Open Access Article
Open Access ArticleNon-invasive diagnosis of bacterial and non-bacterial inflammations using a dual-enzyme-responsive fluorescent indicator†
Yue
He‡
a,
Majun
Yang‡
a,
Jingyi
Cui
c,
Can
Zhao
a,
Bin
Jiang
a,
Jiayun
Guan
a,
Xiaobo
Zhou
a,
Miao
He
a,
Yaya
Zhen
a,
Yuxue
Zhang
a,
Rongrong
Jing
c,
Qi
Wang
*a,
Yuling
Qin
 *a and
Li
Wu
*a and
Li
Wu
 *ab
*ab
aSchool of Public Health, Nantong Key Laboratory of Public Health and Medical Analysis, Nantong University, 9 Seyuan Road, Nantong, 226019, P. R. China. E-mail: wangqi@ntu.edu.cn; ylqin@ntu.edu.cn; wuli8686@ntu.edu.cn
bSchool of Life Science, Nantong University, Nantong 226001, China
cDepartment of Laboratory Medicine, Affiliated Hospital of Nantong University, No. 20, Xisi Road, Nantong 226001, Jiangsu, China
First published on 18th March 2024
Abstract
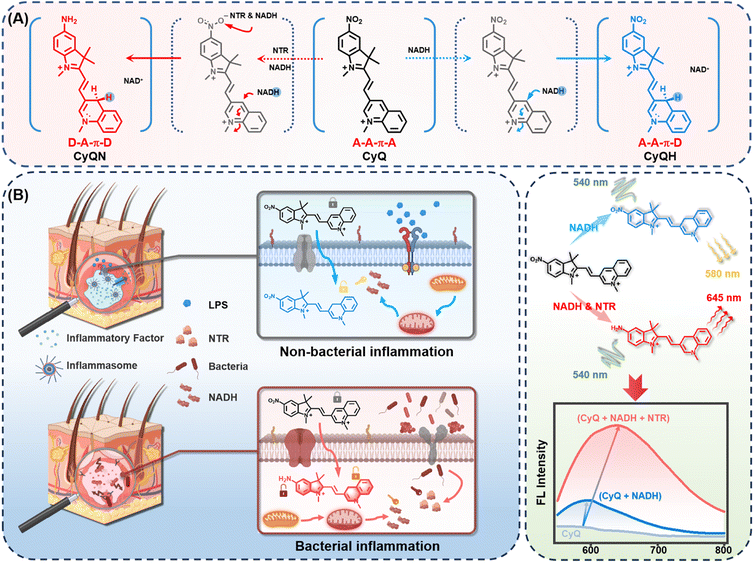
Bacterial infections, as the second leading cause of global death, are commonly treated with antibiotics. However, the improper use of antibiotics contributes to the development of bacterial resistance. Therefore, the accurate differentiation between bacterial and non-bacterial inflammations is of utmost importance in the judicious administration of clinical antibiotics and the prevention of bacterial resistance. However, as of now, no fluorescent probes have yet been designed for the relevant assessments. To this end, the present study reports the development of a novel fluorescence probe (CyQ) that exhibits dual-enzyme responsiveness. The designed probe demonstrated excellent sensitivity in detecting NTR and NAD(P)H, which served as critical indicators for bacterial and non-bacterial inflammations. The utilization of CyQ enabled the efficient detection of NTR and NAD(P)H in distinct channels, exhibiting impressive detection limits of 0.26 μg mL−1 for NTR and 5.54 μM for NAD(P)H, respectively. Experimental trials conducted on living cells demonstrated CyQ's ability to differentiate the variations in NTR and NAD(P)H levels between A. baumannii, S. aureus, E. faecium, and P. aeruginosa-infected as well as LPS-stimulated HUVEC cells. Furthermore, in vivo zebrafish experiments demonstrated the efficacy of CyQ in accurately discerning variations in NTR and NAD(P)H levels resulting from bacterial infection or LPS stimulation, thereby facilitating non-invasive detection of both bacterial and non-bacterial inflammations. The outstanding discriminatory ability of CyQ between bacterial and non-bacterial inflammation positions it as a promising clinical diagnostic tool for acute inflammations.
Introduction
Inflammation is an intricate physiological response that the body initiates in response to damage, infection, or stimulation.1 The immune system primarily orchestrates this process, functioning as a protective mechanism aimed at eradicating pathogens, facilitating tissue repair, and promoting the restoration of health.2 The clinical classification of inflammation includes two main types: bacterial and non-bacterial.3 “Bacterial inflammation” encompasses diverse inflammatory reactions elicited by bacterial infections.4 These encompass conditions such as bacterial pneumonia,5 bacterial arthritis,6 bacterial gastroenteritis (Salmonella and Escherichia coli),7 bacterial meningitis,8 appendicitis caused by bacterial infection within the abdominal cavity,9 and peritonitis.10 “Non-bacterial inflammation” refers to inflammatory reactions induced by non-bacterial agents.11 This category encompasses autoimmune diseases such as rheumatoid arthritis and systemic lupus erythematosus, allergic reactions including allergic rhinitis and asthma, chronic inflammatory bowel diseases like Crohn's disease and ulcerative colitis, drug reactions such as drug allergies or drug-induced vasculitis, and viral infections leading to inflammation such as respiratory tract inflammation triggered by respiratory viruses or skin inflammation caused by herpes virus infections.12–15Accurately distinguishing between bacterial and non-bacterial inflammation holds significant clinical importance in diagnosing and treating inflammatory conditions.16,17 Firstly, the prompt and accurate differentiation of inflammation is crucial in providing clinical guidance for patients' therapeutic strategies. Bacterial inflammation often necessitates the administration of antibiotics, whereas non-bacterial inflammation may require alternative interventions such as antifungal or antiviral medications.18 Secondly, refined clinical diagnoses are essential to reduce antibiotic misuse by accurately distinguishing between bacterial and non-bacterial inflammation, thereby preventing unnecessary antibiotic use and lowering the risk of antibiotic resistance. The third benefit lies in its contribution to disease control. Specifically, the early identification and isolation of the source of infection plays a crucial role in controlling the spread of certain bacterial inflammations such as pneumonia or infectious diarrhea.19 Finally, it also enables physicians to achieve a more precise evaluation of patient prognosis. The management of bacterial inflammation may necessitate more aggressive therapeutic interventions and prolonged recovery periods, whereas non-bacterial inflammation may present a more favourable prognosis. Physicians can appraise patient prognosis with enhanced precision, enabling them to implement appropriate disease management and progress monitoring measures. Hence, the swift and accurate differentiation of inflammation types in clinical practice plays a pivotal role in diagnosing and treating inflammatory disorders.
