Open Access Article
Open Access ArticleIntermittent proton bursts of single lactic acid bacteria†
Jia
Gao
 ,
Kai
Zhou
,
Haoran
Li
,
Yaohua
Li
,
Kairong
Yang
and
Wei
Wang
,
Kai
Zhou
,
Haoran
Li
,
Yaohua
Li
,
Kairong
Yang
and
Wei
Wang
 *
*
State Key Laboratory of Analytical Chemistry for Life Science, School of Chemistry and Chemical Engineering, ChemBIC (Chemistry and Biomedicine Innovation Center), Nanjing University, Nanjing, 210023, China. E-mail: wei.wang@nju.edu.cn
First published on 25th January 2024
Abstract
Lactic acid bacteria are a kind of probiotic microorganisms that efficiently convert carbohydrates to lactic acids, thus playing essential roles in fermentation and food industry. While conventional wisdom often suggests continuous release of protons from bacteria during acidification, here we developed a methodology to measure the dynamics of proton release at the single bacteria level, and report on the discovery of a proton burst phenomenon, i.e., the intermittent efflux of protons, of single Lactobacillus plantarum bacteria. When placing an individual bacterium in an oil-sealed microwell, efflux and accumulation of protons consequently reduced the pH in the confined extracellular medium, which was monitored with fluorescent pH indicators in a high-throughput and real-time manner. In addition to the slow and continuous proton release behavior (as expected), stochastic and intermittent proton burst events were surprisingly observed with a typical timescale of several seconds. It was attributed to the regulatory response of bacteria by activating H+-ATPase to compensate the stochastic and transient depolarizations of membrane potential. These findings not only revealed an unprecedented proton burst phenomenon in lactic acid bacteria, but also shed new lights on the intrinsic roles of H+-ATPase in membrane potential homeostasis, with implications for both fermentation industry and bacterial electrophysiology.
Introduction
Membrane potential plays essential roles in maintaining and regulating the physiological functions of cells and microorganisms.1–5 While the membrane potential of non-excitable mammalian cells is relatively stable, recent studies have shown that the membrane potential of single bacteria exhibited extraordinary fluctuation behavior.6,7 It was because the surface area (∼1 μm2), and thus the membrane capacitance (C), of bacteria were 2–3 orders of magnitude smaller than those of mammalian cells. A tiny amount of transmembrane ion transport (ΔQ) could lead to a relatively large change in the membrane potential (ΔU = ΔQ/C).1,6 In addition, the number of specific membrane proteins in an individual bacterium is sometimes so small that the stochastic gating of a single ion channel or transporter could lead to fluctuations in the intracellular concentration of ions (charges), causing flashes in the membrane potential.5–8Flashing membrane potential of bacteria is not only a feature of bacterial electrophysiology, but also plays electrical signaling roles to regulate different physiological functions.3,9–13 For example, using a genetically encoded dye, the presence of a dynamic flashing behavior of the membrane potential in intact E. coli cells, similar to that of neuronal cells, was reported for the first time in 2011.6 This spontaneous, whole-bacteria flashing behavior was manifested as a rapid depolarization and slow recovery of the membrane potential on a timescale of a few seconds.6,7 Subsequent studies demonstrated that these dynamic changes of bacterial membrane potential are mediated by the random opening or closing of bacterial ion channels in the temporal dimension.7,9,14,15 The discovery of this phenomenon provided unprecedented insights into electrophysiological mechanisms such as bacterial signal transduction and inter-bacterial electrical communication.16,17 It has been reported to be involved in regulating bacterial intracellular pH as well as reactive oxygen levels.18 Our recent studies on bioluminescent bacteria, P. phosphoreum, have shown that the fluctuations in the activity of the Na+/H+ antiporter led to an intermittently pulsed behavior of the membrane potential, as revealed by the blinking bioluminescence emission of a single bacterium.8 Although current research studies have shed light on the membrane potential dynamics of bacteria in multiple dimensions, new perspectives are still expected. For example, it remains unclear whether such fluctuating behavior permeates the metabolite secretion process of bacteria.
In this work, we developed a method for measuring the proton release dynamics of single lactic acid bacteria based on microwell arrays and extracellular pH probes. By encasing individual bacteria in a picoliter-sized microwell sealed with inert oil, protons released from a single bacterium can be effectively confined in the droplets and accumulated to detectable concentrations,19,20 which greatly enhanced the sensitivity and accuracy of proton detection. We chose Lactobacillus plantarum (L. plantarum) as a model microorganism, which, as a member of lactic acid bacteria family, efficiently converted glucose to lactic acid, accompanying a significant decrease in the extracellular pH, i.e., acidification. The proton release rate was measured by introducing a pH-sensitive fluorescent dye in the buffer-free, extracellular medium. It was found that the fermentation acid production process of a single L. plantarum coupled two types of proton efflux kinetics: continuous proton release and intermittent, burst-like proton release with a timescale of several seconds. Further investigations have attributed these two processes to the passive diffusion of electroneutral lactic acid molecules and active pumping of protons by H+-ATPase as a regulatory response to the flashing bacterial membrane potential, respectively.
