Open Access Article
Open Access ArticleA logic-activated nanoswitch for killing cancer cells according to assessment of drug-resistance†
Lihua Zhang ab,
Xiangxi Dengb,
Zhihe Qingb,
Yanli Lei
ab,
Xiangxi Dengb,
Zhihe Qingb,
Yanli Lei b,
Feng Feng
b,
Feng Feng *a,
Ronghua Yang
*a,
Ronghua Yang c and
Zhen Zou
c and
Zhen Zou *c
*c
aCollege of Chemistry and Chemical Engineering, Shanxi Datong University, Datong, 037009, China. E-mail: feng-feng64@263.net
bHunan Provincial Key Laboratory of Cytochemistry, School of Chemistry and Biological Engineering, Changsha University of Science and Technology, Changsha, 410004, China
cKey Laboratory of Chemical Biology & Traditional Chinese Medicine Research Ministry of Education, Institute of Interdisciplinary Studies, College of Chemistry and Chemical Engineering, Hunan Normal University, Changsha, 410081, China. E-mail: kobe474779970@126.com
First published on 30th September 2024
Abstract
A logic-activated nanoswitch that could diagnose the differences between drug-resistant and non-drug-resistant cancer cells and control the release of drugs was developed for enhanced chemo-gene therapy using a standalone system. Compared to traditional treatments, the nanoswitch displayed improved anti-tumor efficiency in vitro.
Heterogeneity is one of the most important characteristics of tumors, and it refers to intrinsic differences among cancer cells in tumor patients, such as cellular morphology, metabolism, gene expression, and metastatic potential.1 Due to this heterogeneity, the bulk tumor may contain genetically distinct tumor-cell subpopulations with various levels of susceptibility to therapy, posing considerable obstacles to the design of treatment strategies.2 Remarkably, heterogeneity provides the fuel for drug resistance in chemotherapy.3 Traditional chemotherapy strategies implement a one-size-fits-all policy to kill drug-resistant cancer cells or non-drug-resistant cancer cells.4 Although those approaches may achieve initial success, subpopulations of cells resistant to the treatment can lead to fast iteration of antineoplastic drugs, causing the recurrence of tumor growth and the dilemma of no drugs available. Therefore, how to distinguish heterogeneous tumor cells and deliver drugs on demand is an urgent issue to be solved.
Biocomputation can sense biochemical signals in a biological system and execute a user program. Boolean logic-based algorithm to generate a functional output.5 Due to the capability of intelligent judgment, biocomputation holds excellent potential for precise definition and innovative treatment of diseases. Designed with disease biomarkers as inputs, stimuli-responsive biomaterials can sense and respond to pathological markers and site-specifically-release therapeutics.6 Up to now, small molecules (e.g., organic7 or organometallic8), biological macromolecules (e.g., DNA,9 peptide,10 and protein11), and various multi-stimuli-responsive materials (e.g., polymer-based particles,12 films and hydrogels13) have been used as scaffolds to construct programmed devices with logic gates, which provide powerful tools for diagnostics, drug delivery and tissue engineering. These findings have provided a strong push for developing more intricate controlled-release nanodevices. For example, Chen et al. fabricated a dual-logic-based hydrogel to match the immune-osteo cascade for diabetic bone regeneration.14 Liu et al. presented a logic-based hydrogel that utilizes diagnostic logic to recognize the pathological cue MMP13 and then control drug release for cartilage regeneration.15 Although the existing works describe impressive designs, there is rarely a report to use programmable logic devices for distinguishing heterogeneous tumor cells.