Upon inflammation, the immune system responds physiologically to injury, infection, or other stimuli, often involving the activation of immune cells and the release of inflammatory factors.20 The disruption of these factors impairs mitochondrial function, thereby affecting cellular energy metabolism and leading to an augmented production and utilization of 1,4-dihydro-nicotinamide adenine dinucleotide (NADH) or its phosphorylated form (NADPH).21 In cases of bacterial inflammation, bacteria utilize nitroreductase (NTR) to reduce nitrate ions (NO3−) to nitrite ions (NO2−) or even further to nitrogen gas (N2) as an alternative electron acceptor for energy production in oxygen-deprived environments, ensuring cellular energy supply without relying on oxygen.22 Consequently, quantifying the NAD(P)H and NTR levels in vivo can establish a clear distinction between bacterial and non-bacterial inflammation. Moreover, NAD(P)H and NTR, as reliable indicators suitable for discerning bacterial and non-bacterial inflammations, wield the potential to diagnose inflammation types and possibly assess their severity differentially. This capability provides a dependable foundation for making therapeutic decisions. A group of scientists has pioneered a range of small-molecule fluorescent probes specifically designed for the individual detection of NAD(P)H and NTR, as exemplified by the work of König, Bhuniya, Tang, Yin, Duan, Chan, Lin, Zhang, Wu, Hu, etc.23–34 Although promising, until now, the field of concurrent dual-enzyme detection using a fluorescent probe has remained unexplored. In this study, we embarked upon an investigation leveraging a synthetically engineered dual-responsive probe to elucidate variations in NAD(P)H levels across HUVEC cells stimulated by varying concentrations of LPS. Additionally, we delved into the fluctuations in NTR concentrations within HUVEC cells infected with bacteria such as Acinetobacter baumannii (A. baumannii), Staphylococcus aureus (S. aureus), Enterococcus faecium (E. faecium) and Pseudomonas aeruginosa (P. aeruginosa), systematically constructing models for the detection of bacterial and non-bacterial inflammations through the discernible shifts in fluorescence signals. Moreover, we validated the effectiveness of these probes in the context of bacterial and non-bacterial inflammations through in vivo experimentation utilizing zebrafish. The establishment of this zebrafish model not only corroborates our findings but also serves as a crucial foundation for the prospective clinical application of these methods in detecting and differentiating bacterial and non-bacterial inflammations.
Results and discussion
Design and synthesis of the NTR and NAD(P)H dual-responsive indicator
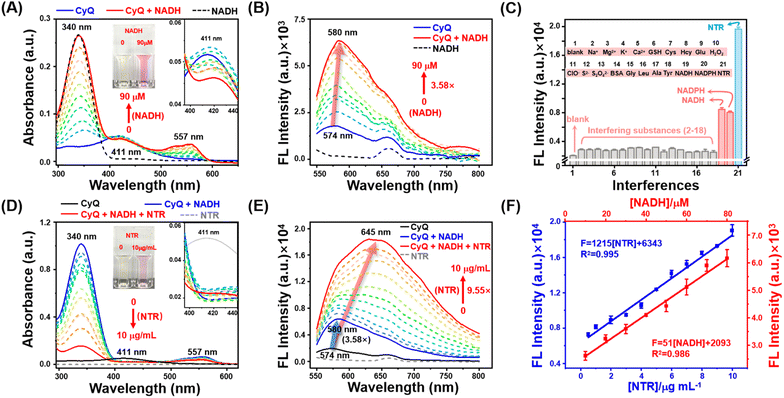
The exceptional stability, excellent photophysical characteristics, and versatile ability of cyanine dyes to target specific organelles have led to their extensive utilization in the development of fluorescent probes.35 Nonetheless, developing dual-enzyme responsive probes based on the cyanine framework that can accurately detect NAD(P)H and NTR with specific reporting signals in vivo remains a formidable challenge. In this study, heterocycle-quinolinium and 5-nitro-indole were selected as specific recognition groups for NAD(P)H and NTR, respectively, based on our previous work.29,36 These recognition groups were linked to corresponding fluorophores through a polymethine chain to form the indicator CyQ (Fig. 1A).The 3-position substituted quinoline salt is well-known for its inherent tendency to undergo two-electron reduction reactions with NAD(P)H analogs.27,29 Upon the NAD(P)H-mediated reaction, a hydride is transferred from NAD(P)H to the acceptor moiety of quinolinium, resulting in the formation of a novel acceptor–π–donor (A–A–π–D) system exhibiting cyanine characteristics (Fig. 1A). On the other hand, another electron-withdrawing moiety (5-nitro-1,2,3,3-tetramethylindolium) present in CyQ can be reduced by NTR, thereby converting the molecule into a 5-amino-1,2,3,3-tetramethylindolium fluorophore with weakened electron-withdrawing properties. Consequently, the altered electron distribution within the molecule results in CyQ adopting a weak acceptor–π–donor (D–A–π–D) configuration, thereby leading to changes in the fluorescence emission spectrum. Moreover, the yellow fluorescence signal emitted by the CyQH (λex = 540 nm, λem = 580 nm, exhibiting a 3.58-fold enhancement, red solid line in Fig. 2B) upon selective activation by NAD(P)H can be differentiated from the red fluorescence signal of CyQN (λex = 540 nm, λem = 645 nm, showing a 9.55-fold enhancement, red solid line in Fig. 2E) that NTR specifically triggers. The efficient discrimination between the two distinct fluorescence channels significantly reduces spectral crosstalk to a considerable extent.
Specifically, the synthesis of CyQ was accomplished via a three-step procedure as depicted in Fig. S1.† First, 2,3,3-trimethyl-5-nitro-3H-indole was subjected to reflux with two equivalents of methyl iodide in acetonitrile for 18 hours, resulting in the formation of 1,2,3,3-tetramethyl-5-nitro-3H-indolium. The 1,2,3,3-tetramethyl-5-nitro-3H-indolium was subjected to a subsequent Knoevenagel condensation reaction with 3-quinolinecarboxaldehyde to afford 1,3,3-trimethyl-5-nitro-2-(2-(quinolin-3-yl)vinyl)-3H-indolium. Finally, the desired product was isolated by precipitation in trichloromethane through a reaction with methyl trifluoromethanesulfonate. The products were comprehensively characterized by high-resolution mass spectrometry, 1H NMR, and 13C NMR (Fig. S2–S10†).
