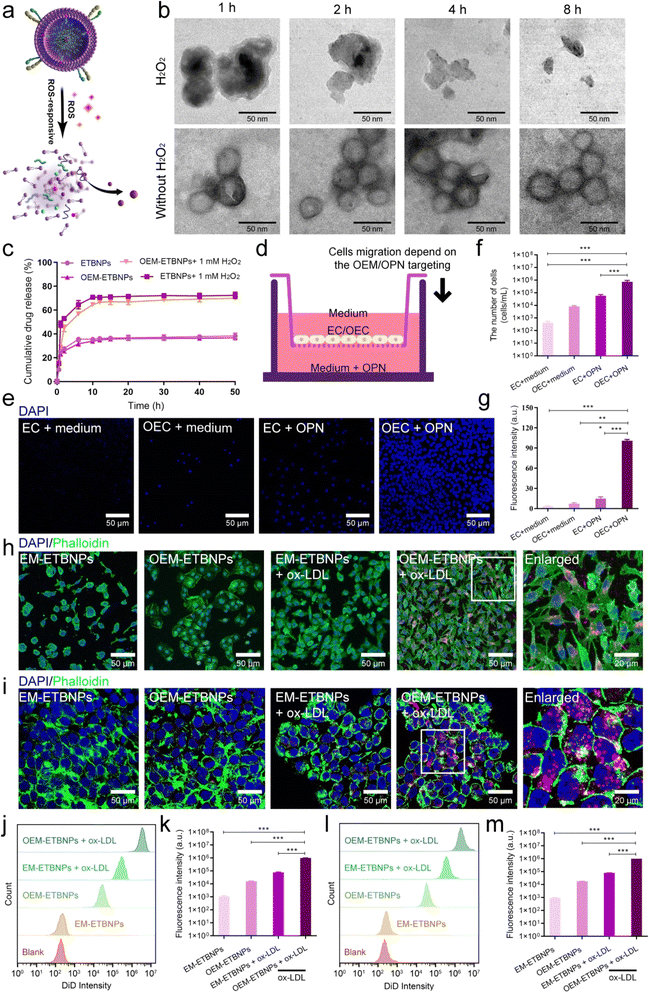
To investigate the release behavior of the ROS-responsive prodrug, ETBNPs and OEM-ETBNPs were exposed to PBS with or without H2O2. As shown in Fig. 4a, b and ESI Fig. 10,† TEM images revealed a stable nanoscale morphology for OEM-ETBNPs in PBS. However, upon exposure to a H2O2 stimulus, a progressive degradation of OEM-ETBNPs was observed, which could be attributed to the loss of thermodynamic stability caused by the cleavage of OC bonds within the OEM-ETBNPs triggered by ROS. Furthermore, a comprehensive evaluation of the prodrug release kinetics was conducted (Fig. 4c). The release profile displayed a rapid release trend within the first 2 h, followed by a relatively slow-release stage extending until 10 h, ultimately reaching a plateau. In response to the ROS stimulus provided by H2O2, the cumulative drug release rate of OEM-ETBNPs was significantly increased from 36.7% to 70.0% in PBS. This remarkable enhancement in drug release further confirmed the destruction of OEM-ETBNPs, resulting in accelerated prodrug activation and subsequent release from the nanoparticles. These findings shed light on the ROS-responsive behavior of the prodrug OEM-ETBNPs and underscored the potential of their controllable drug release capabilities. The dynamic interplay between the ROS stimulus, bond cleavage, and subsequent prodrug activation of the OEM-ETBNPs represented a pivotal mechanism that could be harnessed for targeted drug delivery and on-demand drug release applications.Subsequently, the targeting performance was validated in vitro. OPN has been identified to be highly expressed in FCs localized within AS plaques.19,20 Moreover, it has been established that OPN acts as a ligand for the integrin α9β1 receptor. Thus, a transwell experiment was introduced to ascertain chemotaxis of the overexpression of α9β1 for OPN. As shown in Fig. 4d, the tested cells, EC or OEC, were independently introduced into the upper chamber, while OPN was introduced into the lower chamber. Notably, OEC exhibited a significant migratory capacity towards OPN compared with EC (Fig. 4e–g). As is well known, FCs mainly present two subtypes, i.e., macrophage derived cells (MFCs) and smooth muscle derived cells (SFCs).25–27 Hence, the targeting capability of OEM-ETBNPs towards these two typical FCs was systematically validated. As illustrated in Fig. 4h, SFCs induced by oxidized low-density lipoprotein (ox-LDL) were respectively incubated with EM-ETBNPs and OEM-ETBNPs. The CLSM images revealed that compared with the normal SMCs, SFCs exhibited a significant increase in fluorescence intensity, demonstrating the enhanced uptake of both EM-ETBNPs and OEM-ETBNPs. Moreover, by employing flow cytometry analysis for further quantification, SFCs were found to exhibit a 63.7-fold increased uptake of OEM-ETBNPs compared to the normal SMCs (Fig. 4j and k). Furthermore, MFCs displayed a similar performance in selective uptake as observed in SFCs. As illustrated in Fig. 4i, cellular uptake content of the normal MCs towards EM-ETBNPs and OEM-ETBNPs was relatively weak prior to FC formation. However, after being induced with ox-LDL, the uptake capacity of MFCs experienced a substantial augmentation. Notably, through flow cytometry analysis, MFCs exhibited a 55.0-fold increased uptake of OEM-ETBNPs compared to the normal MCs (Fig. 4l and m).
In vitro cytotoxicity and blood compatibility
