Open Access Article
Open Access ArticlePhytofabricated ZnO-NPs mediated by Hibiscus tiliaceus leaf extract and its potential as a diosgenin delivery vehicle†
Oktavina Kartika Putri ab,
Lina Oktavia Rahayub,
Yuly Kusumawati
ab,
Lina Oktavia Rahayub,
Yuly Kusumawati *a,
Arif Fadlan
*a,
Arif Fadlan a,
Riki Subagyo
a,
Riki Subagyo a and
Mardi Santoso
a and
Mardi Santoso a
a
aDepartment of Chemistry, Institut Teknologi Sepuluh Nopember, Surabaya, 60111, Indonesia. E-mail: y_kusumawati@chem.its.ac.id
bDepartment of Pharmacy, Politeknik Kesehatan Putra Indonesia Malang, Malang, 65122, Indonesia
First published on 23rd July 2024
Abstract
Zinc oxide nanoparticles (ZnO-NPs) have provided promising potential in the biomedical field, including the ability to overcome various health problems. Diosgenin is used to treat multiple health disorders but has very low solubility in water. Using ZnO-NPs as a diosgenin delivery vehicle was expected to increase the solubility of diosgenin, which would affect its bioavailability. This study demonstrates phytofabrication and characterization of ZnO-NPs, loading of diosgenin onto the ZnO-NPs, characterization of the product (ZnO-NPs/diosgenin), and evaluations of diosgenin release. Phytofabrication of the ZnO-NPs was carried out with zinc precursors and Hibiscus tiliaceus leaf extract (HLE) obtained with various extraction solvents. To explore the potential of using the ZnO-NPs as a diosgenin delivery vehicle, diosgenin release from the ZnO-NPs/diosgenin was studied. Based on the X-ray fluorescence (XRF) and X-ray diffraction (XRD) results, ZnO-NPs with high purity have been successfully fabricated. Nano-sized particles were detected using scanning electron microscopy (SEM) and confirmed by transmission electron microscopy (TEM), revealing the smallest particle size of 45.924 ± 27.910 nm obtained from the methanol extract with the zinc acetate precursor. The ZnO-NPs had hexagonal wurtzite and rod-like structures. Diosgenin was successfully added to the ZnO-NPs with loadings of 79.972% for ZnO-HLMEA-D500 (ZnO-NPs/diosgenin produced by doping with a 500 μg mL−1 of diosgenin solution) and 39.775% for ZnO-HLMEA-D1000 (ZnO-NPs/diosgenin produced by doping with a 1000 μg mL−1 of diosgenin solution). The solubilities of diosgenin from ZnO-HLMEA-D500 and ZnO-HLMEA-D1000 were higher than that of free diosgenin, confirming that ZnO-NPs have potential as delivery vehicles for diosgenin and conceivably other water-insoluble drugs.
Introduction
ZnO nanoparticles (ZnO-NPs) have gained widespread favor in biological applications due to their cost-effectiveness, excellent biocompatibility, and minimal toxicity.1 ZnO-NPs have provided promising potential in the biomedical field. The diseases that can be overcome by ZnO-NPs include microbial infection,2–4 diabetes,5–7 inflammation,6,8 cancer,5,6,9,10 thyroid disease,11 anxiety disorders, hepatopulmonary syndrome,6 etc.Several studies have shown that the smaller the ZnO particle size, the better the performance. The antibacterial activity of ZnO-NPs against Staphylococcus epidermidis and Salmonella typhi was better than pharmaceutical-grade ZnO microparticles (ZnO-MPs).3 The regenerative effect of ZnO-NPs on burn wounds was higher than that of ZnO-MPs.12 The antifungal activity of 27 nm sized ZnO-NPs against Aspergillus niger was more potent than that of 84 nm sized ZnO-NPs.13 The cytotoxicity of 26 nm-sized ZnO-NPs was higher than 62 nm and 90 nm against Caco-2 cells.14 Therefore, the synthesis method to produce ZnO-NPs of a small size needs to be carefully carried out.
