Open Access Article
Open Access ArticleTopological effect of an intramolecular split G-quadruplex on thioflavin T binding and fluorescence light-up†
Mengmeng
Lv
ab,
Jiangtao
Ren
 *b and
Erkang
Wang
*b and
Erkang
Wang
 *ab
*ab
aCollege of Chemistry, Jilin University, Changchun, Jilin 130012, China
bState Key Laboratory of Electroanalytical Chemistry, Changchun Institute of Applied Chemistry, Chinese Academy of Sciences, Changchun, Jilin 130022, China. E-mail: jiangtaoren@ciac.ac.cn; ekwang@ciac.ac.cn
First published on 9th February 2024
Abstract
In this work, the topological effect on binding interaction between a G-quadruplex and thioflavin T (ThT) ligand was systematically investigated on a platform of an intramolecular split G-quadruplex (Intra-SG). Distinct fluorescence changes from ThT were presented in the presence of distinct split modes of Intra-SG structures and an intriguing phenomenon of target-induced fluorescence light-up occurred for split modes 2![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 10, 5
10, 5![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 7 and 8
7 and 8![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 4. It was validated that hybridization between the Intra-SG spacer and target did not unfold the G-quadruplex, but facilitated the ThT binding. Moreover, the 3′ guanine-rich fragment of Intra-SG was very susceptible to topology variation produced by the bound target strand. Additionally, a bioanalytical method was developed for ultrasensitive gene detection, confirming the utility of the ThT/Intra-SG complex as a universal signal transducer. It is believed that the results and disclosed rules will inspire researchers to develop many new DNA-based signal transducers in the future.
4. It was validated that hybridization between the Intra-SG spacer and target did not unfold the G-quadruplex, but facilitated the ThT binding. Moreover, the 3′ guanine-rich fragment of Intra-SG was very susceptible to topology variation produced by the bound target strand. Additionally, a bioanalytical method was developed for ultrasensitive gene detection, confirming the utility of the ThT/Intra-SG complex as a universal signal transducer. It is believed that the results and disclosed rules will inspire researchers to develop many new DNA-based signal transducers in the future.
Introduction
A G-quadruplex (G4), as a secondary structure formed by guanine rich sequences,1 widely exists at the end of human telomeres and plays an important biological role.2 G4 is related to cell division, biological aging and tumor cell immortalization,3,4 and numerous organic ligands which are capable of recognizing and stabilizing G4 for the purpose of disease diagnosis and treatment5,6 have been reported, such as PcN4,7 BAMA,8 and TCPP.9 Many G4 ligands reveal a prominent feature of G4-induced luminescence enhancement, and intriguing G4-specific probes, e.g., N-methyl mesoporphyrin IX (NMM)10 and crystal violet (CV),11 have been obtained and introduced into the field of biology and bioanalysis.6 Nevertheless, much effort is required to clarify factors influencing the interaction between G4 and luminescent organic ligands, and explore new types of G4-based molecular probes.Apart from well-known metal ion-dependent properties,12 the topology of G4 including the conformation13,14 the sequence and length of the loop15,16 have been regarded as fundamental and even decisive factors in the binding event of G4 and ligands. For instance, NMM as a classical G4-specific probe is capable of discriminating G4 structures from single-stranded DNAs and duplexes.10 Due to the protection from the quadruplex and loops, CV emitted very intense fluorescence upon binding to antiparallel G4, in contrast to binding to parallel G4, and thus was harnessed for monitoring G4 transition.11 Varieties of analytical methods and logic circuits have been reported on the basis of release, folding or assembly of G4, with utilization of G4-binding ligands (e.g., hemin and other luminescent dyes).17–20 Splitting G4 is one of the attractive strategies for signal transduction. A unimolecular G4 sequence could be dissected into halves at specific sites, yielding intermolecular split G4 (Inter-SG)21 or intramolecular split G4 (Intra-SG).22 Traditional binary probes were obtained by flanking two Inter-SG stands with two sequences which were able to hybridize to one target strand simultaneously.23,24 Intra-SG-based single probes were generated by simply inserting an external DNA sensing sequence at the nicking site.22 Moreover, derived from distinct split modes, multifarious topological G4 structures could be yielded and sensitive quantitation of the target nucleic acid was attained through optimal complexation between split G4 probes and organic ligands (e.g., hemin and NMM).25 The split G4 system is an attractive platform to investigate binding interaction between diverse topological G4 structures and ligands, and select new and effective DNA probes for bioanalysis.
Thioflavin T (ThT) as a cationic benzothiazole dye possesses stable fluorescence properties, cell penetration capability and low toxicity, and was first utilized as a fluorescent probe for monitoring amyloid in tissue sections.26 In the last decade, ThT whose weak fluorescence at 492 nm also can be intensified upon binding to specific nucleic acids,27–29 has been applied for detecting potassium ions, imaging intracellular nucleic acids and monitoring RNA metabolism in vivo.29–31 For example, ThT triggered G4 folding in the 22AG human telomeric DNA, producing 2100-fold enhanced fluorescence, and exhibited high selectivity toward distinct G4 structures.32 Although much progress has been made on nucleic acid recognition by ThT, it is still essential and significant to systematically investigate the topological effect of G4 on ThT binding and its fluorescence response.