Before administration treatment in vivo, ensuring the safety and excellent hemocompatibility of the nanomedicine is crucial for its further application. Therefore, the cytotoxicity of the formulations was systematically assessed on SMCs and MCs. As shown in ESI Fig. 11,† a favorable cell viability was observed after incubating the cells with the formulations at doses ranging from 20 μg mL−1 to 100 μg mL−1 for a duration of 24 h. Notably, even at a high drug concentration of 100 μg mL−1, the cells exhibited robust viability, indicating the exceptional cell compatibility of OEM-ETBNPs. Additionally, the blood compatibility of OEM-ETBNPs was also evaluated through a hemolysis test in vitro. As demonstrated in ESI Fig. 12,† the hemolysis ratios of all drug-contained formulations were found to be below 5%, which is a widely accepted safety standard for further applications in vivo. This finding confirms the excellent hemocompatibility of the formulations.Inhibition of FCs by regulating phenotypic conversion in vitro
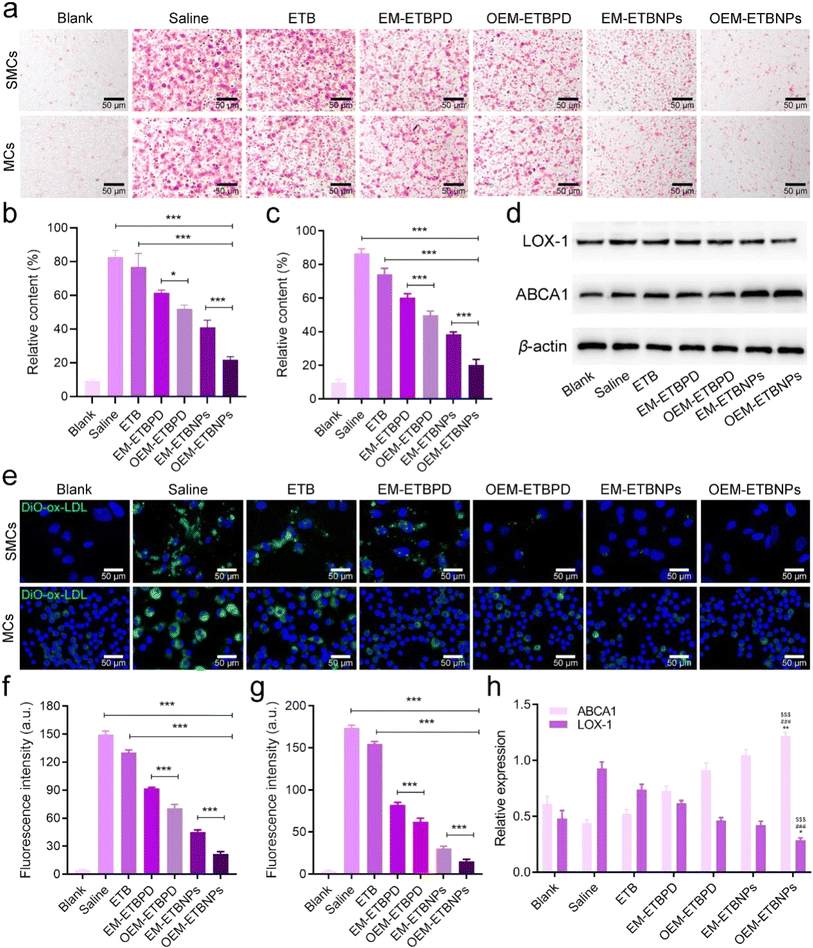
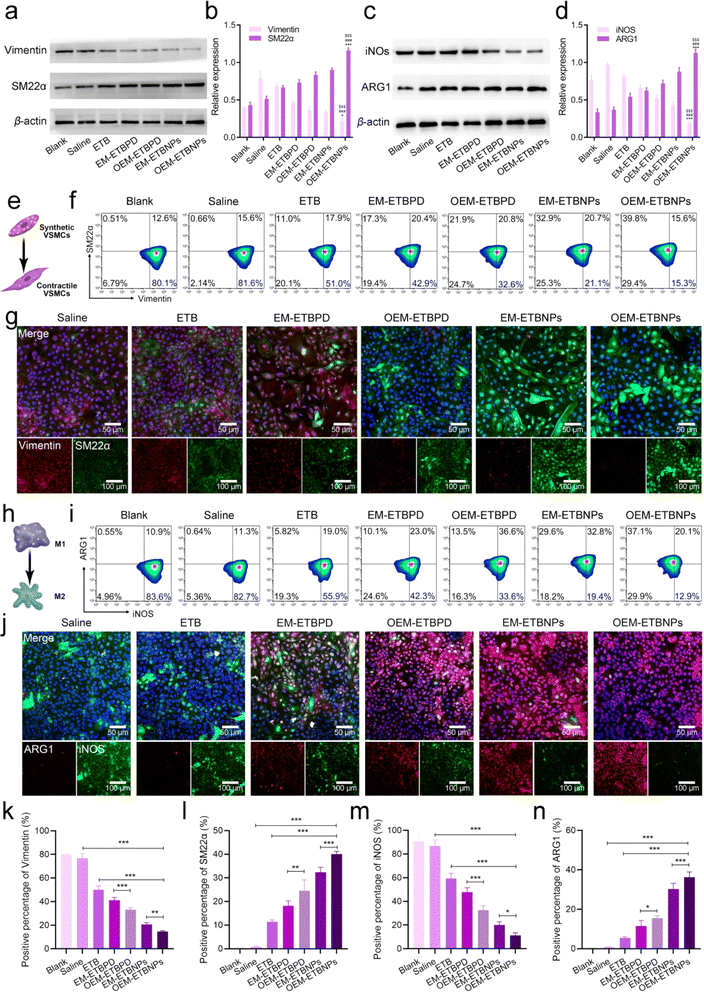
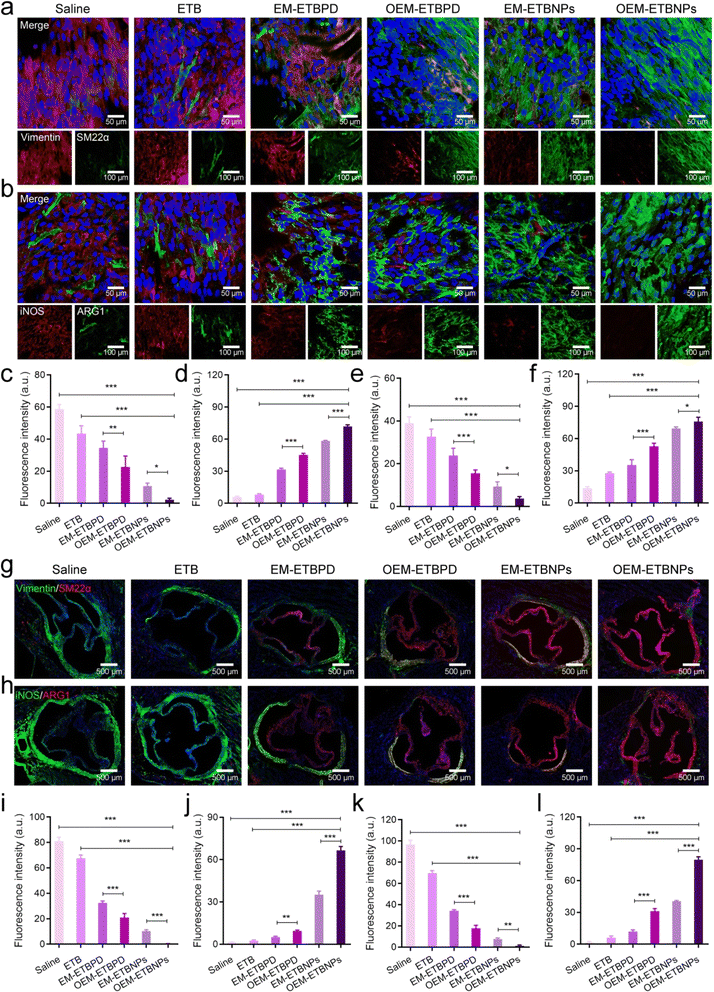
The results of Oil Red O (ORO) staining exhibited remarkable efficiency in inhibiting ox-LDL uptake in both SFCs and MFCs treated with free ETB, EM-ETBPD, OEM-ETBPD, EM-ETBNPs, and OEM-ETBNPs compared with the saline group. Notably, OEM-ETBNPs demonstrated the most profound inhibition of lipid uptake, as evidenced by the lowest relative contents of lipid in both SFCs and MFCs (Fig. 5a–c). Furthermore, the results of WB expression of LOX-1, a critical receptor for ox-LDL internalization, was significantly decreased in MFCs, suggesting a reduction in cholesterol endocytosis. Notably, the expression of the ABCA1 receptor was upregulated in cells treated with the formulations, further underscoring their ability to enhance cholesterol efflux (Fig. 5d and h). Subsequently, after incubating with high-density lipoprotein, OEM-ETBNPs were able to significantly promote lipid efflux, further supporting their critical role in facilitating lipid efflux from FCs (ESI Fig. 13†). Moreover, the cellular uptake of DiO-labeled ox-LDL by SFCs and MFCs was further examined visually supporting the efficacy of OEM-ETBNPs in reducing intracellular lipid accumulation (Fig. 5e–g).To investigate phenotypic conversion resulting from lipid uptake and efflux, the expression of specific markers for SFCs and MFCs was examined. In SFCs, the expression of Vimentin and SM22α was examined to confirm the conversion from a synthetic phenotype to a contractile phenotype. The results of WB and flow cytometry similarly exhibited a gradual decrease in Vimentin expression and a concurrent increase in SM22α (Fig. 6a and b), indicating a conversion from a contractile to a synthetic phenotype in SFCs. Additionally, in the case of SFCs, there was a noticeable reduction in the abundance of lysosomes (ESI Fig. 14†), accompanied by a transition in the cell morphology from spindle-shaped to flat and elongated.28–30 This phenotypic shift was observed as the cells transformed from a synthetic phenotype to a contractile one. Notably, SFCs treated with OEM-ETBNPs exhibited the most pronounced decrease in Vimentin expression and the highest increase in SM22α expression, providing evidence for efficiently promoting conversion from the synthetic phenotype to the contractile phenotype treated with OEM-ETBNPs (Fig. 6e–g, k, l, ESI Fig. 15a and b†).
In MFCs, the expression of iNOS and ARG1 was examined to assess the conversion from a pro-inflammatory M1 phenotype to an anti-inflammatory M2 phenotype. As shown in Fig. 6c and d, a gradual attenuation of iNOS expression along with an elevation in ARG1 expression was observed in WB, revealing a distinct phenotype conversion in the MFCs. Consistently, OEM-ETBNPs exhibited the most diminished expression of iNOS and the highest expression of ARG1, suggesting the efficient cell phenotypic conversion of MFCs from a pro-inflammatory phenotype to an anti-inflammatory one (Fig. 6h–j, m, n, ESI Fig. 15c and d†). Moreover, secretion of inflammatory mediators from M1 MCs was examined, observing a substantial reduction in the expression of TNF-α, IL-6, and IL-1β subsequent to treatments31 (ESI Fig. 16†).
