Open Access Article
Open Access ArticleBiogenic fabrication of a gold nanoparticle sensor for detection of Fe3+ ions using a smartphone and machine learning†
Kim-Phuong T. Dang‡
a,
T. Thanh-Giang Nguyen‡a,
Tien-Dung Cao*b,
Van-Dung Leac,
Chi-Hien Dangac,
Nguyen Phuc Hoang Duya,
Pham Thi Thuy Phuongac,
Do Manh Huya,
Tran Thi Kim Chid and
Thanh-Danh Nguyen *ac
*ac
aInstitute of Chemical Technology, Vietnam Academy of Science and Technology, Ho Chi Minh City, Vietnam. E-mail: ntdanh@ict.vast.vn; danh5463bd@yahoo.com
bSchool of Information Technology, Tan Tao University, Long An, Vietnam. E-mail: dung.cao@ttu.edu.vn
cGraduate University of Science and Technology, Vietnam Academy of Science and Technology, 18 Hoang Quoc Viet, Cau Giay District, Hanoi, Vietnam
dInstitute of Materials Science, Vietnam Academy of Science and Technology, 18 Hoang Quoc Viet, Cau Giay District, Hanoi, Vietnam
First published on 27th June 2024
Abstract
In recent years, smartphones have been integrated into rapid colorimetric sensors for heavy metal ions, but challenges persist in accuracy and efficiency. Our study introduces a novel approach to utilize biogenic gold nanoparticle (AuNP) sensors in conjunction with designing a lightbox with a color reference and machine learning for detection of Fe3+ ions in water. AuNPs were synthesized using the aqueous extract of Eleutherine bulbosa leaf as reductants and stabilizing agents. Physicochemical analyses revealed diverse AuNP shapes and sizes with an average size of 19.8 nm, with a crystalline structure confirmed via SAED and XRD techniques. AuNPs exhibited high sensitivity and selectivity in detection of Fe3+ ions through UV-vis spectroscopy and smartphones, relying on nanoparticle aggregation. To enhance image quality, we developed a lightbox and implemented a reference color value for standardization, significantly improving performance of machine learning algorithms. Our method achieved approximately 6.7% higher evaluation metrics (R2 = 0.8780) compared to non-normalized approaches (R2 = 0.8207). This work presented a promising tool for quantitative Fe3+ ion analysis in water.
1 Introduction
Iron, abundant among transition metal ions of Earth, is predominantly found in its Fe3+ state in rust.1–3 Iron ions (Fe2+ and Fe3+) play vital roles in various metabolic and intracellular activities, serving as cofactors for enzymes and proteins involved in oxygen and electron transport, DNA synthesis, and catalyzing oxido-reductase processes.4–9 While integral to biological functions, iron distribution requires careful regulation due to its potential for harm,10–12 implicated in conditions such as encephalopathy, Parkinson's, Alzheimer's, and Huntington's diseases.13,14 Wastewater discharged from iron and steel industries contributes significantly to water contamination, posing serious health risks to organisms, causing damage at cellular, tissue, and organ levels.15,16Various analytical methods have been developed for detecting iron ions, including spectrophotometry, stripping voltammetry, fluorescent probes, atomic absorption spectroscopy, and luminol flow injection analysis.17–19 Particularly, optical sensors stand out for their cost-effectiveness. However, traditional optical sensors necessitate a well-equipped laboratory with expensive instruments like UV-vis and photoluminescence spectroscopy, as well as skilled personnel, resulting in high costs and time consumption. Consequently, there's a growing interest in easy-to-use, portable optical sensors that can swiftly analyze substances in the field without expert intervention.20–22 The integration of smartphones and new technologies such as Artificial Intelligence, Internet of Things, Big data, holds immense potential for revolutionizing optical analysis, reducing costs, enhancing economic value, and improving quality of life.23 Optical sensors, when coupled with image analysis employing machine learning, have garnered significant attention in research circles worldwide.24,25 This approach leverages smartphone cameras for image analysis to track changes in analyte concentrations interacting with optical sensors.26–28 Moreover, employing machine learning algorithms to process these images enhances the accuracy of analytical methods, promising precise on-site analysis of hazardous chemicals. However, challenges persist in utilizing smartphone cameras, influenced by external factors. These factors can lead to erroneous predictions by machine learning algorithms.29,30
Gold nanoparticles (AuNPs) have seen extensive research and utilization in various applications, including sensing, imaging, therapeutics, diagnostics, drug delivery, catalysis, and surface-enhanced Raman spectroscopy.31–33 Their unique physical and chemical properties, such as morphology-dependent electronics, surface chemistry, and compatibility with surface functionalization, have garnered significant attention.2,34 Morphology-dependent Surface Plasmon Resonance (SPR) absorption of AuNPs has been widely harnessed as an analytical tool for colorimetric sensing across biotechnological and chemical systems, detecting ions, anions, nucleic acids, proteins, and peptides.35–37 In particular, colorimetric sensing of metal ions using AuNPs-based chemosensors shows promise, with numerous studies focusing on designing AuNPs as probes for detecting various compounds and metal ions in environmental samples.38,39 Several studies have explored the use of different AuNPs for colorimetric detection of Fe3+ ions.40–42 However, traditional methods for producing UV-based AuNPs chemosensors often rely on costly and toxic reducing and stabilizing agents. Recently, there has been a growing interest in green synthesis methods for AuNPs in sensing applications. These methods are environmentally friendly, cost-effective, and avoid the use of toxic chemical agents. While some studies have successfully utilized plant extracts for AuNPs synthesis, the application of green-synthesized AuNPs for colorimetric detection of transition metal ions remains underexplored.43,44
Eleutherine bulbosa (EB), a herbaceous plant belonging to the Iridaceae family, is widely cultivated across South America, Africa, and Southeast Asia. Compounds extracted from E. bulbosa include xanthones, fatty acid esters, naphthalenes, phenolics, flavonoids, isoquinolines, anthraquinone, naphthoquinone, tannins, saponins, quinones, steroids, and triterpenoids.45–47 These compounds serve as both reducing and stabilizing agents in the synthesis of metallic nanoparticles.48–50
In this study, we utilized an aqueous extract of E. bulbosa leaves for the green synthesis of AuNPs. These synthesized AuNPs were then used to detect Fe3+ ions through UV-vis spectroscopy and smartphone-based analysis as follows: (i) Fe3+ ions interact with the surface of AuNPs, inducing nanoparticle aggregation and altering their SPR bands and color intensity, which correlate with the Fe3+ ion concentration; (ii) images of the solution cuvette are captured using a smartphone camera; (iii) these images are analyzed to extract color values for a machine learning model; (iv) a linear regression model is employed to correlate these color values with Fe3+ ion concentrations. The method performance is significantly influenced by image quality, affected by factors such as lighting conditions, smartphone camera quality, and user experience. To mitigate these variables, a lightbox with a reference color was designed. Each image is normalized using this reference color, adjusting brightness to standardize color values under experimental conditions. This approach improves the performance of the machine learning algorithm compared to non-normalized methods.
