Open Access Article
Open Access ArticleThe intricate link between membrane lipid structure and composition and membrane structural properties in bacterial membranes
Tzong-Hsien
Lee
 a,
Patrick
Charchar
a,
Patrick
Charchar
 b,
Frances
Separovic
b,
Frances
Separovic
 c,
Gavin E.
Reid
c,
Gavin E.
Reid
 cd,
Irene
Yarovsky
cd,
Irene
Yarovsky
 b and
Marie-Isabel
Aguilar
b and
Marie-Isabel
Aguilar
 *a
*a
aDepartment of Biochemistry and Molecular Biology, Monash University, Clayton, VIC 3800, Australia. E-mail: mibel.aguilar@monash.edu
bSchool of Engineering, RMIT University, Melbourne, Victoria 3001, Australia
cSchool of Chemistry, Bio21 Molecular Science and Biotechnology Institute, University of Melbourne, VIC 3010, Australia
dDepartment of Biochemistry and Pharmacology, University of Melbourne, Parkville, VIC 3010, Australia
First published on 31st January 2024
Abstract
It is now evident that the cell manipulates lipid composition to regulate different processes such as membrane protein insertion, assembly and function. Moreover, changes in membrane structure and properties, lipid homeostasis during growth and differentiation with associated changes in cell size and shape, and responses to external stress have been related to drug resistance across mammalian species and a range of microorganisms. While it is well known that the biomembrane is a fluid self-assembled nanostructure, the link between the lipid components and the structural properties of the lipid bilayer are not well understood. This perspective aims to address this topic with a view to a more detailed understanding of the factors that regulate bilayer structure and flexibility. We describe a selection of recent studies that address the dynamic nature of bacterial lipid diversity and membrane properties in response to stress conditions. This emerging area has important implications for a broad range of cellular processes and may open new avenues of drug design for selective cell targeting.
1. Introduction
Cells in all living organisms are surrounded by a membrane only a few nanometers in thickness. This biomembrane is vital to cell function and the organisation of individual lipids into different structural and physical domains is highly controlled by complex regulatory processes. Advances in high-resolution mass spectrometry technologies for lipidomics and combinatory approaches are beginning to reveal the extraordinary chemical diversity of lipid components in eukaryotic and bacterial membranes.1–6 However, the way in which the myriad of membrane lipids are marshalled together to create a functional biomembrane is poorly understood and represents an enormous challenge.An enduring scientific question with regard to the properties of cell membranes is how the highly complex assortment of membrane lipids, and their tendency to self-organise and segregate into domains of different compositions and properties, determine the constantly changing physical properties that make up the lipid bilayer. Also, why does such diversity exist when most membrane functions can be reconstituted using one or few molecularly defined lipids? The answers to these questions are essential to improve our understanding of the biochemical and biophysical principles that allow the cell to preserve each membrane function within living cells.
It is now evident that the cell manipulates lipid composition to regulate different processes, such as membrane protein insertion, assembly and function.7,8 Although the membrane plays an important protective barrier function, irreversible changes to the integrity of the membrane after exposure to stress can result in the loss of electrochemical gradients across the membrane and lead to cell death.
Moreover, changes in membrane composition, structure, properties occur during growth and differentiation and in response to external stress. These changes are often associated with changes in cell size and shape, and have been related to drug resistance across mammalian species and a range of microorganisms.9–12 Therefore, the significance of characterizing the impact of compositional changes in lipid species on (1) the physical properties and (2) the spatiotemporal structural organization of membranes is enormous. The impact of these changes is even more far-reaching when one considers these changes in different cellular states and the response to external stimuli such as antimicrobial drugs.
The lipid code used by the cell to curate a membrane to perform a specific function remains as a last frontier of science (compared with the nucleotide code for gene structure and the amino acid code for protein structure). While lipidomics is revealing the lipid composition in unprecedented detail,7,13 new techniques are required to allow us to measure the assembly, structure, biophysical properties and biomolecular interactions for complex mixtures of membrane lipids. These multiparametric composition-property characterizations are now being employed, with the ultimate goal to establish a comprehensive all-encompassing model describing the role that lipid composition has on the dynamic structural and mechanical properties and biological activities of membranes.
Membrane lipids display remarkable structural diversity, driven by factors such as a wide variety of chain lengths and headgroups, a multitude of oxidative, reductive, substitutional and ring-forming biochemical transformations, as well as modification with sugar residues and other functional groups of different biosynthetic origin. In addition to the structural diversity of the headgroups and backbone of lipid molecules, the molecular diversity of membrane lipids is much more complex when the different length, degree of saturation, and other structural modifications of the acyl chains are considered.
The compositional difference in lipid chemical structures underpins the functional differences between cell types and growth phases. The lateral interaction between lipids leads to the formation of domains that range between nano- to micrometer in size and this topographical heterogeneity is finely tuned by the bilayer composition.14–17 This fine tuning of the lipid layers also differs between the outer and inner bilayer leaflets. However, little is known about the physical properties of bilayer domains, how they are formed from different lipid compositions, and the compositional differences between bilayer leaflets.
A number of biophysical approaches have been used to explore the nanostructure characteristics of model membranes and cell surfaces and to monitor the structural changes of the membrane bilayer during interaction with peptides and proteins. Optical biosensors, such as surface plasmon resonance spectroscopy (SPR) and dual polarisation interferometry (DPI), have been applied to explore the changes in membrane order and structure during molecule–membrane interactions.18,19 This has allowed the spotlight of cell interactions to be shared by the biomolecule and the cell membrane, whereas previously the focus of membrane-mediated research has been on the interacting molecule and largely ignored the membrane bilayer as an interacting partner.
Finally, in order to be able to manipulate membrane-mediated processes, such as signaling, intracellular drug delivery, cell–material interactions, and to understand the evolution of resistance to cytolytic drugs, we need to have a much more detailed understanding of the factors that regulate the bilayer structure and flexibility. This perspective aims to address this challenge. We describe a selection of recent studies that address the dynamic nature of membranes with a focus on bacterial lipid diversity and membrane properties in response to stress conditions. The intent is to provide a timely contribution to this emerging area which has important implications for a broad range of cellular processes and may open up new avenues of drug design for selective cell targeting.
2. Bacterial membrane lipid profiles
2.1 The main classes of membrane lipids
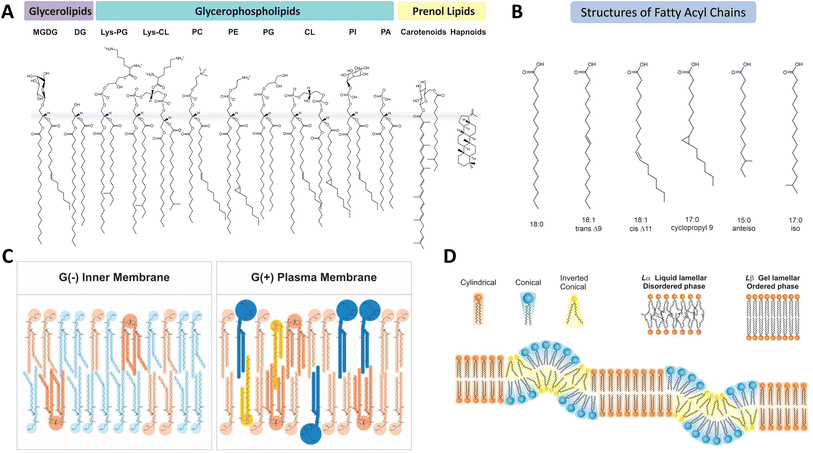
The main classes of lipids in bacterial membranes include glycerophospholipids, glycerolipids and prenol lipids (Fig. 1) with glycerophospholipids being the most abundant. In general, the bacterial membrane contains a mixture of the negatively charged phosphatidylglycerol (PG) and cardiolipin (CL) and zwitterionic phosphatidylethanolamine (PE). While PEs are major phospholipids in Gram-negative (G(−)) bacteria and Gram-positive (G(+)) Enterobacteriaceae and bacilli, PE is not found in G(+) staphylococci, streptococci and enterococci.5,20PE is synthesized via decarboxylation of the precursor PS catalysed by a membrane associated phosphatidylserine decarboxylase (PSD) on the cytoplasmic side of the membrane, which results in an asymmetric distribution of PE in the membrane. Conversion of PS to PE is very fast and efficient considering that the PS is only transiently present in the membrane and is less than 0.1% of the total lipids.21 Accumulation of PS in the membrane can cause growth arrest in E. coli mutants.22E. coli mutant strains unable to synthesise PE adapt by increasing the levels of PG and CL. In addition, these strains require the presence of 10–50 mM Ca2+, Mg2+ or Sr2+, most likely to ion pair with the higher proportion of negatively charged PG and CL in the membrane.23 It is also possible that the divalent ions and CL play a role in the formation of non-lamellar phases replacing the normal function of PE. The mutant strains lacking PE grow 2–3 times slower and become filamentous with multiple genomes and cytolysis quickly occurred without the divalent cations in the culture conditions.24E. coli cells that lack PE progressively change from a filamentous to rod shape by gradually increasing the PE content in the membrane from near zero to 75%.25 Thus the rod shape is associated with the bilayer asymmetry of PE, predominantly in the cytoplasmic leaflet of the inner membrane. Redistribution of PE also influences the distribution of other lipids between the leaflets and the asymmetric transmembrane distribution of PE and CL are tightly regulated to control the membrane order.
PGs are the dominant anionic phospholipid in bacterial membranes, representing 20–25% of total phospholipids in most G(−) bacteria, where PG is restricted to the inner membrane. In G(+) bacteria, PGs can be as high as 60% and are the major phospholipid in addition to lysyl-phosphatidylglycerol, CL and glucosyl-diacylglycerol.2,3 In E. coli membranes, the proportion of the anionic phospholipids to the zwitterionic PE is tightly regulated to balance the membrane surface charge. The negatively charged surface is important in controlling protein-membrane interactions, such as electrostatic interactions with peripheral DnaA proteins and initiating DNA replication, and in mediating the binding of cationic antimicrobial peptides (AMPs).26–31 The high abundance of PGs underlines their importance in modifying membrane properties. For example, systematic biophysical studies of PE/PG mixtures used as natural bacterial membrane mimics showed that PGs decrease the protrusions of PE headgroup into the water phase and restrict the PE headgroup motions along the bilayer normal.32 PGs are important in stabilising the membrane by preventing lipid desorption and decreasing membrane permeability. E. coli mutants lacking PG and CL in the membrane are still viable at temperatures less than 40 °C33 and the functions of PG in a E. faecalis mutant with reduced membrane PG can be compensated by diglucosyl-diacylglycerol lipids.34
CLs are synthesised through the condensation of two molecules of PG by cardiolipin synthases ClsA/B and transfer of a phosphatidyl moiety from PE to PG by ClsC in E. coli, while a stress inducible cls1 and constitutive cls2 genes are expressed in S. aureus for CL synthesis.21 The formation of distinct domains rich in CLs at cell poles and septa are critical during cell division.16
Aminoacyl-PGs are commonly present in the membranes of G(+) bacteria, notably Staphylococcus, Bacillus, Clostridium, Lactobacillus, Listeria, and Streptococcus.26,35 Aminoacyl-PGs are synthesised via esterification of the glycerol headgroup of PG with lysine, alanine and less commonly arginine and ornithine. Aminoacyl-tRNAs are used as amino acid donors catalysed by the enzyme multiple peptide resistance factor (MprF).36 The optimal level of aminoacyl-PGs is regulated by the balance between the aminoacylation and the hydrolysis, which is mediated by the aminoacyl-PG hydrolases in various bacteria. For example, an aminoacyl-PG hydrolase, AhyD, catalysed the hydrolysis of Lys-PG and Ala-PG in the membrane of E. faecium.37 Thus, aminoacyl-PG synthases and hydrolases may act in concert to fine tune aminoacyl-PG levels towards adaptation under changing environmental conditions. The cationic Lys-PGs are the most widely studied in S. aureus, which modulate the membrane negative charges that affect the spectrum of antimicrobial resistance.35 An increased proportion of Lys-PG in S. aureus membranes correlates with resistance to host defensive peptides and membrane-active antibiotics and is related to changes in the membrane interfacial charge and lipid order, which lower AMP binding and membrane insertion.36,38,39
2.2 The structural diversity of membrane lipids
To date, 1862 different glycerophospholipids have been characterised in E. coli based on the Escherichia coli metabolome database (ECMDB).44 The questions we need to address are: (1) which lipids are essential for membrane function; (2) how many structurally distinctive lipids are essential for membrane function; and (3) since the elimination or dramatic changes in a specific lipid level can result in irreversible destabilisation and potentially increase permeability, how tolerant are membranes to changes in lipid composition while maintaining their structure and function without affecting growth? In order to answer these questions, the impact of the head group and the fatty acid chain on membrane properties must first be considered.The proportion of different membrane lipids are fine-tuned for the optimal surface charge in maintaining membrane potential and protein activities. Membrane lipid compositions are under constant surveillance for membrane homeostasis.21,45 Several regulatory pathways constantly monitor and respond to perturbations in chemical structure and compositions of lipids and the subsequent changes in the physical properties of the membrane as a strategy to adapt to changing growth conditions and stress.46 For example, the asymmetric distribution of zwitterionic, anionic and cationic headgroups between the inner and outer membrane leaflet are highly controlled for the optimum surface electrostatic charge to maintain proper protein topologies and assembly.25,47,48 The charged head groups are also important in the selectivity of AMPs for targeting of membranes.
In addition to the diversity of phospholipid head groups in bacterial membranes, the structural diversity of phospholipid acyl chains further increases the complexity of membrane lipids. Structures of lipid acyl chain (Fig. 1B) are highly diverse in length, number and position of double bonds, and branched or cyclic configurations. While technical challenges remain in assessing errors in lipid identification, profiling rare lipid species, quantitation of lipid abundance, sample preparations and asymmetric distribution of lipids between leaflets and lateral domains, the intricate processes by which bacteria constantly modify their lipid membrane are beginning to be understood.