Herein, we proposed a logic-activated nanoswitch for killing cancer cells according to the judgment of drug-resistant or non-drug-resistant cancer cells. As displayed in Fig. 1A, both a triple-helix nucleic acid switch (TNS) and the anticancer drug doxorubicin (DOX) are adsorbed onto a GSH-responsive MnO2 nanocarrier to form MnO2@TNS/DOX nanoswitch. Of note, a TNS probe composed of an Anti-Pgp-sequence-containing external strand (Anti-Pgp-E) (Table S1 and Fig. S1†) in the loop and a stem-forming oligonucleotide let-7a mimics are rationally designed to signal the molecular recognition event of drug-resistant cancer cells. 16 The Anti-Pgp-E sequence is bound to P-glycoprotein messenger RNA, and two arm segments flank the let-7a mimic via Watson–Crick and Hoogsteen base pairing. However, once endocytosed by non-drug-resistant MCF-7 breast cancer cells, the nanoswitch MnO2@TNS/DOX is decomposed by reducing MnO2 nanosheets by overexpressed GSH which plays essential roles in different biological functions,17–19 releasing DOX and TNS that maintains the triplex conformation (Fig. 1B).20 Of note is that TNS is kept in a “silent” state. The triplex formation closes the crucial domain for gene regulation of the let-7a mimic without P-gp mRNA, limiting its therapeutic activity. Only DOX executes the operation of cell killing. Compared with drug-resistant MCF-7/ADR breast cancer cells, the overexpressed GSH is depleted by the nanoswitch MnO2@TNS/DOX to form Mn2+, releasing DOX and TNS that keep the triplex configuration. Attractively, TNS is further activated by the overexpressed P-gp mRNA besides the DOX feature. The TNS undergoes a conformational alteration from a triplex to linear, resulting in the intelligent release of the therapeutic let-7a mimics. These activated sequences can concurrently regulate a wide range of genes related to chemoresistance, particularly those responsible for prosurvival pathways, DNA repair mechanisms, and drug efflux pumps, thereby sensitizing tumor cells to subsequent treatment with traditional chemotherapies and promoting apoptosis.21 Our developed logic-activated nanoswitch is simple in design and sophisticated to control, providing an attractive tool to enhance multiple therapeutics in heterogeneous tumor cells using a single nanodevice.
 | ||
| Fig. 1 Schematic illustration of (A) the fabrication and drug loading process and (B) the intracellular release of logic-activated MnO2@TNS/DOX according to the assessment of drug resistance. | ||
To prepare MnO2@TNS/DOX nanoswitch, we first synthesized uniform, monodispersed, and sheet-like (≈140 nm) (Fig. S2 and S3A†) MnO2 nanosheets with a ζ-potential of about −28.2 mV and a strong UV-vis absorption spectra at 360 nm (Fig. S3B and C†). Then, the assembly and disassembly of TNS in response to a triggering signal were monitored employing a marker fluorescence signal (Fig. S4A†). For this purpose, the Cy5/Cy3 pair was marked to both edges of Anti-Pgp-E. As observed, Cy3 (donor) and Cy5 (acceptor) were close to each other due to the formation of triple-helix stem-loop conformation, thus resulting in the appearance of a fluorescence resonance energy transfer (FRET) signal. The above results were further verified by circular dichroism (CD) spectra and native-PAGE, demonstrating the assembly of TNS and the importance of the anti-Pgp sequence in stimulus recognition (Fig. S4B and C†).
Then the MnO2@TNS/DOX was prepared by the programmed assembly of MnO2 nanosheets, TNS, and DOX. TEM imaging showed that MnO2@TNS/DOXs were nearly shuttle-shaped nanoparticles (Fig. 2A and B), and the transition from MnO2 nanosheets to MnO2@TNS/DOXs was also characterized by AFM (Fig. 2C and D). The UV-vis spectra of MnO2@TNS/DOX showed prominent adsorption peaks at 360 nm and 488 nm, which was assigned to the characteristic absorption of MnO2 and DOX, respectively (Fig. 2E). Furthermore, the composition of MnO2@TNS/DOX was analyzed by EDX (Fig. S5†), where Mn, O, and P were co-existed, indicating that the MnO2 nanosheet was wrapped up TNS. In addition, in contrast to the MnO2 nanosheet with the zeta potential of −28.23 ± 1.15 mV, the value of MnO2@TNS/DOX was changed into −3.42 ± 0.08 mV, further indicating the successful coating process (Fig. 2F). The TNS and DOX loading on each MnO2 was 0.2 and 2.6 (Fig. S6 and S7†). The high loading was attributed to the large surface area. Attractively, when MnO2@TNS/DOX was dispersed in the 10% FBS serum within 2.3 h, negligible fluorescence was changed, indicating excellent stability under physiological conditions (Fig. S8†). Moreover, the hemolysis assay was applied to test the biocompatibility of MnO2@TNS/DOX; the photographs of RBCs after incubation with MnO2@TNS/DOX are shown in Fig. S9,† which revealed its low hemolytic activity. These results suggested that the multidrug-loaded MnO2@TNS/DOX was successfully assembled with favorable stability and held promise for in vivo application.