Transcriptomic and bioinformatics analysis of the mechanism of action of OEM-ETBNPs
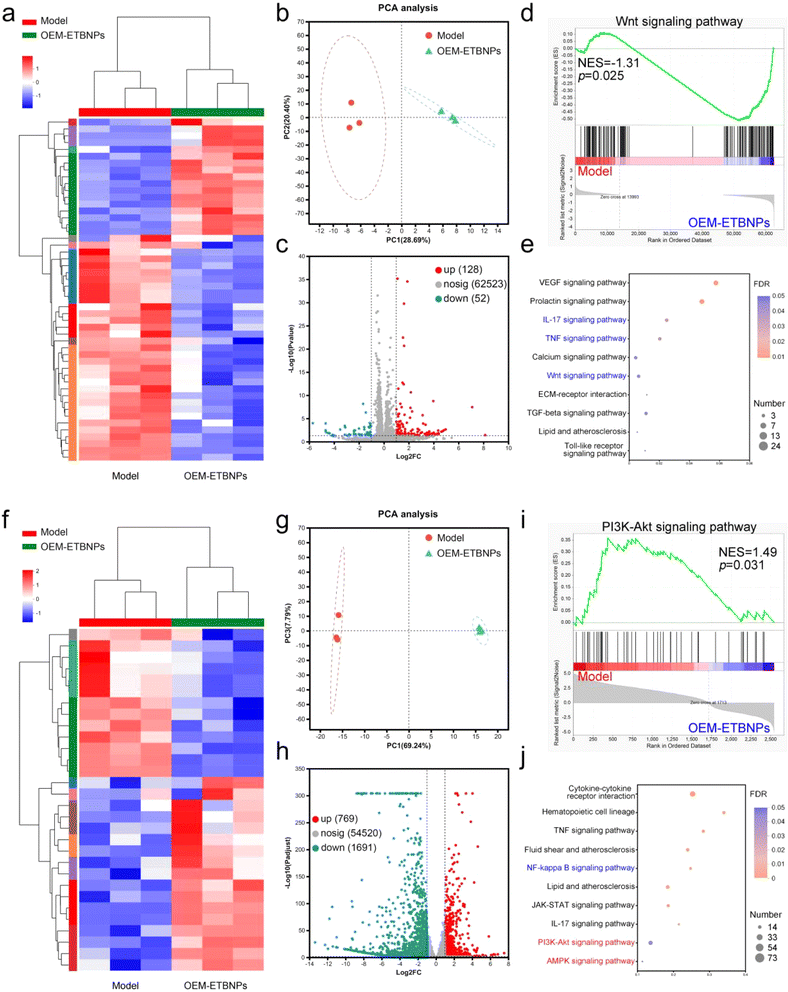
To gain insights into the molecular mechanisms underlying nanotherapeutics utilizing OEM-ETBNPs in SFCs and MFCs, high-throughput transcriptomics analysis was performed. The resulting heat map clearly displayed the substantial differences in the mRNA expression profiles of the two FCs with or without OEM-ETBNP treatment. Specifically, 180 mRNAs with differential expression in SFCs and 2460 mRNAs in MFCs were identified, highlighting significant transcriptomic distinctions (Fig. 7a, b, f and g). Volcano plot analysis demonstrated that in SFCs, OEM-ETBNP treatment led to the upregulation of 128 mRNAs and the downregulation of 52 mRNAs, reflecting substantial changes in gene expression. Furthermore, in MFCs, the same treatment resulted in the significant upregulation of 769 mRNAs and downregulation of 1691 mRNAs, indicating distinct and specific molecular responses to the nanotherapeutics in FCs (Fig. 7c and h). To gain insights into the biological functions of the differentially expressed mRNAs, gene ontology (GO) analysis was performed. The results indicated that OEM-ETBNP treatment could potentially regulate the cell part and cell process (ESI Fig. 17†).To gain a deeper understanding of the biological functions of the differentially expressed genes induced by OEM-ETBNPs, gene set enrichment analysis (GSEA) using the Kyoto encyclopedia of genes and genomes (KEGG) to analyze the signaling pathway was performed. In SFCs, OEM-ETBNPs were able to suppress the Wnt signaling pathway, IL-17 signaling pathway, and TNF signaling pathway (Fig. 7d, e, ESI Fig. 18a and b†). In MFCs, OEM-ETBNPs were capable of activating the PI3K-Akt signaling pathway and AMPK signaling pathway, while inhibiting the NF-κB signaling pathway (Fig. 7i, j, ESI Fig. 18c and d†). Similarly, these findings provide confirmation that OEM-ETBNPs exerted their effects on the positive cell phenotype conversion by regulating lipid uptake and efflux.
Characterization of target drug delivery and therapeutic efficacy in ApoE−/− mice
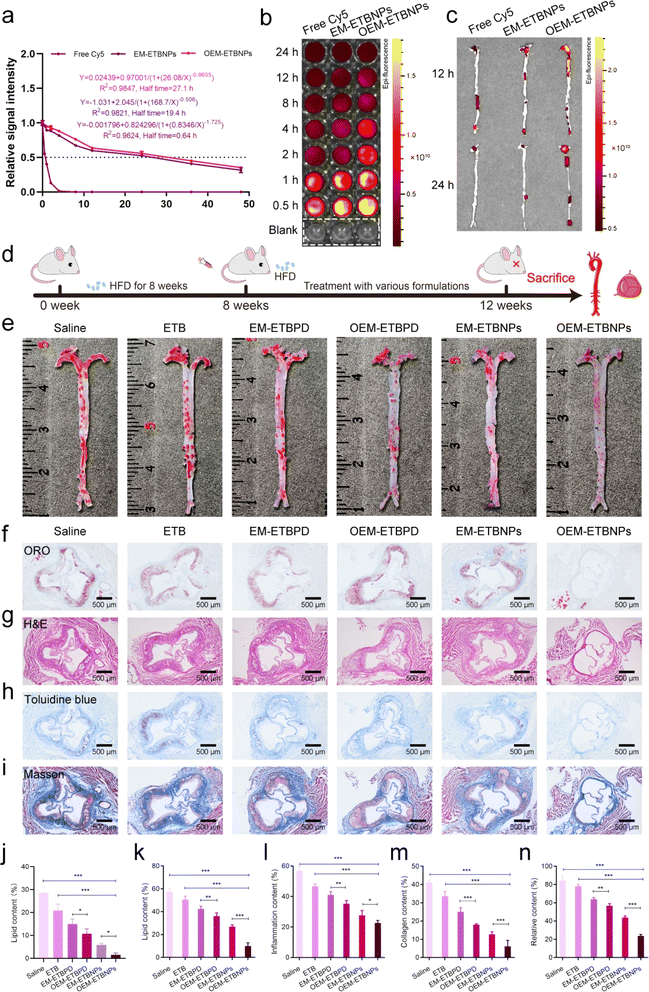
To evaluate the blood distribution of OEM-ETBNPs, free Cy5, Cy5-labeled EM-ETBNPs, and Cy5-labeled OEM-ETBNPs were intravenously injected into ApoE−/− mice. As shown in Fig. 8a, blood circulation measurements confirmed that the OEM-ETBNPs had much longer blood circulation time than the ETBNPs. Furthermore, the fluorescence intensity gradually waned during 24 h investigation. OEM-ETBNPs exhibited a sustained and robust fluorescence signal within 8 h, while the fluorescence signals of the other two groups started to diminish after 4 h (Fig. 8b and ESI Fig. 19a†). These results underscored a significantly prolonged blood circulation of OEM-ETBNPs, facilitating their superior accumulation in the aortic arch and aortic root. In addition, the distribution of the aorta and main organs was evaluated in ApoE−/− mice (Fig. 8c and ESI Fig. 19b†), and OEM-ETBNPs displayed noteworthy accumulation in the aortic arch at 12 h, exhibiting the highest intensity of fluorescence. Even after 24 h, while the fluorescence intensity overall decreased, the OEM-ETBNP group invariably exhibit the strongest signal, indicating their sustained performance in the enhanced target area delivery. In addition, the accumulation of fluorescence intensity in the heart, liver, spleen, lung and kidney gradually diminished in 24 h (ESI Fig. 19c–e†).Moreover, the anti-AS ability of formulations was assessed in ApoE−/− mice as illustrated in Fig. 8d. The results of ORO staining of the aortas showed that formulations were able to reduce the lipid content of plaques compared with the saline group. Quantitative analysis indicated that the free ETB treatment had a limited curative effect, with the lipid content ratio slightly decreasing from 28.6% to 20.9% (Fig. 8e and j). In contrast, the proportion of plaques treated with EM-ETBPD, OEM-ETBPD, EM-ETBNPs, and OEM-ETBNPs decreased to 15.0%, 10.8%, 5.9% and 3.0%, respectively, representing approximately 1.9-fold, 2.6-fold, 4.8-fold and 9.6-fold reduction compared with the saline group, highlighting that OEM-ETBNPs could efficiently enhance therapy for AS. Furthermore, ORO staining of the aortic roots showed a consistent trend with the aortic plaque results. The lipid content of the plaque decreased from 57.3% to 50.6%, 42.3%, 36.1%, 27.0% and 10.0% after treatment with saline, free ETB, EM-ETBPD, OEM-ETBPD, EM-ETBNPs, and OEM-ETBNPs, respectively, which were consistent with the tendency in AS plaque of the aorta (Fig. 8f and k). Moreover, the H&E staining results demonstrated that ApoE−/− mice treated with OEM-ETBNPs showed the least inflammatory response in the aorta (Fig. 8g and l). The necrotic areas in the aortic roots were assessed using Toluidine blue staining. A large necrotic area with substantial cholesterol crystals was observed in the saline group, while the necrotic area was significantly reduced after OEM-ETBNP treatment. Quantitative analysis results showed that compared with the saline group (41.1%), the average necrotic area was decreased to 33.6%, 25.1%, 18.0%, 12.7% and 6.2% for free ETB, EM-ETBPD, OEM-ETBPD, EM-ETBNPs, and OEM-ETBNP treated groups, respectively (Fig. 8h and m). As shown in Fig. 8i and n, Masson's trichrome staining was used to assess the content of collagen in plaque areas. Compared with the saline group, free ETB, EM-ETBPD, OEM-ETBPD, EM-ETBNPs, and OEM-ETBNPs effectively decreased the collagen content, especially for OEM-ETBNP treatment. These results confirmed that OEM-ETBNP treatment showed comprehensive anti-AS effects, i.e., reducing the plaque area, diminishing the inflammatory response, decreasing the necrotic area, and lowering the collagen content in ApoE−/− mice.