The asymmetric distribution of lipids in the outer membrane (OM) of G(−) has been extensively characterised.47,49–51 In general, it has been shown that the outer leaflet of the OM is enriched with highly negatively charged lipopolysaccharide (LPS) while the inner leaflet is composed of mainly glycerophospholipids. The modifications in the structures and composition of LPS also correlated to the changes in the physical properties such as lower the negative surface charge and increasing lipid packing order as the resistance mechanisms to antibiotics and AMPs. In contrast, the functional roles of lipids with predominantly PE in the inner leaflet of OM remains to be explored for their compositional changes in OM stabilisation.
Methods to precisely quantify the asymmetry distribution of lipids in membrane are limited and most methods have been applied to mammalian cells. Various enzymatic, chemical and mechanical treatments have been developed to remove the thick peptidoglycan layer and OM in G(+) and G(−) bacteria, respectively. Ideally, transbilayer asymmetry of lipids should be characterised without labelling although it remains a challenge to examine the lipid components in each membrane leaflet using high-resolution mass spectrometry without labelling. Specific chemical, fluorescence and enzymatic labels are, therefore, used to label mainly PE and CL in bacterial membranes.17,25
The asymmetric distribution of phospholipids in the IM of G(−) bacteria has been characterised using inside-out vesicles (ISOv) prepared from E. coli and Yersinia pseudotuberculosis.25 The localisation and dynamics of PE were characterised by the different amounts of PE in the ISOv that were labelled by either the membrane permeable 1,5-difluoro-2,4-dinitrobenzene (DFDNB) or the membrane impermeable 2,4,6-trinitrobenzenesulfonic acid (TNBS). 75% of PE was mainly localised in the cytoplasmic side of IM in rod-shaped cells while the opposite distribution (75% of PE in the periplasmic leaflet of IM) of PE in E. coli filamentous cells. The redistribution of PE in different stages also influences the distribution of other lipids between the leaflets and regulates lipid order of the bilayer. The bilayer asymmetry is thus tightly controlled and adjusts the physical properties for optimal growth.
Characterizing the global lipid compositions in bacterial membrane are still a main focus in membrane research. Resolving the dynamic spatial distribution of different lipid species of individual lipid molecules in the membrane remains a challenge. Modern super-resolution microscopy combined with specific lipid labelling and complementary modelling approaches would provide more information on the effect of heterogeneous lipid domains on the physical properties and function of membranes.
3. Fundamental membrane properties
Bacterial cells respond to perturbations in membrane structure by various molecular machineries evolved to synthesize new lipids and modify existing membrane lipids.5,52 This regulatory control is critical to remodeling the membrane structures and modifying properties that are potentially detrimental to membrane function. However, understanding the intricate relationship whereby the physical properties of a membrane are modulated by the lipid composition and vice versa is a key challenge to understanding the drivers behind the remodeling. It is extraordinary that cells can actively change the membrane lipid composition to maintain optimal physical properties of the membrane so as to regulate cell size and shape during growth. Consequently, cells constantly respond to membrane destabilization by changing their lipid composition and redistribution of lipids both laterally and transversely.How can these changes in membrane structure be measured and which physical parameters provide the most useful insight into membrane remodeling? Several structural and physical properties have been either defined conceptually or measured by various techniques and are listed in Fig. 2. These parameters can be categorised according to whether they are a fundamental property that defines a static or a dynamic structure. The static properties include thickness, surface charge and intrinsic curvature. Properties that describe a dynamic structure all relate to the concept of membrane “fluidity” and include molecular packing or order, rigidity and stiffness. Specific parameters that can be experimentally determined include rate and amplitude of lipid motion, orientational order parameter in 2H and 13C-NMR, viscosity and Young's modulus, bending stiffness and lateral pressure.
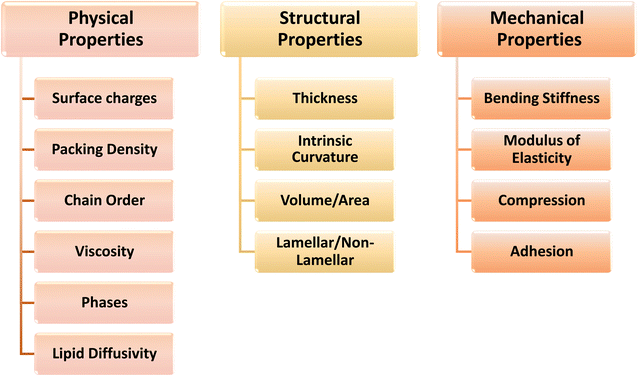 | ||
| Fig. 2 The interrelated physical, structural and mechanical properties of bulk membranes and various parameters for each membrane property. | ||
All these parameters are related to the properties of the individual constituent lipids, the dynamic structure of which can be described by a wide range of diffusion processes, including rotational, translational, short-range, long-range and flip-flop diffusion, which all contribute to the lateral pressure profile of the cell membrane and are modulated in response to various extrinsic environmental factors (see Fig. 2 and 1D). These properties must also be considered together with the lipid saturation index (ratio of SFA to UFA), the cis–trans configuration, number and position of lipid unsaturation sites, the lipid head groups and specific non-phospholipid contents, such as sterols, hopanoids, carotenoids and glycolipids.6,8,53–55
The adaptation of bacterial membranes to changes in extrinsic parameters, such as temperature, hydration, nutrition level, osmosis and chemicals (Fig. 3), involves fine-tuning of these membrane properties through various mechanisms that include synthesis, modification and degradation of lipids and fatty acids and head-group-specific acyl chain remodeling in the membrane lipidome.7 Membranes can adapt to extrinsic stress conditions with changes in only a small fraction of lipid head group species, whereby only a small fraction of the lipidome is required for adaptive membranes while the majority of lipid species do not vary substantially under any stress conditions. Finally, the growth phase and rate at which any of these changes take place within the cell growth cycle can vary depending on the nature of the environmental stress factor.
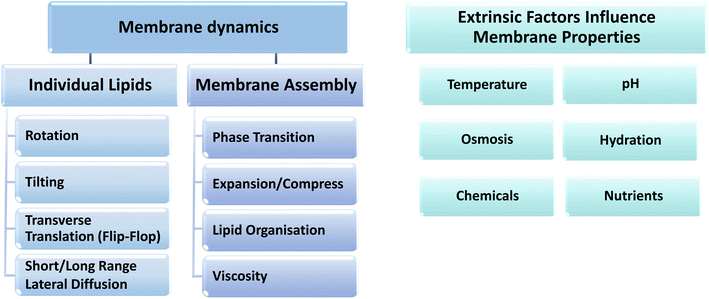 | ||
| Fig. 3 The dynamic parameters for individual lipids and bulk membrane assembly, and the different extrinsic properties that can influence the dynamic behavior of membranes. | ||
Biophysical techniques such as light scattering (X-ray, neutron), nuclear magnetic resonance, electron paramagnetic resonance, fluorescence, infrared and Raman spectroscopies, differential scanning calorimetry, fluorescence, electron, cryo-electronic and atomic force microscopies, ellipsometry and dual polarization interferometry, optical tweezers and micropipette aspiration have widely been used to explore the dynamic physical properties of model and native bacterial membranes.19,56–63
The experimental determination of membrane biophysical parameters, such as surface charge, polarity, viscosity, hydration, tension and micro/nanodomains, can be quantitatively measured by specific environment-sensitive fluorescent probes.64,65 These probes generally reside in a specific location in the bilayer and report on membrane physical properties that are related to the anisotropic properties, which in turn are related to the motional freedom of lipid molecules and the packing or order of the acyl chains. The simultaneous use of different fluorescent probes, that have a well-defined bilayer location and orientation within the lipid structure relative to the interface-carbonyl, polar heads-phosphate, hydrophobic region-acyl chain, provides information on the in-depth landscape of lipid motion/mobility in different membrane structures.
There have been a number of attempts to describe the dynamic complexity of the cell membrane via a unified theory of membrane structure.66–68 As first reported in 1974, one model describes maintenance of the membrane in its liquid-crystalline state through an unique innate mechanism of “homeoviscous adaptation”.69 This concept was built on the observation that E. coli changes lipid composition at different growth temperatures to adapt to these conditions. The lipid changes include synthesis of longer and more saturated fatty acids which then impacts on acyl chain disorder and increased rotational and lateral diffusion of molecules, as increased fluidity is associated with increased temperature.70 This model which describes the membrane as a medium of regions with differing viscosity, therefore, provides a useful model to describe on-going dynamic changes in membrane properties.
However, while technologies continue to improve for the analysis of the changes in lipid compositions, the changes in structural and physical properties in membranes that impact on cellular process are less-well characterized. Our understanding of membrane structure needs to integrate the regulation of membrane phase equilibria in terms of changes in lipid configurations that impact on bilayer formation, membrane thickness and lipid packing. This phase regulation cannot be obtained from only considering the overall membrane viscosity (fluidity).
In addition, advances in theoretical and molecular modelling techniques have proven to be a valuable tool in understanding lipid membrane structure, composition, and function at resolutions scaling from isolated lipid molecules to large (bilayer and other types of) assemblies. While the pioneering lipid simulations of the 1980s were limited to picosecond timescales and overly simple hydrocarbon systems, the current state-of-the-art can now explore complex biological phenomena involving millions of molecules in realistic cellular environments that evolve over microseconds of molecular dynamics.71,72 Through multiple scales of well-designed molecular models, a wide gamut of physical, structural, and mechanical properties (Fig. 2) are able to be systematically explored as a function of dynamics and extrinsic factors (Fig. 3).73,74 Considerable developments in lipid specific interatomic interaction potentials (forcefields) have greatly improved the quality and reliability of molecular dynamics simulations when evaluated against experimental properties, in particular lipid diversity, temperature dependence, and phase behavior.75 It cannot be understated how beneficial and exciting sophisticated visualizations and animated graphics have become, catalyzing the understanding and interpretation of complex membrane systems.76 This is especially true for the qualitative and quantitative analysis of: lipid diffusion and fluidity;77 membrane packing defects;78 and lipid domain phase separations.15,79 Furthermore, there is a diverse range of computational techniques that enable the exploration of realistic simulations of biological membrane curvature and shapes known to be linked to many cellular functions. These encompass both lamellar and non-lamellar membrane models with a range of chemical compositions and intrinsic curvatures, including planar, rippled, local membrane protrusions/tubules, positively and negatively curved assemblies, micelles, bicelles, vesicles/liposomes, and various other shapes (up to and including full viral envelopes).72,80–84
In summary, the physical, structural and mechanical properties described above are all interrelated and influenced by the lipid composition and extrinsic variables. They clearly paint a complex picture of membrane structure in terms of how the structural and physical properties are influenced by changes in composition and extrinsic stress. At the same time, it is also evident that the extent of changes in membrane physical properties can be tolerated by the cell within a threshold level of changes depending on the mechanism, which can occur via de novo synthesis, assimilation of exogeneous lipids (e.g., biofilm, culture media), enzymatic modifications of existing lipids, lipid degradation or production of membrane vesicles. Thus, membrane lipid compositions are tightly regulated to maintain the optimum physical properties and stability for membrane protein function and the connection between lipid composition and the membrane physical and structural properties is remarkably dynamic.
4. The dynamic relationship between lipid chemical structure, lipid composition and the physical properties of membranes
4.1 Role of lipid head groups in modulating membrane properties
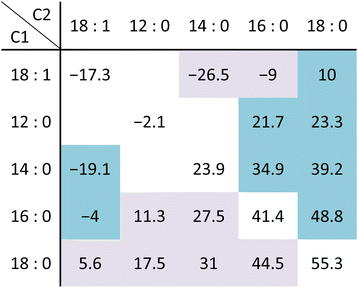
The membrane surface charge and potential are regulated by the relative ratio of charged to neutral head groups. The maintenance of membrane surface charge is critical for the electrical potential of the membrane, which regulates bacterial cellular and membrane behaviour in membrane transport, cell mobility, cell division, environmental sensing and antibiotic resistance.85 The phase behaviour is also modulated by the head groups, although the main phase transitions are closely related to the type of fatty acyl chain.86 The lamellar gel (Lβ) to lamellar liquid-crystalline (Lα) phase transition (Fig. 1D) is the main phase (gel-fluid) transition (Tm) of lipid bilayers, which has been studied in terms of cooperativity, order of lipid molecules, the role of head groups and acyl chain conformation. The Lβ–Lα phase transition is accompanied by a lateral expansion with a decrease in bilayer thickness and an increase in the area per lipid molecules. Furthermore, the phase transition is connected to the acyl chain trans–gauche isomerization, intermolecular interactions, the polar forces between the hydrophilic moieties, the lateral pressure in a bilayer due to steric repulsions, electrostatic interactions, and the hydrophobic effect. All these parameters of membrane order–disorder are also affected by temperature (thermotropic), hydration (lyotropic), pressure (barotropic), pH and ionic strength.The structural contribution of each lipid to membrane order is commonly guided by in vitro trends from individual lipid Tm values and the head groups have a significant role in the thermotropic phase transition (Table 1). For a model membrane composed of the same acyl chain length for different head groups, the Lβ–Lα phase transition temperature decreases in the order of PA > PE > PS > PC = PG > PI. While the Tm values of CL are similar to those of PE, the variation in the ratio of lipid head groups impacts on the phase behavior of a membrane. However, this thermotropic effect of different head groups on the lipid lamellar phase remains to be characterised for specific lipid compositions of bacterial membranes. The exact contribution of individual head groups to the membrane order, lipid packing and phase behavior of the natural membrane in vivo is not well-known and is assessed by the thermotropic phase transition of model membranes prepared from the native bacterial membrane lipid extract. For example, the Tm values of the order-to-disorder phase transition for vesicles of a wild type E. coli lipid extract was 38 °C which increased to 55 °C for a PE-deficient E. coli lipid extract in the presence of 50 mM Ca2+.23,87
18![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) : :![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1/18 1/18![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) : :![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1 1 |
16![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) : :![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 0/18 0/18![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) : :![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1 1 |
12![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) : :![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 0/12 0/12![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) : :![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 0 0 |
14![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) : :![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 0/14 0/14![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) : :![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 0 0 |
16![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) : :![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 0/16 0/16![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) : :![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 0 0 |
18![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) : :![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 0/18 0/18![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) : :![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 0 0 |
20![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) : :![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 0/20 0/20![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) : :![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 0 0 |
22![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) : :![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 0/22 0/22![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) : :![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 0 0 |
24![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) : :![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 0/24 0/24![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) : :![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 0 0 |
|
|---|---|---|---|---|---|---|---|---|---|
| PA | −4 | 28 | 31 | 52 | 65 | 75 | |||
| PE | −16 | 25 | 29 | 50 | 63 | 74 | 86 | ||
| PS | −11 | 14 | 14 | 35 | 54 | 68 | |||
| PC | −17 | −2 | −2 | 24 | 41 | 55 | 66 | 75 | 80 |
| PG | −18 | −2 | −3 | 23 | 41 | 55 | |||
| PI | 21 | 41 | |||||||
| CL | 25 | 52 | 62 | 73 |
Bacterial membranes are highly asymmetric in terms of lipid distribution within the plane and transverse to the bilayer. PE and PG are distributed differently and form domains in E. coli, while CLs are found localized in the cell poles and septa of E. coli and B. subtilis [ref]. Although spontaneous, lipid microphase separation, location-specific lipid synthesis associated with membrane proteins, and intrinsic curvature are all considered important for asymmetric lipid distribution. However, the mechanisms by which CL, PE and PG-rich domains form in the bacterial membrane are uncertain.