Sensing properties of the as-obtained indicator
The spectroscopic investigation of CyQ in response to NADH and NTR under physiological conditions was conducted in a piperazine-1,4-bisethanesulfonic acid (PIPES) buffer (25 mM, 100 mM NaCl, pH 7.4) at a temperature of 37 °C. As shown in Fig. 2A, prior to the addition of NADH, CyQ (5 μM) displayed a weak absorption peak at 411 nm. Upon the titration of NADH (10–90 μM), a prominent absorption peak at 557 nm rapidly emerged, accompanied by a subtle decrease at 411 nm. Meanwhile, the solution underwent a color transition, shifting from a pale yellow hue to a vibrant shade of purple (Fig. 2A inset). The fluorescence emission spectra (Fig. 2B) demonstrated that upon addition of NADH (10–90 μM) to CyQ (5 μM), the fluorescence signal at 580 nm was enhanced by a factor of 3.58, accompanied by a shift in the emission wavelength from 574 nm to 580 nm. The observed phenomenon can be ascribed to the transfer of a hydride from NADH to the acceptor moiety of quinolinium, forming a distinctive acceptor–acceptor–p–donor (A–A–π–D) system. The titration experiment indicated that CyQ exhibited a robust linear response (R2 = 0.986) in the detection of NADH within the concentration range of 10–80 μM, and a similar response (R2 = 0.995) in the detection of NTR (1–10 μg mL−1) (Fig. 2F). The minimum detection limits (LOD) for CyQ in response to NADH (5.54 μM) and NTR (0.26 μg mL−1) were determined by using the regression equation (3σ/k). The fluorescence intensity of CyQ at 580 nm did not show a continuous increase, as demonstrated in Fig. S11,† even with further increases in NADH concentration (100–500 μM). The subsequent measurement of the reaction rate between CyQ and NADH (100 μM) revealed rapid attainment of peak fluorescence intensity at 580 nm within 12 minutes, indicating a high reaction rate (Fig. S11†). The subsequent step involved conducting NTR titration experiments to evaluate the responsiveness of CyQ towards NTR. It is well-known that NTR catalyzes the conversion of nitro compounds into nitroso intermediates, which are further transformed into hydroxylamines and eventually yield amine products.37 The involvement of the cofactor NAD(P)H as an electron donor is essential for this process. Hence, NADH (500 μM) was included as a cofactor in the NTR titration experiments. The absorption spectra in Fig. 2D and the inset figure demonstrate a significant enhancement at 557 nm upon adding NTR (0–10 μg mL−1). The fluorescence emission spectrum (Fig. 2E) revealed a gradual increase in the fluorescence intensity of CyQ (5 μM) at 580 nm, ranging from 3.58 fold to 4.38 fold with the addition of NTR (0 to 2 μg mL−1). Surprisingly, with the increasing concentration of NTR (3–10 μg mL−1), there was a continuous and significant rise in the maximum emission intensity (9.55 fold, Fig. 2E). Additionally, the emission peak exhibited a pronounced redshift, shifting from 580 nm to 645 nm. The underlying cause of this phenomenon lies in the initial conversion of CyQ molecules into CyQH due to an excess of NADH. In the presence of NADH as a coenzyme, the enzyme NTR facilitates the further conversion of the nitro groups in CyQH molecules into amino groups. During this process, the electron distribution of CyQ undergoes a transition from A–A–π–D to D–A–π–D, resulting in a redshift in the emission spectrum.Selectivity is a critical factor to consider when evaluating the stability of fluorescent probes for cellular detection. As illustrated in Fig. 2C, the emission response of CyQ remained unaffected in the presence of various other biologically relevant agents, including different metal ions (Na+, Mg2+, K+, and Ca2+), thiols (GSH, Cys and Hcy), amino acids (Glycine (Gly), leucine (Leu), alanine (Ala), and tyrosine (Tyr)), ROS (H2O2 and ClO−), bioreductants (NADH, NADPH, SO32−, Na2S2O4, and S2−), BSA, glucose, and NTR (in addition with 500 μM NADH). This result underscores the exceptional chemical selectivity of CyQ in detecting NTR or NAD(P)H. Photostability experiments indicated that CyQ, CyQH (CyQ + 100 μM NADH) and CyQN (CyQ + 10 μg mL−1 NTR + 500 μM NADH) maintained stable fluorescence emission even after 30 minutes of exposure to UV irradiation (Fig. S12†). The developed CyQ assay offers a rapid kinetic profile, excellent specificity, good stability and an exceptionally low detection limit for NTR and NAD(P)H. The aforementioned characteristics enable it to accurately measure NTR and NAD(P)H levels exclusively in living cells.
