Open Access Article
Open Access ArticleIntegrative chemoproteomics reveals anticancer mechanisms of silver(I) targeting the proteasome regulatory complex†
Xiaojian
Shao
 ab,
Fangrong
Xing
ab,
Fangrong
Xing
 ab,
Yiwei
Zhang
ab,
Yiwei
Zhang
 ab,
Chun-Nam
Lok
ab,
Chun-Nam
Lok
 ab and
Chi-Ming
Che
ab and
Chi-Ming
Che
 *ab
*ab
aDepartment of Chemistry and State Key Laboratory of Synthetic Chemistry, The University of Hong Kong, Pokfulam Road, Hong Kong, P. R. China. E-mail: cmche@hku.hk
bLaboratory for Synthetic Chemistry and Chemical Biology Limited, Units 1503-1511, 15/F., Building 17W, Hong Kong Science Park, New Territories, Hong Kong, P. R. China
First published on 28th February 2024
Abstract
Silver compounds have favorable properties as promising anticancer drug candidates, such as low side effects, anti-inflammatory properties, and high potential to overcome drug resistance. However, the exact mechanism by which Ag(I) confers anticancer activity remains unclear, which hinders further development of anticancer applications of silver compounds. Here, we combine thermal proteome profiling, cysteine profiling, and ubiquitome profiling to study the molecular mechanisms of silver(I) complexes supported by non-toxic thiourea (TU) ligands. Through the formation of AgTU complexes, TU ligands deliver Ag+ ions to cancer cells and tumour xenografts to elicit inhibitory potency. Our chemical proteomics studies show that AgTU acts on the ubiquitin-proteasome system (UPS) and disrupts protein homeostasis, which has been identified as a main anticancer mechanism. Specifically, Ag+ ions are released from AgTU in the cellular environment, directly target the 19S proteasome regulatory complex, and may oxidize its cysteine residues, thereby inhibiting proteasomal activity and accumulating ubiquitinated proteins. After AgTU treatment, proteasome subunits are massively ubiquitinated and aberrantly aggregated, leading to impaired protein homeostasis and paraptotic death of cancer cells. This work reveals the unique anticancer mechanism of Ag(I) targeting the 19S proteasome regulatory complex and opens up new avenues for optimizing silver-based anticancer efficacy.
Introduction
Platinum-based chemotherapy drugs represented by cisplatin are widely used in cancer treatment.1 However, clinically used anticancer platinum chemotherapy drugs have shortcomings such as adverse side effects and drug resistance, which has triggered interest in the development of other metal-based anticancer drugs.2,3 Compared to platinum compounds, silver complexes have rarely been developed for anticancer therapy but have received widespread attention due to their potent antibacterial and antiviral properties against human infections and their low toxicity to humans.4–8 Although there are sporadic reports on the anticancer effects of silver complexes, most studies are still in the academic trial stage, and few anticancer silver complexes have been approved to enter clinical trials.9 Furthermore, the anticancer activity conferred by Ag(I) complexes remains unclear. Nevertheless, the relatively low toxicity and anti-inflammatory properties of silver complexes in humans make them valuable candidates for developing anticancer therapies.Previous studies have shown that silver complexes and silver nanoparticles have unique anticancer mechanisms.10 After entering cancer cells, Ag+ ions are released from the complex, bind to proteins through coordination or are reduced to Ag0 and aggregate into nanoparticles.11,12 Ag+ ions have been shown to preferentially target the sulfur atoms of cysteine and methionine.13 Ag+ ions are isoelectronic with Cu+ ions and may compete with Cu+ ions to coordinate with copper transporters and deregulate copper homeostasis.14 Possible protein targets for Ag+ ions include cysteine-rich proteins and methionine-containing proteins, such as metallothionein, copper chaperones, high-affinity copper transporter CTR1, α-synuclein, and prion proteins.15 Because silver complexes tend to anchor to enzymes, they can denature oncogenic proteins, thereby exerting anticancer activity.6
The molecular mechanism of anticancer silver complexes is closely related to their ancillary ligands, which determines their water solubility, lipophilicity, redox ability, and Ag+ ion release rate, leading to their different anti-tumour activities.16,17 Ligands with strong σ-donors, such as N-heterocyclic carbenes (NHC), stabilize Ag+ ions and slow their release under cellular conditions.18,19 Many ligands of anticancer silver complexes are derived from anti-inflammatory drugs, including aspirin, naproxen, nimesulide, and disulfiram.20–22 Previously, we reported that AgTU, a silver complex supported by the thiourea (TU) ligand, inhibits the inflammatory TNF-α-stimulated response by targeting cysteine of IκB kinase.23 This complex has been shown to inhibit cancer cell viability at low micromolar concentrations.24 However, the molecular mechanism of AgTU's action in cancer cells has not been studied from a systematic perspective.
In this study, we applied chemical proteomics techniques, including thermal proteome profiling (TPP) and activity-based protein profiling (ABPP), combined with ubiquitome analysis to study protein targets related to the anticancer effects of AgTU. Our results suggest that Ag+ ions released from AgTU act directly on proteasome regulatory subunits, potentially inducing cysteine oxidation and thereby preventing the proteasome from recruiting ubiquitinated proteins for subsequent degradation. AgTU treatment was found to deregulate the ubiquitin-proteasome system (UPS), increase ubiquitinated proteins, and disrupt protein homeostasis in cancer cells. AgTU treatment also impedes the autophagy process and promotes proteasome ubiquitination and aggregation. Disruption of protein homeostasis ultimately triggers paraptosis-like cancer cell death, suggesting the potential of silver(I) complexes containing non-toxic ligands as effective anticancer therapies by targeting 19S proteasome regulatory particles.