Herein, a series of Intra-SG structures derived from a unimolecular G4 were designed (Fig. S1†) for complexation with ThT. Fluorescence variation was observed, especially upon hybridization between the inserted DNA spacer and complementary strand. The topological effect was systematically studied via multiple methods including circular dichroism (CD), UV-vis absorption and thermal denaturation; common rules were disclosed and effective ThT/Intra-SG-based DNA probes were obtained. Meanwhile, ultrasensitive detection of a gene segment from Hepatitis B virus (HBV) was realized through successfully coupling the ThT/Intra-SG transducer with two cascaded DNA amplification techniques including asymmetric polymerase chain reaction (A-PCR)33 and rolling circle amplification (RCA).34
Experimental section
Chemicals and reagents
HPLC-purified oligonucleotides (Table S1†) were synthesized by Sangon Biotech Co., Ltd (Shanghai, China). ThT was purchased from Frontier Scientific, Inc. (Logan, USA). Potassium chloride was purchased from Sinopharm Group Chemical Reagent Co., Ltd (Shanghai, China). The gene segments from Hepatitis B and Ebola viruses and Nova virus (Table S2†) were purchased from Jilin Kumei Biotechnology Co. Ltd. Taq PCR Master Mix for A-PCR, reaction buffers and GelRed were obtained from Sangon Biotech Co., Ltd (Shanghai, China). dNTP mix and enzymes (T4 DNA ligase and phi29 DNA polymerase) for RCA were obtained from New England Biolabs (Beijing, China). All the oligonucleotides were dissolved in Tris–HCl buffer (25 mM, pH 8.0), quantified using UV-vis absorption spectroscopy, stored at −20 °C, and annealed by heating at 95 °C for 3 min and gradually cooling to room temperature before use. ThT stock solutions (5 mM) were prepared in DMSO and stored in the dark at −20 °C. The human serum samples were provided by The First Bethune Hospital of Jilin University. Ultrapure water was utilized throughout.Fluorometric assays
Each fluorescence sample (300 μl, Fig. 1C, 4, S2, S4 and S6–S10†) containing ThT (5 μM) and the indicated DNA strand (300 nM) was prepared in Tris–HCl buffer (25 mM, pH 8.0) containing 300 mM K+, and incubated for 1 h at room temperature. Then, fluorescence spectra were collected on a Cary Eclipse Fluorescence Spectrophotometer (Agilent, USA), with slits of 10/10 nm, a scan rate of 600 nm min−1, excitation at 425 nm and emission at 490 nm.CD measurements
Each CD sample (200 μl, Fig. 2A–C and S5A†) contained 7 μM indicated DNA strand in Tris–HCl buffer (25 mM, pH 8.0) with 300 mM K+. After incubation for 1 h at room temperature, CD measurements from 200 to 350 nm were carried out at room temperature on a JASCO J-820 spectropolarimeter (Tokyo, Japan), and a cuvette of 1 mm light path was utilized.Thermal melting experiments
The samples (750 μl, Fig. 2D and S5B†) containing 2 μM indicated DNA strand were prepared in Tris–HCl buffer (25 mM, pH 8.0) with 300 mM K+. After incubation for 1 h, they were covered with 200 μl paraffin liquid and absorption spectra from 200 to 350 nm were collected as a function of temperature with a ramping rate of 1 °C min−1, on a Cary 60 UV-vis spectrometer (Agilent, USA). Through first derivative analysis of the obtained UV-melting profiles drawn at 295 nm for Intra-SG, and 260 nm for the duplex using the Origin 9.0 software, melting temperature (Tm) of each sample was obtained.UV analysis of ThT–DNA mixtures
Each sample (300 μl, Fig. 3 and S3†) containing 2.5 μM ThT and 5 μM each indicated DNA (Intra-SG or target C) was prepared in Tris–HCl buffer (25 mM, pH 8.0) with 300 mM K+. After incubation for 1 h at room temperature, absorption spectra from 200 to 600 nm were recorded using the mentioned UV-vis spectrophotometer.Results and discussion
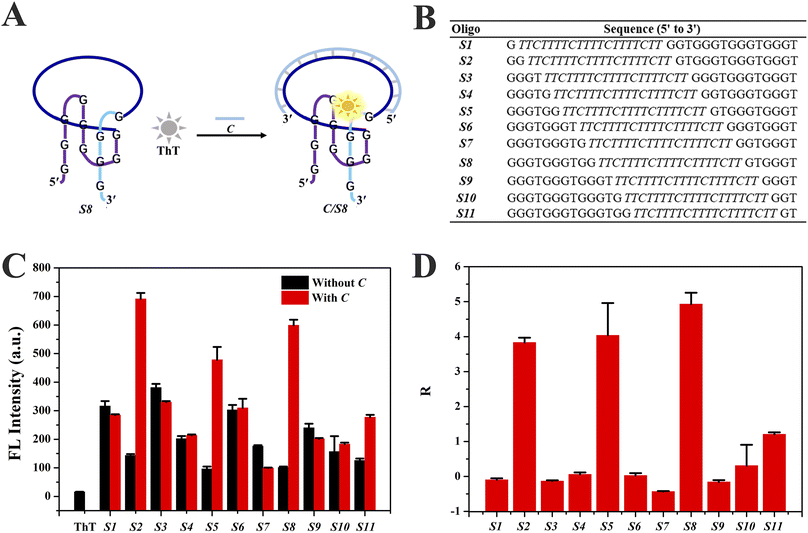
Eleven Intra-SG strands (S1–S11) were generated by splitting a typical unimolecular G4 (T30695, Table S1†) at different sites, namely 1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 11, 2
11, 2![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 10, 3
10, 3![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 9, 4
9, 4![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 8, 5
8, 5![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 7, 6
7, 6![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 6, 7
6, 7![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 5, 8
5, 8![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 4, 9
4, 9![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 3, 10
3, 10![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 2 and 11
2 and 11![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1 based on the number of guanine bases, and inserting a DNA spacer (C′) consisting of a repeated sequence (TTCTT) (Fig. S1†). A representative Intra-SG structure of split mode 8
1 based on the number of guanine bases, and inserting a DNA spacer (C′) consisting of a repeated sequence (TTCTT) (Fig. S1†). A representative Intra-SG structure of split mode 8![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 4 (S8) and detailed sequences of the eleven Intra-SG strands are shown in Fig. 1A and B, respectively. It has been verified that, similar to T30695, all Intra-SG sequences can fold into unimolecular G4 structures in Tris/K buffer (pH 8.0, 300 mM K+).22 As mentioned above, ThT is able to bind to specific G4 structures and emit enhanced emission.32Fig. 1C shows that the fluorescence intensity of ThT increased to varying degrees in the presence of different Intra-SG structures, since the topology of Intra-SG varied with the change in split mode. Drastic fluorescence light-up was observed for S2, S5 and S8, after hybridization between the inserted DNA spacer and its complementary strand (C). A ratio (R) representing the signal change amplitude toward strand C was calculated according to (F0 − FX)/F0, where F0 and FX are fluorescence intensities of the ThT/Intra-SG complex without and with C, respectively (Fig. 1D). High R values were obtained as 3.8, 4.0 and 5.0, respectively, for S2, S5 and S8, and manifest that these ThT/Intra-SG complexes are suitable for exploration as molecular probes for nucleic acid-triggered signal transduction. As for most of the G4-based molecular beacons, the input of the target would trigger unfolding of G4, and lead to release of organic dye and fluorescence change.35 However, via comprehensively analyzing the conformation and thermodynamic stability of Intra-SG structures and their interactions with ThT, a distinct principle was disclosed, which was hybridization-facilitated ThT binding to G4 and fluorescence enhancement (Fig. 1A).
