Open Access Article
Open Access ArticleSynthesis, in vitro and in silico study of novel 1,3-diphenylurea derived Schiff bases as competitive α-glucosidase inhibitors†
Anam Rubbab Pashaab,
Saeed Ullahb,
Ajmal Khanbh,
Sobia Ahsan Halimb,
Javid Hussainc,
Tanzila Rehmand,
Rimsha Taliba,
Rima D. Alharthye,
Hamdy Kashtoh*f,
Magda H. Abdellattif g,
Ahmed Al-Harrasi*b and
Zahid Shafiq
g,
Ahmed Al-Harrasi*b and
Zahid Shafiq *a
*a
aInstitute of Chemical Sciences, Bahauddin Zakariya University, Multan-60800, Pakistan. E-mail: zahidshafiq@bzu.edu.pk
bNatural and Medical Sciences Research Centre, University of Nizwa, P.O. Box 33, PC 616, Birkat Al Mauz, Nizwa, Sultanate of Oman. E-mail: aharrasi@unizwa.edu.om
cDepartment of Biological Sciences and Chemistry, University of Nizwa, Oman
dDepartment of Chemistry, The Women University, Multan-60000, Pakistan
eDepartment of Chemistry, Science & Arts College, Rabigh Branch, King Abdulaziz University, Rabigh 21911, Saudi Arabia
fDepartment of Biotechnology, Yeungnam University, Gyeongsan 38541, Gyeongbuk, Republic of Korea. E-mail: hamdy_kashtoh@ynu.ac.kr
gChemistry Department, College of Sciences, University College of Taraba, Taif University, Taif 21944, Saudi Arabia
hDepartment of Chemical and Biological Engineering, College of Engineering, Korea University, 145 Anam-ro, Seongbuk-gu, Seoul 02841, Republic of Korea
First published on 16th September 2024
Abstract
Diabetes mellitus has become a major global health burden because of several related consequences, including heart disease, retinopathy, cataracts, metabolic syndrome, collapsed renal function, and blindness. In the recent study, thirty Schiff base derivatives of 1,3-diphenylurea were synthesized and their anti-diabetic activity was evaluated by targeting α-glucosidase. The compounds exhibited an overwhelming inhibitory potential for α-glucosidase with higher potency ranging from 2.49–37.16 μM. The most effective compound, 5h, showed competitive inhibition of α-glucosidase (Ki = 3.96 ± 0.0048 μM) in the kinetic analysis and strong binding interactions with key residues α-glucosidase in docking analysis, indicating its potential for better glycemic control in diabetes patients.
1. Introduction
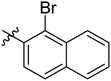
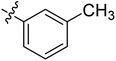
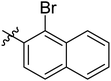
Diabetes mellitus is a chronic condition that has become a major global health concern because of various complications associated with it including heart disease, retinopathy, cataracts, metabolic syndrome, collapsed renal function, and blindness. Globally, >1.31 billion (1.22–1.39) people are estimated to have diabetes by 2050. Age-standardized global diabetes prevalence rates more than 10% are expected for two super-regions: North Africa and the Middle East, at 16.8% (16.1–17.6), and Latin America and the Caribbean, at 11.3% (10.8–11.9).1,2 The primary risk factor for type 2 diabetes (T2DM) is postprandial hyperglycemia, which is linked to the deficiency of insulin or defect in the function of insulin.3,4 The key carbohydrate metabolic enzyme, α-glucosidase (EC 3.2.1.20) is present in brush boarder of small intestine, and converts non-absorbable complex carbohydrates into absorbable monosaccharides, such as glucose molecules. Inhibiting α-glucosidase is a crucial approach to manage conditions linked to the absorption of carbohydrates, such as diabetes, obesity, dental caries, and periodontal illnesses. The α-glucosidase inhibitors block its catalytic activity, thereby slow down carbohydrates digestion and control blood glucose level.5 Therefore, in the current study, new Schiff base of 1,3-dipheny urea derivatives were synthesized and their antidiabetic potential was evaluated by particularly inhibiting α-glucosidase enzyme.Urea scaffold is embedded in a variety of important bioactive compounds and FDA approved drugs like regorafenib and sorafenib which shows its therapeutic importance.6,7 Diarylurea core has gained noteworthy pharmacological interest due to the presence of NH–CO scaffold which binds with diverse range of biological targets8 and consequently produces broad spectrum of biological activities like anti-viral, antitumor, antimalarial9–11 activities. Furthermore, urea derivatives are well known α-glucosidase inhibitors and various studies have explored their potential as antidiabetic agents (Fig. 1).12–17
 | ||
| Fig. 1 The chemical structures of some reported α-glucosidase inhibitors are shown.12–18 | ||
Azomethine functionality has shown anticancer, antioxidant, antifungal, antibacterial, antiviral and antidiabetic activities14,18–24 which makes it a promising pharmacophore to develop new drug candidates. Salicylaldehyde and its derivatives are well known synthetic precursor for the preparation of different drugs like Aspirin, Warfarin, and Salsalate.25 Salicylaldehyde nucleus linked with Schiff bases, have exhibited potential biological activities like antibacterial,26 anticancer,27,28 antiviral,29 tyrosinase,30 antimicrobial,31 antioxidant,32 and antidiabetic33 activities. Notably, the presence of halogen moiety (chloro or bromo) in salicylaldehyde Schiff base nucleus have shown significant biological activity due to more facilitated interactions with the binding sites of biological targets.34
Naphthalene moiety is a typical fluorophore, present in different naturally occurring bioactive phytoconstituents like Patentiflorin A, and Rifampicin. Naphthalene is a crucial building block in the design of new drugs because of its antifungal, antitumor,35 antibacterial,36 and antidiabetic,37 activities. Moreover, various molecules with naphthalene moiety are available as FDA approved drugs.38 Because of the high significance of these moieties, we aimed to join the urea and Schiff base pharmacophore in one molecule along with the salicylaldehyde or naphthaldehyde core to explore the pharmacological profile of urea clubbed imines. For this purpose, 1,3-diphenyl urea analogues were reacted with 5-chlorosalicylaldehyde, 5-bromosalicylaldehyde and 1-bromo-2-naphthaldehyde and Schiff base derivatives of 1,3-dipheny urea were synthesized and screened those analogues against α-glucosidase.
