Open Access Article
Open Access ArticleCreative Commons Attribution 3.0 Unported Licence
Design, synthesis, and antiproliferative activity of new 1,2,3-triazole/quinazoline-4-one hybrids as dual EGFR/BRAFV600E inhibitors†
Amira M. Mohameda,
Ola M. F. Abou-Ghadira,
Yaser A. Mostafaa,
Zainab M. Almarhoon b,
Stefan Bräse
b,
Stefan Bräse *c and
Bahaa G. M. Youssif
*c and
Bahaa G. M. Youssif *a
*a
aPharmaceutical Organic Chemistry Department, Faculty of Pharmacy, Assiut University, Assiut 71526, Egypt. E-mail: bgyoussif2@gmail.com; bahaa.youssif@pharm.aun.edu.eg
bDepartment of Chemistry, College of Science, King Saud University, Riyadh 11451, Saudi Arabia
cInstitute of Biological and Chemical Systems, IBCS-FMS, Karlsruhe Institute of Technology, 76131 Karlsruhe, Germany. E-mail: braese@kit.edu
First published on 5th December 2024
Abstract
A novel series of 1,2,3-triazole/quinazoline-4-one hybrids (8a–t) were designed and synthesized as dual-targeted antiproliferative agents. Compounds 8a–t were evaluated for their antiproliferative efficacy against a panel of four cancer cell lines. The results indicated that most of the evaluated compounds exhibited strong antiproliferative activity, with 8f, 8g, 8h, 8j, and 8l demonstrating the highest potency. These five compounds were investigated as EGFR and BRAFV600E inhibitors. The in vitro tests showed that compounds 8g, 8h, and 8j are strong antiproliferative agents that might work as dual EGFR/BRAFV600E inhibitors. Compounds 8g and 8h were further examined as activators of caspases 3, 8, and Bax and down-regulators of the anti-apoptotic protein Bcl2. The results indicated that the studied compounds had considerable apoptotic antiproliferative action. The investigation of the cell cycle and apoptosis revealed that compound 8g induces cell cycle arrest during the G1 phase transition. Molecular docking experiments are thoroughly examined to validate the binding interactions of the most active hybrids with the active sites of EGFR and BRAFV600E. The data indicated that the examined compounds can efficiently engage with essential amino acid residues in both kinases.
1. Introduction
Cancer is abnormal cellular proliferation resulting from an imbalance between cell division and apoptosis.1,2 Despite the availability of different therapeutic modalities, including surgical intervention, radiation, immunotherapy, and chemotherapy, cancer continues to be a contentious clinical issue. Nevertheless, effective chemotherapeutic agents with minimal side effects have garnered significant interest.3,4 Elucidating and revealing cancer's molecular and cellular roots has significantly contributed to advancing cancer therapies. Currently, cancer therapy identifies membrane receptors of the tyrosine kinase (TK) family as primary targets.5,6 The epidermal growth factor (EGF) family includes important membrane receptors, with the epidermal growth factor receptor (EGFR) getting much attention. It has been found that blocking EGFR signaling is recognized as a potent therapeutic strategy.7,8Clinical studies have shown that combining B-Rapidly Accelerated Fibrosarcoma (B-raf proto-oncogene or BRAF) and tyrosine kinase (TK) inhibitors effectively stops tumor growth and overcomes resistance. Concomitant use of EGFR inhibitors may mitigate resistance to vemurafenib, a mutant BRAF (BRAFV600E) inhibitor, in thyroid cancer.9 This combination has also demonstrated encouraging outcomes in BRAFV600E colorectal cancer.10 Furthermore, researchers have found several compounds in vitro that contain the critical pharmacophoric groups required to inhibit tyrosine kinases, such as epidermal growth factor receptor/vascular endothelial growth factor receptor-2 (EGFR/VEGFR-2) and BRAF.11,12 For example, compound I (Fig. 1) inhibited wild-type BRAF and EGFR, exhibiting IC50 values in the nanomolar range. Furthermore, imidazo[1,2-b]pyridazine II inhibited BRAF and VEGFR-2.
Heterocyclic compounds have yielded a plethora of commercialized medicines and bioactive substances, and their notable cytotoxicity has been essential in the design and production of anticancer agents.13,14 Consequently, quinazolines have garnered significant attention owing to their efficacy and specificity.15,16 Quinazolines' anticancer properties can be traced back to Paganini (vaccine), a naturally occurring quinazoline discovered in 1888.17 Gefitinib, erlotinib, and lapatinib are FDA-approved quinazoline-based anticancer agents that function as EGFR inhibitors18–20 (Fig. 2).
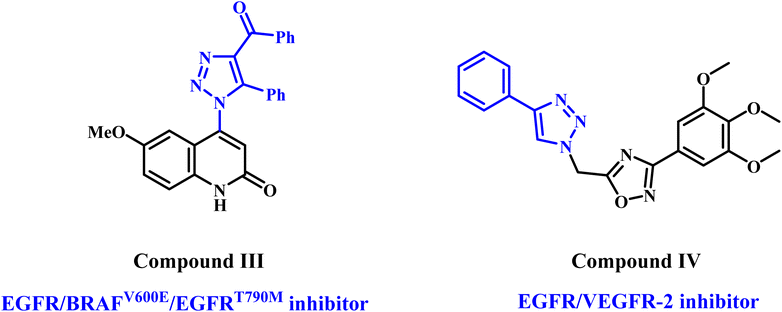
On the other hand, 1,2,3-triazoles are important scaffolds often synthesized using the 1,3-dipolar cycloaddition reaction (click chemistry reaction) between terminal acetylenes and azides.21–23 The anticancer activity of 1,2,3-triazoles has garnered significant attention regarding their therapeutic properties.24–26 In a recent publication from our lab,24 we describe the discovery of a novel class of 1,2,3-triazole/quinoline hybrids as antiproliferative compounds that act as multi-targeted inhibitors. Most novel compounds demonstrated considerable antiproliferative activity against a panel of four cancer cell lines. With IC50 values of 57 nM for EGFR, 68 nM for BRAFV600E, and 9.70 nM for EGFRT790M, compound III (Fig. 3) was the most effective at blocking these three proteins. The apoptotic assay results indicated that compound III functions as caspase-3, 8, and Bax activator while down-regulating the antiapoptotic protein Bcl2.
In another publication,26 we report compound IV (Fig. 3), a 1,2,3-triazole/1,2,4-oxadiazole hybrid, as a potent apoptotic antiproliferative agent that may function as dual inhibitors of EGFR and VEGFR-2.
1.1. Rational design
Recently,27 we reported a series of 1,2,4-oxadiazole/quinazoline-4-one hybrids (Va–o, Fig. 4) designed as antiproliferative agents targeting EGFR/BRAFV600E. The results indicated that most evaluated compounds exhibited strong antiproliferative activity, with Vb, Vc, Vh, Vk, and Vl demonstrating the highest potency. We tested these compounds as EGFR and BRAFV600E inhibitors and discovered they are good at stopping cell growth and may work as dual inhibitors of EGFR and BRAFV600E. | ||
| Fig. 4 Structures of previously published quinazoline-4-one derivatives Va–o and new target compounds 8a–t. | ||
As recently discovered, hybrid molecules have addressed several challenges associated with traditional drugs, including side effects and multidrug resistance (MDR).28 In support of our efforts to develop anticancer drugs with dual or multi-targeted mechanisms,27,29–35 we synthesized new quinazolin-4-ones coupled to 1,2,3-triazoles (8a–t, Fig. 4). We assessed their antiproliferative effectiveness against four different cancer cells. The most potent compounds were further tested as dual inhibitors of EGFR/BRAF. Also, the most potent hybrids were investigated for their apoptotic potential as activators of caspase 3,8 and BAX and down-regulators of antiapoptotic Bcl2. Finally, the most potent derivative was tested for cell cycle arrest and apoptosis.
2. Experimental
2.1. Chemistry
General details: see Appendix A (ESI file).†2.1.1.1. 2-(((1-Phenyl-1H-1,2,3-triazol-4-yl)methyl)thio)-3-phenyl quinazolin-4(3H)-one (8a). Yield: 0.26 g (74%), white solid, m.p: 136–138 °C, Rf. 0.56 (hexane
![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) ethyl acetate, 1
ethyl acetate, 1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 2, v/v). 1H NMR (400 MHz, δ ppm CDCl3): 8.19 (d, J = 7.8 Hz, 1H, Ar–H), 8.01 (s, 1H, triazole CH), 7.70 (t, J = 7.4 Hz, 1H, Ar–H), 7.61 (d, J = 7.4 Hz, 3H, Ar–H), 7.47–7.37 (m, 5H, Ar–H), 7.36–7.30 (m, 2H, Ar–H), 7.24–7.14 (m, 2H, Ar–H), 4.50 (s, 2H, S–CH2). 13C NMR (100 MHz, δ ppm CDCl3): 161.71, 156.87, 147.68, 135.52, 135.19, 134.89, 130.18, 129.80, 129.78, 129.44, 129.16, 128.81, 128.44, 127.50, 126.12, 126.01, 120.59, 120.01, 27.25. LC-MS (m/z) for: C23H17N5OS (exact mass = 411.12); calculated [M + H]+: 412.12; found [M + H]+: 412.00.
2, v/v). 1H NMR (400 MHz, δ ppm CDCl3): 8.19 (d, J = 7.8 Hz, 1H, Ar–H), 8.01 (s, 1H, triazole CH), 7.70 (t, J = 7.4 Hz, 1H, Ar–H), 7.61 (d, J = 7.4 Hz, 3H, Ar–H), 7.47–7.37 (m, 5H, Ar–H), 7.36–7.30 (m, 2H, Ar–H), 7.24–7.14 (m, 2H, Ar–H), 4.50 (s, 2H, S–CH2). 13C NMR (100 MHz, δ ppm CDCl3): 161.71, 156.87, 147.68, 135.52, 135.19, 134.89, 130.18, 129.80, 129.78, 129.44, 129.16, 128.81, 128.44, 127.50, 126.12, 126.01, 120.59, 120.01, 27.25. LC-MS (m/z) for: C23H17N5OS (exact mass = 411.12); calculated [M + H]+: 412.12; found [M + H]+: 412.00.
2.1.1.2. 2-(((1-(4-Chlorophenyl)-1H-1,2,3-triazol-4-yl)methyl)thio)-3-phenyl quinazolin-4(3H)-one (8b). Yield: 0.29 g (77%), white solid, m.p: 146–148 °C, Rf. 0.55 (hexane
![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) ethyl acetate, 1
ethyl acetate, 1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 2, v/v). 1H NMR (400 MHz, δ ppm DMSO-d6): 8.76 (s, 1H, triazole CH), 8.08 (d, J = 7.6 Hz, 1H, Ar–H), 7.87 (d, J = 8.8 Hz, 2H, Ar–H p-Cl C6H4), 7.84 (d, J = 7.8 Hz, 1H, Ar–H), 7.77 (d, J = 8.0 Hz, 1H, Ar–H), 7.63 (d, J = 8.8 Hz, 2H, Ar–H p-Cl C6H4), 7.59–7.52 (m, 3H, Ar–H), 7.51–7.43 (m, 3H, Ar–H), 4.53 (s, 2H, S–CH2). 13C NMR (100 MHz, δ ppm DMSO-d6): 160.86, 156.49, 147.32, 143.95, 135.77, 135.37, 135.05, 133.03, 130.06, 129.92, 129.61, 129.51, 126.64, 126.30, 126.18, 122.29, 121.83, 119.71, 26.66. LC-MS (m/z) for: C23H16ClN5OS (exact mass = 445.08); calculated [M + H]+: 446.08; found [M + H]+: 446.00.