4 (S8) and detailed sequences of the eleven Intra-SG strands are shown in Fig. 1A and B, respectively. It has been verified that, similar to T30695, all Intra-SG sequences can fold into unimolecular G4 structures in Tris/K buffer (pH 8.0, 300 mM K+).22 As mentioned above, ThT is able to bind to specific G4 structures and emit enhanced emission.32Fig. 1C shows that the fluorescence intensity of ThT increased to varying degrees in the presence of different Intra-SG structures, since the topology of Intra-SG varied with the change in split mode. Drastic fluorescence light-up was observed for S2, S5 and S8, after hybridization between the inserted DNA spacer and its complementary strand (C). A ratio (R) representing the signal change amplitude toward strand C was calculated according to (F0 − FX)/F0, where F0 and FX are fluorescence intensities of the ThT/Intra-SG complex without and with C, respectively (Fig. 1D). High R values were obtained as 3.8, 4.0 and 5.0, respectively, for S2, S5 and S8, and manifest that these ThT/Intra-SG complexes are suitable for exploration as molecular probes for nucleic acid-triggered signal transduction. As for most of the G4-based molecular beacons, the input of the target would trigger unfolding of G4, and lead to release of organic dye and fluorescence change.35 However, via comprehensively analyzing the conformation and thermodynamic stability of Intra-SG structures and their interactions with ThT, a distinct principle was disclosed, which was hybridization-facilitated ThT binding to G4 and fluorescence enhancement (Fig. 1A).
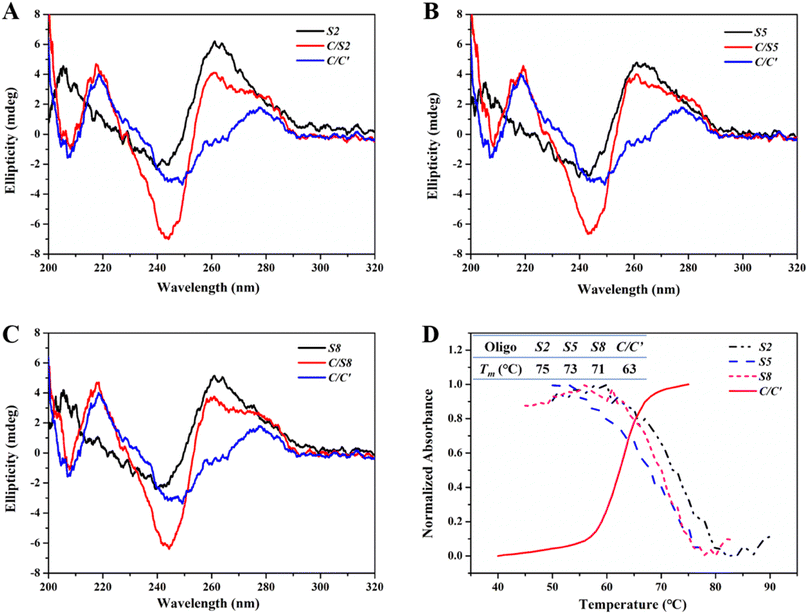
CD spectroscopy was used to confirm possible conformational changes of Intra-SG after spacer hybridization. As described above, the eleven Intra-SG structures revealed the analogous G4 topology to be T30695.10 After mixing these Intra-SG structures (S2, S5 or S8) with strand C, a CD peak at around 280 nm analogous to that of duplex C/C′ appeared, and the characteristic CD signal of G4 at 264 nm still existed (Fig. 2A–C), indicating that hybridization occurred between C and the 20-base spacer but the generated duplex cannot unfold the Intra-SG structures. UV-melting profiles of S2, S5, and S8 at 295 nm and the C/C′ duplex at 260 nm were collected (Fig. 2D) and melting temperature (Tm) was calculated (the inset of Fig. 2D). The results show that Tm values of Intra-SG structures of S2, S5, and S8 are much higher than that of the C/C′ duplex and confirm that the three Intra-SG structures cannot be opened by the generation of a C/C′ duplex. It was validated that duplex C/C′ itself cannot intensify the ThT emission (Fig. S2†). Hence, the fluorescence light-up from ThT/Intra-SG was ascribed to hybridization-facilitated ThT binding to G4.
