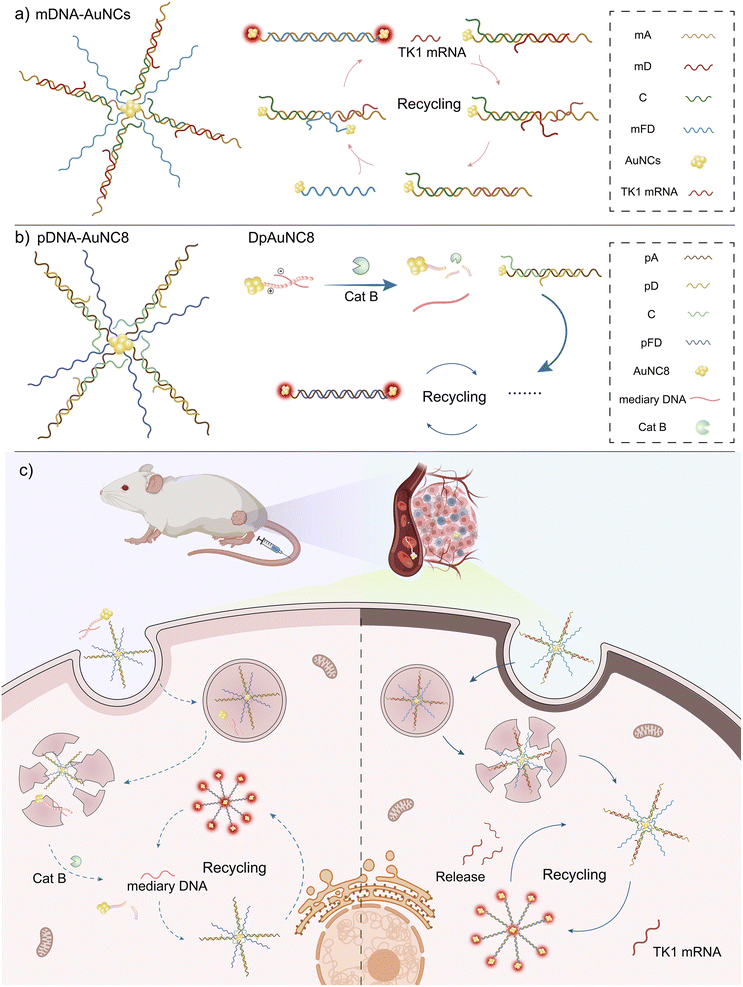
Open Access Article
Open Access ArticleProlonged near-infrared fluorescence imaging of microRNAs and proteases in vivo by aggregation-enhanced emission from DNA-AuNC nanomachines†
Ting
Wang
a,
Kai
Jiang
a,
Yifan
Wang
a,
Limei
Xu
a,
Yingqi
Liu
a,
Shiling
Zhang
a,
Weiwei
Xiong
 a,
Yemei
Wang
a,
Fenfen
Zheng
*a and
Jun-Jie
Zhu
a,
Yemei
Wang
a,
Fenfen
Zheng
*a and
Jun-Jie
Zhu
 *b
*b
aSchool of Environmental & Chemical Engineering, Jiangsu University of Science and Technology, Changhui Rd. 666, Zhenjiang, Jiangsu 212003, China. E-mail: zhengfenfen@just.edu.cn
bState Key Laboratory of Analytical for Life Science, School of Chemistry and Chemical Engineering, Nanjing University, Xianlin Ave 163, Nanjing, Jiangsu 210023, China. E-mail: jjzhu@nju.edu.cn
First published on 21st December 2023
Abstract
Developing a comprehensive strategy for imaging various biomarkers (i.e., microRNAs and proteases) in vivo is an exceptionally formidable task. Herein, we have designed a deoxyribonucleic acid-gold nanocluster (DNA-AuNC) nanomachine for detecting tumor-related TK1 mRNA and cathepsin B in living cells and in vivo. The DNA-AuNC nanomachine is constructed using AuNCs and DNA modules that incorporate a three component DNA hybrid (TD) and a single-stranded fuel DNA (FD). Upon being internalized into tumor cells, the TK1 mRNA initiates the DNA-AuNC nanomachine through DNA strand displacement cascades, leading to the amplified self-assembly and the aggregation-enhanced emission of AuNCs for in situ imaging. Furthermore, with the aid of a protease nanomediator consisting of a mediator DNA/peptide complex and AuNCs (DpAuNCs), the DNA-AuNC nanomachine can be triggered by the protease-activated disassembly of the DNA/peptide complex on the nanomediator, resulting in the aggregation of AuNCs for in vivo protease amplified detection. It is worth noting that our study demonstrates the impressive tumor permeability and accumulation capabilities of the DNA-AuNC nanomachines via in situ amplified self-assembly, thereby facilitating prolonged imaging of TK1 mRNA and cathepsin B both in vitro and in vivo. This strategy presents a versatile and biomarker-specific paradigm for disease diagnosis.