2, v/v). 1H NMR (400 MHz, δ ppm DMSO-d6): 8.76 (s, 1H, triazole CH), 8.08 (d, J = 7.6 Hz, 1H, Ar–H), 7.87 (d, J = 8.8 Hz, 2H, Ar–H p-Cl C6H4), 7.84 (d, J = 7.8 Hz, 1H, Ar–H), 7.77 (d, J = 8.0 Hz, 1H, Ar–H), 7.63 (d, J = 8.8 Hz, 2H, Ar–H p-Cl C6H4), 7.59–7.52 (m, 3H, Ar–H), 7.51–7.43 (m, 3H, Ar–H), 4.53 (s, 2H, S–CH2). 13C NMR (100 MHz, δ ppm DMSO-d6): 160.86, 156.49, 147.32, 143.95, 135.77, 135.37, 135.05, 133.03, 130.06, 129.92, 129.61, 129.51, 126.64, 126.30, 126.18, 122.29, 121.83, 119.71, 26.66. LC-MS (m/z) for: C23H16ClN5OS (exact mass = 445.08); calculated [M + H]+: 446.08; found [M + H]+: 446.00.
2.1.1.3. 2-(((1-(4-Methoxyphenyl)-1H-1,2,3-triazol-4-yl)methyl)thio)-3-phenylquinazolin-4(3H)-one (8c). Yield: 0.29 g (78%), white solid, m.p: 152–154 °C, Rf. 0.58 (hexane
![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) ethyl acetate, 1
ethyl acetate, 1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 2, v/v). 1H NMR (400 MHz, δ ppm DMSO-d6): 8.63 (s, 1H, triazole CH), 8.08 (d, J = 7.6 Hz, 1H, Ar–H), 7.84 (t, J = 7.2 Hz, 1H, Ar–H), 7.76 (d, J = 8.0 Hz, 1H, Ar–H), 7.73 (d, J = 8.6 Hz, 2H, Ar–H p-O–CH3 C6H4), 7.63–7.40 (m, 6H, Ar–H), 7.10 (d, J = 8.6 Hz, 2H, Ar–H p-O–CH3 C6H4), 4.52 (s, 2H, S–CH2), 3.80 (s, 3H, O–CH3). 13C NMR (100 MHz, δ ppm DMSO-d6): 160.88, 159.34, 156.59, 147.34, 143.32, 135.80, 135.07, 130.06, 130.01, 129.61, 129.52, 126.66, 126.28, 126.18, 122.22, 121.83, 119.72, 114.94, 55.63, 26.77. LC-MS (m/z) for: C24H19N5O2S (exact mass = 441.13); calculated [M + H]+: 442.13; found [M + H]+: 442.10.
2, v/v). 1H NMR (400 MHz, δ ppm DMSO-d6): 8.63 (s, 1H, triazole CH), 8.08 (d, J = 7.6 Hz, 1H, Ar–H), 7.84 (t, J = 7.2 Hz, 1H, Ar–H), 7.76 (d, J = 8.0 Hz, 1H, Ar–H), 7.73 (d, J = 8.6 Hz, 2H, Ar–H p-O–CH3 C6H4), 7.63–7.40 (m, 6H, Ar–H), 7.10 (d, J = 8.6 Hz, 2H, Ar–H p-O–CH3 C6H4), 4.52 (s, 2H, S–CH2), 3.80 (s, 3H, O–CH3). 13C NMR (100 MHz, δ ppm DMSO-d6): 160.88, 159.34, 156.59, 147.34, 143.32, 135.80, 135.07, 130.06, 130.01, 129.61, 129.52, 126.66, 126.28, 126.18, 122.22, 121.83, 119.72, 114.94, 55.63, 26.77. LC-MS (m/z) for: C24H19N5O2S (exact mass = 441.13); calculated [M + H]+: 442.13; found [M + H]+: 442.10.
2.1.1.4. 2-(((1-(p-Tolyl)-1H-1,2,3-triazol-4-yl)methyl)thio)-3-phenylquinazolin-4(3H)-one (8d). Yield: 0.27 g (75%), white solid, m.p: 138–140 °C, Rf. 0.57 (hexane
![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) ethyl acetate, 1
ethyl acetate, 1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 2, v/v). 1H NMR (400 MHz, δ ppm DMSO-d6): 8.68 (s, 1H, triazole CH), 8.08 (d, J = 7.2 Hz, 1H, Ar–H), 7.83 (d, J = 8.4 Hz, 1H, Ar–H), 7.76 (d, J = 8.0 Hz, 1H, Ar–H), 7.70 (d, J = 6.0 Hz, 2H, Ar–H p-CH3–C6H4), 7.60–7.41 (m, 6H, Ar–H), 7.35 (d, J = 6.0 Hz, 2H, Ar–H p-CH3 C6H4), 4.52 (s, 2H, S–CH2), 2.34 (s, 3H, CH3). 13C NMR (100 MHz, δ ppm DMSO-d6): 160.86, 156.55, 147.31, 138.42, 135.77, 135.05, 134.33, 130.29, 130.04, 129.59, 129.50, 126.64, 126.27, 126.17, 122.15, 122.14, 120.01, 119.70, 26.73, 20.61. LC-MS (m/z) for: C24H19N5OS (exact mass = 425.13): calculated [M + H]+: 426.13; found [M + H]+: 426.10.
2, v/v). 1H NMR (400 MHz, δ ppm DMSO-d6): 8.68 (s, 1H, triazole CH), 8.08 (d, J = 7.2 Hz, 1H, Ar–H), 7.83 (d, J = 8.4 Hz, 1H, Ar–H), 7.76 (d, J = 8.0 Hz, 1H, Ar–H), 7.70 (d, J = 6.0 Hz, 2H, Ar–H p-CH3–C6H4), 7.60–7.41 (m, 6H, Ar–H), 7.35 (d, J = 6.0 Hz, 2H, Ar–H p-CH3 C6H4), 4.52 (s, 2H, S–CH2), 2.34 (s, 3H, CH3). 13C NMR (100 MHz, δ ppm DMSO-d6): 160.86, 156.55, 147.31, 138.42, 135.77, 135.05, 134.33, 130.29, 130.04, 129.59, 129.50, 126.64, 126.27, 126.17, 122.15, 122.14, 120.01, 119.70, 26.73, 20.61. LC-MS (m/z) for: C24H19N5OS (exact mass = 425.13): calculated [M + H]+: 426.13; found [M + H]+: 426.10.
2.1.1.5. 2-(((1-Phenyl-1H-1,2,3-triazol-4-yl)methyl)thio)-3-(p-tolyl)quinazolin-4(3H)-one (8e). Yield: 0.27 g (75%), white solid, m.p: 139–141 °C, Rf. 0.57 (hexane
![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) ethyl acetate, 1
ethyl acetate, 1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 2, v/v). 1H NMR (400 MHz, δ ppm DMSO-d6): 8.75 (s, 1H, triazole CH), 8.09 (d, J = 7.7 Hz, 1H, Ar–H), 7.91–7.80 (m, 3H, Ar–H), 7.77 (d, J = 8.0 Hz, 1H, Ar–H), 7.58 (t, J = 7.6 Hz, 2H, Ar–H), 7.51–7.45 (m, 2H, Ar–H), 7.36 (d, J = 8.0 Hz, 2H, Ar–H p-CH3 C6H4), 7.33 (d, J = 8.0 Hz, 2H, Ar–H p-CH3 C6H4), 4.53 (s, 2H, S–CH2), 2.40 (s, 3H, CH3). 13C NMR (100 MHz, δ ppm DMSO-d6): 160.90, 156.83, 147.33, 143.68, 139.75, 136.57, 135.00, 133.14, 130.08, 129.95, 129.20, 128.77, 126.64, 126.26, 126.12, 122.22, 120.13, 119.69, 26.73, 20.90. LC-MS (m/z) for: C24H19N5OS (exact mass = 425.13); calculated [M + H]+: 426.13; found [M + H]+: 426.10.
2, v/v). 1H NMR (400 MHz, δ ppm DMSO-d6): 8.75 (s, 1H, triazole CH), 8.09 (d, J = 7.7 Hz, 1H, Ar–H), 7.91–7.80 (m, 3H, Ar–H), 7.77 (d, J = 8.0 Hz, 1H, Ar–H), 7.58 (t, J = 7.6 Hz, 2H, Ar–H), 7.51–7.45 (m, 2H, Ar–H), 7.36 (d, J = 8.0 Hz, 2H, Ar–H p-CH3 C6H4), 7.33 (d, J = 8.0 Hz, 2H, Ar–H p-CH3 C6H4), 4.53 (s, 2H, S–CH2), 2.40 (s, 3H, CH3). 13C NMR (100 MHz, δ ppm DMSO-d6): 160.90, 156.83, 147.33, 143.68, 139.75, 136.57, 135.00, 133.14, 130.08, 129.95, 129.20, 128.77, 126.64, 126.26, 126.12, 122.22, 120.13, 119.69, 26.73, 20.90. LC-MS (m/z) for: C24H19N5OS (exact mass = 425.13); calculated [M + H]+: 426.13; found [M + H]+: 426.10.
2.1.1.6. 2-(((1-(4-Chlorophenyl)-1H-1,2,3-triazol-4-yl)methyl)thio)-3-(p-tolyl)quinazolin-4(3H)-one (8f). Yield: 0.30 g (76%), white solid, m.p: 158–160 °C, Rf. 0.54 (hexane
![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) ethyl acetate, 1
ethyl acetate, 1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 2, v/v). 1H NMR (400 MHz, δ ppm CDCl3): 8.28 (d, J = 7.2 Hz, 1H, Ar–H), 8.05 (s, 1H, triazole CH), 7.76 (t, J = 6.4 Hz, 1H, Ar–H), 7.69 (d, J = 7.6 Hz, 1H, Ar–H), 7.64 (d, J = 7.8 Hz, 2H, Ar–H p-Cl C6H4), 7.48 (d, J = 7.8 Hz, 2H, Ar–H p-Cl C6H4), 7.40 (d, J = 8.0 Hz, 1H, Ar–H), 7.33 (d, J = 7.2 Hz, 2H, Ar–H p-CH3 C6H4), 7.19 (d, J = 7.2 Hz, 2H, Ar–H p-CH3 C6H4), 4.59 (s, 2H, S–CH2), 2.45 (s, 3H, CH3). 13C NMR (100 MHz, δ ppm CDCl3): 161.72, 156.88, 147.65, 139.97, 135.34, 134.86, 134.65, 131.04, 129.95, 129.56, 129.54, 127.54, 126.12, 126.02, 125.92, 121.70, 120.96, 120.03, 27.13, 21.37. LC-MS (m/z) for: C24H18ClN5OS (exact mass = 459.09); calculated [M + H]+: 460.09; found [M + H]+: 460.00.
2, v/v). 1H NMR (400 MHz, δ ppm CDCl3): 8.28 (d, J = 7.2 Hz, 1H, Ar–H), 8.05 (s, 1H, triazole CH), 7.76 (t, J = 6.4 Hz, 1H, Ar–H), 7.69 (d, J = 7.6 Hz, 1H, Ar–H), 7.64 (d, J = 7.8 Hz, 2H, Ar–H p-Cl C6H4), 7.48 (d, J = 7.8 Hz, 2H, Ar–H p-Cl C6H4), 7.40 (d, J = 8.0 Hz, 1H, Ar–H), 7.33 (d, J = 7.2 Hz, 2H, Ar–H p-CH3 C6H4), 7.19 (d, J = 7.2 Hz, 2H, Ar–H p-CH3 C6H4), 4.59 (s, 2H, S–CH2), 2.45 (s, 3H, CH3). 13C NMR (100 MHz, δ ppm CDCl3): 161.72, 156.88, 147.65, 139.97, 135.34, 134.86, 134.65, 131.04, 129.95, 129.56, 129.54, 127.54, 126.12, 126.02, 125.92, 121.70, 120.96, 120.03, 27.13, 21.37. LC-MS (m/z) for: C24H18ClN5OS (exact mass = 459.09); calculated [M + H]+: 460.09; found [M + H]+: 460.00.