What has also not been characterised is the distinct composition of lipid head groups, their role in the formation of domains enriched in certain lipids, and whether they are localized in an ordered or non-lamellar (hexagonal) phase. Moreover, how these clustered lipids and properties are finely tuned for cell-division and resisting the action of AMPs has yet to be established. For example, PE- and PG-enriched domains have been observed in the membranes of both E. coli and B. subtilis and PE-rich domains have a higher degree of membrane order compared to PG-rich domains.17,88
4.2 Role of lipid fatty acid in modulating membrane properties
Acyl chains of lipid molecules have diverse roles in regulating membrane protein activities and maintaining membrane function and integrity. As described in Section 2, the acyl chains of membrane lipids exhibit high levels of structural and compositional complexity, which directly correlates with the pleiotropic behaviour of membrane properties. This complexity arises from the substantial variations in the acyl chain length, modifications in number, position and cis–trans stereoisomers of double bonds, cyclopropane and methyl branching (Fig. 1A and B).Complex systems for modifying lipid fatty acid profiles have evolved in cells to respond to altered growth conditions and various environmental changes, ensuring adjustment of the membrane physical and structural properties within an optimal range. Various innate mechanisms for modifying the lipid fatty acid profiles to modulate both viscosity and phase changes are present in bacteria.
There are a range of basic mechanisms that regulate the degree of unsaturation and length of fatty acid acyl chain in all cells, while cyclisation, branching and isomerization are responsive mechanisms peculiar to specific cell species. The rate of chemical restructure of lipid acyl chains can vary from minutes to days, depending on the regulatory enzymatic mechanisms and the rate and extent of disturbance of the membrane structural integrity and physical properties. The cells can also regulate the fatty acid compositions with more than one mechanism or can switch between different response modes that depend on the growth conditions and environmental factors.
Lipid acyl chains of 12 to 24 carbons in length are generally synthesized and incorporated into a bacterial membrane.6,89–94 Fatty acids with 16–18 carbons are the most abundant in membrane lipids while exceptionally long fatty acids, known as mycolic acids of 60–90 carbons, are present in the outer membrane of Mycobacteria.95 The acyl chain length is the main determinant in modulating the membrane thickness and phase transition. In addition to the acyl chain length, the specific fatty acid structures and the sn-1 and sn-2 position of glycerophospholipids have significant impact on the phase behavior and transition temperature [Table 2] and have different regulatory roles in homeophasic adaptation. Unique chemical structures of lipid fatty acids have been characterized for different bacteria. Even-number straight-chain saturated and unsaturated fatty acids are synthesized by E. coli and S. pneumoniae while odd-number branch fatty acids are predominantly found in many G(+) bacteria, such as B. subtilis and S. aureus.93,96,97
Bacteria are able to introduce unsaturation into the membrane lipids by one of three different mechanisms, which are: (1) de novo synthesis via incorporation of the desired acyl chain into the glycerol backbone, (2) conversion of the saturated chain to the desired unsaturated lipid, or (3) incorporation of an exogenously derived acyl chain (which cannot be synthesized by the bacteria). It is not well understood why certain bacteria utilize a specific mechanism but the rate of changes required for survival will define the mechanism used.
The changes in the SFA/UFA ratio are the primary focus of the homeoviscous response of membranes for cells under temperature stress, such as in E. coli and B. subtilis. For example, in E. coli, the introduction of a double bond at the 10-carbon intermediate by fatty acid synthesis through de novo synthesis produces palmitoleoyl (16![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1 Δ9), and cis-vaccenoyl (18
1 Δ9), and cis-vaccenoyl (18![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1 Δ11) chains as the dominant acyl chain together with saturated palmitoyl (16
1 Δ11) chains as the dominant acyl chain together with saturated palmitoyl (16![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 0) in the membrane lipid extract.93
0) in the membrane lipid extract.93
The physical state of the cell membrane is thus modulated by the incorporation of a mixture of fatty acids into phospholipids with different phase transition temperatures. In E. coli grown at 37 °C, the total UFA content was 45 mole% which then increased to 60 mole% in E. coli grown at 17 °C.93 This increase in UFA was associated with a doubling of the amount of cis-vaccenoyl (18![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1 Δ11) chains and 10% drop in the fully saturated palmitoyl (16
1 Δ11) chains and 10% drop in the fully saturated palmitoyl (16![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 0) chains. However, the fraction of unsaturated palmitoleoyl (16
0) chains. However, the fraction of unsaturated palmitoleoyl (16![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1 Δ9) remained unchanged in the inner membrane with variable temperature. Thus, the extent of unsaturation was accompanied by an increase in the average chain length of the lipid acyl chains under cold stress, which presumably assists in maintaining a more stable membrane at lower temperatures.
1 Δ9) remained unchanged in the inner membrane with variable temperature. Thus, the extent of unsaturation was accompanied by an increase in the average chain length of the lipid acyl chains under cold stress, which presumably assists in maintaining a more stable membrane at lower temperatures.
The incorporation of a cis-double bond introduces a 30° kink in the acyl chain, which decreases the packing order in a lipid bilayer as commonly characterized by 2H solid state (ss)-NMR and the steady-state anisotropy of fluorescence probes such as diphenylhexatriene (DPH). This packing disorder in a lamellar structure is more pronounced when the cis-double bond is located in the middle of the acyl chains.99 Introducing the double bond into the fatty acids not only affects the packing order of hydrophobic core of bilayer, the acyl chain unsaturation can further increase the non-lamellar propensity of PE due to the relatively small head group size versus the lipid length and volume.93 This will impose more negative curvature stress on the membrane creating lipid packing defects.
The degree of unsaturation through de novo synthesis in E. coli and other anaerobes can be modulated by growing cells in the exponential phase when the fatty acids are produced. As the growth of cells enters stationary phase, the production of fatty acids ceases and the double bonds are converted into cyclopropane, with an additional carbon, while still keeping the 30° kink. These cyclic fatty acids exert similar effects to unsaturated fatty acids on the bilayer packing order and are more resistant to oxidation as the acidity is raised in the stationary phase (see Section 4.2.3 below).
The maintenance of optimal viscosity in a membrane is critical for the physiological function of cells. In one study, the membrane viscosity was controlled by the extent of fatty acid unsaturation where a 10-fold decrease in viscosity (20 to 2 poise) occurred when the unsaturation increased from 20% to 60% as estimated by the diffusion coefficient of nitrobenzoxadiazole-conjugated phosphatidylethanolamine (NBD-PE) in E. coli inner membrane vesicles using fluorescence recovery after photobleaching (FRAP).100 Furthermore, membrane viscosity can also be maintained by different combinations of unsaturated, cyclic and straight chain fatty acids at different growth phases. The cellular respiration rates are also regulated by the membrane viscosity and tightly controlled by the unsaturated fatty acid composition.
In contrast to the de novo synthesis of fatty acids, fatty acid unsaturation can be incorporated via the introduction of a double bond into the existing membrane phospholipids (Mechanism 2), which occurs in some bacteria under cold stress. In bacilli, pseudomonads, mycobacteria and cyanobacteria, a cis-double bond is introduced into the saturated fatty acyl chain of membrane phospholipids by a multi-component membrane desaturase under aerobic conditions.101–104 This has been well characterized in B. subtilis where the activation of desaturase is related to changes in membrane structure upon cooling whereby membrane thickness increased with higher packing order.101 The cis-double bond was introduced only at a specific site on the SFA. Insertion at the Δ5 position was exclusively found in B. subtilis and B. megaterium and a Δ10 desaturase additional to Δ5 desaturase inserting a cis-double at Δ10 position was used by B. cereus to generate mono/diunsaturation at Δ5 or/and Δ10.102,105 The proportion of UFA in B. cereus can change from 27% predominantly Δ10-UFA at 37 °C to 45% with both Δ5 and Δ10-UFA at 25 °C due to increased Δ5 desaturase expression and activity.105 As no lipid synthesis is required, this way of modifying the existing saturated acyl chains allows rapid adjustment of the SFA/UFA ratio. However, modulation of membrane properties by desaturation involves activation of desaturase gene transcription and translation and, once the membrane properties have adapted to the cold stress, the desaturase expression is down-regulated. This type of machinery that is sensitive to membrane thickness and packing order highlights the sophisticated regulation of membrane properties via enzyme activity and gene expression in homeoviscous adaptation. As no enzyme induction and synthesis is necessary for the de novo synthesis of UFA, and only the rate of enzyme turnover is changed, the rate of fatty acid synthesis can occur rapidly. However, for anaerobic fatty acid biosynthesis the incorporation of UFA into membrane lipids is the rate-limiting step in modulating the membrane properties. While the unsaturated palmitoleic (16![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1 Δ7) and oleic (18
1 Δ7) and oleic (18![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1 Δ9) acids predominantly synthesized in E. coli growing at low temperature, the different double bond positions (16
1 Δ9) acids predominantly synthesized in E. coli growing at low temperature, the different double bond positions (16![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1 Δ5 and 16
1 Δ5 and 16![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1 Δ10) introduced by the desaturase represent additional mechanisms by which membrane viscosity can be modulated.
1 Δ10) introduced by the desaturase represent additional mechanisms by which membrane viscosity can be modulated.
Membrane lipid unsaturation can also be regulated by uptake of exogeneous unsaturated fatty acids to result in changes in membrane properties (Mechanism 3). The fatty acyl chain compositions of E. coli membrane lipids were strongly affected by the exogeneous fatty acids which made up 45% of the total membrane FA.106 Significantly different Tm values were measured for E. coli cultured with different fatty acid supplements.107 The exogeneous fatty acids are phosphorylated and deposited in the membrane in exchange for a new fatty acid. Exogeneous UFAs can replace only 50% of the membrane lipid fatty acids in S. aureus with a concomitant reduction of branch-chained fatty acids in the membrane.97 In contrast, all membrane lipid fatty acids can be replaced by exogeneous UFA in S. pneumoniae. The types of exogeneous fatty acids in different sources, such as human serum and skin homogenate, can also impact on membrane properties and integrity and alter the resistance to the fatty acid synthesis inhibitors.92,108 For example, the incorporation of oleic acids and serum UFAs increased the membrane order as indicated by an enhanced DPH anisotropy due to an increase in the proportion of SFA and decrease in branched-chain fatty acids (BCFAs).97,109–111 While the UFAs from skin are known to be toxic to S. aureus, the incorporation of serum UFAs into S. aureus membrane increases resistance to FASII inhibitors.112
Incubation of wild-type and methicillin-resistant S. aureus strains with exogenous UFA in serum resulted in 25% UFA being detected in membrane lipids. The presence of UFA would be expected to increase the membrane fluidity. However, the anisotropy of DPH was enhanced in S. aureus incubated with serum, consistent with increased membrane order. This increase in DPH anisotropy can be related to an accompanying increase in the content of carotenoid pigment (staphyloxanthin) in the membrane.111 A similar increase in the DPH anisotropy was also observed for wild-type and fakA mutants of S. aureus incubated in media with 0.01% oleic acids. This was not due to incorporation into phospholipids. Rather, the increased membrane order was likely due to the reduced BCFA/SFA ratio in both the WT and fakA mutant, with a significant amount of UFA incorporated into the WT S. aureus while no UFA was detected in the fakA mutant.110
The isomerisation of UFA from cis to trans configuration plays an important role as an adaptation response to heat stress, the presence of organic solvents, heavy metals, osmotic stress, and exposure to antibiotics and AMPs, by increasing the lipid packing order in membrane and thereby reduced permeability. The cis–trans isomerisation is catalysed by a periplasmic cis–trans isomerase which converts the cis-palmitoleic (C16![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1Δ9cis) and cis-vaccenic acid (C18
1Δ9cis) and cis-vaccenic acid (C18![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1Δ11cis) of phospholipids within the inner membrane of some G(−) bacteria including G(−) Pseudomonas sp., Vibrio sp. and G(+) Enterococcus faecalis FA2-2.113–115 The amount of trans-UFA is low in the membrane of these bacteria under normal conditions. The degree of cis–trans isomerisation in G(−) bacteria varied with the extent of the stressors such as the toxicity, hydrophobicity, and concentration of toxic organic compounds and the duration of nutrient deprivation. The trans-UFA can reach up to 40% of total fatty acid in Pseudomonas sp., and 20% of total fatty acids in Vibrio sp. and Colwellia maris sp. However, although the growth phase and temperature influence the cis–trans isomerisation, the low amount of trans-UFA in E. faecalis is not affected by the extent of stressors and the bulk physical properties are not significantly altered by the low level of trans-UFA. Such a transformation contributes significantly to some G(−) bacteria survival by increasing the lipid order and packing when a rapid response is required to resist environmental stress.