Introduction
The real-time monitoring of dynamic changes in tumor-related biomarkers (i.e., microRNAs or proteases) during tumor progression is of great importance for understanding tumorigenesis mechanisms and advancing clinical theranostics.1–6 In recent years, there has been significant progress in the development of fluorescently labeled DNA/peptide functionalized nanomaterials for imaging microRNAs or proteases within living cells and in vivo, which offers promising means for studying molecular-level cellular activities.7–12 While nanomaterials have shown their utility in delivering DNA or peptide probes to tumor sites through the enhanced permeability and retention (EPR) effect, their imaging effectiveness can sometimes fall short due to the limited penetration or retention. The unique structure and environment of tumor tissues create the paradox that large nanoparticles tend to accumulate around tumor blood vessels rather than penetrating deep into tumor parenchyma, while smaller nanoparticles can penetrate effectively but are quickly cleared from the target site, resulting in poor tumor retention and compromising long-term molecular imaging.13–15 Moreover, conventional fluorophores or quantum dots used to label DNA or peptides often suffer from poor tumor retention and exhibit aggregation-caused quenching in the concentrated intracellular environment.16,17 This may lead to a temporary weakening of emission at the tumor site. Therefore, there is an urgent need for a new strategy to design activatable imaging probes that can combine high tumor permeability with prolonged retention, addressing the challenges of imaging tumor-related biomarkers effectively.In order to effectively target tumor sites, achieving optimal probe concentrations and superior imaging outcomes, it is essential to employ size-tunable nanoprobes capable of altering their sizes at the right moment, ensuring improved penetration and retention within the tumor microenvironment.18,19 Recently, metal nanoclusters (MNCs) have garnered significant attention due to their self-assembly and aggregation-induced enhanced emission (AIE) effect originated from the restriction of the rotation and vibration motions of thiolate ligands.20–25 Compared to organic AIE fluorescent molecules, MNCs offer several advantages, such as ease of synthesis, versatile functionalization, tunable luminescence, exceptional photostability, and outstanding biocompatibility.26,27 The AIE phenomenon in MNCs, induced by metal ions, low pH, peptides, proteins, or polymers, has been harnessed for the in vitro detection of various biomarkers.28–33 Furthermore, by virtue of exceptional sequence specificity, favorable physiochemical properties and programmable supramolecular self-assembly of DNAs, DNA-functionalized gold nanoclusters (AuNCs) have been successfully utilized for probing microRNA or telomerase activity in living cells and in vivo through an in situ self-assembly strategy.34,35 This strategy leverages the enhanced tumor permeability of small sized nanoparticles and the prolonged tumor retention of large sized ones, showing promise for imaging biomolecules at tumor sites with improved accumulation and amplified imaging signals. Nevertheless, there is a limited body of research exploring the potential of this strategy for developing versatile probes with prolonged imaging time and specific amplified responses to microRNAs and proteases in vivo.
Herein, by taking advantage of the toehold-mediated stand displacement (TMSD) reaction and the AIE property of AuNCs triggered by TMSD-driven assembly, we have designed a versatile and activatable DNA-AuNC nanomachine for imaging the microRNAs and proteases in vivo (Scheme 1). To construct the DNA-AuNC nanomachine responsive to target TK1 mRNA (mDNA-AuNCs), we immobilized a three-component DNA hybrid (mTD, consisting of mA, mD and C) and a single-stranded fuel (mFD) on the AuNCs (λem = 605 nm (named mDNA-AuNC6) or 826 nm (named mDNA-AuNC8)) via a Au–S covalent bond. In the presence of TK1 mRNA, the nanomachine was triggered by the cascaded TMSD reaction. In brief, the TK1 mRNA initially binds to the single-stranded toehold domain in mTD, setting off a strand-displacement reaction that forms an intermediate complex and releases “mD”. Subsequently, mFD promptly binds to the newly exposed domain