Open Access Article
Open Access ArticleExposure to polyethylene microplastics exacerbate inflammatory bowel disease tightly associated with intestinal gut microflora
Souvik Ghosal a,
Sagar Bag
a,
Sagar Bag b,
S. R. Rao
b,
S. R. Rao a and
Sudipta Bhowmik
a and
Sudipta Bhowmik *ab
*ab
aMahatma Gandhi Medical Advanced Research Institute (MGMARI), Sri Balaji Vidyapeeth (Deemed to be University), Pondy–Cuddalore Main Road, Pillaiyarkuppam, Pondicherry – 607402, India
bDepartment of Biophysics, Molecular Biology and Bioinformatics, University of Calcutta, 92, A. P. C. Road, Kolkata – 700009, India. E-mail: sbbmbg@caluniv.ac.in
First published on 13th August 2024
Abstract
Polyethylene microplastics (PE MPs) have sparked widespread concern about their possible health implications because of their abundance, pervasiveness in the environment and in our daily life. Multiple investigations have shown that a high dosage of PE MPs may adversely impact gastrointestinal health. In tandem with the rising prevalence of Inflammatory bowel disease (IBD) in recent decades, global plastic manufacturing has risen to more than 300 million tons per year, resulting in a build-up of plastic by-products such as PE MPs in our surroundings. We have explored current advancements in the effect PE MPs on IBD in this review. Furthermore, we compared and summarized the detrimental roles of PE MPs in gut microbiota of different organisms viz., earthworms, super worm's larvae, yellow mealworms, brine shrimp, spring tails, tilapia, gilt-head bream, crucian carp, zebrafish, juvenile yellow perch, European sea bass, c57BL/6 mice and human. According to this review, PE MPs played a significant role in decreasing the diversity of gut microbiota of above-mentioned species which leads to the development of IBD and causes severe intestinal inflammation. Finally, we pinpoint significant scientific gaps, such as the movement of such hazardous PE MPs and the accompanying microbial ecosystems and propose prospective research directions.
1. Introduction
Plastics, synthetic organic polymers, are a major non-natural product made by humans and contribute to Anthropocene stratigraphic markers. By 2050, landfills and the environment will contain 13.2 billion tons of plastic waste. Larger plastics cause physical entanglement and entrapment, while smaller microplastics, less than 5 mm in size, are referred to as microplastics.1–3 Microplastics pollution is a significant global environmental issue, causing significant consequences like climate shifts, ozone depletion, and ocean acidification.4–6 With annual plastic output increasing since the 1960s, poor waste management leads to environmental dumping and microplastics pollution.7–12 Plastic trash releases microplastics, smaller particles between 1 μm and 5 mm in size, which can enter human cells.1,13–16 These particles can cause chronic illness in humans due to their larger diameters, which can be harmful to human health.17,18 MPs, a type of microbead, are a common ingredient in skincare products and are rapidly absorbed by human cells, potentially affecting cellular functions. A marine scientist at the University of Plymouth in the UK called professor Richard Thompson first introduced the phrase “microplastics” in 2004. They are also released into the environment through municipal wastewater, possibly due to larger plastic waste fragmentation.19–22 MPs, found in various water sources, can cause inflammatory and oxidative effects in the colon and gut epithelial permeability, with ingesting being the most dangerous route.23,24 Microplastics contamination in human health is unresolved, but it's crucial to examine its impact on intestinal microbiota and gut inflammation, with common plastic polymers like PE MPs and polypropylene found in tap and freshwater.25 PE, a synthetic petroleum-based plastic, is widely used in disposable containers and agriculture as a plastic mulch since 1938.26 It has been demonstrated by earlier animal studies that microplastics cause intestinal and liver malfunction. For example, Kang et al. discover that fish suffer intestinal injury from microplastics through two distinct routes.27 MPs exhibiting a size of 50 nm is more susceptible to oxidative stress, whereas MPs sized 45 μm significantly disrupts the gut flora. According to Kim et al., fish's ability to digest food is inhibited by microplastics as a result of a microalgae-crustacean-small yellow croaker food chain.28 Additionally, intestinal barrier and metabolic function are compromised in microplastics exposed animals, according to Jin et al.29 In the simulated human gastrointestinal system, Tan et al. show that microplastics dramatically impair lipid digestion, with MPs exhibiting the strongest suppression.30 The size of MPs has no bearing on the reduction in lipid digestion. According to Lu et al., exposure to MPs induces local infections, lipid buildup, and disturbances in fish liver energy metabolism.31 Furthermore, Deng et al. find that mice's metabolites change dramatically following exposure to microplastics and organophosphorus flame retardants (OPFRs). Furthermore, it is evident that microplastics increase the toxicity of OPFRs, underscoring the dangers to one's health posed by exposure to microplastics and other pollutants.32 There are two types of microplastics–primary and secondary microplastics. Primary microplastics are tiny pieces of plastic that are used in cosmetics, face cleansers, and air-blasting media to remove paint and rust from boat hulls and machinery. Secondary microplastics are tiny pieces of plastic that are produced as bigger pieces of plastic waste are broken down by biological, photochemical, and physical wave action. Examples and properties of different microplastics are listed in Table 1.| Microplastics | Properties | Reference |
|---|---|---|
| Polystyrene | The liquid hydrocarbon polystyrene (PS) is produced commercially from petroleum and is derived from the monomer styrene. PS is typically a solid thermoplastic, although it may be liquefied at extremely high temperatures for molding or extrusion, then resolidified at room temperature. Styrene is an aromatic monomer, while PS is an aromatic polymer | 17 |
| Polyethylene terephthalate | The most often utilised thermoplastic polymer resin of the polyester family is polyethylene terephthalate, sometimes referred to as PET, PETE, or the antiquated PETP or PET-P. When mixed with glass fibre, it is also used in industrial operations like thermoforming and engineering resins | 18 |
| Polypropylene | The thermoplastic polymer polypropylene (PP), often called polypropene, is employed in a wide range of applications. Through chain-growth polymerization, it is created from the monomer propylene. Polypropylene, which is a member of the polyolefin family, is non-polar and somewhat crystalline | 19 |
| Poly vinyl chloride | The chemical formula for vinyl chloride is H2C![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) CHCl, and poly (vinyl chloride, or PVC), which is a synthetic resin manufactured from vinyl chloride is (H2C–CHCl)n, where n indicates the degree of polymerization. PVC is a part of a vast family of polymers collectively referred to as “vinyls” CHCl, and poly (vinyl chloride, or PVC), which is a synthetic resin manufactured from vinyl chloride is (H2C–CHCl)n, where n indicates the degree of polymerization. PVC is a part of a vast family of polymers collectively referred to as “vinyls” |
20 |
| Polyamides | Polyamides (PA) (–CO–NH–) contain repeating amide linkages. It is formed by copolymers with different units and condensing identical units. It shows high temperature and electrical resistance, because of its crystalline structure, and PA also shows excellent chemical resistance | 21 |
| Polytrimethylene terephthalate | Polytrimethylene terephthalate is a synthetic polyester that was first invented in 1941. Condensation polymerization or trans esterification are the processes used to make it. Terephthalic acid, also known as dimethyl terephthalate, and 1,3-propanediol are the two monomers needed to create this polymer | 22 |
| Polyphenylene sulphide | Polyphenylene sulphide (PPS) resin is a crystalline heat-resistant polymer with a straightforward chemical composition composed of sulphur and benzene | 23 |
| Low-density polyethylene | Low-density polyethylene (LDPE) is made up of several short branches somewhere between 4000 and 40![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 000 carbon atoms. High pressure (1000–3000 bar; 80–300 °C) and a polymerization with free radicals' technique are used to create it. The two primary methods for producing LDPE are stirred autoclaves and tubular routes 000 carbon atoms. High pressure (1000–3000 bar; 80–300 °C) and a polymerization with free radicals' technique are used to create it. The two primary methods for producing LDPE are stirred autoclaves and tubular routes |
24 |
| High-density polyethylene | Petroleum is used to create high density polyethylene (HDPE), a thermoplastic polymer with the standard chemical formula (C2H4)n. The repeating monomer unit of ethylene was used to build the poly-ethylene molecular chain. Since HDPE is referred to as a “linear” chain, it differs from other kinds of polyethylene in that its side chain branching frequency is lower | 25 |
| Polyoxymethylene | Polyformaldehyde, acetal and polyacetal are some other names for polyoxymethylene. It is an engineering thermoplastic used to make precise components with great dimensional stability, minimal friction, and high stiffness requirements | 26 |
| Polycarbonates | Carbonate groups are a component of a type of thermoplastic polymers known as polycarbonates (PC). It is used in engineering because it is a strong, long-lasting substance, and some grades are optically transparent | 27 |
| Polyurethane | A class of polymers known as “polyurethane” are composed of organic units joined together by carbamate (urethane) bonds. Contrary to other popular polymers like PS and PE, this chemical variety also produces polyurethanes with different chemical structures, along with varnishes and coatings, stiff and flexible foams, electrical potting compounds, adhesives and fibers like polyurethane laminate and spandex | 28 |
Plastic pollution has been proven to have severe environmental and organismal consequences. MPs inevitably find their way into the food chain; convergent investigations have shown them to be present in a variety of foods and drinking water.33,34 This has sparked worries regarding the possible health impacts of MPs after consumption because it has been determined that they are present in human faeces, colonic tissues, and blood. But determining the health danger that MPs pose to people is still a problem on a worldwide scale. Specifically, not much is known about how these particles get up within the human digestive system.35 During their passage, MPs come into contact with the gut barrier, which stops the host from translocating exogenous aggressors like infections and foreign particles.36,37 The gut barrier is made up of intestinal epithelium, mucus, and gut microbiota on its luminal side. The first line of the host's chemical, biological, and physical defense is the mucus layer, a viscoelastic gel that coats and shields the intestinal epithelium. Mucus, which is secreted by goblet cells, is mostly made up of complex glycoproteins known as mucins (or MUC2 in the human gut).38 The vast and diverse collection of microorganisms from nearly every kingdom of life that are carried within the human gut, particularly in the colon, is known as the gut microbiota. These microbes include bacteria, viruses, fungus, archaea, and protozoa.39 These gut-dwelling bacteria interact and cohabit to support a number of vital host physiological processes. For example, they participate in the metabolism of medicines, poisons, and xenobiotics as well as the control of host immunity. Furthermore, they facilitate the digestion of indigestible food (such as dietary fibres), leading to a notable generation of secondary metabolites, including gas, volatile organic compounds (VOCs), short chain fatty acids (SCFAs), and aryl hydrocarbon receptor (AhR) ligands, which are primarily derived from tryptophan metabolisms. There is a connection between gut microbial activity and the maintenance of an efficient intestinal barrier because SCFAs and AhR ligands, in particular, are involved in the preservation of the integrity of intercellular tight junctions in the intestinal epithelium. Deciphering the possible connections between MPs and intestinal mucus, epithelium, and the gut microbiota is therefore a crucial problem that has not received much attention up to this point.40,41
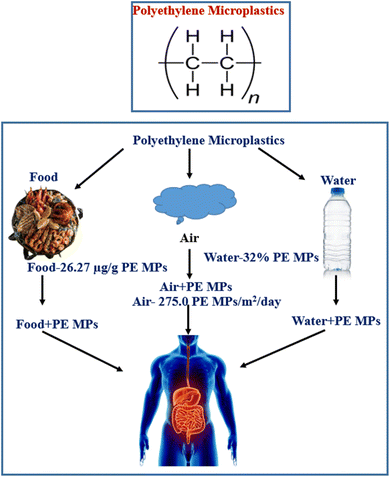
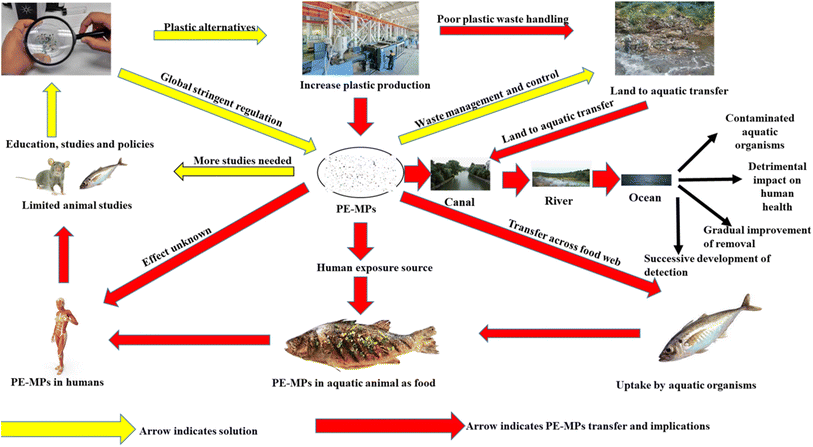
PE MPs, the most common form of microplastics in China, have been found to be a risk factor for bowel illnesses, despite several nations outlawing their sale.42–48 A study found that PE MPs, a synthetic plastic material, can cause changes in gut and serum inflammatory markers and intestinal flora, potentially impacting societal health. PE is abundant in edible shellfish tissues.47–49 Polyethylene is widely used in food packaging for preserving and it is very convenient to use it during transportation and storage, which contributes to the migration from food packaging to food. The gut health will almost certainly be compromised after ingesting PE MPs through contaminated food and drink (Fig. 1).50,51 The most critical aspect in protecting gut and even overall health is an intact gut barrier, and its role is strongly tied to the performance of gut microbiota, intestinal stem cells (ISCs), mucus layer, immune cells, and intestinal epithelium. Studies on earthworm, super worm's larvae, yellow mealworm, brine shrimp, spring tails, tilapia, gilt-head bream, crucian carp, zebrafish, juvenile yellow perch, European sea bass, c57BL/6 mice and human in vitro gut model exposed to PE MPs have revealed a disruption in gut flora. In vitro and in vivo studies have shown that PE MPs have a negative influence on the epithelial barrier and gut microbiota.52–56
This review explores the interactions of PE MPs with the intestinal tracts of several organisms, assesses the impacts of the same materials on healthy bodies, and the detailed molecular framework of occurrence of IBD after interacting with PE MPs. This is the uniqueness and advantage of this study. However, PE MPs and their impact on the onset and development of Inflammatory Bowel Disease (IBD) have not been yet explored. Thus, we are eager to find out more about the impact of PE MPs on beginning and expansion of IBD. Our study focuses on the influence of ubiquitous PE MPs on the progression of chronic disease IBD in a circumstance where people are constantly exposed to PE MPs. Finally, we report new information and future perspectives regarding the toxicity of PE MPs.
