Open Access Article
Open Access ArticlePrecisely modulating the chromatin tracker via substituent engineering: reporting pathological oxidative stress during mitosis†
Jinsong
Li‡
a,
Yingyong
Ni‡
a,
Junjun
Wang
*a,
Yicai
Zhu
a,
Aidong
Wang
c,
Xiaojiao
Zhu
a,
Xianshun
Sun
a,
Sen
Wang
a,
Dandan
Li
 a and
Hongping
Zhou
a and
Hongping
Zhou
 *ab
*ab
aSchool of Chemistry and Chemical Engineering, School of Materials Science and Engineering, Institute of Physical Science and Information Technology, Center of Free Electron Laser & High Magnetic Field, Key Laboratory of Structure and Functional Regulation of Hybrid Materials, Ministry of Education, Key Laboratory of Functional Inorganic Materials Chemistry of Anhui Province, Key Laboratory of Chemistry for Inorganic/Organic Hybrid Functionalized Materials of Anhui Province, Anhui University, Hefei, 230601, P. R. China. E-mail: zhpzhp@263.net; wangjunjun@ahu.edu.cn
bSchool of Chemical and Environmental Engineering, Anhui Polytechnic University, Wuhu, 241000, P. R. China
cKey Laboratory of Drug Design, Huangshan University, Huangshan, 245021, P. R. China
First published on 31st January 2024
Abstract
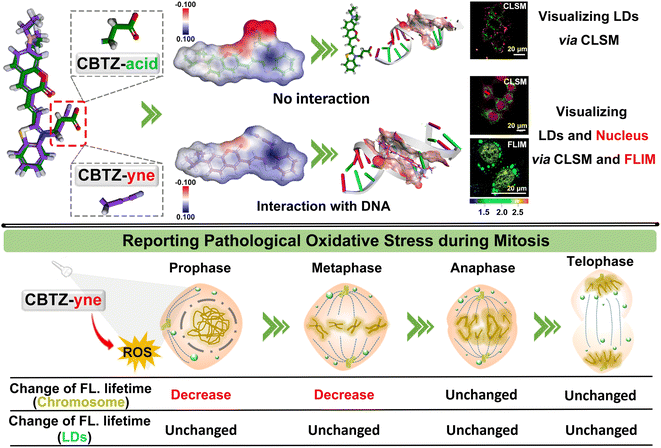
An in-depth understanding of cancer-cell mitosis presents unprecedented advantages for solving metastasis and proliferation of tumors, which has aroused great interest in visualizing the behavior via a luminescence tool. We developed a fluorescent molecule CBTZ-yne based on substituent engineering to acquire befitting lipophilicity and electrophilicity for anchoring lipid droplets and the nucleus, in which the low polarity environment and nucleic acids triggered a “weak–strong” fluorescence and “short–long” fluorescence-lifetime response. Meaningfully, CBTZ-yne visualized chromatin condensation, alignment, pull–push, and separation as well as lipid droplet dynamics, for the first time, precisely unveiling the asynchronous cellular mitosis processes affected by photo-generation reactive oxygen species according to the subtle change of fluorescence-lifetime. Our work suggested a new guideline for tracking the issue of the proliferation of malignant tumors in photodynamic therapy.
Introduction
Cancerous cells have the characteristics of rapid proliferation and metastasis ascribed to uncontrolled mitosis, thus posing a major problem in tumor treatment.1–3 As is known, mitosis involves prophase, metaphase, anaphase, and telophase with the character of chromatin condensation, alignment, pull–push, and separation, which is participated by nucleic acids (including DNA and RNA).4,5 Therefore, visualizing the dynamic physiological processes of DNA at each cycle stage of mitosis was critical during the cell's lifespan.6–11 Certainly, it was imperative to develop fluorescent bio-probe labelling DNA, whose strategies were mainly based on the introduction of positive charge to the probe or biological orthogonal technology.12–14 Until now, the developed small-molecule fluorescent probes capable of detecting, visualizing, and monitoring DNA structures in human cells offered a powerful and direct means to explore its putative roles.15–21 Nevertheless, the subtle variation in cellular microenvironments and complexity of multiple sub-organelles usually expressed the higher requirements for the sensitivity and selectivity of probes using confocal laser scanning microscopy (CLSM).In contrast to CLSM utilizing changes in fluorescence intensity to study the cellular microenvironment, fluorescence lifetime imaging microscopy (FLIM) has been used to provide more accurate messages through changes in fluorescence lifetime.22–27 For example, Liu et al. prepared a polarity-sensitive probe CQPP that monitored lipid droplets and the nucleus, successfully distinguishing between cellular ferroptosis and apoptosis using FLIM technology, which raised the possibility of observing the subtle variation in cellular microenvironments.28 Except for polarity, reactive oxygen species (ROS) was also believed as an important factor in affecting the microenvironments of cancerous cells, which could oxidize DNA leading to mutagenesis and affecting cellular proliferation.29–31 Unfortunately, visualizing the effect of ROS on cellular proliferation was somehow neglected, while was meaningful to fully track the issue of the proliferation of malignant tumors in photodynamic therapy.32
Besides, the cell nucleus and lipid droplets existed in close correlation, for instance, lipid droplet formation affected the nuclear morphology,33 which makes it urgent to develop a dual-specificity and sensitive tracker for observing the process of pathological oxidative stress between them. Compared with the traditional single-mode fluorescent probes, the two points of molecular design were considered to construct a dual-mode fluorescent tracker (CBTZ-yne). Firstly, the excellent lipophilicity of coumarin groups was selected for anchoring lipid droplets. Secondly, the substituent on the N-methyl-benzothiazole site was adjusted to bind DNA, the binding mechanism of which was investigated through 1H nuclear magnetic resonance (NMR) spectra, circular dichroism (CD) experiments, and docking calculation. CBTZ-yne showed a sensitive response of fluorescence and fluorescence lifetime when there is a slight change in the polarity environment or DNA contents, beneficial to observe the subtle variation in cellular microenvironments. Overall, CBTZ-yne first visualized subtle photo-damage on mitosis of cancer cells under photo-production ROS via FLIM, which offered a valuable reference to investigate the conundrum of asynchronous cellular mitosis (Scheme 1).