In vitro detection of NAD(P)H and NTR in bacterial cells
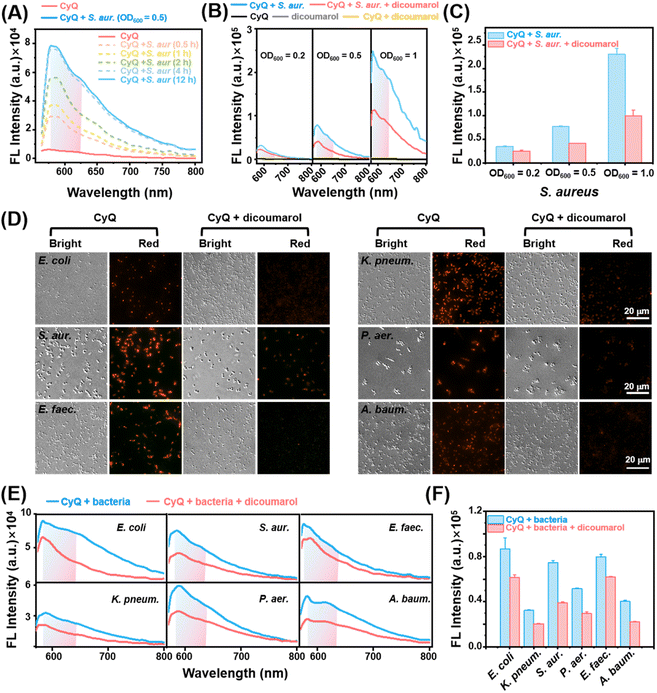
Before conducting cellular experiments, the detection capability of CyQ towards NAD(P)H and NTR in different bacteria strains was evaluated. NTR is an oxidoreductase expressed in a broad spectrum of bacterial species, which reduces a variety of nitroaromatics by simultaneously transferring two electrons from NAD(P)H via a flavin cofactor to substrates. Thus, NTR can be an essential biomarker for accurately diagnosing bacterial infections.38,39S. aureus was initially employed to evaluate the detection capabilities of CyQ for NAD(P)H and NTR within bacterial cells. As depicted in Fig. 3A, the incubation of CyQ (5 μM) with S. aureus (OD600 = 0.5) resulted in a significant enhancement of fluorescence by 4.71 times (pink dashed line) within 30 minutes, reaching its peak emission (increased by 12.58 times, blue dashed line) after 4 hours. The substantial increase in fluorescence at 585 nm indicates the rapid ability of CyQ to penetrate bacterial cell membranes. The phenomenon occurs due to the facile binding of the two positive charges carried by CyQ to the bacterial cell wall, facilitating its entry into bacteria. In contrast to detecting NTR in a solution (Fig. 2E), when utilizing CyQ for S. aureus detection in bacterial culture, the maximum fluorescence emission peak occurs at 585 nm. This is attributed to the inadequate secretion of NTR in the S. aureus culture (OD600 = 0.5), preventing the complete catalysis of CyQ into CyQN and thereby hindering the attainment of fluorescence emission at 645 nm. As depicted in the earlier fluorescence titration graph (Fig. 2E), when NTR concentrations are low (0–4 μg mL−1), CyQN exhibits its maximum fluorescence emission around 580 nm. However, as the concentration of NTR surpasses 4 μg mL−1, the maximum absorption peak of CyQN gradually shifts towards 645 nm (65 nm red shift). Consequently, we employed the maximum fluorescence emission intensity at 585 nm, in conjunction with the fluorescence intensity within the 585–645 nm range, to evaluate the NTR production capabilities in various bacterial samples.The detection capabilities of CyQ were further evaluated on various concentrations of S. aureus. The increase in S. aureus concentration (OD600 = 0.2/0.5/1) significantly elevated CyQ's maximum fluorescence emission intensity, as demonstrated in Fig. 3B (6.58-fold to 43.85-fold increase). We incorporated a control experiment with an NTR inhibitor (dicoumarol, 0.1 mM) to validate the fluorescence enhancement attributed to NTR activity in the bacteria. The results of the control experiment indicated a significant decrease in CyQ's fluorescence intensity at 585 nm upon the addition of dicoumarol, with this difference becoming more pronounced as S. aureus concentrations increased (Fig. 3C). The rationale behind this observation is the specific inhibition exerted by dicoumarol on the activity of NTR, thereby restricting the catalysis function of NTR in reducing the nitro group in CyQ to an amino group.
Subsequently, the detection capability of CyQ towards several clinically prevalent pathogenic bacteria was investigated, including Escherichia coli (E. coli), S. aureus, E. faecium, Klebsiella pneumoniae (K. pneumonia), P. aeruginosa and A. baumannii. Fluorescence imaging and their corresponding mean fluorescence intensities in Fig. 3D and S13† revealed that these six bacteria exhibited evident fluorescence emission in the red channel (570–640 nm) after incubation with CyQ (5 μM for 4 hours).
This observation is consistent with the preliminary findings where NTR catalyzed the conversion of CyQ to CyQN. However, the control group pre-incubated with dicoumarol (0.1 mM) demonstrated a significant fluorescence reduction and the effective inhibition of NTR production by dicoumarol in these six bacteria. Moreover, the varying degrees of fluorescence suppression among the six bacterial strains suggested a significant difference in the expression levels of NTR among these strains. To further assess CyQ's capability in detecting NTR levels across various bacterial solutions, experiments were conducted to evaluate the response of CyQ towards six different bacterial solutions.
The fluorescence spectra in Fig. 3E and F demonstrate the effective detection capability of CyQ for assessing NTR levels in the aforementioned six bacterial strains in solution. When inhibited by dicoumarol (0.1 mM), all six pathogenic bacteria exhibited a significant reduction in fluorescence intensity at 585 nm. Additionally, the fluorescence intensity of S. aureus in both viable and non-viable states was assessed using CyQ to demonstrate its capability in detecting live bacteria. The fluorescence imaging and corresponding mean fluorescence intensities depicted in Fig. S14† demonstrated that S. aureus exhibits prominent fluorescence emission in the red channel (570–640 nm) under normal conditions, while no fluorescence was observed in the deceased state.
The aforementioned experiments have demonstrated the effective detection capability of CyQ for several clinically prevalent pathogenic bacteria, thus laying a foundation for its potential application in identifying bacterial infections in living cells or tissues.