Results and discussion
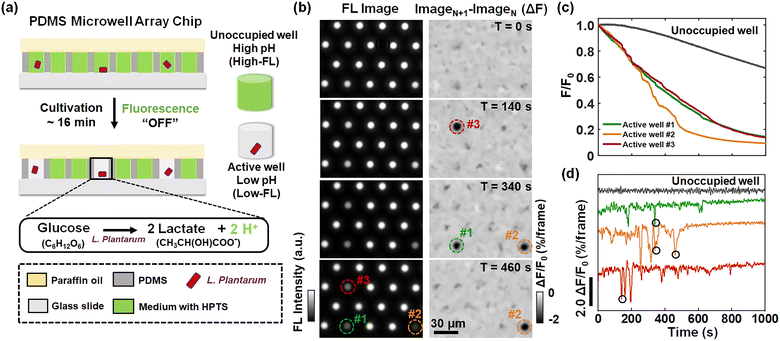
Burst-like proton release of single L. plantarum bacterium
L. plantarum is a kind of commonly used lactic acid bacteria in the fermentation food industry.21,22 The molecular mechanism of lactic acid production by L. plantarum has been extensively investigated and reported.23,24 In an ensemble experiment involving a flask containing billions of bacteria, the continuously declining extracellular pH (pHe) was always observed and used to evaluate the performance of bacterial strains. However, for individual bacteria, it remains largely unexplored whether the proton release process is continuous or it involves discrete steps.In order to address this question, we developed a single-bacterium-in-microwell strategy to continuously monitor the acid production rate of single L. plantarum (Fig. 1a) by using a microwell array and fluorescence (FL) pH indicators. Details on the fabrication of the microwell array25 and the subsequent encapsulation of single bacteria are given in Fig. S1.† In brief, a given number of L. plantarum cells were uniformly dispersed in unbuffered culture medium containing a fluorescent pH indicator, 8-hydroxypyrene-1,3,6-trisulphonic acid, trisodium salt (HPTS). The medium was adjusted to an initial pH of 6.5, under which conditions the bacteria maintained good metabolic activity and HPTS was sensitive to pH drop. The composition of the culture medium is shown in Table S1.† This cell suspension was then loaded into the picolitre wells on the microarray chip. By sealing with a layer of paraffin oil, individual bacteria were randomly and effectively encased within the microwells. In this process, bacteria encapsulated in microwells underwent glycolysis, consuming glucose and converting it into acidic metabolites such as lactic acid. The accumulation of released protons in the environment led to a rapid drop in the pHe of the microwell, as indicated by a decrease in the FL of HPTS. As a result, the FL intensity of the microwells containing bacteria (active wells, as indicated by circles in Fig. 1b) was gradually reduced to nearly zero. In contrast, the FL intensity of the empty microwells without bacteria (unoccupied wells) remained almost unchanged except for a slow and slight decrease due to photobleaching (Fig. 1b).
In a typical experiment, we used an inverted FL microscope equipped with a 20× air objective to collect the emitted light at 505–555 nm under 488 nm excitation. With such configurations, about six hundred microwells can be investigated simultaneously in the view-field, allowing for statistical analysis (Fig. S2†). The occupation ratio of bacteria in microwells followed the Poisson distribution law, where P(x) is the probability of finding a given number of bacteria, x, in a microwell. λ is the average number of bacteria per microwell:26
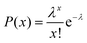 | (1) |
Representative FL images of a small region of interest at different moments are shown in the left panel of Fig. 1b. The circles in the images mark the locations of three active wells whose FL intensity gradually declined over time, indicating the metabolic activity of individual L. plantarum. Comprehensive illustrations on the time-lapsed images are provided in Movie S1.† The FL intensities of the active wells were plotted against the recording time to obtain the acid production kinetic curves of individual bacteria (Fig. 1c). Note that the photobleaching effect has been removed from the curve by subtracting the decreasing FL baselines of the surrounding unoccupied wells (Fig. S5†). As shown in Fig. 1c, while it was expected to see a declining trend of FL intensity (acidification), we surprisingly observed discrete and sudden FL decrease events (‘drops’). Such drops were better illustrated by showing the differential of the FL kinetic curves (Fig. 1d), where the positions circled in black indicated the four FL drop events at the corresponding locations and moments marked in the differential FL images (ImageN+1 − ImageN) shown in the right panel of Fig. 1b. A detailed video of the proton burst events is provided in Movie S2.† These sudden drops were consequences of transient and intensive proton efflux events from an individual encapsulated bacterium to the extracellular medium. We refer to this phenomenon as a burst-like release of protons. In contrast, the gradually decreasing FL intensity is attributed to the constant release of protons. While it was well understood that L. plantarum bacteria produced and secreted protons as a result of metabolism, single bacterium measurements revealed that it involved two processes with significantly distinct kinetics, namely, constant proton release and burst-like proton release.