 | ||
| Scheme 1 Design of the chromatin tracker for reporting pathological oxidative stress during mitosis. | ||
Results and discussion
Synthesis and characterization
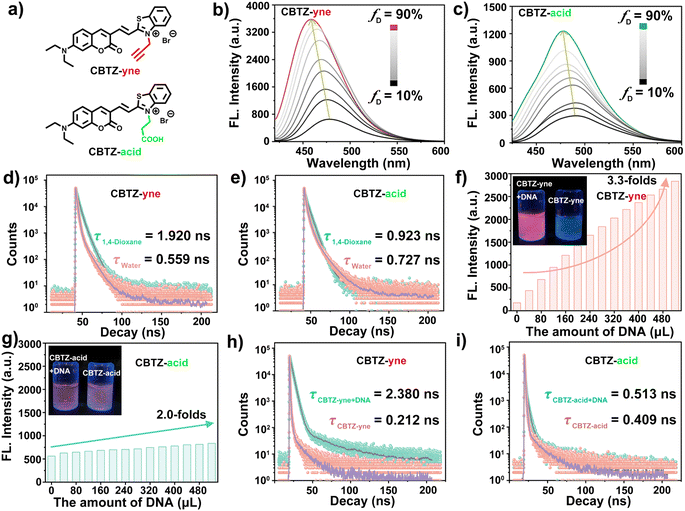
To adjust the lipophilicity and electrophilicity, the coumarin fragment was selected as an electron donor (D) by virtue of its unique lipophilicity, as well as two cationic benzothiazole derivatives as an acceptor (A) with different substituent groups, which provided a prerequisite for designing a fluorescent molecule which showed a response to lipid droplets and DNA. In this regard, the D–π–A system (CBTZ-yne and CBTZ-acid) was easily constructed to achieve excellent fluorescence emission. Among them, the terminal alkyne was used as a functional moiety to modulate the electrophilicity of CBTZ-yne for enhancing DNA binding.34–37 And then, CBTZ-yne and CBTZ-acid were synthesized by the Knoevenagel condensation reaction in one step, whose molecular structures were fully characterized by high-resolution mass spectroscopy (HRMS) as well as 1H NMR and 13C NMR spectroscopy (Fig. S1–S6†).Photophysical properties
The photophysical properties of CBTZ-yne and CBTZ-acid were studied using UV-vis absorption and fluorescence spectra. As we can see in Fig. S7,† the measured absorption maxima of CBTZ-yne and CBTZ-acid presented two absorption peaks in 1,4-dioxane, of which the one around 400 nm was consistent with that of compound 2 (with the same coumarin groups). Besides, the fluorescence spectra centered at 470 nm in Fig. S8† revealed that CBTZ-yne and CBTZ-acid exhibited emission (λex = 405 nm), which might originate from the local excitation (LE) process of coumarin groups.28,32 Also, the emission peaks of CBTZ-yne and CBTZ-acid centered at around 660 nm (λex = 561 nm) were attributed to the intramolecular charge transfer (ICT) process of the D–π–A molecule (Fig. S9†).Given that fluorescent molecules with the D–π–A structure are susceptible to solvent polarity, 1,4-dioxane with low polarity and H2O with high polarity were selected as solvents to investigate the photophysical properties of CBTZ-yne and CBTZ-acid. As displayed in Fig. 1b and c, weak fluorescence emission at around 465 nm was observed in aqueous solution, while the fluorescence intensity increased by degrees with the increasing 1,4-dioxane fraction of the mixture, accompanied by a blue-shift emission peak (Δλ = ca. 21 nm that of CBTZ-yne and Δλ = ca. 13 nm that of CBTZ-acid). Similarly, as shown in Fig. S10,† with the increase of solvent polarity, the fluorescence intensity of CBTZ-yne and CBTZ-acid in the solvent gradually decreases. Among them, the fluorescence intensity of CBTZ-yne has a larger attenuation amplitude, indicating that it is more sensitive to polarity. Subsequently, the transient fluorescence dynamics were also further investigated using a time-correlated single photon counter. As observed in Fig. 1d and e, the measured lifetime of CBTZ-yne in 1,4-dioxane (1.92 ns) was longer than that in aqueous solution (0.559 ns) at λex = ca. 373 nm. In contrast, the fluorescence lifetime of CBTZ-acid was fitted to be 0.923 ns in 1,4-dioxane, and 0.727 ns in aqueous solution, for which the difference (Δτ = 0.196 ns) was smaller than that of CBTZ-yne (Δτ = 1.361 ns). The prolonged lifetime indicated more fractions of excited states relaxing in radiative paths, which was well matched with the above observations of enhanced fluorescence intensity. These results presented that CBTZ-yne and CBTZ-acid showed a response to change in polar environment, especially, CBTZ-yne displayed more susceptibility to changes polarity of the solvent.