Results
AgTU inhibits cancer cell viability in vitro and suppresses tumour growth in vivo
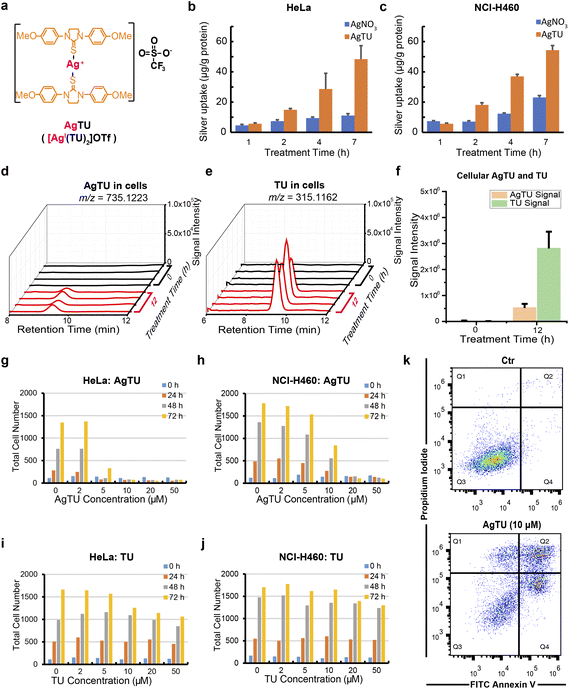
The synthesis of AgTU and TU ligands has been detailed in our previous paper.23 Herein, we measured the silver content in AgTU-treated cancer cells using ICP-MS and compared it with AgNO3-treated cancer cells (Fig. 1a). The results showed that the silver content in AgTU-treated HeLa and NCI-H460 cells was more than 3 times that of AgNO3-treated cells, indicating that TU ligands facilitate the entry of Ag+ into cancer cells (Fig. 1b and c). The silver content in AgTU-treated HeLa cells gradually increased over time, reaching 5.7 and 48.4 μg per g protein at 1 h and 7 h post-treatment, respectively (Fig. 1b). An ammonium bicarbonate aqueous solution of AgTU (10 mM) was stable for 72 h without any changes observed (Fig. S1a†), while metabolite analysis of AgTU in cells showed a significant increase in the free TU ligand within 12 h (Fig. 1d–f). This suggests that AgTU demetallates and releases Ag+ ions in the cellular environment. These results also show that TU ligands act as ionophores to facilitate the uptake of AgTU by cancer cells, while Ag+ ions are the reaction center and are released in the cellular environment.We studied the anti-proliferative and cytotoxic effects of AgTU on various cancer cell lines using MTT assay (Table 1) and high-content microscopy analysis. AgTU was found to inhibit the viability of a broad spectrum of cancer cell lines with half maximal inhibitory concentration values (IC50) ranging from 3.3 to 19.1 μM (Table 1). AgTU exerted antiproliferative effects and cytotoxic effects in HeLa and NCI-H460 cancer cells, as determined by high-content live-dead cell assays (Fig. 1g, h, S1b and c†). In contrast, TU ligand treatment did not have any discernible effect on the viability of a panel of cancer cells (Fig. 1i, j, S1d and e†). These findings show that TU acts as a nontoxic ligand for Ag+ ions, accelerating silver-mediated anticancer activity. Flow cytometry analysis through annexin V and propidium iodide cell staining revealed AgTU-induced apoptosis in cancer cells (Fig. 1k). By directly observing AgTU-induced cell morphological changes in HeLa cells, we confirmed that AgTU induces cell death with paraptosis-like phenotypes,25 including the formation of vacuoles in the cytoplasm, disruption of mitochondria and endoplasmic reticulum structures, and deregulation of mitochondrial respiration (Fig. S2†). AgTU treatment inhibited maximal respiratory capacity as shown in oxygen consumption assay but did not affect glycolysis in extracellular acidification assay (Fig. S2c and d†), revealing that mitochondrial bioenergetics is particularly sensitive to AgTU treatment.
| HeLa (cervical cancer) | NCI-H460 (lung cancer) | A549 (lung cancer) | HCT-116 (colon cancer) | MDA-MB-231 (breast cancer) | MHCC97L (liver cancer) | |
|---|---|---|---|---|---|---|
| AgTU | 3.3 ± 0.8 | 12.2 ± 1.3 | 19.1 ± 1.3 | 4.6 ± 0.6 | 16.1 ± 1.0 | 6.3 ± 1.5 |
| TU | >100 | >100 | >100 | >100 | >100 | >100 |
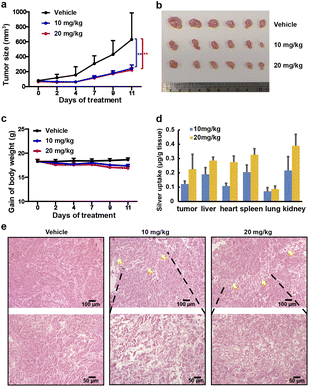
The in vivo anti-tumour effects of AgTU were further investigated in nude mice bearing xenografts derived from human HeLa cervical cancer cells. Nude mice were subcutaneously inoculated with HeLa cells and treated with vehicle control, 10, and 20 mg kg−1 of AgTU, respectively. AgTU treatment every 2–3 days for 11 days inhibited tumour growth, reducing tumour volume by 58.1% and 62.1%, respectively (Fig. 2a and b), and no mouse death or significant body weight loss was observed (Fig. 2c). Determination of the bio-distribution of silver in mice using ICP-MS showed that the accumulation of silver in tumours and organs increased with AgTU concentration (Fig. 2d). At higher doses (20 mg kg−1), silver accumulation in tumours was comparable to that in the liver, heart, spleen, and kidneys, and much higher than accumulation in the lungs. AgTU treatment resulted in tumour necrosis after 11 days, while no significant toxicity to the liver and kidneys was observed using H&E staining and blood biochemistry assays (Fig. 2e and S3†), showing the efficacy and specificity of AgTU against tumours.