A phytofabrication process was used to produce the ZnO-NPs with the help of Hibiscus tiliaceus leaf extract (HLE). Phytofabrication is a plant-mediated metal oxide preparation technique. Compared to other organs of H. tiliaceus, the leaf contains high levels of flavonoid and phenolic compounds,15 which significantly contribute to the formation of metal oxides. Previous studies showed that the bioactivities of metal oxides were increased with the presence of flavonoids in the leaf extract.3,16 Many factors influence the type of compound extracted from plant parts, one of which is its use as an extraction solvent.17,18 The ZnO-NPs synthesized via the reduction with Salvadora persica L. root extract proved that the extraction solvent could affect the morphology of the ZnO-NPs formed.19 Studies showed that the shapes of ZnO particles influence their pharmacological activities.20 This fact prompted the use of three different solvents, including ethanol, methanol, and distilled water, to extract H. tiliaceus leaves in this research.
The shapes and sizes of ZnO-NPs are also influenced by the zinc precursors obtained from the zinc salts. The scanning electron microscopy (SEM) images of ZnO-NPs obtained from Laurus nobilis L. leaf aqueous extract and two zinc precursors, including zinc nitrate and zinc acetate, illustrated that different precursors affected the sizes and shapes of the nanoparticles.21 Differences in sizes and shapes of ZnO-NPs mediated by Justicia adhatoda leaf extract also arose due to using different precursors.22 Studies showed that the sizes of ZnO particles also affect their pharmacological activities.23 Based on these facts, three zinc salts, including zinc chloride, zinc acetate, and zinc nitrate, were compared for use as precursors. Nine variations of ZnO-NPs, generated from three different solvents and precursors, have emerged as intriguing subjects warranting further investigation.
Arthritis, asthma, cancer, cardiovascular disease, and diabetes are health disorders that can be treated with diosgenin,24 which has the International Union of Pure and Applied Chemistry (IUPAC) name (3β,25R)-spirost-5-en-3-ol.25 Diosgenin is also known in the pharmaceutical industry as a starting material for several steroidal drugs, such as cortisone, oral contraceptives, and sex hormones. However, behind those positive features, its low solubility in water is a negative feature of diosgenin. Moreover, diosgenin is reported to have low bioavailability, approximately 6%.26 Studies showed that using nano-sized delivery vehicles changed the drug's biodistribution and pharmacokinetic profiles, boosting the bioavailability and efficacy of hydrophobic active compounds.27 In addition, the use of nano-sized delivery vehicles in drug delivery systems has increased the drug's chemotherapeutic effects, solubilities, and stabilities.28,29 ZnO-NPs are among the candidates used to mitigate the negative features of diosgenin. ZnO-NPs and their combinations have been successfully used as delivery vehicles for several water-insoluble active compounds, such as paclitaxel,29 quercetin,30 and mangiferin.31 This occurs because organic compounds in the phytofabricated ZnO-NPs originate from plant extracts. These organic compounds can bind water-insoluble active compounds and release them in suitable solvents, thereby helping to increase their solubility.
This study was selected because there is a limited article on phytofabricated ZnO-NPs as a delivery vehicle for diosgenin. The best characteristics of ZnO-NPs and diosgenin that have been mentioned are expected to improve the quality of the combination of the two (ZnO-NPs/diosgenin). In ZnO-NPs/diosgenin, ZnO-NPs act as a delivery vehicle for diosgenin. The primary focus of this research was to prepare ZnO-NPs via a phytofabrication technique mediated by HLE from various solvents, including ethanol, methanol, and distilled water with several zinc precursors including zinc chloride, zinc acetate, and zinc nitrate, characterize the phytofabricated ZnO-NPs, produce, and then characterize ZnO-NPs/diosgenin, and evaluate diosgenin release from ZnO-NPs/diosgenin.