2. Results and discussion
2.1. Chemistry
o-Phenylenediamine (1) was reacted with equimolar amount of different substituted isocyanates (2a–t) by constant stirring at room temperature overnight and the resulting mono substituted 1,3-diphenyl ureas (3a–t) were refluxed for 3–4 hours with substituted aldehyde (4) via condensation in methanol to obtain the final products (5a–t) and (6a–j). The scope of reaction was broadened by using a variety of different mono substituted 1,3-diphenyl ureas. The targeted compounds (5a–t) and (6a–j) were obtained in good to excellent yield (Scheme 1). The structures of Schiff base 1,3-dipheny urea derivatives were established using microanalysis (CHN) and spectral data i.e., 1H NMR and 13C NMR. In 1H NMR, phenolic –OH was observed between 11.62–11.91 ppm while Ph–NH–CO proton appeared in the range of 9.99–8.89 ppm as broad singlet. Second NH–R was observed in the range of 8.85–9.22 ppm whereas the imine C![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) N proton showed singlet ranging from 8.23–8.63 ppm. The spectral data of other aromatic and aliphatic protons were in accordance with the structures of anticipated compounds which supports the proposed structure of 1,3-diphenylurea derived Schiff based derivatives. 13C NMR also supported the structure of the synthesized compounds and the carbon peaks were in complete agreement with the structures. CHN analysis corresponds to the molecular formula of the synthesized compounds. In order to determine the purity of compounds, HPLC analysis was carried out using CH3CN
N proton showed singlet ranging from 8.23–8.63 ppm. The spectral data of other aromatic and aliphatic protons were in accordance with the structures of anticipated compounds which supports the proposed structure of 1,3-diphenylurea derived Schiff based derivatives. 13C NMR also supported the structure of the synthesized compounds and the carbon peaks were in complete agreement with the structures. CHN analysis corresponds to the molecular formula of the synthesized compounds. In order to determine the purity of compounds, HPLC analysis was carried out using CH3CN![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) H2O = 80
H2O = 80![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 20 eluent system with 263 nm wavelength. All the compounds exhibited great than 95% purity. QTOF MS was also carried out to find out the molecular mass of the compounds which further supports characterization of our target compounds.
20 eluent system with 263 nm wavelength. All the compounds exhibited great than 95% purity. QTOF MS was also carried out to find out the molecular mass of the compounds which further supports characterization of our target compounds.
The synthesized molecules were in vitro tested against α-glucosidase to reveal their potential in the treatment of diabetes mellitus. All the compounds exhibited potent inhibition of α-glucosidase with IC50 values in the range of 2.49–37.16 μM (Table 1), as compared to the available marketed drug, acarbose (IC50 = 873.34 ± 1.67 μM). The structure activity relationship of these compounds was established by segregating them into three groups based on their R-substituents, namely A, B, and group C.
Group A comprises of ten molecules (5a–5j) with similar R1 (5-chloro 2-hydroxy phenyl) with diverse R-group moieties. Group B contains compounds 5k–5t with similar R1 (5-bromo 2-hydroxy phenyl) and different R-substituents. Due to different R1 and R moieties, groups A and B exhibited varied α-glucosidase inhibitory capability. For instance, compound 5a exhibited potent inhibitory capability (IC50 = 3.26 ± 0.10 μM), while 5k with similar R-substituent exhibited decreased α-glucosidase inhibition (IC50 = 16.38 ± 0.40 μM) as compared to 5a. In contrast, 5b with p-methyl phenyl R group exhibited low inhibitory activity (IC50 = 20.10 ± 0.51 μM) as compared to 5l (IC50 = 5.10 ± 0.13 μM). Compound 5c with phenyl substituent R-group exhibited less potent inhibition (IC50 = 14.20 ± 0.30 μM) as compared to 5m (IC50 = 6.30 ± 0.19 μM). Similarly, compound 5d with m-flouro phenyl R-group displayed less potent inhibitory activity (IC50 = 25.16 ± 0.57 μM), as compared to 5n (IC50 = 7.05 ± 0.13 μM) with similar R moiety. While compound 5e with p-flouro phenyl substituent exhibited higher potent inhibition (IC50 = 3.76 ± 0.11 μM) as compared to 5o (IC50 = 9.15 ± 0.25 μM) with similar substituent (p-flouro phenyl). Compound 5f (IC50 = 20.18 ± 0.36 μM) with o-methyl phenyl substituent exhibited less potent inhibitory activity as compared to 5p (IC50 = 8.24 ± 0.17 μM) with m-chloro phenyl group. Whereas compound 5g (IC50 = 4.03 ± 0.12 μM) with m-methyl phenyl substituent exhibited higher inhibitory activity as compared to 5q (IC50 = 10.60 ± 0.31 μM) with p-methoxy phenyl substitution. Interestingly, compound 5h (IC50 = 2.49 ± 0.10 μM) with m-chloro phenyl substitution exhibited the higher potent inhibitory activity against α-glucosidase as compared to 5r (IC50 = 15.20 ± 0.36 μM) with p-keto phenyl moiety. Compound 5i (IC50 = 18.35 ± 0.47 μM) with p-methoxy phenyl substitution exhibited less potent inhibition as compared to 5s (IC50 = 4.10 ± 0.11 μM) with m-flouro phenyl substitution. Compound 5j (IC50 = 16.35 ± 0.28 μM) with acetophenyl R-substituent exhibited less potent inhibition of α-glucosidase as compared to 5t (IC50 = 12.22 ± 0.30 μM) with phenyl substitution.