It should be noted that there was efficient supply of oxygen within the microwell, i.e., aerobic culture, even though the bacteria were sealed with a thin layer of oil film (∼100 μm). It is well known that L. plantarum, as a kind of facultative anaerobic bacteria, efficiently produces lactic acids under both aerobic and anaerobic conditions. Standard incubation conditions of L. plantarum occurred in the presence of dissolved oxygen.27 In order to further examine the influence of oxygen supply on the FL kinetics, a strictly anaerobic environment in the microwell was achieved by sealing with a 2 cm-thick PDMS (instead of oil film with a typical thickness of 100 microns). Similar constant and burst-like proton release processes were observed (Fig. S6†), indicating that the observed proton bursts represented inherent fluctuations of L. plantarum under both aerobic and anaerobic conditions.
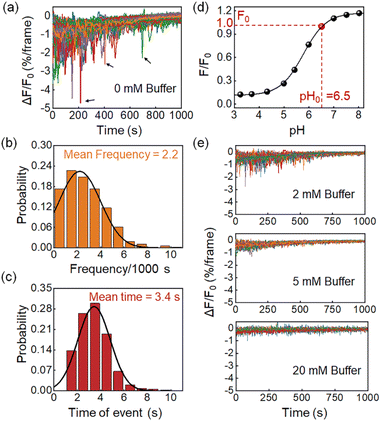
Features of proton burst events
Fig. 2a shows the differential FL kinetic profiles of 94 active microwells, which contain 206 proton burst events, corresponding to an average of 2.2 burst events per bacterium per 1000 s (Fig. 2b). Analysis further revealed an average duration of 3.4 s for the proton burst events (Fig. 2c). Data processing was performed using a home-made MATLAB code based on well-established methods to quantitatively analyze the burst events (Fig. S7†).8,28 Another feature in Fig. 2a is that the proton burst events were mainly distributed in the initial stage of the recording. It was due to the S-type dose response of HPTS to pH (Fig. 2d). As the reaction time progressed and the pHe gradually approached the acidic range, the HPTS probe became less sensitive to the release of proton bursts. In addition, the release of lactate also caused an increase in the buffering capacity of the medium, which further reduced the sensitivity of the probe. To verify this point, we increased the concentration of phosphate buffer salt in the medium from 0 (Fig. 2a) to 2, 5, and 20 mM (Fig. 2e), and found that both the frequency and depth of burst events were significantly reduced as the buffer concentration increased (Fig. 2e). The results confirmed that the FL drops indeed revealed the pH drops caused by the proton efflux, which were significantly suppressed in the presence of the buffer solution.Two proton release processes existed in single L. plantarum
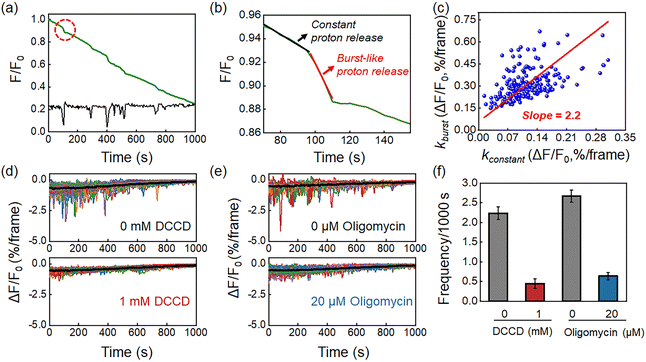
In contrast to measurements from ensemble specimens which showed a continuous decrease in the pHe, the kinetics of individual bacteria consisted of two distinct proton release processes. Fig. 3a shows a representative FL curve, accompanied by several different proton burst events (black curve). As shown in Fig. 3b, zooming into one of these events reveals two distinct processes, which are a constant proton release with a smaller slope (kconstant, black curve) and a burst-like proton release with a larger slope (kburst, red curve). Analysis of 200 events yielded a positive correlation between kconstant and kburst, with a mean slope of 2.2 (Fig. 3c and S8†). This result indicated that the burst proton release state had approximately 2 times more protons compared to the constant proton release state. Occasionally a ratio as large as 10 was observed, suggesting intensive proton efflux during burst events (Fig. S9†). The proton burst events were subsequently attributed to the transient action of H+-ATPase because of the following reasons. Firstly, H+-ATPase is a major proton pump in L. plantarum that extrudes charged H+ by hydrolyzing ATP to regulate intracellular pH homeostasis.29,30 Gene sequencing of the as-studied L. plantarum strain also verified the existence of the H+-ATPase gene (Table S2†). The role of H+-ATPase in the proton burst phenomenon was validated by investigating the influence of N,N′-dicyclohexylcarbodiimide (DCCD), a specific and popular inhibitor of bacterial H+-ATPase.31 As shown in Fig. 4d and f, 1 mM DCCD was effective for suppressing 80% of the proton burst events, which was consistent with literature reports on the inhibition efficiency of DCCD on H+-ATPase at this concentration.32 Another inhibitor, oligomycin,33 exhibited similar inhibition on the burst events (Fig. 3e and f). It was worth noting that both inhibitors (DCCD and oligomycin) had a negligible effect on the constant proton release process (black curve in the profiles, Fig. 3d, e and S10†). These results were consistent with the passive proton efflux (diffusion) pathway, in which intracellular protons passed through the membrane in the form of neutral molecules of lactic acid.34,35 Secondly, L. plantarum inherently lacks an integrated respiratory system, which precludes proton efflux mediated by the electron transport chain complex.8,27 Thirdly, some other electrogenic proton transport proteins, such as Na+/H+ antiporters and K+/H+ antiporters, are responsible for H+ uptake and metal cation efflux, which should cause an increase in medium pH and are therefore opposite to the pH drop in the observed phenomenon.36,37To demonstrate the generality of this proton burst phenomenon, two other Lactobacillus plantarum strains, including Lactobacillus plantarum strain ACCC11095 and Lactobacillus plantarum subsp. plantarum, were further investigated. Similar proton burst phenomena were also observed in these two strains (Fig. S11†).