Result and discussion
Characterization of the ZnO-NPs
The white to cream-colored powders resulting from the phytofabrication process were examined to ascertain the characteristics of the ZnO produced. The morphology of nine ZnO-NPs powders is presented in the ESI file.† The first study, X-ray fluorescence (XRF), was used to determine the chemical compositions of the synthesized compounds. The results showed that the nine ZnO-NPs produced contained more than 97% ZnO, indicating high purity. This value is similar to that of ZnO purchased from US Research Nanomaterials.32 XRF results of 9 types of ZnO-NPs are presented in the ESI file.†The second analysis, X-ray diffraction (XRD), was used to check the shapes and crystallinities of the phytofabricated ZnO-NPs. Fig. 1(a) shows the XRD pattern for the phytofabricated ZnO-NPs. The diffractogram peaks at 2θ = 31.8025°, 34.4457°, 36.3028°, 47.5873°, 56.6412°, 62.9096°, 66.4176°, 67.9934°, 69.0975°, 72.5834°, 76.9642°, 81.3736°, and 89.5855° corresponded to the (100), (002), (101), (102), (110), (103), (200), (112), (201), (004), (202), (104), and (203) lattice planes of the hexagonal wurtzite ZnO-NPs (JCPDS number 36-1451). The exact form was also reported for ZnO-NPs prepared via phytofabrication mediated by Alstonia macrophylla,33 Ficus carica,34 and Tecoma castanifolia leaf extracts.35 The high intensities and sharp peaks of the nine XRD diffraction patterns of phytofabricated ZnO-NPs illustrate the high crystallinity. No peaks representing impurities were found.
Fig. 1(b) presents the wavenumber versus transmittance patterns for the nine phytofabricated ZnO-NPs and H. tiliaceus leaf. Those spectra showed identical peaks at the same wavelengths. The peaks found at 400–500 cm−1 were correlated with the stretching vibrations of the Zn–O–Zn bonds. A broad peak indicated –OH at 3400–3500 cm−1 due to O–H stretching vibrations. This was reinforced by the presence of O–H in-plane bending vibrations, which generated peaks at 1300–1400 cm−1.36 The hydroxyl functional groups on the surfaces of the ZnO-NPs originated from aquo, hydroxy, and oxo species.37 The peak at approximately 890 cm−1 represented C–H bending vibrations. The peak confirmed this at approximately 2800 cm−1 for C–H bending vibrations. The peak at approximately 1500 cm−1 represented C![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) C stretching vibrations in the aromatic ring and polyphenol (C
C stretching vibrations in the aromatic ring and polyphenol (C![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) O). The peak at approximately 1600 cm−1 represented C
O). The peak at approximately 1600 cm−1 represented C![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) C stretching vibrations of the alkyl groups.38 Functional groups identical to those of the organic compounds indicated that the organic compounds in the HLE were attached to ZnO-NPs because the phytofabrication process did not include calcination. Calcination at 350 °C produces ZnO-NPs that still bind several biomolecules, as in the biosynthesis of ZnO-NPs mediated by the Pongamia pinnata extract,39 however, biosynthesis without calcination is more likely to produce the same results. HLE contained the active compounds rutin hydrate, quercetin, ellagic acid, and catechins.15
C stretching vibrations of the alkyl groups.38 Functional groups identical to those of the organic compounds indicated that the organic compounds in the HLE were attached to ZnO-NPs because the phytofabrication process did not include calcination. Calcination at 350 °C produces ZnO-NPs that still bind several biomolecules, as in the biosynthesis of ZnO-NPs mediated by the Pongamia pinnata extract,39 however, biosynthesis without calcination is more likely to produce the same results. HLE contained the active compounds rutin hydrate, quercetin, ellagic acid, and catechins.15
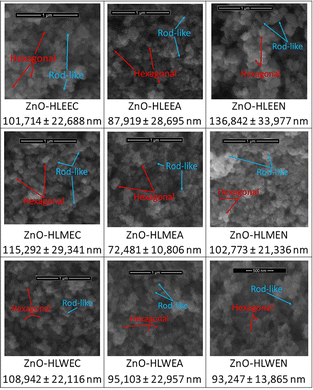
The SEM micrograph of the phytofabricated ZnO-NPs shows that nanosized ZnO was successfully synthesized. The particle sizes were between 57 and 96 nm, with an average particle size of 72.481 ± 10.806 nm. The particle sizes for the nine ZnO-NPs are presented in Fig. 2. ZnO-NPs phytofabricated from Aloe vera leaf extract and Eucalyptus lanceolata leaf litter also showed the same particle size ranges.40,41 The hexagonal particles in the image confirmed the results obtained from the XRD analyses. Rod-like ZnO-NPs were also seen in the SEM image.