Inhibition of FCs by regulating phenotypic conversion in vivo
The phenotypic conversion of SFCs and MFCs in the aortic root was further analyzed using en face staining in an AopE−/− mouse. As shown in Fig. 9, consistent with the cellular outcomes, the treatment with formulations led to notable changes in the phenotype of SFCs. These changes were characterized by a gradual decrease in Vimentin expression and an increase in SM22α expression (Fig. 9a, c and d). Similarly, significant alterations were observed in the cell phenotype of MFCs following treatment with formulations. These changes were characterized by a gradual reduction in the expression of iNOS and a concomitant increase in the expression of ARG1 (Fig. 9b, e and f). Subsequently, immunofluorescence staining of aortic root sections provided valuable insights into the phenotypic conversion of the two FCs. OEM-ETBNPs exhibited the lowest expression of Vimentin and the highest expression of SM22α in SFCs, indicating a profound transition from the synthetic phenotype to the contractile phenotype in SMCs upon treatment with OEM-ETBNPs (Fig. 9g, i and j). Additionally, in MFCs, OEM-ETBNPs displayed the lowest expression of iNOS and the highest expression of ARG1, indicating a positive conversion from the M1 phenotype to the M2 phenotype in MFCs upon treatment with OEM-ETBNPs (Fig. 9h, k and l).Biosafety assessment
To evaluate biosafety in vivo, the typical biochemical markers in plasma and pathological analysis of major organs were used. The body weight of mice treated with OEM-ETBNPs did not show any significant difference compared with the other groups, suggesting that the treatment did not induce any systemic toxicity or adverse effects that could affect the overall health and well-being of the mice (ESI Fig. 20a†). Complete blood count tests were performed to assess the effects of the treatments on blood parameters. The cell contents of white blood cells (WBCs) and RBCs displayed no significant change, which are important components of the blood and play crucial roles in various physiological processes, including immune function (ESI Fig. 20b and c†). Furthermore, the contents of alkaline phosphatase (ALP) and alanine transaminase (ALT) had no significant change after one-month treatments, indicating normal liver function. The blood urea nitrogen (BUN) and creatine kinase (CK) also remained within the normal range (ESI Fig. 20d–g†). Additionally, pathological analysis of the major organs stained with H&E showed no obvious histological toxicity in all the treatment groups (ESI Fig. 21†). Overall, these findings demonstrated that OEM-ETBNPs could be a potential candidate as a safe and effective treatment formulation for chronic vascular diseases.Discussion
In summary, our study unveiled an innovative strategy encompassing the overexpression of integrin α9β1 on the cell membrane and the utilization of a universal assembly approach employing PS mediated right-side-out assembly on the inner leaflet of the cell membrane. This innovative approach was capable of enhancing target drug delivery to FCs while regulating the cellular phenotype, leading to effective treatment of AS.Our previous study had demonstrated the successful spontaneous right-side-out coupling of nanoparticles via specific coupling between the transmembrane protein band 3 and P4.2 peptides. However, this methodology was confined to the use of RBC membranes, necessitating the further exploration of a novel and versatile approach to facilitate the spontaneous right-side-out coating of nanoparticles applicable to universal cell membranes. In this study, a PS-targeted peptide was employed as a feasible targeting moiety, conferring specific binding to PS on the inner leaflet of the cell membranes. Subsequently, through deft utilization of a PEG linker as the functional soft arm, PS-targeted peptides were expertly conjugated with the prodrug to harvest ETBNPs. Remarkably, by using a straightforward co-incubation protocol with a diverse repertoire of cell membranes, devoid of additional treatments such as co-extrusion or ultrasound, we orchestrated the assembly of nanoparticles assuming an enchanting right-side-out orientation. These nanoparticles flawlessly encapsulated and inherited the biological functionality endowed by the source cells.
Based on our findings using extracellular membrane-specific DNA probes, a favorable right-side-out orientated assembly utilizing PS-targeted peptide functionalized nanoparticles was observed compared with the traditional co-extrusion without PS-targeted peptide modified nanoparticles. Notably, cell membrane coated ETBNPs exhibited a thin corona layer, while cell membrane coated ETBPD showed a substantial coating through repeated co-extrusion, resulting in a thick corona layer. Subsequently, the distribution of PS on the cell membrane was validated by incubating OEM-ETBNPs with an Annexin V-Cy5 probe. Annexin V was an affinity-driven protein for PS normally localized on the inner leaflet of the cell membrane. Through the implementation of the Annexin V-Cy5 fluorescent probe, the exposed PS on the cell membrane was able to be specifically labeled and detected. This analysis provided crucial insights into the right-side-out orientation of cell membrane coating on the nanoparticles, introducing a novel method for the efficient and spontaneous harvest of well-defined OEM-ETBNPs using a simple and convenient mixing approach.
According to previous reports,32,33 FCs, including SFCs and MFCs, are formed by the engulfment of a large amount of lipids by either SMCs or MCs in the arterial wall, and are the pathological cells in AS plaques.26,31,34,35 Interestingly, the cell phenotype in SFCs treated with OEM-ETBNPs underwent a conversion from the abnormal synthetic phenotype to the normal contractile phenotype, whereas in MFCs treated with OEM-ETBNPs, a conversion from the pro-inflammatory M1 phenotype to the anti-inflammatory M2 phenotype was exhibited. The therapeutic effects revealed the positive cell phenotypic conversion mediated by lipid metabolism regulation. Transcriptome sequencing was performed on the treated SFCs and MFCs to gain valuable insights into the underlying molecular mechanisms. The analysis showed significant alterations in gene expression profiles in both cell types. In SFCs, OEM-ETBNPs were able to significantly inhibit the Wnt signaling pathway mediated by the binding of ox-LDL receptor-related proteins, thereby regulating SMC proliferation, migration, differentiation, and apoptosis, ultimately facilitating a positive regulation of SMC phenotype conversion.36 Additionally, OEM-ETBNPs demonstrated inhibition of the TNF pathway and IL-17 pathway, thereby suppressing inflammatory responses and reducing proliferation and migration, resulting in the SMCs' phenotype conversion from synthetic to contractile.37,38 In MFCs, the activation of the AMPK signaling pathway was confirmed to inhibit inflammatory responses and the M1 phenotype. Moreover, the PI3K-Akt pathway was found to play a key role in regulating MC polarization and lipid metabolism through regulating the expression of inflammatory factors and nuclear receptors. Following treatment with OEM-ETBNPs, both the AMPK signaling pathway and the PI3K-Akt pathway were activated, suggesting a beneficial effect on MC regulation.39,40 Moreover, after treatment with OEM-ETBNPs, the NF-κB signaling pathway was inhibited, highlighting a potential therapeutic intervention to mitigate MC-associated inflammation.41 These findings further provided valuable molecular evidence supporting the therapeutic potential of OEM-ETBNPs in regulating the positive cellular phenotypes with favorable biocompatibility for safe and efficient AS management.