 | ||
| Fig. 2 TEM image of (A) MnO2 and (B) MnO2@TNS/DOX. AFM image of (C) MnO2 and (D) MnO2@TNS/DOX. (E) UV-vis absorption spectrum and (F) zeta potential of MnO2 nanosheets and MnO2@TNS/DOX. | ||
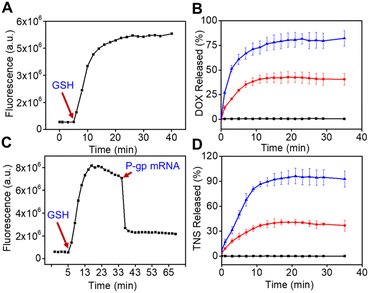
First, the logic-activated behaviors of nanoswitch were investigated using the fluorescence spectrum. The activated effect of GSH on the MnO2@TNS/DOX was then explored. As seen in Fig. 3A and B, the apparent time-dependent fluorescence recovery was observed in 1 mM GSH, when GSH was added into the nanoswitch at 5 min. In contrast, a negligible fluorescence recovery appeared without GSH (Fig. S10†). This was due to the transformation from MnO2@TNS/DOX to Mn2+ by GSH depletion, which was verified by the decrease in absorption and fading of the MnO2 solution (Fig. S11†). Given that the GSH was used to trigger the reduction of MnO2 solution, which was relevant to the concentration of GSH in tumor cells.22 As anticipated, with the expansion of time, the fluorescence gradually recovered with increasing amounts of GSH (Fig. 3C and D). In contrast, the MnO2@TNS/DOX without GSH exposure remained relatively stable and hardly recovered fluorescence during the same period. Additionally, a visible fluorescence of Cy3 at 560 nm recovery and Cy5 at 670 nm reduction was obtained only at a P-gp mRNA concentration of 3 nM, suggesting a high response by the TNS (Fig. S12†). The above results validated that the structure of MnO2@TNS/DOX was decomposed by adding GSH, indicating that the MnO2@TNS/DOX was activated. In addition, significant fluorescence enhancements at 600 and 670 nm were presented with the increase in GSH concentration, suggesting the release of DOX and TNS (Fig. S13†).
Furthermore, the mRNA-dependent activation of TNS was further studied. As displayed in Fig. 3C, upon adding P-gp mRNA, the fluorescence was remarkably decreased, indicating the activation of the TNS and conformational changes of TNS from a folded triplex extended to a linear structure. We thus reasoned that TNS could be stably loaded onto MnO2@TNS/DOX in circulation until MnO2@TNS/DOX would be endocytosed by cells. Collectively, the rapidly activated behavior resulting from the dual-responsive MnO2@TNS/DOX responding to GSH and P-gp mRNA made the MnO2 nanosheets ideal nanocarriers for concurrent delivery of TNS and DOX, which could realize a GSH-responsive to release both DOX and TNS as well as P-gp mRNA-stimulant to release both Anti-Pgp-E and let-7a mimic.
To evaluate the efficacy of nanoswitch cellular uptake and the ability to the logic activation, MCF-7 cells (with low expression of P-gp mRNA) and DOX-treated MCF-7/ADR cells (with high expression of P-gp mRNA) were adopted.23 As such, a noncytotoxic MnO2 nanosheet was examined to evaluate the biocompatibility (Fig. S14†). Then, after incubating the MnO2@TNS/DOX with MCF-7 cells at different times, confocal fluorescent imaging validated that the MnO2@TNS/DOX could be progressively activated in the cytoplasm of MCF-7 cells, indicating that this nanoswitch was activated only by GSH and released DOX (green) and TNS (red) (Fig. S15†).
