Open Access Article
Open Access ArticleThe positional and numerical effect of N6-methyladenosine in tracrRNA on the DNA cleavage activity of Cas9†
Binh Thanh Tran‡
a,
Seulgi Go‡a,
Kyoung-Ran Kim‡a,
Hak Suk Chung ab and
Dae-Ro Ahn
ab and
Dae-Ro Ahn *ab
*ab
aChemical and Biological Integrative Research Center, Korea Institute of Science and Technology (KIST), Hwarangno 14-gil 5, Seongbuk-gu, Seoul 02792, Republic of Korea. E-mail: drahn@kist.re.kr
bDivision of Bio-Medical Science and Technology, KIST School, University of Science and Technology (UST), Seoul 02792, Republic of Korea
First published on 27th June 2024
Abstract
Post-transcriptional modifications on the guide RNAs utilized in the Cas9 system may have the potential to impact the activity of Cas9. In this study, we synthesized a series of tracrRNAs containing N6-methyadenosine (m6A), a prevalent post-transcriptional modification, at various positions. We evaluated the effect of these modifications on the DNA cleavage activity of Cas9. Our results show that multiple m6As in the anti-repeat region of tracrRNA reduce the DNA cleavage activity of Cas9. This suggests that the m6A-modified tracrRNA can be used for Cas9 only when the number and the position of the modified residue are properly chosen in tracrRNA.
Introduction
CRISPR-Cas9, originally reported as an adaptive immune system of archaea and bacteria to defend against viruses, has been extensively employed for gene editing technology.1–3 The gene editing activity of Cas9, a nuclease capable of double strand breakage, is mediated by its complexing with guide RNAs such as crRNA and tracrRNA. When these guide RNAs are introduced into cells for genome engineering, they are susceptible to the intracellular environment, which can impact their stability and the gene editing activity of Cas9. Chemically modified nucleotides have been incorporated to enhance nuclease resistance and the intracellular stability of RNA-based therapeutics.4 In addition to degradation by nucleases, internalized guide RNAs may also elicit an innate immune response. Modifications in RNAs can also contribute to avoiding an undesired immune response as seen in mRNA vaccines modified with N1-methylpseudouridine (m1Ψ), a naturally occurring RNA modification, for decreased innate immune stimulation.5Besides m1Ψ, other endogenously found RNA modifications can also be considered for preparation of low immunogenic guide RNAs.6 In this context, N6-methyladenosine (m6A) is a potential modification for immune evasion, as m6A is the most abundant RNA modification in eukaryotes, accounting for 0.1–0.4% of total adenosines in RNA of mammalian cells.7,8 Indeed, exogenous viral RNAs can be N6-methylated on adenosines by the host RNA methyltransferases for evasion of innate immune responses of host cells.9,10
Recent studies showed that guide RNAs incorporated with modified bases including m6A may compromise the activity of Cas9 while they reduce immune response and cytotoxic effects of guide RNAs.11–13 However, the numerical and positional effect of m6A in guide RNAs still remains elusive as the previous studies used modified single guide (sgRNA) prepared by in vitro transcription in which incorporation of the modified nucleotide at desired positions is not possible. Information on the numerical and positional effect would guide researchers to design m6A-modified sgRNA for reduced immunogenicity without compromising Cas9 activity.
To investigate the numerical and positional effect of m6A on the nuclease activity of Cas9, sequence-specific incorporation of m6A into guide RNAs is necessary. TracrRNA, which has an identical sequence regardless of DNA target sequences, is a more suitable guide RNA for this study than crRNA or sgRNA, which have variable sequences depending on the target. We here synthesized various tracrRNAs containing m6As at different positions via ligation of two RNA oligonucleotides synthesized on solid supports using a DNA/RNA synthesizer. We performed in vitro DNA cleavage reaction of Cas9 complexed with crRNA and the m6A-modified tracrRNAs to assess the positional and numerical effect of m6A in tracrRNA on Cas9 activity. In addition, cellular activity of Cas9 with the m6A-modified tracrRNAs was also verified.