2 Materials and methods
2.1. Materials
All reagents and materials were employed without additional purification. Hydrogen tetrachloroaurate(III) hydrate (HAuCl4·3H2O) were purchased from Acros (Belgium). E. bulbosa leaves were collected in the Thien Cam Son Mountain area, An Giang Province, Vietnam during May–August, 2022. Distilled water was used throughout.2.2. Plant extract preparation
The E. bulbosa leaves were dried in a drying oven for 24 h and finely ground by an electronic blender. The resultant powder (20 g) was refluxed with water (200 mL) for 1.5 h. The mixture was filtered via a whatman paper and the dark green filtrate was storable in the refrigerator at 4 °C for further studies.2.3. Biosynthesis of gold nanoparticles
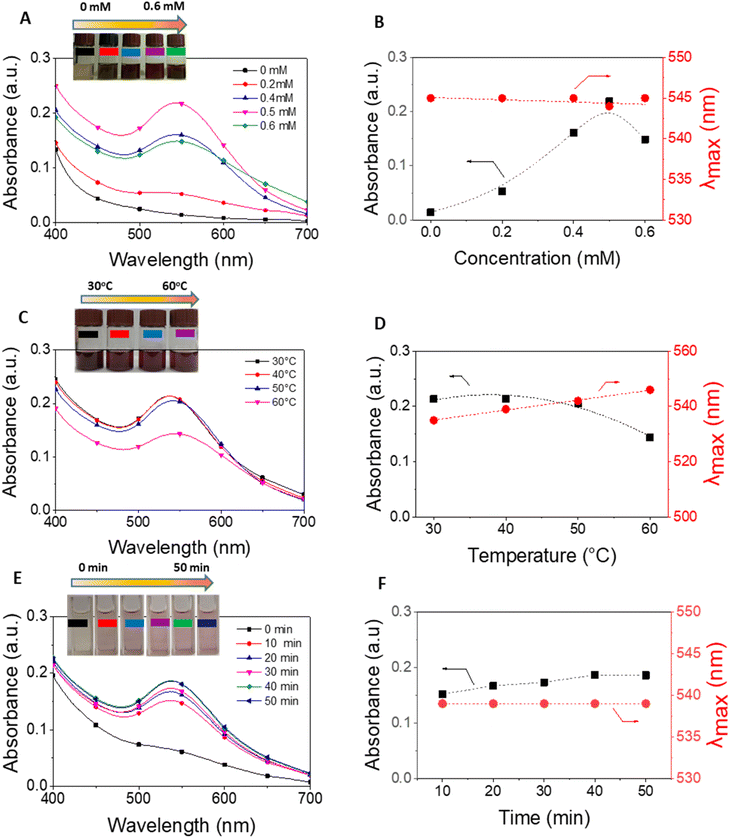
The E. bulbosa extract was stirred in Au3+ ion solution for 30 min at 1200 rpm in a dark condition. The formation of gold nanoparticles was visible due to color changes. UV-vis spectra in range from 200 to 800 nm was used to investigate the optimization of reaction parameters including metallic ion concentration (0.0, 0.2, 0.4, 0.5 and 0.6 mM), reaction temperature (in the range of 30–60 °C), and reaction time (10, 20, 30, 40, 50 min). The reduction of Au3+ ions by the plant extract resulted in an increase in absorbance at around 537 nm. The AuNPs synthesized by E. bulbosa extract (EB-AuNPs) in the optimal circumstances were used for further studies. The crystalline EB-AuNPs were obtained by centrifugation at 8500 rpm for 30 min and then washed twice with water to eliminate the impurities. Finally, the powder of EB-AuNPs was obtained after spreading and drying in the Petri dish under the fume hood for 1.5 h.2.4. Physicochemical characterizations of gold nanoparticles
The optimized samples were employed for exploring the physicochemical characterization and its various applications. UV-vis spectra were measured on a JASCO V-630 spectrophotometer (USA) The Fourier-transform infrared (FTIR) spectra of solid E. bulbosa extract and synthesized AuNPs were captured by FT-IR spectrometer, PerkinElmer, UK at wavelengths spanning from 400 to 4000 cm−1. Chemical elements of the solid AuNPs sample were analyzed by energy-dispersive X-ray (EDX) spectroscopy on JEOL JEM2100. Crystalline characterization was conducted using XRD patterns (Model-D8 Advance, Bruker, Germany) measuring directly powder EB-AuNPs. To examine the morphology of the obtained AuNPs, field emission scanning electron microscopy (FESEM) was utilized, using the HITACHI S-4800 (Japan). The investigation of nano size, crystalline and elemental distribution was conducted using advanced techniques including high-resolution transmission electron microscopy (HRTEM), selected area electron diffraction (SAED), and scanning transmission electron microscopy mapping (STEM-mapping) with the JJEOL JEM2100 instrument. NanoPartica Horiba SZ-100 (Japan), which determined electrical potential at the slipping plane and particle size distribution, was utilized to assess zeta potential and dynamic light scattering (DLS) using gel solutions (1.0 mg mL−1).2.5. Colorimetric detection of Fe3+ ion
The colorimetric detection was performed with as-prepared AuNPs solution (1 mL). To evaluate selectivity, the colorimetric sensing ability of EB-AuNPs was individually examined for a range of environmentally significant metal ions. The metallic ions including Ba2+, Ca2+, Ni2+, Al3+, Zn2+, Fe2+, Fe3+, Pb2+, Mn2+, Cd2+ and Cr2+ were carried out at a consistent concentration of 10 ppm. For UV-vis sensor, AuNPs solution mixed with various concentrations of Fe3+ ion (0.3–30.0 ppm) were measured by UV-vis spectroscopy. The absorption intensity of peaks versus the concentration of Fe3+ ions were plotted.2.6. Smartphone-based sensor
The red – green – blue (RGB) colorimetry method for solution concentrations using smartphones often faces challenges from external factors like varying light intensity, camera resolution, and internal color adjustments. To overcome these obstacles, a novel approach was developed to detect the region of interest (ROI). This involved designing a lightbox as a closed system, incorporating an LED light system within a metal frame, as depicted in Fig. 1. The smartphone and cuvette were positioned at a fixed distance of 6 cm and held in specific orientations. A small green square frame, serving as a reference color value, was placed in front of the cuvette. The region inside this frame, known as the ROI, was identified based on the presence of the green border.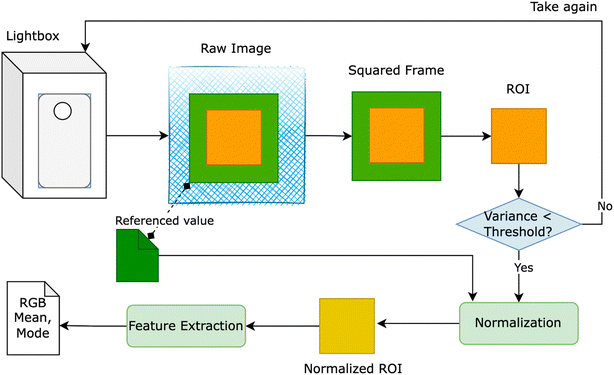 | ||
| Fig. 1 Light box – a closed system for measuring RGB value to extract the features from the raw image taken by smartphone. | ||
Fig. 1 provides an overview of our proposed solution, wherein users were prompted to retake the image if the variance of ROI exceeded a predetermined threshold. Despite the apparent closure of the lightbox system for smartphone image capture, the RGB values of the ROI exhibited inconsistency when multiple images of a solution with identical concentrations were taken using the same smartphone. To rectify this inconsistency, we utilized the reference value of the green squared frame to adjust the overall color of the image. Two normalization methods were devised:
(1) Delta method involved adjusting the mean RGB values of the ROI by adding the difference between the color value of the green squared frame and the RGB reference values (e.g., R = 40, G = 150, B = 90) to the measured mean RGB values in the region of color adjustment. The formula is expressed as eqn (1).
