Open Access Article
Open Access ArticleCreative Commons Attribution 3.0 Unported Licence
A β-barrel-like tetramer formed by a β-hairpin derived from Aβ†
Tuan D.
Samdin
 a,
Chelsea R.
Jones
a,
Chelsea R.
Jones
 a,
Gretchen
Guaglianone
a,
Gretchen
Guaglianone
 a,
Adam G.
Kreutzer
a,
Adam G.
Kreutzer
 a,
J. Alfredo
Freites
a,
J. Alfredo
Freites
 a,
Michał
Wierzbicki
a,
Michał
Wierzbicki
 a and
James S.
Nowick
a and
James S.
Nowick
 *ab
*ab
aDepartment of Chemistry, University of California, Irvine, California 92697-2025, USA. E-mail: jsnowick@uci.edu
bDepartment of Pharmaceutical Sciences, University of California, Irvine, Irvine, California 92697-2025, USA
First published on 28th November 2023
Abstract
β-Hairpins formed by the β-amyloid peptide Aβ are building blocks of Aβ oligomers. Three different alignments of β-hairpins have been observed in the structures of Aβ oligomers or fibrils. Differences in β-hairpin alignment likely contribute to the heterogeneity of Aβ oligomers and thus impede their study at high-resolution. Here, we designed, synthesized, and studied a series of β-hairpin peptides derived from Aβ12–40 in one of these three alignments and investigated their solution-phase assembly and folding. These assays reveal the formation of tetramers and octamers that are stabilized by intermolecular hydrogen bonding interactions between Aβ residues 12–14 and 38–40 as part of an extended β-hairpin conformation. X-ray crystallographic studies of one peptide from this series reveal the formation of β-barrel-like tetramers and octamers that are stabilized by edge-to-edge hydrogen bonding and hydrophobic packing. Dye-leakage and caspase 3/7 activation assays using tetramer and octamer forming peptides from this series reveal membrane-damaging and apoptotic properties. A molecular dynamics simulation of the β-barrel-like tetramer embedded in a lipid bilayer shows membrane disruption and water permeation. The tetramers and octamers described herein provide additional models of how Aβ may assemble into oligomers and supports the hypothesis that β-hairpin alignment and topology may contribute directly to oligomer heterogeneity.
Introduction
The formation and biological activity of β-amyloid oligomers are central events in the pathogenesis and progression of Alzheimer's disease. Dimers of Aβ isolated from Alzheimer's brains have been shown to disrupt neuritic integrity in vitro, and in a separate study impair synaptic structure and function in vivo.1,2 Synthetic trimers of peptides derived from Aβ have been used to raise antibodies that recognize pathological features in tissues isolated from Alzheimer's brains.3 Tetramers and octamers of Aβ have demonstrated pore-forming activity against lipid-bilayers.4 Photo-induced crosslinking studies of Aβ42 have revealed the formation of pentamers and hexamers, while ion mobility-mass spectrometry studies have shown that Aβ42 forms β-barrel shaped hexamers in the presence of membrane mimetic micelles.5,6 Understanding the biophysical and biological activity of Aβ oligomers is key to understanding the molecular basis of neurodegeneration and cognitive decline in Alzheimer's disease.Aβ oligomers, however, are unstable and exhibit significant variation in their stoichiometry and structure—variation that is reflected by the dizzying alphabet soup of terms used to describe them, including ADDLs (Aβ-derived diffusible ligands), AβOs (Aβ oligomers), PFs (protofibrils), PFOs (prefibrillar oligomers), and APFs (annular protofibrils).7–11 Structure–activity relationship studies are often frustrated by the heterogeneous and polydisperse nature of Aβ assemblies, as it equilibrates between monomeric, oligomeric, and fibrillar species.12–16 To overcome these difficulties fragment-based investigations, covalent-stabilization, synthetic Aβ oligomer homologues, and in silico modeling have emerged as tools to aid in the study of more uniform and homogenous models of Aβ oligomers.17 A substantial body of evidence has emerged from these investigations, identifying β-hairpins as a key structural element of Aβ oligomers.
The NMR-based structure of a tetramer formed by full-length Aβ42 reported by Carulla and coworkers is the only atomic-resolution structure of an Aβ oligomer that has been deposited in the Protein Data Bank, (PDB 6RHY).4 The tetramer comprises a six-stranded antiparallel β-sheet, with two β-hairpins of Aβ42 that flank two antiparallel β-strands of Aβ42 (ESI Fig. S1a†). Additional solution-phase studies of this tetramer also provide evidence for the formation of an octamer. Using molecular dynamics (MD) simulations, Carulla and coworkers propose a model in which this tetramer, as well as the octamer, can act to disrupt a lipid membrane and facilitate water permeation. Collectively, these studies have brought into sharp relief the importance of β-hairpins in the structures of Aβ oligomers.
Several additional studies have also established the significance of β-hairpins in Aβ oligomers.18–22 In 2008, Härd and Hoyer reported the NMR structure of a monomer of Aβ40 adopting a β-hairpin conformation when sequestered and stabilized by an affibody.19 Härd and coworkers subsequently stabilized this Aβ β-hairpin using an intermolecular disulfide-bridge and found that the stabilized β-hairpin formed oligomers that mimicked some of the properties of oligomers formed by unmodified Aβ.20 NMR spectroscopic studies revealed that related disulfide-stabilized β-hairpins derived from Aβ16–42 assembled to form a barrel-shaped hexamer stabilized by hydrophobic packing and edge-to-edge hydrogen bonding between β-hairpins (ESI Fig. S1b†).21
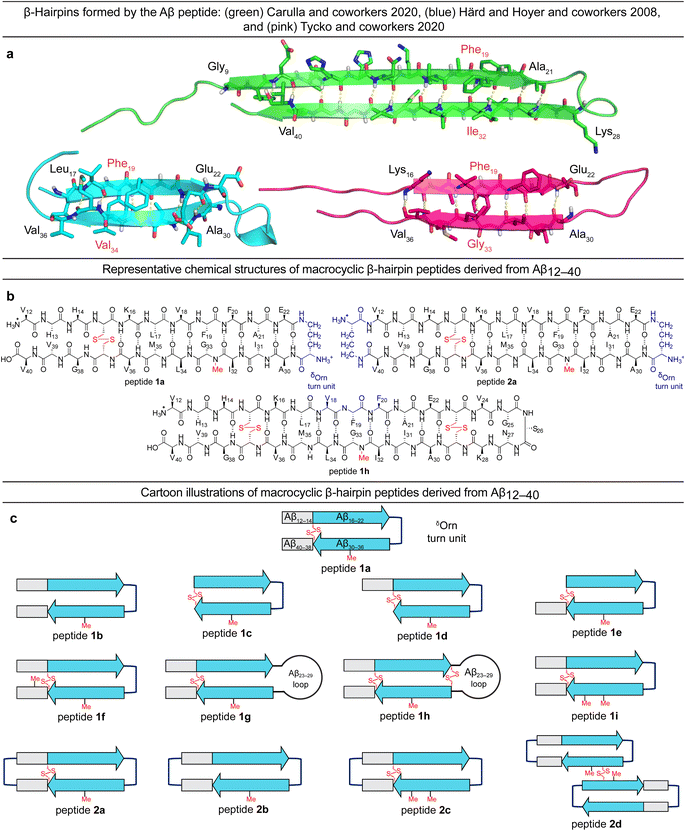
Tycko and coworkers recently reported a β-hairpin as a component in the structure of an atypical Aβ40 fibril structure, in which the characteristic core of parallel in-register β-sheets is coated by an outer layer of β-hairpins formed by Aβ (ESI Fig. S1c†).23 A model of the β-hairpin that fits the cryo-EM and NMR spectroscopic data shows residues 16–22 and 30–36 hydrogen bonding to form a β-sheet, with the intervening residues 23–29 forming a loop (Fig. 1a).