Results and discussion
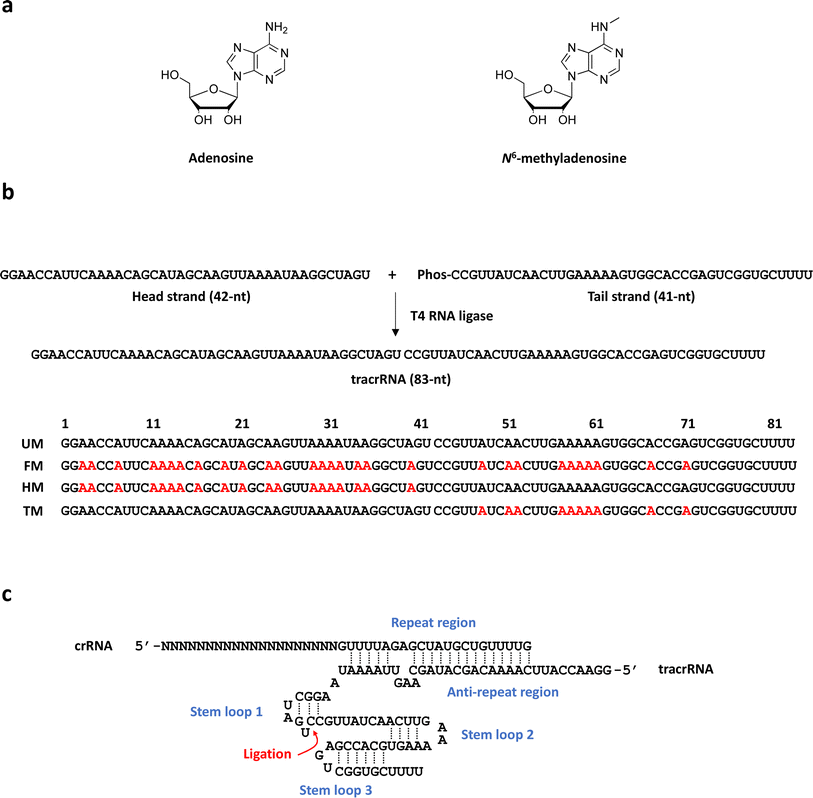
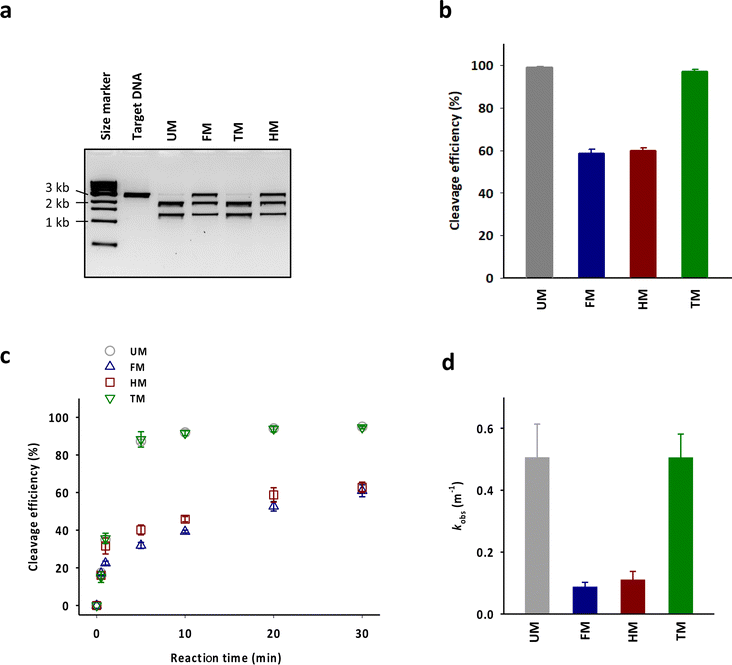
83-mer tracrRNAs were prepared by ligation of two RNA strands, the 42-mer head strand and the 41-mer tail strand that were chemically synthesized using the conventional solid phase-based oligonucleotide synthesis (Fig. 1a and b). Ligating head and tail strands containing m6As produced tracrRNAs modified with m6A at various positions (Table S1 and Fig. S1†). The head strand includes the anti-repeat region, which hybridizes with the repeat region of crRNA and a part of stem loop 1. The tail strand comprises the remaining part of stem loop 1, stem loop 2, and stem loop 3 (Fig. 1c).The cleavage efficiency of a linearized DNA plasmid substrate containing a target sequence by a ribonucleoprotein (RNP) complex composed of Cas9, crRNA, and tracrRNA was studied (Fig. 2a). The results showed that N6-methylation of all 29 adenosines in the tracrRNA (FM) resulted in a significantly lower cleavage efficiency compared to the unmodified tracrRNA (UM) (Fig. 2b). This indicates that m6As in tracrRNA can interfere the in vitro DNA cleavage activity of Cas9, consistent with the previous report.12 To examine the positional effect of m6As on the regulation of the activity, the DNA cleavage activity of Cas9 was tested using tracrRNA with m6As in the head part (HM) or tail part (TM). The results showed that HM reduced the activity, while TM was similarly active as UM (Fig. 2a and b). The cleavage rate (kobs) was also obtained from the time-dependent reaction data and fitting to the equation for single-turnover conditions (Fig. 2c, d, and S2†). The cleavage rate of Cas9 with TM was similar to that with native UM, and the cleavage rate of Cas9 with HM was similar to that with native FM. The reactions with HM and FM were similarly slower than those with UM and TM, with a rate approximately five times lower (Fig. 2d).