Of the nine ZnO-NPs produced, the smallest particle size was obtained from the zinc acetate precursor mediated by HLME (ZnO-HLMEA). Several studies indicate that the smaller the ZnO particle size, the better the antimicrobial, cytotoxicity, and other activities.3,12–14 Therefore, ZnO-HLMEA was chosen for further analysis, starting with TEM analysis. The TEM image corroborated the SEM image, affirming the nanoscale dimensions of the resultant particles. In particular, the average size of the ZnO-NPs recorded with TEM and analyzed with ImageJ software was 45.924 ± 27.910 nm. The ZnO-NPs shown in the TEM image, as presented in Fig. 3(a), had hexagonal and rod-like shapes consistent with the SEM analysis. The particle size distribution can be discerned from the TEM data obtained from ZnO-HLMEA. Fig. 3(b) depicts the histogram of the particle size distribution of these compounds. The data show that most particles have sizes between 20 and 35 nm. Only a tiny portion of particles was found to be more than 100 nm in size.
 | ||
| Fig. 3 TEM images with hexagonal and rod-like shapes (a) and the particle size distribution visualized in the histogram (b) of ZnO-HLMEA. | ||
Previous studies have shown that using zinc acetate as the precursor gives ZnO-NPs rod-like structures. Moreover, the morphology of the ZnO nanopowder produced from zinc acetate is smoother than that of zinc nitrate.42 It can be concluded that the precursor used can influence the structure and size of the ZnO-NPs that are formed.
Preparation and characterization of ZnO-HLMEA-D500 and ZnO-HLMEA-D1000
Two samples of ZnO-NPs/diosgenin were produced by doping with 500 μg mL−1 diosgenin, called ZnO-HLMEA-D500, and 1000 μg mL−1, called ZnO-HLMEA-D1000. ZnO-HLMEA-D500 and ZnO-HLMEA-D1000 were prepared as grayish-white powders. The amounts of diosgenin contained in the ZnO-HLMEA were determined with eqn (1).
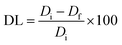 | (1) |
Based on UV-vis spectrophotometry absorbance measurements, the Di value of ZnO-HLMEA-D500 was 0.735 μg mg−1, and the Df value was 0.147 μg mg−1. After calculation using eqn (1), the DL value was 79.972%. So, 0.0500 grams of ZnO-HLMEA-D500 contains 39![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 986.105 μg of diosgenin. Other data obtained include the Di value of ZnO-HLMEA-D1000 was 1.407 μg mg−1, and the Df value was 0.847 μg mg−1. After calculation using eqn (1), the DL value was 39.775%. So, 0.0500 grams of ZnO-HLMEA-D1000 contains 19
986.105 μg of diosgenin. Other data obtained include the Di value of ZnO-HLMEA-D1000 was 1.407 μg mg−1, and the Df value was 0.847 μg mg−1. After calculation using eqn (1), the DL value was 39.775%. So, 0.0500 grams of ZnO-HLMEA-D1000 contains 19![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 887
887![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 285 μg of diosgenin.
285 μg of diosgenin.