The Aβ β-hairpins reported by Carulla and coworkers, Härd and Hoyer and coworkers, and Tycko and coworkers all differ in the alignment of their β-strands (Fig. 1a).4,19,23 In the tetramer reported by Carulla and coworkers, β-strands comprising residues 9–21 and 28–40 hydrogen bond to form an antiparallel β-sheet, with residues 22–27 forming a loop. In the barrel-shaped hexamer reported Härd and coworkers, β-strands comprising residues 17–22 and 30–36 hydrogen bond to form an antiparallel β-sheet, with residues 23–29 forming a loop. These differences in β-strand alignment alter the overall topology of the β-hairpins by shifting residue pairings across the β-strands, the hydrophobicity of the β-hairpin surfaces, and the size of the loop segments between the β-strands. In the β-hairpin reported by Carulla and coworkers, Ile32 is across from Phe19; in the β-hairpin reported by Tycko and coworkers, Gly33 is across from Phe19; and in the β-hairpin reported by Härd and Hoyer and coworkers, Val34 is across from Phe19 (Fig. 1a). These changes in alignment and topology may contribute to the immense variation and heterogeneity observed in the assembly and structures of Aβ fibrils and oligomers.
In the current study, we set out to explore oligomers formed by β-hairpins in the alignment reported by Tycko and coworkers by designing, synthesizing, and studying a series of β-hairpin peptides derived from Aβ12–40.17,24 The structures of these peptides are illustrated in Fig. 1b and c. Four of these peptides (1a, 1d, 1h, and 2a) assemble to form octamers in SDS-PAGE. X-ray crystallographic studies of peptide 2a reveal a hitherto unprecedented β-barrel-like tetramer composed of β-hairpins. The crystallographic tetramers concatenate within the crystal lattice to create an octamer, and thus suggest a structural model for the octamers observed in SDS-PAGE.
Results and discussion
Design of peptides 1a–i and 2a–d
We designed peptide 1a to fold into an Aβ β-hairpin in the alignment reported by Tycko and coworkers to probe the assembly of Aβ β-hairpins into oligomers. Peptide 1a comprises two peptide β-strands of Aβ12–22 and Aβ30–40 linked by a δOrn turn unit connecting residues 22 and 30, a cross-strand disulfide bridge replacing Gln15 and Gly37, and an N-methyl group on Gly33 to prevent uncontrolled aggregation.22,25–33 We designed homologous peptides 1b–i and 2a–d to further explore the effects of the N- and C-terminal residues 12–14 and 38–40, the Aβ23–29 loop, and additional δOrn turn units and disulfide bridges on folding and assembly.Oligomerization of peptides 1a–h and 2a–d
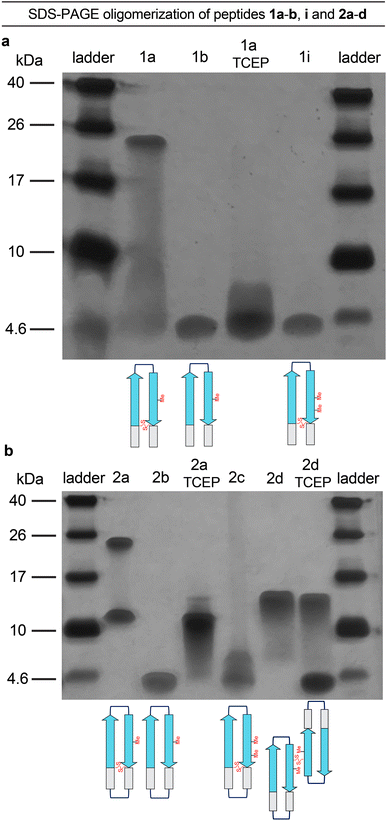
The formation of oligomers that can be observed in SDS-PAGE is a hallmark of Aβ.7,34–37 Peptide 1a (2.5 kDa) runs as an oligomer in SDS-PAGE, migrating at a molecular weight consistent with that of an octamer (ca. 20 kDa; Fig. 2a). The octamer band streaks downward from below the 26 kDa ladder band, suggesting that the octamer is in equilibrium with lower molecular weight species. Unlike the oligomer formed by peptide 1a, the oligomers formed by full-length, unmodified Aβ are heterogeneous in size and typically display a substantial band for the monomer (Fig. 2a).5,34,36 The formation of a well-defined oligomer band by peptide 1a suggests that this constrained Aβ β-hairpin peptide forms a well-defined supramolecular assembly in the membrane-like environment provided by the amphiphilic SDS molecules.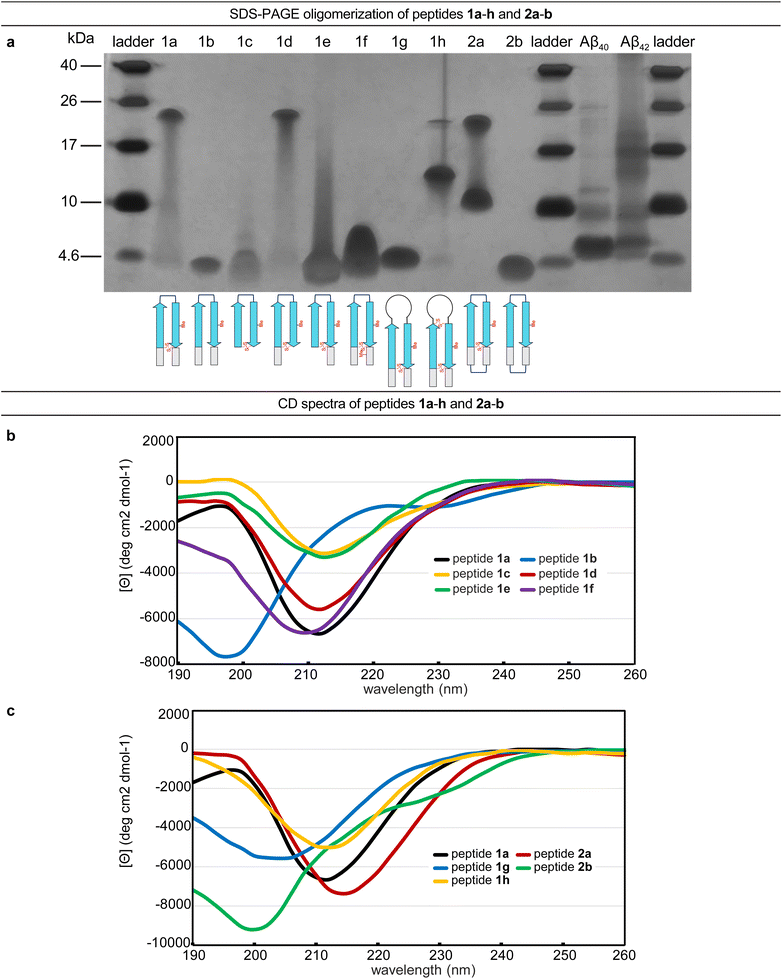 | ||
| Fig. 2 (a) Silver stained SDS-PAGE of peptides 1a–h, 2a–b, and Aβ40 and Aβ42. SDS-PAGE was performed in Tris buffer at pH 6.8 with 2% (w/v) SDS on a 16% polyacrylamide gel with 50 µM solutions of peptide in each lane. (b and c) Circular dichroism (CD) spectra of peptides 1a–h and 2a–b. CD spectra were acquired for each peptide at 50 µM in 10 mM phosphate buffer at pH 7.4; ellipticity was normalized for the number of residues in each peptide. Additional CD spectra for peptides 1i, 2c, and 2d are provided in the (ESI Fig. S2†). | ||
To better understand the assembly of the putative octamer, we prepared and studied peptides 1b–h (Fig. 1c). Peptide 1b (2.5 kDa) lacks a cross-strand disulfide bridge and instead contains the native Gln15 and Gly37 residues. Peptide 1b does not assemble in SDS-PAGE, migrating just below the 4.6 kDa ladder band (Fig. 2a). Peptide 1c (1.8 kDa) maintains the cross-strand disulfide bridge but lacks the N- and C-terminal residues 12–14 and 38–40. Peptide 1c does not assemble, migrating as a downward-streaking band from the 4.6 kDa ladder band. In contrast to peptide 1c, peptide 1d (2.3 kDa) lacks only the C-terminal residues 38–40 and does assemble. Like peptide 1a, peptide 1d migrates as a downward-streaking band from below the just 26 kDa ladder band. The band formed by peptide 1d suggests an octamer in equilibrium with lower molecular weight species. Peptide 1e (2.1 kDa) lacks only the N-terminal residues 12–14 but does not assemble. Instead, peptide 1e migrates as an upward-streaking band from just below the 4.6 kDa ladder band.