on mTD to trigger another strand-displacement reaction, resulting in the aggregation of AuNCs and a noticeable increase in fluorescence. Concurrently, the TK1 mRNA is regenerated to initiate a new cycle for signal amplification. Furthermore, by incorporating protease-responsive DpAuNC8 nanomediators consisting of mediator DNA and protease substrate peptide modified AuNC8 (λem = 826 nm), the pDNA-AuNC8 nanomachine composed of pTD and pFD (pTD and pFD have similar base composition to mTD and mFD that respond to TK1 mRNA, but they respond to mediator DNA that only triggers the TMSD reaction of pTD and pFD) can be employed to detect protease activity in vivo through protease activated mediator DNA-triggered cascade TMSD reactions. Consequently, this versatile DNA-AuNCs nanoplatform demonstrates tremendous potential for long-term dynamic imaging of biomarkers in vivo.
Results and discussion
Characterization of the DNA-AuNC nanomachine
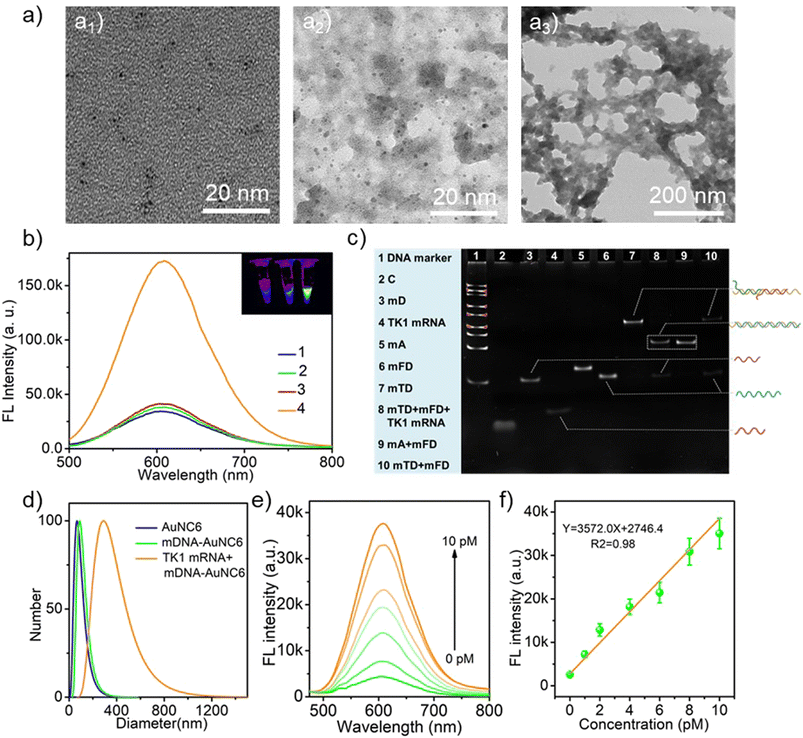
The AuNCs (λem = 605 nm, AuNC6) were first synthesized by using GSH and HAuCl4 as precursors. As shown in Fig. 1a, TEM images confirmed the successful synthesis of well-dispersed spherical AuNC6 with an average diameter of around 2 nm. Remarkably, the surface modification of AuNC6 with DNA molecules did not alter their morphology and dispersibility (Fig. 1a2). Upon incubation with target TK1 mRNA, a noticeable aggregation of mDNA-AuNC6 was observed (Fig. 1a3). As depicted in Fig. S1,† AuNC6 exhibited a typical UV-vis absorption curve, featuring wide absorption from 200 to 500 nm, while mDNA-AuNC6 displayed a characteristic absorption peak at 260 nm, signifying the successful surface modification of DNA on the AuNCs. After incubation with TK1 mRNA, a subtle red-shifted absorption peak was observed, indicating the TMSD reaction induced aggregation of AuNC6. Zeta potential measurements and dynamic light scattering (DLS) further confirmed the aggregation phenomena (Fig. S2† and 1d). Zeta potential measurements demonstrated that the negative surface charge of mDNA-AuNC6 was greater than that of bare AuNC6, and this negative surface charge further increased after TK1 mRNA-induced aggregation (Fig. S2†). DLS provided the dispersibility and size distribution of these nanoparticles. In Fig. 1d, only a single peak was observed for mDNA-AuNC6 that is similar to bare AuNC6, indicating excellent dispersibility in water and expected single-dispersion behavior. After incubation with TK1 mRNA, the size of the nanoparticles increased dramatically, indicating the aggregation of DNA-AuNCs triggered by TK1 mRNA. To further validate the feasibility of the DNA nanomachine reaction, polyacrylamide gel electrophoresis (PAGE) was employed (Fig. 1c). When mTD and mFD were mixed and incubated at 37 °C for 1 h, clear bands were observed (lane 10). Conversely, when the mixture was incubated with TK1 mRNA, new mA/mFD hybrid bands were generated (lane 8), indicating the release of TK1 mRNA, C, and mD. It is worth noting that the short single-stranded C and TK1 mRNA were challenging to stain with bromine ethidium bromide in dilute solution. This underscores the feasibility of the proposed mDNA-AuNC6 nanomachine as a DNA amplifier. All these results establish the viability of the constructed mDNA-AuNC6 nanomachine with specific response to target TK1 mRNA.Next, the performance of mDNA-AuNC6 in detecting TK1 mRNA was studied by using fluorescence spectroscopy (Fig. 1b). Compared with the feeble fluorescence of bare AuNC6, non-target mDNA-AuNC6 (mDNA-AuNC6-R, including the random DNAs has no response to TK1 mRNA) or mDNA-AuNC6 without TK1 mRNA, the fluorescence intensity of mDNA-AuNC6 was significantly increased after incubation with TK1-mRNA, indicating the aggregation-induced enhanced emission of mDNA-AuNC6 nanomachines. To optimize the experimental conditions, we carefully examined the concentrations of mTD and mFD modified on the surface of AuNC6 and the incubation time. As illustrated in Fig. S3,† the fluorescence intensity increased with an increasing mTD/mFD concentration and reached a threefold increase at a concentration of 100 nM. At a fixed mTD/mFD concentration of 100 nM, the fluorescence intensity increased with the extended incubation time, up to 4 h (Fig. S4†). Under these optimal experimental conditions, the TK1 mRNA was determined (Fig. 1e). The fluorescence emissions from mDNA-AuNC6 at 605 nm exhibited a gradual increase with the increasing TK1 concentration from 0 to 10 pM with a detection limit of 0.3 pM (Fig. 1f). The results demonstrate that the aggregation-induced enhanced emission of mDNA-AuNC6 nanomachines can be used to detect TK1 mRNA with high specificity and sensitivity.