2. Compounds that are conjugated with microplastics
2.1 Bacterial community on microplastics
MPs offer a novel microbial habitat, as shown by the composition and functional characteristics of bacterial communities.57–59 Human pathogenic bacteria from the Pseudomonas genus are discovered on microplastics.60 Bacteroidetes, Firmicutes and Proteobacteria are the primary phyla that are colonized on MPs. MPs could be selectively enriched with antibiotic-resistant bacteria or pathogens. Microplastics in a river habitat were niche to the several bacterial species Pseudomonas mendocina, Pseudomonas monteilii and Pseudomonas syringae, Actinobacteria, Proteobacteria and Bacteroidetes, which are accounted for more than 75% of the bacteria on the MPs. Cyanobacteria, Chloroflexi, and Verrucomicrobia were also discovered on the microplastics. Rhodoferax, Flavobacterium, Pseudomonas, Janthinobacterium and Cyanobacteria were among the bacterial populations that made up more than 2% of the microplastics. Klebsiella pneumonia, Streptococcus mitis, Pseudomonas putida, Pseudomonas fluorescens, Pseudomonas savastanoi, Streptococcus mitis, Salmonella enterica, Pseudomonas stutzeri, Pseudomonas entomophila and Aeromonas hydrophila were the top 10 human bacterial pathogens species discovered in microplastics.612.2 Toxic compounds on microplastics
Microplastics collect and concentrate dangerous organic compounds, increasing their toxicity, and represent a major threat to human well-being if consumed or inhaled.62 Prior to being outlawed globally by the Stockholm Convention on Persistent Organic Pollutants in 2001 and in the United States by the Toxic Substances Control Act in 1979, PCBs (Polychlorinated Biphenyl), which are extremely carcinogenic chemical compounds, were used in consumer and industrial products, DDTs (dichlorodiphenyltrichloroethane) which is an odorless, colorless, crystalline, tasteless, chemical compound an organochloride. DDT primarily used as an insecticide over decades has caused serious irreversible damage to human health and environment. The insecticide HCHs (hexachlorocyclohexane) used as a dust, powder, liquid, or concentrate, and PAHs (polycyclic aromatic hydrocarbons) have been associated to serious asthma flare-ups, lung function and an increased risk of obstructive lung and cardiovascular disease. It also has a negative impact on a child's ability to think and behave. Microplastics are known to absorb PBT (polybutylene terephthalate), the persistent chemical in the bio-accumulative hazardous category remain in the environment for long periods of time. When these persistent chemicals and their residues are consumed, they bioaccumulate in the adipose tissues, bones, and brains of creatures.63Chemicals known as additives are added to plastic on purpose to give it properties like colour and transparency. They also improve the performance of plastic products by strengthening their resistance to ozone, temperature, light radiation, mould, bacteria, and humidity, as well as their mechanical, thermal, and electrical resistance. Among them are plasticisers, dyes, UV stabilisers, and so on.64 In order to decrease the forces of physical attraction between molecules and improve their mobility, workability, or distensibility, complex chemical products known as plasticisers are injected between the chains. These products have low vapour pressure, are insoluble in liquids, and are chemically stable. This increases the produced resin's flexibility and plasticity as well as the product's resistance to impact when in use.65 Using phenols and aromatic amines, stabilisers prevent thermal breakdown during processing and oxidation, which breaks the polymeric chains because plastics are particularly sensitive to the degrading effects of heat, light, and UV radiation. They are mostly made up of lead, barium, or organic or inorganic cadmium salts.66 Organic or inorganic materials in the form of fine powders are known as soluble or insoluble dyes, and they give polymers the desired colour. Soluble dyes preserve the plastic's transparency, while insoluble dyes, or pigments, cover it to make it opaque. While organic pigments comprise different chromophoric families such as azo pigments, phthalocyanine pigments, anthraquinone chromophores, and various other chromophores, many inorganic pigments involve heavy metals.67
2.3 Presence of antibiotic resistance gene (ARG) on microplastics
Microplastics increase the dissemination of antibiotic resistance genes among different types of bacteria in the environment.64 The risk of ARGs spreading through transfer was caused by microorganisms creating extensive biofilm formations on microplastics. Additionally, it has been noted that microplastics contain one hundred to five thousand times more antibiotic-resistant bacteria than the water around.65 As possible hosts for ARGs, bacterial pathogens like Flavobacterium and Chryseobacterium were also enriched on microplastics. As the bacterial population on microplastics grows denser, the rate of horizontal gene transfer of ARGs on MGEs (mobile genetic elements) between taxa increases. There is a larger frequency of MGE on microplastics, which is an indicative of a higher rate of horizontal gene transfer of ARGs. These investigations gave concrete proof of the variety of MGEs on microplastics that include ARGs, which may encourage the spread and development of ARGs in bacterial communities on microplastics. Therefore, increased transfer rates and the development of ARGs on microplastics are favored by denser bacterial populations, pollutants and aggregates. As antibiotic resistant gene gets entry into the human body, it reduces or eliminates the utility of antibiotics so it's harder for the immune system to combat illness. Global health is currently severely threatened by this antibiotic-resistant gene.662.4 The impact of the transformation of PE MPs on their effects
The breakdown and surface oxidation of PE MPs via oxidation, which results in fragmentation, mass loss, and changes to the surface characteristics. When oxidised PE MPs are released, the effects on human health and the environment can vary depending on the procedure, and the experimental settings that determined the behaviour and extent of PE MPs oxidation. Concerns regarding the toxicity of nanoplastics should also be addressed, since oxidation processes and oxidative dissolution have the potential to reduce MP sizes and produce nanoplastics. Yong et al. (2020) claim that endocytosis is one way that nanoplastics might penetrate mammalian cells and induce cellular stress. According to their findings, fish's innate immune systems may be stressed by nanoplastics, and mammals, including humans, may also experience this.68Reactive oxygen species (ROS) are known to exist in trace amounts in all plastics because of their polymerization and processing background. The interaction of light with the PE MPs can raise the concentration of free radicals during the weathering process. Zhu et al. (2020) observed improved cytotoxicity of photo-aged phenol-formaldehyde resin microplastics under simulated sunlight irradiation using in vitro human lung epithelial adenocarcinoma cells (A549).69 In comparison to non-photoaged MNPs, the photoaged MNPs displayed some altered physicochemical characteristics, as demonstrated by the increased levels of conjugated carbonyls, ROS, and environmentally persistent free radicals. These changes increased the oxidative potential of the MNPs and increased their cytotoxicity.70
3. An introduction to polyethylene microplastics (PE-MPs): composition & diseases
The most widely produced synthetic, petroleum-based plastic substance has the chemical formula (C2H4)n, and it is known as PE MPs.67 For the production of disposable items like bottles, bags, storage containers, and toys PE MPs are widely utilized.68 Despite the fact that the sale of rinse-off cosmetics containing MPs has been outlawed in many nations, PE MPs microbeads are nevertheless added to cosmetic goods.69 Since 1938, PE MPs has been regularly used in agriculture as a plastic mulch70 and currently the yearly production of polyethylene resin exceeded 100 million tons, or 34% of the worldwide plastics market. There is a significant buildup of PE in the environment as a result of its widespread use, unsuitable trash, inadequate recycling, and the fact that PE MPs is one of the least biodegradable polymers.71 PE MPs can absorb environmental pollutants and has a high persistence.72 The presence of plastic waste in ocean water has been discovered to enable the accumulation of various microbial species that are not typically present in the natural marine substrate, resulting in the formation of a distinct community known as the “plastisphere”. Plastisphere can result in the production of greenhouse gases, the transportation of harmful and invasive species, and a change in animal feeding habits. PE MPs, which makes up 40.5% of all polymers, is the most prevalent plastic polymer discovered in rivers.73 Freshwater and tap water include PE MPs the most often found plastic polymers, while atmospheric fallout contains the second highest concentration of PE MPs.74 Food has been found to contain PE MPs.73 For example, PE MPs ranked as the second most common polymer in edible quahogs or clam tissues that were sold for human consumption (Fig. 1). One of the reasons to transfer PE MPs from food packaging to food itself is caused by the extensive usage of PE MPs in food packaging for preservation facilitating easy handling during transportation and storage.75 Human exposure consequently follows pollution of the environment and food.76 It has been established that PE MPs are the most common type of polymer in human feces.77To understand the effects PE MPs, on people when they enter the body, more research is required. However, based on a growing body of evidence showing negative effects of exposure to PE MPs on the health of marine species, experts have expressed worry that PE MPs exposure can result in:
• Inflammation (associated with rheumatoid arthritis, inflammatory bowel illness, cancer, heart disease, and more).
• Genotoxicity (damage that results in mutations that can cause cancer).
• Chronic conditions (such as diabetes, cardiovascular disease, breast cancer and atherosclerosis).
• Autoimmune conditions.
Taken together, recent studies indicates that PE exposure presents a significant danger to human intestinal health. Furthermore, evidence suggests a favorable connection between fecal MP content and illness activity severity (Harvey–Bradshaw index and Mayo score). Polyethylene has been found in these patients' feces. As a result, the presence of PE in the stool may trigger the emergence of IBD inflammation.