AgTU targets the 19S proteasome regulatory complex
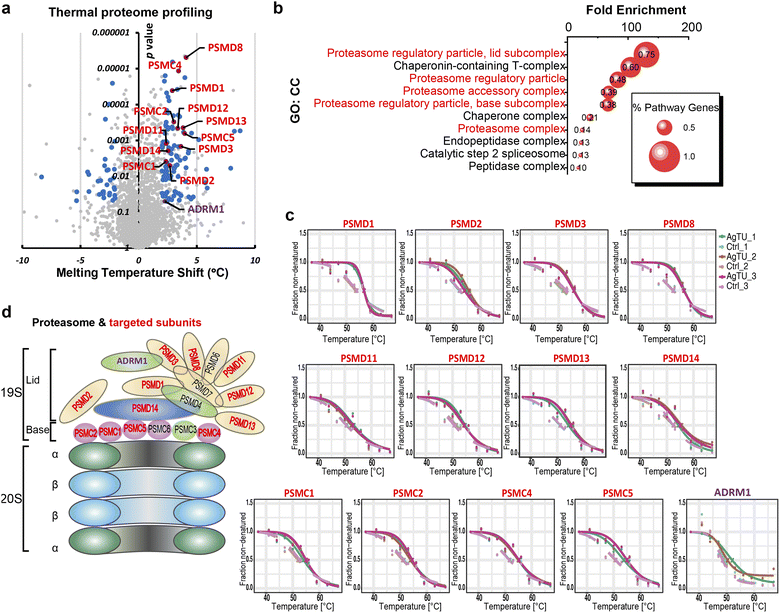
To reveal the anticancer mechanism of AgTU, we utilized thermal proteome profiling (TPP) to identify its potential protein targets. The premise of this strategy is that binding of a compound to a protein results in a change in the thermal stability of the protein.26 In HeLa and NCI-H460 cells, the thermal stability of many subunits of the proteasome 19S complex increased after AgTU treatment (Fig. 3a and S4a†). In HeLa cells, gene ontology (GO) enrichment analysis showed that the most enriched protein targets were the proteasome 19S regulatory subunits (Fig. 3b–d), including its base (PSMC1, PSMC2, PSMC4, and PSMC5) and lid (PSMD1, PSMD2, PSMD3, PSMD8, PSMD11, PSMD12, PSMD13, PSMD14, and ADRM1). In NCI-H460 cells, AgTU treatment resulted in the thermal stabilization of several 19S proteasome subunits, namely, ADRM1, PSMC3, PSMC6, PSMD2, PSMD3, PSMD11, and PSMD13 (Fig. S4b†). The results suggest that AgTU may act on the proteasome regulatory particles, specifically by targeting its ubiquitin receptor (ADRM1) and deubiquitinase (PSMD14). Given that these proteins are involved in recruiting ubiquitinated substrates for deubiquitylation and degradation,27,28 their targeting implies that AgTU may deregulate the ubiquitin-proteasome system (UPS).Cysteines of the 19S proteasome complex are partially oxidized by AgTU
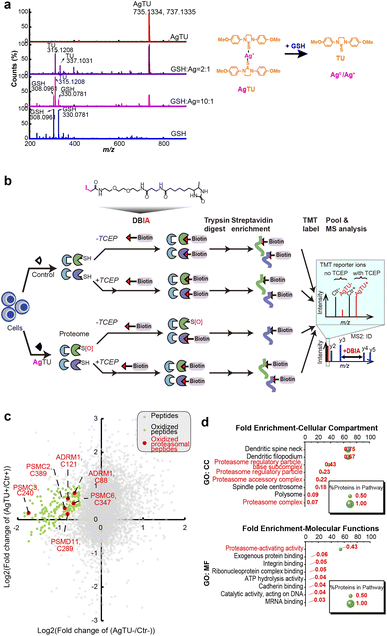
Although AgTU was found to release its TU ligands in the presence of glutathione (GSH) or N-acetyl cysteine (NAC), no interaction between the released Ag+ ions and GSH or NAC was detected in the MS analysis (Fig. 4a and S5†). In-gel fluorescence assays with an iodoacetamide-based fluorescent probe showed reduced cysteine labeling in proteins extracted from the AgTU-treated HeLa cells (Fig. S6†). This may be due to direct targeting of cysteine by AgTU or AgTU-induced cysteine oxidation to form disulfide, sulfenic acid, or sulfinic acid groups.29 Considering the possibility that AgTU induces cysteine oxidation, we then added the reductant tris(2-carboxyethyl)phosphine (TCEP) to reverse the oxidation.30 AgTU treatment did not reduce the cysteine labeling in the presence of TCEP (Fig. S6†). These findings suggest that Ag+ ions released from AgTU may induce protein cysteine oxidation.We analyzed the AgTU-induced cysteine changes from the perspective of the entire proteome (cysteinome), aiming to provide a broader perspective on Ag+-induced protein oxidation and functional changes. We performed activity-based protein profiling (ABPP) by using the cysteine-targeting probe DBIA (desthiobiotinpolyethylene oxide iodoacetamide) combined with the reducing agent TCEP to identify AgTU-mediated cysteinome oxidation (Fig. 4b).31 Our cysteinome study identified 17![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 746 peptides, of which 50% (8630 peptides) were grafted with DBIA and TMT. The high proportion of DBIA labeling confirmed the success of cysteine probe ligation, and the identification of over 8000 peptides was comparable to recent cysteine profiling studies.32,33 We used TMT MS2 ions to quantify the signal intensity of the modified peptides and compared the signal intensities of the control and AgTU groups in the absence and presence of TCEP respectively.
746 peptides, of which 50% (8630 peptides) were grafted with DBIA and TMT. The high proportion of DBIA labeling confirmed the success of cysteine probe ligation, and the identification of over 8000 peptides was comparable to recent cysteine profiling studies.32,33 We used TMT MS2 ions to quantify the signal intensity of the modified peptides and compared the signal intensities of the control and AgTU groups in the absence and presence of TCEP respectively.
By plotting the fold change of cysteine-containing peptides (Fig. 4c), we identified 190 peptides whose DBIA-tagged cysteine was reduced by more than 30% after AgTU treatment. Addition of TCEP also reversed this change, indicating that the AgTU-induced cysteine changes were possibly attributed to cysteine oxidation. Gene ontology (GO) enrichment analysis showed that the most enriched oxidized proteins were proteasome 19S regulatory subunits, including ADRM1 (C88 and C121), PSMC2 (C389), PSMC3 (C240), PSMC6 (C347), and PSMD11 (C289) (Fig. 4d). We then also performed cysteine profiling by using label-free quantification (Fig. S7†), which suggested that ADRM1 (C80 and C88) and PSMC6 (C347) may be oxidized. Cysteine residue C88 of ADRM1 is located in its ubiquitin receptor domain and is responsible for binding ubiquitinated proteins for subsequent deubiquitination and degradation.34 Taken together, the cysteine profiling data support the TPP data, showing that AgTU targets the proteasome system and may induce oxidation of cysteine in the 19S proteasome complex.
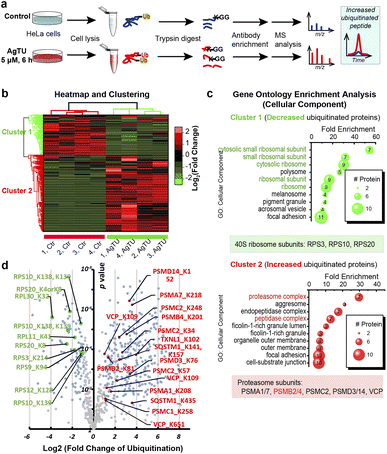
AgTU induces ubiquitination and aggregation of proteasomes
Given that AgTU targets the 19S proteasome complex, we then employed immunoprecipitation and proteomic analysis to study its impact on overall global protein ubiquitination. First, total proteins were extracted from the control and AgTU treatment groups and digested into peptides, allowing the identification and quantification of ubiquitinated proteins with lysine (K) residues converted to a K-ε-GG motif (Fig. 5a). Peptides with the K-ε-GG motif were then pulled down using antibodies immobilized on beads and subsequently analyzed by mass spectrometry. Approximately 800 ubiquitinated peptides were identified, of which about 150 peptides were more than 2-fold more ubiquitinated (Fig. 5b).Heatmap visualization and GO clustering analysis revealed that peptides from proteasomes were highly enriched in cluster 2 (upregulation) and ribosomal proteins were highly enriched in cluster 1 (downregulation) (Fig. 5c and d). Proteasome proteins with upregulated ubiquitination levels include PSMA1 (K208), PSMA7 (K27), PSMC2 (K57), PSMD3 (K76), PSMD14 (K152), and VCP (K112). Given that proteasomes are primarily polyubiquitinated,35 whereas ribosomes may undergo monoubiquitination,36,37 ubiquitome analysis suggests that AgTU may have an impact on both monoubiquitination and polyubiquitination levels. The ubiquitome results are consistent with the above TPP and ABPP data, suggesting that AgTU engages and oxidizes the 19S proteasome, alters its stability, and induces accumulation of ubiquitinated proteins, including the accumulation of polyubiquitinated proteasome subunits.