1Δ11cis) of phospholipids within the inner membrane of some G(−) bacteria including G(−) Pseudomonas sp., Vibrio sp. and G(+) Enterococcus faecalis FA2-2.113–115 The amount of trans-UFA is low in the membrane of these bacteria under normal conditions. The degree of cis–trans isomerisation in G(−) bacteria varied with the extent of the stressors such as the toxicity, hydrophobicity, and concentration of toxic organic compounds and the duration of nutrient deprivation. The trans-UFA can reach up to 40% of total fatty acid in Pseudomonas sp., and 20% of total fatty acids in Vibrio sp. and Colwellia maris sp. However, although the growth phase and temperature influence the cis–trans isomerisation, the low amount of trans-UFA in E. faecalis is not affected by the extent of stressors and the bulk physical properties are not significantly altered by the low level of trans-UFA. Such a transformation contributes significantly to some G(−) bacteria survival by increasing the lipid order and packing when a rapid response is required to resist environmental stress.
Overall, the multiple effects of fatty acid unsaturation on membrane physical properties and physiological functions require further study to explore various aspects of regulatory mechanisms for optimal unsaturation in different cellular states and in response to environmental stress. Moreover, further understanding of the influence of host fatty acids on the bacterial membrane properties is required to further establish the resistance mechanisms to antimicrobial drugs in a host environment.
S. aureus has a deficiency in UFAs and, therefore, utilises predominantly BCFA lipids with an acyl chain length of 15 to modulate the membrane in response to environmental stimuli.111,118,119 Since anteiso-branching perturbs the lateral packing of lipids to a greater extent than iso-branching, for lipid fatty acids with an equivalent carbon number, anteiso-branching lowers the gel to liquid-crystalline phase transition temperature more than iso-branching, while cis-unsaturation in the centre of the hydrocarbon chain is most efficient in lowering the phase transition temperature.99,120 Membranes enriched in anteiso-BCFA lipids are more disordered with a lower DPH anisotropy than those of iso-BCFA lipids in which anteiso-BCFA promotes lower viscosity than the corresponding iso-BCFA in vivo.121 Depletion of BCFA in B. subtilis mutants has been shown to be accompanied by the accumulation of SFAs with a gradual increase in viscosity and leading to growth arrest. The effect of such drastic changes in the SFAs in the membrane properties on cell growth cannot be explained by fluidity alone. The increase in the membrane rigidity can also change the bilayer thickness, the membrane permeability and electron transport chain function.100 In S. aureus, increases in both anteiso- and iso-BCFA in membrane lipids is exploited to maintain optimum molecular diffusion under cold stress, while in B. cereus only the proportion of iso-BCFA decreased in membrane lipids, causing increase in the anteiso/iso ratio and promoting the membrane fluidity for growth at lower temperatures.
Multiple modes of regulating fatty acid compositions are utilized by bacteria to adapt to deleterious environmental effects on membrane structural integrity and physical properties. However, there is no universal mechanism for regulating the compositions of membrane lipid species and fatty acids and switching between different regulation modes has been developed for optimal growth.
Microorganisms are flexible in their fatty acid requirement if the minimum demand of unsaturated fatty acids, for example, are satisfied. Exogenous fatty acids from the host, even those not synthesized by a microorganism, are incorporated into cellular lipids so that variations in the physical properties of phospholipids are minimized under the stress.92,108,110 Although BCFAs and SCUFAs both increase membrane fluidity, these fatty acids impact differently on cellular morphologies,87 and adaptation to cold stress.111,122,123 Expression of virulence factors is significantly different in serum grown organisms,124 and there are global changes in gene expression when S. aureus is grown in blood.125S. aureus grown in serum or blood have different membrane lipid compositions than cells grown in laboratory media and this may have a significant impact on the expression of virulence factors and pathogenesis of the organism. Due to the ability of a pathogen to adapt and undergo dramatic alterations when subjected to a host environment, the membrane properties of bacteria grown in vivo can, therefore, be very different from when it is grown in vitro. This distinction may have a huge impact on critical cellular attributes that control pathogenesis and resistance to antibiotics.126
In addition to the production of CFAs in cells growing under acid stress and at early stationary phase, the biosynthesis of CFAs can also be induced by other conditions such as high temperature, reduction of respiratory components, limitation of ammonium or phosphate, acid pH, anionic detergents at low concentrations, high NaCl and nucleotide (ppGpp, pppGpp) concentrations, low oxygen tension, and adequate levels of Mg2+ ions and sulfate.130 CFAs have biophysical properties similar to UFAs with higher stability towards acidity and oxidation. While the cyclopropane group adopts the same 30° kink in the acyl chain as UFAs, the effects on the membrane order are still not clear as opposite effects on membrane ordering have been shown by determining the phase transition temperatures in model membranes and acyl chain order with NMR.130,131 Depending on the configuration, the conversion of a double bond to a cyclopropane causes pronounced effects by reducing the angular fluctuation along the sn-2 acyl chain but no significant impact on the sn-1 acyl chain as shown by 2H-NMR and molecular dynamic simulation.132,133 Furthermore, the order of the hydrocarbon chain, as reflected in a higher relaxation time, is higher for CFAs than UFAs of the same configuration. Thus, the restricted motion and bulkiness of the cyclopropane moiety prevents the tight packing of the lipid bilayer which maintains the membrane in a more fluid liquid-crystalline state. However, the cyclopropane ring possesses a higher chain order parameter which remains structurally rigid over a broad temperature range. Overall, CFAs may increase membrane fluidity while simultaneously inducing a more ordered state within the hydrocarbon chains compared to UFAs. As a consequence, the cyclopropane moiety might play a dual regulatory role in stabilizing structural and dynamic properties of a membrane in response to environmental stressors.
In E. coli mutants, membranes deficient in CFAs are more permeable to protons, although the role of CFAs in modulating the membrane properties is not clear. A potential function of CFA formation is to stabilize the membrane by increasing the lipid order to avert the enhanced membrane permeability induced by the oxidative and acidic damage.133 Although CFAs contributing an ordering effect to the UFAs counterparts of a model membrane of extracted lipids, it is not known why membranes of whole cells, with a high CFAs content, appear to be more fluid than membranes with a lower CFAs content.134 More thorough studies are required to understand how the membrane properties and structures are modified by the lipid CFAs (Table 3).
| No | Parameters |
|---|---|
| 1 | Changes in the ratio of lipid classes and species in each class |
| 2 | Membrane properties differentially modulated by headgroup-specific acyl chain remodelling |
| 3 | Ratio of UFA/SFA; CFA/SFA; BCFA/SFA |
| 4 | Length of acyl chain |
| 5 | Ratio of different branching structure, iso/anteiso/ω-alicyclic fatty acids |
| 6 | Position of specific acyl chain structure on the glycerol backbone, e.g., unsaturated sn-2 acyl chain |
| 7 | Incorporation of exogeneous FAs |
| 8 | Cis/trans isomerization of UFAs |
| 9 | Proportion of other specific lipids altering lipid packing order, thickness and other molecular interactions |
| 10 | Enzymatic modification of lipid head group; alteration of membrane surface charge |
| 11 | Domains of different size, thickness, packing order and viscosity/diffusivity |
| 12 | Species-selective lipid degradation and production of membrane vesicles |
| 13 | Limiting/inhibiting synthesis of specific lipids |
5. Measuring the structural and mechanical properties of functional membranes
The homeostatic adaptation of bacterial membrane properties to various stress conditions is associated with spatiotemporal modulation of membrane structures including bilayer thickness, area/volume per lipid and lateral phase and domain segregation. The variation in the lipid composition of different membranes, between the membrane leaflets, and the heterogeneous distribution in different membrane areas are all interrelated and contribute to the maintenance of their structural stability. The loss of the integrated control of the dynamic modulation of membrane structure and properties can lead to growth arrest and cell death. The key to fully understanding these complex systems is the ability to measure different physical parameters in native membranes.The bacterial membranes of both G(−) and G(+) bacteria have been visualized as a low electron density layer about 5 nm in thickness by transmission electron microscopy (TEM) and cryo-EM.135–138 In addition to the thickness, changes in the shape and curvature of bacterial membranes have also been observed in relation to changes in lipid compositions, under stress conditions and by antibiotic treatment.135,137
Although the alteration of membrane ultrastructure can be visualised in situ by EM, the low contrast of the plasma membrane relative to the cytoplasm provides limited resolution in order to correlate the heterogeneous lateral structure and domains as molecular adaptation mechanisms in response to stress conditions and antimicrobial peptides (AMPs) and agents. To understand the interrelationship between lipid composition and structural characteristics of membranes, many of the comparative studies of membrane structures have been based on various model membrane systems rather than the native cellular state. The bilayer thickness, area per lipid molecule and effective hydrophobic thickness can be accurately determined by X-ray and neutron scattering which can be correlated with the order of the lipid acyl chains and head group dynamics analysed by NMR to provide both the structure and physical properties of particular lipid compositions.139 The collective in vitro structural characteristics of individual lipids in membranes also provide valuable quantitative information for building in silico models toward understanding the contribution of each different lipid species in response to stress and AMPs. However, the dynamic changes in the membrane structures cannot be obtained from the static structural parameters obtained by scattering analysis of the multi-lipid bilayer stacks.
Understanding the impact of lipid compositions on the dynamic organisation and structural characteristics of membranes, therefore, rely on techniques capable of resolving spatiotemporal changes at nanoscale resolution and at millisecond timescales under physiological conditions. Since first reported in 1986, AFM equipped with various scanning modes is now widely applied to resolve at nanoscale the dynamic changes in bilayer thickness and coexistence of fluid and crystalline domains in membranes so as to explore the relationship between lipid composition and domain formation in regulation of membrane homeostasis.140–146 The effect of different lipid acyl chain length and degree of unsaturation on membrane structure can now be resolved to less than 1 nm differences between liquid-disordered and ordered phases.141,147 The rate of nucleation and size variation of phase-separated domains in multicomponent lipid bilayers, as a function of stress conditions such as temperature, type and concentration of AMPs, can also be correlated to understand the role of lipid composition on the mechanisms of membrane destabilisation.60,148–152 By mapping the surface topographic changes of synthetic lipid membranes and natural bacterial membrane extracts, a broad range of distinctive changes in membrane structure induced by AMPs have been revealed. In addition to pore formation and carpet mechanisms, these include lipid clustering, non-lamellar structure, membrane thinning/thickening, monolayer extraction and nanopitting.153 This wide range of membrane disruptions, which describe unique feature of vertical and lateral changes in bilayer structure, provide new insights into the broad spectrum of action for membrane-active peptides.
Changes in the membrane thickness and the release of membrane vesicles can also be induced by antibacterial agents and have been characterised by high resolution AFM.154–156 These diverse peptide-membrane structural complexes visualised by AFM have shown that the changes in membrane structure induced by peptides are distinctive and complex whereby multiple modes of action can be adopted by one peptide and vary based on lipid composition.60
The structural characteristics and the surface architecture of the bacterial cell surface have also been imaged by AFM and exhibit considerable variation between G(+) and G(−) and on the presence of polysaccharides or S-layers.157–163 A smooth surface was characterised for G(−) bacteria due to the presence of an outer membrane while a rough surface was found for the thick peptidoglycan layer on the surface of G(+). The binding of polymyxin to LPS-containing E. coli outer membranes resulted in reduced membrane thickness, expansion of membrane area and increased stiffness. Polymyxin also rearranged these LPS-containing membranes into hexagonal crystalline structures. Further exploration of the rate dependence of AMP-induced changes on membrane structural, physical and mechanical properties with various lipid compositions should provide quantitative information to elucidate drug-resistance mechanisms at membrane level.
In addition to the resolution of topological nanostructures of membranes, the mechanical properties of membrane and cell surfaces can also be characterised by AFM using different strategies.144,145,164–166 The nanomechanical properties are mostly derived from the force–distance (F–D) curve by recording the deformation of the cantilever as the tip indents with varying amount of force into different depths of the cell surface. The multi-parametric stiffness (Young's modulus), elastic, and adhesive mechanical properties provide further information in relation to the structural and physical properties as a result of the modulation of lipid composition by membrane homeostasis and drug-resistance mechanisms. For example, the role of composition and ultrastructure of the polysaccharide capsule in the resistance to osmotic pressure by K. pneumoniae has been explored by F–D curve analysis of nanomechanical measurements.167,168 The resulting F–D curves were fitted with several physical models representing different stages of probe indentation for various depths of capsule having different properties. These types of F–D curve analysis allow the changes in cell wall elasticity to be mapped in response to antibiotic treatment as shown by the changes in elasticity.169
Although the structure and mechanical properties of a membrane can be obtained from the AFM-based indentation of solid supported lipid bilayers, the quantitative information has not been well related to the bending modulus for membrane deformation with different lipid compositions.157,170,171 The relationship between membrane stiffness and other viscoelastic properties and the bending/deformation of membranes can be studied using lipid vesicles and whole cells.169 Due to the presence of a thick wall of peptidoglycan in G(+) bacteria and an outer membrane in G(−) bacteria, which restricts access to the plasma membrane, enzymatic or mechanical treatment is required to completely remove the cell wall and outer membrane to obtain structural details of bacterial plasma membranes by AFM analysis. Quantitative analysis of size, surface structure, and viscoelastic and bending properties of membrane vesicles (<200 nm in diameter) secreted by bacteria also contribute to understand the role of lipid composition on the membrane mechanical and structural stability.155,156,159,172 Further characterisation of the collective membrane structural, physical, and mechanical properties of bacterial membrane vesicles and cell-wall deficient bacteria (spheroplast/protoplast) will shed light on the resistance mechanisms in relation to the bacterial surface and membranes.