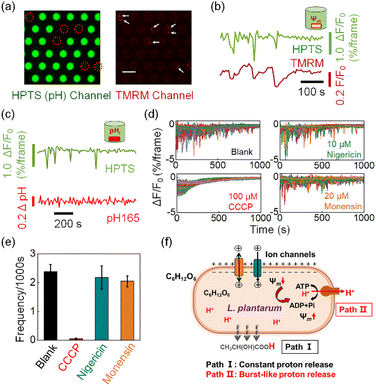
The involvement of burst-like proton release in the recovery of voltage depolarization
We subsequently demonstrate that the transient action of H+-ATPase is a regulatory response to the stochastic depolarization of membrane potential (Ψm). When simultaneously measuring the pHe dynamics in the microwell and the membrane potential of the encapsulated bacterium, excellent synchronization was always observed between the pHe drops (burst events) and the membrane potential depolarization events. In order to achieve high-resolution two-channel FL imaging, we fabricated a new microwell array chip by etching microwells on a 170 μm-thick glass coverslip. The thin coverslip allowed us to use a high numerical aperture (N.A. = 1.49) 60× oil immersion objective for single bacterial imaging. We introduced tetramethylrhodamine methyl ester (TMRM), a membrane permeable fluorescent voltage reporter38 to label the surface-immobilized bacteria, and recorded the membrane potential dynamics of individual bacteria in the microwell. The microscope was programmed to alternatively measure the FL emission from HPTS (pHe channel, 488 nm excitation) and TMRM (Ψm channel, 561 nm excitation). As shown in Fig. 4a, decreasing FL emission in the pHe channel (green) was recorded in 6 microwells, indicating the effective acidification of the medium caused by the encapsulated bacteria. The existence of 8 bacteria within these 6 microwells was successfully validated in the red channel, where a single bacterium is illustrated as red dots because of the staining of TMRM. These results not only proved that the FL decrease was indeed due to the effect of the encapsulated single bacterium, but also demonstrated the technical applicability of simultaneous two-channel FL imaging. Time-lapsed imaging allowed for resolving the trajectories of pHe and Ψm in the same microwell, as shown in Fig. 4b. It was found that the proton burst events (green curve) were in good pace with the depolarization-and-recovery events of the bacterial membrane potential (red curve). More examples are provided in Fig. S12† to show that these two processes were always synchronized with each other, suggesting a mechanistic link between the proton efflux and membrane potential. It was well-documented that the H+-ATPase mediated proton efflux should make the intracellular potential more negative (a polarization process).39,40 Therefore, it was believed that the bacterium underwent stochastic and spontaneous depolarization because of the stochastic gating of ion channels or secondary ion transporters.7,8,41 As a regulatory response, H+-ATPase started to transiently act to pump out protons until the recovery of membrane potential. It is noteworthy that although TMRM and HPTS are both small molecules, their response mechanisms differ significantly. Impermeable HPTS senses the pH change of extracellular medium within 1 second (Fig. S13†). In contrast, TMRM molecules react to membrane potential changes by redistribution across the bacterial membrane according to the Nernst equation.42 This transmembrane process typically lasts for dozens of seconds or several minutes.6,43 Therefore, the signal of TMRM usually requires more time to reach equilibrium after the membrane potential recovery indicated by the finish of proton bursts.By using a similar apparatus, we also introduced a ratiometric pH-sensitive fluorophore RatioWorks™ PH165 NHS to investigate the relationship between intracellular pH (pHi) and bacterial proton burst events.8 As shown in Fig. 4c, while the pHe trajectory showed frequent and distinct fluctuation characteristics, the simultaneously recorded pHi remained unchanged. The translocation of a small number of protons from bacterial cytoplasm to the extracellular medium caused drastic change in pHe but negligible change in pHi because of the distinct buffering capacity. While bacterial cytoplasm exhibited strong buffering capacity due to the presence of the high content of cytoplasmic proteins, the medium was free of buffer solution so as to amplify the effect of proton release, underscoring the strength of pHe recording for studying the proton transport.