Considering that positively charged molecules easily bind with DNA because of their electrostatic interaction, the ability of CBTZ-yne and CBTZ-acid to respond to DNA was assessed via UV-vis absorption/fluorescence spectroscopy. As seen in Fig. S11a and b,† the redshifted absorption (Δλ = ca. 16 nm) of CBTZ-yne (10−5 M) was observed with the addition of DNA in aqueous solution, while that of CBTZ-acid (10−5 M) showed slight redshifts (Δλ = ca. 5 nm). Not only that, the fluorescence spectra in Fig. S12a and b† displayed the weakly emissive behavior of CBTZ-yne and CBTZ-acid in Tris–HCl buffer. Upon increasing the concentration of DNA, the fluorescent color of the CBTZ-yne solution changed from colorless to red, which was observed under a UV lamp (365 nm), along with a gradual increase in the fluorescence intensity. Also, the fluorescence quantum yield of CBTZ-acid measured using an ultrafast time-resolved fluorescence spectrometer was 2.97%, and the fluorescence quantum yield of CBTZ-acid + DNA was 20.16%. Comparatively, the enhanced appearance of CBTZ-acid was less significant than that of CBTZ-yne (Fig. 1f, g and S13†). Subsequently, the response of the tracker to DNA was further explored through transient fluorescence spectra (λem = ca. 665 nm). When DNA was added, the fluorescence lifetime of CBTZ-yne was prolonged from 0.212 ns to 2.38 ns (Δτ = 2.168 ns) (Fig. 1h). In contrast, under the addition of equal DNA, the fluorescence lifetime of CBTZ-acid rarely followed that change (Δτ = 0.104 ns), indicative of the weaker response to DNA (Fig. 1i).
As noted above, various biologically relevant ions and molecules (i.e., PO43−, nucleic acids, amino acids, etc.) were selected to investigate the selectivity of CBTZ-yne and CBTZ-acid to DNA. As shown in Fig. S14a†, the addition of DNA to CBTZ-yne showed the enhancement of fluorescence intensity distinct from those found with the addition of others. Instead, only a slight fluorescence increase at ca. 675 nm was found when CBTZ-acid was added with DNA and other analytes (Fig. S14b†). These observations suggested that the response of CBTZ-yne to DNA was more sensitive than that of CBTZ-acid.
The binding affinity of the tracker for DNA
The different substituent structures of target compounds and the fluorogenic response elicited by DNA prompted us to investigate their binding affinity. As can be seen in Fig. 2a, the 1H NMR titration experiments of the prepared tracker (10−2 M in DMSO-d6, each) with DNA (2 mg mL−1 in D2O-d2) were carried out.38,39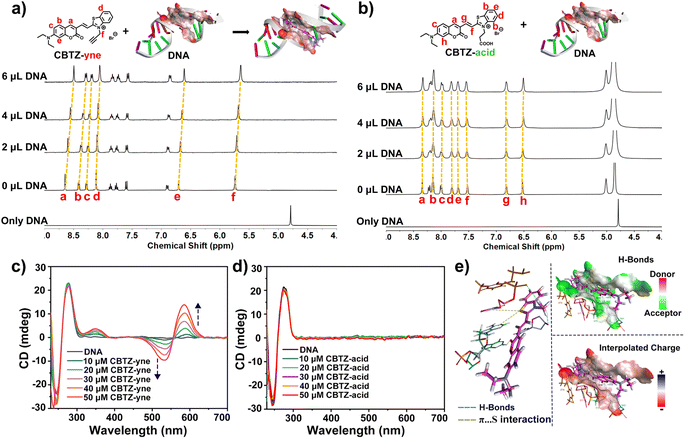 | ||
| Fig. 2 (a and b) 1H NMR spectra of trackers upon titration with 0 μL to 6.0 μL of DNA (2 mg mL−1 in D2O-d2); (c and d) CD spectra of DNA in Tris–HCl buffer (pH 7.4) in the presence of increasing concentrations of trackers; (e) molecular modeling calculations based on CBTZ-yne and DNA (PDB code 5JU4). | ||
With the titration of DNA from 0 μL to 6.0 μL, the Ha–Hf proton resonances of CBTZ-yne shifted up-field by 0.07–0.10 ppm, in part due to the interplay of π–π stacking which increased charge density easily overlapped with the hydrogen orbital to weaken its vibration, reflecting that CBTZ-yne inserted into the groove of the DNA. In contrast, when CBTZ-acid was mixed with DNA in D2O-d2 (0 μL to 6.0 μL), its protons Ha–Hh showed weak up-field shifts only from 0 to 0.02 ppm (Fig. 2b). The above results provided insights into the stronger binding affinity of CBTZ-yne for DNA compared with those of CBTZ-acid, in line with the results of the enhanced fluorescence intensity.