The lack of oligomer formation by peptide 1b reveals that the cross-strand disulfide bridge near the N- and C-terminal residues is necessary for assembly of the putative octamer. It has been noted that SDS is known to contribute to the assembly of small Aβ oligomers.38 The differences in the oligomerization of peptides 1a and 1b suggests that stabilization of the β-hairpin conformation by the disulfide bridge, rather than SDS, induces the formation of the putative octamer. The assembly of peptide 1d, in contrast to peptides 1c and 1e, is surprising and suggests that residues 12–14 participate in intermolecular interactions crucial for assembly of the putative octamer.
To further examine the role of interactions between the N- and C-terminal residues of peptide 1a, 12–14 and 38–40, in the assembly of the octamer, we prepared and studied peptide 1f. In peptide 1f, an N-methyl group on Gly38 is positioned to disrupt hydrogen bonding interactions between residues 12–14 and 38–40. In contrast to peptide 1a, peptide 1f (2.5 kDa) does not assemble as an octamer, but instead migrates as a band between the 10 kDa and 4.6 kDa ladder bands that streaks downward. This result suggests that residues 12–14 and 38–40 hydrogen bond together as part of an extended β-hairpin in the octamer formed by peptide 1a.
To examine whether the putative octamer formed by peptide 1a can accommodate residues 23–29 as a loop, and to better mimic endogenous Aβ β-hairpins and Aβ oligomers, we prepared and studied peptides 1g and 1h. In peptide 1g a loop comprising residues 23–29 replaces the δOrn turn unit. In contrast to peptide 1a, peptide 1g (3.0 kDa) does not assemble and instead migrates just above the 4.6 kDa ladder band. Peptide 1h is a homologue of peptide 1g, that incorporates an additional cross-strand disulfide bridge replacing Asp23 and Gly29 to fortify the β-hairpin conformation of the peptide. Peptide 1h (3.0 kDa) migrates as two bands, a lower molecular weight band consistent with a tetramer and a higher molecular weight band consistent with an octamer. The tetramer band migrates between the 10 and 17 kDa ladder bands and is much greater in intensity than the octamer band, which migrates just below the 26 kDa ladder band. The difference in intensity between the tetramer and octamer bands suggest that peptide 1h favors assembly of the tetramer. Thus, it appears that the octamer formed by peptide 1a can accommodate the Aβ23–29 loop, but that the loop destabilizes the β-hairpin conformation required for assembly—unless an additional stabilizing disulfide bridge is present, as in peptide 1h.
Peptide 2a is a macrocyclic homologue of peptide 1a with a second δOrn turn unit connecting the N- and C-terminal residues 12 and 40. Like peptide 1h, peptide 2a (2.6 kDa) migrates as two oligomers, a lower molecular weight oligomer consistent with a tetramer and a higher molecular weight oligomer consistent with an octamer. The tetramer band migrates just above the 10 kDa ladder band, and the octamer band migrates between the 17 and 26 kDa ladder bands. Both bands are equal in their intensity, with the octamer band streaking downward toward the tetramer band, suggesting that the two species are in equilibrium with each other. Peptide 2b is a homologue of peptide 2a that lacks the cross-strand disulfide bridge and does not assemble, instead migrating just below the 4.6 kDa ladder band.
Collectively, the SDS-PAGE studies of peptides 1b–1h and 2a–2b highlight key factors in the formation of an octamer by peptide 1a. The N-terminal residues 12–14 form critical intermolecular contacts in the octamer. The formation of both tetramers and octamers by peptides 1h and 2a indicates that the tetramers and octamers are in equilibrium and suggests that the tetramers may be components of the octamers, for these peptides as well as for peptide 1a. Peptides 1h and 2a bear additional stabilizing constraints not present in peptide 1a—a second disulfide bridge and a second δOrn turn unit respectively. Rather than stabilizing the octamer, these additional constraints destabilize the octamer and promote the formation of a tetramer, which appears to be a subunit of the octamer. The lack of assembly by peptide 2b, despite the second δOrn turn unit, suggests that the cross-strand disulfide bridge replacing Gln15 and Gly37 is essential for octamer assembly.
Folding of peptides 1a–h and 2a–b
The CD spectrum of peptide 1a is similar to that of a β-sheet, suggesting that the peptide folds as designed into a β-hairpin (Fig. 2b). The CD spectrum of peptide 1a displays a strong negative band centered at ca. 212 nm, with increasing ellipticity at lower wavelengths that reaches a maximum at ca. 198 nm before decreasing once again. The disulfide bridge in peptide 1a appears to be essential for folding—the CD spectrum of peptide 1b indicates that it does not fold and that a random coil conformation predominates. Peptide 1b displays a strong negative band centered at ca. 198 nm and a weak maximum at ca. 220 nm (Fig. 2b). The CD spectra of peptides 1c–f and 1h show that these peptides also fold to adopt β-hairpin-like conformations.Differences between the CD spectra of peptides 1a and 1g reveal that replacing the δOrn turn unit with Aβ residues 23–29 abrogates β-hairpin folding (Fig. 2c). The CD spectrum of peptide 1g displays a broad and shallow negative band centered at ca. 204 nm, which suggests a random-coil-like conformation. Addition of a disulfide linkage between residues 23 and 29 partially restores folding. Thus peptide 1h displays a negative band centered at ca. 212 nm with increasing ellipticity at lower wavelengths.
The CD spectra of peptides 2a and 2b suggest that incorporation of a second δOrn turn unit connecting residues 12 and 40 does not substantially affect folding (Fig. 2c). Peptide 2a displays a strong negative band centered at ca. 214 nm with increasing ellipticity at lower wavelengths, similar to peptide 1a, reflecting a β-hairpin-like conformation. Peptide 2b displays a strong negative band centered at ca. 200 nm, similar to peptide 1b, indicating a random coil conformation. Thus, the constraint of peptide 1b into a macrocycle but without a disulfide bridge is not sufficient to induce β-hairpin folding.
Correlation of these CD studies with the SDS-PAGE studies suggests that folding is necessary for assembly, but that not all folded β-hairpin peptides can assemble. Key residues and contacts also appear to be necessary to achieve the putative octamer that is observed in SDS-PAGE for peptides 1a, 1d, 1h, and 2a.