To investigate the positional impact of the single m6A on the DNA cleavage activity of Cas9, single m6A-modified tracrRNAs were synthesized by replacing one of 19 adenosines in the head part of tracrRNA (SM1 to SM19) (Fig. 3a). The results showed that the DNA cleavage efficiency of Cas9 with all single m6A-modified tracrRNAs was slightly lower than that with UM (Fig. 3b and S3†). The positional effect of the single m6A in tracrRNA was not observed.
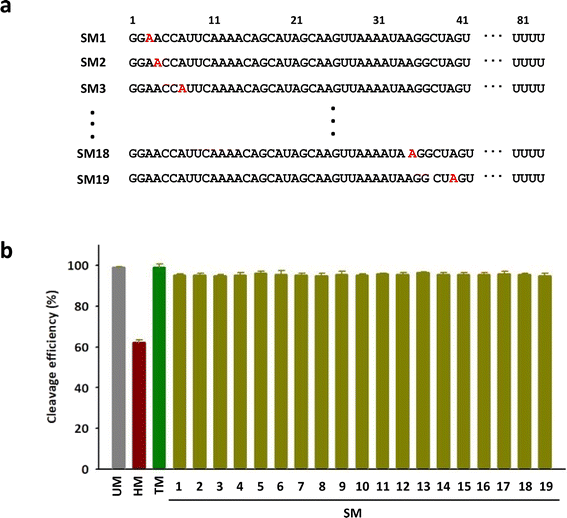 | ||
| Fig. 3 (a) The sequences of tracrRNAs containing a single m6A. (b) The DNA cleavage efficiency of Cas9 with tracrRNAs quantified based on the band intensity in Fig. S3.† The data are an average of three independent experiments with standard deviations. | ||
Next, we attempted to verify the number and the position of m6As in the head part required to exhibit the activity comparable to that of HM (Fig. 4a). We mainly focused on the adenosines in the anti-repeat region of tracrRNA, which play a critical role in forming the RNA duplex with the crRNA repeat region. As shown in Fig. 4b and S4,† the tracrRNAs containing seven m6As in the base-pairing region (M3) showed reduced activity compared to the tracrRNA with four base-paired m6As (M1 and M2). The activity of M5, which contained six m6As (four paired and two unpaired) was similar to that of M3. M4, with two unpaired m6As, showed slightly lower activity than unmodified tracrRNA. M6, with a total of 13 m6As (from M2 and M5), showed activity comparable to that of HM containing 19 m6As. These results suggest that multiple m6As in the anti-repeat region are required to significantly reduce the Cas9 cleavage activity, while the N6-methylation of unpaired adenosines in the head part may be less critical for activity than the modification in paired adenosines. The reduced activity was roughly proportional to the number of m6As, with a decrease in activity observed as the number of m6As increased, indicating that cumulated number of m6As in the anti-repeat region could lead to significant decrease of the cleavage activity of Cas9 (Fig. 4c).