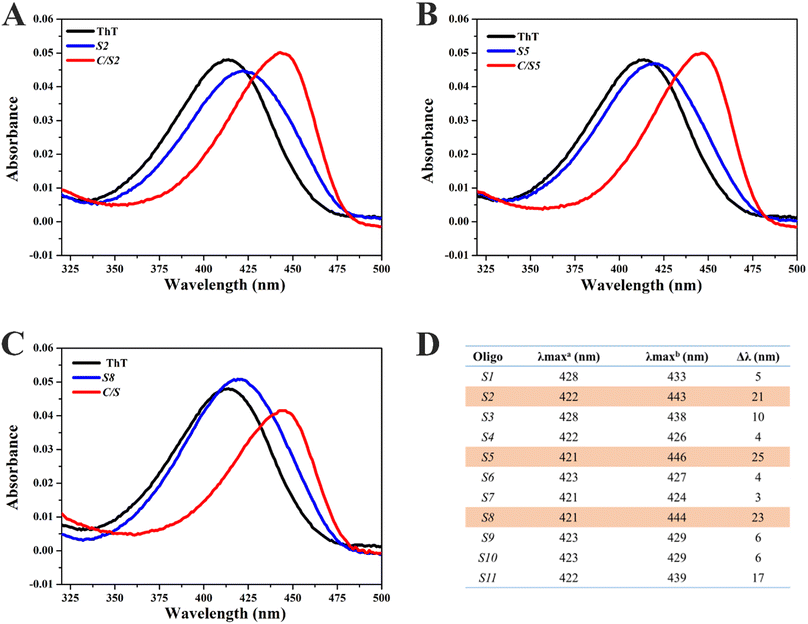
In order to explore the interaction between ThT and Intra-SG, UV-vis absorption spectra of ThT were then obtained in the presence of different Intra-SG structures or C/Intra-SG hybrids (Fig. 3A–C and S3†). ThT in Tris/K buffer exhibited an absorption peak at 412 nm. As DNA (5 μM) was added to a solution containing 2.5 μM ThT, the absorption peak shifted bathochromically and hypochromically, demonstrating apparent binding of ThT to Intra-SG structures. Different degrees of red shifts (Fig. 3D) reflected the differential binding capabilities of ThT with these Intra-SG structures. Particularly, large red shifts (up to 25 nm) were observed for C/S2, C/S5 and C/S8 hybrids, as compared to Intra-SG only, verifying the remarkable binding boost by C hybridization for the three Intra-SG structures. It was postulated that DNA C via loop hybridization can trigger small changes in the G4 microenvironment of S2, S5 and S8, facilitate the binding of ThT and restriction of torsional motion around the C–C bond between the benzothiazole and dimethylaminobenzene groups, and result in a decrease in the nonradiative decay process and remarkable fluorescence enhancement.36–39
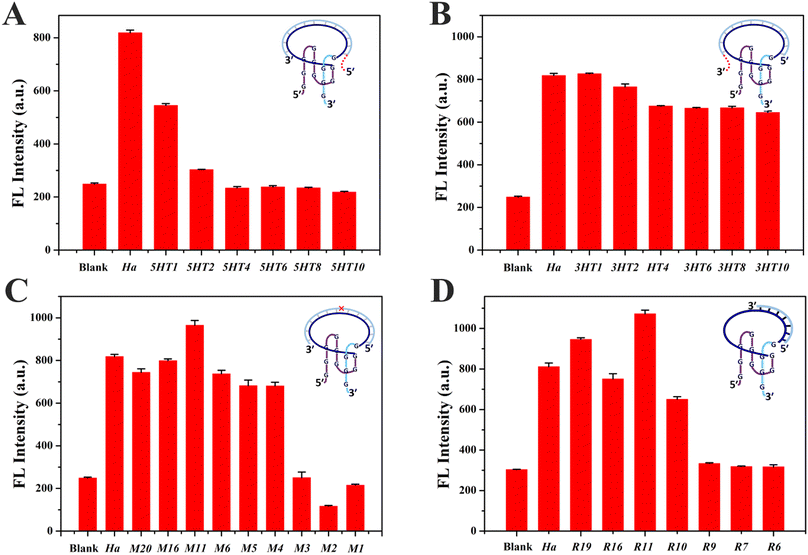
The phenomenon of hybridization-facilitated ThT binding and fluorescence light-up was verified for the simple DNA sequence C. What we were most concerned about was whether it could be applied for random DNA sequences and employed to design a new type of molecular beacon for nucleic acid analysis. Hence, derived from S8, four new Intra-SG structures (S8Ha′, S8Hb′, S8Hc′ and S8Hd′, Table S1†) were designed for recognizing four short single-stranded DNA segments from the HBV gene (Ha, Hb, Hc and Hd).40,41 The HBV has been regarded as a major threat to human health and its infection could lead to liver diseases (e.g., hepatocellular carcinoma).42 Analogous to ThT/S8, all four new ThT/Intra-SG systems emitted enhanced fluorescence in the presence of the respective target strand (Fig. S4†), but different R values were obtained, which indicated that the fluorescence response of the ThT/Intra-SG system correlated with the sequence of the inserted DNA spacer. Thereinto, ThT/S8Ha′ exhibited the highest response toward Ha (R = 2.79, Fig. S4A† and 4A), and a similar response mechanism was also confirmed by CD and UV-melting data (Fig. S5†), namely, typical CD signals corresponding to G4 and higher thermodynamic stability of Intra-SG in Ha/S8Ha′ than the Ha/Ha′ duplex. The ThT/S8Ha′ system was selected for the subsequent study of the topological effect via grafting, mutation and tailoring strategies.
DNA probes usually bind to a small segment in a long nucleic acid strand, which would lead to a complicated sequence environment. It is essential to investigate the influence of the sequence environment on the fluorescence response of ThT/S8Ha′. DNA protrusions consisting of different numbers of thymine (T) bases were grafted at the 5′ or 3′ ends of Ha, resulting in 5HTm and 3HTn strands (m, n = 1, 2, 4, 6, 8 and 10), where m and n represent the number of grafted T bases at the 5′ and 3′ ends of Ha, respectively. ThT/S8Ha′ emitted enhanced fluorescence after mixing with Ha, while remarkable fluorescence decreases were observed for ThT/S8Ha′ in the presence of 5HTm (Fig. 4A), and an overhang with only four T could quench the emission completely (5HT4, Fig. 4A). Relatively, a DNA overhang of 3HTn at the 3′ end exhibited little impact (Fig. 4B). A consistent trend was observed for the strands with protruding polyadenine (5HAm and 3HAn, Fig. S6A and B†), and similar phenomena existed for other Intra-SG with different split modes (S2, S5 and S8) (Fig. S6C–F†). These data suggested that the 5′-protruding strands of the target on Intra-SG structures apparently affected the binding interaction between ThT and Intra-SG and fluorescence emission of ThT.