X-ray crystallographic and REMD studies of peptide 2a
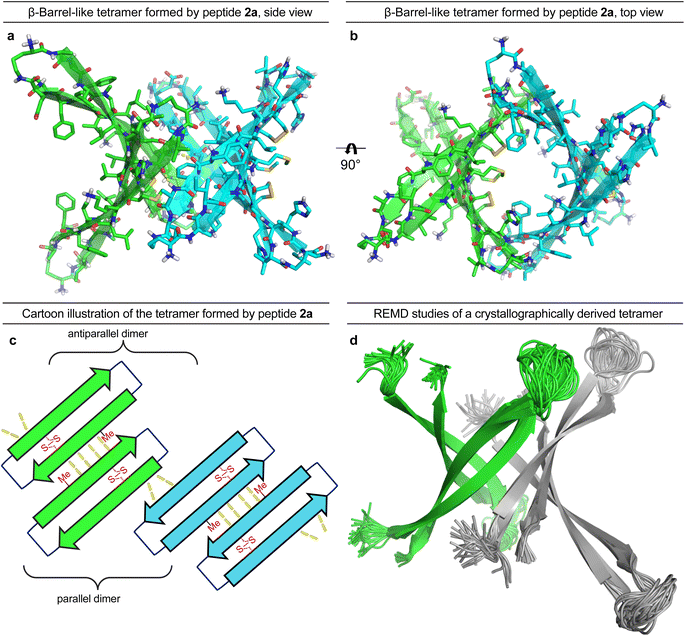
To gain further insights into the structures of the oligomers observed in SDS-PAGE, we grew crystals of peptide 2a and performed X-ray crystallography. None of the other peptides afforded crystals. The X-ray crystallographic structure of peptide 2a reveals that the peptide assembles to form antiparallel β-sheet dimers that further assemble in a face-to-back fashion to form concatenated β-barrel-like tetramers (Fig. 3, PDB 7RTZ). The core of each β-barrel-like tetramer is lined with hydrophobic residues (Cys15, Val18, Phe20, Ile32, Ile34, Met35, Val36 and Cys37), which pack against each other while creating a pore through the center, ca. 7.5 Å in diameter (ESI Fig. S3b†). ESI Table S1† summarizes the crystallographic properties, conditions, data collection, and model refinement statistics for peptide 2a.The β-hairpin subunits comprise residues 12–22 and 30–40 and include a disulfide bridge that replaces Gln15 and Gly37 (ESI Fig. S3a†). The β-hairpin subunits assemble in pairs to form the antiparallel β-sheet dimers (ESI Fig. S3b†), which are stabilized by two pairs of intermolecular hydrogen bonds between Met35 and Cys37. The two dimers further assemble through parallel β-sheet interactions between His14 and Phe19 to form the eight-stranded β-barrel-like tetramer (Fig. 3a–c). Molecular modeling suggests that full-length Aβ can assemble in the same fashion as peptide 2a to form a β-barrel-like tetramer (Fig. 3d). Thus, we observed that Aβ9–42 could form a β-barrel-like tetramer without steric clashes when modeled into the crystallographic coordinates of the tetramer using replica exchange molecular dynamics (REMD) simulations in implicit solvent to generate realistic conformations of the loops and N- and C-terminal regions.
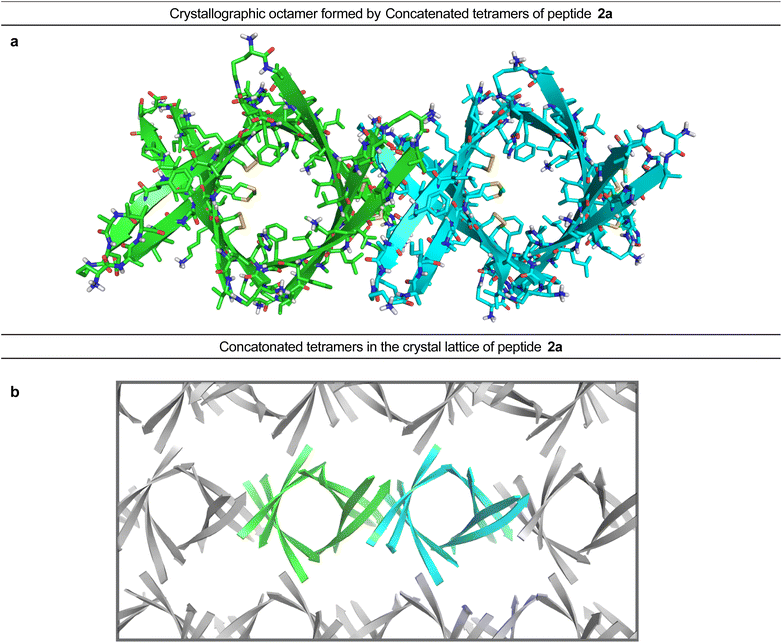
In the crystal lattice, the β-barrel-like tetramers link together to form concatenated networks of β-barrels running the length of the lattice, with the interface between each tetramer also constituting a β-barrel-like tetramer (Fig. 4). Thus, two tetramers may be thought of as composing an octamer consisting of three linked β-barrels. The two tetramers that make up the octamer are parallel to each other, and the tetramer formed at their interface is perpendicular but otherwise identical. Larger oligomers composed of more tetramers can also be envisioned. The crystallographically observed tetramers and octamers may explain the structures of the tetramers and octamers observed in SDS-PAGE for peptides 1a, 1d, 1h, and 2a, with each tetramer consisting of a single β-barrel and two tetramers further assembling to form an octamer consisting of three concatenated β-barrels.
Correlating the crystallographic tetramer and octamer with the oligomers observed in SDS-PAGE
To better understand the tetramers and octamers observed in SDS-PAGE and to correlate these species with the crystallographically observed tetramers and octamers formed by peptide 2a, we performed TCEP reduction, additional N-methylation, and disulfide crosslinking experiments (Fig. 5a and b).39 Reduction of the disulfide bonds of peptides 1a and 2a with tris(2-carboxyethyl)phosphine (TCEP) disrupts octamer formation in SDS-PAGE. Treatment of peptide 1a with TCEP results in a streaky band that is similar in position to that of peptide 1b, which does not assemble (Fig. 5a). Treatment of peptide 2a with TCEP eliminates octamer formation and gives a downward streaking band at a position that corresponds to a tetramer (Fig. 5b). Disruption of the dimerization interface by N-methylation of Met35, in peptides 1i and 2c, also disrupts octamer formation (Fig. 5a and b) while partially or fully retaining propensities to adopt β-hairpin-like conformations (ESI Fig. S2†). These experiments demonstrate that disruption of the hydrogen-bonding interfaces or destabilization of component β-hairpins disrupts octamer or tetramer formation.Covalent crosslinking of the antiparallel β-sheet dimer subunit observed in the X-ray crystallographic structure of peptide 2a stabilizes tetramer formation. Peptide 2d is a homologue of peptide 2b in which Val36 is replaced with cysteine and crosslinked intermolecularly. Peptide 2d runs as a tetramer in SDS-PAGE (Fig. 5b). Upon treatment with TCEP, peptide 2d runs as bands that correspond to monomer and tetramer (Fig. 5b). The formation of the monomer upon reduction further correlates the tetramer assembly observed in SDS-PAGE with that observed in the crystal structure. The CD spectrum of peptide 2d resembles a β-sheet, suggesting that unlike peptide 2b, it folds as designed to adopt a β-hairpin like conformations (ESI Fig. S2†).
We performed dynamic light scattering (DLS) studies to further characterize the oligomers formed by peptides 1a, 2a, and 2b in an aqueous environment without SDS. Under the same pH and buffer conditions of the CD experiments, DLS shows that these peptides form aggregates with hydrodynamic diameters of about 2 µm (ESI Fig. S4†). The aggregates observed by DLS are far larger than the tetramer or octamer observed by SDS-PAGE, or the crystallographic tetramer formed by peptide 2a. These observations suggest that peptides derived from the β-hairpin reported by Tycko and coworkers can assemble to form large assemblies in aqueous environments.