We further examined the effect of three kinds of small molecules that can regulate the membrane potential and pH gradient in different ways. Carbonyl cyanide m-chlorophenylhydrazine (CCCP) is a commonly used proton motive force (PMF) dissipator capable of eliminating both transmembrane voltage (Ψm) and proton gradients (ΔpH).39 The presence of 100 μM CCCP in the medium completely eliminated the burst proton release event (Fig. 4d and e). Two other proton carriers, nigericin and monensin, which are antibiotics that mediate electroneutral H+/K+ and H+/Na+ antiport, can deliver protons without perturbing the membrane potential.44Fig. 4e shows that the addition of 10 μM nigericin (blue column) or 20 μM monensin (yellow column) had negligible effects on the proton burst frequency. These results further confirmed that the proton burst process was related to the bacterial membrane potential rather than the intracellular pHi.
According to these results, we propose a hypothesis to understand the proton release dynamics of single L. plantarum (Fig. 4f). There are two pathways involved in the proton efflux of L. plantarum during acid production. The constant proton release pathway is derived from the continuous and passive transmembrane diffusion of the metabolite lactate as neutral molecules (Path I). It does not involve in membrane potential change and thus contributes to the downward portion of the baseline in the fluorescence trajectory. For the single bacterium encapsulated in the microwell, its membrane potential undergoes stochastic and transient depolarizations because of the extremely small surface area and thus insufficient membrane capacitance to maintain membrane potential against charge fluctuations. As a regulatory mechanism, the depolarized membrane potential activates the function of H+-ATPase to pump out a small number of protons to restore the membrane potential (Path II). The proton burst events resulted in the transient pHe drops in the absence of a buffer system in the extracellular medium. The proton burst event was random and independent among different individuals; this phenomenon was only accessible at the single bacterium level, underscoring the value of the present methodology for studying single bacteria dynamics.45–48
Although the proton bursts and membrane potential dynamics are phenomenologically and mechanistically correlated with each other, the present pHe recording exhibited several important strengths for studying membrane potential dynamics over conventional voltage-sensitive dyes such as TMRM. First, while conventional organic and genetic staining required molecular modification to the bacterium and thus inevitably disturbed its physiological states, the present approach minimized such disturbance because the pHe indicator dyes were distributed in the extracellular medium. It was not available for uptake by bacteria due to its high-water solubility. Second, the uptake efficiency of organic dyes and the expression efficiency of genetic dyes often varied significantly from individual to individual, making the statistical and quantitative analysis challenging.6,49,50 The proposed pHe recording exhibited much better consistency among different bacteria because the FL signal came from a homogeneous solution. Third, while the FL probes for membrane potential are rather limited, there were plenty of pHe indicators available with excellent features on response time, spectral selection, photo- and chemical stability. For example, as shown in Fig. 4b, the width of FL drops in the pHe channel (HPTS) was systematically narrower than that in the Ψm channel (TMRM). These strengths made pHe recording in a microwell a promising strategy for reporting the membrane potential dynamics, with implications for bacterial electrophysiology.
Conclusions
In summary, this study presents a new method based on microwell array technology to detect the proton efflux kinetics of individual bacteria in a non-invasive, real-time and high throughput manner. By using L. plantarum as a model strain, it was found that the proton release from single lactic acid bacteria was not constant. Instead, stochastic and intermittent proton burst events were observed with a typical lasting time of several seconds. The generality of such a phenomenon was verified with two other kinds of strains. Systematic investigations demonstrated that the H+-ATPase mediated proton efflux was responsible for the proton burst event. It acted as a regulatory mechanism to restore the membrane potential homeostasis. These findings not only demonstrated the membrane protein-associated proton burst phenomenon for the first time, but also provided a promising tool for studying the bacterial electrophysiology at the single bacteria level.Data availability
The authors declare that all the data are available within the article and its ESI† or from the corresponding author upon reasonable request.Author contributions
J. G. and W. W. designed the project and wrote the paper. J. G. carried out most of the experiments and analyzed the data. K. Z. and H. R. L. assisted in the fabrication of microarray chip devices. Y. H. L. and K. R. Y. analyzed and discussed the results and helped the treatments on bacteria. W. W. conceived and supervised the project.Conflicts of interest
There are no conflicts to declare.Acknowledgements
We thank the National Natural Science Foundation of China (Grants 21925403, 21874070), and the Excellent Research Program of Nanjing University (Grant ZYJH004) for financial support.References
- J. M. Benarroch and M. Asally, The Microbiologist's Guide to Membrane Potential Dynamics, Trends Microbiol., 2020, 28, 304–314 CrossRef CAS PubMed.