Via single-base mutation of Ha from the 5′ to the 3′ end, a series of strands Mx (x = 1, 2, 3, 4, 5, 6, 11, 16, 19) were generated. As shown in Fig. 4C, low fluorescence responses were observed for ThT/S8Ha′ in the presence of Mx (x = 1, 2 and 3) whose mutation site was close to the 5′ terminus of the target, while the influence disappeared when the mutation site was at or behind the 4th base (M4–M20). Thus, single-base mismatched strands (M1, M2 and M3) could be distinguished very well if their mispairing sites are located close to the 5′ terminus of the target, i.e., the 3′ guanine-rich fragment of S8Ha′, and complete hybridization of the spacer in proximity to the 3′ guanine-rich fragment is decisive for binding interaction between ThT and S8Ha′. Considering the aforementioned apparent proximity effect from 5′-protruding strands of the target, it could be inferred that, in contrast to the 5′ guanine-rich fragment of Intra-SG, its 3′ guanine-rich fragment was much more susceptible to interference from adjacent protruding DNA and mismatches, which impeded hybridization-facilitated ThT binding and fluorescence light-up.
According to the grafting and mutation data, a critical sequence which is the shortest for the target-induced fluorescence enhancement should be obtained in proximity to the 3′ guanine-rich terminus in the DNA spacer of Intra-SG. Thus, different number of bases were cut out from the 3′ end or 5′ end of Ha, yielding different lengths of strands Ry and Ly (y = 19, 16, 11, 10, 9, 7 and 6). Similar to Ha, Ry also triggered fluorescence enhancement from ThT/S8Ha′ (Fig. 4D), until the number of bases of target strands was less than eleven. Obvious emission quenching occurred for Ry (y = 10, 9, 7 and 6). This was not contradictory to the mutation data in Fig. 4C, since a duplex with less than eleven base pairings cannot be stable at room temperature. The eleven-base spacer in S8Ha′ was the critical region which was vital for ThT binding. The R value of ThT/S8Ha′ toward R11 was calculated to be 3.31, and the 3′-protruding DNA consisting of as many as 30 T bases (RT30) cannot affect its fluorescent response obviously (Fig. S7†). Distinguished from Ha and Ry, all Ly stands caused very weak fluorescence response (Fig. S8†), further demonstrating that the target sequence in proximity to the 3′ guanine-rich terminus of Intra-SG is critical for the fluorescence response, and the 3′ guanine-rich fragment of Intra-SG is very susceptible to topology variation produced by the bound target strand.
The fluorescence background and sensitivity are related to the spacer sequence of Intra-SG. To acquire ThT/Intra-SG structures (S8Ra′, S8Rb′, S8Rc′, S8Rd′, S8Re′ and S8Rf′) derived from S8, six random DNA target strands (Ra, Rb, Rc, Rd, Re and Rf) were designed. These DNA sequences are shown in Table S1.† As high as 5.85 of R was obtained for the ThT/S8Ra′ system (Fig. S9†). Through analysis of these Intra-SG sequences via the NUPACK online software suite (https://www.nupack.org/), it could be summed up that the low tendency of duplex formation contributed to the high fluorescence sensitivity. In addition, ThT/S8Ra′ was also employed for topological effect investigation, and relative grafting, mutation and tailoring experimental data in Fig. S10A–G† validated the common rules, namely, the susceptibility of the 3′ guanine-rich fragment of Intra-SG and the existence of a critical recognition sequence. D11 is an eleven-base strand and completely complementary to the recognition sequence of S8Ra′. The R value of ThT/S8Ra′ toward D11 reached 6.04, and the 3′-protruding DNA consisting of as many as 30 T bases (RD30) did not affect the fluorescent response obviously (Fig. S10G†). Finally, ThT/S8Ra′ was selected for the subsequent analytical application in gene detection.
Not only were the intriguing rules for ThT binding interaction with Intra-SG structures disclosed, but the ThT/Intra-SG system can also be selected as a reporter in bioanalytical methods. It is feasible that small single-stranded nucleic acids (e.g., Ha) could be detected directly by ThT/Intra-SG-based molecular beacons. However, apparent limitations, e.g., low sensitivity and inhibition of protruding DNA, should not be ignored. Additionally, it is not applicable for detecting long genes which mostly exist in the double-stranded form in real life. Hence, a label-free amplified bioanalytical platform (LABP) was constructed for the detection of a model gene segment with 360 base pairings (bp) from the HBV (HF, Table S2†),43 by virtue of the ThT/S8Ra′ complex as a signal transducer and in combination with two DNA amplification techniques, A-PCR and RCA. As illustrated in Fig. 5, A-PCR is initiated first with the HF target as a template, producing numerous single-stranded DNA (SC). SC containing the primer sequence (Ha) then acts as a primer for the ligation of a padlock probe (Pa) and triggers RCA. The generated long single-stranded DNA chains containing a repeated sequence (BL′) are able to displace blocker DNA (BL) from conversion probes (Rv/BL), thus leading to the release of Rv which is produced by grafting four bases at the 3′ end of D11 and will finally induce a ThT fluorescence increase after recognition through hybridization with the recognition sequence of S8Ra′.