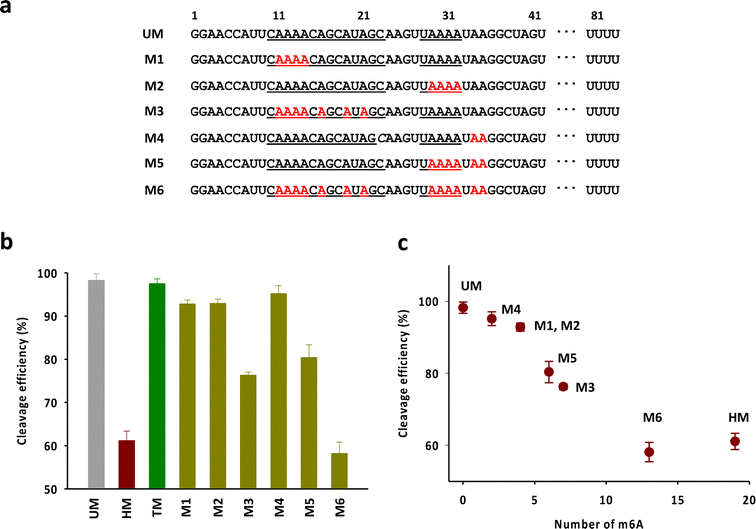 | ||
| Fig. 4 (a) The sequences of tracrRNAs containing multiple m6As in the anti-repeat region. (b) The DNA cleavage efficiency of Cas9 with tracrRNAs quantified based on the band intensity in Fig. S4.† The data are an average of three independent experiments with standard deviations. (c) Correlation between the cleavage efficiency and the number of m6A in the anti-repeat region of tracrRNA. | ||
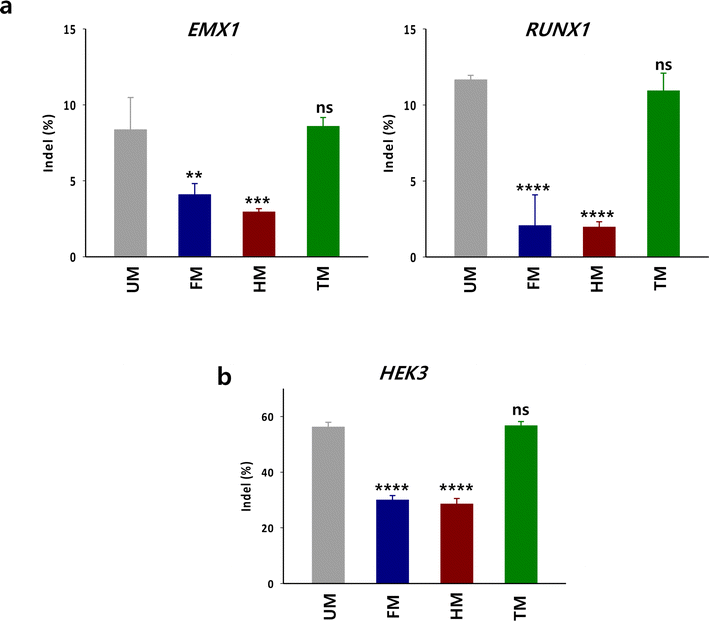
To examine whether the in vitro DNA cleavage activity with TM is also effective for targeted gene disruption by Cas9 in cells, HEK293 cells were treated with the RNP complex composed of Cas9, crRNA targeting endogenous genes (RUNX1 or EMX1) and TM (Fig. 5a). The indel levels in the target sequences were evaluated by Tracking of Indels by Decomposition (TIDE) analysis.14 The indel level in the target by the RNP with TM was comparable to that with unmodified tracrRNA (UM). In contrast, the gene disruption activity of RNP with HM and FM was substantially lower than that with UM. We evaluated gene disruption level by the m6A-modified tracrRNAs in another cell line such as K562 cells. The gene disruption tendency in K562 cells was consistent with the observations in HEK293 cells (Fig. 5b). The indel level at HEK3 by TM was comparable to that by UM and was higher than that by HM and FM in K562 cells. These results illustrate that the m6A-dependent in vitro DNA cleavage activity of Cas9 translates into gene disruption activity within cellular environments. Thus, it is recommended to replace only the 10 adenosines with m6As in the tail part of tracrRNA when designing m6A-modified tracrRNAs.