Group C is comprising of compounds 6a–6j with similar R1 group (2 bromo naphthyl) and diverse R-substituents, these molecules displayed slight variation in the α-glucosidase inhibition. Like compounds 6a and 6c with para and m-chloro phenyl substituents exhibited almost similar potency against α-glucosidase with IC50 values of 20.24 ± 0.38 μM and 18.35 ± 0.40 μM, respectively. Compound 6b (IC50 = 29.42 ± 0.40 μM) with p-methyl phenyl substituent displayed slightly higher inhibition as compared to 6i (IC50 = 34.62 ± 0.85 μM) and 6j (IC50 = 37.16 ± 0.75 μM) with m- and o-methyl phenyl substituents, respectively. Compound 6d (IC50 = 27.29 ± 0.37 μM) with p-methoxy phenyl and 6e (IC50 = 26.17 ± 0.43 μM) with p-aceto phenyl substituents exhibited very close inhibition of α-glucosidase. While 6f (IC50 = 31.48 ± 0.64 μM) with phenyl R group exhibited less inhibition compared to 6d and 6e. The flouro substituted compounds, 6g (IC50 = 17.00 ± 0.42 μM) with p-flouro phenyl substituent exhibited significantly higher potent inhibition as compared to 6h (IC50 = 22.39 ± 0.54 μM) with m-flouro phenyl substituent. Overall, compounds in group A and B displayed higher potency than compounds in group C. This favorable inhibitory effect might be due to the R1 and R substituents.
2.2. Kinetic study
The mechanism of action of the identified inhibitors was deduced in vitro by kinetic analysis of the most potent compound, 5h which showed concentration dependent type of inhibition with Ki value 3.96 ± 0.0048 μM. The mechanistic analysis indicates that 5h binds at the active site of α-glucosidase. Thus, increases Km of the enzyme while Vmax of the enzyme remains constant (Fig. 2).2.3. Molecular docking
Docking was performed to elucidate the mode of binding of 5h at the active site of α-glucosidase, which reflects excellent binding of 5h with active site residues. The urea moiety of 5h mediates a hydrogen bond with Gln279 (2.06 Å) which is one of the residues in catalytic triad. While the hydroxyl group of 5h exhibited a hydrogen bond with the –OH of Thr306 (1.79 Å), and substituted chlorine at phenyl ring forms halogen bond with the side chain of Arg213 (2.47 Å). Moreover, a solvent molecule provided a hydrogen bond to the amino group of 5h (1.96 Å) and Tyr72 creates a hydrophobic interaction with the chloro-substituted phenyl ring of 5h (4.31 Å). These interactions help in fitting of 5h at the active site of enzyme with a highly negative docking score (−6.15 kcal mol−1). The binding mode of 5h is shown in Fig. 3 in 3D and 2D-mode.3. Conclusion
Schiff base diphenyl urea derivatives play a crucial role in medicinal chemistry. The current study demonstrates the synthesis of novel Schiff bases of 1,3-diphenyl urea derivates their evaluation against α-glucosidase to explore their therapeutic potential for diabetes mellitus. Fortunately, all the compounds exhibited several fold potent inhibition of α-glucosidase in the range of 2.49–37.16 μM, as compared to standard drug. In the kinetic analysis, the most potent inhibitor, compound 5h reflected competitive mode of inhibition and demonstrates favorable interactions with the active site residues of α-glucosidase in the molecular docking investigation. The nature of these interactions, such as hydrogen bonds, and hydrophobic interactions provides insight into the molecular basis for the high inhibitory activity of 5h, observed in kinetic studies. These findings highlight the therapeutic potential of the identified inhibitors for the treatment of diabetes mellitus. α-Glucosidase hydrolysis carbohydrates into glucose and subsequently increases blood glucose levels. By inhibiting α-glucosidase, 5h can slow down the absorption of glucose in the intestine, thereby control postprandial blood glucose spikes and reduces the risk of diabetes associated long-term complications. These promising results warrant further preclinical evaluation of 5h for the development of a new class of antidiabetic agent with better glycemic control.4. Materials and method
All the starting materials employed in the synthesis were purchased from Sigma-Aldrich Co. (Germany) and used without purification. Methanol, absolute ethanol, and other solvents were also purchased from different commercial sources in adequate purity and used without purification in the reaction media. To monitor the reaction, thin layer chromatography (TLC) was performed with silica gel 60 aluminum backed plates with suitable solvent system. Spotson TLC plates were visualized by using UV light with 254 nm. The 1H and 13C nuclear magnetic resonance (NMR) spectra were recorded using DMSO-d6 as solvents via Bruker spectrophotometer 600 MHz and 151 MHz as dilute solution at 25 °C. Chemical shifts were reported in parts per million (δ = ppm) and coupling constants (J) were expressed in Hertz (Hz). The signals were described as singlet (s), doublet (d), triplet (t) multiplet (m). HPLC was carried out on Agilent, Germany (Liquid Chromatographic Column 150 mm × 4.6 mm (id) packed with 5-micron C18; 263 nm). Mass spectra (ESI-MS), were recorded by means of Agilent QTOF MS 6530 WITH 1260 HPLC. Thermo Scientific FLASH 2000 CHNS/O analyzer. Melting points were determined using MPS10 melting point apparatus.4.1. Chemistry: general procedure for the synthesis of Schiff base 1,3-dipheny urea derivatives
o-Phenylenediamine (1) (5 mmol) was dissolved in 15–20 mL of chloroform by constant stirring at room temperature. Then equimolar amount of different substituted isocyanates (2a–t) were added carefully dropwise with the help of dropping funnel into this diamine solution. Immediately, solid product precipitated out at stirring that was filtered followed by washing with n-hexane and dried under vacuum. The resulting mono substituted 1,3-diphenyl urea (3a–t) (1 mmol) were refluxed for 3–4 hours with substituted aldehyde (4) (1 mmol) in 8–10 mL of methanol to obtain the final products (5a–t) and (6a–j) that were filtered, washed with cold ethanol, and dried under vacuum.4.2. Experimental data
![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) H2O = 80
H2O = 80![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 20); anal. calcd for C20H15Cl2N3O2: C, 60.02; H, 3.78; N, 10.50; found: C, 60.06; H, 3.81; N, 10.45; QTOF MS ES+ (m/z): [M + H]+, calcd: 400.0619, found: 400.0590.