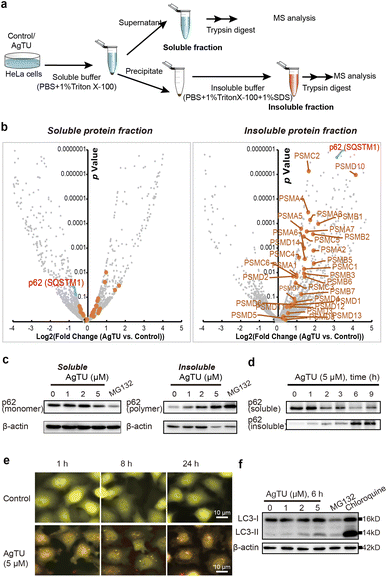
Polyubiquitinated proteins are either directly subject to proteasomal degradation or are degraded by autophagy upon aggregation.35 We studied AgTU-mediated protein aggregation by analyzing the composition of soluble and insoluble protein fractions (Fig. 6a). Proteomics analysis of the insoluble fractions revealed a general increase in proteasome subunits in AgTU-treated cells, indicating AgTU-induced proteasome aggregation (Fig. 6b). In contrast, there was no significant increase of proteasome subunits in the soluble fraction (Fig. 6b). We observed a significant decrease in the soluble fraction of p62/sequestosome-1 (SQSTM1) and a corresponding increase in the insoluble fraction (Fig. 6c, d, S8a and b†). SQSTM1/p62 is an essential protein that promotes the recruitment of ubiquitinated proteins, and its polymerization is a prerequisite for the recruitment of ubiquitinated protein aggregates and autophagy proteins, such as LC3 to autophagosome.38,39 The levels of the p62 monomer and aggregates in the soluble and insoluble fractions can be used as an indicator of protein aggregation.40 After treating HeLa cells with AgTU, we observed a dose- and time-dependent decrease in p62 monomer levels and a corresponding increase in its aggregation degree (Fig. 6c and d). The upregulation of p62 aggregates suggests that AgTU impairs protein homeostasis, leading to accumulation of ubiquitinated proteins and specific dysfunction of the proteasomes and autophagosomes.
Damaged proteasomes have been shown to undergo ubiquitination and autophagic degradation;41 here we examined the effects of AgTU on autophagy. Using the fluorescent reporter tfLC3 (LC3 tagged with GFP and mRFP in tandem),42 we observed AgTU-induced accumulation of autophagosomes (yellow puncta) but the presence of a small number of autolysosomes (red puncta) (Fig. 6e). AgTU treatment was also shown to increase LC3-II levels (Fig. 6f and S8c†). The results show that AgTU impairs autophagic flux, which is associated with the accumulation of damaged proteasomes and ubiquitinated protein aggregates.
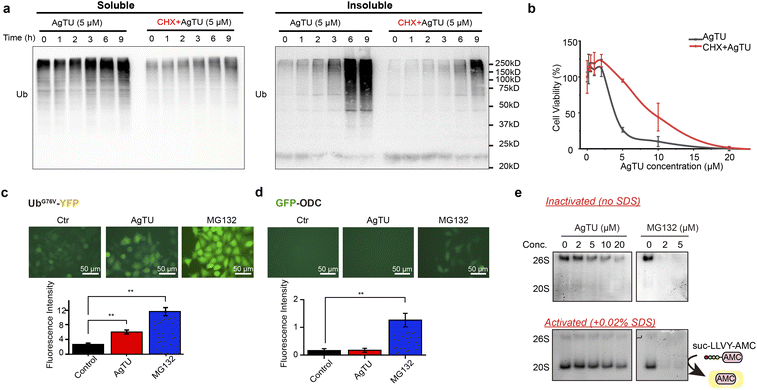
AgTU impairs proteasome activity
Consistent with the results of the proteasome targeting studies mentioned above, AgTU treatment significantly increased the levels of polyubiquitinated proteins in HeLa cancer cells (Fig. 7a and S9a†). Notably, AgTU mainly induces the accumulation of insoluble ubiquitinated proteins, leading to an increase in denatured ubiquitinated protein aggregates and suggesting disruption of protein homeostasis. Our data demonstrate that AgTU targets and inhibits the proteasome, leading to apoptosis- or paraptosis-like cell death, manifested by significant accumulation of cytoplasmic vacuoles, ER morphology changes, and cytoplasmic Ca2+ release (Fig. S2a and b†). We then pretreated cells with CHX before AgTU treatment, as previous reports showed that CHX can impair paraptosis and sometimes attenuate apoptosis.43 In addition, CHX has been shown to attenuate cytotoxicity induced by the proteasome inhibitors MG132 and bortezomib.44,45 Consistent with this, western blot assay showed that CHX impaired AgTU-induced accumulation of polyubiquitinated proteins, possibly due to proteasome inhibition (Fig. 7a). CHX also alleviated AgTU-induced cytotoxicity (Fig. 7b), further demonstrating that AgTU exerts anticancer activities by targeting the proteasome.AgTU-induced accumulation of ubiquitinated proteins was also demonstrated using HeLa cells containing the UbG76V-YFP protein reporter46 as a UPS degradation substrate (Fig. 7c). Accumulation of the UPS reporter protein is triggered by AgTU, albeit to a lesser extent than that by the proteasome inhibitor MG132, which targets the 20S protease activity. Furthermore, we utilized HeLa cells expressing fluorescent GFP-ODC protein47 as a reporter for ubiquitin-independent proteasome 20S protease activity (Fig. 7d). While MG132 induced significant accumulation of the reporter protein, AgTU did not. We also evaluated the effect of AgTU on cellular 26S and 20S proteasome activity by performing in-gel protease activity assay using the switch-on fluorescent proteasome substrate suc-LLVY-AMC (Fig. 7e and S9b†). The results showed that AgTU downregulated the 20S and 26S proteasome protease activities, but not to a different extent compared to MG132, which completely inhibited the activity through the target 20S complex (Fig. 7e). The effect of AgTU on 20S proteasome chymotrypsin-like peptidase activity was also examined using a homogeneous fluorometric assay of suc-LLVY-AMC hydrolysis (Fig. S10†). The results showed that AgTU could partially inhibit the proteasome activity, but not to the same extent as MG132, which completely inhibited the protease activity of the 20S proteasome. In summary, our results indicate that consistent with the TPP and cysteine profiling analysis described above, AgTU preferentially targets the 19S proteasome regulatory subunits rather than the 20S proteolytic core.