| Mean_RGB_ROI = mean_RGB_ROI + (mean_RGB_squared_frame − RGB_predefined) | (1) |
(2) Ratio method: the mean RGB values of the ROI underwent adjustment by multiplying the ratio between the color value of the green squared frame and the RGB reference values (e.g., R = 40, G = 150, B = 90) by the mean RGB values measured in the region of color adjustment. The formula is expressed as eqn (2)
| Mean_RGB_ROI = mean_RGB_ROI × (mean_RGB_squared_frame/RGB_predefined) | (2) |
Finally, six features were extracted from the ROI corresponding to mean and mode of RGB, then use a regression algorithm to learn the correlation between RGB value of images and concentration. The completed solution, i.e., data processing, normalization, visualization, machine learning model, was implemented by Python and its libraries: OpenCV2,51 Seaborn,52 Scikit-learn.53
3 Results and discussion
3.1. Biosynthesis of gold nanoparticles
The study methodology of our work is illustrated in Fig. 2A. Initially, dried E. bulbosa leaves were refluxed with water for 1.5 hours, resulting in a dark green solution post-filtration. To determine the optimal synthetic conditions, various reaction parameters such as metallic ion concentration, reaction temperature, and reaction time were systematically altered to enhance the stability of EB-AuNPs synthesis. Color changes in the colloidal solution, along with UV-vis measurements, confirmed the formation of EB-AuNPs.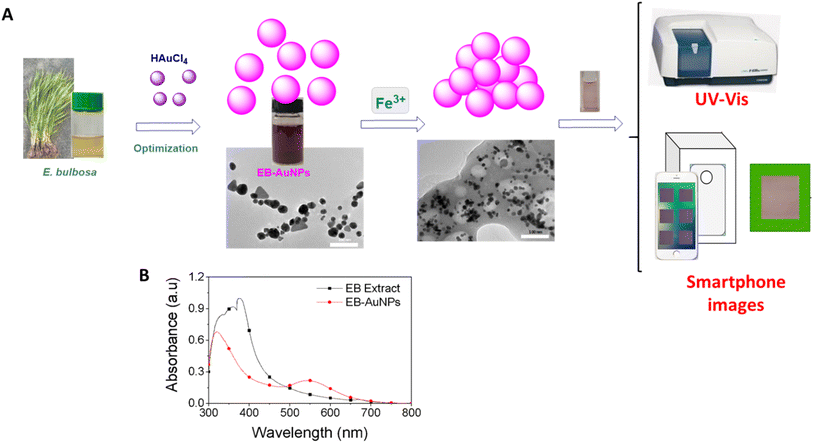 | ||
| Fig. 2 Schematic illustration of the study strategy (A) and UV-vis spectra of the extract and EB-AuNPs (B). | ||
UV-vis spectra revealed an intense peak at 330–400 nm, indicative of aromatic chemical transitions of polyphenols, including π → π* and π → n* (Fig. 2B). This peak signifies the presence of polyphenols, such as flavonoids, responsible for reducing gold ions into nanoparticles. The colloidal solutions of EB-AuNPs exhibited the SPR bands around 537 nm. Interestingly, the disappearance of aromatic chemical transitions in the EB-AuNPs spectrum suggests the conversion of polyphenols and gold ions into AuNPs. Following synthesis, the EB-AuNPs underwent washing and collection via centrifugation. Subsequently, physicochemical analytical techniques were employed to characterize the synthesized nanoparticles and facilitate colorimetric detection of Fe3+ ions in water using UV-vis spectroscopy and smartphone cameras combined with machine learning.
The transformation of metallic ions into nanoparticles exerts a profound influence on the size and morphology of AuNPs stabilized by plant extract. In this study, we delved into optimizing three key parameters including the concentration of gold ions, reaction temperature, and reaction time, to achieve the most effective synthesis of EB-AuNPs, leveraging absorption spectra analysis. Alterations in absorbance intensity and surface plasmon resonance (SPR) peaks serve as valuable indicators of physicochemical changes.54
Fig. 3 illustrates UV-vis spectra and plots depicting absorbance and λmax values against the varied parameters. Our findings underscore the pivotal role of gold ion concentrations in AuNP production. Despite fluctuations in salt concentrations from 0 to 0.6 mM, the maximum absorption values of the SPR band consistently hovered around 545 nm, suggesting negligible influence on the size and morphology of the resultant EB-AuNPs (Fig. 3A and B). However, discernible shifts in absorbance values within the SPR bands were apparent, particularly with increasing salt concentrations. Notably, the peak absorption density peaked at a salt concentration of 0.5 mM, beyond which AuNP agglomeration in the colloidal solution led to decreased absorbance.
Further investigation into the effect of reaction temperature, ranging from 30 to 60 °C, revealed intriguing trends (Fig. 3C and D). At 30 °C, a pronounced absorption of SPR bands was observed, indicative of abundant AuNPs in the colloid solution. However, as the reaction temperature escalated, the absorption intensity declined. Nonetheless, λmax values exhibited a slight increment over the course of the investigation. The heightened mobility of metallic molecules at elevated temperatures likely induced an enlargement in size and alteration in morphology of the formed AuNPs, contributing to their coagulation and subsequent decrease in absorbance values.
UV-vis spectra were measured at 10 minutes intervals to assess the impact of reaction time on EB-AuNP biosynthesis (Fig. 3E and F). Our findings underscore the critical role of reaction time in EB-AuNP formation. Initially, a distinct absorption peak of AuNPs emerged within the first 10 minutes, followed by a gradual synthesis process. The highest absorbance value was attained after 40 minutes of synthesis. Remarkably, despite this temporal evolution, λmax values remained largely constant, suggesting stable morphology of the biosynthesized EB-AuNPs. Based on these observations, the synthesis of EB-AuNPs for further physicochemical characterizations and applications was conducted under optimal conditions including Au3+ ion concentration of 0.5 mM, temperature of 30 °C, and reaction time of 40 minutes.