20); anal. calcd for C20H15Cl2N3O2: C, 60.02; H, 3.78; N, 10.50; found: C, 60.06; H, 3.81; N, 10.45; QTOF MS ES+ (m/z): [M + H]+, calcd: 400.0619, found: 400.0590.![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) H2O = 80
H2O = 80![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 20); anal. calcd for C21H18ClN3O2: C, 66.40; H, 4.78; N, 11.06; found: C, 60.36; H, 3.88; N, 10.55; QTOF MS ES+ (m/z): [M + H]+, calcd: 380.1165, found: 380.1158.
20); anal. calcd for C21H18ClN3O2: C, 66.40; H, 4.78; N, 11.06; found: C, 60.36; H, 3.88; N, 10.55; QTOF MS ES+ (m/z): [M + H]+, calcd: 380.1165, found: 380.1158.![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) H2O = 80
H2O = 80![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 20); anal. calcd for C20H16ClN3O2: C, 65.67; H, 4.41; N, 11.49; found: C, 65.76; H, 4.39; N, 11.54; QTOF MS ES+ (m/z): [M + H]+, calcd: 366.1009, found: 366.0992.
20); anal. calcd for C20H16ClN3O2: C, 65.67; H, 4.41; N, 11.49; found: C, 65.76; H, 4.39; N, 11.54; QTOF MS ES+ (m/z): [M + H]+, calcd: 366.1009, found: 366.0992.![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) H2O = 80
H2O = 80![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 20); anal. calcd for C20H15ClFN3O2: C, 62.59; H, 3.94; N, 10.95; found: C, 62.76; H, 3.89; N, 10.99; N, 11.54; QTOF MS ES+ (m/z): [M + H]+, calcd: 384.0915, found: 384.0900.
20); anal. calcd for C20H15ClFN3O2: C, 62.59; H, 3.94; N, 10.95; found: C, 62.76; H, 3.89; N, 10.99; N, 11.54; QTOF MS ES+ (m/z): [M + H]+, calcd: 384.0915, found: 384.0900.![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) H2O = 80
H2O = 80![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 20); anal. calcd for C20H15ClFN3O2: C, 62.59; H, 3.94; N, 10.95; found: C, 62.66; H, 3.85; N, 11.02; QTOF MS ES+ (m/z): [M + H]+, calcd: 384.0915, found: 384.0901.
20); anal. calcd for C20H15ClFN3O2: C, 62.59; H, 3.94; N, 10.95; found: C, 62.66; H, 3.85; N, 11.02; QTOF MS ES+ (m/z): [M + H]+, calcd: 384.0915, found: 384.0901.![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) H2O = 80
H2O = 80![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 20); anal. calcd for C21H18ClN3O2: C, 66.40; H, 4.78; N, 11.06; found: C, 66.36; H, 4.84; N, 11.11; QTOF MS ES+ (m/z): [M + H]+, calcd: 380.1165, found: 380.1206.
20); anal. calcd for C21H18ClN3O2: C, 66.40; H, 4.78; N, 11.06; found: C, 66.36; H, 4.84; N, 11.11; QTOF MS ES+ (m/z): [M + H]+, calcd: 380.1165, found: 380.1206.![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) H2O = 80
H2O = 80![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 20); anal. calcd for C21H18ClN3O2: C, 66.40; H, 4.78; N, 11.06; found: C, 66.45; H, 4.87; N, 11.21; QTOF MS ES+ (m/z): [M + H]+, calcd: 380.1165, found: 380.1091.
20); anal. calcd for C21H18ClN3O2: C, 66.40; H, 4.78; N, 11.06; found: C, 66.45; H, 4.87; N, 11.21; QTOF MS ES+ (m/z): [M + H]+, calcd: 380.1165, found: 380.1091.![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) H2O = 80
H2O = 80![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 20); anal. calcd for C20 H15Cl2N3O2: C, 60.02; H, 3.78; N, 10.50; found: C, 60.10; H, 3.88; N, 10.45; QTOF MS ES+ (m/z): [M + H]+, calcd: 400.0619, found: 400.0574.
20); anal. calcd for C20 H15Cl2N3O2: C, 60.02; H, 3.78; N, 10.50; found: C, 60.10; H, 3.88; N, 10.45; QTOF MS ES+ (m/z): [M + H]+, calcd: 400.0619, found: 400.0574.![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) H2O = 80
H2O = 80![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 20); anal. calcd for C21H18Cl N3O3: C, 63.72; H, 4.58; N, 10.62; found: C, 63.79; H, 4.68; N, 10.55; QTOF MS ES+ (m/z): [M + H]+, calcd: 396.1114, found: 396.1089.
20); anal. calcd for C21H18Cl N3O3: C, 63.72; H, 4.58; N, 10.62; found: C, 63.79; H, 4.68; N, 10.55; QTOF MS ES+ (m/z): [M + H]+, calcd: 396.1114, found: 396.1089.![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) H2O = 80
H2O = 80![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 20); anal. calcd for C22H18ClN3O3: C, 64.79; H, 4.45; N, 10.30; found: C, 64.88; H, 4.38; N, 10.45.