4. An insight into the diversity and functionalities of gut microbiota
The term “gut microbiota” refers to the complete community of microorganisms that dwell in the gut, which includes archaea, viruses, fungi, and protozoans in addition to bacteria.78,79 In recent years, there has been a surge of interest in the gut microbiota in the scientific community, and it has been linked to a wide range of human diseases, including luminal diseases like IBD (inflammatory bowel disease) and IBS (irritable bowel syndrome), metabolic diseases (diabetes, obesity) and allergic disease, neurodevelopmental illnesses.80,81 The gut microbiota has long been thought to serve a significant functional role in ensuring normal individual and human health and different factors affecting gut microbiota (Fig. 2).4.1 Role of gut microbiota
The gut microbiota interacts symbiotically with the gut mucosa in a healthy individual and performs critical metabolic, immunological and protecting activities for the gut. The gut microbiota, which derives its resources from host dietary components and shed epithelial cells, is an organ in and of itself with substantial functional plasticity and a tremendously high metabolic rate.82This section provides an overview summary of the primary roles of the normal gut microbiota.
5. Inflammatory bowel disease (IBD): an understanding & pathogenetic consequences
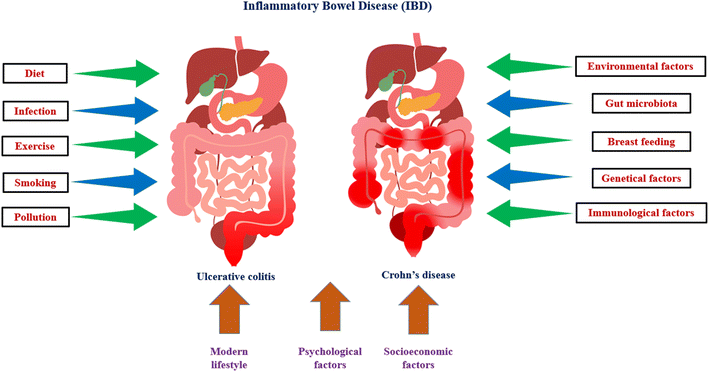
The term “inflammatory bowel disease” (IBD) refers to a collection of several illnesses that are characterised by recurrent, chronic inflammation of the gastrointestinal tract with unknown aetiology and pathophysiology. The host immune system, genetic diversity, and environmental variables may all play a role in the aetiology of IBD. IBD is histopathologically distinguished from Crohn's disease (CD) and ulcerative colitis (UC) based on symptoms (Fig. 3), location of the illness, and histological features. UC results in chronic inflammation and superficial ulcerative disease in the colon, whereas CD is a transmural illness that can affect any portion of the digestive system and is frequently accompanied by granuloma development. IBD can be associated with life-threatening conditions, including primary sclerosing cholangitis, blood clots, and colon cancer. IBD is usually diagnosed between the ages of 20 and 40 years, but can start at any age. IBD shows alternating periods of clinical relapse and remission. Epithelial, Goblet and Paneth, stromal, and immune cells make up the intestinal mucosa. A monolayer of epithelial cells held together by tight connections and sandwiched by immune cells makes up the intestinal epithelium. The intestine is made up of a number of villi, or protrusions, and crypts of Lieberkühn, which are invaginations. The intestinal epithelium acts as a physical barrier to the intestinal lumen's contents while also taking role in nutrient absorption. Additionally, the epithelium communicates with the immune system and gut bacteria, transmitting and receiving signals from both.93,94Goblet and Paneth cells are found in the epithelium, and they generate antimicrobial peptides and mucus, respectively, to stop the spread of luminal bacteria. In Crohn's disease, a loss of mucus layer thickness has been associated to a significant decline in goblet cell counts, and aberrant mucus composition has been described in UC. The lamina propria, which sits beneath the epithelium, is made up of stromal cells such fibroblasts, myofibroblasts, and perivascular pericytes. These cells play a role in fibrosis and wound repair and may be connected to the worsening of UC through their ability to produce chemokines such the immune system modulator interleukin (IL)-33 and the chemokine (C–C motif) ligands CCL19 and CCL21.95,96
Immunoglobulin (Ig)A, which is released by plasma cells, prevents the invasion of pathogenic microbes and aids in maintaining a homeostatic balance between the commensal and host microbiota. The epithelium is a crucial modulator of intestinal homeostasis and the pathogenesis of IBD, as are other non-immune intestinal components.
There are some factors responsible for IBD, which are discussed below.
5.1 Genetical factors
Over the past few decades, there have been considerable advancements in our understanding of the genetic factors that contribute to IBD.97,98 This is due to improvements in DNA analysis and sequencing technology and the use of huge international databases.99 The number of IBD-related gene loci has expanded to 163, of which 110 are linked to both illnesses, 30 with CD, and 23 with UC. Genetic investigations have discovered two autophagy-related genes, ATG16L1 and IRGM, and concluded that autophagy is crucial for immune responses in IBD.100 By destroying and recycling cytosolic components and organelles, as well as enhancing immunity to infection and eradicating intracellular pathogens, autophagy supports intracellular homeostasis.101 All forms of autophagy need ATG16L1, and the coding variant T300A has been associated with a higher risk of developing CD. The p47 immunity-related GTPase family includes IRGM. Reduced protein expression is brought on by IRGM polymorphisms linked with CD. Epithelial and dendritic cells with ATG16L1 and NOD2 polymorphisms have defective antibacterial autophagy.1025.2 Environment
Environment related factors are very important in the pathophysiology of IBD.103,104 IBD risk factors include smoking, poor nutrition, prescription drugs, geography, social stress, and psychological problems.105 The normal understanding of vitamin D's effects focuses on bone health and calcium metabolism. The immunological benefits of vitamin D are becoming more well acknowledged. Recent studies have shown that vitamin D plays a diverse role in a range of illnesses, including IBD.106 Leslie et al. found that low vitamin D levels increased the incidence of IBD and that vitamin D deficiency was common in IBD patients.107 Aspirin and nonsteroidal anti-inflammatory drugs (NSAIDs) are well known for their effects on the digestive system.108 Stress has long been implicated in the development of CD and UC.109 It has been found that people who are less stressed are less likely to develop the illness. Recent ecological and epidemiological research suggest that air pollution may be a factor in the occurrence of CD and UC. In rising countries, CD and UC are becoming more common, which is consistent with the expansion of industry.1105.3 Immunological factors
Investigations into mucosal immunity, especially the T cell response, have long dominated studies into the pathophysiology of IBD.111 There is proof that aberrant intestinal inflammatory responses occur in IBD patients as a result of dysfunctions in the innate and adaptive immune pathways. Over the past two decades, the majority of research has focused on the role of abnormal adaptive immune responses in the pathophysiology of IBD. The focus on the adaptive immune response eventually gave rise to the hypothesis that the two primary types of IBD represent two unique types of intestinal inflammation: CD has long been believed to be driven by a Th1 response, whereas UC has been linked to an unconventional Th2 response.112,1136. Exploring the interplaying relationship between IBD and gut microbiota
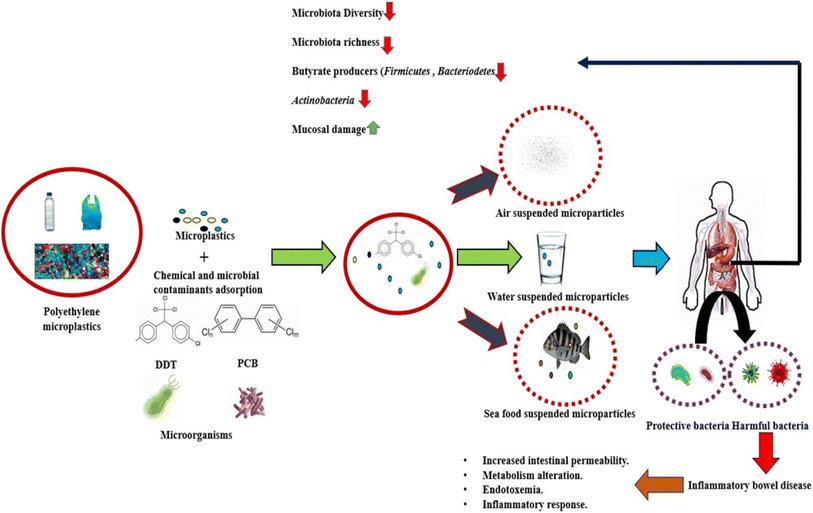
Intestinal dysbiosis is unquestionably related to IBD.114 When an individual is susceptible due to genetics and other concurrent environmental factors come into play, changes in the microbiome are crucial in deciding when a pathology may manifest.115 According to an applicable parameter, IBD patients have a general loss of biodiversity in addition to a drop in specific taxa including Firmicutes and Bacteroidetes, Lactobacillus, and Eubacterium. Additionally, the species that make butyrate, a short-chain fatty acid that favorably regulates intestinal homeostasis and decreases inflammation, are decreased in IBD patients. With the relative growth of Enterobacteriaceae, which includes Escherichia coli and Fusobacterium, a parallel taxonomic change has been seen.116 Increased Ruminococcus gnavus, decreased Bifidobacterium adolescentis, Dialister invisus, Faecalibacterium prausnitzii and an undefined Clostridium cluster XIVa were found in CD patients by Joossens et al. The variety and overall number of species in the microbiome linked to IBD should be reduced, it is generally agreed.117 Additionally, it has been observed that Enterobacteria are overrepresented in CD patients, whereas Firmicutes and Bacteroidetes are relatively rare. In contrast, Clostridium spp. is less common and Escherichia coli are more prevalent in UC patients.118In the early stages of IBD, the gut microbiota's makeup can shift. IBD patients experience more variation in the composition of their gut microbiota than do healthy people.119 According to certain research, CD patients have a higher level of dysbiosis than people with UC. Bifidobacterium longum, Eubacterium rectale, Faecalibacterium prausnitzii, Roseburia intestinalis, and other helpful bacteria levels in CD and UC were significantly lower than in healthy controls, while Bacteroides fragilis relative abundance and growth rate increased.120 At the time the disease first manifests, CD and UC are also enriched in Ruminococcus torques and Ruminococcus. The considerable variations in the abundance of Clostridium hathewayi, Clostridium bolteae and Ruminococcus gnavus indicate that a small number of strains also exhibit increased transcriptional activity. In CD patients, the groups Christensenellaceae, Coriobacteriaceae, and particularly Clostridium leptum decline, whereas Actinomyces species, Veillonella and Escherichia coli rise. Eubacterium rectum is enriched in UC patients, while levels of Akkermansia muciniphila decline and E. coli levels rise.121 According to comparison research, Coprococcus spp. abundance considerably reduces in CD while Intestinibacter spp. abundance rises in both CD and UC. R. gnavus is noticeably more prevalent in IBD patients, according to Hall et al.122 A. muciniphila was shown to be a pathobiont that fosters the growth of IBD and NOD-like receptor 6 (NLRP6) and was discovered to be a major regulator of the organism's abundance. The low abundance in the genus Roseburia is significantly correlated with the IBD-related genes Caspase recruitment domain family member 9 (CARD9), nucleotide binding oligomerization domain containing 2 (NOD2), Autophagy related 16 like 1 (ATG16L), Immunity related GTPase M (IRGM), and Fucosyltransferase 2 (FUT2). Compared to healthy people, patients with active IBD had lower Blastocystis spp. prevalence. Weersma et al. identified variants in several genes, including Myelin gene regulatory factor (MYRF), SEC16 homolog A (SEC16A), Interleukin 17 receptor Elike (IL17REL) and WD repeat domain 78 (WDR78), which were associated with IBD using 12 exome wide microbial quantitative trait loci (mbQTL) analyses.123 In the etiology of IBD, the immune system-affecting genetic variations have a significant impact on the microbiota. It has been noted that the pathophysiology of IBD is also influenced by the environment and genetic susceptibility of the gut flora and their interaction with PE-MPs (Fig. 4).