6. Changes in physical properties of membranes associated with resistance to AMPs
The increasing incidence of drug-resistant microbial infections is one of the most significant threats to global health. Solutions to control and reduce the emergence and spread of drug-resistant bacterial infections remain as major challenges. Understanding the mechanisms of antimicrobial resistance related to the emergence, transmission, bacterial fitness, persistence, and potential resistance-relapse upon withdrawing the drug provides essential information for the design and development of antibacterial agents and technologies complementary to conventional antibiotics for clinical use.173 Among the various antimicrobial agents, AMPs with broad-spectrum activity show great promise against drug-resistant bacteria. Classic mechanisms of bactericidal action for several AMPs have been generally related to the formation of transmembrane peptide pores and micellization of membranes. These mechanisms mainly focus on the final structure, orientation, and assembly of peptides in the membrane environment. However, the changes in the physical properties and geometrical parameters of membranes associated with the peptide binding are less well characterised for each mechanism of action, but are essential to development of new antimicrobial strategies. The perturbation of a membrane upon exposure to AMPs can cause changes in different physical properties of membranes, including thickening/thinning, membrane expansion, specific domain formation and redistribution, membrane curvature deformation, and non-lamellar phases and lateral phase segregation, which can all lead to destabilizing the membrane, increased permeability, and loss of membrane potential.174–176The selective antibacterial activity of AMPs is strongly influenced by the lipid compositions of bacterial membranes where the negatively charged phospholipids play a prominent role in promoting peptide binding and insertion into the membrane, while other peptides tend to bind primarily at the interfacial region of membrane consisting solely of zwitterionic lipids. Following exposure to AMPs, the phenotypic changes in membranes associated with alterations in lipid composition and structure, and subsequently impacting on the physical properties of the membrane, are emerging as potential mechanisms to counteract the action of AMPs. These physical changes involve a change in the net negative surface charge, thickening of the cell wall, modification of the membrane fluidity and order, and changes in the membrane thickness, which all are interlinked and result in changes to other physicochemical properties of the membrane as discussed below.
6.1 Changes in surface charge
Changes in lipid fraction and specific modification of the lipid head group can impact on the surface charge of membranes. For example, decreases in the percentage of PG/CL reduce the negative charge. Aminoacylation of the PG and CL head group with lysine or alanine and translocation from the inner to outer leaflet of a membrane, is catalysed by the multiple peptide resistance factor (MprF) via catalysis of the transfer of the aminoacyl moiety from Lys-tRNALys to the free distal hydroxyl group of the glycerol moiety of phosphatidylglycerol (PG).26 This reaction is an intrinsic resistance mechanism to AMPs first identified in Staphylcocci and later in other bacteria such as Bacilli, Pseudomonas, Listeria and Mycobacteria except for enterobacteria.35 This pathway of aminoacyl PG and CL biosynthesis with concomitant reduction in PG molecules thereby reduces the overall surface negative charge and generates an asymmetric distribution of lipids between the inner and outer membrane leaflets and thus reduces the ability of anionic AMPs to bind to the membrane.It remains unclear if the presence of Lys-PG or Ala-PG has different consequences for cationic AMP resistance. While Lys-PG has a positive net charge, Ala-PG is a zwitterionic lipid with a less pronounced impact on the net charge of the membrane surface and on cationic AMP repulsion than Lys-PG. Nevertheless, the fact that bacteria produce Ala-PG either alone or in combination with Lys-PG indicates that it should have a distinct impact on membrane properties and may be a means to fine-tune the charge of the membrane in response to environmental challenges as recently suggested.26,177 However, there is no consistent correlation between lipid composition in terms of the levels of PG, Lysyl-PG, PE and CL and AMP resistance, which indicates that more than one mechanism evolves to confer the apparent AMP resistance.
6.2 Impact of alteration of lipid acyl chains on molecular packing and bilayer thickness
In addition to the changes in membrane surface charge, the structure and composition of the acyl chains of bacterial membrane lipids can vary greatly in response to environmental pressures and impact on the mechanism of action of, and resistance to, AMPs. Current understanding of the role of acyl chain composition on AMP activity remains obscure, as changes in the chain length, the degree of unsaturation, branched and cyclic groups in the lipid acyl chain all have complex effects on the physical properties of a membrane. Although changes in the fatty acyl chain length and unsaturation alter membrane thickness and fluidity, how these changes affect AMP activity and resistance depends on the specific mechanism by which the AMP disrupts the membrane.Increased levels of longer and unsaturated acyl chains in membrane lipids have been reported for S. aureus following exposure to platelet-derived peptide and class IIa bacteriocin.178,179 In contrast, increased amounts of longer SFA and the ratio of SFA/UFA were identified in various strains of E. faecalis displayed colinear cross-resistance to pediocin, nisin and alamethicin.180,181 A similar decrease in the UFA with increased membrane rigidity was identified in daptomycin resistance E. faecium. However, it is not clear from these studies whether the altered acyl chain lengths correlated with any changes in the hydrophobic thickness of the membrane.
Due to the lack of probes and methods that are sufficiently sensitive to measure the membrane thickness changes in bacterial cells, the effects of acyl chain length and membrane thickness on AMP activity are generally explored by use of model membranes composed of defined mixtures of lipids with various acyl chain lengths. For example, a linear dependence in increased dye efflux time induced by δ-lysin dependent on increasing bilayer thickness was observed for liposomes composed a homologous series of di-monounsaturated PCs, namely di(14![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1)PC, di(16
1)PC, di(16![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1)PC, di(18
1)PC, di(18![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1)PC, di(20
1)PC, di(20![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1)PC, and di(22
1)PC, and di(22![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1)PC.182 However, there was no clear dependence of dye efflux time on the membrane thickness for liposomes composed of PCs with polyunsaturated and asymmetric (POPC, SOPC) acyl chains whose phase transition temperatures are well below ambient. Similar equilibrium binding constants were observed for δ-lysin binding to POPC, di(18
1)PC.182 However, there was no clear dependence of dye efflux time on the membrane thickness for liposomes composed of PCs with polyunsaturated and asymmetric (POPC, SOPC) acyl chains whose phase transition temperatures are well below ambient. Similar equilibrium binding constants were observed for δ-lysin binding to POPC, di(18![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1)PC, di(22
1)PC, di(22![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1)PC and di(18
1)PC and di(18![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 3)PC liposomes. As it is harder for δ-lysin to insert into thicker membranes, the membrane thickness is one of the critical determinants in modulating the peptide insertion into membrane.
3)PC liposomes. As it is harder for δ-lysin to insert into thicker membranes, the membrane thickness is one of the critical determinants in modulating the peptide insertion into membrane.
A mismatch between the bilayer thickness and the peptide length was shown to reduce the helical content of the AMP, maculatin 1.1 (Mac1.1), in the presence of model membranes composed of PC with different acyl chain lengths.183 Changes in membrane thickness may also impact on the possible oligomeric state of Mac1.1 inserted into the membrane, with a higher number of peptide monomers found in thicker membranes. In this study, the thickness and lipid order of each PC bilayer showed a linear dependence on the acyl chain length. The binding of Mac1.1 exhibited a biphasic dependence between the amount of bound Mac1.1 and bilayer thickness, whereby the mass of bound peptide increased for PC with C14 to C16 chain length and then decreased from C16 to C22. Significant perturbation of 31P chemical shift anisotropy (CSA) values was only observed for DOPC (C18) and DEPC (C22), respectively. In the case of DEPC, the greater range in CSA indicated different headgroup conformations or environments in the presence of Mac1.1. Overall, the results indicated that there is a significant change in the bilayer order upon binding of Mac1.1 and this occurred in a co-operative manner at higher concentrations of Mac1.1 with increasing bilayer thickness and order. An optimal bilayer thickness and lipid order was required for effective membrane perturbation by Mac1.1 and an increase in bilayer thickness and order may counteract the action of Mac1.1 and play a role in antimicrobial resistance to AMPs.
6.3 Bilayer fluidity and phase separation
The variation in the acyl chain length not only impacts on the bilayer hydrophobic thickness, but the length and changes in the position and number of double bonds and the presence of branched and cyclic moieties also cause changes in the fluidity and phase separation in membranes. For example, membrane fluidity has been shown to be an important property in determining the activity of a number of AMPs, including daptomycin, a non-pore forming cyclic hexapeptide, cWFW, thrombocidin-derived peptides and the synthetic bactericidal peptide 2.184–186 The incubation of live B. subtilis and model membranes with a non-pore forming synthetic cyclic hexapeptide cWFW led to an increase in Laurdan generalized polarization as a result of a substantial increase in membrane rigidity and formation of discrete membrane domains for antibacterial activity. A rapid reduction in membrane fluidity (i.e., more rigid) followed by lipid phase separation was observed in liposomes containing CL, which was independent of the specific fatty acid or lipid head group.186 A reduction in membrane fluidity has also been associated with daptomycin resistance in both E. faecalis and E. faecium clinical isolates and nisin resistance in L. monocytogenes.181,187 In addition, the UFAs were significantly reduced in the membrane lipids isolated from the mundticin KS (a model of class IIa bacteriocins)-resistant E. faecium together with an increase in the zwitterionic amino-containing PLs and cyclopropanyl(9)-C19![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 0 lipids.188
0 lipids.188
This is in contrast to an increase in membrane fluidity reported in the resistance to DAP in S. aureus, and an increase in the ratios of UFA/SFA and short- to long-acyl chains of PG in the clinical isolates of leucocin A-resistant L. monocytogenes.189 The decrease in membrane fluidity in daptomycin-resistant E. faecalis could not be linked to changes in the degree of acyl chain saturation and proportion of cyclic fatty acid.181 In contrast, a significant decrease in unsaturated fatty acids and an increase in cyclopropane fatty acids were identified in DAP-resistant E. faecium.
Factors other than the degree of saturation can also be important for the decrease in membrane fluidity. The changes in membrane fluidity with the variation in the ratio of saturated/saturated acyl chains can also be accompanied by changes in the composition of cyclic and branched acyl chains. The effect of cyclopropane fatty acyl chains on the gel–liquid crystalline phase transition of bilayers depends on lipid composition.130 Higher phase transition temperatures were found for more rigid bilayers when cyclopropane was present in both the sn-1 and sn-2 acyl chains, while a lower transition temperature was observed for more fluid bilayers with cyclopropane present only in the sn-2 acyl chain. However, the influence of the steric configuration of cyclopropane acyl chains on membrane fluidity is still not clear. It has been suggested that cyclopropane fatty acids pack less tightly than the unsaturated fatty acyl chains in a membrane and enhance the fluidity,133 while others have shown that the presence of cyclopropane fatty acids leads to more rigid membranes with increases in the phase transition temperature.131 Overall therefore, regulation of cyclopropane fatty acid content is an important mechanism by which bacteria can modify the structural and dynamic properties to control membrane fluidity and survive various environmental stresses.
6.4 Lipid packing and bilayer order
In view of the role played by chemical composition and geometrical properties of membranes in regulating the activity of AMPs and the emergence of resistance to AMPs, changes in the lateral density and packing/order of lipids induced upon AMPs binding also induce transcriptional changes, which modify the lipid compositions, surface charge, fluidity, and organisation of membrane to reduce the AMP activity. The self-assembly and alignment of lipids into highly ordered structures generates an anisotropic system with an unique optical property and the degree of molecular order and lipid acyl chain packing can be evaluated by the measurement of birefringence values. This is possible using DPI which allows the real-time changes in lipid bilayer birefringence to be measured as a function of peptide mass bound to the membrane to reveal the kinetic perturbation in membrane structure associated with distinctive mechanisms of AMP action on membranes of different compositions.19It has also been reported that higher lipid packing/order is related to increases in Lysyl-PG and Lysyl-PE content in the membrane, whilst peptide binding was reduced minimally to membranes containing 30% aminoacylated lipids. Although anticipated that Lysyl-PG/PE would reduce the electrostatic binding of cationic AMPs to bacterial membranes, these findings suggested that the presence of Lysyl-lipids also stabilise the membranes rich in anionic PG by increasing the packing order and reducing membrane perturbation by AMPs.38,190 Increased bacterial resistance to antimicrobial agents with noticeable increases in cis-vaccenic acid and cyclopropane fatty acids content in the membrane is possible by limiting membrane permeability and insertion through lipid order.191–193
6.5 Curvature and domains
The intrinsic curvature propensities of lipids are of particular importance for stabilizing and maintaining cell shape and morphology during the cell division process. Among various bacterial lipids, the physicochemical properties of inverted conical shape lipids, PE and CL, by clustering into intrinsic negative curvature structures (Fig. 1D), can lower the energetic barrier to membrane curvature changes for cell division intermediates.The membrane physical properties can also be modified without substantial changes in lipid composition for AMP-resistant bacteria. Redistribution of lipids through either in-plane diffusion or inner-outer flipping can result in the formation of specific lipid domains and asymmetric localization of specific lipids in the outer or inner leaflet of the membrane bilayer. This asymmetric distribution can then impact on the membrane curvature due to different packing parameters of lipid structures once they are localised into domains of different size.
For example, the transfer of CL from the inner to the outer leaflet upon the interaction of antibacterial agents can also induce a change in membrane curvature. The appearance of these highly curved microscopic regions could be the consequence of CL redistribution, fluidity changes, and enhancement in lipid phase separation. The formation of CL domains and their preferential septal and polar localization have been observed in E. coli, B. subtilis, and P. putida staining with the CL-specific fluorescence dye 10-N-nonyl acridine orange (NAO).16 The increased CL and reduced PG content in the nucleoid-free minicell divided from the cell pole of a minCDE null mutant analysed by mass spectrometry also supported the localization of CL-domain in the E. coli cell pole.194
Relocation and clustering of CL into microdomains has been observed in the P. aeruginosa membrane upon interaction of the CL-binding aminoglycoside antibiotics 3′,6-dinonyl neamine (diNn). The insertion of diNn into the membrane of spheroplasts resulted in decreased fluidity, increased permeability and loss of bacterial rod shape through a decrease in length and increase in curvature,195 which coincided with an enrichment of CL in the membrane outer layer, leading to an increase in membrane curvature.