Overall, our study presented a comprehensive and innovative approach for target delivery and treatment of FCs within AS plaques. By exploiting the overexpression of α9β1 integrin and utilizing the inner leaflet assembly with a PS-targeted peptide, OEM-ETBNPs were able to significantly regulate the aberrant lipid metabolism and subsequently promote positive cell phenotypic conversion. These findings had significant implications for the future development for targeting FC therapies and provided valuable insights into the underlying mechanisms of AS treatment. Further research in preclinical investigation is warranted to further optimize and translate this strategy with the potential to revolutionize the precise AS management.
Materials and methods
The PS-targeted peptides were purchased from Nanjing Taiye Co. Ltd (Nanjing, China), while HO-PEG2K-Mal and HO-PEG2K–OH were obtained from Pengshuo Co. Ltd (Shanghai, China). OC was purchased from Macklin Co. Ltd (Shanghai, China). Ezetimibe (ETB) was sourced from Yuanye Co. Ltd (Shanghai, China). The Annexin V-Cy5 apoptosis detection kit, 1,1′-dioctadecyl-3,3,3′,3′-tetramethylindodicarbocyanine, 4-chlorobenzenesulfonate (DiD) dyes, and 3,3′-dioctadecyloxacarbocyanine perchlorate (DiO) dyes were obtained from Biotium Inc (Fremont, USA). Fluorescein isothiocyanate (FITC), Cy5-N-hydroxysuccinimide (NHS), and D,L-dithiothreitol were provided by Sigma-Aldrich (MO, USA). NAP-5 columns were procured from GE Healthcare (Pittsburgh, PA). DAPI and cell total protein extraction kits were provided by Beyotime Institute of Biotechnology (Jiangsu, China). 22-(N(7-Nitrobenz-2-oxa-1,3-diazol-4-yl) amino)-23,24-bisnor-5-cholen3β-ol (22-NBD cholesterol) was obtained from J&K Scientific Ltd (Shanghai, China). The endothelial cell line stably transfected with mouse integrin alpha-9 (α9) lentivirus was purchased from Yueyongdajiang Co., Ltd (Chengdu, China). Antibodies against ARG1 (ab239731), iNOS (ab178945), Vimentin (ab8978), SM22α (ab14106), LOX-1 (ab60718), ABCA1 (ab307534), goat anti-rabbit IgG H&L (Alexa Fluor® 488) (ab150077), and goat anti-mouse IgG H&L (Alexa Fluor® 555) (ab150118) were sourced from Abcam. Fetal bovine serum (FBS) and DMEM high glucose were purchased from Biological Industries (Chongqing, China). The membrane and cytosol protein extraction kit were purchased from Beyotime (Shanghai, China).Synthetic of the ROS-sensitive polymeric prodrug (PS-Mal-PEG2K-ETB-PEG2K-Mal-PS)
ETB (41 mg, 1 mmol) and HO-PEG2K-OH (400 mg) were dissolved in 5 mL of methylene chloride (CH2Cl2) and added to OC (63.4 mg, 5 mmol) along with triethylamine (50 μL). The reaction mixture was initially stirred at 4 °C for 1 h and then the temperature was raised to 30 °C for 5 h with continuous stirring. After the excess OC was removed by reduced pressure distillation, the mixture was supplemented with Mal-PEG2K-OH (80 mg), and the reaction was allowed to proceed overnight at room temperature to obtain Mal-PEG2K-ETB-PEG2K-Mal. Subsequently, after Mal-PEG2K-ETB-PEG2K-Mal re-dissolved in DMSO, the PS-targeted peptide (17 mg) was introduced, and reacted at 40 °C for 12 h. Following this, the resulting mixture underwent freeze-drying, and the lyophilized products were suspended in DMSO. The products were dialyzed (MWCO: 3500 Da) against deionized water to remove DMSO. Finally, the dialyzed samples were subjected to a 48 h lyophilization process, resulting in the formation of ETBNPs.Overexpression of integrin α9 on ECs by lentivirus
The integrin α9 lentiviruses were transfected into a normal EC following the guidelines outlined in the Lentivirus Operation Manual to obtain an OEC. After the transfection process, WB analysis was employed to confirm the successful expression of the target protein.Preparation of biomimetic ROS-sensitive nanoparticles (OEM-ETBNPs)
OEMs were prepared as previously described.23 OEMs and ETBNPs were co-cultured for 30 min at 37 °C to harvest OEM-ETBNPs. For OEM-ETBPD synthesis, OEMs and ETBPD were co-extruded using an Avestin mini-extruder (Avestin, LF-1, Canada, 100 nm polycarbonate porous membrane) for 10 times to harvest OEM-ETBPD.General characterization of OEM-ETBNPs
Mal-PEG2K-ETB-PEG2K-Mal and PS-Mal-PEG2K-ETB-PEG2K-Mal-PS were dissolved in DMSO-d6 or CDCl3, and then 400 μL of solution was transferred into NMR tubes. The structure characterization of the samples was conducted using 1H NMR and 13C NMR spectroscopy. The relative molecular weight and its distribution of PS-Mal-PEG2K-ETB-PEG2K-Mal-PS were determined using GPC.The Dh and zeta potentials of ETBNPs, ETBPD, EM/OEM-ETBPD and EM/OEM-ETBNPs were measured using a Malvern Zetasizer Nano ZS unit (Nano ZS 90, Malvern, UK) equipped with a He–Ne laser (λ = 633 nm) at a scattering angle of 90° at 25 °C. The stability of free ETB, ETBNPs, ETBPD, OEM/EM-ETBNPs and OEM/EM-ETBPD dispersed in 1× PBS and 10% serum was evaluated by monitoring the changes in particle size after 60 h of incubation.
The membrane proteins of OECs, OEMs and OEM-ETBNPs were extracted using the membrane and cytosol protein extraction kit and subjected to SDS-PAGE. The expression of integrin α9 was detected by WB.
The morphologies of ETBNPs, cell membrane-ETBPD, and cell membrane-ETBNPs were visualized using TEM at 200 kV (JEM-2100F, JEOL, Japan). For the preparation of the TEM samples, ETBNPs, cell membrane-ETBPD, and cell membrane-ETBNP suspension droplets were dripped on copper-coated mesh grids for 2 min and then negatively stained with 2% (wt/vol) sodium phosphotungstate for 1 min and rinsed with filtered 1× PBS three times.
Preparation of the Annexin V-Cy5 probe for OEM-ETBNPs
The Annexin V-Cy5 probe was utilized to assess the distribution of PS in OEM-ETBNPs. Initially, under normal conditions, OEM-ETBNPs and the Annexin V-Cy5 probe were co-incubated for 1 h, followed by centrifugation at 6500×g for 5 min to wash the samples. Subsequently, after the addition of cell lysate and H2O2, another 1 h co-incubation of OEM-ETBNPs with the Annexin V-Cy5 probe was carried out, followed by a second round of centrifugation at 6500×g for 5 min to wash the samples again. Through this experimental procedure, the distribution of PS in OEM-ETBNPs during the encapsulation process could be evaluated.Preparation of the ssDNA probe for OEM-ETBNPs
The ssDNA probe was prepared following a previously reported method.42 Initially, the ssDNA probe (50 μM) was reduced in DTT (0.1 M, 0.17 M phosphate buffer solution, pH = 8.0) for 1 h. Subsequently, the ssDNA probe was purified using a NAP-5 column with 1× PBS as the eluent. A different cell membrane was incubated with the ssDNA probe at a concentration of 25 μM for 1 h at room temperature, and then sonicated and washed with 1× PBS to obtain the probe-cell membrane.To produce probe-cell membrane-ETBNPs, the probe-cell membrane co-incubation ETBNPs were kept at 4 °C for 1 h. Alternatively, to obtain probe-cell membrane-ETBPD, the probe-cell membrane and ETBPD were co-extruded using an Avestin mini-extruder 10 times.