Moreover, the activation of P-gp mRNA was further confirmed. As seen in Fig. 4, after treating MCF-7/ADR cells with MnO2@TNS/DOX, blue and green fluorescence were observed but no red fluorescence, implying that the release of Anti-Pgp-E and let-7a mimic from the TNS was only reliant on P-gp mRNA. In contrast to the MnO2@TNS/DOX, we found that ConL-MnO2@TNS/DOX (a ConL-TNS with loop region randomly altered to be unable to recognize P-gp mRNA, as a control nanoswitch) (Table S1 and Fig. S1†), which was deprived of the ability to identify P-gp mRNA, failed to be activated in MCF-7/ADR cells. In addition, similar results have been observed in MCF-7 cells pretreated with MnO2@TNS/DOX, indicating that the release of DOX and TNS from the nanoswitch was only dependent on GSH. The above results were attributed to the structural transformation of TNS, confirming that TNS can be activated in drug-resistant cells in response to P-gp mRNA. The hybridization of P-gp mRNA in MCF-7/ADR cells changed the stem-loop conformation of TNS, and then, the two fluorophores of TNS were separated with low FRET efficiency.
The above perspective results inspired us to further study the regulatory effect of let-7a mimic after recovery from the P-gp mRNA-induced allosteric responses. To this end, western blot assay was tested to evaluate the protein expression levels of P-gp (target mRNA of P-gp mRNA) and HMGA2 (target mRNA of let-7a) in MCF-7/ADR cells.23,24 As expected, for MCF-7 cells treated with MnO2@TNS/DOX, little down-regulating effect of P-gp and HMGA2 protein expression was exhibited(Fig. S16†). Similarly, owing to the lack of response to P-gp mRNA, ConL-MnO2@TNS/DOX failed to recognize the stimulus signals in MCF-7/ADR cells. They maintained that let-7a mimic impeded, thereby exhibiting a weak inhibitory effect on the expression of HMGA2. In contrast, MnO2@TNS/DOX-treated MCF-7/ADR cells showed 90.0% and 61.0% reduction of P-gp and HMGA2 expression, indicating that the HMGA2 protein knockdown was logic-activated. The above results confirmed that MnO2@TNS/DOX could effectively knock down the target gene on the target cells.
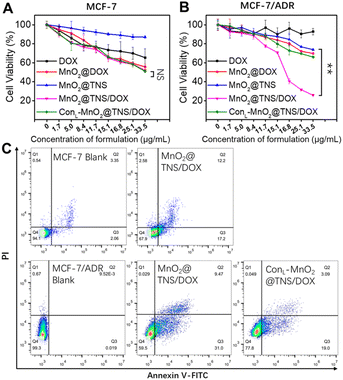
Subsequently, the cell-killing activity of nanoswitch was explored. MTT assay showed that MnO2@TNS/DOX displayed dose-dependent cytotoxicity against MCF-7 cells. Actually, at 48 h post-treatment, the utilization of the MnO2@TNS/DOX resulted in a viability population of 52.0% at the highest concentration of 33.5 μg per mL MnO2, but no significant difference among other nanoswitches, indicative of their cleavage only by GSH (Fig. 5A). The above results indicated that the cytotoxic effects on MCF-7 cells were attributed to the function of DOX, validating our previous hypothesis. Then, high cytotoxicity was obtained after treating MCF-10A cells (normal breast cells in humans) with the MnO2@TNS/DOX. These results indicated that activation of the MnO2@TNS/DOX could not only generate in cancer cells but also result in the release of DOX from MnO2@TNS/DOX in normal cells due to the GSH microenvironment (Fig. S17†).