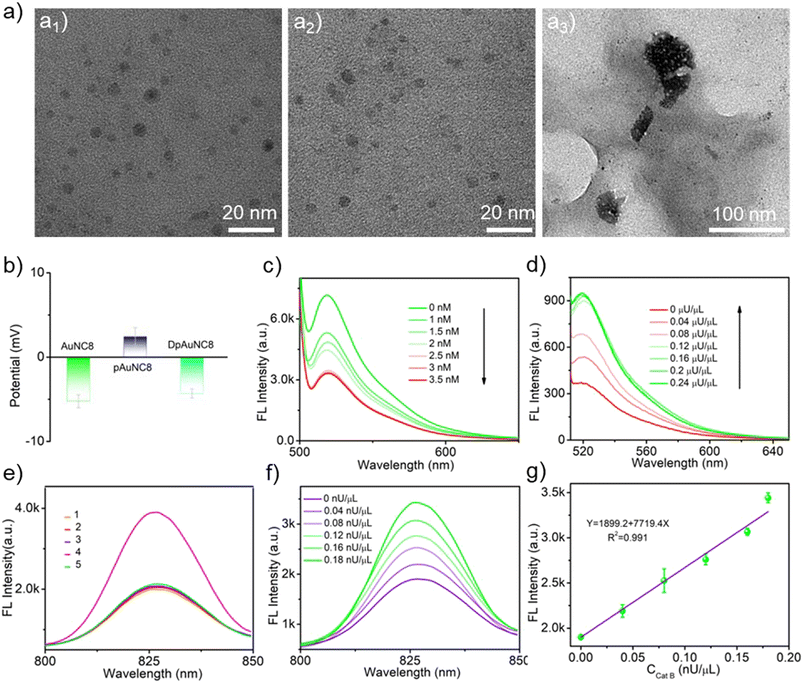
Characterization of the Cat B-responsive DpAuNC8 nanomediator and pDNA-AuNC8 nanomachine
To engineer mediator DNA molecules capable of triggering downstream cascade TMSD reactions of DNA-AuNC nanomachines, we devised a DpAuNC8 nanomediator to respond to Cat B cleavage by assembling the electronegative mediator DNAs on the surface of electropositive Cat B substrate peptide modified AuNCs with emission at 826 nm (AuNC8) via electrostatic interaction. The successful preparation of DpAuNC8 was proved by zeta potential and UV-vis absorption measurements (Fig. 2b and S5†). To evaluate the responsiveness of DpAuNC8 to Cat B, DNA molecules were equipped with FAM fluorophores. The fluorescence intensity of DNA-FAM decreased with increasing concentrations of pAuNC8 due to the successful assembly of DNA-FAM on pAuNC8 (Fig. 2c). In the presence of Cat B (Fig. 2d), the fluorescence intensity of DpAuNC8 increased gradually with increasing Cat B concentrations, implying the release of mediator DNA trigged by Cat B induced peptide cleavage.To construct the pDNA-AuNC8 nanomachines responsible for relaying the mediator DNA strands, the “mA”, “mD” and “mFD” were replaced by “pA”, “pD” and “pFD” that respond to mediator DNA and thus initiate the TMSD reaction to amplify the Cat B detection signal. The feasibility of the pDNA-AuNC8 nanomachine comprising “pA”, “pD”, and “pFD” was confirmed through PAGE assay (Fig. S6†). TEM images vividly depicted the aggregation upon the mixture of the DpAuNC8 nanomediators and the pDNA-AuNC8 nanomachines in the presence of Cat B, indicating the Cat B triggered mediator DNA release and thus inducing the aggregation of AuNC8 through the TMSD reaction (Fig. 2a3). Dynamic light scattering (DLS) also confirmed the aggregation phenomenon (Fig. S7†). To further assess the cascade responsiveness of DpAuNC8 and pDNA-AuNC8 nanomachines, various probe formulations were incubated in different reaction buffers. In the presence of excess Cat B (1 μU μL−1), the cascade of DpAuNC8 and pDNA-AuNC8 nanomachines yielded an approximately 2-fold increase in fluorescence intensity, unlike the unchanged fluorescence when the Cat B inhibitor (antipain hydrochloride, AH) was introduced or when either DpAuNC8 or pDNA-AuNC8 was used alone (Fig. 2e). Since the fluorescence of DNA-AuNC8 showed a positive correlation with the mediator DNA concentration (Fig. S8†), the cascade system for sensing Cat B activity was accomplished by sequentially integrating DpAuNC8 with pDNA-AuNC8. As shown in Fig. 2f, the fluorescence emissions of cascade pDNA-AuNC8 nanomachines at 826 nm displayed a linear relationship with the Cat B activity in the range of 0 to 0.18 nU μL−1 (Fig. 2g). Notably, the AuNCs with emission at 605 nm could also be employed for Cat B detection (named pDNA-AuNC6 and DpAuNC6, Fig. S9 and S10†), while AuNC8 were suitable for TK1 detection (named mDNA-AuNC8, Fig. S11 and S12†).
Fluorescence imaging in living cells
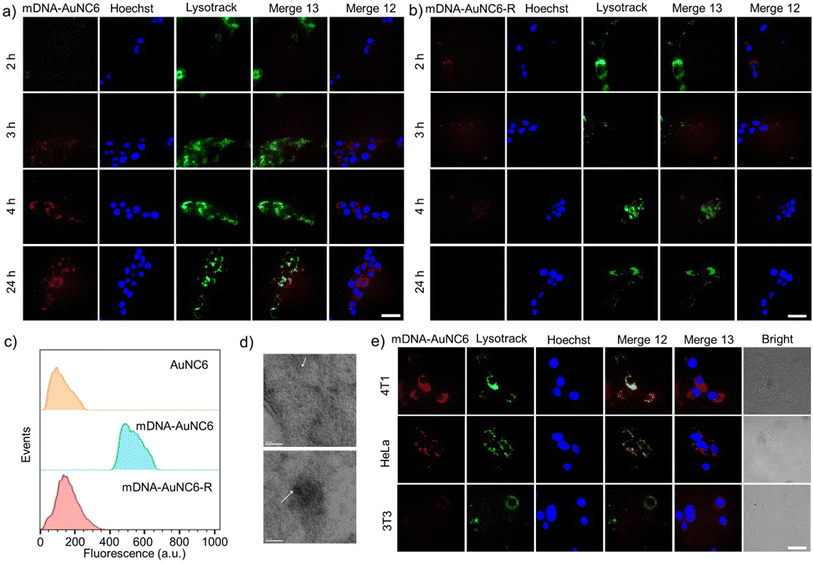
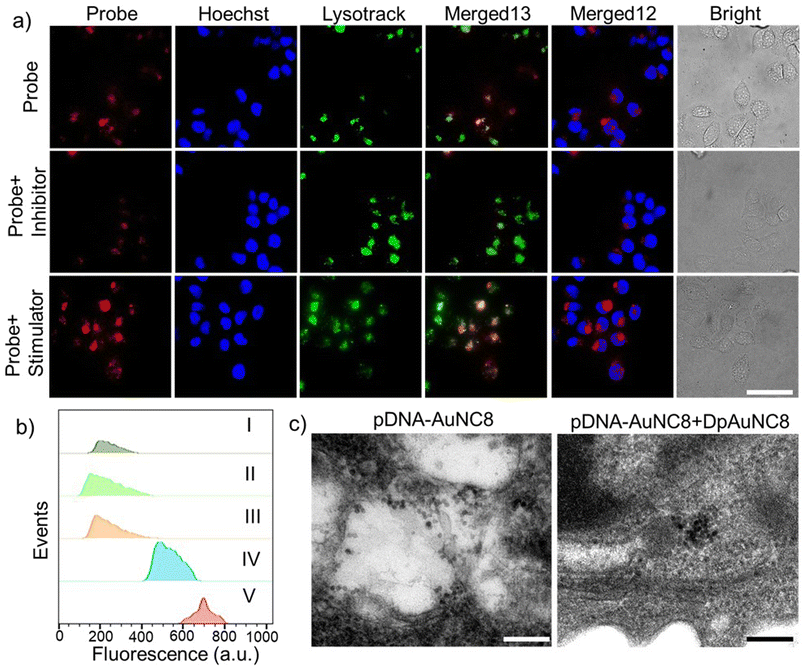
Before delving into the performance of the DNA-AuNCs nanoplatform for imaging TK1 mRNA or Cat B activity in living cells, we conducted a biocompatibility assessment of the nanomachines on 4T1 cells, a crucial step in biomedical application. Following a 24-h incubation of the nanomachines with 4T1 cells, no distinct cytotoxicity was detected. Even at a high concentration of 100 μg mL−1, cell viability exceeded 80%, underscoring their exceptional biocompatibility (Fig. S13†). Additionally, the live/dead staining assay using calcein and PI confirmed the outstanding biocompatibility of DNA-AuNC nanomachines (Fig. S14†). Subsequently, the cell uptake of the nanomachines was evaluated by ICP-OES. The DNA-AuNC nanomachines were incubated with 4T1 cells for varying durations (0.5, 1, 2, 4, 8, and 24 h), and the Au content in the lysed cells was quantified by ICP-OES (Fig. S15†). The optimal incubation time was