2.1.1.7. 2-(((1-(4-Methoxyphenyl)-1H-1,2,3-triazol-4-yl)methyl)thio)-3-(p-tolyl)quinazolin-4(3H)-one (8g). Yield: 0.31 g (80%), white solid, m.p: 168–170 °C, Rf. 0.58 (hexane
![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) ethyl acetate, 1
ethyl acetate, 1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 2, v/v). 1H NMR (400 MHz, δ ppm CDCl3): 8.19 (d, J = 7.4 Hz, 1H, Ar–H), 7.89 (s, 1H, triazole CH), 7.70 (t, J = 7.2 Hz, 1H, Ar–H), 7.60 (d, J = 8.0 Hz, 1H, Ar–H), 7.50 (d, J = 8.8 Hz, 2H, Ar–H p-O–CH3 C6H4), 7.35 (t, J = 7.5 Hz, 1H, Ar–H), 7.25 (d, J = 8.0 Hz, 2H, Ar–H p-CH3 C6H4), 7.11 (d, J = 8.0 Hz, 2H, Ar–H p-CH3 C6H4), 6.91 (d, J = 8.8 Hz, 2H, Ar–H p-O–CH3 C6H4), 4.49 (s, 2H, S–CH2), 3.77 (s, 3H, O–CH3), 2.35 (s, 3H, CH3). 13C NMR (100 MHz, δ ppm CDCl3): 161.82, 159.88, 157.12, 147.70, 140.43, 134.78, 132.82, 130.58, 130.48, 130.41, 130.15, 128.81, 127.51, 126.01, 125.99, 122.23, 120.01, 114.77, 55.65, 27.27, 21.43. LC-MS (m/z) for: C25H21N5O2S (exact mass = 455.14); calculated [M + H]+: 456.14; found [M + H]+: 456.10.
2, v/v). 1H NMR (400 MHz, δ ppm CDCl3): 8.19 (d, J = 7.4 Hz, 1H, Ar–H), 7.89 (s, 1H, triazole CH), 7.70 (t, J = 7.2 Hz, 1H, Ar–H), 7.60 (d, J = 8.0 Hz, 1H, Ar–H), 7.50 (d, J = 8.8 Hz, 2H, Ar–H p-O–CH3 C6H4), 7.35 (t, J = 7.5 Hz, 1H, Ar–H), 7.25 (d, J = 8.0 Hz, 2H, Ar–H p-CH3 C6H4), 7.11 (d, J = 8.0 Hz, 2H, Ar–H p-CH3 C6H4), 6.91 (d, J = 8.8 Hz, 2H, Ar–H p-O–CH3 C6H4), 4.49 (s, 2H, S–CH2), 3.77 (s, 3H, O–CH3), 2.35 (s, 3H, CH3). 13C NMR (100 MHz, δ ppm CDCl3): 161.82, 159.88, 157.12, 147.70, 140.43, 134.78, 132.82, 130.58, 130.48, 130.41, 130.15, 128.81, 127.51, 126.01, 125.99, 122.23, 120.01, 114.77, 55.65, 27.27, 21.43. LC-MS (m/z) for: C25H21N5O2S (exact mass = 455.14); calculated [M + H]+: 456.14; found [M + H]+: 456.10.
2.1.1.8. 2-(((1-(p-Tolyl)-1H-1,2,3-triazol-4-yl)methyl)thio)-3-(p-tolyl)quinazolin-4(3H)-one (8h). Yield: 0.28 g (75%), white solid, m.p: 143–145 °C, Rf. 0.56 (hexane
![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) ethyl acetate, 1
ethyl acetate, 1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 2, v/v). 1H NMR (400 MHz, δ ppm CDCl3): 8.18 (d, J = 7.2 Hz, 1H, Ar–H), 7.92 (s, 1H, triazole CH), 7.67 (t, J = 6.4 Hz, 1H, Ar–H), 7.59 (d, J = 7.6 Hz, 1H, Ar–H), 7.46 (d, J = 6.8 Hz, 2H, p-CH3 C6H4), 7.34 (t, J = 6.6 Hz, 1H, Ar–H), 7.23 (d, J = 8.0 Hz, 2H, p-CH3 C6H4), 7.14 (d, J = 8.0 Hz, 2H, p-CH3 C6H4), 7.09 (d, J = 6.8 Hz, 2H, p-CH3 C6H4), 4.49 (s, 2H, S–CH2), 2.34 (s, 3H, CH3), 2.31 (s, 3H, CH3). 13C NMR (100 MHz, δ ppm CDCl3): 161.83, 157.13, 147.70, 140.42, 138.97, 134.80, 134.68, 132.84, 130.47, 130.24, 130.15, 128.81, 127.50, 126.02, 126.00, 121.10, 120.48, 120.01, 27.29, 21.44, 21.12. LC-MS (m/z) for: C25H21N5OS (exact mass = 439.15); calculated [M + H]+: 440.15; found [M + H]+: 440.20.
2, v/v). 1H NMR (400 MHz, δ ppm CDCl3): 8.18 (d, J = 7.2 Hz, 1H, Ar–H), 7.92 (s, 1H, triazole CH), 7.67 (t, J = 6.4 Hz, 1H, Ar–H), 7.59 (d, J = 7.6 Hz, 1H, Ar–H), 7.46 (d, J = 6.8 Hz, 2H, p-CH3 C6H4), 7.34 (t, J = 6.6 Hz, 1H, Ar–H), 7.23 (d, J = 8.0 Hz, 2H, p-CH3 C6H4), 7.14 (d, J = 8.0 Hz, 2H, p-CH3 C6H4), 7.09 (d, J = 6.8 Hz, 2H, p-CH3 C6H4), 4.49 (s, 2H, S–CH2), 2.34 (s, 3H, CH3), 2.31 (s, 3H, CH3). 13C NMR (100 MHz, δ ppm CDCl3): 161.83, 157.13, 147.70, 140.42, 138.97, 134.80, 134.68, 132.84, 130.47, 130.24, 130.15, 128.81, 127.50, 126.02, 126.00, 121.10, 120.48, 120.01, 27.29, 21.44, 21.12. LC-MS (m/z) for: C25H21N5OS (exact mass = 439.15); calculated [M + H]+: 440.15; found [M + H]+: 440.20.
2.1.1.9. 2-(((1-Phenyl-1H-1,2,3-triazol-4-yl)methyl)thio)-3-(m-tolyl)quinazolin-4(3H)-one (8i). Yield: 0.25 g (70%), white solid, m.p: 155–157 °C, Rf. 0.57 (hexane
![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) ethyl acetate, 1
ethyl acetate, 1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 2, v/v). 1H NMR (400 MHz, δ ppm DMSO-d6): 8.75 (s, 1H, triazole CH), 8.08 (d, J = 7.8 Hz, 1H, Ar–H), 7.87–7.81 (m, 3H, Ar–H), 7.77 (d, J = 8.40 Hz, 1H, Ar–H), 7.56 (t, J = 7.6 Hz, 2H, Ar–H), 7.51–7.40 (m, 3H, Ar–H), 7.35 (d, J = 7.5 Hz, 1H, Ar–H), 7.25 (d, J = 8.0 Hz, 2H, Ar–H), 4.53 (s, 2H, S–CH2), 2.36 (s, 3H, CH3). 13C NMR (100 MHz, δ ppm DMSO-d6): 160.77, 156.56, 147.27, 143.58, 139.15, 136.54, 135.64, 134.96, 130.64, 129.90, 129.66, 129.33, 128.71, 126.58, 126.44, 126.24, 126.09, 122.20, 120.08, 119.66, 26.70, 20.78. LC-MS (m/z) for: C24H19N5OS (exact mass = 425.13); calculated [M + H]+: 426.13; found [M + H]+: 426.10.
2, v/v). 1H NMR (400 MHz, δ ppm DMSO-d6): 8.75 (s, 1H, triazole CH), 8.08 (d, J = 7.8 Hz, 1H, Ar–H), 7.87–7.81 (m, 3H, Ar–H), 7.77 (d, J = 8.40 Hz, 1H, Ar–H), 7.56 (t, J = 7.6 Hz, 2H, Ar–H), 7.51–7.40 (m, 3H, Ar–H), 7.35 (d, J = 7.5 Hz, 1H, Ar–H), 7.25 (d, J = 8.0 Hz, 2H, Ar–H), 4.53 (s, 2H, S–CH2), 2.36 (s, 3H, CH3). 13C NMR (100 MHz, δ ppm DMSO-d6): 160.77, 156.56, 147.27, 143.58, 139.15, 136.54, 135.64, 134.96, 130.64, 129.90, 129.66, 129.33, 128.71, 126.58, 126.44, 126.24, 126.09, 122.20, 120.08, 119.66, 26.70, 20.78. LC-MS (m/z) for: C24H19N5OS (exact mass = 425.13); calculated [M + H]+: 426.13; found [M + H]+: 426.10.
2.1.1.10. 2-(((1-(4-Chlorophenyl)-1H-1,2,3-triazol-4-yl)methyl)thio)-3-(m-tolyl)quinazolin-4(3H)-one (8j). Yield: 0.28 g (72%), white solid, m.p: 177–179 °C, Rf. 0.54 (hexane
![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) ethyl acetate, 1
ethyl acetate, 1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 2, v/v). 1H NMR (400 MHz, δ ppm CDCl3): 8.19 (d, J = 7.7 Hz, 1H, Ar–H), 7.96 (s, 1H, triazole CH), 7.70 (t, J = 7.3 Hz, 1H, Ar–H), 7.60 (d, J = 8.0 Hz, 1H, Ar–H), 7.55 (d, J = 8.4 Hz, 2H, Ar–H p-Cl C6H4), 7.39 (d, J = 8.4 Hz, 2H, Ar–H p-Cl C6H4), 7.35 (d, J = 7.2 Hz, 1H, Ar–H), 7.32 (d, J = 8.0 Hz, 1H, Ar–H), 7.25 (d, J = 7.6 Hz, 1H, Ar–H), 7.02 (d, J = 6.4 Hz, 2H, Ar–H), 4.49 (s, 2H, S–CH2), 2.33 (s, 3H, CH3). 13C NMR (100 MHz, δ ppm CDCl3): 161.77, 157.03, 147.66, 145.10, 140.48, 135.41, 134.83, 134.58, 132.79, 130.49, 130.14, 129.93, 128.79, 128.06, 127.53, 126.08, 125.94, 121.69, 120.93, 120.01, 27.16, 21.44. LC-MS (m/z) for: C24H18ClN5OS (exact mass = 459.09); calculated [M + H]+: 460.09; found [M + H]+: 460.10.
2, v/v). 1H NMR (400 MHz, δ ppm CDCl3): 8.19 (d, J = 7.7 Hz, 1H, Ar–H), 7.96 (s, 1H, triazole CH), 7.70 (t, J = 7.3 Hz, 1H, Ar–H), 7.60 (d, J = 8.0 Hz, 1H, Ar–H), 7.55 (d, J = 8.4 Hz, 2H, Ar–H p-Cl C6H4), 7.39 (d, J = 8.4 Hz, 2H, Ar–H p-Cl C6H4), 7.35 (d, J = 7.2 Hz, 1H, Ar–H), 7.32 (d, J = 8.0 Hz, 1H, Ar–H), 7.25 (d, J = 7.6 Hz, 1H, Ar–H), 7.02 (d, J = 6.4 Hz, 2H, Ar–H), 4.49 (s, 2H, S–CH2), 2.33 (s, 3H, CH3). 13C NMR (100 MHz, δ ppm CDCl3): 161.77, 157.03, 147.66, 145.10, 140.48, 135.41, 134.83, 134.58, 132.79, 130.49, 130.14, 129.93, 128.79, 128.06, 127.53, 126.08, 125.94, 121.69, 120.93, 120.01, 27.16, 21.44. LC-MS (m/z) for: C24H18ClN5OS (exact mass = 459.09); calculated [M + H]+: 460.09; found [M + H]+: 460.10.