The percentage of probe-cell membrane-ETBNPs/ETBPD with a right-side-out orientation, denoted as P, was calculated using the following equation:
| p = (F0 − F1)/(F0 − F2) | (1) |
In vitro drug release from nanotherapeutics
The drug release from ETBNPs and OEM-ETBNPs was investigated using the dialysis method.43 Solutions of ETBNPs and OEM-ETBNPs, prepared at a concentration of 1 mg mL−1, were introduced into dialysis bags with a MWCO of 3500 Da. Subsequently, the dialysis bags, one containing ETBNPs and the other containing OEM-ETBNPs, were immersed in 50 mL of release medium. To evaluate the influence of H2O2 on drug release, one set of dialysis bags was supplemented with 1 mM H2O2, while the other set served as the control without H2O2. At predetermined time intervals, 2 mL of the release medium from each dialysis bag was withdrawn, and an equivalent volume of fresh medium was replenished. The concentration of released drug was determined using a UV-Vis spectrophotometer at 280 nm, employing a standard curve for quantification.Chemotaxis assays
To assess the targeting capability of OEMs to OPN, chemotaxis assays were performed as previously described.44 In brief, a total of 1 × 105 EC/OEC containing serum-free media were added to the upper chamber. Subsequently, 500 μL of culture media containing either pure medium or OPN was added to the lower well and cultured at 37 °C with 5% CO2 for 12 h. A membrane with a pore size of 8.0 μm was used to facilitate the migration of cells from the upper chamber to the lower chamber. After the 12 h incubation period, the cells that had migrated to the lower chamber were collected, and their nuclei were stained with DAPI for subsequent counting.FC formation and lipid uptake inhibition
SMCs and MCs were cultured in DMEM supplemented with 10% FBS and 100 μg mL−1 penicillin/streptomycin at 37 °C and 5% CO2. Once the cells reached an approximate confluence of 80%, they were transitioned to serum-free DMEM. Subsequently, the MCs and SMCs were pretreated with various formulations, including saline, free ETB, EM-ETBPD, OEM-ETBPD, EM-ETBNPs, and OEM-ETBNPs, each at a concentration of 1 mg mL−1, for a period of 12 h. Concurrently, SMCs were exposed to a culture medium containing 80 μg mL−1 of ox-LDL, and the MCs were treated with 100 ng mL −1 of LPS and 80 μg mL−1 of ox-LDL for a duration of 24 h to assess their ability to inhibit lipid uptake.45–47 The blank group was only treated with serum-free DMEM.Cellular uptake
To further assess the targeting capability of OEM-ETBNPs, SMCs and MCs, SFCs and MFCs were selected for cell uptake studies.48 These cells were seeded in 6-well plates at a density of 1 × 105 cells per well in 2 mL of DMEM medium containing 10% FBS and incubated at 37 °C with 5% CO2 for 24 h. Subsequently, DiD-labeled EM-ETBNPs and OEM-ETBNPs were added to each well, and the samples were incubated for 4 h. For fluorescence imaging, the cells were washed three times with 1 × PBS and fixed with 4% paraformaldehyde (PFA) at room temperature for 30 min. To prevent non-specific binding, the cells were then blocked with 5% bovine serum albumin before incubation with green-phalloidin for 12 h, respectively. Finally, the samples were counter-stained with DAPI to visualize the cell nuclei and imaged using a CLSM (Leica).In vitro toxicity assays
Cell viability was assessed using the MTS assay. SMCs and MCs were seeded in a 96-well plate. These cells were then treated with various concentrations of free ETB, EM-ETBPD, OEM-ETBPD, EM-ETBNPs, and OEM-ETBNPs in serum-free medium for a duration of 24 h.In vitro cholesterol efflux quantification
SFCs and MFCs were incubated with different samples, including free ETB, EM-ETBPD, OEM-ETBPD, EM-ETBNPs, and OEM-ETBNPs at a concentration of 1 mg mL−1 in serum-free DMEM for a duration of 12 h. Subsequently, the cells were incubated with 22-NBD cholesterol (5 μg mL−1) for 12 h after washing twice with 1× PBS and incubated in serum-free DMEM containing high-density lipoprotein (50 μg mL−1) for an additional 6 h. The amount of cholesterol efflux was determined by calculating the percentage of fluorescence in the medium relative to the total fluorescence detected in both the cells and the medium. To quantify cholesterol efflux, the medium was collected, and the fluorescence associated with the released cholesterol was measured using a microplate reader (FlexStation II, NanoDrop, USA).Hemolysis assay
RBCs were obtained from whole blood of healthy C57BL/6 mice and subsequently washed three times with saline (150×g, 10 min, and 4 °C). Subsequently, different groups suspended in normal saline were incubated with RBC suspension at 37 °C for 1 h. As controls, pure water was used as the positive control, with normal saline as the negative control. After centrifugation at 1000×g for 5 min, the absorbance of the supernatants from the collected each group was measured at 545 nm using a UV-Vis spectrophotometer.The hemolytic activity was calculated as a percentage using the following equation:
| Hemolytic activity (%) = (ODT − ODNC)/(ODPC − ODNC) × 100% | (2) |
Oil red O staining
To assess the extent of AS, the cells or aorta were subjected to ORO staining, following a previously established protocol.49 A stock ORO solution (Sigma-Aldrich) was prepared by dissolving 150 mg of ORO powder in 50 mL of isopropanol (Sigma-Aldrich). Firstly, the cells or aorta were fixed with 4% PFA to preserve their structural integrity, and washed with 60% isopropanol to remove any residual fixative and prepare them for staining. The fixed cells or aorta were stained for at least 1 h to ensure sufficient dye uptake and proper visualization of lipid-rich areas associated with AS.Flow cytometry
A total of 1 × 105 cells were seeded in advance onto 6-well plates and treated with various labeled fluorescence groups. The cells were digested with trypsin, followed by centrifugation at 150×g for 5 min. followed by centrifugation. A cell filter was employed to ensure uniform dispersion of the cell population. Flow cytometry data were obtained and analyzed using FlowJo software (Treestar, Ashland, USA). The blank group was only treated with serum-free DMEM.To assess the phenotypic transition of SMCs and MCs, SMCs and MCs were co-cultured with different treatments for 12 h. Cell surface receptors were fluorescently labeled with antibodies against ARG1, iNOS, Vimentin, and SM22α. The percentages represent SM22α/ARG1 and Vimentin/iNOS double negative (lower left quadrant), double positive (upper right quadrant), solely Vimentin/iNOS positive (lower right quadrant), and solely SM22α/ARG1 positive (upper left quadrant), respectively.
Transcriptomic analysis
Cell samples were collected MFCs, MFCs treated with OEM-ETBNPs, SFCs, and SFCs treated with OEM-ETBNPs. The above samples were subjected to rapid freezing in liquid nitrogen. To analyze the genetic information of these cell samples, high-throughput sequencing was performed by Meiji Co., Ltd located in Shanghai, China. The sequencing data generated from the samples were then processed and analyzed using the Meiji Cloud platform, an online analysis tool.Enzyme-linked immunosorbent assay
MFCs were cultured in 24-well plates and incubated with serum-free DMEM containing different samples, including free ETB, EM-ETBPD, OEM-ETBPD, EM-ETBNPs, and OEM-ETBNPs for 12 h. The blank group was treated only with serum-free DMEM. After the incubation period, the cell culture supernatants were collected and subjected to centrifugation at 13![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 000×g for 2 min to remove any cellular debris. The concentrations of TNF-α, IL-6 and IL-1β were determined using specific ELISA kits, which allow for the quantification of these inflammatory cytokines.
000×g for 2 min to remove any cellular debris. The concentrations of TNF-α, IL-6 and IL-1β were determined using specific ELISA kits, which allow for the quantification of these inflammatory cytokines.