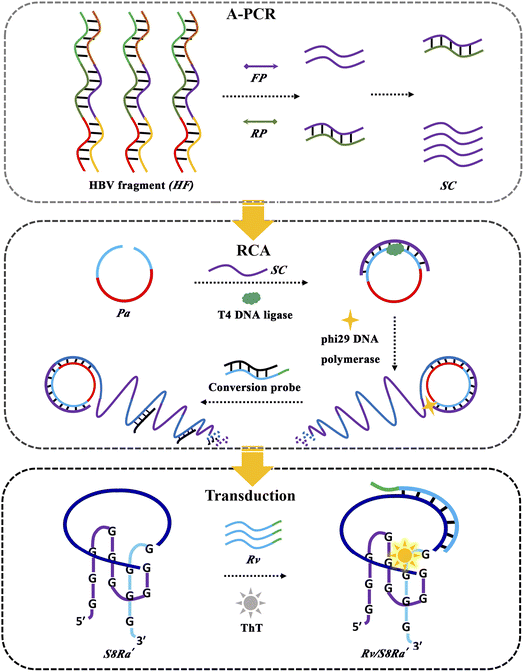 | ||
| Fig. 5 Schematical illustration of a label-free amplified bioanalytical platform (LABP) for HF detection by integrating A-PCR, RCA and ThT/S8Ra′-based signal transduction. | ||
Systematical experimental optimization and characterization were performed for the LABP. The molar ratio of the forward and reverse primer (FP and RP) in A-PCR varied and an optimum molar ratio of 10![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1 was selected, since the agarose gel electrophoresis image in Fig. 6A showed that SC of high yield was obtained (a red rectangle in lane 4). Pa cyclization with Ha as a primer was confirmed by denaturing polyacrylamide gel electrophoresis (a red rectangle in lane 4, Fig. 6B). The native polyacrylamide gel electrophoresis image in Fig. 6C shows that a large number of long DNA chains (lane 2) were produced after incubation of the cyclized Pa with phi29 DNA polymerase, and a displacement reaction between the RCA products and conversion probes (Rv/BL) occurred, leading to the release of Rv (indicated by a red rectangle in lane 4). Additionally, ThT/S8Ra′displayed high fluorescence response toward Rv (Fig. S10H†). Consequently, an ultrasensitive method for HF determination was successfully obtained through integrating A-PCR, RCA, a conversion probe and the ThT/S8Ra′ transducer together. Fig. 6D shows the fluorescence increase for the LABP after titration of HF. A linear relationship was obtained between fluorescence intensity and the logarithm of HF molecule numbers from 1 to 103 copies, and the regression equation is y = 324.59512x + 576.59311 (R2 = 0.991). The detection limit was calculated to be 1.0375 copies at a signal-to-noise of 3. Two gene segments from the Ebola virus (EF) and Nova virus (NF) were used to verify its specificity. Significant responses were observed only for HF, both in buffer and in 2% serum (Fig. 6E), demonstrating the excellent selectivity and applicability of the LABP for ultrasensitive gene detection.
1 was selected, since the agarose gel electrophoresis image in Fig. 6A showed that SC of high yield was obtained (a red rectangle in lane 4). Pa cyclization with Ha as a primer was confirmed by denaturing polyacrylamide gel electrophoresis (a red rectangle in lane 4, Fig. 6B). The native polyacrylamide gel electrophoresis image in Fig. 6C shows that a large number of long DNA chains (lane 2) were produced after incubation of the cyclized Pa with phi29 DNA polymerase, and a displacement reaction between the RCA products and conversion probes (Rv/BL) occurred, leading to the release of Rv (indicated by a red rectangle in lane 4). Additionally, ThT/S8Ra′displayed high fluorescence response toward Rv (Fig. S10H†). Consequently, an ultrasensitive method for HF determination was successfully obtained through integrating A-PCR, RCA, a conversion probe and the ThT/S8Ra′ transducer together. Fig. 6D shows the fluorescence increase for the LABP after titration of HF. A linear relationship was obtained between fluorescence intensity and the logarithm of HF molecule numbers from 1 to 103 copies, and the regression equation is y = 324.59512x + 576.59311 (R2 = 0.991). The detection limit was calculated to be 1.0375 copies at a signal-to-noise of 3. Two gene segments from the Ebola virus (EF) and Nova virus (NF) were used to verify its specificity. Significant responses were observed only for HF, both in buffer and in 2% serum (Fig. 6E), demonstrating the excellent selectivity and applicability of the LABP for ultrasensitive gene detection.
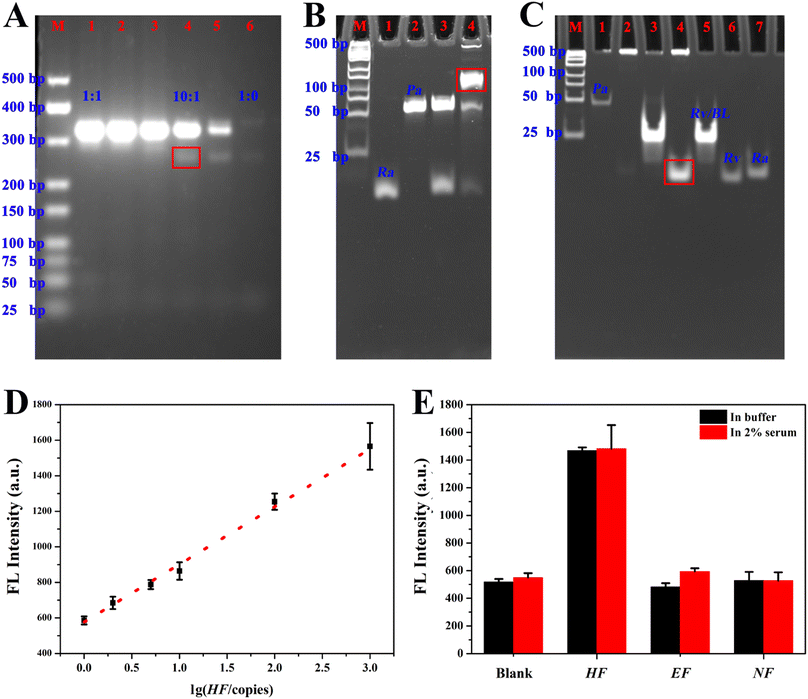 | ||
Fig. 6 (A) Agarose gel electrophoresis for optimizing the molar ratio of FP and HP in A-PCR. The ratios from lane 1 to lane 6 were 1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) : :![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1, 2 1, 2![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) : :![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1, 5 1, 5![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) : :![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1, 10 1, 10![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) : :![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1, 50 1, 50![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) : :![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1 and 1 1 and 1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) : :![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 0, respectively. The band corresponding to single-stranded A-PCR products (SC) with a selected ratio of 10 0, respectively. The band corresponding to single-stranded A-PCR products (SC) with a selected ratio of 10![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) : :![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1 is indicated by a red rectangle. (B) Denaturing polyacrylamide gel electrophoresis demonstrating DNA ligation (i.e., Pa cyclization) with Ha as a primer in the presence of T4 DNA ligase. The band corresponding to cyclized Pa is indicated by a red rectangle. (C) Native polyacrylamide gel electrophoresis confirming RCA polymerization (lane 2) and the displacement reaction between RCA products and the conversion probe (Rv/BL) (lane 3 and 4), and the released sequence Rv was indicated by a red rectangle in lane 4. Lane M represents the DNA marker ladder (25–500 bp). (D) A linear calibration curve for HF detection with a LABP. (E) Fluorescence responses of the LABP toward gene segments from different viruses, in buffer or in 2% serum. The final amount of target (HF, EF and NF) was 105 copies. The experimental details are presented in the ESI.