Discussion
It has been well documented for decades that silver complexes offer alternative anticancer drug candidates that circumvent anticancer drug resistance and are less cytotoxic to humans.48 However, the anticancer mechanism action of silver complexes needs to be studied from a systematic perspective to facilitate their optimization and development. In this work, we combine several chemoproteomic strategies to identify protein targets of silver complexes to elucidate silver-based anticancer mechanisms.By developing the silver thiourea complex AgTU and examining its cytotoxicity, our research shows that the coordination of silver ions with nontoxic TU ligands can promote the uptake of silver by cancer cells, making it more effective against cancer than silver nitrate (Fig. 1). The anticancer activity was significantly enhanced. In the cellular environment, AgTU was found to release Ag+ ions to exert its anticancer effects. AgTU demonstrated potent antiproliferative activity against a panel of cancer cell lines and anti-tumour activity in mice xenograft models (Fig. 2).
To reveal its anticancer mechanism, we combined several chemoproteomics strategies to resolve its protein targets. Thermal proteome profiling revealed that AgTU acts directly on many subunits of the 19S proteasome complex, which has a total of 18 subunits, 13 of which were identified as potential targets in HeLa cells (Fig. 3). The proteasome is a promising anticancer target because it is highly expressed in cancer cells and is responsible for clearing damaged proteins in cells.49,50 Therefore, proteasome inhibitors are considered to be valuable compounds for the treatment of various cancers.51,52 Many proteasome inhibitors are in clinical trials for anticancer therapy, and older drugs are being repurposed to evaluate their inhibitory effects against proteasome.53,54 Several proteasome inhibitors in clinical trials have shown broad anticancer properties and have different primary targets, including the targeting proteasome 20S protease and 19S proteasome-associated deubiquitinases, PSMD14, UCHL5, and USP14, as well as the ubiquitin receptors ADRM1 and PSMD4.47,55–58 In the proteasome, the 19S complex serves to recognize ubiquitin-tagged substrates, cleave their polyubiquitin chains, unfold the protein, and transfer it to the catalytic 20S in the core.53 Our study found that AgTU specifically acts on the 19S proteasome regulatory complex, and the components engaged by AgTU include PSMD14, which breaks down polyubiquitin chains, and ADRM1, which recruits ubiquitinated proteins. Directly targeting the 19S proteasome with the silver complex has the potential to be developed as a novel cancer therapy.
Silver(I) ions have a d10 electronic configuration, are considered “soft” Lewis acids, and exhibit strong reactivity toward cysteine.59,60 Cysteine residues often appear in the catalytic center of enzymes, affecting their activity and interactions with potential inhibitors.61–64 In addition, cysteine can be oxidized to form disulfide bonds, sulfenic acid, sulfinic acid, or sulfonic acid, greatly affecting protein structure and function.65 Previous studies have shown that targeting proteasome subunits can alter the structure and function of proteasome, leading to the accumulation of polyubiquitinated proteins and impairing protein homeostasis.28 Through cysteine profiling, we found that AgTU treatment induced changes in the oxidation state of the cysteinome sulphur atom. Since thermal proteome profiling (Fig. 3) and ubiquitome data (Fig. 5) indicate that AgTU targets proteasome regulatory complexes, we performed GO analysis of the cysteine profiling showing that the cysteine oxidized protein profile in AgTU-treated cells was enriched in proteasome proteins (Fig. 4c and d). AgTU may induce cysteine oxidation in vitro (Fig. 4a and S5†), so we focused on how AgTU treatment affects protein cysteine oxidation in the cysteine profiling analysis, especially on those proteasome subunits that contain reactive cysteines. AgTU targets cysteines of proteasome regulatory subunits, such as the C88 and C121 residues of ADRM1 (Fig. 4), both of which are located in the ubiquitin receptor domain and are responsible for recruiting ubiquitinated proteins for degradation. Previous studies reported that proteasome activity can be regulated through oxidation of proteasome subunits,66 and therefore oxidation of the ADRM1 cysteine may interfere with the degradation process. However, overall proteasome oxidation levels ranged from 49% to 74%, indicating that AgTU treatment induced mild oxidation. Several other cysteines in the 19S proteasome regulatory complex, including PSMC2 (C389), PSMC3 (C240), PSMC6 (C347), and PSMD11 (C289), are also mildly oxidized. Partial oxidation of cysteines in the 19S complex may deregulate proteasome functions, leading to aggregation of denatured proteins and accumulation of ubiquitinated proteins.
Since AgTU-induced cysteine oxidation of the 19S complex is expected to impair proteasome functions, we performed ubiquitome profiling in AgTU-treated cells. Conjugation of monoubiquitin and polyubiquitin to substrate proteins can modulate their subcellular localization, signal regulation, and protein levels and activities through the UPS and autophagy systems.67 Consistent with its targeting of the 19S proteasome complex, AgTU upregulates the levels of ubiquitinated and aggregated proteins. Further proteomic study of ubiquitinated proteins after AgTU treatment showed increased ubiquitination levels of proteasome subunits (Fig. 5), including 3 subunits from 19S regulatory particles, i.e., PSMC2 (K57), PSMD3 (K51), and PSMD14 (K152), as well as 2 subunits of the 20S core catalytic particle: PSMA1 (K208), and PSMA7 (K27). Furthermore, several reported monoubiquitinated sites of the ribosome, including RPS3 (K214), RPS10 (K138, K139), and RPS20 (K4, K8),36,37 were observed to be reduced upon AgTU treatment. While AgTU treatment may affect ubiquitinated protein levels, the identity of monoubiquitin versus polyubiquitin remains to be determined, as ubiquitinated protein pull-down experiments cannot distinguish the number of protein ubiquitin units.
Proteomics analysis of AgTU-induced protein aggregation confirmed an increase in proteasome aggregates after AgTU treatment (Fig. 6). Ubiquitinated proteasomes are known to be subject to autophagy-mediated degradation.35 In this work, AgTU impaired the autophagy process, which may be responsible for the accumulation of ubiquitinated proteasomes and their aggregates.
Analysis of proteasome protease activity showed that proteasome activity was deregulated after AgTU treatment (Fig. 7). In western blot analysis, accumulation of ubiquitinated proteins was also observed. Previous studies have shown that metal complexes, including compounds derived from silver, copper, gold, and rhodium, have the potential to target the proteasome and inhibit proteasome activity.22,68–70 Specifically, silver ions were found to inhibit the protease activity of the 20S proteasome and the deubiquitinase activity of the 19S proteasome, while the silver diethyldithiocarbamate complex mainly targets the deubiquitinase subunits of the 19S proteasome.22,71 Our studies demonstrate that AgTU has a distinct mode of targeting proteasome protease activity, leading to possible oxidation of proteasome subunits and an increase in ubiquitinated and aggregated proteasomes in cancer cells. It has been reported that impaired proteasome activity triggers paraptosis which is morphologically characterized by cytoplasmic vacuolation.72,73 Here, AgTU impairs proteasome activity, which is associated with increased protein aggregates and paraptosis in cancer cells. Proteomic analysis showed that AgTU inhibits proteasome activity by targeting and deregulating the 19S regulatory complex, which is different from the typical proteasome inhibitor MG132 that directly inhibits the proteasome proteolytic center of the 20S catalytic complex.