The N6-methylation on adenosines is known to weaken the A–U base pair possibly by removing one of the two hydrogens on the exocyclic amine alternatively participating in the base pair with uridines and thereby destabilizes the duplex hybridization.15,16 An increased number of paired m6As up to 11 in the head part would in general lower the stability of the crRNA-tracrRNA complex formed via an intermolecular duplex, which could lead to decreased DNA cleavage activity of Cas9. In contrast, destabilization of the stem loops by 4 paired m6As (three in stem loop 2 and one in stem loop 3) in the tail part may not be effective enough to change the stem loop structures as the intramolecular hairpins have relatively high stability. In addition, all adenosines in the tail part of tracrRNA, with the exception of one, do not engage in interactions with Cas9.17 This suggests that m6As have a minimal effect on the interaction between the tail part and Cas9, and therefore on the DNA cleavage activity of Cas9.
The previously known consensus RNA sequence for cellular m6A modifications is DRACH where D, R, and H respectively stand for A/G/U, A/G, and A/C/U.18,19 Among the adenosines in the anti-repeat region, only two residues fall within the DRACH motif. Therefore, it is unlikely that intracellularly delivered tracrRNA significantly decrease Cas9 activity via endogenous N6-methylation on adenines in tracrRNA.
Incorporating methylated residues into RNA-based therapeutics, such as mRNA and siRNA, has been shown to improve their intracellular stability and immune evasion.20,21 The methylation on 2′-hydroxyl group enhances nuclease resistance while that on nucleobases including m6A decreases the immunogenicity of RNAs.22 Identifying the numerical and positional effect of m6As in tracrRNA on the activity of Cas9 is important for clinical application of Cas9 to avoid the potential trade-off between gene editing activity and immune evasion. Incorporating as many m6As as possible into positions that do not interfere the DNA cleavage activity of Cas9 could suppress undesired immune stimulation without compromising the in vivo therapeutic efficacy of the gene editing system.
Conclusion
In this study, we demonstrated that there is the positional and numerical effect of m6As in tracrRNA on the DNA cleavage activity of Cas9. Our findings indicate that multiple m6As in the anti-repeat region of tracrRNA, which hybridizes with the repeat region of crRNA, decrease the DNA cleavage activity of Cas9 whereas m6As in other positions of tracrRNA has little impact on the activity. The cellular activity of Cas9 could be maintained when the number and the position of m6As are properly chosen in tracrRNA. These results could be utilized for design of tracrRNAs for gene therapy applications, as they provide information on how to modulate the cellular activity and immune evasion properties of guide RNAs without compromising the gene editing efficacy of Cas9.Data availability
The data supporting this article have been included as part of the ESI.†Author contributions
B. T. T. and K.-R. K. synthesized m6A-modified tracrRNAs. B. T. T. performed in vitro DNA cleavage experiments. H. S. C. expressed and purified recombinant Cas9. K.-R. K. and S. G. performed cellular gene disruption experiments. S. G. analyzed indel frequencies. D.-R. A. designed the study, and wrote the manuscript.Conflicts of interest
There are no conflicts to declare.Acknowledgements
We thank Dr Gayeong Lim for providing the K562 cells, which were used to evaluate cellular activity of RNP complexes. This study was supported by intramural research programs of the Korea Institute of Science and Technology (KIST) (2E33132) and the Bio & Medical Technology Development Program of the National Research Foundation (NRF) funded by the Korean government (MSIT) (2022M3E5F1018118, RS-2024-00396224, RS-2024-00397042).References
- J. A. Doudna and E. Charpentier, Science, 2014, 346, 1258096 CrossRef PubMed.
- P. D. Hsu, E. S. Lander and F. Zhang, Cell, 2014, 157, 1262 CrossRef CAS PubMed.
- H. Kim and J.-S. Kim, Nat. Rev. Genet., 2014, 15, 321 CrossRef CAS PubMed.