 | ||
| Fig. 4 Schematic representation of interplaying relationship between polyethylene microplastics, IBD and gut microbiota. | ||
7. Polyethylene microplastics alters the gut microflora: biological impact
7.1 Earthworm (Metaphire guillelmi)
Earthworm, also called angleworm, any one of more than 1800 species of terrestrial worms of the class oligochaeta (phylum Annelida)—in particular, members of the genus Lumbricus. Earthworms occur in virtually all soils of the world in which the moisture and organic content are sufficient to sustain them. In earthworm after being exposed to PE MPs, the relative abundance of Sphingomonadaceae in their gut significantly decreased. In the experimental groups, the relative abundance of Firmicutes increased dramatically from the control group's 11.16% to 59.47, 47.04, and 62.27% (low, medium, and high PE-MPs groups, respectively). As opposed to the control group, Fusobacteria and Bacteroidetes dramatically decreased in the PE MPs treatment groups (Fig. 5).123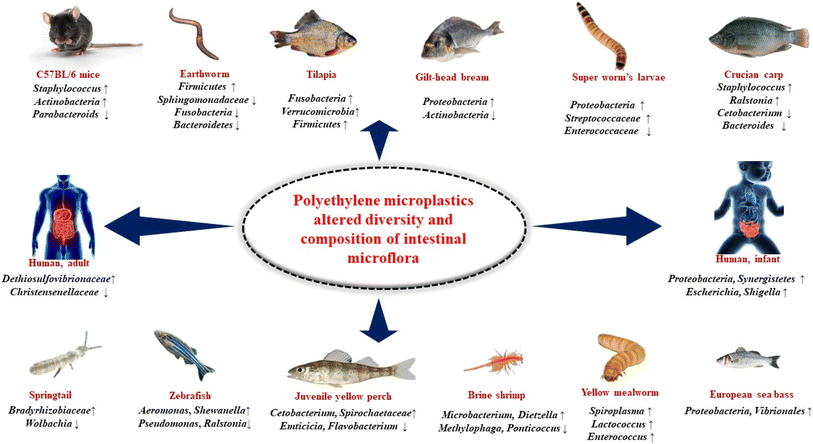 | ||
| Fig. 5 Polyethylene microplastics altered intestinal microflora in various organisms. ‘↑’ indicates increasing, and ‘↓’ indicates decreasing. | ||
7.2 Super worm's larvae (Zophobas atratus)
Zophobas morio (phylum Arthopoda) of the class Insecta, members of the genus Zophobas is a species of darkling beetle, whose larvae are known by the common name superworm, kingworm, morio worm or simply zophobas. Relative abundance analysis indicated that the gut microbiota in LDPE-fed larvae mostly comprised of six distinct phyla: Proteobacteria, Firmicutes, Bacteroidetes, Tenericutes, Fusobacteria and Actinobacteria. Firmicutes and Proteobacteria were dominant phyla. After ingestion of plastics, the relative abundance of Firmicutes in LDPE-fed larvae significantly decreased from 65.91% to 43.67%, while the relative abundance of Proteobacteria significantly increases from 30.01% to 48.26%. The LDPE diet in super worm's larvae resulted in decrease in the abundance of Enterococcaceae from 44.73% to 36.80% and increase in the relative abundance of Streptococcaceae from 2.50% to 3.50% (Fig. 5).1247.3 Yellow mealworm (Tenebrio molitor)
Mealworms are the larval form of the yellow mealworm beetle, Tenebrio molitor, a species of darkling beetle. It is under phylum Arthopoda, class Insecta and genus Tenebrio. In the gut of mealworms, the major phyla reported were Lactococcus, Enterococcus, and Spiroplasma indicating that these may be necessary for the mealworms' digestion and survival. After feeding a meal containing PE MPs, although not harmful, a significant abundance of Spiroplasma in the gut microbiome reported which is not harmful. Additionally, mealworms fed with diet containing PE MPs Lactococcus and Enterococcus were frequent genera, with Lactococcus being present in every area of the gut while Enterococcus was absent in the foregut and anterior midgut (Fig. 5).1257.4 Brine shrimp (Artemia salina)
Artemia is a genus of aquatic crustaceans also known as brine shrimp. It is under phylum Arthopoda, class Branchiopoda and genus Artemia. Artemia populations are found worldwide, typically in inland saltwater lakes, but occasionally in oceans. In case of brine shrimp, after exposure of PE MPs, it has been noted that the proportion of Microbacterium and Dietzella is high and Methylophaga and Ponticoccus is low and the dominant phyla are Proteobacteria, Actinomycetes and Bacteriodaceae.1267.5 Springtails (Folsomia candida)
Springtails (Collembola) form the largest of the three lineages of modern hexapods that are no longer considered insects (the other two are the Protura and Diplura). It is under phylum Arthopoda, sub-phylum Hexapoda, and subclass Collembola. PE MPs exerted a significant toxic effect on spring tails and change their gut microbial community. Wolbachia's relative abundance greatly dropped, but Bradyrhizobiaceae, Ensifer, and Stenotrophomonas's relative abundance significantly rose (Fig. 5).1277.6 Tilapia (Oreochromis niloticus)
Tilapia, common name used for certain species of fishes belonging to the family Cichlidae (order Perciformes), represented by numerous, mostly freshwater species native to Africa. Tilapias are perhaps best known because of their potential as an easily raised and harvested food fish. In case of tilapia the composition of the gut microbiota was altered as a result of ingesting PE MPs with Verrucomicrobia, Fusobacteria, and Firmicutes being the overrepresented bacterial taxa (Fig. 5).1287.7 Gilt-head bream (Sparus aurata)
The gilt-head (sea) bream (Sparus aurata) is a fish of the bream family Sparidae found in the Mediterranean Sea and the eastern coastal regions of the North Atlantic Ocean. In case of gilt-head bream, after exposure of low-density polyethylene (LDPE) microplastics the relative abundance of certain bacteria is changed (Fig. 5). In case of 200–500 μm LDPE, the relative proportion of microbiota are Actinobacteria – 0.4%, Bacteroidetes – 8.9%, Firmicutes – 36.9%, Proteobacteria – 51.6%, Verrucomicrobia – 1.3%. In case of 501–1000 μm LDPE, the relative proportion of microbiota are Actinobacteria – 1.3%, Bacteroidetes – 9.3%, Firmicutes – 34.0%, Proteobacteria –51.8%, Verrucomicrobia – 2.6%.1297.8 Crucian carp (Carassius carassius)
The crucian carp is a medium-sized member of the common carp family Cyprinidae. It occurs widely in northern European regions. In case of crucian carp after exposure to PE MPs, Staphylococcus and Ralstonia grew whereas Cetobacterium and Bacteroides shrank. Additionally, Pseudomonas and other bacteria that were not present in the control group but showed up in the PE MPs exposed groups (Fig. 5).1307.9 Zebrafish (Danio rerio)
The zebrafish is a freshwater fish belonging to the minnow family of the order Cypriniformes. Native to India and South Asia, it is a popular aquarium fish. The phyla Firmicutes, Bacteroidetes, Proteobacteria and Verrucomicrobia significantly changed in zebrafish after exposed to PE MPs. Furthermore, 16S RNA gene sequencing showed that the alpha diversity of the control and 1000 mg L−1 PE MPs treated groups differed significantly. Microbacterium, Shewanella, Methyloversatili, Nevskia, and Aeromonas have all shown notable increases in abundance at the genus level. On the other hand, after exposure to PE MPs, there were a significant decrease in the abundance of Ralstonia, Stenotrophomonas and Pseudomonas.1317.10 Juvenile yellow perch (Perca flavescens)
Juvenile yellow perch is a freshwater perciform fish native to much of North America. In case of juvenile yellow perch, after exposure of high-density PE MPs, it has been noted that the proportion of Cetobacterium is >75%, Spirochaetaceae is 50–75%, Saprosipraceae is 0.05–0.10%, Emticicia is 0.05–0.10%, Luteolibacter is 0.50–1.0%, Flavobacterium is 0.10–0.50%, Enterobacteriaceae is 5–10%, Pleslomonas is 5–10% and Cetobacterium is 10–25%.1327.11 European sea bass (Dicentrarchus labrax)
The European seabass is a primarily ocean-going fish native to the waters off Europe's western and southern and Africa's northern coasts, though it can also be found in shallow coastal waters and river mouths during the summer months and late autumn. It is one of only six species in its family, Moronidae, collectively called the temperate basses. In case of European sea bass after exposed to PE MPs beneficial bacterial genera are decreased and Proteobacteria, Vibrionales are increased which is an indicator of intestinal dysbiosis (Fig. 5).767.12 C57BL/6 mice (Mus musculus)
C57BL/6, often referred to as “C57 black 6”, “B6”, “C57” or “black 6”, is a common inbred strain of laboratory mouse. It is the most widely used “genetic background” for genetically modified mice for use as models of human disease. When C57BL/6 mice exposed to PE MPs, it has been seen that in their gut Staphylococcus, Firmicutes, Melainabacteria, Actinobacteria is increased and parabacteroids is decreased. It has also seen that in exposed mice, the Gastranaerophilales were more prevalent and the Verrucomicrobiales were less prevalent (Fig. 5).1337.13 Human, infant in vitro gut model
In an infant in vitro gut model, exposure to PE MPs changes the makeup of the gut microbiota by changing the ratio of potentially pathogenic Enterobacteriaceae, Oscillospiraceae, Dethiosulfovibrionaceae, and Moraxellaceae to Monoglobaceae populations. A higher-diversity was obtained by adding PE MPs repeatedly. It's interesting to note that the human in vitro study by Tamargo and colleagues on the exposure to PET MPs revealed an increase in Proteobacteria and Synergistetes (phylum of Dethiosulfovibrionaceae), which suggests that these populations could be thought of as biomarkers of exposure to MPs in the human gut microbiota. The impact of PE MPs on the adult gut microbiota, which reveals an increase in Dethiosulfovibrionaceae and Enterobacteriaceae luminal and mucus-associated microbiota, supports this theory. Dethiosulfovibrionaceae has been linked to colorectal cancer, but Enterobacteriaceae, which includes well-known enteric pathogens including Escherichia coli, Campylobacter, Salmonella, and Shigella may be more prevalent in human intestine inflammatory illnesses (Fig. 5).467.14 Human, adult in vitro gut model
Exposure to PE MPs in a human, adult in vitro gut model results in an increase in the well-known gut pathobionts Desulfovibrionaceae, Dethiosulfovibrionaceae and Enterobacteriaceae as well as a decrease in the populations of Akkermansiaceae and Christensenellaceae which are thought to be indicators of a healthy state of gut (Fig. 5).678. Uncovering the possible role of PE MPs and altered gut microflora on IBD
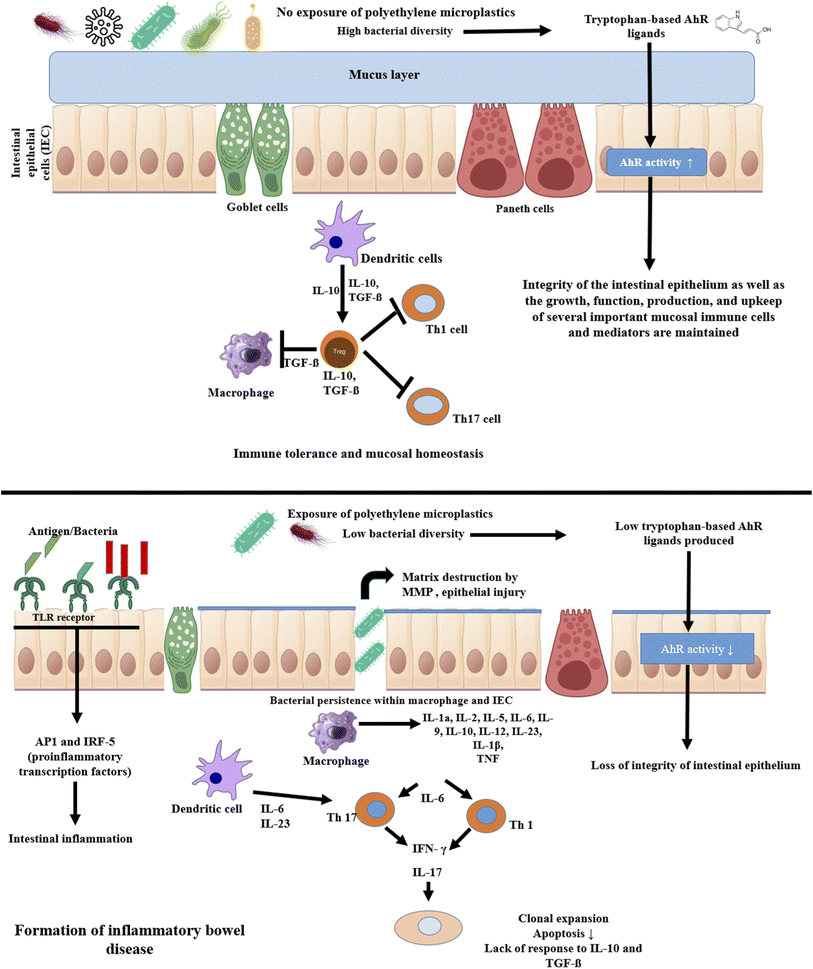