3.2. Physicochemical characterization
FTIR analysis of both E. bulbosa leaf extract and the synthesized EB-AuNPs is used to determine the role of organic compounds from the plant extract in nanoparticle synthesis, as depicted in Fig. 4. The absorption bands of the leaf extract revealed peaks at 3409, 2943, 2702, 2128, 1606, 1411, 1328, 1286, 1077, 618, and 533 cm−1. In particular, the peaks at 3409 cm−1 and 1411 cm−1 corresponded to the stretching and bending vibrations of hydroxyl (OH) groups, respectively, characteristic of polyphenols.55 Additionally, peaks at 2943 cm−1 and 1077 cm−1 were assigned to C–H and C–O–C stretching vibrations, while the peak at 1606 cm−1 related to carboxyl group (COO−) stretching vibration. Following the reduction of Au3+ ions into AuNPs, these characteristic spectra manifested as new peaks at 3294, 2918, 2851, 2356, 1653, 1242, 1012, 720, and 511 cm−1. Notably, the marked decrease in intensity at the peak of 3409 cm−1 suggested the involvement of polyphenol compounds in the aqueous extract for the reduction of gold ions.56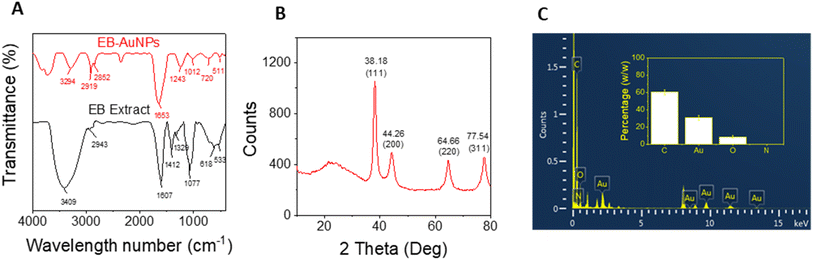 | ||
| Fig. 4 (A) FTIR spectra of E. bulbosa leaf extract and EB-AuNPs; (B) XRD pattern of synthesized EB-AuNPs and (C) EDX pattern and elemental percentage (inset) of EB-AuNPs. | ||
Powder XRD analysis was employed to assess the crystallinity of the synthesized AuNPs. Fig. 4B illustrates the XRD pattern of EB-AuNPs, revealing distinct diffraction peaks at 38.18°, 44.26°, 64.66°, and 77.54°, corresponding to the face-centered cubic (fcc) crystal planes of (1 1 1), (2 0 0), (2 2 0), and (3 1 1), respectively. These peaks confirmed the crystalline nature of EB-AuNPs, with the most intense signal indicating preferential growth in the (1 1 1) direction.57 The average crystalline size ‘D’ of the AuNPs crystal was calculated from the XRD pattern using the Debye–Scherrer equation: D = 0.9λ/β![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) cos
cos![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) θ. In this formula, λ represents the wavelength of the X-rays employed for diffraction, and β denotes the full width at half maximum (FWHM) of the diffraction peak. XRD data yielded a calculated crystal size of 6.4 nm and an estimated cell volume of approximately 68.0 Å3, further affirming the presence of AuNPs in the samples (Fig. S1†).58
θ. In this formula, λ represents the wavelength of the X-rays employed for diffraction, and β denotes the full width at half maximum (FWHM) of the diffraction peak. XRD data yielded a calculated crystal size of 6.4 nm and an estimated cell volume of approximately 68.0 Å3, further affirming the presence of AuNPs in the samples (Fig. S1†).58
EDX analysis of the EB-AuNPs revealed a prominent peak of Au near 2.2 keV,59 confirming the characteristic spectrum of AuNPs (Fig. 4C). Signals from C, O, and N elements corroborated the presence of stabilizing organic components around the AuNPs surface, such as polysaccharides and proteins. The mean content of C, Au, O, and N elements was estimated to be 60.47 ± 2.87, 30.69 ± 2.88, 8.63 ± 1.53, and 0.20 ± 0.08%, respectively. The presence of these organic components alongside a significant AuNPs content in the samples suggests promising potential for sensing applications in the detection of heavy metallic ions.
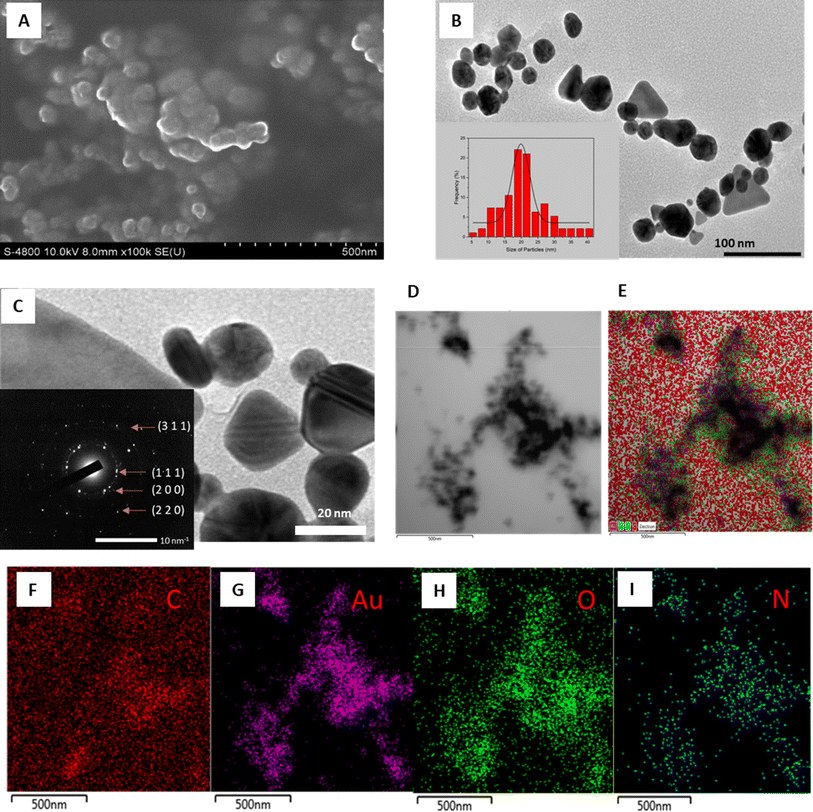
The morphological characteristics of AuNPs are thoroughly examined through various microscopic techniques, including FESEM, HRTEM, and STEM-mapping, as illustrated in Fig. 5. The FESEM image depicts densely packed AuNPs with estimated diameters below 50 nm, indicative of their compact nature (Fig. 5A). Meanwhile, the TEM image reveals a diverse array of morphologies, ranging from spherical to triangular and octagonal shapes, within a size range of 5.4–37.8 nm, with an average size of 19.8 nm (Fig. 5B). Discrepancies in reduction potentials and phytochemical compositions among different stabilizing agents of nanoparticles may account for the observed variations in size and shape of AuNPs.60
The crystalline structure of the biosynthesized AuNPs was further elucidated through SAED pattern analysis. HRTEM imaging unveils a well-defined crystal lattice, with the SAED image exhibiting bright cycles indicative of the crystalline nature of AuNPs, with preferential crystal growth occurring along the (111) plane (Fig. 5C). The observed rings correspond to reflections from planes (111), (200), (220), and (311), affirming the face-centered cubic structure inherent in the gold crystal.61
STEM-mapping images of the synthesized EB-AuNPs are presented in Fig. 5D–I, revealing the presence of elemental constituents such as Au, C, N, and O. The black particles signify gold elements, thus confirming the presence of AuNPs in the samples (Fig. 5G). Notably, the overlapping distribution of N and O elements with AuNPs suggests the presence of organic components with high solubility in aqueous medium such as proteins and polysaccharides, binding to the surface of AuNPs, serving as capping and stabilizing agents.62,63
3.3. Detection of Fe3+ ions using UV-vis spectroscopy