In daptomycin (DAP)-susceptible E. faecalis, cell membrane structure is preserved. However, in the absence of activation of the LiaFSR system, CL-enriched domains are localized at sites of high membrane curvature of the cell septa and poles. At the sub-MIC level of Ca2+-DAP, the peptide binds to the cell membrane sparing the septum. As the concentration of DAP increases to or above the MIC, the peptide accumulates at the septum, which induces changes in membrane architecture and impairment of cell division, leading to cell death. In DAP-resistant E. faecalis, activation of the LiaSR two-component regulatory system results in redistribution of CL-enriched domains away from the septum. In combination with a drastic reduction in membrane PG content, the calcium-complexed DAP cannot effectively bind to the septum and is diverted to planner membrane sites which are rich in non-PG negatively charged lipids. The alteration in lipid homeostasis together with redistribution of lipid domain-inhibiting AMP oligomerisation, failing to fully distort membrane structure also confers AMP resistance mechanisms.196,197
7. Conclusions and future prospects
There is increasing interest in the properties of biomembranes and how their physical properties are manipulated by the cell in response to environmental stressors. Most studies have focussed on either the molecular composition and lipidomics to give insight into the changes in physical properties or direct measurement of the physical properties and spatial organisation of the membrane. It is timely now to combine these different approaches to delineate in more detail the relationship between all these factors and how these changes in membrane properties impact on the binding of membrane-active molecules, e.g., AMPs.The overall approaches of combining lipidomics, nanostructure imaging and modelling, and quantitative physical and mechanical properties can generate more comprehensive mechanisms for formation of membrane domains and the key molecular factors that can lead to bacterial resistance to membrane-active compounds. While these phenomena are likely to occur across all species and cell types, the focus of this perspective has been the existence and relevance of bacterial membrane domains on the development of resistance to AMP drugs in terms of the compositional effects on membrane structural properties.
Various issues remain to be determined for the molecular mechanisms of membrane remodelling, including:
• How do changes in physical properties affect membrane lipid composition and membrane physiological function or vice versa?
• How do bacteria sense membrane properties and lipid composition and relay the signals to regulate the activity of enzymes and the transcriptional levels of genes involved in lipid remodelling?
• What are the molecular identities of various enzymes (two/three component systems) involved in lipid acyl chain remodelling?
• What are the roles of glycerolipids (MGDG/DGDG) and minor lipids (prenol, carotenoids) in membrane remodelling under stress?
• What are the roles of membrane vesicles in membrane lipid remodelling?
• What are the mechanisms of PE, PI, PS, and CL remodelling of membranes?
• What is the abundance and role of minor lipids in modulating of membrane structural and physical properties?
The premier challenge is to understand the role of physicochemical properties of the membrane bilayer in the regulation of protein activities and cell–cell interactions and how the constantly changing structure impacts on processes such as AMP action, amyloid formation and cell signalling.
Biophysical characterization and nanoimaging analyses of membranes are now poised to make significant advances in understanding the tight regulation of dynamic changes in lipid composition on the homeostatic control of membrane structure and function. The implementation of multiparameter approaches will have enormous impact on a broad range of areas such as membrane protein structure–function, drug design, cell signaling, and biomaterial science.
Author contributions
All authors contributed to the drafting and editing of this perspective.Conflicts of interest
There are no conflicts to declare.Acknowledgements
The authors acknowledge the support of the National Health and Medical Research Council project grant APP1142750 and Ideas Grant ID: 2011990.References
- X. Han and R. W. Gross, J. Lipid Res., 2022, 63, 100164 CrossRef CAS PubMed
.
- K. L. F. Hilton, C. Manwani, J. E. Boles, L. J. White, S. Ozturk, M. D. Garrett and J. R. Hiscock, Chem. Sci., 2021, 12, 13273–13282 RSC
.
- L. R. Joyce and K. S. Doran, PLoS Pathog., 2023, 19, e1011026 CrossRef CAS PubMed
.
- A. Shevchenko and K. Simons, Nat. Rev. Mol. Cell Biol., 2010, 11, 593–598 CrossRef CAS PubMed
.
- C. Sohlenkamp and O. Geiger, FEMS Microbiol. Rev., 2016, 40, 133–159 CrossRef CAS PubMed
.
- Y. M. Zhang and C. O. Rock, Nat. Rev. Microbiol., 2008, 6, 222–233 CrossRef CAS PubMed
.
- G. Chwastek, M. A. Surma, S. Rizk, D. Grosser, O. Lavrynenko, M. Rucinska, H. Jambor and J. Saenz, Cell Rep., 2020, 32, 108165 CrossRef CAS PubMed
.
- T. Harayama and H. Riezman, Nat. Rev. Mol. Cell Biol., 2018, 19, 281–296 CrossRef CAS PubMed
.
- V. I. Band and D. S. Weiss, Antibiotics, 2015, 4, 18–41 CrossRef PubMed
.
- A. Peschel and H. G. Sahl, Nat. Rev. Microbiol., 2006, 4, 529–536 CrossRef CAS PubMed
.
- H. G. Sahl and Y. Shai, Biochim. Biophys. Acta, 2015, 1848, 3019–3020 CrossRef CAS PubMed
.
- T. Dingjan and A. H. Futerman, Bioessays, 2021, 43, e2100021 CrossRef PubMed
.
- E. Ryan and G. E. Reid, Acc. Chem. Res., 2016, 49, 1596–1604 CrossRef CAS PubMed
.
-
A. F. Alvarez and D. Georgellis, in Biogenesis of Fatty Acids, Lipids and Membranes, ed. O. Geiger, Springer International Publishing, Cham, 2019, pp. 575–592, DOI:10.1007/978-3-319-50430-8_39
.
- A. Bochicchio, A. F. Brandner, O. Engberg, D. Huster and R. A. Bockmann, Front. Cell Dev. Biol., 2020, 8, 601145 CrossRef PubMed
.
- E. Mileykovskaya and W. Dowhan, Biochim. Biophys. Acta, 2009, 1788, 2084–2091 CrossRef CAS PubMed
.
- H. Strahl and J. Errington, Annu. Rev. Microbiol., 2017, 71, 519–538 CrossRef CAS PubMed
.
- T. H. Lee, D. J. Hirst and M. I. Aguilar, Biochim. Biophys. Acta, 2015, 1848, 1868–1885 CrossRef CAS PubMed
.
- T. H. Lee, D. J. Hirst, K. Kulkarni, M. P. Del Borgo and M. I. Aguilar, Chem. Rev., 2018, 118, 5392–5487 CrossRef CAS PubMed
.
- I. M. Lopez-Lara and O. Geiger, Biochim. Biophys. Acta, Mol. Cell Biol. Lipids, 2017, 1862, 1287–1299 CrossRef CAS PubMed
.
- J. B. Parsons and C. O. Rock, Prog. Lipid Res., 2013, 52, 249–276 CrossRef CAS PubMed
.
- E. Hawrot and E. P. Kennedy, J. Biol. Chem., 1978, 253, 8213–8220 CrossRef CAS PubMed
.
- A. G. Rietveld, J. A. Killian, W. Dowhan and B. de Kruijff, J. Biol. Chem., 1993, 268, 12427–12433 CrossRef CAS
.
- E. Mileykovskaya and W. Dowhan, J. Bacteriol., 2000, 182, 1172–1175 CrossRef CAS
.
- M. Bogdanov, K. Pyrshev, S. Yesylevskyy, S. Ryabichko, V. Boiko, P. Ivanchenko, R. Kiyamova, Z. Guan, C. Ramseyer and W. Dowhan, Sci. Adv., 2020, 6, eaaz6333 CrossRef CAS PubMed
.
- C. M. Ernst and A. Peschel, Mol. Microbiol., 2011, 80, 290–299 CrossRef CAS PubMed
.
- T. H. Lee, K. N. Hall and M. I. Aguilar, Curr. Top. Med. Chem., 2016, 16, 25–39 CrossRef CAS PubMed
.
- K. Matsumoto, Mol. Microbiol., 2001, 39, 1427–1433 CrossRef CAS PubMed
.
- K. Hall, T. H. Lee, A. I. Mechler, M. J. Swann and M. I. Aguilar, Sci. Rep., 2014, 4, 5479 CrossRef CAS
.
- T. H. Lee, K. N. Hall, M. J. Swann, J. F. Popplewell, S. Unabia, Y. Park, K. S. Hahm and M. I. Aguilar, Biochim. Biophys. Acta, 2010, 1798, 544–557 CrossRef CAS PubMed
.
- T. H. Lee, C. Heng, M. J. Swann, J. D. Gehman, F. Separovic and M. I. Aguilar, Biochim. Biophys. Acta, 2010, 1798, 1977–1986 CrossRef CAS
.
- W. Zhao, T. Rog, A. A. Gurtovenko, I. Vattulainen and M. Karttunen, Biochimie, 2008, 90, 930–938 CrossRef CAS
.
- S. Kikuchi, I. Shibuya and K. Matsumoto, J. Bacteriol., 2000, 182, 371–376 CrossRef CAS PubMed
.
- R. Rashid, Z. J. Nair, D. M. H. Chia, K. K. L. Chong, A. Cazenave Gassiot, S. A. Morley, D. K. Allen, S. L. Chen, S. S. Chng, M. R. Wenk and K. A. Kline, mBio, 2023, 14, e0307322 CrossRef PubMed
.
- C. Slavetinsky, S. Kuhn and A. Peschel, Biochim. Biophys. Acta, Mol. Cell Biol. Lipids, 2017, 1862, 1310–1318 CrossRef CAS PubMed
.
- A. Peschel, R. W. Jack, M. Otto, L. V. Collins, P. Staubitz, G. Nicholson, H. Kalbacher, W. F. Nieuwenhuizen, G. Jung, A. Tarkowski, K. P. van Kessel and J. A. van Strijp, J. Exp. Med., 2001, 193, 1067–1076 CrossRef CAS PubMed
.
- A. M. Smith, J. S. Harrison, K. M. Sprague and H. Roy, J. Biol. Chem., 2013, 288, 22768–22776 CrossRef CAS PubMed
.
- E. Kilelee, A. Pokorny, M. R. Yeaman and A. S. Bayer, Antimicrob. Agents Chemother., 2010, 54, 4476–4479 CrossRef CAS PubMed
.
- R. P. Rehal, H. Marbach, A. T. M. Hubbard, A. A. Sacranie, F. Sebastiani, G. Fragneto and R. D. Harvey, Chem. Phys. Lipids, 2017, 206, 60–70 CrossRef CAS PubMed
.
- H. Goldfine, J. Lipid Res., 1984, 25, 1501–1507 CrossRef CAS
.
- G. Lindblom, J. B. Hauksson, L. Rilfors, B. Bergenstahl, A. Wieslander and P. O. Eriksson, J. Biol. Chem., 1993, 268, 16198–16207 CrossRef CAS PubMed
.
- A. R. Niemi, L. Rilfors and G. Lindblom, Biochim. Biophys. Acta, 1995, 1239, 186–194 CrossRef PubMed
.
- L. Rilfors, A. Wieslander and G. Lindblom, Subcell. Biochem., 1993, 20, 109–166 CAS
.
- T. Sajed, A. Marcu, M. Ramirez, A. Pon, A. C. Guo, C. Knox, M. Wilson, J. R. Grant, Y. Djoumbou and D. S. Wishart, Nucleic Acids Res., 2016, 44, D495–D501 CrossRef CAS
.
- R. Ernst, C. S. Ejsing and B. Antonny, J. Mol. Biol., 2016, 428, 4776–4791 CrossRef CAS
.
- R. Ernst, S. Ballweg and I. Levental, Curr. Opin. Cell Biol., 2018, 53, 44–51 CrossRef CAS PubMed
.
- J. E. Horne, D. J. Brockwell and S. E. Radford, J. Biol. Chem., 2020, 295, 10340–10367 CrossRef CAS PubMed
.
- L. M. Mitchison-Field and B. J. Belin, Curr. Opin. Microbiol., 2023, 74, 102315 CrossRef CAS PubMed
.
- J. C. Henderson, S. M. Zimmerman, A. A. Crofts, J. M. Boll, L. G. Kuhns, C. M. Herrera and M. S. Trent, Annu. Rev. Microbiol., 2016, 70, 255–278 CrossRef CAS PubMed
.
- B. Lugtenberg and L. Van Alphen, Biochim. Biophys. Acta, 1983, 737, 51–115 CrossRef CAS
.
- E. Lundstedt, D. Kahne and N. Ruiz, Chem. Rev., 2021, 121, 5098–5123 CrossRef CAS
.
- J. R. Willdigg and J. D. Helmann, Front. Mol. Biosci., 2021, 8, 634438 CrossRef CAS
.
- B. J. Belin, N. Busset, E. Giraud, A. Molinaro, A. Silipo and D. K. Newman, Nat. Rev. Microbiol., 2018, 16, 304–315 CrossRef CAS PubMed
.
- M. Bloom, E. Evans and O. G. Mouritsen, Q. Rev. Biophys., 1991, 24, 293–397 CrossRef CAS PubMed
.
- W. Seel, D. Baust, D. Sons, M. Albers, L. Etzbach, J. Fuss and A. Lipski, Sci. Rep., 2020, 10, 330 CrossRef CAS PubMed
.
- B. Bechinger and S. U. Gorr, J. Dent. Res., 2017, 96, 254–260 CrossRef CAS PubMed
.
- D. Roversi, C. Troiano, E. Salnikov, L. Giordano, F. Riccitelli, M. De Zotti, B. Casciaro, M. R. Loffredo, Y. Park, F. Formaggio, M. L. Mangoni, B. Bechinger and L. Stella, Biophys. Chem., 2023, 300, 107060 CrossRef CAS PubMed
.
- M. A. Sani and F. Separovic, Chemistry, 2018, 24, 286–291 CrossRef CAS
.
- F. Separovic, D. W. Keizer and M. A. Sani, Front. Biomed. Biotechnol., 2020, 2, 610203 Search PubMed
.
- K. Hammond, M. G. Ryadnov and B. W. Hoogenboom, Biochim. Biophys. Acta, Biomembr., 2021, 1863, 183447 CrossRef CAS PubMed
.
- C. Aisenbrey, A. Marquette and B. Bechinger, Adv. Exp. Med. Biol., 2019, 1117, 33–64 CrossRef CAS PubMed
.