- N. D. Cook, G. B. Carvalho and A. Damasio, From membrane excitability to metazoan psychology, Trends Neurosci., 2014, 37, 698–705 CrossRef CAS PubMed.
- H. Strahl and L. W. Hamoen, Membrane potential is important for bacterial cell division, Proc. Natl. Acad. Sci. U. S. A., 2010, 107, 12281–12286 CrossRef CAS PubMed.
- D. D. Lee, A. Prindle, J. Liu and G. M. Suel, SnapShot: Electrochemical Communication in Biofilms, Cell, 2017, 170, 214 CrossRef CAS PubMed.
- H. Z. Zhang, Y. P. Pan, L. Y. Hu, M. A. Hudson, K. S. Hofstetter, Z. C. Xu, M. Q. Rong, Z. Wang, B. V. V. Prasad, S. W. Lockless, W. Chiu and M. Zhou, TrkA undergoes a tetramer-to-dimer conversion to open TrkH which enables changes in membrane potential, Nat. Commun., 2020, 11, 547 CrossRef CAS PubMed.
- J. M. Kralj, D. R. Hochbaum, A. D. Douglass and A. E. Cohen, Electrical spiking in Escherichia coli probed with a fluorescent voltage-indicating protein, Science, 2011, 333, 345–348 CrossRef CAS PubMed.
- G. N. Bruni, R. A. Weekley, B. J. T. Dodd and J. M. Kralj, Voltage-gated calcium flux mediates Escherichia coli mechanosensation, Proc. Natl. Acad. Sci. U. S. A., 2017, 114, 9445–9450 CrossRef CAS PubMed.
- Y. Li, H. Li, J. Gao, B. Niu, H. Wang and W. Wang, Visualizing the Intermittent Gating of Na(+)/H(+) Antiporters in Single Native Bioluminescent Bacteria, Angew. Chem., Int. Ed., 2023, 62, e202215800 CrossRef CAS PubMed.
- A. Prindle, J. Liu, M. Asally, S. Ly, J. Garcia-Ojalvo and G. M. Suel, Ion channels enable electrical communication in bacterial communities, Nature, 2015, 527, 59–63 CrossRef CAS PubMed.
- J. Humphries, L. Y. Xiong, J. T. Liu, A. Prindle, F. Yuan, H. A. Arjes, L. Tsimring and G. M. Suel, Species-Independent Attraction to Biofilms through Electrical Signaling, Cell, 2017, 168, 200–209 CrossRef CAS PubMed.
- K. Kikuchi, L. Galera-Laporta, C. Weatherwax, J. Y. Lam, E. C. Moon, E. A. Theodorakis, J. Garcia-Ojalvo and G. M. Suel, Electrochemical potential enables dormant spores to integrate environmental signals, Science, 2022, 378, 43–49 CrossRef CAS PubMed.
- T. Sirec, J. M. Benarroch, P. Buffard, J. Garcia-Ojalvo and M. Asally, Electrical Polarization Enables Integrative Quality Control during Bacterial Differentiation into Spores, iScience, 2019, 16, 378–389 CrossRef CAS PubMed.
- M. Hennes, N. Bender, T. Cronenberg, A. Welker and B. Maier, Collective polarization dynamics in bacterial colonies signify the occurrence of distinct subpopulations, PLoS Biol., 2023, 21, e3001960 CrossRef CAS PubMed.
- L. Galera-Laporta, C. J. Comerci, J. Garcia-Ojalvo and G. M. Suel, IonoBiology: The functional dynamics of the intracellular metallome, with lessons from bacteria, Cell Syst., 2021, 12, 497–508 CrossRef CAS PubMed.
- B. Martinac, Y. Saimi and C. Kung, Ion channels in microbes, Physiol. Rev., 2008, 88, 1449–1490 CrossRef CAS PubMed.
- P. V. Krasteva, Bacterial electrophysiology brought to light, Nat. Methods, 2011, 8, 714 CrossRef PubMed.
- J. M. Jones and J. W. Larkin, Toward Bacterial Bioelectric Signal Transduction, Bioelectricity, 2021, 3, 116–119 CrossRef PubMed.