The selectivity of the EB-AuNPs probe was thoroughly investigated by examining their interactions with several environmentally relevant metallic ions, including Ba2+, Ca2+, Ni2+, Al3+, Zn2+, Fe2+, Fe3+, Pb2+, Mn2+, Cd2+, and Cr2+, each at a concentration of 10 ppm, as depicted in Fig. 6. Upon the addition of metallic ions such as Ba2+, Ca2+, Ni2+, Zn2+, Mn2+, Cd2+, and Cr2+ the colloidal solution exhibited minimal discoloration, with insignificant changes observed in the intensity of the SPR peak of AuNPs. However, with the introduction of Fe2+, Pb2+ and Fe3+ ions, a noticeable decrease in the absorbance value of the SPR band was observed. Of particular interest, the absorbance value of AuNPs in the presence of Fe3+ ions exhibited a five-fold and nine-fold decrease compared to that observed with Fe2+ and Pb2+ ions, respectively. This phenomenon can be attributed to the high chelating ability of Fe3+ ions with EB-AuNPs compared to other ions, thereby indicating the superior selectivity of EB-AuNPs towards Fe3+ ions in an aqueous medium.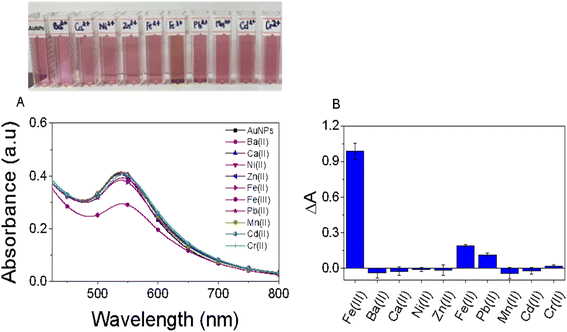 | ||
| Fig. 6 The influence of diverse metal ions on alterations in color and absorption spectra of the AuNPs test solution (A) and the colorimetric detection of AuNPs toward a range of metal ions (B). | ||
Changes in the intensity of the SPR peak and the color of the AuNPs solution in response to the target analyte are proposed to occur via a mechanism involving nanoparticle aggregation/agglomeration. Understanding the morphology of the samples is crucial for studying the detection mechanism of heavy metallic ions. In this study, we conducted analysis of DLS size distribution and TEM images for EB-AuNPs before and after the addition of Fe3+ ions, as illustrated in Fig. 7. DLS measurements revealed that the particle size distribution of EB-AuNPs exhibited a monodispersity index in both the samples, with the particle size significantly increasing (500–1000 nm) after the addition of the analyte compared to the EB-AuNP sample before analyte addition (120–400 nm). In particular, the analysis of TEM images before the addition of Fe3+ ions showed individual separated particles, whereas agglomeration of the synthesized nanoparticles was clearly observed after treatment with Fe3+ ions. Thus, the mechanism for detecting Fe3+ ions can be proposed to involve chelating interactions between Fe3+ ions and capping functional groups such as NH2 and COO− attached to the surface of AuNPs. The enhanced attraction between the nanoparticles results in their subsequent agglomeration, thereby reducing the distance between the AuNPs. Additionally, TEM images reveal a decrease in the size of individual particles, which can be attributed to the oxidation of Au(0) atoms by Fe3+ ions.64 Based on the experimental data from this study, a plausible mechanism for colorimetric sensing of Fe3+ ions using EB-AuNPs is proposed in Fig. 7E.
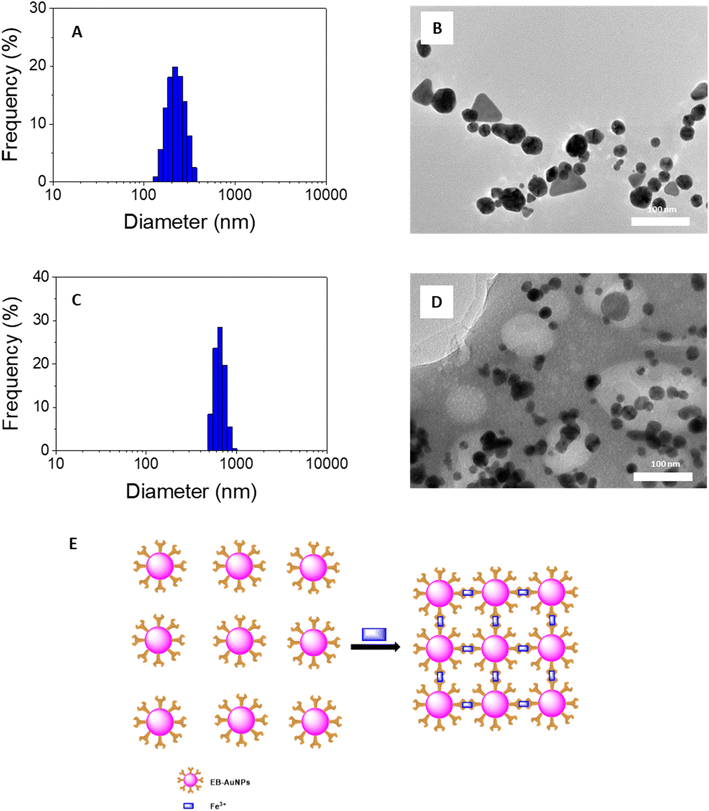 | ||
| Fig. 7 DLS spectra (left) and TEM images (right) of EB-AuNPs before (A and B) and after (C and D) adding Fe3+ ion and (E) proposed mechanism for colorimetric detection of Fe3+ ion in aqueous ion. | ||
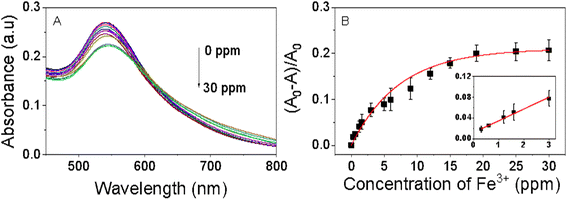
The colorimetric analysis aimed at detecting Fe3+ ions was conducted under ambient air conditions at room temperature. The findings of this investigation are depicted in Fig. 8. As the concentration of Fe3+ ions increases, the purple color of the EB-AuNPs solution diminishes, transforming the dispersion solution to a white appearance, indicative of gradual nanocomposite aggregation. This observed color change correlates with a progressive decrease in absorbance of AuNPs, as revealed by UV-vis spectra analysis. Ratios of absorption intensity rise with increase of different Fe3+ ion concentrations in range of 0.3–30 ppm. Across the range of measured Fe3+ ion concentrations, the absorption intensity demonstrates a linear trend in a narrow range of 0.3–3.0 ppm, as determined by the regression equation (A0 − A)/A0 = 0.00226CFe + 0.011 with an R2 value of 0.99 (Fig. 7C). The limit of detection (LOD) was determined using the equation LOD = 3σ/s, where σ represents the standard deviation of the blank signal and s denotes the slope of the regression equation. The computed LOD value is established to be 0.118 ppm. The LOD value of EB-AuNPs is relatively lower than the US National Secondary Drinking Water Regulations for iron (0.3 ppm) as well as previous reports as predicted in Table 1. The results demonstrated that EB-AuNPs are an effective probe for detection of Fe3+ ion.
| Probes | Detection mechanism | Linear range (ppm) | LOD (ppm) | References |
|---|---|---|---|---|
| Kojate motifs | Coordination | — | 2.01 | 65 |
| AgNPs@N-acetyl-L-cysteine | Reduction | 0.005–4.48 | 0.05 ppm | 66 |
| AuNPs@ pyrophosphate | Aggregation | 0.56–3.36 | 0.314 | 67 |
| GQD | Coordination | 0–4.48 | 0.404 | 68 |
| Green tea extract | Coordination | 2.5–17.5 | 0.90 | 69 |
| AuNCs@L-DOPA | Aggregation | 0.28–71.7 | 0.196 | 70 |
| EB-AuNPs | Aggregation | 0.3–3.0 | 0.118 | This work |
3.4. Detection of Fe3+ ions using smartphone images
In the experiment involving CuSO4 solutions, we conducted five separate trials, capturing ten images at each concentration range using two distinct techniques (Table 2): (1) for experiments 1–3, the technician utilized the focus button on the camera to focus on the Region of Interest (ROI); (2) for experiments 4 and 5, the technician refrained from using the focus button. Following the extraction of the ROI from each image, we applied a variance threshold to filter out images exhibiting high discrepancies in pixel values within the ROI. However, all images captured in the laboratory under the supervision of a single technician, aided by our lightbox, were deemed acceptable. Table 2 provides an overview of our dataset, highlighting that all experiments and image acquisitions were conducted under consistent conditions by the same technician.