- B. Bechinger, J. M. Resende and C. Aisenbrey, Biophys. Chem., 2011, 153, 115–125 CrossRef CAS PubMed
.
-
Solid-State NMR: Applications in Biomembrane Structure, ed. F. Separovic and M. A. Sani, IOP Publishing Ltd, 2020 Search PubMed
.
- A. S. Klymchenko and R. Kreder, Chem. Biol., 2014, 21, 97–113 CrossRef CAS
.
- Y. Niko and A. S. Klymchenko, J. Biochem., 2021, 170, 163–174 CrossRef CAS PubMed
.
- J. Bernardino de la Serna, G. J. Schütz, C. Eggeling and M. Cebecauer, Front. Cell Dev. Biol., 2016, 4, 106 CrossRef PubMed
.
- J. Lombard, Biol. Direct, 2014, 9, 32 CrossRef PubMed
.
- G. L. Nicolson and G. Ferreira de Mattos, Biomedicines, 2022, 10, 1711 CrossRef CAS PubMed
.
- M. Sinensky, Proc. Natl. Acad. Sci. U. S. A., 1974, 71, 522–525 CrossRef CAS PubMed
.
- D. Chapman, Q. Rev. Biophys., 1975, 8, 185–235 CrossRef CAS PubMed
.
- G. Enkavi, M. Javanainen, W. Kulig, T. Rog and I. Vattulainen, Chem. Rev., 2019, 119, 5607–5774 CrossRef CAS PubMed
.
- S. J. Marrink, V. Corradi, P. C. T. Souza, H. I. Ingolfsson, D. P. Tieleman and M. S. P. Sansom, Chem. Rev., 2019, 119, 6184–6226 CrossRef CAS
.
- A. Blanco-Gonzalez, A. Pineiro and R. Garcia-Fandino, Comput. Struct. Biotechnol. J., 2022, 20, 2798–2806 CrossRef CAS PubMed
.
- S. Moradi, A. Nowroozi and M. Shahlaei, RSC Adv., 2019, 9, 4644–4658 RSC
.
- A. N. Leonard, E. Wang, V. Monje-Galvan and J. B. Klauda, Chem. Rev., 2019, 119, 6227–6269 CrossRef CAS PubMed
.
- R. A. Corey, M. Baaden and M. Chavent, Front. Bioinform., 2023, 3, 1149744 CrossRef CAS PubMed
.
- M. Chavent, T. Reddy, J. Goose, A. C. Dahl, J. E. Stone, B. Jobard and M. S. Sansom, Faraday Discuss., 2014, 169, 455–475 RSC
.
- M. Tripathy, S. Thangamani and A. Srivastava, J. Chem. Theory Comput., 2020, 16, 7800–7816 CrossRef CAS PubMed
.
- R. X. Gu, S. Baoukina and D. P. Tieleman, J. Am. Chem. Soc., 2020, 142, 2844–2856 CrossRef CAS PubMed
.
- A. H. Larsen, Int. J. Mol. Sci., 2022, 23, 8098 CrossRef CAS PubMed
.
- W. Pezeshkian and S. J. Marrink, Curr. Opin. Cell Biol., 2021, 71, 103–111 CrossRef CAS PubMed
.
- M. Pohnl, C. Kluge and R. A. Bockmann, J. Chem. Theory Comput., 2023, 19, 1908–1921 CrossRef PubMed
.
- M. Ramezanpour, M. L. Schmidt, B. Y. M. Bashe, J. R. Pruim, M. L. Link, P. R. Cullis, P. E. Harper, J. L. Thewalt and D. P. Tieleman, Langmuir, 2020, 36, 6668–6680 CrossRef CAS PubMed
.
- M. Sonora, L. Martinez, S. Pantano and M. R. Machado, J. Chem. Inf. Model., 2021, 61, 408–422 CrossRef CAS PubMed
.
- J. M. Benarroch and M. Asally, Trends Microbiol., 2020, 28, 304–314 CrossRef CAS PubMed
.
-
R. Koynova and B. Tenchov, in Encyclopedia of Biophysics, ed. G. C. K. Roberts, Springer Berlin Heidelberg, Berlin, Heidelberg, 2013, pp. 1841–1854, DOI:10.1007/978-3-642-16712-6_542
.
- S. Legendre, L. Letellier and E. Shechter, Biochim. Biophys. Acta, 1980, 602, 491–505 CrossRef CAS PubMed
.
- S. Vanounou, D. Pines, E. Pines, A. H. Parola and I. Fishov, Photochem. Photobiol., 2002, 76, 1–11 CrossRef CAS PubMed
.
- E. Chmiel, C. E. Galuska, P. Koper, B. Kowalczyk, T. Urbanik-Sypniewska, M. Palusinska-Szysz and B. Fuchs, Metabolites, 2022, 12, 418 CrossRef CAS PubMed
.
- S. E. Diomande, C. Nguyen-The, M. H. Guinebretiere, V. Broussolle and J. Brillard, Front. Microbiol., 2015, 6, 813 Search PubMed
.
- J. Gidden, J. Denson, R. Liyanage, D. M. Ivey and J. O. Lay, Int. J. Mass Spectrom., 2009, 283, 178–184 CrossRef CAS PubMed
.
- K. M. Hines, G. Alvarado, X. Chen, C. Gatto, A. Pokorny, F. Alonzo III, B. J. Wilkinson and L. Xu, mSphere, 2020, 5 DOI:10.1128/msphere.00339-20
.
- S. Morein, A. Andersson, L. Rilfors and G. Lindblom, J. Biol. Chem., 1996, 271, 6801–6809 CrossRef CAS PubMed
.
- M. Suutari and S. Laakso, Crit. Rev. Microbiol., 1994, 20, 285–328 CrossRef CAS PubMed
.
-
M. Daffé, A. Quémard and H. Marrakchi, in Biogenesis of Fatty Acids, Lipids and Membranes, ed. O. Geiger, Springer International Publishing, Cham, 2017, pp. 1–36, DOI:10.1007/978-3-319-43676-0_18-1
.
- S. E. Diomande, M. H. Guinebretiere, B. De Sarrau, C. Nguyen-the, V. Broussolle and J. Brillard, BMC Res. Notes, 2015, 8, 329 CrossRef PubMed
.
- J. B. Parsons, M. W. Frank, C. Subramanian, P. Saenkham and C. O. Rock, Proc. Natl. Acad. Sci. U. S. A., 2011, 108, 15378–15383 CrossRef CAS
.
-
D. Marsh, Handbook of lipid bilayers, CRC Press, 2nd edn, 2013 Search PubMed
.
- C. Huang, Lipids, 2001, 36, 1077–1097 CrossRef CAS PubMed
.
- I. Budin, T. de Rond, Y. Chen, L. J. G. Chan, C. J. Petzold and J. D. Keasling, Science, 2018, 362, 1186–1189 CrossRef CAS PubMed
.
- E. Saita, D. Albanesi and D. de Mendoza, Biochim. Biophys. Acta, 2016, 1861, 837–846 CrossRef CAS PubMed
.
-
S. G. Altabe, M. C. Mansilla and D. de Mendoza, in Stearoyl-CoA Desaturase Genes in Lipid Metabolism, ed. P. D. J. M. Ntambi, Springer New York, New York, NY, 2013, pp. 209–231, DOI:10.1007/978-1-4614-7969-7_15
.
- M. C. Mansilla and D. de Mendoza, Arch. Microbiol., 2005, 183, 229–235 CrossRef CAS PubMed
.
- K. Zhu, K. H. Choi, H. P. Schweizer, C. O. Rock and Y. M. Zhang, Mol. Microbiol., 2006, 60, 260–273 CrossRef CAS PubMed
.
- L. Chazarreta Cifre, M. Alemany, D. de Mendoza and S. Altabe, Appl. Environ. Microbiol., 2013, 79, 6271–6279 CrossRef PubMed
.
- E. Shechter, L. Letellier and G. Gulik-Krzywicki, Eur. J. Biochem., 1974, 49, 61–76 CrossRef CAS PubMed
.
- M. Esfahani, A. R. Limbrick, S. Knutton, T. Oka and S. J. Wakil, Proc. Natl. Acad. Sci. U. S. A., 1971, 68, 3180–3184 CrossRef CAS PubMed
.
- G. Kenanian, C. Morvan, A. Weckel, A. Pathania, J. Anba-Mondoloni, D. Halpern, M. Gaillard, A. Solgadi, L. Dupont, C. Henry, C. Poyart, A. Fouet, G. Lamberet, K. Gloux and A. Gruss, Cell Rep., 2019, 29, 3974–3982 CrossRef CAS PubMed
.
- J. B. Parsons, T. C. Broussard, J. L. Bose, J. W. Rosch, P. Jackson, C. Subramanian and C. O. Rock, Proc. Natl. Acad. Sci. U. S. A., 2014, 111, 10532–10537 CrossRef CAS PubMed
.
- Z. DeMars, V. K. Singh and J. L. Bose, J. Bacteriol., 2020, 202 DOI:10.1128/jb.00128-20
.
- S. Sen, S. Sirobhushanam, S. R. Johnson, Y. Song, R. Tefft, C. Gatto and B. J. Wilkinson, PLoS One, 2016, 11, e0165300 CrossRef PubMed
.
- C. N. Krute, M. J. Ridder, N. A. Seawell and J. L. Bose, Microbiology, 2019, 165, 197–207 CrossRef CAS PubMed
.
- C. Eberlein, T. Baumgarten, S. Starke and H. J. Heipieper, Appl. Microbiol. Biotechnol., 2018, 102, 2583–2593 CrossRef CAS PubMed
.
- H. Keweloh and H. J. Heipieper, Lipids, 1996, 31, 129–137 CrossRef CAS PubMed
.
- T. Kondakova, S. Kumar and J. E. Cronan, Chem. Phys. Lipids, 2019, 222, 23–35 CrossRef CAS PubMed
.
- T. Kaneda, Microbiol. Rev., 1991, 55, 288–302 CrossRef CAS PubMed
.
- T. Kaneda, Biochim. Biophys. Acta, 1972, 270, 32–39 CrossRef CAS PubMed
.
- J. C. Kaiser, S. Sen, A. Sinha, B. J. Wilkinson and D. E. Heinrichs, Mol. Microbiol., 2016, 102, 850–864 CrossRef CAS PubMed
.
- V. K. Singh, D. S. Hattangady, E. S. Giotis, A. K. Singh, N. R. Chamberlain, M. K. Stuart and B. J. Wilkinson, Appl. Environ. Microbiol., 2008, 74, 5882–5890 CrossRef CAS PubMed
.
- R. N. Lewis, B. D. Sykes and R. N. McElhaney, Biochemistry, 1987, 26, 4036–4044 CrossRef CAS PubMed
.
- M. Gohrbandt, A. Lipski, J. W. Grimshaw, J. A. Buttress, Z. Baig, B. Herkenhoff, S. Walter, R. Kurre, G. Deckers-Hebestreit and H. Strahl, EMBO J., 2022, 41, e109800 CrossRef CAS PubMed
.
- D. F. Silbert, R. C. Ladenson and J. L. Honegger, Biochim. Biophys. Acta, 1973, 311, 349–361 CrossRef CAS PubMed
.
- L. H. Wang, M. S. Wang, X. A. Zeng and Z. W. Liu, Biochim. Biophys. Acta, 2016, 1858, 1791–1800 CrossRef CAS PubMed
.
- Y. Oogai, M. Matsuo, M. Hashimoto, F. Kato, M. Sugai and H. Komatsuzawa, Appl. Environ. Microbiol., 2011, 77, 8097–8105 CrossRef CAS PubMed
.
- N. Malachowa, A. R. Whitney, S. D. Kobayashi, D. E. Sturdevant, A. D. Kennedy, K. R. Braughton, D. W. Shabb, B. A. Diep, H. F. Chambers, M. Otto and F. R. DeLeo, PLoS One, 2011, 6, e18617 CrossRef CAS PubMed
.
- A. Mu, W. P. Klare, S. L. Baines, C. N. Ignatius Pang, R. Guerillot, N. Harbison-Price, N. Keller, J. Wilksch, N. T. K. Nhu, M. D. Phan, B. Keller, B. Nijagal, D. Tull, S. Dayalan, H. H. C. Chua, D. Skoneczny, J. Koval, A. Hachani, A. D. Shah, N. Neha, S. Jadhav, S. R. Partridge, A. J. Cork, K. Peters, O. Bertolla, S. Brouwer, S. J. Hancock, L. Alvarez-Fraga, D. M. P. De Oliveira, B. Forde, A. Dale, W. Mujchariyakul, C. J. Walsh, I. Monk, A. Fitzgerald, M. Lum, C. Correa-Ospina, P. Roy Chowdhury, R. G. Parton, J. De Voss, J. Beckett, F. Monty, J. McKinnon, X. Song, J. R. Stephen, M. Everest, M. I. Bellgard, M. Tinning, M. Leeming, D. Hocking, L. Jebeli, N. Wang, N. Ben Zakour, S. A. Yasar, S. Vecchiarelli, T. Russell, T. Zaw, T. Chen, D. Teng, Z. Kassir, T. Lithgow, A. Jenney, J. N. Cole, V. Nizet, T. C. Sorrell, A. Y. Peleg, D. L. Paterson, S. A. Beatson, J. Wu, M. P. Molloy, A. E. Syme, R. J. A. Goode, A. A. Hunter, G. Bowland, N. P. West, M. R. Wilkins, S. P. Djordjevic, M. R. Davies, T. Seemann, B. P. Howden, D. Pascovici, S. Tyagi, R. B. Schittenhelm, D. P. De Souza, M. J. McConville, J. R. Iredell, S. J. Cordwell, R. A. Strugnell, T. P. Stinear, M. A. Schembri and M. J. Walker, Nat. Commun., 2023, 14, 1530 CrossRef CAS PubMed
.
- A. Y. Wang and J. E. Cronan Jr, Mol. Microbiol., 1994, 11, 1009–1017 CrossRef CAS PubMed
.
- Y. Y. Chang and J. E. Cronan Jr, Mol. Microbiol., 1999, 33, 249–259 CrossRef CAS PubMed
.