2.1.1.11. 2-(((1-(4-Methoxyphenyl)-1H-1,2,3-triazol-4-yl)methyl)thio)-3-(m-tolyl)quinazolin-4(3H)-one (8k). Yield: 0.28 g (73%), white solid, m.p: 186–188 °C, Rf. 0.58 (hexane
![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) ethyl acetate, 1
ethyl acetate, 1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 2, v/v). 1H NMR (400 MHz, δ ppm CDCl3): 8.28 (d, J = 5.6 Hz, 1H, Ar–H), 8.00 (s, 1H, triazole CH), 7.78 (s, 1H, Ar–H), 7.71 (s, 1H, Ar–H), 7.58 (d, J = 5.8 Hz, 2H, Ar–H p-OCH3 C6H4), 7.43 (d, J = 6.4 Hz, 2H, Ar–H), 7.34 (s, 1H, Ar–H), 7.12 (s, 2H, Ar–H), 6.99 (d, J = 5.8 Hz, 2H, Ar–H p-OCH3 C6H4), 4.61 (s, 2H, S–CH2), 3.86 (s, 3H, OCH3), 2.42 (s, 3H, CH3). 13C NMR (100 MHz, δ ppm CDCl3): 161.76, 159.87, 157.00, 147.69, 139.93, 135.39, 134.82, 131.00, 130.38, 129.55, 129.54, 129.27, 127.47, 126.03, 126.01, 125.88, 125.34, 122.21, 120.00, 114.76, 55.65, 27.31, 21.38. LC-MS (m/z) for: C25H21N5O2S (exact mass = 455.14); calculated [M + H]+: 456.14; found [M + H]+: 456.10.
2, v/v). 1H NMR (400 MHz, δ ppm CDCl3): 8.28 (d, J = 5.6 Hz, 1H, Ar–H), 8.00 (s, 1H, triazole CH), 7.78 (s, 1H, Ar–H), 7.71 (s, 1H, Ar–H), 7.58 (d, J = 5.8 Hz, 2H, Ar–H p-OCH3 C6H4), 7.43 (d, J = 6.4 Hz, 2H, Ar–H), 7.34 (s, 1H, Ar–H), 7.12 (s, 2H, Ar–H), 6.99 (d, J = 5.8 Hz, 2H, Ar–H p-OCH3 C6H4), 4.61 (s, 2H, S–CH2), 3.86 (s, 3H, OCH3), 2.42 (s, 3H, CH3). 13C NMR (100 MHz, δ ppm CDCl3): 161.76, 159.87, 157.00, 147.69, 139.93, 135.39, 134.82, 131.00, 130.38, 129.55, 129.54, 129.27, 127.47, 126.03, 126.01, 125.88, 125.34, 122.21, 120.00, 114.76, 55.65, 27.31, 21.38. LC-MS (m/z) for: C25H21N5O2S (exact mass = 455.14); calculated [M + H]+: 456.14; found [M + H]+: 456.10.
2.1.1.12. 2-(((1-(p-Tolyl)-1H-1,2,3-triazol-4-yl)methyl)thio)-3-(m-tolyl)quinazolin-4(3H)-one (8l). Yield: 0.26 g (70%), white solid, m.p: 158–160 °C, Rf. 0.56 (hexane
![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) ethyl acetate, 1
ethyl acetate, 1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 2, v/v). 1H NMR (400 MHz, δ ppm CDCl3): 8.17 (s, 1H, Ar–H), 7.93 (s, 1H, triazole CH), 7.67 (s, 1H, Ar–H), 7.60 (s, 1H, Ar–H), 7.45 (s, 2H, Ar–H p-CH3 C6H4), 7.32 (s, 2H, Ar–H), 7.19 (s, 3H, Ar–H), 7.02 (s, 2H, Ar–H p-CH3 C6H4), 4.48 (s, 2H, S–CH2), 2.31 (s, 6H, 2CH3). 13C NMR (100 MHz, δ ppm CDCl3): 161.78, 157.01, 147.71, 139.93, 138.93, 135.49, 135.41, 135.34, 134.82, 134.72, 130.99, 130.24, 129.56, 129.55, 127.47, 126.19, 126.04, 125.99, 120.47, 120.01, 27.30, 21.38, 21.12. LC-MS (m/z) for: C25H21N5OS (exact mass = 439.15); calculated [M + H]+: 440.15; found [M + H]+: 440.00.
2, v/v). 1H NMR (400 MHz, δ ppm CDCl3): 8.17 (s, 1H, Ar–H), 7.93 (s, 1H, triazole CH), 7.67 (s, 1H, Ar–H), 7.60 (s, 1H, Ar–H), 7.45 (s, 2H, Ar–H p-CH3 C6H4), 7.32 (s, 2H, Ar–H), 7.19 (s, 3H, Ar–H), 7.02 (s, 2H, Ar–H p-CH3 C6H4), 4.48 (s, 2H, S–CH2), 2.31 (s, 6H, 2CH3). 13C NMR (100 MHz, δ ppm CDCl3): 161.78, 157.01, 147.71, 139.93, 138.93, 135.49, 135.41, 135.34, 134.82, 134.72, 130.99, 130.24, 129.56, 129.55, 127.47, 126.19, 126.04, 125.99, 120.47, 120.01, 27.30, 21.38, 21.12. LC-MS (m/z) for: C25H21N5OS (exact mass = 439.15); calculated [M + H]+: 440.15; found [M + H]+: 440.00.
2.1.1.13. 2-(((1-Phenyl-1H-1,2,3-triazol-4-yl)methyl)thio)-3-ethyl quinazolin-4(3H)-one (8m). Yield: 0.21 g (67%), white solid, m.p: 120–122 °C, Rf. 0.51 (hexane
![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) ethyl acetate, 1
ethyl acetate, 1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 2, v/v). 1H NMR (400 MHz, δ ppm DMSO-d6): 8.82 (s, 1H, triazole CH), 8.08 (d, J = 7.7 Hz, 1H, Ar–H), 7.86 (d, J = 7.0 Hz, 1H, Ar–H), 7.68–7.60 (m, 4H, Ar–H), 7.48 (t, J = 7.2 Hz, 1H, Ar–H), 7.30 (d, J = 8.4 Hz, 2H, Ar–H), 4.70 (s, 2H, S–CH2), 4.08 (q, J = 6.8 Hz, 2H, N–CH2), 1.16 (t, J = 6.8 Hz, 3H, CH3). 13C NMR (100 MHz, δ ppm DMSO-d6): 161.29, 155.65, 147.31, 143.74, 138.96, 135.66, 134.42, 130.33, 127.32, 125.95, 125.83, 123.12, 121.58, 119.85, 39.85, 26.68, 13.29.
2, v/v). 1H NMR (400 MHz, δ ppm DMSO-d6): 8.82 (s, 1H, triazole CH), 8.08 (d, J = 7.7 Hz, 1H, Ar–H), 7.86 (d, J = 7.0 Hz, 1H, Ar–H), 7.68–7.60 (m, 4H, Ar–H), 7.48 (t, J = 7.2 Hz, 1H, Ar–H), 7.30 (d, J = 8.4 Hz, 2H, Ar–H), 4.70 (s, 2H, S–CH2), 4.08 (q, J = 6.8 Hz, 2H, N–CH2), 1.16 (t, J = 6.8 Hz, 3H, CH3). 13C NMR (100 MHz, δ ppm DMSO-d6): 161.29, 155.65, 147.31, 143.74, 138.96, 135.66, 134.42, 130.33, 127.32, 125.95, 125.83, 123.12, 121.58, 119.85, 39.85, 26.68, 13.29.
2.1.1.14. 2-(((1-(4-Chlorophenyl)-1H-1,2,3-triazol-4-yl)methyl)thio)-3-ethyl quinazolin-4(3H)-one (8n). Yield: 0.23 g (69%), white solid, m.p: 132–134 °C, Rf. 0.53 (hexane
![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) ethyl acetate, 1
ethyl acetate, 1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 2, v/v). 1H NMR (400 MHz, δ ppm CDCl3): 8.15 (dd, J = 8.0, 1.1 Hz, 1H, Ar–H), 7.99 (s, 1H, triazole CH), 7.69–7.61 (m, 1H, Ar–H), 7.55 (d, J = 8.8 Hz, 2H, Ar–H p-Cl C6H4), 7.52 (s, 1H, Ar–H), 7.38 (d, J = 8.8 Hz, 2H, Ar–H p-Cl C6H4), 7.35–7.30 (m, 1H, Ar–H), 4.63 (s, 2H, S–CH2), 4.10 (q, J = 7.1 Hz, 2H, N–CH2), 1.29 (t, J = 7.1 Hz, 3H, CH3). 13C NMR (100 MHz, δ ppm CDCl3): 161.32, 155.62, 147.21, 135.45, 134.59, 134.49, 132.99, 129.93, 127.19, 125.95, 125.63, 124.92, 121.71, 119.56, 39.87, 26.55, 13.27. LC-MS (m/z) for: C19H16ClN5OS (exact mass = 397.08); calculated [M + H]+: 398.08; found [M + H]+: 398.00.
2, v/v). 1H NMR (400 MHz, δ ppm CDCl3): 8.15 (dd, J = 8.0, 1.1 Hz, 1H, Ar–H), 7.99 (s, 1H, triazole CH), 7.69–7.61 (m, 1H, Ar–H), 7.55 (d, J = 8.8 Hz, 2H, Ar–H p-Cl C6H4), 7.52 (s, 1H, Ar–H), 7.38 (d, J = 8.8 Hz, 2H, Ar–H p-Cl C6H4), 7.35–7.30 (m, 1H, Ar–H), 4.63 (s, 2H, S–CH2), 4.10 (q, J = 7.1 Hz, 2H, N–CH2), 1.29 (t, J = 7.1 Hz, 3H, CH3). 13C NMR (100 MHz, δ ppm CDCl3): 161.32, 155.62, 147.21, 135.45, 134.59, 134.49, 132.99, 129.93, 127.19, 125.95, 125.63, 124.92, 121.71, 119.56, 39.87, 26.55, 13.27. LC-MS (m/z) for: C19H16ClN5OS (exact mass = 397.08); calculated [M + H]+: 398.08; found [M + H]+: 398.00.
2.1.1.15. 2-(((1-(4-Methoxyphenyl)-1H-1,2,3-triazol-4-yl)methyl)thio)-3-ethyl quinazolin-4(3H)-one (8o). Yield: 0.23 g (69%), white solid, m.p: 142–144 °C, Rf. 0.49 (hexane
![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) ethyl acetate, 1
ethyl acetate, 1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 2, v/v). 1H NMR (400 MHz, δ ppm CDCl3): 8.15 (d, J = 7.6 Hz, 1H, Ar–H), 7.92 (s, 1H, triazole CH), 7.64 (t, J = 7.2 Hz, 1H, Ar–H), 7.49 (d, J = 8.5 Hz, 2H, Ar–H p-OCH3 C6H4), 7.32 (t, J = 7.4 Hz, 1H, Ar–H), 7.21 (d, J = 8.0 Hz, 1H, Ar–H), 6.90 (d, J = 8.5 Hz, 2H, Ar–H p-OCH3 C6H4), 4.62 (s, 2H, S–CH2), 4.10 (q, J = 6.8 Hz, 2H, N–CH2), 3.76 (s, 3H, OCH3), 1.28 (t, J = 6.8 Hz, 3H, CH3). 13C NMR (100 MHz, δ ppm CDCl3): 161.38, 159.85, 155.66, 149.57, 147.31, 134.42, 128.64, 127.13, 125.85, 123.33, 122.22, 119.56, 115.26, 114.75, 55.63, 39.83, 26.68, 13.26. LC-MS (m/z) for: C20H19N5O2S (exact mass = 393.13); calculated [M + H]+: 394.13; found [M + H]+: 394.00.
2, v/v). 1H NMR (400 MHz, δ ppm CDCl3): 8.15 (d, J = 7.6 Hz, 1H, Ar–H), 7.92 (s, 1H, triazole CH), 7.64 (t, J = 7.2 Hz, 1H, Ar–H), 7.49 (d, J = 8.5 Hz, 2H, Ar–H p-OCH3 C6H4), 7.32 (t, J = 7.4 Hz, 1H, Ar–H), 7.21 (d, J = 8.0 Hz, 1H, Ar–H), 6.90 (d, J = 8.5 Hz, 2H, Ar–H p-OCH3 C6H4), 4.62 (s, 2H, S–CH2), 4.10 (q, J = 6.8 Hz, 2H, N–CH2), 3.76 (s, 3H, OCH3), 1.28 (t, J = 6.8 Hz, 3H, CH3). 13C NMR (100 MHz, δ ppm CDCl3): 161.38, 159.85, 155.66, 149.57, 147.31, 134.42, 128.64, 127.13, 125.85, 123.33, 122.22, 119.56, 115.26, 114.75, 55.63, 39.83, 26.68, 13.26. LC-MS (m/z) for: C20H19N5O2S (exact mass = 393.13); calculated [M + H]+: 394.13; found [M + H]+: 394.00.