| Exp no. | Range | Focus on the ROI | No of images/concentration | Total images |
|---|---|---|---|---|
| 1 | 0.25–2 M | Y | 10 | 80 (10 × 8) |
| 2 | 0.25–2 M | Y | 10 | 80 (10 × 8) |
| 3 | 0.25–2 M | Y | 10 | 80 (10 × 8) |
| 4 | 0.25–2 M | N | 10 | 80 (10 × 8) |
| 5 | 0.25–2 M | N | 10 | 80 (10 × 8) |
| Total | 400 | |||
Our study aims to investigate the influence of environmental factors on predicting the outcomes of a machine learning method by employing our proposed normalization techniques on the dataset, comparing the results with those obtained from the raw data without normalization. For each approach, we randomly split the dataset into two subsets: a training set and a test set, maintaining a ratio of 70![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 30 and then utilize linear regression to construct the machine learning models. Table 3 presents the average and standard deviation of prediction results on the test data after conducting five iterations.
30 and then utilize linear regression to construct the machine learning models. Table 3 presents the average and standard deviation of prediction results on the test data after conducting five iterations.
| Normalization approach | Metrics | |
|---|---|---|
| R2 | Root mean squared error | |
| Raw | 0.9716 ± 0.0023 | 0.0965 ± 0.0039 |
| Delta | 0.9724 ± 0.0026 | 0.0950 ± 0.0054 |
| Ratio | 0.9744 ± 0.0031 | 0.0915 ± 0.0048 |
The results indicate that while both normalization approaches (delta and ratio) enhanced the performance of the machine learning model, the improvements were modest, amounting to only 0.8% for the R2 metric (delta) and 0.28% for the ratio when compared to using raw data. This can be attributed to the stability of CuSO4 characteristics over time and the controlled conditions of our designed lightbox, which mitigates environmental influences during image capture. However, real-world applications present numerous variables affecting prediction accuracy, including evolving characteristics of iron and variations in smartphone camera quality among end users. In our subsequent experiment, we focus on Fe3+ ion to demonstrate the efficacy of the normalization approaches in enhancing prediction outcomes.
In detection of Fe3+ ions, we conducted trials across three distinct instances within the concentration range of 0.3 to 30 ppm, as detailed previously. Each concentration was accompanied by the capture of 10 images, with no specific focus on the ROI. Consequently, our dataset for Fe3+ ions comprises 390 images, calculated from 13 concentrations, each with 10 images, across the three experimental runs. Employing a methodology akin to that of CuSO4, we partitioned the dataset into training and testing sets using a 70![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 30 ratio, both with and without applying normalization techniques. The performance of our linear regression model on the test dataset, following five iterations, is presented in Table 4. The result showed that the normalization approaches enhanced the R2 metrics (5.73% for delta and 5.26% for ratio) compared to approach without normalization, i.e., they make the smaller errors. This result indicated that normalization approaches are useful.
30 ratio, both with and without applying normalization techniques. The performance of our linear regression model on the test dataset, following five iterations, is presented in Table 4. The result showed that the normalization approaches enhanced the R2 metrics (5.73% for delta and 5.26% for ratio) compared to approach without normalization, i.e., they make the smaller errors. This result indicated that normalization approaches are useful.
| Normalization approach | Metrics | |
|---|---|---|
| R2 | Root mean squared error | |
| Raw | 0.8207 ± 0.0196 | 3.9872 ± 0.2205 |
| Delta | 0.8780 ± 0.0077 | 3.2913 ± 0.1057 |
| Ratio | 0.8733 ± 0.0068 | 3.3548 ± 0.0902 |
| Parameters | CuSO4 | Fe3+ ion |
|---|---|---|
| B | −11.67194794 | 258.03961 |
| A1 | 0.1749725058769363 | 18.918431692320038 |
| A2 | −0.8659853255560485 | 2.3887664791957213 |
| A3 | 0.09558946035564549 | 3.018478950326897 |
| A4 | 0.25728815135808075 | −12.756884811298555 |
| A5 | 0.14611200674974775 | 1.0676268307155141 |
| A6 | 0.6633898919692821 | −16.484926316415734 |
| A7 | −0.0029225413551885746 | 0.5362284337472314 |
| A8 | −0.02300907284385169 | −1.8121682896367453 |
| A9 | 0.023310157395674134 | −1.625810063272214 |
| A10 | 0.0044665653250014845 | −1.050626267391651 |
| A11 | −0.022848946883847206 | 1.8510867458015794 |
| A12 | 0.0223940371249181 | 1.4279649782837482 |
| A13 | 0.01868896020592669 | 2.051594167885635 |
| A14 | −0.034996010159643945 | −0.1163897021906569 |
| A15 | 0.03250105403235272 | 1.9882705177771514 |
| A16 | 0.03644820329411289 | −0.9739092392704912 |
| A17 | −0.03845392009176146 | −3.1062507819007705 |
| A18 | −0.10714238706694611 | 0.556601441321064 |
| A19 | −0.020293607203989122 | 1.9732038182084808 |
| A20 | 0.21716408527928446 | −0.9955983036842209 |
| A21 | 0.03025398359255365 | −0.24395525445843377 |
| A22 | −0.003966909407306939 | 0.6050224958128079 |
| A23 | 0.018379714598367163 | −2.284755734470818 |
| A24 | −0.03128443711780003 | −1.678320747941534 |
| A25 | −0.10925068557748736 | 0.5307519637416704 |
| A26 | −0.03317336821218308 | 1.1676053587550543 |
| A27 | 0.020511778996520742 | 1.2271310727686446 |
For detection of Fe3+ ions, the optimal parameters determined through cross-validation were as follows: an R2 score of 0.885621 and an RMSE of 3.189249. These results were achieved by utilizing all features, employing the Delta normalization approach, and employing polynomial regression of degree 2. The resulting equation for the Fe3+ ion model, trained with these refined parameters, is denoted as generic equation form (eqn (3)) with parameters described in Table 5.
| y = B + A1 × meanR + A2 × meanG + A3 × meanB + A4 × modeR + A5 × modeB + A6 × modeG + A7 × meanR2 + A8 × meanR × meanG + A9 × meanR × meanB + A10 × meanR × modeR + A11 × meanR × modeB + A12 × meanR × modeG + A13 × meanG2 + A14 × meanG × meanB + A15 × meanG × modeR + A16 × meanG × modeB + A17 × meanG × modeG + A18 × meanB2 + A19 × meanB × modeR + A20 × meanB × modeB + A21 × meanB × modeG + A22 × modeR2 + A23 × modeR × modeB + A24 × modeR × modeG + A25 × modeB2 + A26 × modeB × modeG + A27 × modeG2 | (3) |
Utilizing the two polynomial regression functions mentioned above, we are able to accurately measure the concentrations of CuSO4 and Fe3+ ions in water using a smartphone camera. Notably, employing machine learning techniques allows for the determination of Fe3+ concentration over a significantly broader range (0–30 ppm) compared to UV-vis spectroscopy. Despite employing hyper-parameter tuning in this study to optimize model performance, our analysis was restricted to the use of polynomial regression due to the limited size of our experimental dataset. Moving forward, our research would encompass a wider range of concentration levels and finer increments to acquire more comprehensive datasets. Furthermore, we plan to explore alternative machine learning algorithms such as random forest, neural networks, and ensemble methods to develop more robust models.