determined to be 2 h. To investigate the nanomachines' specific response to the target mRNA in living cells, mDNA-AuNC6 (responds towards TK1 mRNA) and mDNA-AuNC6-R (including the random DNAs that have no response to TK1 mRNA) were incubated with 4T1 cells and observed by fluorescence microscopy (Fig. 3a and b). Compared with the 4T1 cells treated with mDNA-AuNC6-R, a substantial increase in fluorescence intensity was observed in mDNA-AuNC6 treated cells after a 4-h incubation, indicating the specific aggregation of AuNC6 triggered by overexpressed TK1 mRNA in 4T1 cells.36–39 Moreover, the strong red fluorescence of AuNC6 is located at the nuclear periphery by staining the nucleus and lysosome for colocalization, signifying that the TK1 mRNA is principally distributed in the cytoplasm. Importantly, the fluorescence of the nanomachines remained unfading over 24 h, indicating their excellent long-term imaging capability. Flow cytometry analysis also demonstrated an increased fluorescence intensity in the mDNA-AuNC6 treated group compared to the AuNC6 or mDNA-AuNC6-R treated group, consistent with the results from fluorescence imaging (Fig. 3c). The specific state of AuNC6 in 4T1 cells after incubation with DNA-AuNC6 or DNA-AuNC6-R was further confirmed by TEM (Fig. 3d). As expected, a significant aggregation of AuNC6 was observed in mDNA-AuNC6 treated 4T1 cells, induced by TK1 mRNA via cascade TMSD reactions. Conversely, no particle aggregation was observed in mDNA-AuNC6-R treated cells. To further assess the selectivity of mDNA-AuNC6 nanomachines in living cells, HeLa cells and NIH-3T3 cells were also exposed to the nanomachines (Fig. 3e). In contrast to 4T1 cells, no enhanced fluorescent signals were observed in either HeLa cells or NIH-3T3 cells due to their lower expression level of TK1 mRNA. These results collectively demonstrate the excellent specificity of the nanomachines for targeting cancer cells and their long-term fluorescence imaging capability in living cells.Next, we assessed the real-time imaging capability of the cascade DpAuNC8 and pDNA-AuNC8 nanomachines for monitoring Cat B in living cells. The 4T1 cells were first treated with AH to down-regulate Cat B activity or cysteine (Cys, Cat B stimulator) to up-regulate it, followed by incubation with the cascade nanomachines (Fig. 4a). Compared to the untreated group, the fluorescence intensity decreased in the inhibitor-treated group and increased in the stimulator-treated one, aligning with the findings from flow cytometry analysis (Fig. 4b). The aggregation of AuNC8 in 4T1 cells after incubation with pDNA-AuNC8 or pDNA-AuNC8 plus DpAuNC8 was further validated by TEM (Fig. 4c). As expected, a substantial aggregation of AuNC8 was observed in 4T1 cells treated with pDNA-AuNC8 plus DpAuNC8, triggered by Cat B mediated cascade TMSD reactions, whereas no particle aggregation was observed in cells treated with pDNA-AuNC8 alone, indicating the specific aggregation of AuNC8 triggered by Cat B. Therefore, the cascade nanomachines are capable of monitoring Cat B activity in cancer cells via fluorescence imaging, owing to their effective amplification performance through the Cat B mediated TMSD reaction.
In vivo fluorescence imaging
Motivated by the outstanding performance of the nanomachines in the aforementioned in vitro experiments, we extended our investigation to their in vivo behavior in mice. Mice harboring subcutaneous 4T1 xenograft tumors received doses of mDNA-AuNC6 or mDNA-AuNC6-R via intratumoral injection and were subsequently subjected to whole-body fluorescence imaging at specific time intervals (Fig. 5a).There is an observation of the mouse head being bright, which is probably caused by the hair on the head that is not barbered (Fig. S16†). Time-dependent fluorescence images revealed a significant decrease in fluorescence intensity within 1 h in the mDNA-AuNC6-R treated group (Fig. S18b and e†), a pattern similar to that observed in the group injected with AuNC6 (Fig. S17 and S18c, f†). Conversely, mice treated with mDNA-AuNC6 exhibited a more pronounced fluorescence increase within 2 h, followed by a gradual decline in fluorescence (Fig. S18a and d†), owing to the TK1 mRNA triggered aggregation of AuNC6. This comparison also underscored the imaging specificity of our nanomachines for target TK1 mRNA in vivo.