2.1.1.16. 2-(((1-(p-Tolyl)-1H-1,2,3-triazol-4-yl)methyl)thio)-3-ethylquinazolin-4(3H)-one (8p). Yield: 0.22 g (68%), white solid, m.p: 125–127 °C, Rf. 0.52 (hexane
![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) ethyl acetate, 1
ethyl acetate, 1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 2, v/v). 1H NMR (400 MHz, δ ppm CDCl3): 8.14 (dd, J = 7.9, 0.9 Hz, 1H, Ar–H), 7.95 (s, 1H, triazole CH), 7.66–7.59 (m, 1H, Ar–H), 7.52 (d, J = 8.0 Hz, 1H, Ar–H), 7.46 (d, J = 8.4 Hz, 2H, Ar–H p-CH3 C6H4), 7.34–7.27 (m, 1H, Ar–H), 7.19 (d, J = 8.4 Hz, 2H, Ar–H p-CH3 C6H4), 4.62 (s, 2H, S–CH2), 4.09 (q, J = 7.1 Hz, 2H, N–CH2), 2.31 (s, 3H, benzylic CH3), 1.28 (t, J = 7.1 Hz, 3H, CH3). 13C NMR (100 MHz, δ ppm CDCl3): 161.37, 155.65, 147.31, 144.63, 138.97, 134.66, 134.42, 130.23, 127.12, 125.84, 125.74, 121.11, 120.48, 119.56, 39.82, 26.66, 21.10, 13.26. LC-MS (m/z) for: C20H19N5OS (exact mass = 377.13); calculated [M + H]+: 378.13; found [M + H]+: 378.10.
2, v/v). 1H NMR (400 MHz, δ ppm CDCl3): 8.14 (dd, J = 7.9, 0.9 Hz, 1H, Ar–H), 7.95 (s, 1H, triazole CH), 7.66–7.59 (m, 1H, Ar–H), 7.52 (d, J = 8.0 Hz, 1H, Ar–H), 7.46 (d, J = 8.4 Hz, 2H, Ar–H p-CH3 C6H4), 7.34–7.27 (m, 1H, Ar–H), 7.19 (d, J = 8.4 Hz, 2H, Ar–H p-CH3 C6H4), 4.62 (s, 2H, S–CH2), 4.09 (q, J = 7.1 Hz, 2H, N–CH2), 2.31 (s, 3H, benzylic CH3), 1.28 (t, J = 7.1 Hz, 3H, CH3). 13C NMR (100 MHz, δ ppm CDCl3): 161.37, 155.65, 147.31, 144.63, 138.97, 134.66, 134.42, 130.23, 127.12, 125.84, 125.74, 121.11, 120.48, 119.56, 39.82, 26.66, 21.10, 13.26. LC-MS (m/z) for: C20H19N5OS (exact mass = 377.13); calculated [M + H]+: 378.13; found [M + H]+: 378.10.
2.1.1.17. 2-(((1-Phenyl-1H-1,2,3-triazol-4-yl)methyl)thio)-3-allyl quinazolin-4(3H)-one (8q). Yield: 0.21 g (66%), white solid, m.p: 124–126 °C, Rf. 0.50 (hexane
![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) ethyl acetate, 1
ethyl acetate, 1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 2, v/v). 1H NMR (400 MHz, δ ppm DMSO-d6): 8.70 (s, 1H, triazole CH), 8.06 (d, J = 7.8 Hz, 1H, Ar–H), 7.96 (d, J = 6.8 Hz, 2H, Ar–H), 7.74 (t, J = 7.5 Hz, 1H, Ar–H), 7.61–7.50 (m, 3H, Ar–H), 7.45 (t, J = 7.5 Hz, 1H, Ar–H), 7.35 (d, J = 8.1 Hz, 1H, Ar–H), 5.96–5.82 (m, 1H,
2, v/v). 1H NMR (400 MHz, δ ppm DMSO-d6): 8.70 (s, 1H, triazole CH), 8.06 (d, J = 7.8 Hz, 1H, Ar–H), 7.96 (d, J = 6.8 Hz, 2H, Ar–H), 7.74 (t, J = 7.5 Hz, 1H, Ar–H), 7.61–7.50 (m, 3H, Ar–H), 7.45 (t, J = 7.5 Hz, 1H, Ar–H), 7.35 (d, J = 8.1 Hz, 1H, Ar–H), 5.96–5.82 (m, 1H, ![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) CH), 5.21 (d, J = 10.0 Hz, 1H,
CH), 5.21 (d, J = 10.0 Hz, 1H, ![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) CH2), 5.11 (d, J = 16.0 Hz, 1H,
CH2), 5.11 (d, J = 16.0 Hz, 1H, ![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) CH2), 4.90 (s, 2H, S–CH2), 4.74 (d, J = 4.0 Hz, 2H, N–CH2); 13C NMR (100 MHz, δ ppm DMSO-d6): 160.21, 154.96, 146.46, 138.97, 134.94, 131.63, 131.26, 129.31, 126.96, 126.54, 126.36, 126.02, 125.71, 120.62, 118.74, 117.62, 45.96, 26.80.
CH2), 4.90 (s, 2H, S–CH2), 4.74 (d, J = 4.0 Hz, 2H, N–CH2); 13C NMR (100 MHz, δ ppm DMSO-d6): 160.21, 154.96, 146.46, 138.97, 134.94, 131.63, 131.26, 129.31, 126.96, 126.54, 126.36, 126.02, 125.71, 120.62, 118.74, 117.62, 45.96, 26.80.
2.1.1.18. 2-(((1-(4-Chlorophenyl)-1H-1,2,3-triazol-4-yl)methyl)thio)-3-allyl quinazolin-4(3H)-one (8r). Yield: 0.25 g (71%), white solid, m.p: 137–139 °C, Rf. 0.54 (hexane
![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) ethyl acetate, 1
ethyl acetate, 1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 2, v/v). 1H NMR (400 MHz, δ ppm CDCl3): 8.15 (d, J = 7.6 Hz, 1H, Ar–H), 7.97 (s, 1H, triazole CH), 7.65 (t, J = 7.4 Hz, 1H, Ar–H), 7.54 (d, J = 8.4 Hz, 2H, Ar–H p-Cl C6H4), 7.52 (s, 1H, Ar–H), 7.38 (d, J = 8.4 Hz, 2H, Ar–H p-Cl C6H4), 7.33 (d, J = 7.6 Hz, 1H, Ar–H), 5.91–5.76 (m, 1H,
2, v/v). 1H NMR (400 MHz, δ ppm CDCl3): 8.15 (d, J = 7.6 Hz, 1H, Ar–H), 7.97 (s, 1H, triazole CH), 7.65 (t, J = 7.4 Hz, 1H, Ar–H), 7.54 (d, J = 8.4 Hz, 2H, Ar–H p-Cl C6H4), 7.52 (s, 1H, Ar–H), 7.38 (d, J = 8.4 Hz, 2H, Ar–H p-Cl C6H4), 7.33 (d, J = 7.6 Hz, 1H, Ar–H), 5.91–5.76 (m, 1H, ![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) CH), 5.20 (d, J = 12.0 Hz, 1H,
CH), 5.20 (d, J = 12.0 Hz, 1H, ![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) CH2), 5.17 (d, J = 6.0 Hz, 1H,
CH2), 5.17 (d, J = 6.0 Hz, 1H, ![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) CH2), 4.67 (d, J = 5.1 Hz, 2H, N–CH2), 4.63 (s, 2H, S–CH2). 13C NMR (100 MHz, δ ppm CDCl3): 161.35, 155.75, 147.26, 144.87, 135.41, 134.63, 130.26, 129.94, 127.32, 126.25, 126.04, 125.75, 121.70, 121.07, 119.47, 118.59, 46.39, 26.72. LC-MS (m/z) for: C20H16ClN5OS (exact mass = 409.08); calculated [M + H]+: 410.08; found [M + H]+: 410.00.
CH2), 4.67 (d, J = 5.1 Hz, 2H, N–CH2), 4.63 (s, 2H, S–CH2). 13C NMR (100 MHz, δ ppm CDCl3): 161.35, 155.75, 147.26, 144.87, 135.41, 134.63, 130.26, 129.94, 127.32, 126.25, 126.04, 125.75, 121.70, 121.07, 119.47, 118.59, 46.39, 26.72. LC-MS (m/z) for: C20H16ClN5OS (exact mass = 409.08); calculated [M + H]+: 410.08; found [M + H]+: 410.00.
2.1.1.19. 2-(((1-(4-Methoxyphenyl)-1H-1,2,3-triazol-4-yl)methyl)thio)-3-allyl quinazolin-4(3H)-one (8s). Yield: 0.25 g (72%), white solid, m.p: 148–150 °C, Rf. 0.48 (hexane
![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) ethyl acetate, 1
ethyl acetate, 1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 2, v/v). 1H NMR (400 MHz, δ ppm DMSO-d6): 8.70 (s, 1H, triazole CH), 8.08 (s, 1H, Ar–H), 7.81–7.70 (m, 4H, Ar–H), 7.48 (s, 1H, Ar–H), 7.12 (s, 2H, Ar–H), 5.95–5.85 (m, 1H,
2, v/v). 1H NMR (400 MHz, δ ppm DMSO-d6): 8.70 (s, 1H, triazole CH), 8.08 (s, 1H, Ar–H), 7.81–7.70 (m, 4H, Ar–H), 7.48 (s, 1H, Ar–H), 7.12 (s, 2H, Ar–H), 5.95–5.85 (m, 1H, ![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) CH), 5.16 (d, J = 16.0 Hz, 1H,
CH), 5.16 (d, J = 16.0 Hz, 1H, ![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) CH2), 5.12 (d, J = 8.0 Hz, 1H,
CH2), 5.12 (d, J = 8.0 Hz, 1H, ![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) CH2), 4.68 (s, 4H, N–CH2 & S–CH2), 3.81 (s, 3H, OCH3). 13C NMR (100 MHz, δ ppm DMSO-d6): 161.63, 159.95, 154.98, 149.57, 146.73, 134.82, 131.24, 128.64, 126.89, 126.13, 125.74, 122.25, 120.22, 119.28, 117.56, 114.69, 55.37, 45.93, 26.70. LC-MS (m/z) for: C21H19N5O2S (exact mass = 405.13); calculated [M + H]+: 406.13; found [M + H]+: 406.10.
CH2), 4.68 (s, 4H, N–CH2 & S–CH2), 3.81 (s, 3H, OCH3). 13C NMR (100 MHz, δ ppm DMSO-d6): 161.63, 159.95, 154.98, 149.57, 146.73, 134.82, 131.24, 128.64, 126.89, 126.13, 125.74, 122.25, 120.22, 119.28, 117.56, 114.69, 55.37, 45.93, 26.70. LC-MS (m/z) for: C21H19N5O2S (exact mass = 405.13); calculated [M + H]+: 406.13; found [M + H]+: 406.10.