Cytotoxicity and membrane disruption of peptides 1a and 2a
Aβ oligomers have been shown to activate apoptotic pathways in vivo and damage artificial membranes in vitro.40–44 Because the crystallographic β-barrel-like tetramer formed by peptide 2a resembles a pore, we speculated that peptides 1a and 2a may damage cell membranes through a pore-like mechanism of action. To investigate the cytotoxic and membrane disrupting potential of Aβ derived peptides stabilized in the alignment reported by Tycko and coworkers, we carried out a caspase-3/7 activation assay and a dye-leakage assay using peptides 1a and 2a. Peptides 1a and 2a were thus found to activate caspase 3/7, an apoptotic marker, at 12.5–25 µM in SH-SY5Y cells (ESI Fig. S5†). The dye-leakage assay was performed using negatively charged large unilamellar vesicles (LUVs) and peptides 1a and 2a (ESI Fig. S6†). In these experiments, peptides 1a and 2a disrupt anionic lipid membranes at 0.6–2.7 µM. Together, these data suggest that peptides 1a and 2a have limited potential to mimic some of the apoptotic and membrane-damaging behaviors of oligomers formed by full-length Aβ.Molecular dynamics simulation of an Aβ tetramer in a lipid bilayer membrane
We performed molecular dynamics (MD) simulations using the X-ray crystallographic structure of peptide 2a to explore the consequences of inserting the β-barrel-like tetramer into a membrane.4,45–49 We thus generated a model of an Aβ9–42 β-barrel-like tetrameric assembly embedded in a fully hydrated POPC lipid bilayer from the crystallographic structure of peptide 2a and the REMD simulation and performed a 500 ns all-atom MD simulation (Fig. 6 and ESI S7†). The incorporation of the tetramer in the lipid bilayer results in a reduction of the lipid bilayer thickness around the β-barrel-like assembly (ESI Fig. S8†) as lipid phosphate groups and water molecules interact with polar moieties in the β-hairpins connecting loops and termini (Fig. 6 and ESI S9†). Water molecules cross the membrane through the tetramer pore, and also interact with polar moieties on the outer surface of the tetramer (Fig. 6 and ESI S10 and S11†). Although these results indicate that it is possible for a tetrameric assembly of peptide 2a to disrupt the membrane by forming pores, they cannot establish whether the caspase 3/7 activation and dye leakage observed for peptides 1a and 2a are caused by pore formation.50,51 The caspase 3/7, dye-leakage, and simulation data suggest that peptides 1a and 2a, and other peptides stabilized in the β-hairpin alignment described by Tycko and coworkers can interact with lipid membranes in a manner that our laboratory is beginning to explore.52,53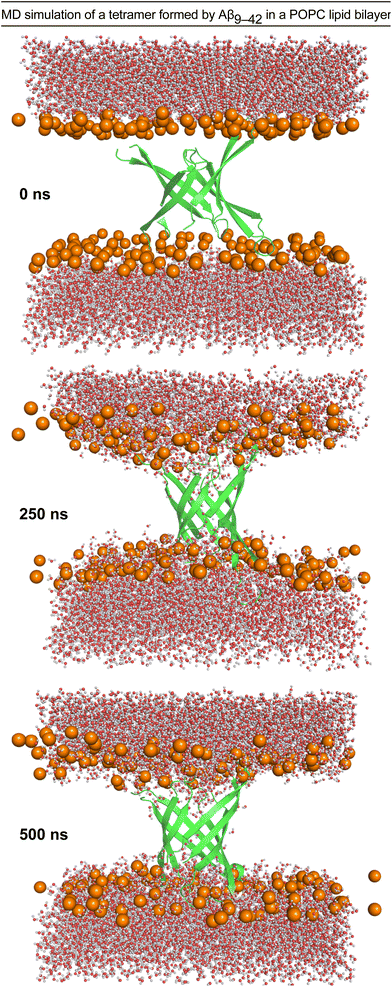 | ||
| Fig. 6 Configuration snapshots from an all-atom molecular dynamics simulation of a β-barrel-like tetramer formed by Aβ9–42 in a POPC lipid bilayer at 0 ns, 250 ns, and 500 ns. The Aβ9–42 tetramer is shown as a secondary structure representation. The lipid phosphate P atoms are shown as orange spheres, and the water molecules are shown in ball-and-stick representations colored by atom (O, red; H, white). The rest of the system is omitted for clarity. The simulation was set up using CHARMM-GUI,54,55 and run for 500 ns at constant temperature and pressure using NMAD 2.14 (ref. 56) with the CHARMM36 forcefield.57,58 Molecular graphics and simulation analyses were generated with VMD 1.9.3.59 | ||
Conclusion
β-Hairpins are a key structural component of Aβ oligomers and may contribute directly to oligomer heterogeneity through variation in hairpin alignment and topology. In this investigation, we studied the assembly and structure of oligomers formed by peptides derived from Aβ12–40 that were stabilized in the β-hairpin alignment reported by Tycko and coworkers. SDS-PAGE and X-ray crystallography reveal that Aβ derived β-hairpins in this alignment assemble to form β-barrel-like tetramers that concatenate to form octamers. We envision that Aβ β-hairpins in other alignments might form other oligomers, with different structures, stoichiometries, and stabilities—and that all of these β-hairpins and oligomers exist in equilibrium. Earlier investigations from our laboratory on related Aβ derived peptide systems have revealed the assembly of dimers, trimers, tetramers, hexamers, octamers, and dodecamers that differ substantially in assembly and structure. Further, mutations in familial Alzheimer's disease that alter the biophysical properties of the Aβ peptide may have a significant impact on Aβ folding and assembly.33,60 These findings would suggest that not all β-hairpins formed by the Aβ peptide have an equal propensity to fold and assemble.It is noteworthy that peptide 2a assembles to form oligomers of the same stoichiometry as the tetramer and octamer reported by Carulla and coworkers, but with significant differences in structure.4 The tetramer reported by Carulla and coworkers is planar and does not form a β-barrel-like assembly; further the octamer is proposed to adopt a β-sandwich conformation. Neither the planar tetramer nor the β-sandwich contain a pore. Instead, the β-barrel-like tetramer formed by peptide 2a more closely resembles the hexameric “cylindrin” formed by peptide fragments of αB crystallin, reported by Eisenberg and coworkers.61 Cylindrin-like assemblies from fragments of Aβ have also been reported.62–64 The tetramers and octamers described herein provide additional models of how Aβ may assemble into oligomers in membrane-like environments and the crystal state.
Data availability
Data analyzed in this study are available from the authors on request. Crystallographic data for peptide 2a has been deposited at the Protein Data Bank under PDB number 7RTZ and can be obtained from https://doi.org/10.2210/pdb7RTZ/pdb.Author contributions
Tuan D. Samdin: conceptualization, methodology, investigation, validation, formal analysis, writing – original draft, writing – reviewing & editing, visualization. Chelsea R. Jones: investigation. Gretchen Guaglianone: investigation. Adam G. Kreutzer: conceptualization, methodology, investigation. J. Alfredo Freites: methodology, software, validation, formal analysis, resources, visualization, supervision. Michał Wierzbicki: investigation. James S. Nowick: conceptualization, methodology, resources, writing – reviewing & editing, supervision, funding acquisition.Conflicts of interest
The authors declare no competing financial interest.Acknowledgements
We thank the National Institutes of Health (NIH) National Institute of General Medical Sciences (NIGMS) and the National Institute on Aging (NIA) for funding (GM097562) and (AG072587). We also thank Dr Dimitry Fishman and the Laser Spectroscopy Laboratories in the UCI Department of Chemistry for assistance with circular dichroism measurements, and the Stanford Synchrotron Radiation Lightsource (SSRL) Structural Molecular Biology Program (SMBP) for synchrotron data collection. The SMBP is supported in part by the DOE Office of Biological and Environmental Research, the NIH, and NIGMS. The SSRL is supported by the U.S. Department of Energy, Office of Science, Office of Basic Energy Sciences under contract no. DE-AC02-76SF00515. The authors thank Dr Robert Tycko for providing the coordinates of his Aβ β-hairpin model (Fig. 1a). M. W. acknowledges the support from the Ministry of Science and Higher Education, Republic of Poland (Mobility Plus grant no. 1647/MOB/V/2017/0). T. D. S. acknowledges the support from the University of California, Irvine, for funding through the Graduate Dean's Dissertation Year Fellowship, and thanks Alberto Smith and Victoria Sahrai for their assistance with peptide synthesis.References
- G. M. Shankar, S. Li, T. H. Mehta, A. Garcia-Munoz, N. E. Shepardson, I. Smith, F. M. Brett, M. A. Farrell, M. J. Rowan, C. A. Lemere, C. M. Regan, D. M. Walsh, B. L. Sabatini and D. J. Selkoe, Amyloid-β Protein Dimers Isolated Directly from Alzheimer's Brains Impair Synaptic Plasticity and Memory, Nat. Med., 2008, 14(8), 837–842, DOI:10.1038/nm1782
.
- G. Brinkmalm, W. Hong, Z. Wang, W. Liu, T. T. O'Malley, X. Sun, M. P. Frosch, D. J. Selkoe, E. Portelius, H. Zetterberg, K. Blennow and D. M. Walsh, Identification of Neurotoxic Cross-Linked Amyloid-β Dimers in the Alzheimer's Brain, Brain, 2019, 142(5), 1441–1457, DOI:10.1093/brain/awz066
.
- A. G. Kreutzer, C. M. T. Parrocha, S. Haerianardakani, G. Guaglianone, J. T. Nguyen, M. N. Diab, W. Yong, M. Perez-Rosendahl, E. Head and J. S. Nowick, Antibodies Raised Against an Aβ Oligomer Mimic Recognize Pathological Features in Alzheimer's Disease and Associated Amyloid-Disease Brain Tissue, bioRxiv, 2023, preprint, 2023.05.11.540404, DOI: DOI:10.1101/2023.05.11.540404.