Multiple investigations using animal models have demonstrated that exposure to PE MPs can cause dysbiosis of the intestinal flora. To allow appropriate development and prevent uncontrolled expansion, the balance between proliferation and differentiation of intestinal stem cells is rigorously managed. Overproduction of paneth cells and goblet cells in the intestine can lead to inflammatory conditions (ulcerative colitis, Crohn's disease) and even malignancy. Several studies suggest that after exposure of PE MPs number of goblet cells decreased.134 Colonic epithelium crypt number and depth are elevated. Intestinal stem cell markers' gene expression levels are also elevated. A key player in cell proliferation and the development of cancer, c-myc and PCNA are markedly elevated in colonic crypts. The notch signaling system is the main regulator of intestinal stem cell fate and the generation of goblet cells. It was further demonstrated that the influence of PE MPs on the imbalanced proliferation and differentiation of crypt was controlled by the overactivation of the notch signaling pathway in intestinal organoids.73 These results showed that colonic epithelium after exposure to PE MPs is in an extremely active state of proliferation. Exposure to PE MPs most dramatically changed the balance between ISC self-renewal and differentiation in the colon epithelium, further demonstrating the disruption of intestinal homeostasis. The incidence and susceptibility of sickness are frequently increased by intestinal homeostasis imbalance. IBD are regarded as chronic conditions triggered by environmental factors, and the incidence of these conditions is correlated with an imbalance in intestinal homeostasis.135 Additionally, studies have revealed that persons with IBD had a significantly higher concentration of PE MPs in their faeces than the general population.1 Consequently, exposure to PE MPs accelerated the development of colitis, which was accompanied by a rapid loss of body weight, diarrhea and bloody stools, macroscopic and pathological damage, and increased levels of inflammation. Microplastics exposure exacerbated liver pathological damage and inflammation in mice with colitis. According to a number of research, colonic epithelial self-renewal and differentiation are in a delicate balance, and microplastics may upset this equilibrium, increasing the risk of developing disorders linked to the environment.136Studies have revealed that Staphylococcus overgrowth is linked to IBD because of the inflammation that its superantigen causes.137,138 It was discovered that ulcerative colitis patients had significantly lower levels of Parabacteroides expression than healthy individuals. Reduced intestinal barrier function, inflammation and an increased risk of weight gain were all linked to lower levels of Akkermansia in the intestine. It has been demonstrated that serum levels of cytokines (such as IL-1a, G-CSF, IL-2, IL-5, IL-6, IL-9, IP-10, and RANTES) and the percentages of Th1 and Th17 rise after consumption of different quantities of PE MPs. The pro-inflammatory cytokine IL-6 is one of the primary regulators of sporadic and inflammatory bowel disease.139 In the gut microbiota, PE MPs therapy may increase the number of Staphylococcus genera, and an increase in Staphylococcus abundance may cause an increase in IL-1. G-CSF is a key player in chronic inflammation and can absorb neutrophils from the circulation into inflammatory tissues.140 It has been shown that B. fragilis in the intestines promotes CD4+ T cell differentiation into Treg cells and inhibits the expansion of Th17 cells. It has been shown that Clostridium stimulation of Treg production is crucial for gut immunological homeostasis. Therefore, PE MPs may be able to regulate the frequency of Th17 and Treg cells by altering the variety of the gut microbiota.141 Gut microbiota can develop intestinal defense mechanisms to prevent the introduction of foreign substances. These mechanisms have the ability to activate and regulate signal transduction pathways, such as the TLR pathway, that are connected to intestinal mucosal immune function.142 TLRs, which seem to be significant regulators, activate the inflammatory component of the mucosal immune response. Increased TLR4 protein levels have been seen in the colonic mucosa of children with IBD, which is dependent on inflammation.143 AP-1 and IRF5 are pro-inflammatory transcription factors that are connected to TLR4. It has been established that TLR4/AP-1 and TLR4/IRF5 signaling activation are significant in intestinal inflammation because the expression of TLR4, AP-1 and IRF5 in the experimental group was noticeably higher than that in the control group. In order to explain this, the authors hypothesized that exposure to PE MPs would change the diversity of gut flora, bacterial abundance and species counts, which would change cytokine release and the ratio of Th1, Th17 and Treg cells among CD4+ cells.133 It has been specifically proposed that changed gut microbiota would result in intestinal inflammation by activating TLR4 signaling (Fig. 6).
It has been examined whether the gut microbiota can activate AhR (aryl hydrocarbon receptor) in response to PE MPs exposure.144 Numerous environmental contaminants have been discovered to induce pro-inflammatory responses in various cell types via AhR. The gut microbiota can produce tryptophan-based AhR ligands such as indole-3-acrylic acid, indole-3-acetic acid, tryptamine, indole-3-aldehyde or indole-3-acetaldehyde, which can affect the integrity of the intestinal epithelium as well as the development, operation, production, and maintenance of several critical mucosal immune cells and mediators. Inflammatory bowel disease has been reported to have a shortfall in the gut microbiota's production of AhR ligands when compared to individuals without the disorder.67 In the gut, when the microbiota exposed to PE MPs, AhR activity was typically lower than the comparable control (Fig. 6).
After being exposed to PE MPs, C57BL/6 mice have shown an increase in mucosal and mucin regions as well as an elevation of Muc2, Vil1, and Chga transcripts in the colon, which indicates dysregulation of colon mucosa differentiation. Additionally, an increase in Ocln and F11r expression was noted, which may indicate barrier breakdown. Increased expression of Ifng and Il6 was brought about by the PE bead, supporting a pro-inflammatory state in the colon. Significant changes in the intestinal immune response were also demonstrated by the modulation of the frequency of CD4+ T lymphocytes, CD8+ T lymphocytes, dendritic cells, and inflammatory monocytes in the proximal small intestine, NK cells in the distal small intestine, and anti-inflammatory macrophages in the colon. Furthermore, exposure to the PE beads reduced the quantity of protective Lactobacillales bacteria. Finally, the PE increased the Rhodospirillales' frequency. In this sense, PE microbeads changed the gut microbiota and caused IBD, which had a more detrimental effect on the homeostasis of intestinal tissues.145,146
In the human digestive tract, exposure to PE MPs dramatically changed the microbial volatile profile, which is defined by five main discriminant volatile organic compounds. After being exposed to PE MPs, the quantity of indole, 3-methyl-, increased significantly while the abundance of three hydrocarbon and one alcohol molecule dropped. Also referred to as skatole, this substance is a tryptophan-derived metabolite that is absorbed by the gut epithelium and is produced through a decarboxylation process from indole-3-acetic acid by the gut microbiota in the small intestine and colon. Its level is often low since it is produced in two phases by at least two distinct bacterial species (Bacteroides, Clostridium, or Escherichia coli and Lactobacillus, Clostridium, Desulfovibrio, or Bacteroides). It's interesting to note that our analysis indicates an increase in the Desulfovibrio population, which may contribute to the creation of skatoles and, at the very least, partially account for the increased skatole production. Skatole was not found in the large intestine during fermentation in healthy people, but it was found in significant concentrations in the guts of IBD patients. This rapid rise in the abundance of indole, 3-methyl-, or skatole following exposure to PE MPs suggests that gastrointestinal dysregulation may be mediated by the microbiota. As such, more research is necessary.147,148
9. Future perspective and conclusions
As emerging contaminants, MPs are widely dispersed in a variety of environmental media. MPs pollution must therefore be continuously monitored. MPs in the environment is anticipated to increase with increased global production and usage of plastics and might seriously impact ecosystems. Following consumption of PE MPs, various organisms of the gut microbiota changed in the diversity and composition. As a result of the occurrence and severity of IBD was accelerated due to imbalanced colonic mucosal epithelial proliferation and differentiation, as well as intestinal homeostasis. PE MPs exposure caused over proliferation of ISCs and the loss of goblet cells. Different PE MPs concentrations may stimulate the serum release of the pro-inflammatory cytokine IL-1. The proportion of Th17 and Treg cells among CD4+ cells was reduced by PE MPs, but the Th17/Treg cell ratio was unaffected. Through the activation of TLR4 signaling, high-concentration PE MPs have a tendency to cause intestinal inflammation. The altered gut microbiota caused by PE MPs decreases the production of AhR ligands, which in turn compromises the integrity of the intestinal epithelium and raises the risk of inflammatory bowel disease. The ability of PE MPs to induce inflammatory bowel illness provides a theoretical framework for the therapy and prevention of problems associated with microplastics. As an environmental risk factor for chronic intestinal disease, this should serve as a wake-up call for lawmakers. It is essential to evaluate the ecological ramifications of PE MPs over a longer time period given their long-term stability. Major research holes still exist in environmental microplastics, nevertheless. More study is required, specifically to address the consequences of microplastics on terrestrial environments and human health.More detailed research on the toxicity of PE MPs for humans and other organisms is necessary. Thus, we recommend that future efforts concentrate on the following aspects:
• In the future, investigators ought to develop a larger variety of study models. Animal model studies have difficulty in recognizing dynamic alterations of specific tissues and organs. As the body is a complex entity, the impact of alterations in specific target systems may not be identified immediately. Effectiveness a single cell model is severely restricted. Attention should be paid to research into numerous cell combinations models and organoid models. This form of model may better emulate and target specific tissues or organs, giving a novel experimental viewpoint for toxicological assessment.
• In vivo toxicological studies should be used to determine the toxicity of PE MPs with various chemical formulations or properties; animal model experiments (e.g., rats, mice, zebra fish) should be used to determine the impact of alterations in the physical and chemical attributes (e.g., shape, size, surface charge) of microplastics on human health; and animal model investigations should be used to investigate the transport processes, accumulation, and metabolism of PE MPs within the human body.
• Experiments must be conducted to determine the distribution as well as the accumulation of PE MPs in human tissues. Additional information is needed for in vivo microplastics localization research. Except for the characterization of microplastics themselves, information on the interactions of materials with the body is still constrained. Furthermore, limited investigations have revealed the build-up of PE MPs in the surroundings or organisms. This circumstance does not imply that PE MPs do not exist, but rather that detection technology is insufficient. As a result, more precise microplastics characterization approaches will be required in the future, particularly for the co-localization of PE MPs in vivo.