In MCF-7/ADR cells, MTT assay revealed that MnO2@DOX, MnO2@TNS, and ConL-MnO2@TNS/DOX yielded significantly low cell viability(Fig. 5B). In contrast, an improved effect was achieved upon employing the MnO2@TNS/DOX, indicating its importance in suppressing the cytotoxicity of MCF-7/ADR cells. We thus reasoned that MnO2@TNS/DOX was selectively activated within tumor cells, allowing for programmable drug release and reversing the drug resistance. The above results confirmed that the nanoswitch could precisely differentiate drug-resistant and non-drug-resistant cancer cells and further achieve the corresponding cell-killing activity.
Finally, the killing mechanism of nanoswitch was also studied using flow cytometry. As shown in Fig. 5C, for the MCF-7 cells, an apoptotic population of 29.4% was observed when MnO2@TNS/DOX exhibited cytotoxic effects. For comparison, an apoptotic rate of 40.5% was observed after treating MCF-7/ADR cells with the MnO2@TNS/DOX. In contrast, the value was reduced when the MnO2@TNS/DOX was replaced with a control therapeutic element, proving the importance of targeting. These confirmed toxicity results from the GSH and P-gp mRNA logic-activated release of active DOX, Anti-Pgp-E, and let-7a mimic through induced cell apoptosis, synchronized with blue and green fluorescence imaging.
In summary, we have successfully developed a logic-activated nanoswitch that distinguishes drug-resistant and non-drug-resistant cancer cells and executes corresponding cell-killing activity. Designed with cancer biomarkers as inputs, the nanoswitch releases Anti-Pgp-E, let-7a mimic and DOX in a programmable manner, achieving optimal therapeutic effects. The ingeniously designed system integrates multiple responses and logic gate operations into a single nanodevice, illustrating its potential in treating complex diseases.
Data availability
The authors confirm that the data supporting the findings of this study are available within the article [and/or its ESI†].Author contributions
Lihua Zhang, Zhen Zou, Zhihe Qing, and Ronghua Yang conceived the idea of the study; Lihua Zhang and Zhen Zou performed the research, analyzed data, and wrote the paper. The remaining authors contributed to refining the ideas, conducting additional analyses, and finalizing this paper.Conflicts of interest
There are no conflicts to declare.Acknowledgements
This work was supported in part by the National Natural Science Foundation of China (21705010), the Hunan Provincial Natural Science Foundation of China (2022JJ20038), the Fundamental Research Program of Datong City (2023058), and the Natural Science Foundation of Changsha City (kq2202189).Notes and references
- C. E. Meacham and S. J. Morrison, Nature, 2013, 501, 328–337 CrossRef CAS PubMed.
- I. Dagogo-Jack and A. T. Shaw, Nat. Rev. Clin. Oncol., 2018, 15, 81–94 CrossRef CAS PubMed.
- H. E. C. Bhang, D. A. Ruddy, V. K. Radhakrishna, J. X. Caushi, R. Zhao, M. M. Hims, A. P. Singh, I. Kao, D. Rakiec, P. Shaw, M. Balak, A. Raza, E. Ackley, N. Keen, M. R. Schlabach, M. Palmer, R. J. Leary, D. Y. Chiang, W. R. Sellers, F. Michor, V. G. Cooke, J. M. Korn and F. Stegmeier, Nat. Med., 2015, 21, 440–448 CrossRef CAS PubMed.
- N. Vasan, J. Baselga and D. M. Hyman, Nature, 2019, 575, 299–309 CrossRef CAS PubMed.
- J. Li, A. A. Green, H. Yan and C. Fan, Nat. Chem., 2017, 9, 1056–1067 CrossRef CAS PubMed.
- P. Zhang, D. Gao, K. An, Q. Shen, C. Wang, Y. Zhang, X. Pan, X. Chen, Y. Lyv, C. Cui, T. Liang, X. Duan, J. Liu, T. Yang, X. Hu, J. J. Zhu, F. Xu and W. Tan, Nat. Chem., 2020, 12, 381–390 CrossRef CAS PubMed.