- S. Ciudad, E. Puig, T. Botzanowski, M. Meigooni, A. S. Arango, J. Do, M. Mayzel, M. Bayoumi, S. Chaignepain, G. Maglia, S. Cianferani, V. Orekhov, E. Tajkhorshid, B. Bardiaux and N. Carulla, Aβ(1-42) Tetramer and Octamer Structures Reveal Edge Conductivity Pores as a Mechanism for Membrane Damage, Nat. Commun., 2020, 11(1), 1–14, DOI:10.1038/s41467-020-16566-1
.
- G. B. Marina, D. Kirkitadze, A. Lomakin, S. S. Vollers, G. B. Benedek and D. B. Teplow, Amyloid β-Protein (Aβ) Assembly: Aβ40 and Aβ42 Oligomerize through Distinct Pathways, Proc. Natl. Acad. Sci. U. S. A., 2003, 100(1), 330–335, DOI:10.1073/pnas.222681699
.
- R. Moons, L. L. Ilag and F. Sobott, Native Ion Mobility-Mass Spectrometry Reveals the Formation of β - Barrel Shaped Amyloid - β Hexamers in a Membrane-Mimicking Environment, J. Am. Chem. Soc., 2019, 141, 10440–10450, DOI:10.1021/jacs.9b04596
.
- E. N. Cline, M. A. Bicca, K. L. Viola and W. L. Klein, The Amyloid-β Oligomer Hypothesis: Beginning of the Third Decade, J. Alzheimer's Dis., 2018, 64, S567–S610, DOI:10.3233/JAD-179941
.
- I. Benilova, E. Karran and B. De Strooper, The Toxic Aβ Oligomer and Alzheimer's Disease: An Emperor in Need of Clothes, Nat. Neurosci., 2012, 15(3), 349–357, DOI:10.1038/nn.3028
.
- D. J. Selkoe and J. Hardy, The Amyloid Hypothesis of Alzheimer's Disease at 25 Years, EMBO Mol. Med., 2016, 8(6), 595–608, DOI:10.15252/emmm.201606210
.
- C. M. Dobson, T. P. J. Knowles and M. Vendruscolo, The Amyloid Phenomenon and Its Significance in Biology and Medicine, Cold Spring Harbor Perspect. Biol., 2020, 12(2), a033878, DOI:10.1101/cshperspect.a033878
.
- A. J. Dear, T. C. T. Michaels, G. Meisl, D. Klenerman, S. Wu, S. Perrett, S. Linse, C. M. Dobson and T. P. J. Knowles, Kinetic Diversity of Amyloid Oligomers, Proc. Natl. Acad. Sci. U. S. A., 2020, 117(22), 28–31, DOI:10.1073/pnas.1922267117
.
- T. C. T. Michaels, A. Šarić, S. Curk, K. Bernfur, P. Arosio, G. Meisl, A. J. Dear, S. I. A. Cohen, C. M. Dobson, M. Vendruscolo, S. Linse and T. P. J. Knowles, Dynamics of Oligomer Populations Formed during the Aggregation of Alzheimer's Aβ42 Peptide, Nat. Chem., 2020, 12(5), 445–451, DOI:10.1038/s41557-020-0452-1
.
- J. Jeon, W. M. Yau and R. Tycko, Early Events in Amyloid-β Self-Assembly Probed by Time-Resolved Solid State NMR and Light Scattering, Nat. Commun., 2023, 14, 2964, DOI:10.1038/s41467-023-38494-6
.
- T. C. T. Michaels, D. Qian, A. Šarić, M. Vendruscolo, S. Linse and T. P. J. Knowles, Amyloid Formation as a Protein Phase Transition, Nat. Rev. Phys., 2023, 5(7), 379–397, DOI:10.1038/s42254-023-00598-9
.
- K. Kulenkampff, M. W. Perez, P. Sormanni, J. Habchi and M. Vendruscolo, Quantifying Misfolded Protein Oligomers as Drug Targets and Biomarkers in Alzheimer and Parkinson Diseases, Nat. Rev. Chem, 2021, 5, 277–294, DOI:10.1038/s41570-021-00254-9
.
- S. De, D. R. Whiten, F. S. Ruggeri, C. Hughes, M. Rodrigues, D. I. Sideris, C. G. Taylor, F. A. Aprile, S. Muyldermans, T. P. J. Knowles, M. Vendruscolo, C. Bryant, K. Blennow, I. Skoog, S. Kern, H. Zetterberg and D. Klenerman, Soluble Aggregates Present in Cerebrospinal Fluid Change in Size and Mechanism of Toxicity during Alzheimer's Disease Progression, Acta Neuropathol. Commun., 2019, 7(1), 120, DOI:10.1186/s40478-019-0777-4
.
- T. D. Samdin, A. G. Kreutzer and J. S. Nowick, Exploring Amyloid Oligomers with Peptide Model Systems, Curr. Opin. Chem. Biol., 2021, 64, 106–115, DOI:10.1016/j.cbpa.2021.05.004
.
- L. Yu, R. Edalji, J. E. Harlan, T. F. Holzman, A. P. Lopez, B. Labkovsky, H. Hillen, S. Barghorn, U. Ebert, P. L. Richardson, L. Miesbauer, L. Solomon, D. Bartley, K. Walter, R. W. Johnson, P. J. Hajduk and E. T. Olejniczak, Structural Characterization of a Soluble Amyloid β-Peptide Oligomer, Biochemistry, 2009, 48(9), 1870–1877, DOI:10.1021/bi802046n
.
- W. Hoyer, C. Grönwall, A. Jonsson, S. Ståhl and T. Härd, Stabilization of a β-Hairpin in Monomeric Alzheimer's Amyloid-β Peptide Inhibits Amyloid Formation, Proc. Natl. Acad. Sci. U. S. A., 2008, 105(13), 5099–5104, DOI:10.1073/pnas.0711731105
.
- A. Sandberg, L. M. Luheshi, S. Söllvander, T. Pereira de Barros, B. Macao, T. P. J. Knowles, H. Biverstål, C. Lendel, F. Ekholm-Petterson, A. Dubnovitsky, L. Lannfelt, C. M. Dobson and T. Härd, Stabilization of Neurotoxic Alzheimer Amyloid-β Oligomers by Protein Engineering, Proc. Natl. Acad. Sci. U.S.A., 2010, 107(35), 15595–15600, DOI:10.1073/pnas.1001740107
.
- C. Lendel, M. Bjerring, A. Dubnovitsky, R. T. Kelly, A. Filippov, O. N. Antzutkin, N. C. Nielsen and T. Härd, A Hexameric Peptide Barrel as Building Block of Amyloid-β Protofibrils, Angew. Chem., Int. Ed., 2014, 53, 12756–12760, DOI:10.1002/anie.201406357
.
- A. G. Kreutzer and J. S. Nowick, Elucidating the Structures of Amyloid Oligomers with Macrocyclic β-Hairpin Peptides: Insights into Alzheimer's Disease and Other Amyloid Diseases, Acc. Chem. Res., 2018, 51, 706–718, DOI:10.1021/acs.accounts.7b00554
.
- U. Ghosh, K. R. Thurber, W.-M. Yau and R. Tycko, Molecular Structure of a Prevalent Amyloid-β Fibril Polymorph from Alzheimer's Disease Brain Tissue, Proc. Natl. Acad. Sci. U.S.A., 2021, 118(4), e2023089118, DOI:10.1073/pnas.2023089118
.
- A. G. Kreutzer and J. S. Nowick, Elucidating the Structures of Amyloid Oligomers with Macrocyclic β-Hairpin Peptides: Insights into Alzheimer's Disease and Other Amyloid Diseases, Acc. Chem. Res., 2018, 51(3), 706–718, DOI:10.1021/acs.accounts.7b00554
.