- Y. Xu, Z. Zhao, W. Tong, Y. Ding, B. Liu, Y. Shi, J. Wang, S. Sun, M. Liu, Y. Wang, Q. Qi, M. Xian and G. Zhao, Nat. Commun., 2020, 11, 1496 CrossRef CAS PubMed
.
- D. W. Grogan and J. E. Cronan Jr, Microbiol. Mol. Biol. Rev., 1997, 61, 429–441 CAS
.
- H. Velly, M. Bouix, S. Passot, C. Penicaud, H. Beinsteiner, S. Ghorbal, P. Lieben and F. Fonseca, Appl. Microbiol. Biotechnol., 2015, 99, 907–918 CrossRef CAS PubMed
.
- E. J. Dufourc, I. C. P. Smith and H. C. Jarrell, Biochemistry, 1984, 23, 2300–2309 CrossRef CAS
.
- D. Poger and A. E. Mark, J. Phys. Chem. B, 2015, 119, 5487–5495 CrossRef CAS PubMed
.
- N. Loffhagen, C. Härtig, W. Geyer, M. Voyevoda and H. Harms, Eng. Life Sci., 2007, 7, 67–74 CrossRef CAS
.
- A. Grigor’eva, A. Bardasheva, A. Tupitsyna, N. Amirkhanov, N. Tikunova, D. Pyshnyi and E. Ryabchikova, Microorganisms, 2020, 8, 1991 CrossRef PubMed
.
- V. R. Matias and T. J. Beveridge, J. Bacteriol., 2006, 188, 1011–1021 CrossRef CAS PubMed
.
- H. T. Nguyen, L. A. O’Donovan, H. Venter, C. C. Russell, A. McCluskey, S. W. Page, D. J. Trott and A. D. Ogunniyi, Antibiotics, 2021, 10, 307 CrossRef CAS PubMed
.
- J. Ubbink and P. Schar-Zammaretti, Micron, 2005, 36, 293–320 CrossRef CAS PubMed
.
- B. A. Cornell and F. Separovic, Biochim. Biophys. Acta, 1983, 733, 189–193 CrossRef CAS PubMed
.
- G. Binnig, C. F. Quate and C. Gerber, Phys. Rev. Lett., 1986, 56, 930–933 CrossRef PubMed
.
- S. D. Connell and D. A. Smith, Mol. Membr. Biol., 2006, 23, 17–28 CrossRef CAS PubMed
.
- Y. F. Dufrene, T. Ando, R. Garcia, D. Alsteens, D. Martinez-Martin, A. Engel, C. Gerber and D. J. Muller, Nat. Nanotechnol., 2017, 12, 295–307 CrossRef CAS PubMed
.
- E. I. Goksu, J. M. Vanegas, C. D. Blanchette, W. C. Lin and M. L. Longo, Biochim. Biophys. Acta, 2009, 1788, 254–266 CrossRef CAS PubMed
.
- L. Picas, P. E. Milhiet and J. Hernández-Borrell, Chem. Phys. Lipids, 2012, 165, 845–860 CrossRef CAS PubMed
.
- R. M. Sullan, J. K. Li and S. Zou, Langmuir, 2009, 25, 7471–7477 CrossRef CAS PubMed
.
- M. Krieg, G. Fläschner, D. Alsteens, B. M. Gaub, W. H. Roos, G. J. L. Wuite, H. E. Gaub, C. Gerber, Y. F. Dufrêne and D. J. Müller, Nat. Rev. Phys., 2019, 1, 41–57 CrossRef
.
- A. Aufderhorst-Roberts, U. Chandra and S. D. Connell, Biophys. J., 2017, 112, 313–324 CrossRef CAS PubMed
.
- A. Alessandrini, H. M. Seeger, T. Caramaschi and P. Facci, Biophys. J., 2012, 103, 38–47 CrossRef CAS PubMed
.
- K. El Kirat, S. Morandat and Y. F. Dufrene, Biochim. Biophys. Acta, 2010, 1798, 750–765 CrossRef CAS PubMed
.
- K. Hammond, G. Benn, I. Bennett, E. S. Parsons, M. G. Ryadnov, B. W. Hoogenboom and A. L. B. Pyne, Methods Mol. Biol., 2021, 2208, 225–235 CrossRef CAS PubMed
.
- A. Melcrova, S. Maity, J. Melcr, N. A. W. de Kok, M. Gabler, J. van der Eyden, W. Stensen, J. S. M. Svendsen, A. J. M. Driessen, S. J. Marrink and W. H. Roos, Nat. Commun., 2023, 14, 4038 CrossRef CAS PubMed
.
- B. Bechinger, J. Pept. Sci., 2015, 21, 346–355 CrossRef CAS PubMed
.
- M. A. Sani and F. Separovic, Acc. Chem. Res., 2016, 49, 1130–1138 CrossRef CAS PubMed
.
- T. H. Lee, V. Hofferek, M. A. Sani, F. Separovic, G. E. Reid and M. I. Aguilar, Faraday Discuss., 2021, 232, 399–418 RSC
.
- Y. Kikuchi, N. Obana, M. Toyofuku, N. Kodera, T. Soma, T. Ando, Y. Fukumori, N. Nomura and A. Taoka, Nanoscale, 2020, 12, 7950–7959 RSC
.
- M. Toyofuku, S. Schild, M. Kaparakis-Liaskos and L. Eberl, Nat. Rev. Microbiol., 2023, 21, 415–430 CrossRef CAS PubMed
.
- Y. F. Dufrene, A. Viljoen, J. Mignolet and M. Mathelie-Guinlet, Cell. Microbiol., 2021, 23, e13324 CrossRef CAS PubMed
.
- S. Ishii, S. Yoshimoto and K. Hori, J. Colloid Interface Sci., 2022, 606, 628–634 CrossRef CAS PubMed
.
- S. Manioglu, S. M. Modaresi, N. Ritzmann, J. Thoma, S. A. Overall, A. Harms, G. Upert, A. Luther, A. B. Barnes, D. Obrecht, D. J. Muller and S. Hiller, Nat. Commun., 2022, 13, 6195 CrossRef CAS PubMed
.
- L. Pasquina-Lemonche, J. Burns, R. D. Turner, S. Kumar, R. Tank, N. Mullin, J. S. Wilson, B. Chakrabarti, P. A. Bullough, S. J. Foster and J. K. Hobbs, Nature, 2020, 582, 294–297 CrossRef CAS PubMed
.
- R. D. Turner, S. Mesnage, J. K. Hobbs and S. J. Foster, Nat. Commun., 2018, 9, 1263 CrossRef PubMed
.
- A. Viljoen, S. J. Foster, G. E. Fantner, J. K. Hobbs and Y. F. Dufrene, mBio, 2020, 11 DOI:10.1128/mbio.03020-19
.
- A. Viljoen, E. Rath, J. D. McKinney, G. E. Fantner and Y. F. Dufrene, J. Bacteriol., 2021, 203 DOI:10.1128/jb.00547-20
.
- S. Garcia-Manyes, L. Redondo-Morata, G. Oncins and F. Sanz, J. Am. Chem. Soc., 2010, 132, 12874–12886 CrossRef CAS PubMed
.
- M. Majewska, D. Mrdenovic, I. S. Pieta, R. Nowakowski and P. Pieta, Biochim. Biophys. Acta, Biomembr., 2020, 1862, 183347 CrossRef CAS PubMed
.
- E. R. Rojas, G. Billings, P. D. Odermatt, G. K. Auer, L. Zhu, A. Miguel, F. Chang, D. B. Weibel, J. A. Theriot and K. C. Huang, Nature, 2018, 559, 617–621 CrossRef CAS PubMed
.
- A. Mularski, J. J. Wilksch, E. Hanssen, R. A. Strugnell and F. Separovic, Biochim. Biophys. Acta, 2016, 1858, 1091–1098 CrossRef CAS PubMed
.
- A. Mularski, J. J. Wilksch, H. Wang, M. A. Hossain, J. D. Wade, F. Separovic, R. A. Strugnell and M. L. Gee, Langmuir, 2015, 31, 6164–6171 CrossRef CAS PubMed
.
- J. W. Goss and C. B. Volle, ACS Appl. Bio Mater., 2020, 3, 143–155 CrossRef CAS PubMed
.
- G. K. Auer and D. B. Weibel, Biochemistry, 2017, 56, 3710–3724 CrossRef CAS PubMed
.
- M. LeClaire, J. Gimzewski and S. Sharma, Nano Sel., 2021, 2, 1–15 CrossRef CAS
.
- D. Vorselen, M. C. Piontek, W. H. Roos and G. J. L. Wuite, Front. Mol. Biosci., 2020, 7, 139 CrossRef CAS PubMed
.
- A. H. Holmes, L. S. Moore, A. Sundsfjord, M. Steinbakk, S. Regmi, A. Karkey, P. J. Guerin and L. J. Piddock, Lancet, 2016, 387, 176–187 CrossRef CAS PubMed
.
- T. H. Lee, V. Hofferek, F. Separovic, G. E. Reid and M. I. Aguilar, Curr. Opin. Chem. Biol., 2019, 52, 85–92 CrossRef CAS PubMed
.
- E. Salnikov, C. Aisenbrey and B. Bechinger, Biochim. Biophys. Acta, Biomembr., 2022, 1864, 183844 CrossRef CAS PubMed
.
- E. Salnikov and B. Bechinger, Biochim. Biophys. Acta, Biomembr., 2022, 1864, 184001 CrossRef CAS PubMed
.
- H. Roy and M. Ibba, Proc. Natl. Acad. Sci. U. S. A., 2008, 105, 4667–4672 CrossRef CAS PubMed
.
- A. S. Bayer, R. Prasad, J. Chandra, A. Koul, M. Smriti, A. Varma, R. A. Skurray, N. Firth, M. H. Brown, S. P. Koo and M. R. Yeaman, Infect. Immun., 2000, 68, 3548–3553 CrossRef CAS PubMed
.
- P. Lather, A. K. Mohanty, P. Jha, A. K. Garsa and S. K. Sood, Eur. Food Res. Technol., 2015, 240, 101–107 CrossRef CAS
.
- R. Kumariya, S. K. Sood, Y. S. Rajput, N. Saini and A. K. Garsa, Biochim. Biophys. Acta, 2015, 1848, 1367–1375 CrossRef CAS PubMed
.
- N. N. Mishra, A. S. Bayer, T. T. Tran, Y. Shamoo, E. Mileykovskaya, W. Dowhan, Z. Guan and C. A. Arias, PLoS One, 2012, 7, e43958 CrossRef CAS PubMed
.
- A. Pokorny, E. M. Kilelee, D. Wu and P. F. Almeida, Biophys. J., 2008, 95, 4748–4755 CrossRef CAS PubMed
.
- T. H. Lee, M. A. Sani, S. Overall, F. Separovic and M. I. Aguilar, Biochim. Biophys. Acta, Biomembr., 2018, 1860, 300–309 CrossRef CAS PubMed
.
- A. Muller, M. Wenzel, H. Strahl, F. Grein, T. N. V. Saaki, B. Kohl, T. Siersma, J. E. Bandow, H. G. Sahl, T. Schneider and L. W. Hamoen, Proc. Natl. Acad. Sci. U. S. A., 2016, 113, E7077–E7086 CrossRef PubMed
.
- S. Omardien, J. W. Drijfhout, F. M. Vaz, M. Wenzel, L. W. Hamoen, S. A. J. Zaat and S. Brul, Biochim. Biophys. Acta, Biomembr., 2018, 1860, 2404–2415 CrossRef CAS PubMed
.
- K. Scheinpflug, M. Wenzel, O. Krylova, J. E. Bandow, M. Dathe and H. Strahl, Sci. Rep., 2017, 7, 44332 CrossRef PubMed
.
- J. Li, M. L. Chikindas, R. D. Ludescher and T. J. Montville, Appl. Environ. Microbiol., 2002, 68, 5904–5910 CrossRef CAS PubMed
.
- Y. Sakayori, M. Muramatsu, S. Hanada, Y. Kamagata, S. Kawamoto and J. Shima, Microbiology, 2003, 149, 2901–2908 CrossRef CAS PubMed
.
- V. Vadyvaloo, J. W. Hastings, M. J. van der Merwe and M. Rautenbach, Appl. Environ. Microbiol., 2002, 68, 5223–5230 CrossRef CAS PubMed
.
- E. Cox, A. Michalak, S. Pagentine, P. Seaton and A. Pokorny, Biochim. Biophys. Acta, 2014, 1838, 2198–2204 CrossRef CAS PubMed
.
- J. E. Cronan and T. Luk, Microbiol. Mol. Biol. Rev., 2022, 86, e0001322 CrossRef PubMed
.
- C. de Carvalho and M. J. Caramujo, Molecules, 2018, 23, 2583 CrossRef PubMed
.
- X. Jiang, Y. Duan, B. Zhou, Q. Guo, H. Wang, X. Hang, L. Zeng, J. Jia and H. Bi, J. Bacteriol., 2019, 201 DOI:10.1128/jb.00374-19
.
- C. M. Koppelman, T. Den Blaauwen, M. C. Duursma, R. M. Heeren and N. Nanninga, J. Bacteriol., 2001, 183, 6144–6147 CrossRef CAS PubMed
.
- M. El Khoury, J. Swain, G. Sautrey, L. Zimmermann, P. Van Der Smissen, J. L. Decout and M. P. Mingeot-Leclercq, Sci. Rep., 2017, 7, 10697 CrossRef PubMed
.
- T. T. Tran, J. M. Munita and C. A. Arias, Ann. N. Y. Acad. Sci., 2015, 1354, 32–53 CrossRef PubMed
.
- T. T. Tran, D. Panesso, N. N. Mishra, E. Mileykovskaya, Z. Guan, J. M. Munita, J. Reyes, L. Diaz, G. M. Weinstock, B. E. Murray, Y. Shamoo, W. Dowhan, A. S. Bayer and C. A. Arias, mBio, 2013, 4 DOI:10.1128/mbio.00281-13
.
| This journal is © The Royal Society of Chemistry 2024 |