In vitro discrimination of bacterial and non-bacterial inflammation
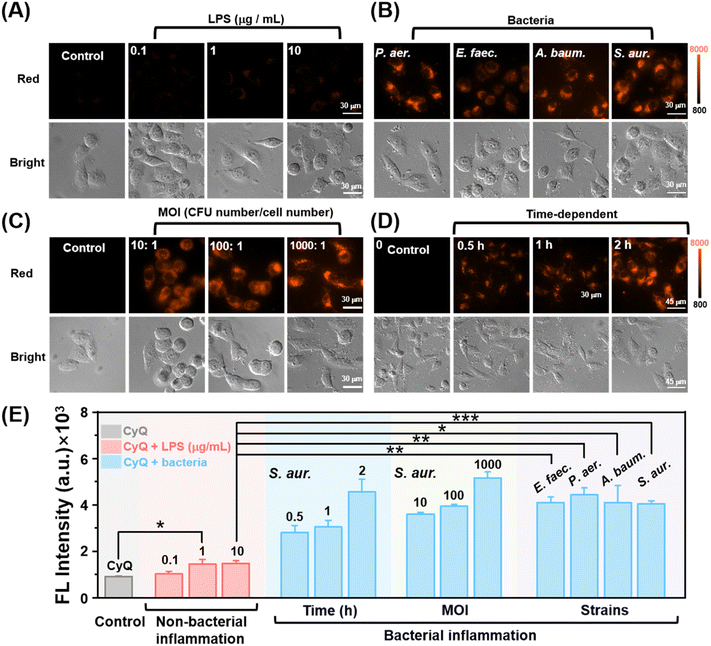
Prior to conducting bacteria and live cell imaging, the cytotoxicity of CyQ on HUVEC cells was evaluated using the MTT assay. Additionally, the bacteria, including E. coli, S. aureus, E. faecium, K. pneumoniae, P. aeruginosa, and A. baumannii, were tested. As illustrated in Fig. S15 and S16,† after incubating HUVEC cells (24 h) and the bacteria (12 h) with various concentrations of CyQ (0–15 μM), the cell viability of HUVEC remained above 85% and the OD600 of the bacteria increased significantly (1.95–2.8 fold). This result indicated that CyQ exhibited good biocompatibility. Colocalization studies were performed in HUVEC cells utilizing Lyso-Tracker Green, Mito-Tracker Green, and DAPI (4′,6-diamidino-2-phenylindole) as co-staining probes. As illustrated in Fig. S17,† HUVEC cells stained with Lyso-Tracker Green, Mito-Tracker Green, and CyQ displayed fluorescence emissions in the green and red channels, respectively. The high Pearson's colocalization coefficient (P = 0.93) between Mito-Tracker Green and CyQ indicated a predominant mitochondrial localization of CyQ in HUVEC cells, whereas the low colocalization with Lyso-Tracker (P = 0.5) and DAPI (P = 0.21) suggested minimal presence in lysosomes and nuclei.To verify the discriminative ability of CyQ in distinguishing between bacterial and non-bacterial inflammation, we established models for both types of inflammation in HUVEC cells by inducing lipopolysaccharide (LPS) stimulation and employing various pathogenic bacteria (S. aureus, E. faecium, P. aeruginosa and A. baumannii), respectively. The detection efficacy of CyQ in relation to LPS-induced non-bacterial inflammation was initially evaluated. LPS is a crucial constituent in the outer membrane of Gram-negative bacteria. The binding of LPS to toll-like receptor 4 (TLR4) and the myeloid differentiation factor (MD2) complex triggers a cascade of intracellular signaling molecules, leading to a wide range of inflammatory responses within cells.40 The fluorescence microscopy images demonstrated that HUVEC cells, after pretreatment with LPS (0.1/1/10 μg mL−1, 24 h) and subsequent incubation with CyQ (5 μM, 2 h), exhibited a slight fluorescence signal enhancement in the red channel compared to the control group, ranging from a 1.12-fold (0.1 μg mL−1) to a 1.62-fold (10 μg mL−1) increment (Fig. 4A). The increased emission observed in the LPS group resulted from the inflammatory response triggered by cellular oxidative stress induced by LPS. The LPS-induced mitochondrial damage subsequently led to aberrant intracellular accumulation of NAD(P)H. The excessive NAD(P)H caused the transformation of CyQ molecules from the A–A–π–A conformation to the A–A–π–D conformation, thereby resulting in enhanced fluorescence emission in the red channel. It's worth noting that HUVEC cells, when stimulated with LPS (1 μg mL−1), exhibited a 1.6-fold increase in fluorescence intensity. However, the fluorescence intensity did not increase further when the LPS concentration was raised to 10 μg mL−1. This observation indicates that more severe non-bacterial inflammation does not further enhance the fluorescence of CyQ. The reason may be that CyQ reacts with NAD(P)H and ultimately transforms into CyQH. Establishing the fluorescence threshold for non-bacterial inflammation is a fundamental basis for effectively distinguishing bacterial inflammation. The bacterial infection inflammation model was based on using the clinically prevalent pathogen S. aureus to infect HUVEC cells. As illustrated in Fig. 4D, HUVEC cells that were pre-incubated with S. aureus (106 CFU mL−1) displayed a significant increase in red fluorescence (3.31 fold) upon incubation with CyQ (5 μM) for 0.5 hours. Following an additional incubation period of 2 hours, the fluorescence intensity reached its zenith, escalating by 4.95 fold. The fluorescence enhancement in cells infected with S. aureus was more robust compared to the non-bacterial inflammation induced by LPS (1 μg mL−1), showing a remarkable increase as early as 30 minutes (more than twice that of the LPS group) and further escalating by 212% after 2 hours of incubation. Even when LPS was increased to 10 μg mL−1, the fluorescence intensity of HUVEC cells infected with S. aureus and incubated with CyQ (5 μM) for 30 minutes remained significantly higher than non-bacterial inflammation (2.05 fold, Fig. 4D). The underlying cause of this phenomenon lies in the bacterial infection-induced redox imbalance within cells, accompanied by mitochondrial dysfunction, leading to an excessive accumulation of NAD(P)H. Moreover, intracellular nitrate conversion to nitrite for energy supply is facilitated by the substantial production of NTR by invading bacteria. The NTR enzyme, utilizing NAD(P)H as a coenzyme, concurrently catalyzes the reduction of –NO2 in CyQ to –NH2, leading to the transformation of the A–A–π–A CyQ form into the D–A–π–D CyQN form. The rapidly intensifying fluorescence signals indicate that CyQ can swiftly detect the bacterial inflammatory status of cells. The capability of CyQ to distinguish various concentrations of S. aureus infection in HUVEC cells was further evaluated. The fluorescence intensity of HUVEC cells significantly increased after 2 hours of incubation with CyQ (5 μM) under various multiplicity of infection (MOI, 1 × 106/107/108 CFU mL−1) conditions, as demonstrated in Fig. 4C. The fluorescence intensity in HUVEC cells increased by 292%, even at a low bacterial concentration with an MOI of 10 (Fig. 4C). The relative fluorescence intensity changes of HUVEC cells incubated with CyQ and DAPI were further compared in the context of bacterial and non-bacterial inflammation. As shown in Fig. S18a,† HUVEC cells pretreated with LPS (0.1/10 μg mL−1) exhibited weaker fluorescence emission in the red fluorescence channel, while HUVEC cells pretreated with different MOIs of S. aureus (1 × 106/107 CFU mL−1) showed stronger fluorescence, with a significant increase in fluorescence intensity as the bacterial concentration increased. Considering that DAPI is a fluorescent stain utilized for DNA labeling, its blue fluorescence intensity remains relatively consistent across different types of inflammation. Therefore, we further compared the ratio of mean fluorescence intensities (MFIs) in the red and blue channels for both types of inflammation. As shown in Fig. S18b,† the ratio (MFICyQ – MFIDAPI/MFIDAPI) of HUVEC cells pretreated with S. aureus was significantly higher (1.55–3.29 fold, Fig. S18b†) than that of LPS-treated cells, indicating that CyQ exhibits a more pronounced enhancement in relative fluorescence intensity in bacterial inflammation compared to non-bacterial inflammation. The rapid and sensitive discriminatory ability of CyQ towards bacterial infection suggests its potential as an effective tool for promptly distinguishing between bacterial and non-bacterial inflammations in clinical settings.