- R. K. Spencer, H. Li and J. S. Nowick, X-Ray Crystallographic Structures of Trimers and Higher-Order Oligomeric Assemblies of a Peptide Derived from Aβ17-36, J. Am. Chem. Soc., 2014, 136(15), 5595–5598, DOI:10.1021/ja5017409
.
- A. G. Kreutzer, I. L. Hamza, R. K. Spencer and J. S. Nowick, X-Ray Crystallographic Structures of a Trimer, Dodecamer, and Annular Pore Formed by an Aβ17-36 β-Hairpin, J. Am. Chem. Soc., 2016, 138(13), 4634–4642, DOI:10.1021/jacs.6b01332
.
- A. G. Kreutzer, S. Yoo, R. K. Spencer and J. S. Nowick, Stabilization, Assembly, and Toxicity of Trimers Derived from Aβ, J. Am. Chem. Soc., 2017, 139(2), 966–975, DOI:10.1021/jacs.6b11748
.
- A. G. Kreutzer, R. K. Spencer, K. J. McKnelly, S. Yoo, I. L. Hamza, P. J. Salveson and J. S. Nowick, A Hexamer of a Peptide Derived from Aβ16-36, Biochemistry, 2017, 56(45), 6061–6071, DOI:10.1021/acs.biochem.7b00831
.
- P. J. Salveson, R. K. Spencer, A. G. Kreutzer and J. S. Nowick, X-Ray Crystallographic Structure of a Compact Dodecamer from a Peptide Derived from Aβ16–36, Org. Lett., 2017, 19(13), 3462–3465, DOI:10.1021/acs.orglett.7b01445
.
- T. D. Samdin, M. Wierzbicki, A. G. Kreutzer, W. J. Howitz, M. Valenzuela, A. Smith, V. Sahrai, N. L. Truex, M. Klun and J. S. Nowick, Effects of N-Terminal Residues on the Assembly of Constrained β-Hairpin Peptides Derived from Aβ, J. Am. Chem. Soc., 2020, 142(26), 11593–11601, DOI:10.1021/jacs.0c05186
.
- S. Haerianardakani, A. G. Kreutzer, P. J. Salveson, T. D. Samdin, G. E. Guaglianone and J. S. Nowick, Phenylalanine Mutation to Cyclohexylalanine Facilitates Triangular Trimer Formation by β-Hairpins Derived from Aβ, J. Am. Chem. Soc., 2020, 142, 20708–20716, DOI:10.1021/jacs.0c09281
.
- A. G. Kreutzer, T. D. Samdin, G. Guaglianone, R. K. Spencer and J. S. Nowick, X-Ray Crystallography Reveals Parallel and Antiparallel β-Sheet Dimers of a β-Hairpin Derived from Aβ16-36 That Assemble to Form Different Tetramers, ACS Chem. Neurosci., 2020, 11(15), 2340–2347, DOI:10.1021/acschemneuro.0c00290
.
- K. J. McKnelly, A. G. Kreutzer, W. J. Howitz, K. Haduong, S. Yoo, C. Hart and J. S. Nowick, Effects of Familial Alzheimer's Disease Mutations on the Assembly of a β-Hairpin Peptide Derived from Aβ16−36, Biochemistry, 2022, 61, 446–454, DOI:10.1021/acs.biochem.1c00664
.
- C. L. Masters, G. Simms, N. A. Weinman, G. Multhaup, B. L. McDonald and K. Beyreuther, Amyloid Plaque Core Protein in Alzheimer Disease and Down Syndrome, Proc. Natl. Acad. Sci. U.S.A., 1985, 82(12), 4245–4249, DOI:10.1073/pnas.82.12.4245
.
- D. J. Selkoe, C. R. Abraham, M. B. Podlisny and L. K. Duffy, Isolation of Low‐molecular‐weight Proteins from Amyloid Plaque Fibers in Alzheimer's Disease, J. Neurochem., 1986, 46(6), 1820–1834, DOI:10.1111/j.1471-4159.1986.tb08501.x
.
- M. B. Podlisny, B. L. Ostaszewski, S. L. Squazzo, E. H. Koo, R. E. Rydell, D. B. Teplow and D. J. Selkoe, Aggregation of Secreted Amyloid β-Protein into Sodium Dodecyl Sulfate-Stable Oligomers in Cell Culture, J. Biol. Chem., 1995, 9564–9570, DOI:10.1074/jbc.270.16.9564
.
- S. Yoo, S. Zhang, A. G. Kreutzer and J. S. Nowick, An Efficient Method for the Expression and Purification of Aβ(M1-42), Biochemistry, 2018, 57(26), 3861–3866, DOI:10.1021/acs.biochem.8b00393
.
- A. D. Watt, K. A. Perez, A. Rembach, N. A. Sherrat, L. W. Hung, T. Johanssen, C. A. McLean, W. M. Kok, C. A. Hutton, M. Fodero-Tavoletti, C. L. Masters, V. L. Villemagne and K. J. Barnham, Oligomers, Fact or Artefact? SDS-PAGE Induces Dimerization of β-Amyloid in Human Brain Samples, Acta Neuropathol., 2013, 125(4), 549–564, DOI:10.1007/s00401-013-1083-z
.
- S. Zhang, S. Yoo, D. T. Snyder, B. B. Katz, A. Henrickson, B. Demeler, V. H. Wysocki, A. G. Kreutzer and J. S. Nowick, A Disulfide-Stabilized Aβ That Forms Dimers but Does Not Form Fibrils, Biochemistry, 2022, 61(4), 252–264, DOI:10.1021/acs.biochem.1c00739
.
- A. Awasthi, Y. Matsunaga and T. Yamada, Amyloid-Beta Causes Apoptosis of Neuronal Cells via Caspase Cascade, Which Can Be Prevented by Amyloid-Beta-Derived Short Peptides, Exp. Neurol., 2005, 196(2), 282–289, DOI:10.1016/j.expneurol.2005.08.001
.
- S. Söllvander, E. Nikitidou, R. Brolin, L. Söderberg, D. Sehlin, L. Lannfelt and A. Erlandsson, Accumulation of Amyloid-β by Astrocytes Result in Enlarged Endosomes and Microvesicle-Induced Apoptosis of Neurons, Mol. Neurodegener., 2016, 11(1), 1–19, DOI:10.1186/s13024-016-0098-z
.
- M. F. M. Sciacca, S. A. Kotler, J. R. Brender, J. Chen, D. K. Lee and A. Ramamoorthy, Two-Step Mechanism of Membrane Disruption by Aβ through Membrane Fragmentation and Pore Formation, Biophys. J., 2012, 103(4), 702–710, DOI:10.1016/j.bpj.2012.06.045
.
- K. J. Korshavn, C. Satriano, Y. Lin, R. Zhang, M. Dulchavsky, A. Bhunia, M. I. Ivanova, Y. H. Lee, C. La Rosa, M. H. Lim and A. Ramamoorthy, Reduced Lipid Bilayer Thickness Regulates the Aggregation and Cytotoxicity of Amyloid-β, J. Biol. Chem., 2017, 292(11), 4638–4650, DOI:10.1074/jbc.M116.764092
.
- S. A. Kotler, P. Walsh, J. R. Brender and A. Ramamoorthy, Differences between Amyloid-β Aggregation in Solution and on the Membrane: Insights into Elucidation of the Mechanistic Details of Alzheimer's Disease, Chem. Soc. Rev., 2014, 43(19), 6692–6700, 10.1039/c3cs60431d
.
- M. Serra-Batiste, M. Ninot-Pedrosa, M. Bayoumi, M. Gairí, G. Maglia and N. Carulla, Aβ42 Assembles into Specific β-Barrel Pore-Forming Oligomers in Membrane-Mimicking Environments, Proc. Natl. Acad. Sci. U. S. A., 2016, 113(39), 10866–10871, DOI:10.1073/pnas.1605104113
.
- N. Kandel, T. Zheng, Q. Huo and S. A. Tatulian, Membrane Binding and Pore Formation by a Cytotoxic Fragment of Amyloid β Peptide, J. Phys. Chem. B, 2017, 121(45), 10293–10305, DOI:10.1021/acs.jpcb.7b07002
.