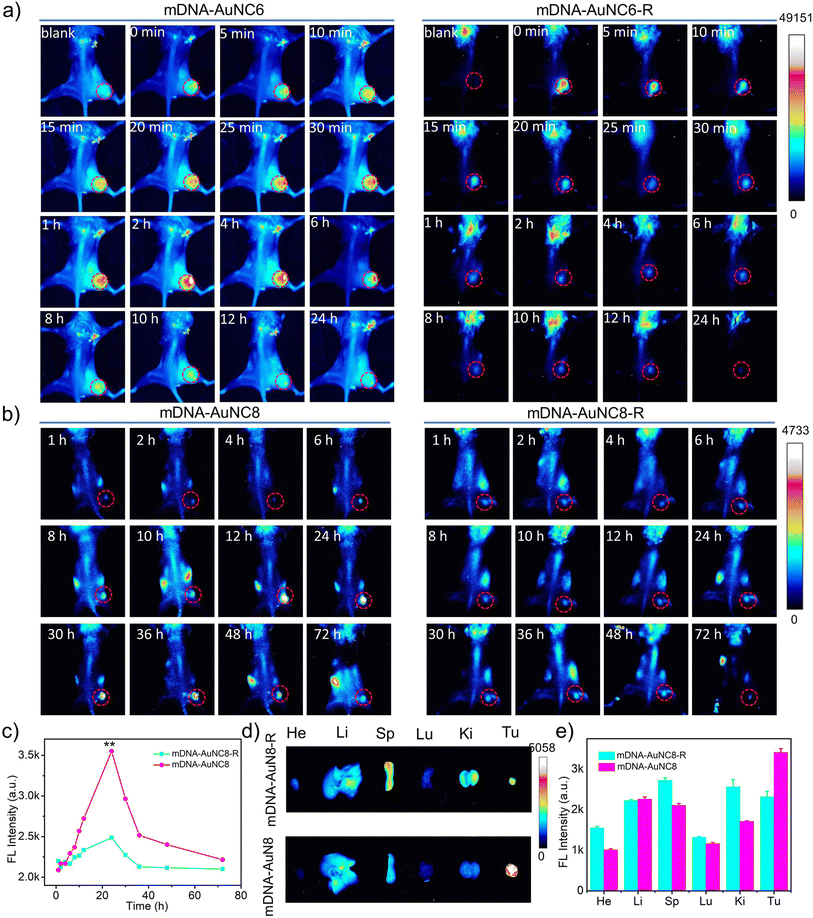
Subsequently, we assessed the imaging performance of the mDNA-AuNC6 nanomachine at tumor sites following intravenous administration. Unfortunately, we failed to obtain ideal whole-body fluorescence images probably due to the inadequate imaging resolution of AuNC6. Only ex vivo fluorescence images of tumors and major organs were attained from the mice at 24 h after intravenous injection of mDNA-AuNC6 or mDNA-AuNC6-R (Fig. S19†), showing that mDNA-AuNC6 has higher fluorescence intensity at the tumor site than mDNA-AuNC6-R attributable to the TK1 mRNA induced aggregation, which could also be evidenced by slower renal clearance of Au in the mDNA-AuNC6 treated group in contrast to the mDNA-AuNC6-R or AuNC6 treated groups (Fig. S20†). In order to visualize the TK1 mRNA level in situ, we further evaluated the imaging performance of the mDNA-AuNC8 nanomachine through intravenous administration. The fluorescence imaging ability of mDNA-AuNC8 was exemplified by comparing the fluorescence at tumors between mice treated with mDNA-AuNC8 and those treated with mDNA-AuNC8-R at specific time intervals (Fig. 5b). This comparison revealed a 1.4-fold enhancement in fluorescence intensity after 24 h post-injection and slower fluorescence decrease within 72 h, ascribed to the aggregation of AuNC8 induced by TK1 mRNA (Fig. 5c and S21†). Notably, 72 h after injection, the signal in the tumor of the mice treated with mDNA-AuNC8 was still clearly visible, proving prominent long-term retention of the nanomachines in vivo due to the aggregation of AuNC8. The ex vivo fluorescence imaging of tumors and major organs collected from the mice at 72 h post-injection exhibited similar results with AuNC6 as shown in Fig. S19† (Fig. 5d and e). All these findings confirmed that the nanomachines could accumulate at tumor sites and persist for a longer duration compared to the control group, owing to the TK1 mRNA induced aggregation.