Encouraged by the promising results obtained from the S. aureus infection model experiment, we proceeded to investigate further CyQ's potential in distinguishing cells infected with A. baumannii, P. aeruginosa (Gram-negative bacteria), S. aureus and E. faecium (Gram-positive bacteria). We employed promptly and inoculated HUVEC cells with A. baumannii, P. aeruginosa, S. aureus, and E. faecium at a density of 106 CFU mL−1 to establish non-bacterial and bacterial inflammation models. As shown in Fig. 4B, after incubation with CyQ (5 μM) for 2 hours, the fluorescence intensity in cells affected by the four pathogenic bacteria in the bacterial inflammation model exhibited a significantly enhanced signal compared to the non-bacterial inflammation group (increasing by 2.72–2.97 fold, Fig. 4B and E). These experiments collectively demonstrated the robust discriminatory ability of CyQ in detecting bacterial inflammation induced by different bacterial infections.
In vivo differentiation between bacterial and non-bacterial inflammation
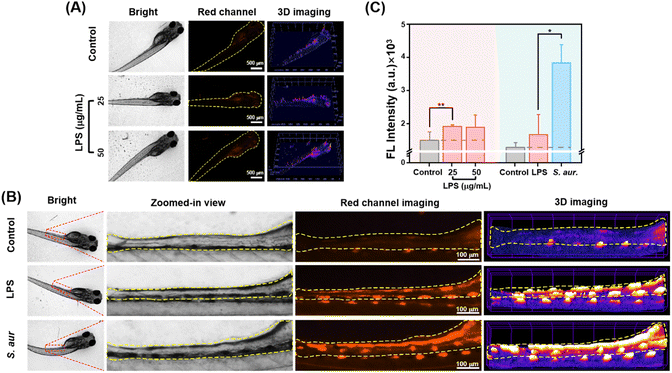
Encouraged by the findings from in vitro experiments, CyQ was further investigated to distinguish between bacterial and non-bacterial inflammation in the zebrafish model. Zebrafish possess both innate and acquired immune systems that closely resemble those found in mammals, and they exhibit rapid natural immune responses to infections and tissue damage, rendering them a widely used animal model for inflammation research.41 The transparency of zebrafish embryos during early development renders them highly suitable for in vivo imaging of inflammation using fluorescent probes. In this study, at 5 days post-fertilization (5 dpf), zebrafish were selected and subjected to sterile inflammation models induced by LPS stimulation and bacterial inflammation models induced by S. aureus, all maintained at a temperature of 28.5 °C.The zebrafish LPS-induced non-bacterial inflammation model is a systemic inflammation model induced by LPS, an effective activator of the innate immune response.42 In this study, the zebrafish were stimulated with varying concentrations of LPS (25/50/100 μg mL−1) to induce an inflammatory response. The results of the LPS-induced inflammation experiments indicated that zebrafish incubated with 25 μg mL−1 and 50 μg mL−1 LPS exhibited typical inflammatory changes, such as sluggish behavior and curved spines, with an overall mortality rate of <40%, making them suitable for the study of non-bacterial inflammation. Therefore, the zebrafish stimulated with LPS concentrations of 25 μg mL−1 and 50 μg mL−1 were selected for in vivo imaging analysis. As shown in Fig. 5A, zebrafish larvae incubated with 25 μg mL−1 LPS and co-incubated with CyQ (5 μM) for 2 hours exhibited an average fluorescence intensity in the fish that was 1.28-fold higher than that of the control group. The fluorescence enhancement primarily arose from cellular damage induced by oxidative stress at the site of inflammation, which subsequently triggered immune cell activation and the release of inflammatory factors. These inflammatory factors hinder the proper functioning of mitochondria, thereby impacting cellular energy metabolism and resulting in an elevated production and utilization of NAD(P)H. NAD(P)H reacts with the quinoline moiety of CyQ, leading to an enhanced fluorescence signal. Surprisingly, the continuous increase in LPS concentration did not significantly enhance the fluorescence signal. The possible reason is that the fluorescent probe CyQ exhibited a near-complete reaction in the presence of overexpressed NADH, resulting in saturation of the fluorescent signal.
The probe's diagnostic efficacy for bacterial inflammation was investigated further. The establishment of the zebrafish model for bacterial inflammation primarily involved two methods: natural infection and microinjection. In this study, we employed the oral infection method by immersing zebrafish larvae in a highly concentrated suspension of S. aureus bacteria (107 CFU mL−1). This method facilitated the entry of bacteria into the organism through the digestive tract, followed by the crossing of the intestinal barrier, ultimately resulting in fish infection. Initially, healthy zebrafish larvae at 5 dpf were immersed in a culture medium containing S. aureus at 28.5 °C for 12 hours, followed by an additional incubation period of 24 hours in a sterile culture medium. Following the incubation of S. aureus, zebrafish exhibited characteristic inflammatory symptoms, including decreased mobility and spinal curvature, confirming the successful establishment of the bacterial inflammation model. Subsequently, after 2 hours of incubation of zebrafish with CyQ (5 μM), fluorescence imaging revealed notable morphological changes in the fish, including enlarged yolk sacs and pericardial edema (Fig. 5B). The zebrafish infected with S. aureus orally displayed a pronounced red fluorescence in the intestinal region, similar to what has been reported in the literature (Fig. 5B and C). Compared to the aseptic inflammation group stimulated with LPS (50 μg mL−1), the fluorescence intensity in the zebrafish intestinal cavity affected by bacterial inflammation increased 2.29 fold. This phenomenon can be attributed to the accumulation of S. aureus within the zebrafish intestinal cavity, disrupting the zebrafish intestinal microenvironment. The bacteria consume a substantial amount of oxygen and release inflammatory factors, resulting in a severe inflammatory response in the gut. These inflammatory factors disrupt the functionality of intestinal epithelial cells and vascular endothelial cell mitochondria, thereby impacting cellular energy metabolism. This ultimately resulted in an elevation in NAD(P)H production and utilization. Additionally, under anaerobic conditions, bacteria cannot rely on oxygen as the final electron acceptor for energy generation. Bacteria employ nitrate reductase to convert nitrate (NO3−) to nitrite (NO2−), thereby generating energy to maintain intracellular energy supply. The generated NTR further reduced the nitro group in CyQ, transforming it from the CyQH form to the CyQN form (D–A–π–D), thereby significantly enhancing fluorescence and causing a redshift in emission wavelength compared to non-bacterial inflammation.