In addition, the binding affinity of the tracker and DNA was further studied by CD spectroscopy.40 As shown in Fig. 2c, the characteristic CD signals of DNA with a strong positive band at ca. 275 nm due to the base pair stacking and a negative band at ca. 236 nm indicated the right-handed helicity of DNA.41 With increasing concentrations of the tracker, the Cotton effect signal at 245 nm and 275 nm was retained. And, the induced CD signals centered at 585 nm (positive) and 530 nm (negative) gradually appeared upon the addition of CBTZ-yne, which was aligned with the absorption maximum (Fig. 2c), while those of CBTZ-acid were not.42 These measurements were suggestive of a chiral helical structure for the CBTZ-yne with DNA, where the handedness was induced by chiral DNA, which further supported the proposed binding affinity of CBTZ-yne upon interaction with DNA. By contrast, the induced CD signals of CBTZ-acid were not observed upon the introduction of DNA (Fig. 2d).
Besides, the density functional theory (DFT) method was used to evaluate the electrostatic potential of the tracker (Fig. S15†).43 The electrostatic potential of the entire molecule of CBTZ-yne was positive, while the electrostatic potential of the carboxyl group of CBTZ-acid was negative. These findings suggested that the weak binding affinity of CBTZ-acid might be due to the negative electrical properties of the carboxyl group, which was further verified by the docking calculation. As shown in Fig. 2e and Table S1†, CBTZ-acid with the carboxyl group was significantly acidic, showing a negative charge, which would not help interact with DNA bearing anionic charge on the surface. Note that, CBTZ-yne could bind with DNA through H-bond and π⋯S interaction.
Co-localization and fluorescence lifetime imaging
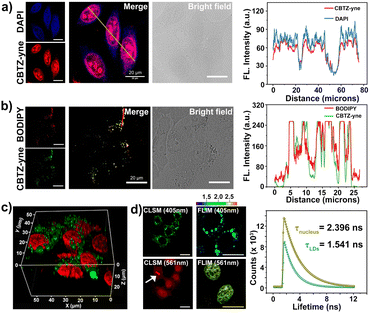
Given the prospective extracellular results, the suitability of CBTZ-yne for fluorescence imaging at the cellular level was then evaluated using various cell lines including HepG2 (human hepatocellular carcinoma cells), HeLa (human cervical cancer cells), A293T (human renal epithelial cells), and HUVEC (human umbilical vein endothelial cells). Confocal fluorescence microscope images in Fig. S16† showed punctate green fluorescence (λex: 405 nm) and irregular red fluorescence (λex: 561 nm) scattered in the cytoplasm within 20 min of incubation with CBTZ-acid. By contrast, the four types of cells incubated with CBTZ-yne exhibited punctate green fluorescence (λex: 405 nm) scattered in the cytoplasm as well as red fluorescence signal (λex: 561 nm) lighting up the nucleus that might be associated with the binding of CBTZ-yne with DNA, which was also suggestive of the good nuclear membrane permeability of CBTZ-yne (Fig. S17†).44Tracker staining lipid droplets and nucleus was examined with colocalization assays in live cells (taking HepG2 cells as an example) using commercial dyes BODIPY and DAPI. As can be seen from Fig. S18a,† the green fluorescence of CBTZ-acid occurred mainly in the lipid droplets, as confirmed by excellent overlap (highly matched fluorescence curve in Fig. S18b†) with the lipid droplet-specific marker, BODIPY. However, the red fluorescence response of CBTZ-acid occurred in the cytoplasm without overlapping with DAPI (Fig. S16c and d†). Therefore, these results suggest that CBTZ-acid was only selective for lipid droplets, not the nucleus. Importantly, the green fluorescence of CBTZ-yne coincided almost exactly with the red fluorescence associated with BODIPY, as well as the NIR fluorescence of CBTZ-yne was consistent with the blue fluorescence of DAPI (Fig. 3a and b), indicating that CBTZ-yne could visualize the lipid droplets and nucleus by two-color fluorescence imaging. It was noted that the avoidable fluorescence spectral crosstalk of the green and red dual-channel emission (Δλ = ca. 160 nm) provided conditions for the three-dimensional (3D) imaging of lipid droplets and the nucleus by CBTZ-yne (Fig. 3c).
In accordance, fluorescence lifetime imaging microscopy (FLIM) analysis of CBTZ-yne showed a short lifetime (τavg = 1.541 ns) in the region of lipid droplets with a cyan pseudo-color and a long lifetime in the region of the nucleus with a golden pseudo-color (τavg = 2.396 ns), respectively, under the same scale bar of lifetime (Fig. 3d). These observations showed that CBTZ-yne could image lipid droplets and the nucleus through dual-channel CLSM and FLIM, which provided a multi-functional luminescence tool for visualizing the behavior activity of cancer cells.