Western blotting
SFCs and MFCs were treated with saline, free ETB, EM-ETBPD, OEM-ETBPD, EM-ETBNPs, and OEM-ETBNPs for 12 h, respectively. SFCs were collected and Vimentin and SM22α were quantified by WB, respectively. MFCs were collected and ABCA1, LOX-1, ARG1, and iNOS were quantified by WB, respectively. The blank group was only treated with serum-free DMEM.Animals
Eight-week-old male AopE−/− mice were procured from Beijing HFK Bioscience Co., Ltd (Beijing, China). All experimental procedures and animal care adhered to institutional and national guidelines for the care and use of laboratory animals. The Laboratory Animal Welfare and Ethics Committee of Chongqing University reviewed and approved all aspects of animal care and experimental protocols (IACUC issue no.: CQU-IACUC-RE-202308-003). The AopE−/− mice were randomly divided into six groups, each consisting of five mice. These mice were fed a western diet comprising 21.0% anhydrous milk fat, 34% sucrose, 20.0% protein and 0.15% cholesterol for a duration of eight weeks. Additionally, the mice received intravenous injections via the tail vein every three days for one month, with a volume of 150 μL. The injected substances varied among the groups as follows: (a) saline, (b) free ETB, (c) EM-ETBPD, (d) OEM-ETBPD, (e) EM-ETBNPs, and (f) OEM-ETBNPs. The injection dosage administered to each mouse was 20 mg kg−1. After the completion of the treatment period, the mice were sacrificed.Ex vivo target delivery
To assess the ex vivo target delivery, 150 μL of free Cy5, Cy5-labeled EM-ETBNPs and OEM-ETBNPs were injected through the tail vein of the mice. After 12 and 24 h, the aorta and organs were collected and washed three times with 1× PBS to remove the blood residues. The imaging and fluorescence quantification of vessels and organs were performed using an in vivo small animal optical imaging system.In vivo blood circulation measurement
To investigate the blood circulation behavior of OEM-ETBNPs, 150 μL of free Cy5, Cy5-labeled EM-ETBNPs and OEM-ETBNPs were intravenously injected into the tail vein of the mice. Blood samples were collected at specific time points: 0.5, 1, 2, 4, 8, 12, and 24 h post-injection. The collected blood samples were diluted with 40 μL of 1× PBS containing 0.2 × 10−3 M EDTA2K and transferred to 96-well plates. The fluorescence intensity of the samples was measured using both a fluorescence microplate reader and an in vivo small animal optical imaging system.Immunofluorescence
The cells or frozen sections were washed with 1× PBS three times to remove any debris or residual substances and fixed in 4% PFA for 15 min at room temperature, ensuring the preservation of cellular structures. To minimize non-specific binding, a blocking step was performed using 5% bovine serum albumin for a duration of 1 h and then incubated overnight at 4 °C with antibodies against ARG1, iNOS, Vimentin, and SM22α. On the next day, the cells or frozen sections were washed three times to remove any unbound primary antibodies and incubated with fluorescence-coupled secondary antibodies for a duration of at least two hours. The nuclei were stained with DAPI. The stained cells or frozen sections were imaged using a laser confocal microscope, which provides high-resolution images and enables the visualization of specific antigens within the cells or tissues. The fluorescence intensity was quantified using Image J software.Histology and immunohistochemistry staining
For histological analysis, the aortic root sections were fixed with 4% PFA for 1 h and stained with ORO, H&E, Toluidine blue, and Masson. Furthermore, H&E staining was performed on the sections of the principal organs, encompassing the heart, liver, spleen, lung, and kidney.Blood and serum biochemistry analysis
Blood samples were subjected to comprehensive analysis to determine the levels of various parameters, including RBC, WBC, ALT, ALP, BUN, and CK. This analysis was performed using a highly advanced and automated analyzer platform (Sysmex KX-21, Sysmex Co., Japan).Statistical analysis
Statistical analyses were conducted using the Statistical Package for Social Sciences, a widely recognized software for data analysis. The results were presented as mean ± SD, providing a measure of central tendency and variability in the data. Each experiment was performed with a minimum of three replications to ensure the robustness and reliability of the findings. Tukey's multiple comparison test, the Mann–Whitney U test, and Student's t-test were used to identify significant differences where appropriate. Significance was indicated as no significance (ns), or *P < 0.05, **P < 0.01, and ***P < 0.0001.Data availability
The data that support the findings of this study are available from the corresponding author upon reasonable request.Author contributions
Xian Qin: designed the research, conducted experiments, and analyzed data, writing original draft. Li Zhu: designed the research, conducted experiments. Yuan Zhong: conducted experiments and analyzed data. Yi Wang: conducted experiments and analyzed data. Xiaoshan Luo: conducted experiments and analyzed data. Jiawei Li: conducted experiments and analyzed data. Fei Yan: conducted experiments and analyzed data. Guicheng Wu: conducted experiments and analyzed data. Juhui Qiu: conducted experiments and analyzed data. Guixue Wang: conducted experiments and analyzed data. Kai Qu: designed the research, Supervision. Kun Zhang: designed the research, conducted experiments and analyzed data. Wei Wu: project administration, supervision, conceptualization, methodology, visualization, investigation.Conflicts of interest
The authors declare no conflict of interest.Acknowledgements
This work was supported by grants from the National Key Research and Development Program of China (2022YFF0710700), National Natural Science Foundation of China (31971301, 32171324, 12102072, and 52303146), Natural Science Foundation of Chongqing (cstc2021jcyj-msxmX0149, CSTB2022NSCQ-BHX0724, CSTB2022NSCQ-BHX0718, and CSTB2023NSCQ-BHX0067), JinFeng Laboratory of Chongqing (jfkyjf202203001), Fundamental Research Funds for Central Universities (2023CDJXY-051, 2022CDJXY-026, 2020CDJQY-A061, and 2022CDJYGRH-011), Chongqing Wanzhou District PhD Direct-Access Research Project (wzstc20220314, wzstc20230408, and wzstc20220410). In addition, some large instruments and equipment were provided by the Analysis and Testing Center of Chongqing University.References
- X. Qin, et al., Spontaneously Right-Side-Out-Orientated Coupling-Driven ROS-Sensitive Nanoparticles on Cell Membrane Inner Leaflet for Efficient Renovation in Vascular Endothelial Injury, Adv. Sci., 2023, 10, 2205093 CrossRef CAS PubMed.
- J. Xie, et al., Oriented Assembly of Cell-Mimicking Nanoparticles via a Molecular Affinity Strategy for Targeted Drug Delivery, ACS Nano, 2019, 13, 5268–5277 CrossRef CAS PubMed.
- C. M. J. Hu, et al., Nanoparticle biointerfacing by platelet membrane cloaking, Nature, 2015, 526, 118–121 CrossRef CAS PubMed.
- A. H. Lystad, et al., Distinct functions of ATG16L1 isoforms in membrane binding and LC3B lipidation in autophagy-related processes, Nat. Cell Biol., 2019, 21, 372–383 CrossRef CAS PubMed.
- Y. Wang, et al., Biomimetic Nanotherapies: Red Blood Cell Based Core-Shell Structured Nanocomplexes for Atherosclerosis Management, Adv. Sci., 2019, 6, 1900172 CrossRef PubMed.
- M. Sancho-Albero, et al., Efficient encapsulation of theranostic nanoparticles in cell-derived exosomes: leveraging the exosomal biogenesis pathway to obtain hollow gold nanoparticle-hybrids, Nanoscale, 2019, 11, 18825–18836 RSC.
- L. Rao, et al., Hybrid cellular membrane nanovesicles amplify macrophage immune responses against cancer recurrence and metastasis, Nat. Commun., 2020, 11, 4909 CrossRef PubMed.
- J. Kranich, et al., In vivo identification of apoptotic and extracellular vesicle-bound live cells using image-based deep learning, J. Extracell. Vesicles, 2020, 9, 1792683 CrossRef CAS PubMed.
- K. Yang, et al., Peptide ligand-SiO2 microspheres with specific affinity for phosphatidylserine as a new strategy to isolate exosomes and application in proteomics to differentiate hepatic cancer, Bioact. Mater., 2022, 15, 343–354 CAS.
- S. Mura, et al., Stimuli-responsive nanocarriers for drug delivery, Nat. Mater., 2013, 12, 991–1003 CrossRef CAS PubMed.
- K. Y. Choi, et al., Hyaluronic Acid-Based Activatable Nanomaterials for Stimuli-Responsive Imaging and Therapeutics: Beyond CD44-Mediated Drug Delivery, Adv. Mater., 2019, 31, e1803549 CrossRef PubMed.
- Q. Yao, et al., Synergistic enzymatic and bioorthogonal reactions for selective prodrug activation in living systems, Nat. Commun., 2018, 9, 5032 CrossRef PubMed.
- S. Ouerd, et al., Endothelium-restricted endothelin-1 overexpression in type 1 diabetes worsens atherosclerosis and immune cell infiltration via NOX1, Cardiovasc. Res., 2020, 117, 1144–1153 CrossRef PubMed.