2.1.1.20. 2-(((1-(p-Tolyl)-1H-1,2,3-triazol-4-yl)methyl)thio)-3-allyl quinazolin-4(3H)-one (8t). Yield: 0.23 g (69%), white solid, m.p: 130–132 °C, Rf. 0.53 (hexane
![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) ethyl acetate, 1
ethyl acetate, 1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 2, v/v). 1H NMR (400 MHz, δ ppm CDCl3): 8.15 (d, J = 7.6 Hz, 1H, Ar–H), 7.94 (s, 1H, triazole CH), 7.64 (t, J = 7.4 Hz, 1H, Ar–H), 7.53 (d, J = 8.0 Hz, 1H, Ar–H), 7.46 (d, J = 8.4 Hz, 2H, Ar–H p-CH3 C6H4), 7.32 (t, J = 7.6 Hz, 1H, Ar–H), 7.18 (d, J = 8.4 Hz, 2H, Ar–H p-CH3 C6H4), 5.94–5.73 (m, 1H,
2, v/v). 1H NMR (400 MHz, δ ppm CDCl3): 8.15 (d, J = 7.6 Hz, 1H, Ar–H), 7.94 (s, 1H, triazole CH), 7.64 (t, J = 7.4 Hz, 1H, Ar–H), 7.53 (d, J = 8.0 Hz, 1H, Ar–H), 7.46 (d, J = 8.4 Hz, 2H, Ar–H p-CH3 C6H4), 7.32 (t, J = 7.6 Hz, 1H, Ar–H), 7.18 (d, J = 8.4 Hz, 2H, Ar–H p-CH3 C6H4), 5.94–5.73 (m, 1H, ![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) CH), 5.20 (d, J = 14.0 Hz, 1H,
CH), 5.20 (d, J = 14.0 Hz, 1H, ![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) CH2), 5.16 (d, J = 6.8 Hz, 1H,
CH2), 5.16 (d, J = 6.8 Hz, 1H, ![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) CH2), 4.67 (d, J = 5.2 Hz, 2H, N–CH2), 4.62 (s, 2H, S–CH2), 2.31 (s, 3H, CH3). 13C NMR (100 MHz, δ ppm CDCl3): 161.38, 155.87, 147.30, 144.50, 138.96, 134.67, 134.58, 130.54, 130.23, 127.27, 125.96, 125.81, 121.12, 120.47, 119.46, 118.78, 46.37, 26.85, 21.10. LC-MS (m/z) for: C21H19N5OS (exact mass = 389.13); calculated [M + H]+: 390.13; found [M + H]+: 390.10.
CH2), 4.67 (d, J = 5.2 Hz, 2H, N–CH2), 4.62 (s, 2H, S–CH2), 2.31 (s, 3H, CH3). 13C NMR (100 MHz, δ ppm CDCl3): 161.38, 155.87, 147.30, 144.50, 138.96, 134.67, 134.58, 130.54, 130.23, 127.27, 125.96, 125.81, 121.12, 120.47, 119.46, 118.78, 46.37, 26.85, 21.10. LC-MS (m/z) for: C21H19N5OS (exact mass = 389.13); calculated [M + H]+: 390.13; found [M + H]+: 390.10.
2.2. Biology
2.3. Docking study
Molecular docking simulations within EGFR (PDB ID: 1M17)42 and BRAFV600E (PDB ID: 4RZV).43 Were validated with a redocking test, whereby the test proteins' structures had remained fixed while the co-crystallized ligands (Erlotinib for EGFR and Vemurafenib for BRAFV600E) were redocked into their crystal-binding pocket. See Appendix A† for more details.3. Results and discussion
3.1. Chemistry
Scheme 1 depicts the synthetic methods for the novel target hybrids (8a–t). We refluxed anthranilic acid (1) in ethanol with isothiocyanate derivatives (2a–e) for 8 h. After the reaction was complete (as determined by TLC), the resulting white precipitate was filtered and recrystallized from an ethanol/dioxane mixture (1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1), yielding quinazoline-4-ones (3a–e) in 90–95% yields.44 The quinazoline derivatives (3a–e) were then alkylated with propargyl bromide (4) in the presence of anhydrous K2CO3 by stirring in DMF, yielding S-propargyl derivatives (5a–e) in 70–83% yields that were purified by ethanol recrystallization.45 FTIR spectrum of 5c, as an example, revealed the appearance of the characteristic peak of
1), yielding quinazoline-4-ones (3a–e) in 90–95% yields.44 The quinazoline derivatives (3a–e) were then alkylated with propargyl bromide (4) in the presence of anhydrous K2CO3 by stirring in DMF, yielding S-propargyl derivatives (5a–e) in 70–83% yields that were purified by ethanol recrystallization.45 FTIR spectrum of 5c, as an example, revealed the appearance of the characteristic peak of ![[triple bond, length as m-dash]](https://www.rsc.org/images/entities/char_e002.gif) C–H stretching at ν 3245 cm−1, 3066 (
C–H stretching at ν 3245 cm−1, 3066 (![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) C–H stretching), 2968, 2928 (sp3C–H stretching), 2116 (C
C–H stretching), 2968, 2928 (sp3C–H stretching), 2116 (C![[triple bond, length as m-dash]](https://www.rsc.org/images/entities/char_e002.gif) C) and 1687 (C
C) and 1687 (C![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) O). The diazotization of aniline derivatives (6a–d) with NaNO2/HCl at 0–5 °C, followed by the addition of sodium azide, resulted in the formation of the corresponding aromatic azides (7a–d) with a yield of 70–75%.46 The FTIR spectrum of 7c confirmed the appearance of the characteristic peak of the azide group at 2109 cm−1.
O). The diazotization of aniline derivatives (6a–d) with NaNO2/HCl at 0–5 °C, followed by the addition of sodium azide, resulted in the formation of the corresponding aromatic azides (7a–d) with a yield of 70–75%.46 The FTIR spectrum of 7c confirmed the appearance of the characteristic peak of the azide group at 2109 cm−1.
1,3-Dipolar cycloaddition reactions between propargyl derivatives (5a–e) and aromatic azides (7a–d) were carried out by refluxing in THF in the presence of CuSO4·5H2O/sodium ascorbate as a catalyst, yielding 1,4-disubstituted 1,2,3-triazoles (8a–t) in good yields (86–90%). The structures of 8a–t were elucidated using 1H NMR, 13C NMR spectroscopy, and LC-MS. The 1H NMR spectra of compound 8g, as an example, confirmed the presence of three characteristic singlet signals in the aliphatic range: first one at δ 2.36 (s, 3H, CH3), second one at δ 3.77 (s, 3H, O–CH3) and third one at δ 4.49 (s, 2H, S–CH2). Additionally, the spectrum revealed another characteristic singlet signal in the aromatic range at δ 7.89 of the triazole CH (s, 1H, triazole CH).
Also, the spectrum revealed two pairs of doublets of the aromatic ring's p-disubstituted patterns and extra signals for the aromatic protons in the quinazoline moiety. The 13C NMR spectrum of 8g indicated the presence of peaks of methoxy, methylene, and methyl groups at δ 55.65, δ 27.27, and δ 21.43 ppm, respectively. The 13C NMR spectrum of 8g also indicated a peak at δ 161.82 ppm of the amidic carbonyl group. LC-MS spectrum of compound 8g (C25H21N5O2S, M. Wt = 455.14) showed a signal at 456.10 m/z [M + H]+.
3.2. Biology
| Comp. | Cell viability % | R1 | R2 | Antiproliferative activity IC50 ± SEM (nM) | ||||
|---|---|---|---|---|---|---|---|---|
| A-549 | MCF-7 | Panc-1 | HT-29 | Average (GI50) | ||||
| 8a | 89 | Phenyl | H | 61 ± 5 | 56 ± 5 | 64 ± 5 | 64 ± 5 | 61 |
| 8b | 90 | Phenyl | Cl | 40 ± 3 | 36 ± 3 | 42 ± 3 | 44 ± 3 | 41 |
| 8c | 86 | Phenyl | OMe | 47 ± 3 | 42 ± 3 | 49 ± 3 | 49 ± 3 | 47 |
| 8d | 89 | Phenyl | Me | 53 ± 4 | 48 ± 4 | 56 ± 4 | 56 ± 4 | 53 |
| 8e | 90 | p-Tolyl | H | 88 ± 8 | 85 ± 8 | 92 ± 8 | 92 ± 8 | 89 |
| 8f | 87 | p-Tolyl | Cl | 37 ± 3 | 34 ± 3 | 38 ± 3 | 39 ± 3 | 37 |
| 8g | 88 | p-Tolyl | OMe | 22 ± 1 | 20 ± 1 | 24 ± 1 | 24 ± 1 | 22 |
| 8h | 92 | p-Tolyl | Me | 25 ± 1 | 22 ± 1 | 26 ± 1 | 27 ± 1 | 25 |
| 8i | 90 | m-Tolyl | H | 65 ± 5 | 59 ± 5 | 68 ± 6 | 68 ± 6 | 65 |
| 8j | 87 | m-Tolyl | Cl | 29 ± 2 | 26 ± 2 | 31 ± 2 | 30 ± 2 | 29 |
| 8k | 85 | m-Tolyl | OMe | 42 ± 3 | 39 ± 3 | 46 ± 3 | 45 ± 3 | 43 |
| 8l | 87 | m-Tolyl | Me | 33 ± 2 | 29 ± 2 | 36 ± 2 | 35 ± 2 | 33 |
| 8m | 90 | Ethyl | H | 72 ± 6 | 68 ± 6 | 74 ± 6 | 74 ± 6 | 72 |
| 8n | 92 | Ethyl | Cl | 75 ± 6 | 71 ± 6 | 78 ± 6 | 79 ± 6 | 76 |
| 8o | 89 | Ethyl | OMe | 69 ± 5 | 63 ± 5 | 74 ± 6 | 74 ± 6 | 70 |
| 8p | 86 | Ethyl | Me | 82 ± 7 | 77 ± 7 | 84 ± 7 | 85 ± 7 | 82 |
| 8q | 87 | Allyl | H | 93 ± 8 | 89 ± 8 | 96 ± 8 | 96 ± 8 | 94 |
| 8r | 90 | Allyl | Cl | 79 ± 6 | 75 ± 6 | 81 ± 7 | 84 ± 7 | 80 |
| 8s | 91 | Allyl | OMe | 57 ± 5 | 53 ± 4 | 59 ± 5 | 59 ± 5 | 57 |
| 8t | 90 | Allyl | Me | 84 ± 7 | 80 ± 7 | 87 ± 7 | 87 ± 7 | 85 |
| Erlotinib | ND | — | — | 30 ± 3 | 40 ± 3 | 30 ± 3 | 30 ± 3 | 33 |
Compared to Erlotinib, which had a GI50 of 33 nM, compounds 8a–t had substantial antiproliferative action, with GI50 values ranging from 22 nM to 94 nM versus the four cancer cell lines evaluated. In that order, the most potent derivatives were 8f, 8g, 8h, 8j, and 8l, with GI50 values of 37, 22, 25, 29, and 33 nM. This means that 8g, 8h, and 8j were stronger than Erlotinib (GI50 = 33 nM). The most potent of the newly synthesized hybrids 8a–t was compound 8g (R1 = p-tolyl, R2 = OMe), which had a GI50 value of 22 nM, which is 1.5 times stronger than the standard Erlotinib (GI50 = 33 nM). The nature of the aryl/alkyl substituents at position 3 of the quinazoline moiety appears to be critical for the 8a–t hybrids' antiproliferative activity. The GI50 values of compounds 8h (R1 = p-tolyl, R2 = Me) and 8j (R1 = m-tolyl, R2 = Cl) were 25 nM and 29 nM, respectively. These values were lower than compound 8g's (GI50 = 22 nM) but higher than the reference erlotinib. Also, compounds 8k (R1 = m-tolyl, R2 = OMe) and 8l (R1 = m-tolyl, R2 = Me) had GI50 values of 43 and 33 nM, respectively. These were less potent than compounds 8g, 8h, and 8j, but 8l performed similarly to erlotinib, while 8k is less effective than erlotinib. These data demonstrated that the p-tolyl group at position 3 of the quinazoline moiety is more tolerated for antiproliferative action than the m-tolyl one. Additionally, the GI50 values for compounds 8c (R1 = phenyl, R2 = OMe), 8o (R1 = ethyl, R2 = OMe), and 8s (R1 = allyl, R2 = OMe) were 47, 70, and 57 nM, respectively. These values are lower than those for compound 8g (R1 = p-tolyl, R2 = OMe) and even Erlotinib. The results highlighted the importance of the substitution pattern at position three of the quinazoline moiety in antiproliferative activity, with efficacy increasing in the following order: p-tolyl > m-tolyl > phenyl > allyl > ethyl.