- D. Wu, W. Qi, W. Nie, Z. Lu, Y. Ye, J. Li, T. Sun, Y. Zhu, H. Cheng and X. Wang, BacFlash signals acid-resistance gene expression in bacteria, Cell Res., 2021, 31, 703–712 CrossRef CAS PubMed.
- F. J. Hol and C. Dekker, Zooming in to see the bigger picture: microfluidic and nanofabrication tools to study bacteria, Science, 2014, 346, 1251821 CrossRef PubMed.
- K. X. Zhang, S. S. Qin, S. X. Wu, Y. Liang and J. H. Li, Microfluidic systems for rapid antibiotic susceptibility tests (ASTs) at the single-cell level, Chem. Sci., 2020, 11, 6352–6361 RSC.
- F. De Filippis, E. Pasolli and D. Ercolini, The food-gut axis: lactic acid bacteria and their link to food, the gut microbiome and human health, FEMS Microbiol. Rev., 2020, 44, 454–489 CrossRef PubMed.
- S. Wuyts, W. Van Beeck, C. N. Allonsius, M. F. L. van den Broek and S. Lebeer, Applications of plant-based fermented foods and their microbes, Curr. Opin. Biotechnol., 2020, 61, 45–52 CrossRef CAS PubMed.
- C. P. Tseng and T. J. Montville, Enzymatic Regulation of Glucose Catabolism by Lactobacillus-Plantarum in Response to Ph Shifts in a Chemostat, Appl. Microbiol. Biotechnol., 1992, 36, 777–781 CrossRef CAS.
- P. D. Cotter and C. Hill, Surviving the acid test: Responses of gram-positive bacteria to low pH, Microbiol. Mol. Biol. Rev., 2003, 67, 429–453 CrossRef CAS PubMed.
- L. Du, H. Liu and J. Zhou, Picoliter droplet array based on bioinspired microholes for in situ single-cell analysis, Microsyst. Nanoeng., 2020, 6, 33 CrossRef CAS PubMed.
- H. H. Gorris and D. R. Walt, Analytical Chemistry on the Femtoliter Scale, Angew. Chem., Int. Ed., 2010, 49, 3880–3895 CrossRef CAS PubMed.
- A. Guidone, R. G. Ianniello, A. Ricciardi, T. Zotta and E. Parente, Aerobic metabolism and oxidative stress tolerance in the Lactobacillus plantarum group, World J. Microbiol. Biotechnol., 2013, 29, 1713–1722 CrossRef CAS PubMed.
- Y. M. Fang, Z. M. Li, Y. Y. Jiang, X. Wang, H. Y. Chen, N. J. Tao and W. Wang, Intermittent photocatalytic activity of single CdS nanoparticles, Proc. Natl. Acad. Sci. U. S. A., 2017, 114, 10566–10571 CrossRef CAS PubMed.
- H. Kobayashi, A Proton-Translocating Atpase Regulates Ph of the Bacterial Cytoplasm, J. Biol. Chem., 1985, 260, 72–76 CrossRef CAS PubMed.
- B. J. Koebmann, D. Nilsson, O. P. Kuipers and P. R. Jensen, The membrane-bound H+-ATPase complex is essential for growth of Lactococcus lactis, J. Bacteriol., 2000, 182, 4738–4743 CrossRef CAS PubMed.
- L. Karapetyan, G. Mikoyan, A. Vassilian, A. Valle, J. Bolivar, A. Trchounian and K. Trchounian, Escherichia coli Dcu C(4)-dicarboxylate transporters dependent proton and potassium fluxes and F(O)F(1)-ATPase activity during glucose fermentation at pH 7.5, Bioelectrochemistry, 2021, 141, 107867 CrossRef CAS PubMed.
- J. C. McEntire, G. M. Carman and T. J. Montville, Increased ATPase activity is responsible for acid sensitivity of nisin-resistant Listeria monocytogenes ATCC 700302, Appl. Environ. Microbiol., 2004, 70, 2717–2721 CrossRef CAS PubMed.
- J. Symersky, D. Osowski, D. E. Walters and D. M. Mueller, Oligomycin frames a common drug-binding site in the ATP synthase, Proc. Natl. Acad. Sci. U. S. A., 2012, 109, 13961–13965 CrossRef CAS PubMed.
- D. D. Axe and J. E. Bailey, Transport of Lactate and Acetate through the Energized Cytoplasmic Membrane of Escherichia-Coli, Biotechnol. Bioeng., 1995, 47, 8–19 CrossRef CAS PubMed.
- A. L. Carvalho, D. L. Turner, L. L. Fonseca, A. Solopova, T. Catarino, O. P. Kuipers, E. O. Voit, A. R. Neves and H. Santos, Metabolic and Transcriptional Analysis of Acid Stress in Lactococcus lactis, with a Focus on the Kinetics of Lactic Acid Pools, PLoS One, 2013, 8, e68470 CrossRef CAS PubMed.