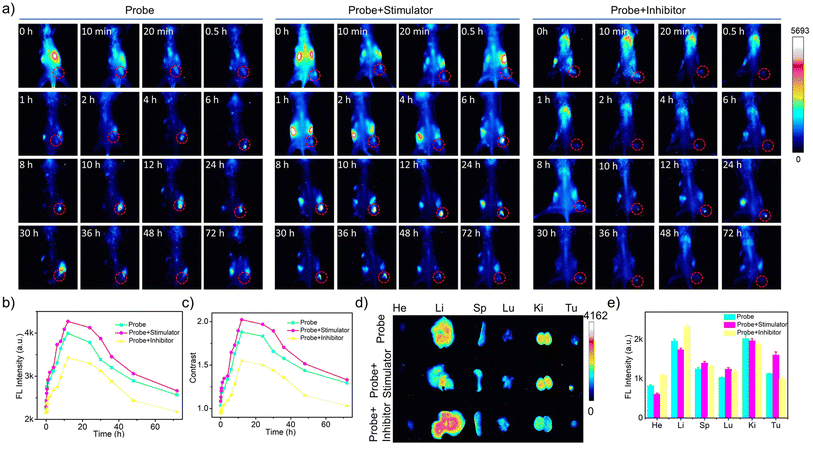
To enable the dynamic monitoring of Cat B activities in vivo, we modulated Cat B activities within tumors by intratumoral injection of AH or Cys prior to both intratumoral and intravenous administration of the cascade pDNA-AuNC8 plus DpAuNC8 nanomachines. The results from both intratumoral (Fig. S22a†) and intravenous (Fig. 6a) administrations showed that the two groups pretreated with the stimulator and inhibitor displayed intensified and diminished fluorescence at each time point in comparison to the control group (Fig. 6b, c and S22b†). Ex vivo fluorescence imaging was conducted on tumor and major organs gathered from the mice at 72 h post-injection. Accordingly, the two groups pretreated with the stimulator and inhibitor, respectively, exhibited increased and weakened fluorescence at the tumor site (Fig. 6d and e). These results provide confirmation that the cascade nanomachines were proficient at discerning Cat B activation levels in the tumors of live animals with acceptable sensitivity and accuracy. Moreover, there was no obvious pathological abnormality in the tissues of the heart, liver, spleen, lung, and kidney of the mice during the imaging (Fig. S23†), demonstrating the superior biocompatibility of the nanomachines.
Conclusions
In summary, we have successfully developed a DNA-AuNC nanomachine for long-term imaging of mRNA and protease, harnessing the TMSD reaction-driven AIE of AuNCs. Leveraging the TMSD reaction-driven AIE of AuNCs with emission at 605 nm or 826 nm, the mDNA-AuNC nanomachine exhibits remarkable sensitivity in TK1 mRNA detection. For Cat B imaging, we introduced cascade pDNA-AuNC8 plus DpAuNC8 nanomachines, skillfully translating intracellular protease-regulated cleavage into TMSD reaction-driven aggregation of AuNC8. What's more, these DNA-AuNC nanomachines offer a label-free feature and outstanding photostability, making them exceptionally suited for long-term imaging owing to their AIE characteristics. In vivo investigations have demonstrated that the size tunability of these DNA-AuNC nanomachines ensured high accumulation and long-term retention at tumor sites. Accordingly, NIR fluorescence imaging using the DNA-AuNC nanomachines proves invaluable for long-term observations in vivo, holding substantial promise for visualizing novel biological phenomena in organisms. In addition, the utility of the DNA-AuNC nanomachines can be extended to probe other mRNAs or proteases by pre-installation of corresponding substrates. The proposed DNA-AuNC nanomachines hold immense potential for future applications in disease diagnosis.Data availability
All experimental supporting data and procedures are availabe in the ESI.†Author contributions
Ting Wang performed the experiments and collected the data. Kai Jiang analyzed the data. Ting Wang and Kai Jiang contributed equally. Yifan Wang, Limei Xu, Yingqi Liu, and Shiling Zhang helped with the data collection and analysis of animal experiments. Weiwei Xiong and Yemei Wang guided part of the experiments. Fenfen Zheng and Jun-Jie Zhu conceived and supervised the project. All authors discussed the results and commented on the manuscript. All authors approved the final version of the manuscript.Conflicts of interest
There are no conflicts to declare.Acknowledgements
This work was financially supported by the National Natural Science Foundation of China (21804059 and 21701059) and the Natural Science Foundation of Jiangsu Province (BK20180974 and BK20210876). All animal studies were performed in accordance with the Jiangsu Animal Care and Use Committee (SYXK(Su)2023).Notes and references
- W. Ma, P. Fu, M. Sun, L. Xu, H. Kuang and C. Xu, J. Am. Chem. Soc., 2017, 139, 11752–11759 CrossRef CAS PubMed.
- H. Dong, J. Lei, L. Ding, Y. Wen, H. Ju and X. Zhang, Chem. Rev., 2013, 113, 6207–6233 CrossRef CAS PubMed.
- J. Conde, N. Oliva, M. Atilano, H. S. Song and N. Artzi, Nat. Mater., 2016, 15, 353–363 CrossRef CAS PubMed.
- T. Ma, Y. Hou, J. Zeng, C. Liu, P. Zhang, L. Jing, D. Shangguan and M. Gao, J. Am. Chem. Soc., 2018, 140, 211–218 CrossRef CAS PubMed.
- S. Yu, Y. Zhou and Y. Sun, et al. , Angew. Chem., Int. Ed., 2021, 60, 5948–5958 CrossRef CAS.
- J. Sun, K. Jiang, Y. Wang, Y. Liu, T. Wang, S. Ding, X. Zhang, W. Xiong, F. Zheng, H. Yang and J. J. Zhu, Adv. Healthcare Mater., 2023, 2302016 CrossRef CAS.
- F. Zheng, T. Meng, D. Jiang, J. Sun, H. Yao, J. J. Zhu and Q. Min, Angew. Chem., Int. Ed., 2021, 60, 21565–21574 CrossRef CAS PubMed.
- D. Jiang, Y. Pan, H. Yao, J. Sun, W. Xiong, L. Li, F. Zheng, S. Sun and J. J. Zhu, Anal. Chem., 2022, 94, 9074–9080 CrossRef CAS PubMed.
- H. Yao, D. Jiang, G. Dong, J. Sun, S. Sun, L. Li, F. Zheng and W. Xiong, Analyst, 2021, 146, 5115–5123 RSC.
- K. Zhang, X. Meng, Z. Yang, Y. Cao, Y. Cheng, D. Wang, H. Lu, Z. Shi, H. Dong and X. Zhang, Adv. Mater., 2019, 31, 1807888 CrossRef PubMed.
- N. Yan, X. Wang, L. Lin, T. Song, P. Sun, H. Tian, H. Liang and X. Chen, Adv. Funct. Mater., 2018, 28, 1800490 CrossRef.