• Currently, researchers are primarily concerned with the impacts of microplastics on the intestinal mechanical barriers and immune responses of IBD patients, while overlooking their impact on the gut microflora. Microplastics have been shown in studies to cause intestinal or extraintestinal organ injury by disrupting gut microbiota. As a result, subsequent studies should focus on PE MPs for the microbiota of the gut and metabolomic alterations in high-risk groups, in order to identify microbial strategies for preventing illness.
More systematic and detailed large-scale studies in various ecosystems is needed to understand role of PE MPs contamination in environment (Fig. 7). The consequences on the plant, animals and other living organisms must be investigated. The major influence of PE MPs on society the scientific community has yet to study. PE MPs research must take an approach that is more realistic. An experimental setting is essential for producing cohesive and reliable research results. For the advancement of microplastic removal procedures, interdisciplinary methods must be promoted. Understanding environmental elements and anthropogenic activity requires a combination of sociological, epidemiological, engineering, biological, and technical techniques. To reduce human health concerns, it is critical to assess and improve treatment technologies for eliminating microplastics from polluted areas (Fig. 7).
Due to the complicated mix of diverse microplastic particles, the present difficulty that has to be addressed is creating technology to regulate the pollutant or impose universal, countrywide or even statewide rules. Until these problems are addressed, every attempt should be undertaken to minimize PE MPs production and consumption, as well as improve recycling and ecologically safe plastic disposal, in tandem with the development of technology that eliminate microplastics from the surroundings.
Declaration
All the photos/artwork that appear in figures in this paper, including those in the TOC graphic, were created by the authors of this manuscript and have not been published elsewhere.Data availability
No primary research results, software or code have been included, and no new data were generated or analyzed as part of this review. All information cited within this review is derived from existing literature and publicly available sources.Conflicts of interest
The authors declare they have no potential or actual competing financial or personal interests.Acknowledgements
Mr Sagar Bag thanks UGC, Govt. of India for providing fellowship and research grant (UGC-Senior Research Fellowship, NTA reference number: 201610001623). Sudipta Bhowmik thanks “Intramural Seed Money Research Committee, SBV” for “SBV-Seed money” research grant (SBV/IRC/SEED MONEY/134/2022).References
- S. Ghosal, S. Bag, M. D. Burman and S. Bhowmik, J. Phys. Chem. Lett., 2023, 14, 10328–10332, DOI:10.1021/acs.jpclett.3c02632.
- S. Ghosal, S. Bag and S. Bhowmik, J. Phys. Chem. Lett., 2024, 15(25), 6560–6567, DOI:10.1021/acs.jpclett.4c00731.
- Y. Cheng, S. Yang, L. Yin, Y. Pu and G. Liang, Ecotoxicol. Environ. Saf., 2023, 249, 114385, DOI:10.1016/j.ecoenv.2022.114385.
- M. T. Khan, I. A. Shah, M. F. Hossain, N. Akther, Y. Zhou, M. S. Khan, M. Al-Shaeli, M. S. Bacha and I. Ihsanullah, Sci. Total Environ., 2023, 860, 160322, DOI:10.1016/j.scitotenv.2022.160322.
- I. Gambino, F. Bagordo, T. Grassi, A. Panico and A. De Donno, Int. J. Environ. Res. Publ. Health, 2022, 19, 5283, DOI:10.3390/ijerph19095283.
- A. Luqman, Environments, 2021, 8, 138 CrossRef.
- P. Schwabl, S. Köppel, P. Königshofer, T. Bucsics, M. Trauner, T. Reiberger and B. Liebmann B, Ann. Intern. Med., 2019, 171, 453–457, DOI:10.7326/M19-0618.
- R. Dris, J. Gasperi, M. Saad, C. Mirande and B. Tassin, Mar. Pollut. Bull., 2016, 104, 290–293, DOI:10.1016/j.marpolbul.2016.01.006.
- M. Klein and E. K. Fischer, Sci. Total Environ., 2019, 685, 96–103, DOI:10.1016/j.scitotenv.2019.05.405.
- K. Liu, X. Wang, T. Fang, P. Xu, L. Zhu and D. Li, Sci. Total Environ., 2019, 675, 462–471, DOI:10.1016/j.scitotenv.2019.04.110.
- E. Gaston, M. Woo, C. Steele, S. Sukumaran and S. Anderson, Appl. Spectrosc., 2020, 74, 1079–1098, DOI:10.1177/0003702820920652.
- S. L. Wright, J. Ulke, A. Font, K. L. A. Chan and F. J. Kelly, Environ. Int., 2020, 136, 105411, DOI:10.1016/j.envint.2019.105411.
- R. Dris, J. Gasperi, C. Mirande, C. Mandin, M. Guerrouache, V. Langlois and B. Tassin, Environ. Pollut., 2017, 221, 453–458, DOI:10.1016/j.envpol.2016.12.013.
- Q. Zhang, Y. Zhao, F. Du, H. Cai, G. Wang and H. Shi, Environ. Sci. Technol., 2020, 54, 6530–6539, DOI:10.1021/acs.est.0c00087.
- L. C. Jenner, Atmos. Environ., 2021, 259, 118512 CrossRef CAS.
- Z. Liao, X. Ji, Y. Ma, B. Lv, W. Huang, X. Zhu, M. Fang, Q. Wang, X. Wang, R. Dahlgren and X. Shang, J. Hazard. Mater., 2021, 417, 126007, DOI:10.1016/j.jhazmat.2021.126007.
- M. Celik, H. Nakano, K. Uchida, A. Isobe and H. Arakawa, Mar. Pollut. Bull., 2023, 190, 114818, DOI:10.1016/j.marpolbul.2023.114818.
- C. Amaneesh, S. Anna Balan, P. S. Silpa, J. W. Kim, K. Greeshma, A. Aswathi Mohan, A. Robert Antony, H. P. Grossart, H. S. Kim and R. Ramanan, Environ. Sci. Technol., 2023, 57, 5–24, DOI:10.1021/acs.est.2c05817.
- C. Wang, W. Wu, Z. Pang, J. Liu, J. Qiu, T. Luan, J. Deng and Z. Fang, J. Hazard. Mater., 2023, 446, 130617, DOI:10.1016/j.jhazmat.2022.130617.
- S. Haldar, Y. Muralidaran, D. Míguez, S. I. Mulla and P. Mishra, Sci. Total Environ., 2023, 861, 160571, DOI:10.1016/j.scitotenv.2022.160571.
- R. Geyer, J. R. Jambeck and K. L. Law, Sci. Adv., 2017, 3, 1700782, DOI:10.1126/sciadv.1700782.
- P. Liu, J. Dai, K. Huang, Z. Yang, Z. Zhang and X. Guo, Crit. Rev. Environ. Sci. Technol., 2023, 53, 865–885 CrossRef CAS.
- A. A. Mamun, T. A. E. Prasetya, I. R. Dewi and M. Ahmad, Sci. Total Environ., 2023, 858, 159834, DOI:10.1016/j.scitotenv.2022.159834.
- K. Grote, F. Brüstle and A. K. Vlacil, Materials, 2023, 16, 3123, DOI:10.3390/ma16083123.
- T. A. Kurniawan, A. Haider, H. M. Ahmad, A. Mohyuddin, H. M. Umer Aslam, S. Nadeem, M. Javed, M. H. D. Othman, H. H. Goh and K. W. Chew, Chemosphere, 2023, 325, 138367, DOI:10.1016/j.chemosphere.2023.138367.
- N. Khalid, M. Aqeel, A. Noman and Z. Fatima Rizvi, J. Hazard. Mater., 2023, 445, 130455, DOI:10.1016/j.jhazmat.2022.130455.
- H. M. Kang, E. Byeon, H. Jeong, M. S. Kim, Q. Q. Chen and J. S. Lee, J. Hazard. Mater., 2021, 405, 124207 CrossRef CAS.
- L. Kim, R. X. Cui, J. Il Kwak and Y. J. An, J. Hazard. Mater., 2022, 440, 129715 CrossRef CAS.
- Y. X. Jin, L. Lu, W. Q. Tu, T. Luo and Z. W. Fu, Sci. Total Environ., 2019, 649, 308–317 CrossRef CAS PubMed.
- H. Tan, T. Yue, Y. Xu, J. Zhao and B. Xing, Environ. Sci. Technol., 2020, 54(19), 12285–12294 CrossRef CAS PubMed.
- Y. Lu, Y. Zhang, Y. Deng, W. Jiang, Y. Zhao, J. Geng, L. Ding and H. Ren, Environ. Sci. Technol., 2016, 50(7), 4054–4060 CrossRef CAS PubMed.
- Y. Deng, Y. Zhang, R. Qiao, M. M. Bonilla, X. Yang, H. Ren and B. Lemos, J. Hazard. Mater., 2018, 357, 348–354 CrossRef CAS.
- J. Kokarakis, E. J. Kokarakis, E. Ladakis and H. Petrakakos, Microplastics and their Impact on the Marine Environment, Paper presented at the SNAME 8th International Symposium on Ship Operations, Management and Economics, OnePetro, Athens, Greece, 2023, DOI:10.5957/SOME-2023-040.
- C. Pironti, M. Ricciardi, O. Motta, Y. Miele, A. Proto and L. Montano, Toxics, 2021, 9, 224, DOI:10.3390/toxics9090224.
- M. A. Bhat, K. Gedik and E. O. Gaga, Air Qual., Atmos. Health, 2023, 16, 233–262, DOI:10.1007/s11869-022-01272-2.
- F. Avilés-Jiménez, G. Yu, K. Torres-Poveda, V. Madrid-Marina and J. Torres, Arch. Med. Res., 2017, 48, 754–765, DOI:10.1016/j.arcmed.2017.11.008.
- G. Nardone and D. Compare D, United Eur. Gastroenterol. J., 2015, 3, 255–260, DOI:10.1177/2050640614566846.
- M. A. Engevik, A. Hickerson, G. E. Shull and R. T. Worrell, Cell. Physiol. Biochem., 2013, 32, 111–128, DOI:10.1159/000356632.
- R. Monroy-Torres, M. A. Hernández-Luna, X. S. Ramírez-Gómez and S. López-Briones, In Prebio. and Probio. Potent. Benefits in Nutrit. and Heal., 2019 Search PubMed.
- G. W. Tannock, Gut Flo. Nutrit. Immun. and Healt., 2003, 1–23 Search PubMed.
- M. Wang, S. Ahrné, B. Jeppsson and G. P. Molin, FEMS Microbiol. Ecol., 2005, 54, 219–231, DOI:10.1016/j.femsec.2005.03.012.
- K. Grote, F. Brüstle and A. K. Vlacil, Materials, 2023, 16, 3123, DOI:10.3390/ma16083123.
- T. A. Kurniawan, A. Haider, H. M. Ahmad, A. Mohyuddin, H. M. Umer Aslam, S. Nadeem, M. Javed, M. H. D. Othman, H. H. Goh and K. W. Chew, Chemosphere, 2023, 325, 138367, DOI:10.1016/j.chemosphere.2023.138367.
- N. Khalid, M. Aqeel, A. Noman and Z. Fatima Rizvi, J. Hazard. Mater., 2023, 445, 130455, DOI:10.1016/j.jhazmat.2022.130455.
- Y. Qiu, S. Zhou, C. Zhang, W. Qin, C. Lv and M. Zou, Sci. Total Environ., 2023, 877, 162891, DOI:10.1016/j.scitotenv.2023.162891.
- E. Fournier, M. Leveque, P. Ruiz, J. Ratel, C. Durif, S. Chalancon, F. Amiard, M. Edely, V. Bezirard, E. Gaultier, B. Lamas, E. Houdeau, F. Lagarde, E. Engel, L. Etienne-Mesmin, S. Blanquet-Diot and M. Mercier-Bonin, J. Hazard. Mater., 2023, 442, 130010, DOI:10.1016/j.jhazmat.2022.130010.