Visualizing different periods of mitosis in cancer cells
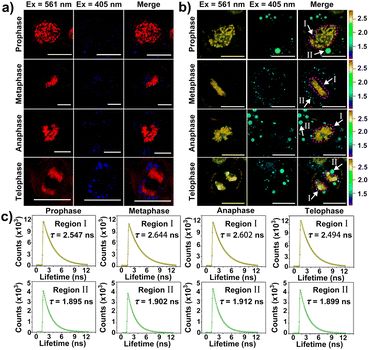
Of note, according to the observation shown in Fig. 3d with white arrows, CBTZ-yne imaged chromosome in the metaphase during mitosis, as well as the fluorescence signals from the green channel occurred in lipid droplets. This potential application made it a preferred candidate as a multi-functional tracker for real-time visualization of chromosomes and lipid droplets in different periods of mitosis of live cells. As shown in the higher resolution micrographs of single cells, when asynchronous cells were imaged, CBTZ-yne stained chromosomes in the red-channel and lipid droplets in the green-channel as cells progress through the cell cycle, and fluorescence images in living cells undergoing prophase, metaphase, anaphase, and telophase can be obtained and displayed in Fig. 4a (Fig. S19†). Moreover, the fluorescence lifetime of CBTZ-yne in cells was examined by FLIM analysis, and shown as pseudo-color at the same scale bar of lifetime. As indicated in Fig. 4b (Fig. S19†), the average lifetime distributed in the nucleus was 2.4–2.6 ns, represented by a golden pseudo-color, in line with the fluorescence lifetime of CBTZ-yne binding with DNA. By contrast, the fitted lifetime of lipid droplets was 1.7–1.8 ns represented by the cyan pseudo-color of II-region, which was matched with CBTZ-yne in low polarity (Fig. 4c and S19†). These results indicate that CBTZ-yne could be used to observe mitosis processes of the asynchronous cells by CLSM and FLIM.Feasibility of damage to DNA in the nucleus induced by photo-generation ROS of CBTZ-yne
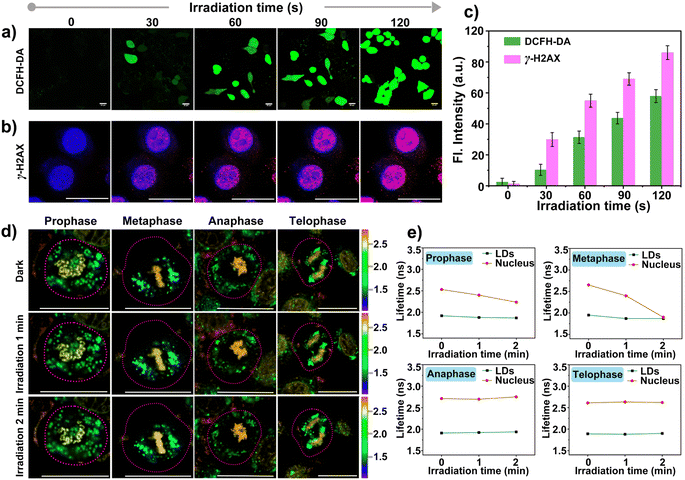
The higher steady-state levels of ROS played pivotal roles in the effect on metabolic activity in cancer cells because of the increased oxidative stress. Before the investigation of self-monitoring the damage to DNA in the nucleus induced by ROS, the abilities of photo-generation ROS by CBTZ-yne were evaluated via UV-vis absorption and fluorescence spectra as well as electron paramagnetic resonance (EPR) spectra with the commercial indicators including 2′,7′-dichlorodihydrofluorescein diacetate (DCFH-DA) for ROS, 9,10-anthracenediyl bis(methylene)dimalonic acid (ABDA) for 1O2, dihydrorhodamine 123 (DHR 123) for O2−˙, 5,5-dimethyl-1-pyrroline N-oxide (DMPO) for O2−˙.45,46 Firstly, with the continuous illumination of LED light, the fluorescence intensity of DCFH-DA increased rapidly within 35 s in the presence of CBTZ-yne (Fig. S20†), showing superior ROS generation of CBTZ-yne. Secondly, compared with the rose bengal (RB) group (commercial photosensitizer), the absorbance spectra in Fig. S21† showed that the absorption of ABDA was basically unchanged in CBTZ-yne aqueous solution with the irradiation of LED light for 80 s, indicating that the type of ROS generated by CBTZ-yne was not 1O2 (type II ROS). However, as shown in Fig. S22,† with the continuous illumination of LED light, the fluorescence intensity of DHR 123 increased more rapidly within 50 s in the presence of CBTZ-yne than that of CBTZ-acid, showing superior O2−˙ generation of CBTZ-yne. Meanwhile, the ESR signal of O2−˙ with DMPO in CBTZ-yne (red line) was detected in EPR spectra (Fig. S23†), indicative of the generation of type I ROS (O2−˙) of CBTZ-yne.Meanwhile, the ROS production ability was evaluated in HepG2 cells incubated with CBTZ-yne (10 μM) and DCFH-DA (2 μM). As displayed in Fig. 5a, under LED-light irradiation, the green fluorescence signal of DCFH-DA within the cells gradually increased, indicating photo-generation ROS of CBTZ-yne at the cellular levels. After that, the photodynamic cell suppression efficiency of CBTZ-yne and CBTZ-acid was tested through MTT (3-(4,5-dimethylthiazol-2-yl)-2,5-diphenyltetrazolium bromide) assay. In the dark, CBTZ-yne and CBTZ-acid showed negligible cytotoxicity and optimal biocompatibility (Fig. S24a†). When the cells incubated by using CBTZ-yne and CBTZ-acid were irradiated with an LED lamp, the cell survival rate decreased to varying degrees. The phototoxicity of CBTZ-yne was significantly better than that of CBTZ-acid at the same dose (Fig. S24b†). Benefiting from the oxidizability of ROS to biological macromolecules, the feasibility of damage to DNA in the nucleus of CBTZ-yne was verified using the commercial dye (γ-H2AX) immunofluorescence with the DAPI marker under irradiation of LED light from 0 s to 120 s.47 As indicated in Fig. 5b, when the HepG2 cells were treated with CBTZ-yne, DAPI, and γ-H2AX without irradiation, the confocal fluorescence image only showed the blue fluorescence of DAPI. However, the red fluorescence of phosphorylated H2AX Ser139 products in HepG2 cells was gradually observed as the irradiation time increased, suggestive of DNA damage after photogenerated ROS. At the same time, Fig. 5c showed the quantification of the fluorescence intensity of DCFH-DA and γ-H2AX in Fig. 5a and b under illumination from 0 s to 120 s.