- T. A. Krulwich, G. Sachs and E. Padan, Molecular aspects of bacterial pH sensing and homeostasis, Nat. Rev. Microbiol., 2011, 9, 330–343 CrossRef CAS PubMed.
- J. Stautz, Y. Hellmich, M. F. Fuss, J. M. Silberberg, J. R. Devlin, R. B. Stockbridge and I. Haenelt, Molecular Mechanisms for Bacterial Potassium Homeostasis, J. Mol. Biol., 2021, 433, 166968 CrossRef CAS PubMed.
- S. Creed and M. McKenzie, Measurement of Mitochondrial Membrane Potential with the Fluorescent Dye Tetramethylrhodamine Methyl Ester (TMRM), Methods Mol. Biol., 2019, 1928, 69–76 CrossRef CAS PubMed.
- G. N. Bruni and J. M. Kralj, Membrane voltage dysregulation driven by metabolic dysfunction underlies bactericidal activity of aminoglycosides, eLife, 2020, 9, e58706 CrossRef CAS PubMed.
- J. Falhof, J. T. Pedersen, A. T. Fuglsang and M. Palmgren, Plasma Membrane H(+)-ATPase Regulation in the Center of Plant Physiology, Mol. Plant, 2016, 9, 323–337 CrossRef CAS PubMed.
- W. A. Catterall, Ion channel voltage sensors: structure, function, and pathophysiology, Neuron, 2010, 67, 915–928 CrossRef CAS PubMed.
- B. Ehrenberg, V. Montana, M. D. Wei, J. P. Wuskell and L. M. Loew, Membrane-Potential Can Be Determined in Individual Cells from the Nernstian Distribution of Cationic Dyes, Biophys. J., 1988, 53, 785–794 CrossRef CAS PubMed.
- D. L. Farkas, M. D. Wei, P. Febbroriello, J. H. Carson and L. M. Loew, Simultaneous Imaging of Cell and Mitochondrial-Membrane Potentials, Biophys. J., 1989, 56, 1053–1069 CrossRef CAS PubMed.
- X. Wang, X. Zhang, Z. Huang, D. Wu, B. Liu, R. Zhang, R. Yin, T. Hou, C. Jian, J. Xu, Y. Zhao, Y. Wang, F. Gao and H. Cheng, Protons Trigger Mitochondrial Flashes, Biophys. J., 2016, 111, 386–394 CrossRef CAS PubMed.
- L. A. Genova, M. F. Roberts, Y. C. Wong, C. E. Harper, A. G. Santiago, B. Fu, A. Srivastava, W. Jung, L. M. Wang, L. Krzeminski, X. W. Mao, X. H. Sun, C. Y. Hui, P. Chen and C. J. Hernandez, Mechanical stress compromises multicomponent efflux complexes in bacteria, Proc. Natl. Acad. Sci. U. S. A., 2019, 116, 25462–25467 CrossRef CAS PubMed.
- X. D. Cheng, K. C. Chen, B. Dong, M. Yang, S. L. Filbrun, Y. Myoung, T. X. Huang, Y. Gu, G. F. Wang and N. Fang, Dynamin-dependent vesicle twist at the final stage of clathrin-mediated endocytosis, Nat. Cell Biol., 2021, 23, 859–869 CrossRef CAS PubMed.
- Y. Q. Sang, X. D. Wen and Y. He, Single-cell/nanoparticle trajectories reveal two-tier Levy-like interactions across bacterial swarms, View, 2022, 3, 20220047 CrossRef CAS.
- Y. H. Li, S. Wang, X. Y. He, S. J. Li, T. H. Zheng, Y. P. Chen, H. Cui and W. Wang, Imaging the oxygen wave with a single bioluminescent bacterium, Chem. Sci., 2021, 12, 12400–12406 RSC.
- A. Miyawaki, Fluorescence imaging of physiological activity in complex systems using GFP-based probes, Curr. Opin. Neurobiol., 2003, 13, 591–596 CrossRef CAS PubMed.
- X. Deng, X. Q. Yao, K. Berglund, B. Dong, D. Ouedraogo, M. A. Ghane, Y. Zhuo, C. McBean, Z. Z. Wei, S. Gozem, S. P. Yu, L. Wei, N. Fang, A. M. Mabb, G. Gadda, D. Hamelberg and J. J. Yang, Tuning Protein Dynamics to Sense Rapid Endoplasmic-Reticulum Calcium Dynamics, Angew. Chem., Int. Ed., 2021, 60, 23289–23298 CrossRef CAS PubMed.
Footnote |
| † Electronic supplementary information (ESI) available: Materials and methods, Fig. S1–S13, Tables S1 and S2, description of supplementary movies. See DOI: https://doi.org/10.1039/d3sc06238d |
| This journal is © The Royal Society of Chemistry 2024 |