In vivo experiments demonstrate that CyQ can effectively differentiate between zebrafish bacterial and non-bacterial inflammation by inducing significant fluorescence intensity changes and emission wavelength alterations. Furthermore, the results indicate that CyQ exhibits high specificity in distinguishing between these two types of inflammation. The fluorescence intensity does not increase as non-bacterial inflammation progresses when detected (Fig. 5C). This distinction differs from the significant increase observed in bacterial inflammation, establishing a foundation for the further clinical application of the probe.
Conclusions
The present study presents a novel dual-enzyme-responsive indicator named CyQ, which exhibits specific responsiveness to NAD(P)H and NTR at low molecular concentrations. In the PIPES buffer, CyQ showed a “turn-on” fluorescence emission (3.58 fold) at wavelengths of 580 nm in the presence of NAD(P)H and a significant enhancement (9.55 fold) around 645 nm upon catalysis with NTR. Due to its exceptional optical capabilities, the CyQ indicator emerges as an ideal choice for on-site bioimaging of NAD(P)H and NTR levels. As NTR stands as a distinctive marker for discerning between bacterial and non-bacterial infections, the CyQ indicator was further employed for distinguishing bacterial and non-bacterial inflammations in vitro and in vivo. Fluorescence imaging demonstrated that the bacterial inflammation induced by S. aureus exhibited a higher fluorescence enhancement threshold than the non-bacterial inflammation induced by LPS. Further application of CyQ to detect Gram-positive and Gram-negative bacteria-induced inflammation showed that CyQ has a broad spectrum of detection capability in discriminating bacterial inflammation.From the perspective of clinical inflammation management, it is highly desirable to have an effective diagnostic technique for in vivo visualization of both bacterial and non-bacterial inflammation. It can be applied in various scenarios, such as detecting drug-resistant bacteria, facilitating rational antibiotic usage, and monitoring disease progression. The presented method represents a pioneering example of in vivo imaging and differentiation between bacterial and non-bacterial inflammation. We firmly believe that this dual-enzyme responsive probe will serve as an invaluable tool for the selective detection of bacterial inflammation in various clinical applications.
Ethical statement
All animal experiments were performed in compliance with the relevant laws and institutional guidelines for the Care and Use of Research Animals established by the Nantong University Animal Studies Committee, and the experiments have been approved by the committee.Data availability
Synthetic procedure, optical properties, details of cell imaging, and NMR and HRMS spectra for the products can be found in the ESI.†Author contributions
Q. W., L. W. and Y. L. Q. conceived the project. Y. H. and M. J. Y. performed the experimental work. J. Y. C. and C. Z. carried out the computational work. B. J., J. Y. G., M. H., Y. Y. Z., Y. X. Z., and X. B. Z. contributed to the biological experiments. R. R. J. and Y. L. Q. analyzed the data. The manuscript was written by Q. W. and L. W. with contributions from Y. H. and commented on by all authors.Conflicts of interest
There are no conflicts to declare.Acknowledgements
This work was financially supported by the National Natural Science Foundation of China (32171452, 82373456, and 62005132), Excellent Youth Foundation of Jiangsu Scientific Committee (BK20220060), Jiangsu Specially-Appointed Professor (06200048, 06200053), “Postgraduate Research & Practice Innovation Program of Jiangsu Province” (KYCX22 3375), Sichuan Association of Thousand Talent Plan Experts, and the Large Instruments Open Foundation of Nantong University.Notes and references
- L. M. Coussens and Z. Werb, Nature, 2002, 420, 860–867 CrossRef CAS PubMed.
- D. Furman, J. Campisi, E. Verdin, P. Carrera-Bastos, S. Targ, C. Franceschi, L. Ferrucci, D. W. Gilroy, A. Fasano, G. W. Miller, A. H. Miller, A. Mantovani, C. M. Weyand, N. Barzilai, J. J. Goronzy, T. A. Rando, R. B. Effros, A. Lucia, N. Kleinstreuer and G. M. Slavich, Nat. Med., 2019, 25, 1822–1832 CrossRef CAS PubMed.
- Z. Lin, Z. Deng, J. Liu, Z. Lin, S. Chen, Z. Deng and W. Li, J. Inflammation Res., 2022, 15, 953–964 CrossRef CAS PubMed.
- J. J. Woodward, A. T. Iavarone and D. A. Portnoy, Science, 2010, 328, 1703–1705 CrossRef CAS PubMed.
- G. Adhanom, D. Gebreegziabiher, Y. Weldu, A. Gebreyesus Wasihun, T. Araya, H. Legese, B. S. Lopes and M. Saravanan, BioMed Res. Int., 2019, 2019, 8768439 Search PubMed.
- D. Kobayashi, O. Takahashi, H. Arioka, S. Koga and T. Fukui, Am. J. Geriatr. Psychiatry, 2013, 21, 957–962 CrossRef PubMed.
- A. Ternhag, A. Törner, A. Svensson, K. Ekdahl and J. Giesecke, Emerging Infect. Dis., 2008, 14, 143–148 CrossRef PubMed.
- GBD 2016 Causes of Death Collaborators, Lancet, 2017, vol. 390, pp. 1151–1210 Search PubMed.
- E. Arnbjörnsson and S. Bengmark, Am. J. Surg., 1984, 147, 390–392 CrossRef PubMed.
- S. W. Biggins, P. Angeli, G. Garcia-Tsao, P. Ginès, S. C. Ling, M. K. Nadim, F. Wong and W. R. Kim, Hepatology, 2021, 74, 1014–1048 CrossRef PubMed.
- S. Shi, Y. Chen, Z. Luo, G. Nie and Y. Dai, Cell Commun. Signaling, 2023, 21, 61 CrossRef CAS PubMed.
- M. Kiriakidou and C. L. Ching, Ann. Intern. Med., 2020, 172, itc81–itc96 CrossRef PubMed.
- R. C. Strunk, A. L. Sternberg, L. B. Bacharier and S. J. Szefler, J. Allergy Clin. Immunol., 2002, 110, 395–403 CrossRef PubMed.