Tracking oxidative stress during mitosis via FLIM
Given that ROS can damage lipid droplets and DNA to cause oxidative stress in cells, the effect of ROS on mitosis processes in asynchronous cells was further investigated through the analysis of the fluorescence lifetime of CBTZ-yne in lipid droplets and the nucleus via FLIM. As can be seen from Fig. 5d and S25,† when HepG2 cells were in different mitotic phases (prophase, metaphase, anaphase, and telophase) under dark conditions, the fitted fluorescence lifetime in lipid droplets and the nucleus was in accordance with the findings in Fig. 4. Nevertheless, the fluorescence lifetime showed changes with different degrees in the different periods of asynchronous cells under irradiation. In detail, while the average fluorescence lifetime (τavg) of CBTZ-yne in the prophase and metaphase of HepG2 cells was 2.53 ns, 2.65 ns, respectively, the values of τavg notably dropped by 0.30–0.75 ns due to the different DNA damage inducers by the production of photoexcited ROS. By contrast, the τavg of the anaphase and telophase of HepG2 cells retained 2.70 ns, 2.55 ns, respectively, suggesting that the effect of ROS on DNA damage was more obvious in the prophase and metaphase of cellular mitosis. To further reveal the mechanism, ROS-induced damage of different-concentration DNA under light was assessed by observing the change in the fluorescence lifetime of CBTZ-yne with different content (30 μL, 300 μL, 500 μL) of DNA (aqueous solution, 2 mg mL−1) via ultrafast time-resolved fluorescence spectroscopy. As shown in Fig. S26,† the fluorescence lifetime of CBTZ-yne binding with high-concentration DNA (300 μL or 500 μL, 2 mg mL−1) decreased more obviously than that with low-concentration DNA (30 μL, 2 mg mL−1) with the increasing time of illumination. The above results SHOWED that (1) DNA of different concentrations affected by photo-generated ROS different, (2) the condensed degree of DNA in the prophase and metaphase might be higher than that in the anaphase and telophase. At the same time, the fluorescence lifetime of the lipid droplets was unchanged significantly in each period, indicative of no effect on the activities of lipid droplets (Fig. 5e). In a word, CBTZ-yne would be used as a tracker for visualizing the effects induced by ROS on lipid droplets and chromosomes during asynchronous cell mitosis according to the subtle change of fluorescence lifetime.Conclusions
In summary, based on substituent engineering, the coumarin derivative (CBTZ-yne) was precisely modulated for anchoring the lipid droplets and nucleus. Mechanically, the LE process in low-polarity environments and the limited intramolecular rotation after binding to DNA resulted in CBTZ-yne showcasing the more significant “weak–strong” fluorescence and “short–long” fluorescence-lifetime response in the two subcellular organelles. Benefiting from self-generated ROS under light, the tailored CBTZ-yne first reported pathological oxidative stress during the entire process of asynchronous cell mitosis via FLIM, which also represented a new paradigm for exploring cellular malignant proliferation to deal with the problem of cancer treatment.Data availability
The datasets supporting this article have been uploaded as part of the ESI.†Author contributions
Jinsong Li prepared the compounds, determined the test methods, and wrote the original draft. Yingyong Ni supplemented the relevant experiment in the revised manuscript and ESI.† Yicai Zhu provided cell culture. Aidong Wang analyzed the test data of docking calculation. Xiaojiao Zhu, Xianshun Sun, Dandan Li, Sen Wang and Hongping Zhou were responsible for funding acquisition. Junjun Wang and Hongping Zhou conceptualized the project, guided the project, and revised the manuscript.Conflicts of interest
There are no conflicts to declare.Acknowledgements
This work was supported by the National Natural Science Foundation of China (52372073, 51972001, 22171001, 22375003, 22305001, and 52103111) and Anhui University Scientific Research Start-up Fund (No. S202418001/112, S020318008/026, and S020318008/028).References
- J. Medlock, A. A. K. Das, L. A. Madden, D. J. Allsup and V. N. Paunov, Chem. Soc. Rev., 2017, 46, 5110–5127 RSC
.
- L. Zhang, N. Montesdeoca, J. Karges and H. Xiao, Angew. Chem., Int. Ed., 2023, 62, e202300662 CrossRef CAS PubMed
.
- P. Zhang, Y. Zhai, Y. Cai, Y. Zhao and Y. Li, Adv. Mater., 2019, 31, 1904156 CrossRef CAS PubMed
.