20); anal. calcd for C22H18ClN3O3: C, 64.79; H, 4.45; N, 10.30; found: C, 64.88; H, 4.38; N, 10.45.![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) H2O = 80
H2O = 80![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 20); anal. calcd for C20H15BrClN3O2: C, 54.02; H, 3.40; N, 9.45; found: C, 54.11; H, 3.48; N, 9.55; QTOF MS ES+ (m/z): [M + 2H]+, calcd: 444.0114, found: 445.9910.
20); anal. calcd for C20H15BrClN3O2: C, 54.02; H, 3.40; N, 9.45; found: C, 54.11; H, 3.48; N, 9.55; QTOF MS ES+ (m/z): [M + 2H]+, calcd: 444.0114, found: 445.9910.![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) H2O = 80
H2O = 80![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 20); anal. calcd for C21H18BrN3O2: C, 59.45; H, 4.28; N, 9.90; found: C, 59.40; H, 4.38; N, 9.95; QTOF MS ES+ (m/z): [M + H]+, calcd: 424.0660, found: 424.0620.
20); anal. calcd for C21H18BrN3O2: C, 59.45; H, 4.28; N, 9.90; found: C, 59.40; H, 4.38; N, 9.95; QTOF MS ES+ (m/z): [M + H]+, calcd: 424.0660, found: 424.0620.![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) H2O = 80
H2O = 80![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 20); anal. calcd for C20H15BrFN3O2: C, 56.09; H, 3.53; N, 9.81; found: C, 56.22; H, 3.58; N, 9.85; QTOF MS ES+ (m/z): [M + H]+, calcd: 428.0409, found: 428.0278.
20); anal. calcd for C20H15BrFN3O2: C, 56.09; H, 3.53; N, 9.81; found: C, 56.22; H, 3.58; N, 9.85; QTOF MS ES+ (m/z): [M + H]+, calcd: 428.0409, found: 428.0278.![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) H2O = 80
H2O = 80![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 20); anal. calcd for C21H18BrN3O2: C, 59.45; H, 4.28; N, 9.90; found: C, 59.51; H, 4.31; N, 9.85; QTOF MS ES+ (m/z): [M + 2H]+, calcd: 424.0660, found: 426.0672.
20); anal. calcd for C21H18BrN3O2: C, 59.45; H, 4.28; N, 9.90; found: C, 59.51; H, 4.31; N, 9.85; QTOF MS ES+ (m/z): [M + 2H]+, calcd: 424.0660, found: 426.0672.![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) H2O = 80
H2O = 80![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 20); anal. calcd for C21H18BrN3O2: C, 59.45; H, 4.28; N, 9.90; found: C, 59.50; H, 4.37; N, 9.98; QTOF MS ES+ (m/z): [M + H]+, calcd: 424.0660, found: 424.0556.
20); anal. calcd for C21H18BrN3O2: C, 59.45; H, 4.28; N, 9.90; found: C, 59.50; H, 4.37; N, 9.98; QTOF MS ES+ (m/z): [M + H]+, calcd: 424.0660, found: 424.0556.![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) H2O = 80
H2O = 80![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 20); anal. calcd for C20H15BrClN3O2: C, 54.02; H, 3.40; N, 9.45; found: C, 54.21; H, 3.38; N, 9.51; QTOF MS ES+ (m/z): [M + 2H]+, calcd: 444.0114, found: 445.9989.
20); anal. calcd for C20H15BrClN3O2: C, 54.02; H, 3.40; N, 9.45; found: C, 54.21; H, 3.38; N, 9.51; QTOF MS ES+ (m/z): [M + 2H]+, calcd: 444.0114, found: 445.9989.![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) H2O = 80
H2O = 80![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 20); anal. calcd for C21H18BrN3O3: C, 57.29; H, 4.12; N, 9.54; found: C, 57.41; H, 4.08; N, 9.58.
20); anal. calcd for C21H18BrN3O3: C, 57.29; H, 4.12; N, 9.54; found: C, 57.41; H, 4.08; N, 9.58.![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) H2O = 80
H2O = 80![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 20); anal. calcd for C22H18BrN3O3: C, 58.42; H, 4.01; N, 9.29; found: C, 58.55; H, 4.08; N, 9.35; QTOF MS ES+ (m/z): [M + 2H]+, calcd: 452.0609, found: 454.0568.
20); anal. calcd for C22H18BrN3O3: C, 58.42; H, 4.01; N, 9.29; found: C, 58.55; H, 4.08; N, 9.35; QTOF MS ES+ (m/z): [M + 2H]+, calcd: 452.0609, found: 454.0568.![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) H2O = 80
H2O = 80![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 20); anal. calcd for C20H15BrFN3O2: C, 56.09; H, 3.53; N, 9.81; found: C, 56.19; H, 3.65; N, 9.95; QTOF MS ES+ (m/z): [M + H]+, calcd: 428.0409, found: 428.0403.
20); anal. calcd for C20H15BrFN3O2: C, 56.09; H, 3.53; N, 9.81; found: C, 56.19; H, 3.65; N, 9.95; QTOF MS ES+ (m/z): [M + H]+, calcd: 428.0409, found: 428.0403.![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) H2O = 80
H2O = 80![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 20); anal. calcd for C20H16Br N3O2: C, 58.55; H, 3.93; N, 10.24; found: C, 58.70; H, 3.88; N, 10.35; QTOF MS ES+ (m/z): [M + H]+, calcd: 410.0504, found: 410.0533.
20); anal. calcd for C20H16Br N3O2: C, 58.55; H, 3.93; N, 10.24; found: C, 58.70; H, 3.88; N, 10.35; QTOF MS ES+ (m/z): [M + H]+, calcd: 410.0504, found: 410.0533.![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) H2O = 80
H2O = 80![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 20); anal. calcd for C24H17BrClN3O: C, 60.21; H, 3.58; N, 8.78; found: C, 60.30; H, 3.50; N, 8.90; QTOF MS ES+ (m/z): [M + 2H]+, calcd: 478.0321, found: 480.0301.