Also, the pattern of substitution at the fourth position of the phenyl group within the 1,2,3-triazole moiety (R2) may significantly impact how effectively 8a–t hybrids inhibit cell proliferation. The GI50 values for compounds 8e (R1 = p-tolyl, R2 = H), 8f (R1 = p-tolyl, R2 = Cl), and 8h (R1 = p-tolyl, R2 = Me) were 89, 37, and 25 nM, respectively, demonstrating lower potency than 8g (R1 = p-tolyl, R2 = OMe), which exhibited a GI50 value of 22 nM. The results show that the antiproliferative activity of these hybrids is affected by the pattern of substitutions at the fourth position of the phenyl group in the 1,2,3-triazole moiety. The activity decreases from OMe to Me to Cl, with compound 8e, the unsubstituted derivative (R2 = H), having the least potency. It is 4-fold less potent than compound 8g, the methoxy derivative.
Another significant feature is that all tested compounds exhibited heightened sensitivity to the breast cancer (MCF-7) cell line compared to the other cell lines investigated. For example, compound 8g (R1 = p-tolyl, R2 = OMe) had the most activity, with IC50 values of 22, 20, 24, and 24 nM against lung cancer-A-549, breast cancer-MCF-7, pancreatic cancer-Panc-1 pancreatic, and colon cancer-HT-29 cancer cell lines, respectively. It was more effective than erlotinib against all four cancer cell lines and twice as effective against the MCF-7 breast cancer cell line. The same rule applies to all derivatives, regardless of the characteristics of (R2) or how the quinazoline moiety is substituted at position 3.
| Compound | EGFR inhibition IC50 ± SEM (nM) | BRAFV600E inhibition IC50 ± SEM (nM) |
|---|---|---|
| 8f | 89 ± 5 | 69 ± 5 |
| 8g | 68 ± 4 | 47 ± 3 |
| 8h | 74 ± 5 | 55 ± 5 |
| 8j | 78 ± 5 | 61 ± 5 |
| 8l | 83 ± 5 | 64 ± 5 |
| Erlotinib | 80 ± 5 | 60 ± 5 |
| Vemurafenib | ND | 30 ± 3 |
We further investigated the ability of compounds 8g and 8h, the most effective in all in vitro studies, to activate apoptotic markers.
3.2.5.1. Caspase-3 activation assay. Compounds 8g and 8h were evaluated as caspase-3 activators against the MCF-7 breast cancer cell line,40 with results presented in Table 3. It was found that derivatives 8g and 8h had significantly higher levels of caspase-3 protein (695 ± 5 and 650 ± 5 pg mL−1, respectively) compared to the standard substance staurosporine (503 ± 4 pg mL−1). The compounds 8g and 8h increased the concentration of caspase-3 protein in the MCF-7 cancer cell line, showing levels that were 11 and 10 times higher than the untreated control and higher than staurosporine. These data indicate that apoptosis may play a role in the antiproliferative effects of the investigated compounds, possibly due to caspase-3 upregulation.
| Compound number | Caspase-3 | |
|---|---|---|
| Conc. (pg mL−1) | Fold change | |
| 8g | 695 ± 5 | 11 |
| 8h | 650 ± 5 | 10 |
| Staurosporine | 503 ± 4 | 8 |
| Control | 63 | 1 |
3.2.5.2. Assay for caspase-8, Bax and Bcl-2 levels. We further examined the effects of compounds 8g and 8h on caspase-8, Bax, and the anti-apoptotic Bcl2 levels in the MCF-7 cancer cell line, using staurosporine as a reference.40 Table 4 displays the findings. The results indicated that 8g and 8h markedly elevated Bax and caspase-8 levels in comparison to staurosporine. Compound 8g had the most overexpression of caspase-8 (2.60 ng mL−1), followed by compound 8h (2.10 ng mL−1) and staurosporine as a control (1.80 ng mL−1). Furthermore, 8g and 8h demonstrated greater Bax induction (315 and 295 pg mL−1, respectively) relative to staurosporine (280 pg mL−1), exhibiting a 39-fold and 37-fold increase compared to untreated control MCF-7 cancer cells.
| Compound number | Caspase-8 | Bax | Bcl-2 | |||
|---|---|---|---|---|---|---|
| Conc. (ng mL−1) | Fold change | Conc. (pg mL−1) | Fold change | Conc. (ng mL−1) | Fold reduction | |
| 8g | 2.60 ± 0.20 | 28 | 315 ± 9 | 39 | 0.70 | 7 |
| 8h | 2.10 ± 0.20 | 23 | 295 ± 9 | 37 | 0.90 | 6 |
| Staurosporine | 1.80 ± 0.10 | 20 | 280 ± 7 | 35 | 1.10 | 5 |
| Control | 0.09 | 1 | 8 | 1 | 5 | 1 |
Finally, compound 8g caused a marketed drop in the concentration of Bcl-2 protein (0.70 ng mL−1), followed by compound 8h (0.90 ng mL−1) in the MCF-7 cell line compared to staurosporine (1.10 ng mL−1). The apoptosis assay showed that compounds 8g and 8h have dual inhibitory effects against EGFR and BRAFV600E, showing strong apoptotic antiproliferative effect.
3.2.6.1. Cell cycle analysis. The effects of 8g on cell cycle progression and apoptosis induction in A-549 cells were examined. A lung cancer (A-549) cell line was subjected to treatment for 24 hours with an IC50 concentration of 8g. We stained the cell line with PI/annexin V and performed flow cytometry using a BD FACS Caliber.41 The results (Fig. 5) showed that A-549 cells treated with compound 8g had a significant accumulation of 86% in the G0/G1 phase after 24 hours of incubation. This means a cell cycle arrest at the G1 transition.
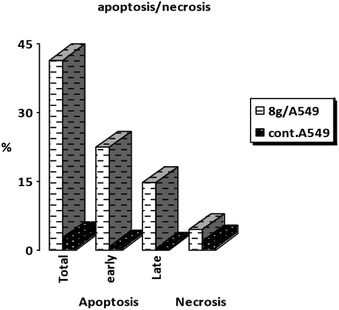
3.2.6.2. Apoptosis induction assay. A-549 cells was stained with annexin V/PI, cultured for 24 hours, and analyzed them to examine 8g's capacity to cause apoptosis. Analysis of early and late apoptosis revealed that compound 8g induced significant apoptosis, with a necrosis percentage of 4.39 (Fig. 6 and 7).
3.3. In silico docking simulation
This study conducted a thorough computer docking analysis to determine the binding relationships between chemicals 8f, 8g, 8h, 8j, and 8l and EGFR-TK. The present study employed the Discovery Studio program to conduct a comprehensive analysis of the interaction mechanism.55,56 To improve this analysis, we incorporated the crystallographic structure of the EGFR-erlotinib complex, as documented in the Protein Data Bank (PDB ID: 1M17),42,57 to give the study a strong structural base, as well as the crystallographic configuration of EGFR complexed with Erlotinib. To test the docking technique's efficacy, we re-docked the co-crystallized ligand erlotinib into the active site of the EGFR protein. The technique produced an S score of −7.30 kcal mol−1, indicating the procedure's precision. A strong hydrogen bond interaction was made between the pyrimidine nitrogen in Erlotinib and the amino acid residue Met769 in the EGFR structure, which made the docking result even more clear. This interaction is crucial for stabilizing the ligand in the active site, underscoring the significance of molecular interactions in the binding process. We docked compounds 8f, 8g, 8h, 8j, and 8l with EGFR kinase to determine the binding affinity of derivatives with the best in vitro activity against EGFR, and Table 5 shows their docking scores.All the tested compounds had a strong and similar ability to bind to erlotinib at the EGFR enzyme's active site. A visual analysis of the optimal docking pose was conducted to determine potential interactions between the test compounds and the amino acid residues constituting the active site. Fig. 8 illustrates how two H-bond interactions with MET 769 and a pi–H interaction with LEU 694 stabilized the structure of compound 8g, which had the highest docking score among its congeners, within EGFR binding site.
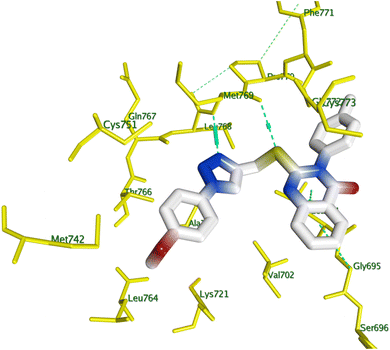 | ||
| Fig. 8 3D closest interactions between active site amino acid residues of EGFR kinase (PDB ID: 1M17) and best docking score compound 8g. | ||
The study's most effective antiproliferative hybrids, 8g and 8h, was looked at in more detail using in silico docking to find out how it binds to the active site of BRAFV600E. This exploratory method utilized the crystal structure of BRAFV600E in complex with Vemurafenib (PDB ID: 4RZV) as a reference point.43,58 Compounds 8g and 8h showed the highest binding affinity (S = −7.97 and −7.23 kcal mol; respectively) among their test congeners, and visual inspection of their docking poses revealed a number of pi-H interactions via –N-p-tolyl moiety and quinazoline ring with LYS 483 and VAL 471, in addition to triazole ring with SER 535, as shown in Fig. 9.
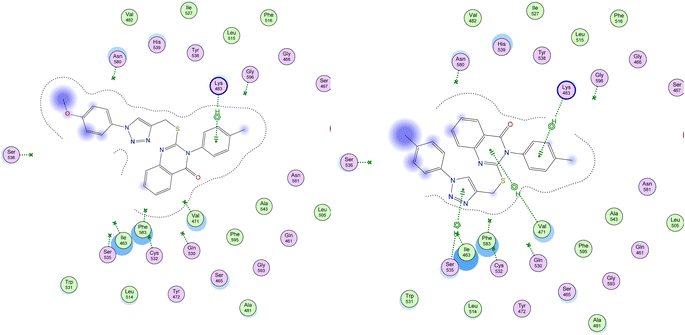 | ||
| Fig. 9 Depicted 2D figures of binding interactions of compounds 8g and 8h within active site of BRAFV600E (PDB ID: 4RZV) showing pi–H bonds as green-dotted lines with LYS 483, VAL 471, and SER 535. | ||
In summary, these docking experiments provide good insights into the potential inhibitory activity of these new quinazolines against EGFR and BRAF kinases, and their high binding affinity within test crystal structures suggesting that they can effectively interact with crucial amino acid residues of both kinases.
4. Conclusion
In search of novel antiproliferative scaffolds, we designed and synthesized twenty new 1,2,3-oxadiazole/quinazoline-4-one hybrids (8a–t) that inhibit both EGFR and BRAFV600E. The novel hybrids showed promising antiproliferative activities. We investigated the inhibitory effects of derivatives 8f, 8g, 8h, 8j, and 8l on EGFR and BRAFV600E. In vitro experiments revealed that compounds 8g, 8h, and 8l are effective cancer-fighting medicines that inhibit both EGFR and BRAFV600E. Additionally, compounds 8g and 8h may have apoptotic action because they activated caspase 3, 8, and Bax while inhibiting Bcl2. Cell cycle analysis and apoptosis induction assays on 8g revealed cell cycle arrest in the G1 phase. Docking experiments revealed structural insights, with compound 8g exhibiting strong binding affinities for both EGFR and BRAFV600E, as indicated by its high docking scores. Upon optimization, these novel hybrids may have the potential to act as anticancer agents.Data availability
Samples of compounds 8a–t are available from the authors.Conflicts of interest
The author reported no potential conflicts of interest(s).Acknowledgements
This work was funded by the Researchers Supporting Project Number (RSPD2024R603) King Saud University, Riyadh, Saudi Arabia. The authors also acknowledge support from the KIT-Publication Fund of the Karlsruhe Institute of Technology.References
- A. Letai, Annu. Rev. Cancer Biol., 2017, 1, 275–294 CrossRef.
- E. J. Lee, Cystogenesis, 2016, 25–34 CAS.
- S. Zhu, T. Zhang, L. Zheng, H. Liu, W. Song, D. Liu, Z. Li and C.-x. Pan, J. Hematol. Oncol., 2021, 14, 156 CrossRef PubMed.
- N. Lu, J. Wu, M. Tian, S. Zhang, Z. Li and L. Shi, Eur. J. Med. Chem., 2024, 116233 CrossRef CAS PubMed.