- D. Li, X. Tian, A. Wang, L. Guan, J. Zheng, F. Li, S. Li, H. Zhou, J. Wu and Y. Tian, Chem. Sci., 2016, 7, 2257–2263 RSC
.
- S. Pillai, J. Nguyen, J. Johnson, E. Haura, D. Coppola and S. Chellappan, Nat. Commun., 2015, 6, 10072 CrossRef CAS PubMed
.
- S. Nam and O. Chaudhuri, Nat. Phys., 2018, 14, 621–628 Search PubMed
.
- S. Nam, V. K. Gupta, H.-p. Lee, J. Y. Lee, K. M. Wisdom, S. Varma, E. M. Flaum, C. Davis, R. B. West and O. Chaudhuri, Sci. Adv., 2019, 5, eaaw6171 Search PubMed
.
- V. Roukos, G. Pegoraro, T. C. Voss and T. Misteli, Nat. Protoc., 2015, 10, 334–348 Search PubMed
.
- Y.-H. Wen, F. Yang, S. Li, X. Yao, J. Song and L.-Z. Gong, J. Am. Chem. Soc., 2023, 145, 4199–4207 CrossRef CAS PubMed
.
- M.-Y. Wu, J.-K. Leung, L. Liu, C. Kam, K. Y. K. Chan, R. A. Li, S. Feng and S. Chen, Angew. Chem., Int. Ed., 2020, 59, 10327–10331 CrossRef CAS PubMed
.
- F. Yi, Y. Zhang, Z. Wang, Z. Wang, Z. Li, T. Zhou, H. Xu, J. Liu, B. Jiang, X. Li, L. Wang, N. Bai, Q. Guo, Y. Guan, Y. Feng, Z. Mao, G. Fan, S. Zhang, C. Wang, L. Cao, B. P. O’Rourke, Y. Wang, Y. Wu, B. Wu, S. You, N. Zhang, J. Guan, X. Song, Y. Sun, S. Wei and L. Cao, Sci. Adv., 2021, 7, eabe5518 CrossRef CAS PubMed
.
- S. Berndl and H.-A. Wagenknecht, Angew. Chem., Int. Ed., 2009, 48, 2418–2421 CrossRef CAS PubMed
.
- P. Gaur, A. Kumar, R. Dalal, S. Bhattacharyya and S. Ghosh, Chem. Commun., 2017, 53, 2571–2574 RSC
.
- Z. Li, S. Sun, Z. Yang, S. Zhang, H. Zhang, M. Hu, J. Cao, J. Wang, F. Liu, F. Song, J. Fan and X. Peng, Biomaterials, 2013, 34, 6473–6481 CrossRef CAS PubMed
.
- G. Lukinavičius, C. Blaukopf, E. Pershagen, A. Schena, L. Reymond, E. Derivery, M. Gonzalez-Gaitan, E. D'Este, S. W. Hell, D. W. Gerlich and K. Johnsson, Nat. Commun., 2015, 6, 8497 CrossRef PubMed
.
- R. T. Michenfelder, L. Delafresnaye, V. X. Truong, C. Barner-Kowollik and H.-A. Wagenknecht, Chem. Commun., 2023, 59, 4012–4015 RSC
.
- A. Nakamura, K. Takigawa, Y. Kurishita, K. Kuwata, M. Ishida, Y. Shimoda, I. Hamachi and S. Tsukiji, Chem. Commun., 2014, 50, 6149–6152 RSC
.
- X. Peng, T. Wu, J. Fan, J. Wang, S. Zhang, F. Song and S. Sun, Angew. Chem., Int. Ed., 2011, 50, 4180–4183 CrossRef CAS PubMed
.
- A. T. Szczurek, K. Prakash, H.-K. Lee, D. J. Żurek-Biesiada, G. Best, M. Hagmann, J. W. Dobrucki, C. Cremer and U. Birk, Nucleus, 2014, 5, 331–340 CrossRef PubMed
.
- M. Tian, J. Sun, B. Dong and W. Lin, Angew. Chem., Int. Ed., 2018, 57, 16506–16510 CrossRef CAS PubMed
.
- K.-N. Wang, Q. Cao, L.-Y. Liu, Z.-J. Zhao, W. Liu, D.-J. Zhou, C.-P. Tan, W. Xia, L.-N. Ji and Z.-W. Mao, Chem. Sci., 2019, 10, 10053–10064 RSC
.
- A. Goujon, A. Colom, K. Straková, V. Mercier, D. Mahecic, S. Manley, N. Sakai, A. Roux and S. Matile, J. Am. Chem. Soc., 2019, 141, 3380–3384 CrossRef CAS PubMed
.
- X. Peng, Z. Yang, J. Wang, J. Fan, Y. He, F. Song, B. Wang, S. Sun, J. Qu, J. Qi and M. Yan, J. Am. Chem. Soc., 2011, 133, 6626–6635 CrossRef CAS PubMed
.
- M. Priessner, P. A. Summers, B. W. Lewis, M. Sastre, L. Ying, M. K. Kuimova and R. Vilar, Angew. Chem., Int. Ed., 2021, 60, 23148–23153 CrossRef CAS PubMed
.
- J. A. Robson, M. Kubánková, T. Bond, R. A. Hendley, A. J. P. White, M. K. Kuimova and J. D. E. T. Wilton-Ely, Angew. Chem., Int. Ed., 2020, 59, 21431–21435 CrossRef CAS PubMed
.