The FTIR spectra obtained for diosgenin, ZnO-HLMEA-D500, ZnO-HLMEA-D1000, ZnO-HLMEA-D500, and ZnO-HLMEA-D1000 after the release of diosgenin at pH 5.5, ZnO-HLMEA-D500, and ZnO-HLMEA-D1000 after release of diosgenin at pH 7.4 and ZnO-HLMEA are shown in Fig. 4. The presence of peaks at 3454 cm−1, 2951 cm−1, 1456 cm−1, 1172 cm−1, and 1053 cm−1 strengthens the uniqueness of the diosgenin spectrum. The bands at 2951 cm−1 and 1456 cm−1 correlate to CH2 stretching and scissoring vibrations, and a band at 3454 cm−1 indicates –OH stretching vibrations. The band at 897 cm−1 is associated with CH2 twist vibrations, and strong characteristic peaks at 1172 cm−1 and 1053 cm−1 can be connected to –C–O stretching vibrations. The vibrational peaks observed for diosgenin were in perfect agreement with previous reports.43,44
ZnO-HLMEA-D500 and ZnO-HLMEA-D1000 showed identical peaks, indicating that the concentration of diosgenin used in the doping process did not change the way diosgenin interacted with the ZnO-NPs, as shown in Fig. 4(c) and (d). The appearance of a peak around 1630 cm−1 in both spectra indicates the presence of the C![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) C group. This functional group is identical to diosgenin (double bond in the cyclic ring).45 However, when compared with the spectrum of pure diosgenin, there is a wavenumber shift, indicating a bond's formation. These peaks still appeared after the release of diosgenin at pH 5.5 and 7.4, indicating the presence of organic compounds with double bonds that remained bound to the ZnO-NPs due to the extract-mediated phytofabrication process. The hydrogen bonds were stronger in ZnO-HLMEA-D500 than in ZnO-HLMEA-D1000, (e) to (f) and (g) to (h), as indicated by the intensities of the broad absorption peaks (approximately 3400–3500 cm−1) caused by O–H stretching vibrations of the hydroxyl groups.46 It was concluded that the intermolecular O–H⋯O–H interactions between the hydroxyl groups and the solvent increased with increasing diosgenin concentration in the doping process.47
C group. This functional group is identical to diosgenin (double bond in the cyclic ring).45 However, when compared with the spectrum of pure diosgenin, there is a wavenumber shift, indicating a bond's formation. These peaks still appeared after the release of diosgenin at pH 5.5 and 7.4, indicating the presence of organic compounds with double bonds that remained bound to the ZnO-NPs due to the extract-mediated phytofabrication process. The hydrogen bonds were stronger in ZnO-HLMEA-D500 than in ZnO-HLMEA-D1000, (e) to (f) and (g) to (h), as indicated by the intensities of the broad absorption peaks (approximately 3400–3500 cm−1) caused by O–H stretching vibrations of the hydroxyl groups.46 It was concluded that the intermolecular O–H⋯O–H interactions between the hydroxyl groups and the solvent increased with increasing diosgenin concentration in the doping process.47
Identical peaks were obtained for ZnO-HLMEA-D500 and ZnO-HLMEA-D1000 after the release of diosgenin at pH 5.5. This showed that regardless of the amount of diosgenin bound to ZnO-NPs, the release process at pH 5.5 followed the exact mechanism. Similar behavior also occurred at pH 7.4. A peak at approximately 1109 cm−1 indicated a C–O stretching vibration in an amino acid,48 as shown by spectra (c) to (f). These amino acids probably came from natural compounds in the HLE that were still bound to ZnO-NPs/diosgenin. These amino acids were still bound after releasing the diosgenin with DPBS at pH 5.5. However, the amino acids were no longer found after releasing the diosgenin with DPBS at pH 7.4, possibly because they were degraded or dissolved. The high levels of amino acids and proteins in H. tiliaceus leaves indicate it could be used as a ruminant feed additive.49