20); anal. calcd for C24H17BrClN3O: C, 60.21; H, 3.58; N, 8.78; found: C, 60.30; H, 3.50; N, 8.90; QTOF MS ES+ (m/z): [M + 2H]+, calcd: 478.0321, found: 480.0301.![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) H2O = 80
H2O = 80![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 20); anal. calcd for C25H20BrN3O: C, 65.51; H, 4.40; N, 9.17; found: C, 65.66; H, 4.48; N, 9.09; QTOF MS ES+ (m/z): [M + 2H]+, calcd: 458.0867, found: 460.1058.
20); anal. calcd for C25H20BrN3O: C, 65.51; H, 4.40; N, 9.17; found: C, 65.66; H, 4.48; N, 9.09; QTOF MS ES+ (m/z): [M + 2H]+, calcd: 458.0867, found: 460.1058.![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) H2O = 80
H2O = 80![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 20); anal. calcd for C24H17BrClN3O: C, 60.21; H, 3.58; N, 8.78; Found; C, 60.32; H, 3.71; N, 8.90; QTOF MS ES+ (m/z): [M + 2H]+, calcd: 478.0321, found: 480.0355.
20); anal. calcd for C24H17BrClN3O: C, 60.21; H, 3.58; N, 8.78; Found; C, 60.32; H, 3.71; N, 8.90; QTOF MS ES+ (m/z): [M + 2H]+, calcd: 478.0321, found: 480.0355.![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) H2O = 80
H2O = 80![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 20); anal. calcd for C25H20BrN3O2: C, 63.30; H, 4.25; N, 8.86; found: C, 63.39; H, 4.28; N, 8.95; QTOF MS ES+ (m/z): [M + 2H]+, calcd: 474.0817, found: 476.1053.
20); anal. calcd for C25H20BrN3O2: C, 63.30; H, 4.25; N, 8.86; found: C, 63.39; H, 4.28; N, 8.95; QTOF MS ES+ (m/z): [M + 2H]+, calcd: 474.0817, found: 476.1053.![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) H2O = 80
H2O = 80![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 20); anal. calcd for C26H20BrN3O2: C, 64.21; H, 4.15; N, 8.64; found: C, 64.30; H, 4.18; N, 8.55; QTOF MS ES+ (m/z): [M + 2H]+, calcd: 486.0817, found: 488.0739.
20); anal. calcd for C26H20BrN3O2: C, 64.21; H, 4.15; N, 8.64; found: C, 64.30; H, 4.18; N, 8.55; QTOF MS ES+ (m/z): [M + 2H]+, calcd: 486.0817, found: 488.0739.![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) H2O = 80
H2O = 80![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 20); anal. calcd for C24H18BrN3O: C, 64.88; H, 4.08; N, 9.46; found: C, 66.79; H, 4.18; N, 9.54; QTOF MS ES+ (m/z): [M + 2H]+, calcd: 444.0711, found: 446.0777.
20); anal. calcd for C24H18BrN3O: C, 64.88; H, 4.08; N, 9.46; found: C, 66.79; H, 4.18; N, 9.54; QTOF MS ES+ (m/z): [M + 2H]+, calcd: 444.0711, found: 446.0777.![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) H2O = 80
H2O = 80![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 20); anal. calcd for C24H17BrFN3O: C, 62.35; H, 3.71; N, 9.09; found: C, 62.44; H, 3.68; N, 9.21; QTOF MS ES+ (m/z): [M + 2H]+, calcd: 462.0617, found: 464.0687.
20); anal. calcd for C24H17BrFN3O: C, 62.35; H, 3.71; N, 9.09; found: C, 62.44; H, 3.68; N, 9.21; QTOF MS ES+ (m/z): [M + 2H]+, calcd: 462.0617, found: 464.0687.![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) H2O = 80
H2O = 80![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 20); anal. calcd for C24H17BrFN3O: C, 62.35; H, 3.71; N, 9.09; found: C, 62.44; H, 3.69; N, 9.18; QTOF MS ES+ (m/z): [M + 2H]+, calcd: 462.0617, found: 464.0687.
20); anal. calcd for C24H17BrFN3O: C, 62.35; H, 3.71; N, 9.09; found: C, 62.44; H, 3.69; N, 9.18; QTOF MS ES+ (m/z): [M + 2H]+, calcd: 462.0617, found: 464.0687.![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) H2O = 80
H2O = 80![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 20); anal. calcd for C25H20BrN3O: C, 65.51; H, 4.40; N, 9.17; found: C, 65.55; H, 4.38; N, 9.25; QTOF MS ES+ (m/z): [M + 2H]+, calcd: 458.0867, found: 460.1104.
20); anal. calcd for C25H20BrN3O: C, 65.51; H, 4.40; N, 9.17; found: C, 65.55; H, 4.38; N, 9.25; QTOF MS ES+ (m/z): [M + 2H]+, calcd: 458.0867, found: 460.1104.![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) H2O = 80
H2O = 80![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 20); anal. calcd for C25H20BrN3O: C, 65.51; H, 4.40; N, 9.17; found: C, 65.57; H, 4.37; N, 9.21; QTOF MS ES+ (m/z): [M + H]+, calcd: 458.0867, found: 460.0945.