- S. Sudhesh Dev, S. A. Zainal Abidin, R. Farghadani, I. Othman and R. Naidu, Front. Pharmacol, 2021, 12, 772510 CrossRef PubMed.
- J. Esteban-Villarrubia, J. J. Soto-Castillo, J. Pozas, M. San Román-Gil, I. Orejana-Martín, J. Torres-Jiménez, A. Carrato, T. Alonso-Gordoa and J. Molina-Cerrillo, Int. J. Mol. Sci., 2020, 21, 8529 CrossRef CAS PubMed.
- L. H. Al-Wahaibi, A. M. Elshamsy, T. F. Ali, B. G. Youssif, S. Bräse, M. Abdel-Aziz and N. A. El-Koussi, ACS Omega, 2024, 9(32), 34358–34369 CrossRef CAS PubMed.
- L. H. Al-Wahaibi, A. F. Mohammed, F. E.-Z. S. Abdel Rahman, M. H. Abdelrahman, X. Gu, L. Trembleau and B. G. Youssif, J. Enzyme Inhib. Med. Chem., 2023, 38, 2218602 CrossRef PubMed.
- T. Notarangelo, L. Sisinni, V. Condelli and M. Landriscina, Cancer Cell Int., 2017, 17, 1–9 CrossRef PubMed.
- S. Mondaca, M. Lacouture, J. Hersch and R. Yaeger, JCO Precis. Oncol., 2018, 2, 1800088 Search PubMed.
- M. Okaniwa, M. Hirose, T. Imada, T. Ohashi, Y. Hayashi, T. Miyazaki, T. Arita, M. Yabuki, K. Kakoi and J. Kato, J. Med. Chem., 2012, 55, 3452–3478 CrossRef CAS PubMed.
- Q. Zhang, Y. Diao, F. Wang, Y. Fu, F. Tang, Q. You and H. Zhou, MedChemComm, 2013, 4, 979–986 RSC.
- K. Laxmikeshav, P. Kumari and N. Shankaraiah, Med. Res. Rev., 2022, 42, 513–575 CrossRef CAS PubMed.
- B. K. Banik and B. Banerjee, Heterocyclic Anticancer Agents, Walter de Gruyter GmbH & Co KG, 2022 Search PubMed.
- M. Moradi, A. Mousavi, Z. Emamgholipour, J. Giovannini, S. Moghimi, F. Peytam, A. Honarmand, S. Bach and A. Foroumadi, Eur. J. Med. Chem., 2023, 115626 CrossRef CAS PubMed.
- S. Sharma, K. Sharma, S. Pathak, M. Kumar and P. K. Sharma, Open Med. Chem. J., 2020, 14, 108–121 CrossRef CAS.
- K. Nepali, S. Sharma, R. Ojha and K. L. Dhar, Med. Chem. Res., 2013, 22, 1–15 CrossRef CAS.
- V. Panwar, K. Mukherji, M. Ghate, D. K. Jindal and D. Kumar, in Biomedical Translational Research: Drug Design and Discovery, Springer, 2022, pp. 387–399 Search PubMed.
- H. T. Abdel-Mohsen, M. M. Anwar, N. S. Ahmed, S. S. Abd El-Karim and S. H. Abdelwahed, Molecules, 2024, 29, 875 CrossRef CAS PubMed.
- R. V. Patel, B. M. Mistry, A. V. Gujarati, A. B. Patel and D. K. Patel, ChemistrySelect, 2023, 8, e202301053 CrossRef CAS.
- H. C. Kolb, M. Finn and K. B. Sharpless, Angew. Chem., Int. Ed., 2001, 40, 2004–2021 CrossRef CAS PubMed.
- M. Akter, K. Rupa and P. Anbarasan, Chem. Rev., 2022, 122, 13108–13205 CrossRef CAS PubMed.
- Z. Y. Wang, J. Li, N. Wang, H. Liu and K. K. Wang, Asian J. Org. Chem., 2023, 12, e202300105 CrossRef CAS.
- E. M. El-Sheref, S. Bräse, H. N. Tawfeek, F. A. Alasmary and B. G. Youssif, Int. J. Mol. Sci., 2023, 24, 13300 CrossRef CAS PubMed.
- E. M. El-Sheref, M. A. Elbastawesy, A. B. Brown, A. M. Shawky, H. A. Gomaa, S. Bräse and B. G. Youssif, Molecules, 2021, 26, 6798 CrossRef CAS PubMed.
- M. A. Mahmoud, A. F. Mohammed, O. I. Salem, T. M. Almutairi, S. Bräse and B. G. Youssif, J. Enzyme Inhib. Med. Chem., 2024, 39, 2305856 CrossRef PubMed.
- A. M. Mohamed, O. M. Abou-Ghadir, Y. A. Mostafa, K. A. Dahlous, S. Bräse and B. G. Youssif, Front. Chem., 2024, 12, 1447618 CrossRef PubMed.
- E. Kucuksayan and T. Ozben, Curr. Top. Med. Chem., 2017, 17, 907–918 CrossRef CAS PubMed.
- H. A. Abou-Zied, E. A. Beshr, H. A. Gomaa, Y. A. Mostafa, B. G. Youssif, A. M. Hayallah and M. Abdel-Aziz, Arch. Pharm., 2023, 356, 2200464 CrossRef CAS PubMed.
- M. B. Alshammari, A. A. Aly, B. G. Youssif, S. Bräse, A. Ahmad, A. B. Brown, M. A. Ibrahim and A. H. Mohamed, Front. Chem., 2022, 10, 1076383 CrossRef CAS PubMed.
- L. H. Al-Wahaibi, H. A. Abou-Zied, E. A. Beshr, B. G. Youssif, A. M. Hayallah and M. Abdel-Aziz, Int. J. Mol. Sci., 2023, 24, 9104 CrossRef CAS PubMed.
- L. H. Al-Wahaibi, E. M. El-Sheref, M. M. Hammouda and B. G. Youssif, Pharmaceuticals, 2023, 16, 467 CrossRef CAS PubMed.
- L. H. Al-Wahaibi, A. M. Gouda, O. F. Abou-Ghadir, O. I. Salem, A. T. Ali, H. S. Farghaly, M. H. Abdelrahman, L. Trembleau, H. H. Abdu-Allah and B. G. Youssif, Bioorg. Chem., 2020, 104, 104260 CrossRef CAS PubMed.
- L. H. Al-Wahaibi, M. Hisham, H. A. Abou-Zied, H. A. Hassan, B. G. Youssif, S. Bräse, A. M. Hayallah and M. Abdel-Aziz, Pharmaceuticals, 2023, 16, 1522 CrossRef CAS PubMed.
- L. H. Al-Wahaibi, M. A. Mahmoud, Y. A. Mostafa, A. E. Raslan and B. G. Youssif, J. Enzyme Inhib. Med. Chem., 2023, 38, 376–386 CrossRef CAS PubMed.
- R. A. Mekheimer, S. M. Allam, M. A. Al-Sheikh, M. S. Moustafa, S. M. Al-Mousawi, Y. A. Mostafa, B. G. Youssif, H. A. Gomaa, A. M. Hayallah and M. Abdelaziz, Bioorg. Chem., 2022, 121, 105693 CrossRef CAS PubMed.
- M. Hisham, H. A. Hassan, H. A. Gomaa, B. G. Youssif, A. M. Hayallah and M. Abdel-Aziz, J. Mol. Struct., 2022, 1254, 132422 CrossRef CAS.
- S. A. El-Kalyoubi, H. A. Gomaa, E. M. Abdelhafez, M. Ramadan, F. Agili and B. G. Youssif, Pharmaceuticals, 2023, 16, 716 CrossRef CAS PubMed.
- L. H. Al-Wahaibi, A. F. Mohammed, M. H. Abdelrahman, L. Trembleau and B. G. Youssif, Molecules, 2023, 28, 1269 CrossRef CAS PubMed.
- B. G. Youssif, A. M. Mohamed, E. E. A. Osman, O. F. Abou-Ghadir, D. H. Elnaggar, M. H. Abdelrahman, L. Treamblu and H. A. Gomaa, Eur. J. Med. Chem., 2019, 177, 1–11 CrossRef CAS PubMed.
- H. A. El-Sherief, B. G. Youssif, S. N. A. Bukhari, A. H. Abdelazeem, M. Abdel-Aziz and H. M. Abdel-Rahman, Eur. J. Med. Chem., 2018, 156, 774–789 CrossRef CAS PubMed.
- M. A. Bhat, B. Tüzün, N. A. Alsaif, A. A. Khan and A. M. Naglah, J. Mol. Struct., 2022, 1257, 132600 CrossRef CAS.
- A. K. Singh, A. Kumar, S. Thareja and P. Kumar, Anti-Cancer Agents Med. Chem., 2023, 23, 278–297 CrossRef CAS PubMed.
- H. S. ElZahabi, M. S. Nafie, D. Osman, N. H. Elghazawy, D. H. Soliman, A. A. H. El-Helby, R. K. Arafa and E. J. of, Med. Chem., 2021, 222, 113609 CAS.
- G. Moussa, R. Alaaeddine, L. M. Alaeddine, R. Nassra, A. S. Belal, A. Ismail, A. F. El-Yazbi, Y. S. Abdel-Ghany and A. Hazzaa, Eur. J. Med. Chem., 2018, 144, 635–650 CrossRef CAS PubMed.
- M. I. Mangione, R. A. Spanevello and M. Anzardi, RSC Adv., 2017, 7, 47681–47688 RSC.
- H. A. El-Sherief, B. G. Youssif, A. H. Abdelazeem, M. Abdel-Aziz and H. M. Abdel-Rahman, Anti-Cancer Agents Med. Chem., 2019, 19, 697–706 CrossRef CAS PubMed.
- A. S. Borude, S. R. Deshmukh, S. V. Tiwari, S. H. Kumar and S. R. Thopate, Eur. J. Med. Chem., 2024, 276, 116727 CrossRef CAS PubMed.
- S. Elmore, Toxicol. Pathol., 2007, 35, 495–516 CrossRef CAS PubMed.
- M. S. D'arcy, Cell Biol. Int., 2019, 43, 582–592 CrossRef PubMed.
- G. W. Foight, Determinants of protein-peptide interaction specificity in the Bcl-2 and TRAF families, Massachusetts Institute of Technology, 2015 Search PubMed.
- Y.-F. Chen, A.-S. Lee, W.-Y. Chen, C.-H. Lin, C.-L. Kuo and J.-G. Chung, Am. J. Chin. Med., 2020, 48, 719–736 CrossRef CAS PubMed.
- N. Naumova and R. Šachl, Membranes, 2020, 10, 299 CrossRef CAS PubMed.
- K. G. Ponder and L. H. Boise, Cell Death Discovery, 2019, 5, 56 CrossRef PubMed.
- T. S. Ibrahim, R. M. Bokhtia, A. M. Al-Mahmoudy, E. S. Taher, M. A. AlAwadh, M. Elagawany, E. H. Abdel-Aal, S. Panda, A. M. Gouda and H. Z. Asfour, Bioorg. Chem., 2020, 99, 103782 CrossRef CAS PubMed.
- M. S. Shaykoon, A. A. Marzouk, O. M. Soltan, A. S. Wanas, M. M. Radwan, A. M. Gouda, B. G. Youssif and M. Abdel-Aziz, Bioorg. Chem., 2020, 100, 103933 CrossRef CAS PubMed.
- V. Jakhmola, T. Parashar, P. Ghildiyal, A. Ansori, R. K. Sharma, N. R. Rao, K. Kalra, N. Singh, N. Nainwal and R. K. Singh, Pharmacogn. J., 2022, 14, 4609–4629 Search PubMed.
- T. Kircher, T. Pantsar, A. Oder, J. P. von Kries, M. Juchum, B. Pfaffenrot, P. Kloevekorn, W. Albrecht, R. Selig and S. Laufer, Eur. J. Med. Chem., 2021, 209, 112901 CrossRef CAS PubMed.
Footnote |
| † Electronic supplementary information (ESI) available. See DOI: https://doi.org/10.1039/d4ra06694d |
| This journal is © The Royal Society of Chemistry 2024 |