- A. Hendel, R. Bak, J. Clark, A. Kennedy, D. Ryan, S. Roy, I. Steinfeld, B. Lunstad, R. Kaiser, A. Wilkens, R. Bacchetta, A. Tsalenko, D. Dellinger, L. Bruhn and M. Porteus, Nat. Biotechnol., 2015, 33, 985 CrossRef CAS PubMed.
- D. Nance and J. L. Meier, ACS Cent. Sci., 2021, 7, 748 CrossRef PubMed.
- P. J. McCown, A. Ruszkowska, C. N. Kunkler, K. Breger, J. P. Hulewicz, M. C. Wang, N. A. Springer and J. A. Brown, Wiley Interdiscip. Rev.: RNA, 2020, 11, e1595 CrossRef CAS PubMed.
- D. T. Dubin and R. H. Taylor, Nucleic Acids Res., 1975, 2, 1653 CrossRef CAS PubMed.
- C. M. Wei, A. Gershowitz and B. Moss, Cell, 1975, 4, 379 CrossRef CAS PubMed.
- D. G. Courtney, E. M. Kennedy, R. E. Dumm, H. P. Bogerd, K. Tsai, N. S. Heaton and B. R. Cullen, Cell Host Microbe, 2017, 22, 377 CrossRef CAS PubMed.
- N. Li, H. Hui, B. Bray, G. M. Gonzalez, M. Zeller, K. G. Anderson, R. Knight, D. Smith, Y. Wang, A. F. Carlin and T. M. Rana, Cell Rep., 2021, 35, 109091 CrossRef CAS PubMed.
- H. Yang, E. Eremeeva, M. l. Abramov, M. Jacquemyn, E. Groaz, D. Daelemans and P. Herdewijn, Nucleic Acid Res., 2023, 51, 1501 CrossRef CAS PubMed.
- D. V. Prokhorova, I. P. Vokhtantsev, P. O. Tolstova, E. S. Zhuravlev, L. M. Kulishova, D. O. Zharkov and G. A. Stepanov, CRISPR J., 2022, 5, 799 CrossRef CAS PubMed.
- D. Prokhorova, A. Matveeva, A. Zakabunin, A. Ryabchenko and G. Stepanov, Int. J. Mol. Sci., 2023, 24, 17116 CrossRef CAS PubMed.
- E. K. Brinkman, T. Chen, M. Amendola and B. V. Steensel, Nucleic Acids Res., 2014, 42, e168 CrossRef PubMed.
- E. Kierzek and R. Kierzek, Nucleic Acid Res., 2003, 31, 4472 CrossRef CAS PubMed.
- C. Roost, S. Lynch, P. J. Batista, K. Qu, H. Y. Chang and E. T. Kool, J. Am. Chem. Soc., 2015, 137, 2107 CrossRef CAS PubMed.
- H. Nishimasu, F. A. Ran, P. D. Hsu, S. Konermann, S. I. Shehata, N. Dohmae, R. Ishitani, F. Zhang and O. Nureki, Cell, 2014, 156, 935 CrossRef CAS PubMed.
- T. Csepany, A. Lin, C. J. Baldick Jr and K. Beemon, J. Biol. Chem., 1990, 265, 20117 CrossRef CAS PubMed.
- J. E. Harper, S. M. Miceli, R. J. Roberts and J. L. Manley, Nucleic Acids Res., 1990, 18, 5735 CrossRef CAS PubMed.
- S. C. Devarkar, C. Wang, M. T. Miller and J. Marcotrigiano, Proc. Natl. Acad. Sci. U.S.A., 2016, 113, 596 CrossRef CAS PubMed.
- A. D. Judge, G. Bola, A. C. H. Lee and I. MacLachlan, Mol. Ther., 2006, 13, 494 CrossRef CAS PubMed.
- X. Lou, J. Wang, Y. Wei and J. Sun, Cell Death Dis., 2021, 12, 300 CrossRef CAS PubMed.
Footnotes |
| † Electronic supplementary information (ESI) available. See DOI: https://doi.org/10.1039/d4ra03957b |
| ‡ These authors contributed equally. |
| This journal is © The Royal Society of Chemistry 2024 |