- A. Tamargo, N. Molinero, J. J. Reinosa, V. Alcolea-Rodriguez, R. Portela, M. A. Bañares, J. F. Fernández and M. V. Moreno-Arribas, Sci. Rep., 2022, 12, 528 CrossRef CAS PubMed.
- L. R. Jones, S. J. Wright and T. W. Gant, Toxicol. Lett., 2023, 385, 51–60 CrossRef CAS PubMed.
- T. W. L. Lam, Y. C. J. Tsui, Y. L. Cheng, A. T. H. Ma and L. Fok, Sci. Total Environ., 2023, 875, 162576, DOI:10.1016/j.scitotenv.
- R. C. Thompson, C. J. Moore, F. S. vom Saal and S. H. Swan, Philos. Trans. R. Soc. Lond. B Biol. Sci., 2009, 364, 2153–2166, DOI:10.1098/rstb.2009.0053.
- C. Jiménez-Arroyo, A. Tamargo, N. Molinero and M. V. Moreno-Arribas, Microb. Biotechnol., 2023, 16, 34–53, DOI:10.1111/1751-7915.14182.
- M. Vancamelbeke and S. Vermeire, Expet Rev. Gastroenterol. Hepatol., 2017, 11, 821–834, DOI:10.1080/17474124.
- R. Ding, Y. Ma, T. Li, M. Sun, Z. Sun and J. Duan, Sci. Total Environ., 2023, 878, 163144, DOI:10.1016/j.scitotenv.
- A. O. Adeniji, K. Okaiyeto, J. N. Mohammed, M. Mabaleha, E. B. Tanor and M. J. George, Int. J. Environ. Sci. Technol., 2023, 1–28, DOI:10.1007/s13762-023-04916-7.
- J. Zalasiewicz, C. N. Waters, J. A. Do Sul, P. L. Corcoran, A. D. Barnosky, A. Cearreta, M. Edgeworth, A. Gałuszka, C. Jeandel, R. Leinfelder and J. R. McNeill, Anthropocene, 2016, 13, 4–17 CrossRef.
- H. Kye, J. Kim, S. Ju, J. Lee, C. Lim and Y. Yoon, Heliyon, 2023, 9, E14359 CrossRef CAS PubMed.
- C. Pironti, M. Ricciardi, O. Motta, Y. Miele, A. Proto and L. Montano, Toxics, 2021, 9, 224, DOI:10.3390/toxics9090224.
- M. A. Bhat, K. Gedik and E. O. Gaga, Air Qual., Atmos. Health, 2023, 16, 233–262, DOI:10.1007/s11869-022-01272-2.
- H. Yu, J. Shao, H. Jia, D. Gang, B. Ma and C. Hu, Engineering, 2023, 37, 69–77 CrossRef.
- H. Liang, W. P. de Haan, M. Cerdà-Domènech, J. Méndez, F. Lucena, C. García-Aljaro, A. Sanchez-Vidal and E. Ballesté, Environ. Pollut., 2023, 319, 120983, DOI:10.1016/j.envpol.2022.120983.
- H. Hu, D. Jin, Y. Yang, J. Zhang, C. Ma and Z. Qiu, Environ. Res., 2021, 200, 111363, DOI:10.1016/j.envres.2021.111363.
- A. I. Osman, M. Hosny, A. S. Eltaweil, S. Omar, A. M. Elgarahy, M. Farghali, P. S. Yap, Y. S. Wu, S. Nagandran, K. Batumalaie, S. C. B. Gopinath, O. D. John, M. Sekar, T. Saikia, P. Karunanithi, M. H. M. Hatta and K. A. Akinyede, Environ. Chem. Lett., 2023 DOI:10.1007/s10311-023-01593-3.
- F. Wang, C. S. Wong, D. Chen, X. Lu, F. Wang and E. Y. Zeng, Water Res., 2018, 139, 208–219, DOI:10.1016/j.watres.2018.04.003.
- N. J. Hahladakis, A. V. Costas, R. Weber, E. Iacovidou and P. Purnell, J. Hazard. Mater., 2018, 344, 179–199 CrossRef PubMed.
- V. Marturano, P. Cerruti and V. Ambrogi, Phys. Sci. Rev., 2017, 2, 20170138, DOI:10.1515/psr-2017-0138.
- E. Hansen, N. H. Nilsson, D. Lithner and C. Lassen, Hazardous substances in plastic materials, COWI and the Danish Technological Institute, Denmark, 2013, pp. 7–8 Search PubMed.
- V. R. Sastri, Plastics in Medical Devices: Properties, Requirements, and Applications, Elsevier, 3rd edn, 2021, pp. 55–72 Search PubMed.
- C. Q. Y. Yong, S. Valiyaveetill and B. L. Tang, Int. J. Environ. Res. Public Health, 2020, 17, 1509 CrossRef CAS PubMed.
- K. Zhu, H. Jia, Y. Sun, Y. Dai, C. Zhang, X. Guo, T. Wang and L. Zhu, Environ. Int., 2020, 145, 106137 CrossRef CAS PubMed.
- S. Jeon, D. K. Lee, J. Jeong, S. I. Yang, J. S. Kim, J. Kim and W. S. Cho, Environ. Pollut., 2021, 281, 117006 CrossRef CAS PubMed.
- N. Khalid, M. Aqeel, A. Noman and Z. Fatima Rizvi, J. Hazard Mater., 2023, 445, 130455, DOI:10.1016/j.jhazmat.2022.130455.
- L. Hu, Y. Zhao and H. Xu, J. Hazard. Mater., 2022, 439, 129652, DOI:10.1016/j.jhazmat.
- M. Djouina, C. Vignal, A. Dehaut, S. Caboche, N. Hirt, C. Waxin, C. Himber, D. Beury, D. Hot, L. Dubuquoy, D. Launay, G. Duflos and M. Body-Malapel, Environ. Res., 2022, 212, 113230, DOI:10.1016/j.envres.2022.113230.
- M. Revel, A. Châtel and C. Mouneyrac, Curr Opin Environ Sci Health., 2018, 1, 17–23 CrossRef.
- Q. Liu, Z. Chen, Y. Chen, F. Yang, W. Yao and Y. Xie, J. Agric. Food Chem., 2021, 69, 10450–10468, DOI:10.1021/acs.jafc.1c04199.
- D. Montero, S. Rimoldi, S. Torrecillas, J. Rapp, F. Moroni, A. Herrera, M. Gómez, A. Fernández-Montero and G. Terova, Sci. Total Environ., 2022, 805, 150402, DOI:10.1016/j.scitotenv.2021.150402.
- P. Kang, Y. Zhao, C. Zuo, Y. Cai, C. Shen, B. Ji and T. Wei, Sci. Total Environ., 2023, 878, 163028, DOI:10.1016/j.scitotenv.2023.163028.
- E. Scarpellini, G. Ianiro, F. Attili, C. Bassanelli, A. De Santis and A. Gasbarrini, Dig. Liver Dis., 2015, 47, 1007–1012, DOI:10.1016/j.dld.
- A. Weiner, S. Turjeman and O. Koren, Neuropharmacology, 2023, 227, 109453, DOI:10.1016/j.neuropharm.
- H. Bisgaard, N. Li, K. Bonnelykke, B. L. Chawes, T. Skov, G. Paludan-Müller, J. Stokholm, B. Smith, K. A. Krogfelt and J. Aller, Clin. Immunol., 2011, 128, 646–652, DOI:10.1016/j.jaci.2011.04.060.
- Z. Li, T. Lu, M. Li, M. Mortimer and L. H. Guo, Chemosphere, 2023, 329, 138692, DOI:10.1016/j.chemosphere.2023.138692.
- S. M. Jandhyala, R. Talukdar, C. Subramanyam, H. Vuyyuru, M. Sasikala and D. Nageshwar Reddy, World J. Gastroenterol., 2015, 21, 8787–8803, DOI:10.3748/wjg.v21.i29.8787.
- E. Mutuyemungu, M. Singh, S. Liu and D. J. Rose, J. Funct.Foods, 2023, 100, 105367 CrossRef.
- J. L. Sonnenburg, J. Xu, D. D. Leip, C. H. Chen, B. P. Westover, J. Weatherford, J. D. Buhler and J. I. Gordon, Science, 2005, 307, 1955–1959, DOI:10.1126/science.1109051.
- A. Conz, M. Salmona and L. Diomede, Nutrients, 2023, 15, 1869, DOI:10.3390/nu15081869.
- T. A. Clayton, D. Baker, J. C. Lindon, J. R. Everett and J. K. Nicholson, Proc. Natl. Acad. Sci., 2009, 106, 14728–14733 CrossRef CAS PubMed.
- B. D. Wallace, H. Wang, K. T. Lane, J. E. Scott, J. Orans, J. S. Koo, M. Venkatesh, C. Jobin, L. A. Yeh, S. Mani and M. R. Redinbo, Science, 2010, 330, 831–835, DOI:10.1126/science.1191175.
- C. Thoda and M. Touraki, Appl. Sci., 2023, 13, 4726 CrossRef CAS.
- C. L. Maynard, C. O. Elson, R. D. Hatton and C. T. Weaver, Nature, 2012, 489, 231–241, DOI:10.1038/nature11551.
- C. L. Maynard, C. O. Elson, R. D. Hatton and C. T. Weaver, Nature, 2012, 489, 231–241, DOI:10.1038/nature11551.
- E. Cario, G. Gerken and D. K. Podolsky, Gastroentero, 2007, 132, 1359–1374, DOI:10.1053/j.gastro.2007.02.056.
- F. Yan, H. Cao, T. L. Cover, M. K. Washington, Y. Shi, L. Liu, R. Chaturvedi, R. M. Jr. Peek, K. T. Wilson and D. B. Polk, J. Clin. Invest., 2011, 121, 2242–2253, DOI:10.1172/JCI44031.
- R. J. Xavier and D. K. Podolsky, Nature, 2007, 448, 427–434, DOI:10.1038/nature06005.
- M. Barreiro-de Acosta, A. Molero, E. Artime, S. Díaz-Cerezo, L. Lizán, H. D. de Paz and M. D. Martín-Arranz, Adv. Ther., 2023, 40, 1975–2014, DOI:10.1007/s12325-023-02473-6.
- C. Abraham and J. H. Cho, N. Engl. J. Med., 2009, 361, 2066–2078, DOI:10.1056/NEJMra0804647.
- Y. Zurba, B. Gros and M. Shehab, Biomedicines, 2023, 11, 747, DOI:10.3390/biomedicines11030747.
- D. R. Gaya, R. K. Russell, E. R. Nimmo and J. Satsangi, Lancet, 2006, 367, 1271–1284, DOI:10.1016/S0140-6736(06)68345-1.
- O. A. Kriger-Sharabi and U. Kopylov, J. Clin. Med., 2023, 12, 2696, DOI:10.3390/jcm12072696.
- R. H. Duerr, Gastroenterology, 2007, 132, 2045–2049, DOI:10.1053/j.gastro.
- S. A. McCarroll, A. Huett, P. Kuballa, S. D. Chilewski, A. Landry, P. Goyette, M. C. Zody, J. L. Hall, S. R. Brant, J. H. Cho, R. H. Duerr, M. S. Silverberg, K. D. Taylor, J. D. Rioux, D. Altshuler, M. J. Daly and R. J. Xavier, Nat. Genet., 2008, 40, 1107–1112, DOI:10.1038/ng.215.
- B. Khor, A. Gardet and R. J. Xavier, Nature, 2011, 474, 307–317, DOI:10.1038/nature10209.
- P. Kuballa, A. Huett, J. D. Rioux, M. J. Daly and R. J. Xavier, PLoS One, 2008, 3, 3391, DOI:10.1371/journal.pone.0003391.