4 Conclusions
This study combined biogenic nanometals with machine learning techniques to quantify Fe3+ ions in water. The aqueous extract of E. bulbosa leaf proved highly effective as both a reducing and stabilizing agent in synthesizing gold nanoparticles (AuNPs). By optimizing synthesis conditions, the morphology of the AuNPs was optimized, and monitoring their characteristics using various analytical techniques. The resultant pure crystalline gold nanoparticles boasted an average diameter of 19.8 nm. Furthermore, our investigation showcased the potential of AuNPs as a colorimetric probe for detecting Fe3+ ions. Through a combination of UV-vis analysis and smartphone camera images, integrated with machine learning algorithms, we devised an efficient method for analyzing Fe3+ ions. Consequently, this study lays the groundwork for developing novel colorimetric sensors capable of accurately and swiftly detecting toxic contaminants in water.Data availability
The data supporting this article have been included as part of the ESI.†Conflicts of interest
No potential conflict of interest was reported by the authors.Acknowledgements
This research is funded by Vietnam Academy of Science and Technology under grant number VAST03.04/23-24.References
- R. R. Crichton and J. R. Boelaert, Inorganic Biochemistry of Iron Metabolism: from Molecular Mechanisms to Clinical Consequences, John Wiley & Sons, 2001 Search PubMed.
- N. Elahi, M. Kamali and M. H. Baghersad, Talanta, 2018, 184, 537–556 CrossRef CAS PubMed.
- S. K. Sahoo, D. Sharma, R. K. Bera, G. Crisponi and J. F. Callan, Chem. Soc. Rev., 2012, 41, 7195–7227 RSC.
- N. Abbaspour, R. Hurrell and R. Kelishadi, J. Res. Med. Sci., 2014, 19, 164 Search PubMed.
- X. Bai, Y. Wang, Z. Song, Y. Feng, Y. Chen, D. Zhang and L. Feng, Int. J. Mol. Sci., 2020, 21, 2480 CrossRef CAS PubMed.
- J. Bartl, L. Reinke, M. Koch and S. Kubik, Chem. Commun., 2020, 56, 10457–10460 RSC.
- D.-Q. Feng, G. Liu, Z. Chen, H. Lu, Y. Gao and X. Fang, Microchem. J., 2020, 157, 104977 CrossRef CAS.
- A. Robin, G. Vansuyt, P. Hinsinger, J. M. Meyer, J.-F. Briat and P. Lemanceau, Adv. Agron., 2008, 99, 183–225 CAS.
- The Ministry of Health, Promulgating National Technical Regulation And Regulations On Inspection And Monitoring Of Domestic Water Quality, 2018, No. 41/2018/TT-BYT Search PubMed.
- S. J. Dixon and B. R. Stockwell, Nat. Chem. Biol., 2014, 10, 9–17 CrossRef CAS PubMed.
- P. Ponka, Kidney Int., 1999, 55, S2–S11 CrossRef PubMed.
- D. R. Richardson and P. Ponka, Biochim. Biophys. Acta, Rev. Biomembr., 1997, 1331, 1–40 CrossRef CAS PubMed.
- J.-L. Liu, Z.-Y. Wang and C. Guo, Front. Neurosci., 2018, 12, 411985 Search PubMed.
- K. Wojtunik-Kulesza, A. Oniszczuk and M. Waksmundzka-Hajnos, Biomed. Pharmacother., 2019, 111, 1277–1289 CrossRef CAS PubMed.
- V. Masindi and K. L. Muedi, Heavy Met., 2018, 10, 115–133 Search PubMed.
- M. Jaishankar, T. Tseten, N. Anbalagan, B. B. Mathew and K. N. Beeregowda, Interdiscip. Toxicol., 2014, 7, 60–72 CrossRef PubMed.
- S. Toyokuni, Cancer Sci., 2009, 100, 9–16 CrossRef CAS PubMed.
- T. Hirayama and H. Nagasawa, J. Clin. Biochem. Nutr., 2017, 60, 39–48 CrossRef CAS PubMed.
- M. J. Hopwood, A. J. Birchill, M. Gledhill, E. P. Achterberg, J. K. Klar and A. Milne, Front. Mar. Sci., 2017, 4, 192 CrossRef.
- S. R. Ahmed, R. Chand, S. Kumar, N. Mittal, S. Srinivasan and A. R. Rajabzadeh, TrAC, Trends Anal. Chem., 2020, 131, 116006 CrossRef CAS.
- K. Lal, S. A. Jaywant and K. M. Arif, Sensors, 2023, 23, 7099 CrossRef CAS PubMed.
- H. Sohrabi, A. Hemmati, M. R. Majidi, S. Eyvazi, A. Jahanban-Esfahlan, B. Baradaran, R. Adlpour-Azar, A. Mokhtarzadeh and M. de la Guardia, TrAC, Trends Anal. Chem., 2021, 143, 116344 CrossRef CAS.
- M. Xu, Z. Chen, J. Zheng, Q. Zhao and Z. Yuan, Semin. Cancer Biol., 2023, 94, 62–80 CrossRef CAS PubMed.
- D. K. Iyer, A. Shaji, S. P. Singh, A. Tripathi, A. Hazra, S. Mandal and P. Ghosh, Coord. Chem. Rev., 2023, 495, 215371 CrossRef CAS.
- C. Wu, P. Tan, X. Chen, H. Chang, Y. Chen, G. Su, T. Liu, Z. Lu, M. Sun and Y. Wang, ACS Appl. Mater. Interfaces, 2023, 15, 48506–48518 CrossRef CAS PubMed.
- J. Nelis, A. Tsagkaris, M. Dillon, J. Hajslova and C. Elliott, TrAC, Trends Anal. Chem., 2020, 115934 CrossRef CAS PubMed.
- H. N. Baghban, K. Ghaseminasab and M. Hasanzadeh, Anal. Methods, 2023, 15, 3549–3561 RSC.
- E. Ozdemir, Y. Alcay, O. Yavuz, M. S. Yildirim, H. Aribuga, U. Ertugral, K. Kaya and I. Yilmaz, Talanta, 2023, 261, 124660 CrossRef CAS PubMed.
- T. H. Bui, B. Thangavel, M. Sharipov, K. Chen and J. H. Shin, Chemosensors, 2023, 11, 468 CrossRef CAS.
- Z.-J. Huang, H. Li, J.-Y. Luo, S. Li and F. Liu, Anal. Chem., 2023, 95, 6156–6162 CrossRef CAS PubMed.
- H. Daraee, A. Eatemadi, E. Abbasi, S. Fekri Aval, M. Kouhi and A. Akbarzadeh, Artif. Cells, Nanomed., Biotechnol., 2016, 44, 410–422 CrossRef CAS PubMed.
- M. H. Jazayeri, H. Amani, A. A. Pourfatollah, H. Pazoki-Toroudi and B. Sedighimoghaddam, Sens. Bio-Sens. Res., 2016, 9, 17–22 CrossRef.