- P. Demoly, N. F. Adkinson, K. Brockow, M. Castells, A. M. Chiriac, P. A. Greenberger, D. A. Khan, D. M. Lang, H. S. Park, W. Pichler, M. Sanchez-Borges, T. Shiohara and B. Y. Thong, Allergy, 2014, 69, 420–437 CrossRef CAS PubMed.
- C. Tschöpe, E. Ammirati, B. Bozkurt, A. L. P. Caforio, L. T. Cooper, S. B. Felix, J. M. Hare, B. Heidecker, S. Heymans, N. Hübner, S. Kelle, K. Klingel, H. Maatz, A. S. Parwani, F. Spillmann, R. C. Starling, H. Tsutsui, P. Seferovic and S. Van Linthout, Nat. Rev. Cardiol., 2021, 18, 169–193 CrossRef.
- S. Gutkin, R. Tannous, Q. Jaber, M. Fridman and D. Shabat, Chem. Sci., 2023, 14, 6953–6962 RSC.
- C. Wang, S. Liu, Z. Wang, M. Wang, H. Pang, Y. Liu, H. Chang and Z. Sui, Anal. Chem., 2024, 96, 1093–1101 CrossRef CAS.
- M. Yin, Y. Chen, X. Liu, S. Tian, L. Zhao, Y. Bai, H. Wang, J. Lin, D. Jiang, Z. Lei, F. Meng, D. Tian and L. Luo, ACS Nano, 2023, 17, 3873–3888 CrossRef CAS PubMed.
- C. Vazquez Guillamet and M. H. Kollef, Clin. Infect. Dis., 2018, 67, 1455–1462 CrossRef PubMed.
- G. M. Barton, J. Clin. Invest., 2008, 118, 413–420 CrossRef CAS PubMed.
- T. Gaber, C. Strehl and F. Buttgereit, Nat. Rev. Rheumatol., 2017, 13, 267–279 CrossRef PubMed.
- D. J. Nicholas, Biol. Rev. Cambridge Philos. Soc., 1963, 38, 530–568 CrossRef CAS.
- S. Luo, R. Zou, J. Wu and M. P. Landry, ACS Sens., 2017, 2, 1139–1145 CrossRef CAS PubMed.
- Y. X. Fu, W. Y. Guo, N. Wang, Y. J. Dai, Z. Y. Zhang, X. L. Sun, W. C. Yang and G. F. Yang, Anal. Chem., 2022, 94, 17692–17699 CrossRef CAS PubMed.
- R. Peng, J. Yuan, D. Cheng, T. Ren, F. Jin, R. Yang, L. Yuan and X. Zhang, Anal. Chem., 2019, 91, 15974–15981 CrossRef CAS PubMed.
- S. H. Gardner, C. J. Brady, C. Keeton, A. K. Yadav, S. C. Mallojjala, M. Y. Lucero, S. Su, Z. Yu, J. S. Hirschi, L. M. Mirica and J. Chan, Angew Chem. Int. Ed. Engl., 2021, 60, 18860–18866 CrossRef CAS PubMed.
- Y. Jiao, L. Zhang, X. Gao, W. Si and C. Duan, Angew Chem. Int. Ed. Engl., 2020, 59, 6021–6027 CrossRef CAS PubMed.
- M. Li, X. Kong, Y. Yin, Y. Zhang, X. Dai, J. Wang and W. Lin, J. Photochem. Photobiol., A, 2022, 424, 113657–113662 CrossRef CAS.
- C. Yang, C. Jiang, M. Yang, Q. Bai, Y. Zhen, Y. Zhang, W. Yin, J. Wang, X. Zhou, G. Li, M. Wu, Y. Qin, Q. Wang, H. Ji and L. Wu, Chem. Biomed. Imaging, 2023, 1, 738–749 CrossRef CAS.
- S. Xu, Q. Wang, Q. Zhang, L. Zhang, L. Zuo, J. D. Jiang and H. Y. Hu, Chem. Commun., 2017, 53, 11177–11180 RSC.
- M. A. Fomin, R. I. Dmitriev, J. Jenkins, D. B. Papkovsky, D. Heindl and B. König, ACS Sens., 2016, 1, 702–709 CrossRef CAS.
- A. Podder, V. P. Murali, S. Deepika, A. Dhamija, S. Biswas, K. K. Maiti and S. Bhuniya, Anal. Chem., 2020, 92, 12356–12362 CrossRef CAS PubMed.
- Y. Zhao, K. Wei, F. Kong, X. Gao, K. Xu and B. Tang, Anal. Chem., 2018, 91, 1368–1374 CrossRef PubMed.
- K. Ma, H. Yang, X. Wu, F. Huo, F. Cheng and C. Yin, Angew. Chem., Int. Ed. Engl., 2023, 62, e202301518 CrossRef CAS PubMed.
- D. Oushiki, H. Kojima, T. Terai, M. Arita, K. Hanaoka, Y. Urano and T. Nagano, J. Am. Chem. Soc., 2010, 132, 2795–2801 CrossRef CAS PubMed.
- M. Yang, W. Zhu, Y. Lv, B. Jiang, C. Jiang, X. Zhou, G. Li, Y. Qin, Q. Wang, Z. Chen and L. Wu, Chem. Sci., 2023, 14, 12961–12972 RSC.
- Y. Li, Y. Sun, J. Li, Q. Su, W. Yuan, Y. Dai, C. Han, Q. Wang, W. Feng and F. Li, J. Am. Chem. Soc., 2015, 137, 6407–6416 CrossRef CAS PubMed.
- L. L. Wu, Q. Wang, Y. Wang, N. Zhang, Q. Zhang and H. Y. Hu, Chem. Sci., 2020, 11, 3141–3145 RSC.
- Y. Ji, Y. Wang, N. Zhang, S. Xu, L. Zhang, Q. Wang, Q. Zhang and H. Y. Hu, J. Org. Chem., 2019, 84, 1299–1309 CrossRef CAS PubMed.
- E. M. Pålsson-McDermott and L. A. O'Neill, Immunology, 2004, 113, 153–162 CrossRef PubMed.
- B. Novoa and A. Figueras, Adv. Exp. Med. Biol., 2012, 946, 253–275 CrossRef CAS PubMed.
- K. H. Park and K. H. Cho, Fish Shellfish Immunol., 2011, 31, 904–910 CrossRef CAS PubMed.
Footnotes |
| † Electronic supplementary information (ESI) available. See DOI: https://doi.org/10.1039/d3sc06866h |
| ‡ These authors contributed equally to this work. |
| This journal is © The Royal Society of Chemistry 2024 |