- W. Feng, et al., 2D vanadium carbide MXenzyme to alleviate ROS-mediated inflammatory and neurodegenerative diseases, Nat. Commun., 2021, 12, 2203 CrossRef CAS PubMed.
- C. Gao, et al., Treatment of atherosclerosis by macrophage-biomimetic nanoparticles via targeted pharmacotherapy and sequestration of proinflammatory cytokines, Nat. Commun., 2020, 11, 2622 CrossRef CAS PubMed.
- J. He, et al., Reactive oxygen species (ROS)-responsive size-reducible nanoassemblies for deeper atherosclerotic plaque penetration and enhanced macrophage-targeted drug delivery, Bioact. Mater., 2022, 19, 115–126 Search PubMed.
- W. Chen, et al., Macrophage-targeted nanomedicine for the diagnosis and treatment of atherosclerosis, Nat. Rev. Cardiol., 2022, 19, 228–249 CrossRef PubMed.
- Z. S. Y. Lok, et al., Osteopontin in Vascular Disease, Arterioscler., Thromb., Vasc. Biol., 2019, 39, 613–622 CrossRef CAS PubMed.
- Z. Zhu, et al., Plasma osteopontin levels and adverse clinical outcomes after ischemic stroke, Atherosclerosis, 2021, 332, 33–40 CrossRef CAS PubMed.
- S. Zhang, et al., Targeting the Microenvironment of Vulnerable Atherosclerotic Plaques: An Emerging Diagnosis and Therapy Strategy for Atherosclerosis, Adv. Mater., 2022, 34, 2110660 CrossRef CAS PubMed.
- D. C. M. Simoes, et al., An integrin axis induces IFN-β production in plasmacytoid dendritic cells, J. Cell Biol., 2022, 221, e202102055 CrossRef CAS PubMed.
- S. Kon, et al., The role of α9β1 integrin and its ligands in the development of autoimmune diseases, J. Cell Commun. Signaling, 2018, 12, 333–342 CrossRef PubMed.
- Y. Zhong, et al., Overexpressed VLA-4 on endothelial cell membrane camouflaging the pathological reactive oxygen species responsive prodrug to enhance target therapy for atherosclerosis, Chem. Eng. J., 2022, 442, 136198 CrossRef CAS.
- J. H. Park, et al., Genetically engineered cell membrane–coated nanoparticles for targeted delivery of dexamethasone to inflamed lungs, Sci. Adv., 2021, 7, eabf7820 CrossRef CAS PubMed.
- G. Miao, et al., Vascular smooth muscle cell c-Fos is critical for foam cell formation and atherosclerosis, Metabolism, 2022, 132, 155213 CrossRef CAS PubMed.
- S. Allahverdian, et al., A. Contribution of intimal smooth muscle cells to cholesterol accumulation and macrophage-like cells in human atherosclerosis, Circulation, 2014, 129, 1551–1559 CrossRef CAS PubMed.
- Y. Wang, et al., Smooth Muscle Cells Contribute the Majority of Foam Cells in ApoE (Apolipoprotein E)-Deficient Mouse Atherosclerosis, Arterioscler., Thromb., Vasc. Biol., 2019, 39, 876–887 CrossRef CAS PubMed.
- J. H. Campbell, et al., Lipid accumulation in arterial smooth muscle cells. Influence of phenotype, Atherosclerosis, 1983, 47, 279–295 CrossRef CAS PubMed.
- J. Thyberg, et al., J. Role of caveolae in cholesterol transport in arterial smooth muscle cells exposed to lipoproteins in vitro and in vivo, Lab. Invest., 1998, 78, 825–837 CAS.
- S. Allahverdian, et al., Smooth muscle cell fate and plasticity in atherosclerosis, Cardiovasc. Res., 2018, 114, 540–550 CrossRef CAS PubMed.
- H. Xu, et al., Duplex Responsive Nanoplatform with Cascade Targeting for Atherosclerosis Photoacoustic Diagnosis and Multichannel Combination Therapy, Adv. Mater., 2023, 35(21), 2300439 CrossRef CAS PubMed.
- G. Tumurkhuu, et al., Chlamydia pneumoniae hijacks a host auto-regulatory IL-1β loop to drive foam cell formation and accelerate atherosclerosis, Cell Metab., 2018, 28, 432–448 CrossRef CAS PubMed.
- C. Kontos, et al., Designed CXCR4 mimic acts as a soluble chemokine receptor that blocks atherogenic inflammation by agonist-specific targeting, Nat. Commun., 2020, 11, 5981 CrossRef CAS PubMed.
- M. O. J. Grootaert, et al., Vascular smooth muscle cells in atherosclerosis: time for a re-assessment, Cardiovasc. Res., 2021, 117, 2326–2339 CrossRef CAS PubMed.
- S. Allahverdian, et al., Contribution of monocyte-derived macrophages and smooth muscle cells to arterial foam cell formation, Cardiovasc. Res., 2012, 95, 165–172 CrossRef CAS PubMed.
- C. Mill, et al., Wnt signalling in smooth muscle cells and its role in cardiovascular disorders, Cardiovasc. Res., 2012, 95, 233–240 CrossRef CAS PubMed.
- P. Kong, et al., circ-Sirt1 controls NF-κB activation via sequence-specific interaction and enhancement of SIRT1 expression by binding to miR-132/212 in vascular smooth muscle cells, Nucleic Acids Res., 2019, 47, 3580–3593 CrossRef CAS PubMed.
- M. J. Butcher, et al., Smooth Muscle Cell-Derived Interleukin-17C Plays an Atherogenic Role via the Recruitment of Proinflammatory Interleukin-17A+ T Cells to the Aorta, Arterioscler., Thromb., Vasc. Biol., 2016, 36, 1496–1506 CrossRef CAS PubMed.
- F. Xu, et al., Astragaloside IV inhibits lung cancer progression and metastasis by modulating macrophage polarization through AMPK signaling, J. Exp. Clin. Cancer Res., 2018, 37, 207 CrossRef PubMed.
- H. Y. Zhao, et al., M2 macrophages, but not M1 macrophages, support megakaryopoiesis by upregulating PI3K-AKT pathway activity, Signal Transduction Targeted Ther., 2021, 6, 234 CrossRef CAS PubMed.
- S. Wang, et al., Macrophage Polarization Modulated by NF-κB in Polylactide Membranes-Treated Peritendinous Adhesion, Small, 2022, 18, e2104112 CrossRef PubMed.
- Z. Fan, et al., Structural elucidation of cell membrane-derived nanoparticles using molecular probes, J. Mater. Chem. B, 2014, 2, 8231–8238 RSC.
- X. Xue, et al., Trojan Horse nanotheranostics with dual transformability and multifunctionality for highly effective cancer treatment, Nat. Commun., 2018, 9, 3653 CrossRef PubMed.
- L. Luo, et al., Bioengineering CXCR4-overexpressing cell membrane functionalized ROS-responsive nanotherapeutics for targeting cerebral ischemia-reperfusion injury, Theranostics, 2021, 11, 8043–8056 CrossRef CAS PubMed.
- S. Pi, et al., The P2RY12 receptor promotes VSMC-derived foam cell formation by inhibiting autophagy in advanced atherosclerosis, Autophagy, 2021, 17, 980–1000 CrossRef CAS PubMed.
- S. Ma, et al., Melatonin Ameliorates the Progression of Atherosclerosis via Mitophagy Activation and NLRP3 Inflammasome Inhibition, Oxid. Med. Cell. Longevity, 2018, 2018, 9286458 Search PubMed.
- L. Izem, et al., Plasminogen Facilitates Foam Cell Formation by Macrophages Utilizing H2B and PAR1, Circulation, 2019, 140, A11411 Search PubMed.
- X. Qin, et al., Uptake of oxidative stress-mediated extracellular vesicles by vascular endothelial cells under low magnitude shear stress, Bioact. Mater., 2022, 9, 397–410 CAS.
- K. Qu, et al., TET1s deficiency exacerbates oscillatory shear flow-induced atherosclerosis, Int J Biol Sci, 2022, 18, 2163–2180 CrossRef CAS PubMed.
Footnotes |
| † Electronic supplementary information (ESI) available. See DOI: https://doi.org/10.1039/d4sc00761a |
| ‡ X. Qin and L. Zhu contributed equally to this work. |
| This journal is © The Royal Society of Chemistry 2024 |