Conclusions
In summary, our study demonstrates that AgTU acts on the 19S proteasome complex to impair proteasome protease activity by participating in proteasome cysteine oxidation. Proteasome damage disrupts protein homeostasis in cancer cells, leading to accumulation of misfolded proteins as well as aggregated and ubiquitinated proteins, thereby triggering cancer cell death. Furthermore, our findings demonstrate that AgTU has potent anti-tumour effects in mouse models and cytotoxic effects against a panel of cancer cell lines. This article highlights the importance of integrating multiple chemoproteomics approaches to study the anticancer effects of silver-based compounds and provides a comprehensive strategy to elucidate their anticancer mechanisms.Ethical statement
All animal experiments were conducted under the guidelines approved by the Committee on the Use of Live Animals in Teaching and Research of the University of Hong Kong.Data availability
All proteome data are uploaded to ProteomeXchange Datasets (PXD042400, PXD043636, PXD048202, PXD042465, PXD042467). Part of experimental supporting data and procedures are available in the ESI and Supporting Data 1.†Author contributions
Chi-Ming Che: conceptualization, funding acquisition, investigation, methodology, supervision, writing – review & editing. Xiaojian Shao: conceptualization, data curation, investigation, methodology, software, writing – original draft, writing – review & editing. Fangrong Xing: investigation, methodology. Chun-Nam Lok: data curation, investigation, writing – review & editing. Yiwei Zhang: data curation, investigation, methodology.Conflicts of interest
There are no conflicts to declare.Acknowledgements
We acknowledge the funding support from the Laboratory for Synthetic Chemistry and Chemical Biology under the Health@InnoHK Program launched by the Innovation and Technology Commission, The Government of Hong Kong Special Administrative Region of the People's Republic of China, and the financial support from the State Key Laboratory of Synthetic Chemistry. We thank Dr Qian Zhao of The Hong Kong Polytechnic University for initial proteomics analysis. We also thank Dr Kwan Ming Ng for his comment on the mass spectrometry data analysis.References
- K. Peng, Y. Zheng, W. Xia and Z. W. Mao, Chem. Soc. Rev., 2023, 52, 2790–2832 RSC.
- L. Messori and A. Merlino, Coord. Chem. Rev., 2016, 315, 67–89 CrossRef CAS.
- X. Wang, X. Wang, X. Jin, N. Muhammad and Z. Guo, Chem. Rev., 2019, 119, 1138–1192 CrossRef CAS PubMed.
- C. N. Lok, T. Zou, J. J. Zhang, I. W. Lin and C. M. Che, Adv. Mater., 2014, 26, 5550–5557 CrossRef CAS PubMed.
- S. Medici, M. Peana, G. Crisponi, V. M. Nurchi, J. I. Lachowicz, M. Remelli and M. A. Zoroddu, Coord. Chem. Rev., 2016, 327–328, 349–359 CrossRef CAS.
- S. Medici, M. Peana, V. M. Nurchi and M. A. Zoroddu, J. Med. Chem., 2019, 62, 5923–5943 CrossRef CAS PubMed.
- M. Cao, S. Wang, J. H. Hu, B. H. Lu, Q. Y. Wang and S. Q. Zang, Adv. Sci., 2022, 9, e2103721 CrossRef PubMed.
- D. Panacek, L. Hochvaldova, A. Bakandritsos, T. Malina, M. Langer, J. Belza, J. Martincova, R. Vecerova, P. Lazar, K. Polakova, J. Kolarik, L. Valkova, M. Kolar, M. Otyepka, A. Panacek and R. Zboril, Adv. Sci., 2021, 8, 2003090 CrossRef CAS PubMed.
- Z. Xu, X. Zha, R. Ji, H. Zhao and S. Zhou, ACS Appl. Mater. Interfaces, 2023, 15, 13983–13992 CAS.
- L. Qin, P. Wang, Y. Guo, C. Chen and M. Liu, Adv. Sci., 2015, 2, 1500134 CrossRef PubMed.
- S. Tang and J. Zheng, Adv. Healthcare Mater., 2018, 7, e1701503 CrossRef PubMed.
- K. Kluska, J. Adamczyk and A. Krężel, Coord. Chem. Rev., 2018, 367, 18–64 CrossRef CAS.
- H. Kozlowski, S. Potocki, M. Remelli, M. Rowinska-Zyrek and D. Valensin, Coord. Chem. Rev., 2013, 257, 2625–2638 CrossRef CAS.
- L. Chen, J. Min and F. Wang, Signal Transduction Targeted Ther., 2022, 7, 378 CrossRef CAS PubMed.
- A. Hecel, P. Kolkowska, K. Krzywoszynska, A. Szebesczyk, M. Rowinska-Zyrek and H. Kozlowski, Curr. Med. Chem., 2019, 26, 624–647 CrossRef CAS PubMed.
- W. Liu and R. Gust, Chem. Soc. Rev., 2013, 42, 755–773 RSC.
- T. J. Siciliano, M. C. Deblock, K. M. Hindi, S. Durmus, M. J. Panzner, C. A. Tessier and W. J. Youngs, J. Organomet. Chem., 2011, 696, 1066–1071 CrossRef CAS.
- C. Hemmert, A. Fabie, A. Fabre, F. Benoit-Vical and H. Gornitzka, Eur. J. Med. Chem., 2013, 60, 64–75 CrossRef CAS PubMed.
- K. A. Ali, M. M. Abd-Elzaher and K. Mahmoud, Int. J. Med. Chem., 2013, 2013, 256836 Search PubMed.
- M. Poyraz, C. N. Banti, N. Kourkoumelis, V. Dokorou, M. J. Manos, M. Simčič, S. Golič-Grdadolnik, T. Mavromoustakos, A. D. Giannoulis, I. I. Verginadis, K. Charalabopoulos and S. K. Hadjikakou, Inorg. Chim. Acta, 2011, 375, 114–121 CrossRef CAS.
- C. N. Banti, A. D. Giannoulis, N. Kourkoumelis, A. M. Owczarzak, M. Poyraz, M. Kubicki, K. Charalabopoulos and S. K. Hadjikakou, Metallomics, 2012, 4, 545–560 CrossRef CAS PubMed.