† 1 is indicated by a red rectangle. (B) Denaturing polyacrylamide gel electrophoresis demonstrating DNA ligation (i.e., Pa cyclization) with Ha as a primer in the presence of T4 DNA ligase. The band corresponding to cyclized Pa is indicated by a red rectangle. (C) Native polyacrylamide gel electrophoresis confirming RCA polymerization (lane 2) and the displacement reaction between RCA products and the conversion probe (Rv/BL) (lane 3 and 4), and the released sequence Rv was indicated by a red rectangle in lane 4. Lane M represents the DNA marker ladder (25–500 bp). (D) A linear calibration curve for HF detection with a LABP. (E) Fluorescence responses of the LABP toward gene segments from different viruses, in buffer or in 2% serum. The final amount of target (HF, EF and NF) was 105 copies. The experimental details are presented in the ESI.† | ||
Conclusion
Systematic investigation of the topological effect on binding interaction between G4 and the ThT ligand was implemented with the universal Intra-SG platform, through fluorescence, UV-vis absorption, CD and thermal denaturation. ThT incubated with Intra-SG structures of eleven split modes revealed different fluorescence variations, and particularly, target-induced fluorescence enhancement was observed for three Intra-SG structures (S2, S5 and S8) whose split modes were 2![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 10, 5
10, 5![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 7 and 8
7 and 8![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 4, respectively. Hybridization-facilitated ThT binding to G4 was corroborated with different Intra-SG sequences. Moreover, it was found that the 3′ guanine-rich fragment of Intra-SG was quite susceptible to interference from adjacent protruding DNA or mismatches, and thus the sequence close to the 3′ guanine-rich terminus was decisive for the target-induced fluorescence enhancement. On the basis of these systems and rules, a well-performing method for ultrasensitive gene detection was constructed with an optimum ThT/Intra-SG system, further confirming its utility as a signal transducer in bioanalytical strategies. Taken together, in this work, critical factors for the interaction between Intra-SG and the ThT ligand were disclosed, and a universal and distinctive DNA-based signal transducer was provided, which will contribute to expanding the repertoire of DNA probes.
4, respectively. Hybridization-facilitated ThT binding to G4 was corroborated with different Intra-SG sequences. Moreover, it was found that the 3′ guanine-rich fragment of Intra-SG was quite susceptible to interference from adjacent protruding DNA or mismatches, and thus the sequence close to the 3′ guanine-rich terminus was decisive for the target-induced fluorescence enhancement. On the basis of these systems and rules, a well-performing method for ultrasensitive gene detection was constructed with an optimum ThT/Intra-SG system, further confirming its utility as a signal transducer in bioanalytical strategies. Taken together, in this work, critical factors for the interaction between Intra-SG and the ThT ligand were disclosed, and a universal and distinctive DNA-based signal transducer was provided, which will contribute to expanding the repertoire of DNA probes.
Data availability
Additional experimental and characterisation data are available in the ESI.†Author contributions
Mengmeng Lv: conceptualization, investigation, methodology, validation, visualization, writing – original draft and review & editing. Jiangtao Ren: conceptualization, investigation, methodology, validation, visualization, supervision, funding acquisition, writing – original draft and review & editing. Erkang Wang: supervision, funding acquisition, writing – review & editing.Conflicts of interest
There are no conflicts to declare.Acknowledgements
This work was supported by the National Natural Science Foundation of China (22004119 and 22374145), Jilin Province Science Technology Development Plan Project (20230508075RC) and Youth Innovation Promotion Association CAS (2021224).References
- Y. Xu, Chem. Soc. Rev., 2011, 40, 2719–2740 RSC.
- Z.-L. Liu, C.-A. Tao and J.-F. Wang, Chin. J. Anal. Chem., 2020, 48, 153–163 CAS.
- A. Bernal and L. Tusell, Int. J. Mol. Sci., 2018, 19, 294 CrossRef PubMed.
- F. Tameire, I. I. Verginadis and C. Koumenis, Semin. Cancer Biol., 2015, 33, 3–15 CrossRef CAS PubMed.
- G. Miglietta, J. Marinello, M. Russo and G. Capranico, Mol. Cancer, 2022, 21, 180 CrossRef PubMed.
- E. Ruggiero and S. N. Richter, Nucleic Acids Res., 2018, 46, 3270–3283 CrossRef CAS PubMed.
- X. Li, Y.-H. Jeon, N. Kwon, J.-G. Park, T. Guo, H.-R. Kim, J.-D. Huang, D.-S. Lee and J. Yoon, Biomaterials, 2021, 266, 120430 CrossRef CAS PubMed.
- W. Chen, Y. Zhang, H.-B. Yi, F. Wang, X. Chu and J.-H. Jiang, Angew. Chem., Int. Ed., 2023, 62, e202300162 CrossRef CAS PubMed.
- Y.-B. Su, X. Zhao, L.-J. Chen, H.-L. Qian and X.-P. Yan, Talanta, 2021, 233, 122567 CrossRef CAS PubMed.
- J. Ren, J. Wang, J. Wang and E. Wang, Biosens. Bioelectron., 2014, 51, 336–342 CrossRef CAS PubMed.
- S. Wang, S. Lu, J. Zhao and X. Yang, ACS Appl. Mater. Interfaces, 2019, 11, 25066–25073 CrossRef CAS PubMed.
- H. Lu, S. Li, J. Chen, J. Xia, J. Zhang, Y. Huang, X. Liu, H.-C. Wu, Y. Zhao, Z. Chai and Y. Hu, Metallomics, 2015, 7, 1508–1514 CrossRef CAS PubMed.