- F. Yang, Q. Yang, L. Yang, J. Li, Y. Zhang, H. Lu, H. Dong and X. Zhang, Anal. Chem., 2022, 94, 6599–6606 CrossRef CAS PubMed.
- E. A. Sykes, J. Chen, G. Zheng and W. C. W. Chan, ACS Nano, 2014, 8, 5696–5706 CrossRef CAS PubMed.
- H. S. Choi, W. Liu, P. Misra, E. Tanaka, J. P. Zimmer, B. I. Ipe, M. G. Bawendi and J. V. Frangioni, Nat. Biotechnol., 2007, 25, 1165–1170 CrossRef CAS PubMed.
- W. Jiang, B. Y. S. Kim, J. T. Rutka and W. C. W. Chan, Nat. Nanotechnol., 2008, 3, 145–150 CrossRef CAS.
- X. Lou, Y. Zhuang, X. Zuo, Y. Jia, Y. Hong, X. Min, Z. Zhang, X. Xu, N. Liu, F. Xia and B. Z. Tang, Anal. Chem., 2015, 87, 6822–6827 CrossRef CAS PubMed.
- Y. Zhuang, M. Zhang, B. Chen, R. Duan, X. Min, Z. Zhang, F. Zheng, H. Liang, Z. Zhao, X. Lou and F. Xia, Anal. Chem., 2015, 87, 9487–9493 CrossRef CAS PubMed.
- X. Liu, Y. Chen, H. Li, N. Huang, Q. Jin, K. Ren and J. Ji, ACS Nano, 2013, 7, 6244–6257 CrossRef CAS PubMed.
- W. Yu, R. Liu, Y. Zhou and H. Gao, ACS Cent. Sci., 2020, 6, 100–116 CrossRef CAS PubMed.
- Z. Luo, X. Yuan, Y. Yu, Q. Zhang, D. Leong, J. Y. Lee and J. Xie, J. Am. Chem. Soc., 2012, 134, 16662–16670 CrossRef CAS PubMed.
- K. Zheng, X. Yuan, K. Kuah, Z. Luo, Q. Yao, Q. Zhang and J. Xie, Chem. Commun., 2015, 51, 15165–15168 RSC.
- F. Semcheddine, N. E. l. Guissi, W. Liu, Tayyaba, L. Gang, H. Jiang and X. Wang, Mater. Horiz., 2021, 8, 2771–2784 RSC.
- A. Yahia-Ammar, D. Sierra, F. Merola, N. Hildebrandt and X. L. Guevel, ACS Nano, 2016, 10, 2591–2599 CrossRef CAS PubMed.
- N. Goswami, Q. F. Yao, Z. T. Luo, J. G. Li, T. K. Chen and J. P. Xie, J. Phys. Chem. Lett., 2016, 7, 962–975 CrossRef CAS PubMed.
- Z. Wu, Q. Yao, O. J. H. Chai, N. Ding, W. Xu, S. Zang and J. Xie, Angew. Chem., Int. Ed., 2020, 59, 9934–9939 CrossRef CAS PubMed.
- Y. Zheng, J. Wu, H. Jiang and X. Wang, Coord. Chem. Rev., 2021, 431, 0010–8545 CrossRef.
- Y. Huang, J. Ji, J. Zhang, F. Wang and J. Lei, Chem. Commun., 2019, 56, 313–316 RSC.
- J. G. You, C. Y. Lu, A. S. K. Kumar and W. L. Tseng, Nanoscale, 2018, 10, 17691–17698 RSC.
- J. Wang, X. Lin, T. Shu, L. Su, F. Liang and X. Zhang, Int. J. Mol. Sci., 2019, 20, 1891 CrossRef CAS PubMed.
- W. X. Wang, Y. Q. Wu and H. W. Li, J. Colloid Interface Sci., 2017, 505, 577–584 CrossRef CAS.
- A. Dutta, U. Goswami and A. Chattopadhyay, ACS Appl. Mater. Interfaces, 2018, 10, 19459–19472 CrossRef CAS.
- F. Qu, Z. Wang, C. Li, D. Jiang and X. E. Zhao, Sens. Actuators, B, 2022, 359, 131610 CrossRef CAS.
- Y. Zi, D. Xu, C. Li, F. Qu and X. E. Zhao, Sens. Actuators, B, 2021, 345, 130243 CrossRef CAS.
- X. Ran, Z. Wang, F. Pu, E. Ju, J. Ren and X. Qu, Mater. Horiz., 2021, 8, 1769–1775 RSC.
- W. Shu, X. Zhang, H. Tang, L. Wang, M. Cheng, J. Xu, R. Li and X. Ran, Anal. Chim. Acta, 2023, 1268, 341372 CrossRef CAS PubMed.
- N. Benjamina, A. Tanirb, K. Lunaa, M. Bellac, G. Simond, H. Tamara and P. Tamara, Cancer Biomarkers, 2010, 7, 65–72 Search PubMed.
- N. Ibnat and E. H. Chowdhury, Sci. Rep., 2023, 536, 2045–2322 Search PubMed.
- S. Barbara, B. Sebastian, A. Christian, B. Thomas, B. Valesca, H. Christoph, R. Christoph, M. Katja, T. Arbel D, V. Mathias, S. Ugur and L. Martin, Front. Oncol., 2020, 10, 2234 Search PubMed.
- G. N. Fanelli, R. Scarpitta, P. Cinacch, B. Fuochi, A. Szumera-Ciećkiewicz, I. K. De, P. Ferrari, A. Fontana, M. Miccoli and A. G. Naccarato, et al. , J. Clin. Med., 2021, 10, 5416 CrossRef CAS.
Footnote |
| † Electronic supplementary information (ESI) available. See DOI: https://doi.org/10.1039/d3sc05887e |
| This journal is © The Royal Society of Chemistry 2024 |