- X. Chen, Q. Yang, J. Chen, P. Zhang, Q. Huang, X. Zhang, L. Yang, D. Xu, C. Zhao, X. Wang and J. Liu, Cell. Physiol. Biochem., 2018, 49, 780–797 CrossRef CAS PubMed.
- I. W. Lin, C. N. Lok, K. Yan and C. M. Che, Chem. Commun., 2013, 49, 3297–3299 RSC.
- K. Yan, C. N. Lok, K. Bierla and C. M. Che, Chem. Commun., 2010, 46, 7691–7693 RSC.
- F. Fontana, M. Raimondi, M. Marzagalli, A. Di Domizio and P. Limonta, Biochim. Biophys. Acta, Rev. Cancer, 2020, 1873, 188338 CrossRef CAS PubMed.
- H. Franken, T. Mathieson, D. Childs, G. M. Sweetman, T. Werner, I. Togel, C. Doce, S. Gade, M. Bantscheff, G. Drewes, F. B. Reinhard, W. Huber and M. M. Savitski, Nat. Protoc., 2015, 10, 1567–1593 CrossRef CAS PubMed.
- C. W. Liu and A. D. Jacobson, Trends Biochem. Sci., 2013, 38, 103–110 CrossRef CAS PubMed.
- D. J. Trader, S. Simanski and T. Kodadek, J. Am. Chem. Soc., 2015, 137, 6312–6319 CrossRef CAS PubMed.
- A. A. Saei, H. Gullberg, P. Sabatier, C. M. Beusch, K. Johansson, B. Lundgren, P. I. Arvidsson, E. S. J. Arner and R. A. Zubarev, Redox Biol., 2020, 32, 101491 CrossRef CAS PubMed.
- E. A. Kisty, J. A. Falco and E. Weerapana, Cell Chem. Biol., 2023, 30, 321–336 CrossRef CAS PubMed.
- E. K. Kemper, Y. Zhang, M. M. Dix and B. F. Cravatt, Nat. Methods, 2022, 19, 341–352 CrossRef CAS PubMed.
- M. Kuljanin, D. C. Mitchell, D. K. Schweppe, A. S. Gikandi, D. P. Nusinow, N. J. Bulloch, E. V. Vinogradova, D. L. Wilson, E. T. Kool, J. D. Mancias, B. F. Cravatt and S. P. Gygi, Nat. Biotechnol., 2021, 39, 630–641 CrossRef CAS PubMed.
- F. Yang, G. Jia, J. Guo, Y. Liu and C. Wang, J. Am. Chem. Soc., 2022, 144, 901–911 CrossRef CAS PubMed.
- P. Dickson, D. Abegg, E. Vinogradova, J. Takaya, H. An, S. Simanski, B. F. Cravatt, A. Adibekian and T. Kodadek, Cell Chem. Biol., 2020, 27, 1371–1382 CrossRef CAS PubMed.
- W. Li, P. He, Y. Huang, Y. F. Li, J. Lu, M. Li, H. Kurihara, Z. Luo, T. Meng, M. Onishi, C. Ma, L. Jiang, Y. Hu, Q. Gong, D. Zhu, Y. Xu, R. Liu, L. Liu, C. Yi, Y. Zhu, N. Ma, K. Okamoto, Z. Xie, J. Liu, R. R. He and D. Feng, Theranostics, 2021, 11, 222–256 CrossRef CAS PubMed.
- A. Garzia, S. M. Jafarnejad, C. Meyer, C. Chapat, T. Gogakos, P. Morozov, M. Amiri, M. Shapiro, H. Molina, T. Tuschl and N. Sonenberg, Nat. Commun., 2017, 8, 16056 CrossRef CAS PubMed.
- A. Garzia, C. Meyer and T. Tuschl, Cell Rep., 2021, 36, 109468 CrossRef CAS PubMed.
- B. Carroll, E. G. Otten, D. Manni, R. Stefanatos, F. M. Menzies, G. R. Smith, D. Jurk, N. Kenneth, S. Wilkinson, J. F. Passos, J. Attems, E. A. Veal, E. Teyssou, D. Seilhean, S. Millecamps, E. L. Eskelinen, A. K. Bronowska, D. C. Rubinsztein, A. Sanz and V. I. Korolchuk, Nat. Commun., 2018, 9, 256 CrossRef PubMed.
- Y. Zhang, S. R. Mun, J. F. Linares, J. Ahn, C. G. Towers, C. H. Ji, B. E. Fitzwalter, M. R. Holden, W. Mi, X. Shi, J. Moscat, A. Thorburn, M. T. Diaz-Meco, Y. T. Kwon and T. G. Kutateladze, Nat. Commun., 2018, 9, 4373 CrossRef PubMed.
- D. Sun, R. Wu, J. Zheng, P. Li and L. Yu, Cell Res., 2018, 28, 405–415 CrossRef CAS PubMed.
- D. Hoeller and I. Dikic, Proc. Natl. Acad. Sci. U.S.A., 2016, 113, 13266–13268 CrossRef CAS PubMed.
- S. Kimura, T. Noda and T. Yoshimori, Autophagy, 2007, 3, 452–460 CrossRef CAS PubMed.
- D. Nedungadi, A. Binoy, N. Pandurangan, B. G. Nair and N. Mishra, Cell Biol. Int., 2021, 45, 164–176 CrossRef CAS PubMed.
- J. Lavie, H. De Belvalet, S. Sonon, A. M. Ion, E. Dumon, S. Melser, D. Lacombe, J. W. Dupuy, C. Lalou and G. Benard, Cell Rep., 2018, 23, 2852–2863 CrossRef CAS PubMed.
- S. T. Nawrocki, J. S. Carew, K. Dunner Jr, L. H. Boise, P. J. Chiao, P. Huang, J. L. Abbruzzese and D. J. McConkey, Cancer Res., 2005, 65, 11510–11519 CrossRef CAS PubMed.
- V. Menendez-Benito, L. G. Verhoef, M. G. Masucci and N. P. Dantuma, Hum. Mol. Genet., 2005, 14, 2787–2799 CrossRef CAS PubMed.
- B. H. Lee, M. J. Lee, S. Park, D. C. Oh, S. Elsasser, P. C. Chen, C. Gartner, N. Dimova, J. Hanna, S. P. Gygi, S. M. Wilson, R. W. King and D. Finley, Nature, 2010, 467, 179–184 CrossRef CAS PubMed.
- Y. H. Lim, K. M. Tiemann, G. S. Heo, P. O. Wagers, Y. H. Rezenom, S. Y. Zhang, F. W. Zhang, W. J. Youngs, D. A. Hunstad and K. L. Wooley, ACS Nano, 2015, 9, 1995–2008 CrossRef CAS PubMed.