- G. Bergamaschi, F. Castiglione, R. D'Incà, M. Astegiano, W. Fries, M. Milla, C. Ciacci, F. Rizzello, S. Saibeni, R. Ciccocioppo, A. Orlando, F. Bossa, M. Principi, P. Vernia, C. Ricci, M. L. Scribano, G. Bodini, D. Mazzucco, G. Bassotti, G. Riegler, A. Buda, M. Neri, F. Caprioli, F. Monica, A. Manca, E. Villa, G. Fiorino, M. Comberlato, N. Aronico, C. Della Corte, R. Caccaro, P. Gionchetti, P. Giuffrida, P. Iovino, M. V. Lenti, C. Mengoli, L. Pellegrini, A. Pieraccini, D. Ribaldone, A. Testa, C. Ubezio, A. Viola, M. Vecchi, C. Klersy and A. Di Sabatino, Inflamm. Bowel Dis., 2023, 29, 76–84, DOI:10.1093/ibd/izac054.
- A. R. Bourgonje, D. Kloska, A. Grochot-Przęczek, M. Feelisch, A. Cuadrado and H. van Goor, Redox Biol., 2023, 60, 102603, DOI:10.1016/j.redox.2023.102603.
- E. V. Jr. Loftus, Gastroenterology, 2004, 126, 1504–1517, DOI:10.1053/j.gastro.2004.01.063.
- M. Garg, J. S. Lubel, M. P. Sparrow, S. G. Holt and P. R. Gibson, Aliment. Pharmacol. Ther., 2012, 36, 324–344, DOI:10.1111/j.1365-2036.
- W. D. Leslie, N. Miller, L. Rogala and C. N. Bernstein, Am. J. Gastroenterol., 2008, 103, 1451–1459, DOI:10.1111/j.1572-0241.2007.01753.
- A. N. Ananthakrishnan, L. M. Higuchi, E. S. Huang, H. Khalili, J. M. Richter, C. S. Fuchs and A. T. Chan, Ann. Intern. Med., 2012, 156, 350–359, DOI:10.7326/0003-4819-156-5-201203060-00007.
- J. E. Mawdsley and D. S. Rampton, Neuroimmunomodulation, 2006, 13, 327–336, DOI:10.1159/000104861.
- K. T. Thia, E. V. Jr. Loftus, W. J. Sandborn and S. K. Yang, Am. J. Gastroenterol., 2008, 103, 3167–3182, DOI:10.1111/j.1572-0241.
- S. Cao, M. Colonna and P. Deepak, J. Crohns. Colit., 2023, 17, 1010–1022, DOI:10.1093/ecco-jcc/jjad008.
- G. M. Cobrin and M. T. Abreu, Immunol. Rev., 2005, 206, 277–295, DOI:10.1111/j.0105-2896.
- S. R. Targan and L. C. Karp, Immunol. Rev., 2005, 206, 296–305, DOI:10.1111/j.0105-2896.
- N. M. Liyanage, D. P. Nagahawatta, T. U. Jayawardena and Y. J. Jeon, Life, 2023, 13, 1026, DOI:10.3390/life13041026.
- E. R. Lane, T. L. Zisman and D. L. Suskind, J. Inflamm. Res., 2017, 10, 63–73, DOI:10.2147/JIR.
- C. Lupp, M. L. Robertson, M. E. Wickham, I. Sekirov, O. L. Champion, E. C. Gaynor and B. B. Finlay, Cell Host Microbe, 2007, 2, 119–129, DOI:10.1016/j.chom.2007.06.010.
- M. Joossens, G. Huys, M. Cnockaert, V. De Preter, K. Verbeke, P. Rutgeerts, P. Vandamme and S. Vermeire, Gut, 2011, 60, 631–637, DOI:10.1136/gut.2010.223263.
- C. Martinez, M. Antolin, J. Santos, A. Torrejon, F. Casellas, N. Borruel, F. Guarner and J. R. Malagelada, Am. J. Gastroenterol., 2008, 103, 643–648, DOI:10.1111/j.1572-0241.2007.
- Y. Zhang, R. Chen, D. Zhang, S. Qi and Y. Liu, Biomed. Pharmacother., 2023, 160, 114295, DOI:10.1016/j.biopha.2023.114295.
- A. Vich Vila, F. Imhann, V. Collij, S. A. Jankipersadsing, T. Gurry, Z. Mujagic, A. Kurilshikov, M. J. Bonder, X. Jiang, E. F. Tigchelaar, J. Dekens, V. Peters, M. D. Voskuil, M. C. Visschedijk, H. M. van Dullemen, D. Keszthelyi, M. A. Swertz, L. Franke, R. Alberts, E. A. M. Festen, G. Dijkstra, A. A. M. Masclee, M. H. Hofker, R. J. Xavier, E. J. Alm, J. Fu, C. Wijmenga, D. M. A. E. Jonkers, A. Zhernakova and R. K. Weersma, Sci. Transl. Med., 2018, 10, 8914, DOI:10.1126/scitranslmed.
- I. R. Pittayanon, J. T. Lau, G. I. Leontiadis, F. Tse, Y. Yuan, M. Surette and P. Moayyedi, Gastroenterology, 2020, 158, 930–946, DOI:10.1053/j.gastro.2019.11.294.
- A. B. Hall, M. Yassour, J. Sauk, A. Garner, X. Jiang, T. Arthur, G. K. Lagoudas, T. Vatanen, N. Fornelos, R. Wilson, M. Bertha, M. Cohen, J. Garber, H. Khalili, D. Gevers, A. N. Ananthakrishnan, S. Kugathasan, E. S. Lander, P. Blainey, H. Vlamakis, R. J. Xavier and C. Huttenhower, Genome Med., 2017, 9, 103, DOI:10.1186/s13073-017-0490-5.
- Y. Cheng, W. Song, H. Tian, K. Zhang, B. Li, Z. Du, W. Zhang, J. Wang, J. Wang and L. Zhu, Chemosphere, 2021, 267, 129219, DOI:10.1016/j.chemosphere.2020.129219.
- B. Y. Peng, Y. Sun, Z. Wu, J. Chen, Z. Shen, X. Zhou, W. M. Wu and Y. Zhang, J. Clean. Prod., 2022, 367, 132987 CrossRef CAS.
- Y. Lou, Y. Li, B. Lu, Q. Liu, S. S. Yang, B. Liu, N. Ren, W. M. Wu and D. Xing, J. Hazard. Mater., 2021, 416, 126222, DOI:10.1016/j.jhazmat.2021.126222.
- B. Li, Y. Ding, X. Cheng, D. Sheng, Z. Xu, Q. Rong, Y. Wu, H. Zhao, X. Ji and Y. Zhang, Chemosph, 2020, 244, 125492, DOI:10.1016/j.chemosphere.2019.125492.
- H. Ju, D. Zhu and M. Qiao, Environ. Pollut., 2019, 247, 890–897, DOI:10.1016/j.envpol.
- X. Lu, J. X. Z. L. Zhang, D. Wu, J. Tian, L. J. Yu, L. He, S. Zhong, H. Du, D. F. Deng, Y. Z. Ding, H. Wen and M. Jiang, Sci. Total Environ., 2022, 841, 156571, DOI:10.1016/j.scitotenv.
- I. Varo, K. Osorio, I. Estensoro, F. Naya-Catala, A. Sitja-Bobadilla, J. C. Navarro, J. Perez-Sanchez, A. Torreblanca and M. C. Piazzon, Aquacult, 2021, 545, 737245 CrossRef CAS.
- J. Hu, J. Zuo, J. Li, Y. Zhang, X. Ai, J. Zhang, D. Gong and D. Sun, Sci. Total Environ., 2022, 802, 149736, DOI:10.1016/j.scitotenv.
- Y. Zhao, Z. Qin, Z. Huang, Z. Bao, T. Luo and Y. Jin, Environ. Pollut., 2021, 282, 117039, DOI:10.1016/j.envpol.
- X. Lu, D. F. Deng, F. Huang, F. Casu, E. Kraco, R. J. Newton, M. Zohn, S. J. Teh, A. M. Watson, B. Shepherd, Y. Ma, M. A. O. Dawood and L. M. R. Mendoza, Anim. Nutr., 2022, 9, 143–158, DOI:10.1016/j.aninu.
- W. Li, R. Wufuer, J. Duo, S. Wang, Y. Luo, D. Zhang and X. Pan, Sci. Total Environ., 2020, 749, 141420, DOI:10.1016/j.scitotenv.
- K. K. Gkouskou, C. Deligianni, C. Tsatsanis and A. G. Eliopoulos, Front. Cell. Infect. Microbiol., 2014, 4, 28, DOI:10.3389/fcimb.2014.00028.
- C. Chang and H. Lin, Best Pract. Res. Clin. Gastroenterol., 2016, 30, 3–15, DOI:10.1016/j.bpg.
- S. Xie, R. Zhang, Z. Li, C. Liu, Y. Chen and Q. Yu, Environ. Res., 2023, 217, 114861, DOI:10.1016/j.envres.
- M. C. Collado, E. Isolauri, K. Laitinen and S. Salminen, Am. J. Clin. Nutr., 2008, 88, 894–899, DOI:10.1093/ajcn/88.4.894.
- M. F. Isler, S. J. Coates and M. D. Boos, Int. J. Dermatol., 2023, 62, 337–345, DOI:10.1111/ijd.16297.
- A. Kasza, Cytok, 2013, 62, 22–33, DOI:10.1016/j.cyto.2013.02.007.
- A. M. Wengner, S. C. Pitchford, R. C. Furze and S. M. Rankin, Blood, 2008, 111, 42–49, DOI:10.1182/blood-2007-07-099648.
- K. Atarashi, T. Tanoue, K. Oshima, W. Suda, Y. Nagano, H. Nishikawa, S. Fukuda, T. Saito, S. Narushima, K. Hase, S. Kim, J. V. Fritz, P. Wilmes, S. Ueha, K. Matsushima, H. Ohno, B. Olle and K. Honda, Nature, 2013, 500, 232–236, DOI:10.1038/nature12331.
- D. Dahiya and P. S. Nigam, Int. J. Mol. Sci., 2023, 24, 5748, DOI:10.3390/ijms24065748.
- K. J. Jung, E. K. Lee, J. Y. Kim, Y. Zou, B. Sung, H. S. Heo, M. K. Kim, J. Lee, N. D. Kim, B. P. Yu and H. Y. Chung, Inflamm. Res., 2009, 58, 143–150, DOI:10.1007/s00011-008-7227-2.
- B. L. Holloman, A. Cannon, K. Wilson, P. Nagarkatti and M. Nagarkatti, mBio, 2023, 14, 0313722, DOI:10.1128/mbio.03137-22.
- A. Bartley, T. Yang, R. Arocha, W. L. Malphurs, R. Larkin, K. L. Magee, T. W. Vickroy and J. Zubcevic, Front. Physiol., 2018, 9, 1593 CrossRef PubMed.
- M. Djouina, C. Vignal, A. Dehaut, S. Caboche, N. Hirt, C. Waxin, C. Himber, D. Beury, D. Hot, L. Dubuquoy and D. Launay, Environ. Res., 2022, 212, 113230 CrossRef CAS PubMed.
- E. Fournier, M. Leveque, P. Ruiz, J. Ratel, C. Durif, S. Chalancon, F. Amiard, M. Edely, V. Bezirard, E. Gaultier, B. Lamas, E. Houdeau, F. Lagarde, E. Engel, L. Etienne-Mesmin, S. Blanquet-Diot and M. Mercier-Bonin, J. Hazard. Mater., 2023, 442, 130010, DOI:10.1016/j.jhazmat.2022.130010.
- R. K. Weersma, A. Zhernakova and J. Fu, Gut, 2019, 69(8), 1510–1519 CrossRef.
| This journal is © The Royal Society of Chemistry 2024 |