20); anal. calcd for C25H20BrN3O: C, 65.51; H, 4.40; N, 9.17; found: C, 65.57; H, 4.37; N, 9.21; QTOF MS ES+ (m/z): [M + H]+, calcd: 458.0867, found: 460.0945.4.3. In vitro α-glucosidase inhibitory assay and statistical analysis
The α-glucosidase inhibitory activity was performed by spectrophotometric assay as published earlier.2 Briefly, in a total of 200 μL per well reaction volume, 135 μL of sodium phosphate buffer (50 mM, pH 7.0), 20 μL of test compounds (0.5 mM in DMSO), and 20 μL of α-glucosidase solution (0.02 U per well) were added into 96-well plate. Blank contained 20 μL 7% DMSO only, while acarbose was used as the positive control. The reaction mixture was incubated at 37 °C for 15 min. 25 μL of substrate solution, p-nitro phenyl α-D-glucopyranoside (0.7 mM) was added and change in absorbance was recorded continuously at 400 nm for 30 min through 96-well plate reader (xMark™ Microplate Spectrophotometer, BIO-RAD). All the reactions were performed in triplicates in 96-well microplates. The kinetic study was performed for 5h through a similar experimental procedure with one modification (i.e., four different concentrations of the substrate were used including 0.1, 0.2, 0.4 and 0.8 mM).SoftMax Pro package and Excel were utilized to analyze the results of biological activity. Grafit 7 software was used for kinetics analysis. The percent inhibition was calculated using the formula given below:
 | (1) |
EZ-FIT (Perrella Scientific, Inc., USA) was used to calculate IC50 of all compounds. To overcome the expected errors, all experiments were performed in triplicate, and variations in the results are reported in Standard Error of Mean values (SEM).
 | (2) |
4.4. Docking analysis
The docking of 5h was performed in 3A4A by Molecular Operating Environment (MOE v2022.02). The enzyme file was prepared by MOE's protonate-3D that added hydrogen atoms in the enzyme and calculated partial charges with Amber12:EHT force field. The chemical structure of 5h was drawn on MOE, AM1-BCC charges were added on it and the structure was energy minimized with MMFF94x force field (RMS gradient = 0.5 kcal mol−1 Å−1). The Triangle-Matcher docking algorithm of MOE and London dG scoring function was scrutinized initially by re-docking of co-crystallized ligand in the active of 3A4A, which showed RMSD of 0.17 Å and docking score of −7.30 kcal mol−1. Therefore, the same parameters were used in the docking of 5h and thirty conformations of 5h were saved. Later, each conformation was analyzed by conformational sampling and the best pose was selected based on the docking score and optimal binding interactions.Data availability
The data supporting this article have been included as part of the ESI.†Conflicts of interest
The authors have declared no conflict of interest.Acknowledgements
The research was funded by Taif University, Saudi Arabia through project number TU-DSPP-2024-19. Z. S. is thankful to the Alexander von Humboldt Foundation for the award of Return Fellowship.References
- A. M. Dirir, M. Daou, A. F. Yousef and L. F. Yousef, Phytochem. Rev., 2022, 21, 1049–1079 CrossRef CAS.
- A. Alam, M. Ali, N. U. Rehman, S. Ullah, S. A. Halim, A. Latif, Zainab, A. Khan, O. Ullah and S. Ahmad, Pharmaceuticals, 2022, 15, 672 CrossRef CAS PubMed.
- S. Ullah, M. Waqas, S. A. Halim, I. Khan, A. Khalid, A. N. Abdalla, H. A. Makeen, A. Ibrar, A. Khan and A. Al-Harrasi, Int. J. Biol. Macromol., 2023, 250, 126227 CrossRef CAS.
- N. Kausar, S. Ullah, M. A. Khan, H. Zafar, M. I. Choudhary and S. Yousuf, Bioorg. Chem., 2021, 106, 104499 CrossRef CAS PubMed.
- S. Akhter, S. Ullah, S. Yousuf, H. Siddiqui and M. I. Choudhary, Bioorg. Chem., 2021, 107, 104531 CrossRef CAS PubMed.
- R. Ronchetti, G. Moroni, A. Carotti, A. Gioiello and E. Camaioni, RSC Med. Chem., 2021, 12, 1046–1064 RSC.
- A. K. Ghosh and M. Brindisi, J. Med. Chem., 2019, 63, 2751–2788 CrossRef PubMed.
- V. B. Bregović and N. Basarić, Coord. Chem. Rev., 2015, 295, 80–124 CrossRef.
- A. Catalano, Curr. Med. Chem., 2022, 29, 4302–4306 CrossRef CAS.
- L. N. Solano, G. L. Nelson, C. T. Ronayne, S. Jonnalagadda, S. K. Jonnalagadda, K. Kottke, R. Chitren, J. L. Johnson, M. K. Pandey and S. C. Jonnalagadda, Sci. Rep., 2020, 10, 17969 CrossRef CAS.
- Y. Zhang, M. Anderson, J. L. Weisman, M. Lu, C. J. Choy, V. A. Boyd, J. Price, M. Sigal, J. Clark and M. Connelly, ACS Med. Chem. Lett., 2010, 1, 460–465 CrossRef CAS PubMed.
- A. R. Pasha, A. Khan, S. Ullah, S. A. Halim, J. Hussain, M. Khalid, M. M. Naseer, A. F. El-Kott, S. Negm and A. Al-Harrasi, Sci. Rep., 2023, 13, 1877 CrossRef CAS.
- L. M. Aroua, A. H. Alosaimi, F. M. Alminderej, S. Messaoudi, H. A. Mohammed, S. A. Almahmoud, S. Chigurupati, A. E. Albadri and N. H. Mekni, Pharmaceutics, 2023, 15, 457 CrossRef CAS PubMed.
- H. Gezegen, M. B. Gürdere, A. Dinçer, O. Özbek, Ü. M. Koçyiğit, P. Taslimi, B. Tüzün, Y. Budak and M. Ceylan, Arch. Pharm., 2021, 354, 2000334 CrossRef CAS.
- J. Y. Kim, J. W. Lee, Y. S. Kim, Y. Lee, Y. B. Ryu, S. Kim, H. W. Ryu, M. J. Curtis-Long, K. W. Lee and W. S. Lee, ChemBioChem, 2010, 11, 2125–2131 CrossRef CAS.
- S. Shamim, K. M. Khan, N. Ullah, M. Mahdavi, M. A. Faramarzi, B. Larijani, U. Salar, R. Rafique, M. Taha and S. Perveen, J. Mol. Struct., 2021, 1242, 130826 CrossRef.