- H. S. Kim and D. Y. Lee, Polymers, 2018, 10, 961 CrossRef PubMed.
- S. C. Gopinath, T. Lakshmipriya and K. Awazu, Biosens. Bioelectron., 2014, 51, 115–123 CrossRef CAS PubMed.
- S. Lepinay, A. Staff, A. Ianoul and J. Albert, Biosens. Bioelectron., 2014, 52, 337–344 CrossRef CAS PubMed.
- T. Yang, X.-X. Zhang, J.-Y. Yang, Y.-T. Wang and M.-L. Chen, Talanta, 2018, 177, 212–216 CrossRef CAS PubMed.
- J. Zong, S. L. Cobb and N. R. Cameron, Biomater. Sci., 2017, 5, 872–886 RSC.
- V. N. Mehta, R. K. Singhal and S. K. Kailasa, RSC Adv., 2015, 5, 33468–33477 RSC.
- A. Safavi, R. Ahmadi and Z. Mohammadpour, Sens. Actuators, B, 2017, 242, 609–615 CrossRef CAS.
- P. Buduru and S. R. Reddy, Sens. Actuators, B, 2016, 237, 935–943 CrossRef CAS.
- S. S. Memon, A. Nafady, A. R. Solangi, A. M. Al-Enizi, M. R. Shah, S. T. Sherazi, S. Memon, M. Arain, M. I. Abro and M. I. Khattak, Sens. Actuators, B, 2018, 259, 1006–1012 CrossRef CAS.
- M. Yu, Z. Zhu, H. Wang, L. Li, F. Fu, Y. Song and E. Song, Biosens. Bioelectron., 2017, 91, 143–148 CrossRef CAS PubMed.
- S. Lokina, R. Suresh, K. Giribabu, A. Stephen, R. L. Sundaram and V. Narayanan, Spectrochim. Acta, Part A, 2014, 129, 484–490 CrossRef CAS PubMed.
- M.-T. Tran, L.-P. Nguyen, D.-T. Nguyen, T. Le Cam-Huong, C.-H. Dang, T. T. K. Chi and T.-D. Nguyen, Res. Chem. Intermed., 2021, 47, 4613–4633 CrossRef CAS.
- T. M. A. Alves, H. Kloos and C. L. Zani, Mem. Inst. Oswaldo Cruz, 2003, 98, 709–712 CrossRef CAS PubMed.
- W. Munaeni, M. Yuhana, M. Setiawati and A. T. Wahyudi, Asian Pac. J. Trop. Biomed., 2019, 9, 397–404 CrossRef CAS.
- W. Munaeni, A. Pariakan, L. B. Abidin and M. Yuhana, Microbiology Indonesia, 2017, 11, 1 CrossRef.
- H. A. Alhadrami, R. Orfali, A. A. Hamed, M. M. Ghoneim, H. M. Hassan, A. S. Hassane, M. E. Rateb, A. M. Sayed and N. M. Gamaleldin, Antibiotics, 2021, 10, 968 CrossRef CAS PubMed.
- J. Wang, N. Zhou, Z. Zhu, J. Huang and G. Li, Anal. Bioanal. Chem., 2007, 388, 1199–1205 CrossRef CAS PubMed.
- T. Ahmad, M. A. Bustam, M. Irfan, M. Moniruzzaman, H. M. A. Asghar and S. Bhattacharjee, Biotechnol. Appl. Biochem., 2019, 66, 698–708 CrossRef CAS PubMed.
- OpenCV2, https://pypi.org/project/opencv-python/, accessed March 01, 2024.
- Seaborn, https://seaborn.pydata.org/, accessed March 01, 2024.
- Scikit-learn, https://scikit-learn.org/stable/, accessed March 01, 2024.
- V. Amendola, R. Pilot, M. Frasconi, O. M. Maragò and M. A. Iatì, J. Phys.: Condens. Matter, 2017, 29, 203002 CrossRef PubMed.
- A. Derenne, V. Van Hemelryck, D. Lamoral-Theys, R. Kiss and E. Goormaghtigh, Biochim. Biophys. Acta, Mol. Basis Dis., 2013, 1832, 46–56 CrossRef CAS PubMed.
- A. Martínez-Cuazitl, M. d. C. Gómez-García, S. Pérez-Mora, M. Rojas-López, R. J. Delgado-Macuil, J. Ocampo-López, G. J. Vázquez-Zapién, M. M. Mata-Miranda and D. G. Pérez-Ishiwara, Int. J. Mol. Sci., 2023, 24, 17138 CrossRef PubMed.
- W. Yan, V. Petkov, S. M. Mahurin, S. H. Overbury and S. Dai, Catal. Commun., 2005, 6, 404–408 CrossRef CAS.
- V. Petkov, Y. Peng, G. Williams, B. Huang, D. Tomalia and Y. Ren, Phys. Rev. B: Condens. Matter Mater. Phys., 2005, 72, 195402 CrossRef.
- D. Mahl, J. Diendorf, S. Ristig, C. Greulich, Z.-A. Li, M. Farle, M. Köller and M. Epple, J. Nanopart. Res., 2012, 14, 1–13 CrossRef.
- I. Hussain, N. Singh, A. Singh, H. Singh and S. Singh, Biotechnol. Lett., 2016, 38, 545–560 CrossRef CAS PubMed.
- S. Sathiyaraj, G. Suriyakala, A. D. Gandhi, R. Babujanarthanam, K. S. Almaary, T.-W. Chen and K. Kaviyarasu, J. Infect. Public Health, 2021, 14, 1842–1847 CrossRef PubMed.
- A. Shavandi, P. Saeedi, M. A. Ali and E. Jalalvandi, in Functional Polysaccharides for Biomedical Applications, Elsevier, 2019, pp. 267–304 Search PubMed.
- C. Wang, X. Gao, Z. Chen, Y. Chen and H. Chen, Polymers, 2017, 9, 689 CrossRef PubMed.
- A. S. Andreani, E. S. Kunarti, T. Hashimoto, T. Hayashita and S. J. Santosa, J. Environ. Chem. Eng., 2021, 9, 105962 CrossRef CAS.
- S. Vallejos, A. Muñoz, F. C. García, R. Colleoni, R. Biesuz, G. Alberti and J. M. García, Sens. Actuators, B, 2016, 233, 120–126 CrossRef CAS.
- X. Gao, Y. Lu, S. He, X. Li and W. Chen, Anal. Chim. Acta, 2015, 879, 118–125 CrossRef CAS PubMed.
- S.-P. Wu, Y.-P. Chen and Y.-M. Sung, Analyst, 2011, 136, 1887–1891 RSC.
- A. Ananthanarayanan, X. Wang, P. Routh, B. Sana, S. Lim, D. H. Kim, K. H. Lim, J. Li and P. Chen, Adv. Funct. Mater., 2014, 24, 3021–3026 CrossRef CAS.
- J. I. Ballesteros, H. J. R. Caleja-Ballesteros and M. C. Villena, Microchem. J., 2021, 160, 105652 CrossRef CAS.
- J.-a. Annie Ho, H.-C. Chang and W.-T. Su, Anal. Chem., 2012, 84, 3246–3253 CrossRef CAS PubMed.
Footnotes |
| † Electronic supplementary information (ESI) available. See DOI: https://doi.org/10.1039/d4ra03265a |
| ‡ These authors contributed equally to this work and share first authorship. |
| This journal is © The Royal Society of Chemistry 2024 |