- S. Suarasan, A.-M. Craciun, E. Licarete, M. Focsan, K. Magyari and S. Astilean, ACS Appl. Mater. Interfaces, 2019, 11, 7812–7822 CrossRef CAS PubMed
.
- Y. Zhao, H. S. Kim, X. Zou, L. Huang, X. Liang, Z. Li, J. S. Kim and W. Lin, J. Am. Chem. Soc., 2022, 144, 20854–20865 CrossRef CAS PubMed
.
- K.-N. Wang, L.-Y. Liu, D. Mao, S. Xu, C.-P. Tan, Q. Cao, Z.-W. Mao and B. Liu, Angew. Chem., Int. Ed., 2021, 60, 15095–15100 CrossRef CAS PubMed
.
- X. Renaudin, Int. Rev. Cell Mol. Biol., 2021, 364, 139–161 CAS
.
- L. A. Loeb, D. C. Wallace and G. M. Martin, Proc. Natl. Acad. Sci. U. S. A., 2005, 102, 18769–18770 CrossRef CAS PubMed
.
- L. B. Sullivan and N. S. Chandel, Cancer Metabol., 2014, 2, 17 CrossRef PubMed
.
- H. Tian, A. C. Sedgwick, H.-H. Han, S. Sen, G.-R. Chen, Y. Zang, J. L. Sessler, T. D. James, J. Li and X.-P. He, Coord. Chem. Rev., 2021, 427, 213577 CrossRef CAS
.
- I. L. Ivanovska, M. P. Tobin, T. Bai, L. J. Dooling and D. E. Discher, J. Cell Biol., 2023, 222, e202208123 CrossRef CAS PubMed
.
- E. M. Kohn, K. Konovalov, C. A. Gomez, G. N. Hoover, A. K. Yik, X. Huang and J. D. Martell, ACS Chem. Biol., 2023, 18(9), 1976 CrossRef CAS PubMed
.
- Z. L. Salie, K. A. Kirby, E. Michailidis, B. Marchand, K. Singh, L. C. Rohan, E. N. Kodama, H. Mitsuya, M. A. Parniak and S. G. Sarafianos, Proc. Natl. Acad. Sci. U.S.A., 2016, 113, 9274 CrossRef CAS PubMed
.
- P. Giastas, M. Neu, P. Rowland and E. Stratikos, ACS Med. Chem. Lett., 2019, 10, 708 CrossRef CAS PubMed
.
- T. T. Talele, J. Med. Chem., 2020, 63, 5625 CrossRef CAS PubMed
.
- I. Crnolatac, I. Rogan, B. Majić, S. Tomić, T. Deligeorgiev, G. Horvat, D. Makuc, J. Plavec, G. Pescitelli and I. Piantanida, Anal. Chim. Acta, 2016, 940, 128–135 CrossRef CAS PubMed
.
- W. Liu, Y.-F. Zhong, L.-Y. Liu, C.-T. Shen, W. Zeng, F. Wang, D. Yang and Z.-W. Mao, Nat. Commun., 2018, 9, 3496 CrossRef PubMed
.
- J. E. F. Alves, J. F. de Oliveira, T. R. C. de Lima Souza, R. O. de Moura, L. B. de Carvalho Júnior, M. d. C. Alves de Lima and S. M. V. de Almeida, Int. J. Biol. Macromol., 2021, 170, 622–635 CrossRef CAS PubMed
.
- V. I. Ivanov, L. E. Minchenkova, A. K. Schyolkina and A. I. Poletayev, Biopolymers, 1973, 12, 89–110 CrossRef CAS PubMed
.
- V. Uma, M. Kanthimathi, T. Weyhermuller and B. U. Nair, J. Inorg. Biochem., 2005, 99, 2299–2307 CrossRef CAS PubMed
.
- Z. Feng, D. Zhang, H. Guo, W. Su, Y. Tian and X. Tian, Nanoscale, 2023, 15, 5486–5493 RSC
.
- K. N. Wang, X. J. Chao, B. Liu, D. J. Zhou, L. He, X. H. Zheng, Q. Cao, C. P. Tan, C. Zhang and Z.-W. Mao, Chem. Commun., 2018, 54, 2635–2638 RSC
.
- X. Li, J. Wu, L. Wang, C. He, L. Chen, Y. Jiao and C. Duan, Angew. Chem., Int. Ed., 2020, 59, 6420–6427 CrossRef CAS PubMed
.
- H. Ma, Y. Lu, Z. Huang, S. Long, J. Cao, Z. Zhang, X. Zhou, C. Shi, W. Sun, J. Du, J. Fan and X. Peng, J. Am. Chem. Soc., 2022, 144, 3477–3486 CrossRef CAS PubMed
.
- X. Zhu, J. Wu, R. Liu, H. Xiang, W. Zhang, Q. Chang, S. Wang, R. Jiang, F. Zhao, Q. Li, L. Huang, L. Yan and Y. Zhao, ACS Nano, 2022, 16, 18849–18862 CrossRef CAS PubMed
.
Footnotes |
| † Electronic supplementary information (ESI) available. See DOI: https://doi.org/10.1039/d3sc06342a |
| ‡ Co-first author. |
| This journal is © The Royal Society of Chemistry 2024 |