- H. Ullah, F. Rahim, E. Ullah, S. Hayat, H. Zada, F. Khan, A. Wadood, F. Nawaz, Z. U. Rehman and S. A. A. Shah, Chem. Data Collect., 2023, 43, 100987 CrossRef CAS.
- S. S. Mukhtar, A. S. Hassan, N. M. Morsy, T. S. Hafez, H. M. Hassaneen and F. M. Saleh, Egypt. J. Chem., 2021, 64, 6541–6554 Search PubMed.
- A. R. Pasha, A. Khan, S. Ullah, S. A. Halim, J. Hussain, M. M. Naseer, A. F. El-Kott, S. Negm, A. Al-Harrasi and Z. Shafiq, Sci. Rep., 2023, 13(1), 1877 CrossRef CAS PubMed.
- K. V. Sashidhara, J. N. Rosaiah, G. Bhatia and J. Saxena, Eur. J. Med. Chem., 2008, 43, 2592–2596 CrossRef CAS PubMed.
- Y. Toubi, F. Abrigach, S. Radi, F. Souna, A. Hakkou, A. Alsayari, A. Bin Muhsinah and Y. N. Mabkhot, Molecules, 2019, 24, 3250 CrossRef.
- S. Ullah, A. Ullah, M. Waqas, S. A. Halim, A. R. Pasha, Z. Shafiq, S. N. Mali, R. D. Jawarkar, A. Khan, A. Khalid, A. N. Abdalla, H. Kashtoh and A. Al-Harrasi, Sci. Rep., 2024, 14(1), 12588 CrossRef CAS.
- H. R. Afzal, N. u. H. Khan, K. Sultana, A. Mobashar, A. Lareb, A. Khan, A. Gull, H. Afzaal, M. T. Khan and M. Rizwan, ACS Omega, 2021, 6, 4470–4479 CrossRef CAS.
- A. Aispuro-Pérez, J. López-Ávalos, F. García-Páez, J. Montes-Avila, L. A. Picos-Corrales, A. Ochoa-Terán, P. Bastidas, S. Montaño, L. Calderón-Zamora and U. Osuna-Martínez, Bioorg. Chem., 2020, 94, 103491 CrossRef PubMed.
- C. R. Sahoo, S. K. Paidesetty, B. Dehury and R. N. Padhy, J. Biomol. Struct. Dyn., 2023, 1–11 Search PubMed.
- Y. Belay, A. Muller, D. T. Ndinteh, O. A. Kolawole, A. S. Adeyinka and T. Y. Fonkui, J. Mol. Struct., 2023, 1275, 134623 CrossRef CAS.
- M. Ishaq, P. Taslimi, Z. Shafiq, S. Khan, R. E. Salmas, M. M. Zangeneh, A. Saeed, A. Zangeneh, N. Sadeghian, A. Asari and H. Mohamad, Synthesis, bioactivity and binding energy calculations of novel 3-ethoxysalicylaldehyde based thiosemicarbazone derivatives, Bioorg. Chem., 2020, 100, 103924 CrossRef CAS.
- S. D. Gupta, B. Revathi, G. I. Mazaira, M. D. Galigniana, C. Subrahmanyam, N. Gowrishankar and N. Raghavendra, Bioorg. Chem., 2015, 59, 97–105 CrossRef PubMed.
- S. Y. Abbas, W. M. Basyouni, K. A. El-Bayouki, R. M. Dawood, T. H. Abdelhafez and M. K. Elawady, Synth. Commun., 2019, 49, 2411–2416 CrossRef CAS.
- A. R. Pasha, M. Khan, A. Khan, J. Hussain, M. Al-Rashida, T. Islam, Z. Batool, H. Kashtoh, M. H. Abdellattif, A. Al-Harrasi and Z. Shafiq, Synthesis, in vitro, and in silico study of novel pyridine based 1, 3-diphenylurea derivatives as tyrosinase inhibitors, Bioorg. Chem., 2024, 107724 CrossRef.
- M. A. Salem, S. Y. Abbas, M. H. Helal and A. Y. Alzahrani, Synth. Commun., 2021, 51, 2984–2990 CrossRef CAS.
- C. Yorur-Goreci, N. Altas-Kiymaz, A. Peksel, B. Bilgin-Eran and M. Sonmez, Sci. Pharm., 2014, 82, 735–748 CrossRef CAS.
- S. Hashmi, S. Khan, Z. Shafiq, P. Taslimi, M. Ishaq, N. Sadeghian, H. S. Karaman, N. Akhtar, M. Islam and A. Asari, Bioorg. Chem., 2021, 107, 104554 CrossRef CAS PubMed.
- E. Hejchman, H. Kruszewska, D. Maciejewska, B. Sowirka-Taciak, M. Tomczyk, A. Sztokfisz-Ignasiak, J. Jankowski and I. Młynarczuk-Biały, Monatsh. Chem., 2019, 150, 255–266 CrossRef CAS.
- M. D. Altıntop, Ö. Atlı, S. Ilgın, R. Demirel, A. Özdemir and Z. A. Kaplancıklı, Eur. J. Med. Chem., 2016, 108, 406–414 CrossRef.
- D. B. Rajamma, G. C. R. Iyer, I. H. Madar and P. Karunakar, Indian J. Pharm. Educ. Res., 2019, 53, 276–284 CrossRef CAS.
- E. Bokor, S. Kun, D. Goyard, M. Toth, J.-P. Praly, S. Vidal and L. Somsak, Chem. Rev., 2017, 117, 1687–1764 CrossRef CAS PubMed.
- S. Makar, T. Saha and S. K. Singh, Eur. J. Med. Chem., 2019, 161, 252–276 CrossRef CAS PubMed.
Footnote |
| † Electronic supplementary information (ESI) available. See DOI: https://doi.org/10.1039/d4ra05767h |
| This journal is © The Royal Society of Chemistry 2024 |