- H. H. Huang, I. D. Ferguson, A. M. Thornton, P. Bastola, C. Lam, Y. T. Lin, P. Choudhry, M. C. Mariano, M. D. Marcoulis, C. F. Teo, J. Malato, P. J. Phojanakong, T. G. Martin 3rd, J. L. Wolf, S. W. Wong, N. Shah, B. Hann, A. N. Brooks and A. P. Wiita, Nat. Commun., 2020, 11, 1931 CrossRef CAS PubMed.
- X. Lu, U. Nowicka, V. Sridharan, F. Liu, L. Randles, D. Hymel, M. Dyba, S. G. Tarasov, N. I. Tarasova, X. Z. Zhao, J. Hamazaki, S. Murata, T. R. Burke Jr and K. J. Walters, Nat. Commun., 2017, 8, 15540 CrossRef PubMed.
- R. K. Anchoori, B. Karanam, S. Peng, J. W. Wang, R. Jiang, T. Tanno, R. Z. Orlowski, W. Matsui, M. Zhao, M. A. Rudek, C. F. Hung, X. Chen, K. J. Walters and R. B. Roden, Cancer Cell, 2013, 24, 791–805 CrossRef CAS PubMed.
- A. L. Goldberg, J. Cell Biol., 2012, 199, 583–588 CrossRef CAS PubMed.
- J. E. Park, Z. Miller, Y. Jun, W. Lee and K. B. Kim, Transl. Res., 2018, 198, 1–16 CrossRef CAS PubMed.
- H. Yang, X. Chen, K. Li, H. Cheaito, Q. Yang, G. Wu, J. Liu and Q. P. Dou, Semin. Cancer Biol., 2021, 68, 105–122 CrossRef CAS PubMed.
- Q. P. Dou and J. A. Zonder, Curr. Cancer Drug Targets, 2014, 14, 517–536 CrossRef CAS PubMed.
- Y. Song, T. Du, A. Ray, K. Chauhan, M. Samur, N. Munshi, D. Chauhan and K. C. Anderson, Blood Cancer J., 2021, 11, 13 CrossRef PubMed.
- T. Du, Y. Song, A. Ray, X. Wan, Y. Yao, M. K. Samur, C. Shen, J. Penailillo, T. Sewastianik, Y. T. Tai, M. Fulciniti, N. C. Munshi, H. Wu, R. D. Carrasco, D. Chauhan and K. C. Anderson, Blood, 2023, 141, 2599–2614 CAS.
- J. A. Harrigan, X. Jacq, N. M. Martin and S. P. Jackson, Nat. Rev. Drug Discovery, 2018, 17, 57–78 CrossRef CAS PubMed.
- C. Gabbiani, F. Scaletti, L. Massai, E. Michelucci, M. A. Cinellu and L. Messori, Chem. Commun., 2012, 48, 11623–11625 RSC.
- L. V. Puchkova, M. Broggini, E. V. Polishchuk, E. Y. Ilyechova and R. S. Polishchuk, Nutrients, 2019, 11, 1364 CrossRef CAS PubMed.
- M. Marzi, M. K. Vakil, M. Bahmanyar and E. Zarenezhad, BioMed Res. Int., 2022, 2022, 7341493 Search PubMed.
- K. Senkane, E. V. Vinogradova, R. M. Suciu, V. M. Crowley, B. W. Zaro, J. M. Bradshaw, K. A. Brameld and B. F. Cravatt, Angew. Chem., Int. Ed., 2019, 58, 11385–11389 CrossRef CAS PubMed.
- X. Shao, F. Ji, Y. Wang, L. Zhu, Z. Zhang, X. Du, A. C. K. Chung, Y. Hong, Q. Zhao and Z. Cai, Anal. Chem., 2018, 90, 11092–11098 CrossRef CAS PubMed.
- C. M. Forester, Q. Zhao, N. J. Phillips, A. Urisman, R. J. Chalkley, J. A. Oses-Prieto, L. Zhang, D. Ruggero and A. L. Burlingame, Proc. Natl. Acad. Sci. U.S.A., 2018, 115, 2353–2358 CrossRef CAS PubMed.
- M. C. Montasell, P. Monge, S. Carmali, L. M. D. Loiola, D. G. Andersen, K. B. Lovschall, A. B. Sogaard, M. M. Kristensen, J. M. Putz and A. N. Zelikin, Nat. Commun., 2022, 13, 4861 CrossRef CAS PubMed.
- M. Qiu, J. Chen, X. Li and J. Zhuang, Int. J. Mol. Sci., 2022, 23, 12197 CrossRef CAS PubMed.
- Z. A. Ronai, Proc. Natl. Acad. Sci. U.S.A., 2016, 113, 8894–8896 CrossRef CAS PubMed.
- Z. Skrott, M. Mistrik, K. K. Andersen, S. Friis, D. Majera, J. Gursky, T. Ozdian, J. Bartkova, Z. Turi, P. Moudry, M. Kraus, M. Michalova, J. Vaclavkova, P. Dzubak, I. Vrobel, P. Pouckova, J. Sedlacek, A. Miklovicova, A. Kutt, J. Li, J. Mattova, C. Driessen, Q. P. Dou, J. Olsen, M. Hajduch, B. Cvek, R. J. Deshaies and J. Bartek, Nature, 2017, 552, 194–199 CrossRef CAS PubMed.
- F.-M. Siu, I. W.-S. Lin, K. Yan, C.-N. Lok, K.-H. Low, T. Y.-C. Leung, T.-L. Lam and C.-M. Che, Chem. Sci., 2012, 3, 1785–1793 RSC.
- N. N. Liu, X. F. Li, H. B. Huang, C. Zhao, S. Y. Liao, C. S. Yang, S. T. Liu, W. B. Song, X. Y. Lu, X. Y. Lan, X. Chen, S. G. Yi, L. Xu, L. L. Jiang, C. G. Zhao, X. X. Dong, P. Zhou, S. J. Li, S. Q. Wang, X. P. Shi, P. Q. Dou, X. J. Wang and J. B. Liu, Oncotarget, 2014, 5, 5453–5471 CrossRef PubMed.
- A. Filippi, M. Zancani, E. Petrussa and E. Braidot, J. Plant Physiol., 2019, 233, 42–51 CrossRef CAS PubMed.
- D. Lee, I. Y. Kim, S. Saha and K. S. Choi, Pharmacol. Ther., 2016, 162, 120–133 CrossRef CAS PubMed.
- H. J. Lee, D. M. Lee, M. J. Seo, H. C. Kang, S. K. Kwon and K. S. Choi, Int. J. Mol. Sci., 2022, 23, 2648 CrossRef CAS PubMed.
Footnote |
| † Electronic supplementary information (ESI) available: Instrumentation, experimental procedures, synthesis and characterization data, details of biological studies, and supplementary figures. See DOI: https://doi.org/10.1039/d3sc04834a |
| This journal is © The Royal Society of Chemistry 2024 |