- I. Prislan, J. Lah and G. Vesnaver, J. Am. Chem. Soc., 2008, 130, 14161–14169 CrossRef CAS PubMed.
- Y. Ma, K. Iida and K. Nagasawa, Biochem. Biophys. Res. Commun., 2020, 531, 3–17 CrossRef CAS PubMed.
- F. Hao, Y. Ma and Y. Guan, Molecules, 2019, 24, 1863 CrossRef CAS PubMed.
- W. Wong, J. Zhuang, S. Ng, L. L. New, S. Hiew, J. Guo, Z. Yang and T. Li, Bioorg. Med. Chem. Lett., 2010, 20, 4689–4692 CrossRef CAS PubMed.
- Y. Guo, Q. Wang, Z. Wang, X. Chen, L. Xu, J. Hu and R. Pei, Sens. Actuators, B, 2015, 214, 50–55 CrossRef CAS.
- N. Li, R. Li, X. Sun, Y. Yang and Z. Li, Microchim. Acta, 2020, 187, 158 CrossRef CAS PubMed.
- H. Wu, Y. Liu, H. Wang, J. Wu, F. Zhu and P. Zou, Biosens. Bioelectron., 2016, 81, 303–308 CrossRef CAS PubMed.
- D. Zhou, W. Wu, Q. Li, J. Pan and J. Chen, Anal. Methods, 2019, 11, 3546–3551 RSC.
- J. Zhu, L. Zhang, S. Dong and E. Wang, Chem. Sci., 2015, 6, 4822–4827 RSC.
- M. Lv, Y. Guo, J. Ren and E. Wang, Nucleic Acids Res., 2019, 47, 9502–9510 CrossRef CAS PubMed.
- J. Ren, T. Wang, E. Wang and J. Wang, Analyst, 2015, 140, 2556–2572 RSC.
- D. Fan, X. Zhu, S. Dong and E. Wang, ChemPhysChem, 2017, 18, 1767–1772 CrossRef CAS PubMed.
- H. Chen, X. Sun, R. Cai, Y. Tian and N. Zhou, Mikrochim. Acta, 2019, 186, 843 CrossRef CAS PubMed.
- M. Biancalana and S. Koide, Biochim. Biophys. Acta, Proteins Proteomics, 2010, 1804, 1405–1412 CrossRef CAS PubMed.
- S. Liu, P. Peng, H. Wang, L. Shi and T. Li, Nucleic Acids Res., 2017, 45, 12080–12089 CrossRef CAS PubMed.
- Q. Xu, M. Yang, Y. Chang, S. Peng, D. Wang, X. Zhou and Y. Shao, Nucleic Acids Res., 2022, 50, 10249–10263 CrossRef CAS PubMed.
- L. Liu, Y. Shao, J. Peng, C. Huang, H. Liu and L. Zhang, Anal. Chem., 2014, 86, 1622–1631 CrossRef CAS PubMed.
- S. Sugimoto, K. Arita-Morioka, Y. Mizunoe, K. Yamanaka and T. Ogura, Nucleic Acids Res., 2015, 43, e92 CrossRef PubMed.
- X. Lan, L. Zhu, Y. Zhang, K. Chen, J. Wang, Z. Du, S. Li, X. Chen and W. Xu, Nucleic Acids Res., 2023, 51, 3556–3572 CrossRef CAS PubMed.
- J. Mohanty, N. Barooah, V. Dhamodharan, S. Harikrishna, P. I. Pradeepkumar and A. C. Bhasikuttan, J. Am. Chem. Soc., 2013, 135, 367–376 CrossRef CAS PubMed.
- T. Nakagawa, J. Tanaka, K. Harada, A. Shiratori, Y. Shimazaki, T. Yokoi, C. Uematsu and Y. Kohara, Anal. Chem., 2020, 92, 11705–11713 CrossRef CAS PubMed.
- J. Ge, Y. Hu, R. Deng, Z. Li, K. Zhang, M. Shi, D. Yang, R. Cai and W. Tan, Anal. Chem., 2020, 92, 13588–13594 CrossRef CAS PubMed.
- B. Guo, Y. Sheng, K. Zhou, Q. Liu, L. Liu and H.-C. Wu, Angew. Chem., Int. Ed., 2018, 57, 3602–3606 CrossRef CAS PubMed.
- V. I. Stsiapura, A. A. Maskevich, V. A. Kuzmitsky, V. N. Uversky, I. M. Kuznetsova and K. K. Turoverov, J. Phys. Chem. B, 2008, 112, 15893–15902 CrossRef CAS PubMed.
- J. Zhu, Z. Yan, W. Zhou, C. Liu, J. Wang and E. Wang, ACS Sens., 2018, 3, 1118–1125 CrossRef CAS PubMed.
- S. Tang, Y. Liu, Y. Yan, O. S. Yannick, X. Zhou, X. Xiang and C. Ma, Microchem. J., 2022, 181, 107655 CrossRef CAS.
- P. K. Singh, M. Kumbhakar, H. Pal and S. Nath, J. Phys. Chem. B, 2009, 113, 8532–8538 CrossRef CAS PubMed.
- F. Galibert, E. Mandart, F. Fitoussi, P. Tiollais and P. Charnay, Nature, 1979, 281, 646–650 CrossRef CAS PubMed.
- A. Panduro, S. Roman, S. Laguna-Meraz and A. Jose-Abrego, Viruses, 2023, 15, 2186 CrossRef PubMed.
- X. Peng, Y. Shi, B. Zhang, C. Xu and J. Lang, J. Med. Virol., 2023, 95, e29084 CrossRef CAS PubMed.
- D. C. Nyan, L. E. Ulitzky, N. Cehan, P. Williamson, V. Winkelman, M. Rios and D. R. Taylor, Clin. Infect. Dis., 2014, 59, 16–23 CrossRef CAS PubMed.
Footnote |
| † Electronic supplementary information (ESI) available. See DOI: https://doi.org/10.1039/d3sc06862e |
| This journal is © The Royal Society of Chemistry 2024 |