- Y. Sun, A. Kakinen, X. Wan, N. Moriarty, C. P. J. Hunt, Y. Li, N. Andrikopoulos, A. Nandakumar, T. P. Davis, C. L. Parish, Y. Song, P. C. Ke and F. Ding, Spontaneous Formation of β-Sheet Nano-Barrels during the Early Aggregation of Alzheimer's Amyloid Beta, Nano Today, 2021, 38, 101125, DOI:10.1016/j.nantod.2021.101125
.
- P. H. Nguyen, A. Ramamoorthy, B. R. Sahoo, J. Zheng, P. Faller, J. E. Straub, L. Dominguez, J.-E. Shea, N. V. Dokholyan, A. De Simone, B. Ma, R. Nussinov, S. Najafi, S. T. Ngo, A. Loquet, M. Chiricotto, P. Ganguly, J. McCarty, M. S. Li, C. Hall, Y. Wang, Y. Miller, S. Melchionna, B. Habenstein, S. Timr, J. Chen, B. Hnath, B. Strodel, R. Kayed, S. Lesné, G. Wei, F. Sterpone, A. J. Doig and P. Derreumaux, Amyloid Oligomers: A Joint Experimental/Computational Perspective on Alzheimer’s Disease, Parkinson’s Disease, Type II Diabetes and Amyotrophic Lateral Sclerosis, Chem. Rev., 2021, 121(4), 2545–2647, DOI:10.1021/acs.chemrev.0c01122
.
- D. Mrdenovic, I. S. Pieta, R. Nowakowski, W. Kutner, J. Lipkowski and P. Pieta, Amyloid β Interaction with Model Cell Membranes – What Are the Toxicity-Defining Properties of Amyloid β?, Int. J. Biol. Macromol., 2022, 200(January), 520–531, DOI:10.1016/j.ijbiomac.2022.01.117
.
- W. B. Kauffman, T. Fuselier, J. He and W. C. Wimley, Mechanism Matters: A Taxonomy of Cell Penetrating Peptides, Trends Biochem. Sci., 2015, 40(12), 749–764, DOI:10.1016/j.tibs.2015.10.004.Mechanism
.
- S. Guha, J. Ghimire, E. Wu and W. C. Wimley, Mechanistic Landscape of Membrane-Permeabilizing Peptides, Chem. Rev., 2019, 119(119), 6040–6085, DOI:10.1021/acs.chemrev.8b00520
.
- G. Guaglianone, B. Torrado, Y. F. Lin, M. C. Watkins, V. H. Wysocki, E. Gratton and J. S. Nowick, Elucidating the Oligomerization and Cellular Interactions of a Trimer Derived from Aβ through Fluorescence and Mass Spectrometric Studies, ACS Chem. Neurosci., 2022, 13(16), 2473–2482, DOI:10.1021/acschemneuro.2c00313
.
- A. G. Kreutzer, G. Guaglianone, S. Yoo, C. M. T. Parrocha, S. M. Ruttenberg, R. J. Malonis, K. Tong, Y.-F. Lin, J. T. Nguyen, W. J. Howitz, M. N. Diab, I. L. Hamza, J. R. Lai, V. H. Wysocki and J. S. Nowick, Probing Differences Among Aβ Oligomers with Two Triangular Trimers Derived from Aβ, Proc. Natl. Acad. Sci. U. S. A., 2023, 120, 1–12, DOI:10.1073/pnas
.
- S. Jo, T. Kim, V. G. Iyer and W. Im, CHARMM-GUI: A Web-Based Graphical User Interface for CHARMM, J. Comput. Chem., 2008, 29(11), 1859–1865, DOI:10.1002/jcc
.
- E. L. Wu, X. Cheng, S. Jo, H. Rui, K. C. Song, E. M. Dávila-Contreras, Y. Qi, J. Lee, V. Monje-Galvan, R. M. Venable, J. B. Klauda and W. Im, CHARMM-GUI Membrane Builder toward Realistic Biological Membrane Simulations, J. Comput. Chem., 2014, 35(27), 1997–2004, DOI:10.1002/jcc.23702
.
- J. C. Phillips, D. J. Hardy, J. D. C. Maia, J. E. Stone, J. V. Ribeiro, R. C. Bernardi, R. Buch, G. Fiorin, J. Hénin, W. Jiang, R. McGreevy, M. C. R. Melo, B. K. Radak, R. D. Skeel, A. Singharoy, Y. Wang, B. Roux, A. Aksimentiev, Z. Luthey-Schulten, L. V. Kalé, K. Schulten, C. Chipot and E. Tajkhorshid, Scalable Molecular Dynamics on CPU and GPU Architectures with NAMD, J. Chem. Phys., 2020, 153(4), 044130, DOI:10.1063/5.0014475
.
- J. Huang, S. Rauscher, G. Nawrocki, T. Ran, M. Feig, B. L. De Groot, H. Grubmüller and A. D. MacKerell, CHARMM36m: An Improved Force Field for Folded and Intrinsically Disordered Proteins, Nat. Methods, 2016, 14(1), 71–73, DOI:10.1038/nmeth.4067
.
- J. B. Klauda, R. M. Venable, J. A. Freites, J. W. O'Connor, D. J. Tobias, C. Mondragon-Ramirez, I. Vorobyov, A. D. MacKerell and R. W. Pastor, Update of the CHARMM All-Atom Additive Force Field for Lipids: Validation on Six Lipid Types, J. Phys. Chem. B, 2010, 114(23), 7830–7843, DOI:10.1021/jp101759q
.
- W. Humphrey, A. Dalke and K. Schulten, VMD: Visual Molecular Dynamics, J. Mol. Graphics, 1996, 14(October 1995), 33–38 CrossRef CAS PubMed
.
- A. Hatami, S. Monjazeb, S. Milton and C. G. Glabe, Familial Alzheimer's Disease Mutations within the Amyloid Precursor Protein Alter the Aggregation and Conformation of the Amyloid-β Peptide, J. Biol. Chem., 2017, 292(8), 3172–3185, DOI:10.1074/jbc.M116.755264
.
- A. Laganowsky, C. Liu, M. R. Sawaya, J. P. Whitelegge, J. Park, M. Zhao, A. Pensalfini, A. B. Soriaga, M. Landau, P. K. Teng, D. Cascio, C. Glabe and D. Eisenberg, Atomic View of a Toxic Amyloid Small Oligomer, Science, 2012, 191(March), 1228–1232 CrossRef PubMed
.
- P. N. Cheng, C. Liu, M. Zhao, D. Eisenberg and J. S. Nowick, Amyloid β-Sheet Mimics That Antagonize Protein Aggregation and Reduce Amyloid Toxicity, Nat. Chem., 2012, 4(11), 927–933, DOI:10.1038/nchem.1433
.
- C. Liu, M. Zhao, L. Jiang, P. N. Cheng, J. Park, M. R. Sawaya, A. Pensalfini, D. Gou, A. J. Berk, C. G. Glabe, J. Nowick and D. Eisenberg, Out-of-Register β-Sheets Suggest a Pathway to Toxic Amyloid Aggregates, Proc. Natl. Acad. Sci. U. S. A., 2012, 109(51), 20913–20918, DOI:10.1073/pnas.1218792109
.
- T. D. Do, N. E. Lapointe, R. Nelson, P. Krotee, E. Y. Hayden, B. Ulrich, S. Quan, S. C. Feinstein, D. B. Teplow, D. Eisenberg, J. E. Shea and M. T. Bowers, Amyloid β-Protein C-Terminal Fragments: Formation of Cylindrins and β-Barrels, J. Am. Chem. Soc., 2016, 138(2), 549–557, DOI:10.1021/jacs.5b09536
.
Footnote |
| † Electronic supplementary information (ESI) available: Procedures for the synthesis of peptides 1a–i and 2a–d, SDS-PAGE and silver staining, circular dichroism studies, molecular dynamics studies, and caspase 3/7 activation assay; X-ray crystallographic data collection, processing, and refinement (PDF); characterization data for peptide 2a were deposited in the Protein Data Bank (PDB) with code 7RTZ (CIF). See DOI: https://doi.org/10.1039/d3sc05185d |
| This journal is © The Royal Society of Chemistry 2024 |