- S. Steiner, J. Wolf, S. Glatzel, A. Andreou, J. M. Granda, G. Keenan, T. Hinkley, G. Aragon-Camarasa, P. J. Kitson, D. Angelone and L. Cronin, Science, 2019, 363, eaav2211 CrossRef CAS PubMed.
- Y. Shen, T. Pan, L. Wang, Z. Ren, W. Zhang and F. Huo, Adv. Mater., 2021, 33, 2007442 CrossRef CAS PubMed.
- M. You, G. Zhu, T. Chen, M. J. Donovan and W. Tan, J. Am. Chem. Soc., 2015, 137, 667–674 CrossRef CAS PubMed.
- Y. Li, S. Sun, L. Fan, S. Hu, Y. Huang, K. Zhang, Z. Nie and S. Yao, Angew. Chem., Int. Ed., 2017, 56, 14888–14892 CrossRef CAS PubMed.
- X. J. Gao, L. S. Chong, M. S. Kim and M. B. Elowitz, Science, 2018, 361, 1252–1258 CrossRef CAS PubMed.
- N. H. Dashti, R. S. Abidin and F. Sainsbury, ACS Nano, 2018, 12, 4615–4623 CrossRef CAS PubMed.
- J. Song, M. H. Rizvi, B. B. Lynch, J. Ilavsky, D. Mankus, J. B. Tracy and G. H. McKinley, ACS Nano, 2020, 14, 17018–17027 CrossRef CAS PubMed.
- D. Li, K. Chen, H. Tang, S. Hu, L. Xin, X. Jing, Q. He, S. Wang, J. Song, L. Mei, R. D. Cannon, P. Ji, H. Wang and T. Chen, Adv. Mater., 2022, 34, 2108430 CrossRef CAS PubMed.
- T. Y. Zhao, H. Y. Deng, J. W. Li, S. L. He, X. Li, H. Li, Z. Yang, H. T. Deng, P. Q. Li, X. Sui, S. P. Jiang, Q. Y. Guo and S. Y. Liu, Adv. Funct. Mater., 2023, 33, 2213019 CrossRef CAS.
- X. Chen, Y. Zhou, X. Peng and J. Yoon, Chem. Soc. Rev., 2010, 39, 2120–2135 RSC.
- Q. Y. Cai, J. Li, J. Ge, L. Zhang, Y. L. Hu, Z. H. Li and L. B. Qu, Biosens. Bioelectron., 2015, 72, 31–36 CrossRef CAS.
- D. Fan, C. Shang, W. Gu, E. Wang and S. Dong, ACS Appl. Mater. Interfaces, 2017, 9, 25870–25877 CrossRef CAS.
- L. H. Zhang, Z. Zou, Y. L. Lei, Z. H. Qing, Y. Zeng, H. Y. Sun, F. Feng and R. H. Yang, Chem. Commun., 2021, 57(91), 12131–12134 RSC.
- J. Chen, H. Meng, Y. Tian, R. Yang, D. Du, Z. Li, L. Qu and Y. Lin, Nanoscale Horiz., 2019, 4, 321–338 RSC.
- F. H. Sarkar, Y. Li, Z. Wang, D. Kong and S. Ali, Drug Resistance Updates, 2010, 13, 57–66 CrossRef CAS PubMed.
- W. Zhu, Z. Dong, T. Fu, J. Liu, Q. Chen, Y. Li, R. Zhu, L. Xu and Z. Liu, Adv. Funct. Mater., 2016, 26, 5490–5498 CrossRef CAS.
- L. Zhu, Y. Guo, Q. Qian, D. Yan, Y. Li, X. Zhu and C. Zhang, Angew. Chem., Int. Ed., 2020, 59, 17944–17950 CrossRef CAS PubMed.
- C. Zhu, Z. Zeng, H. Li, F. Li, C. Fan and H. Zhang, J. Am. Chem. Soc., 2013, 135, 5998–6001 CrossRef CAS PubMed.
Footnote |
| † Electronic supplementary information (ESI) available. See DOI: https://doi.org/10.1039/d4ra04651j |
| This journal is © The Royal Society of Chemistry 2024 |