The weight losses from ZnO-HLMEA-D500 and ZnO-HLMEA-D1000 with increasing temperatures are presented in Fig. 5(a). The curves showed weight losses for ZnO-HLMEA-D1000 of approximately 6% (w/w) and 6.5% (w/w) for ZnO-HLMEA-D500 at temperatures up to 1000 °C. This occurred due to dehydration and the loss of organic active compounds from the HLE attached to the ZnO-NPs. These data also showed that the weight loss of ZnO-HLMEA-D500 was higher than that of ZnO-HLMEA-D1000, which indicated that more diosgenin was attached to ZnO-HLMEA-D500 than to ZnO-HLMEA-D1000. This confirmed the amounts of diosgenin bound to ZnO calculated with eqn (1), which showed that the amount of diosgenin contained in ZnO-HLMEA-D500 was 79.972% and that in ZnO-HLMEA-D1000 was 39.775%. One weight loss for diosgenin was seen in the temperature range 0 to 600 °C, consistent with previous studies.50
 | ||
| Fig. 5 TGA curves of ZnO-HLMEA-D (a) and curves of Vero cells viability treated with diosgenin and ZnO-HLMEA-D1000 (b). | ||
Cell viability assay
The cell viability assay of normal Vero cells exposed to ZnO-HLMEA-D1000 and diosgenin was carried out using the 3-(4,5-dimethylthiazol-2-yl)-2,5-diphenyl-2H-tetrazolium bromide (MTT) method. The MTT method is one of the most frequently used colorimetric test methods for assessing cytotoxicity or cell viability. This assay primarily determines cell viability by measuring the activity of mitochondrial enzymes, such as succinate dehydrogenase. In this test, MTT is reduced to purple formazan by NADH.51 When cells die, they lose the ability to convert MTT to formazan.52 According to Fig. 5(b), exposure to approximately 40 μg mL−1 of ZnO-HLMEA-D1000 can result in the death of 50% of normal Vero cells during a 24 hours incubation period. Diosgenin appears safer for normal Vero cells because it can maintain cell viability at around 62% at concentrations of 50 μg mL−1 or more. However, an article states that phytofabricated ZnO-NPs have an excellent biocompatibility profile, showing reduced cytotoxicity compared to chemically synthesized ZnO-NPs on Vero cells.53 This shows that phytofabricated ZnO-NPs still have significant potential to be developed as drug-delivery vehicles.Evaluation of diosgenin release
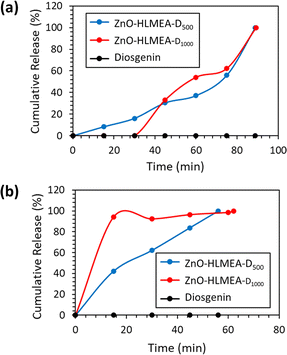
Fig. 6 depicts the amounts of diosgenin released from ZnO-HLMEA-D500 and ZnO-HLMEA-D1000. The amount of diosgenin contained in 0.0500 g of ZnO-HLMEA-D500 was 39![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 986.105 μg, and in 0.0500 g of ZnO-HLMEA-D1000 was 19
986.105 μg, and in 0.0500 g of ZnO-HLMEA-D1000 was 19![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 887.285 μg. The solubilities of diosgenin in DPBS at pH 5.5 and 7.4 were tested and showed that diosgenin did not dissolve completely in DPBS at pH 5.5 or 7.4, confirming that diosgenin has poor bioavailability.26 However, the diagrams show that diosgenin was successfully released into DPBS at pH 5.5 and 7.4, indicating that the ZnO-NPs increased its solubility. The amounts of diosgenin released were not significantly affected by the pH. The pattern for releasing active compounds bound to ZnO-NPs was the same as seen in several previous studies: the longer the time, the greater the number of compounds released.30,31 However, diosgenin release was more spontaneous in the early time range (before 20 minutes) at pH 7.4 compared to pH 5.5. This difference can be attributed to the increased ionic strength at pH 7.4.54
887.285 μg. The solubilities of diosgenin in DPBS at pH 5.5 and 7.4 were tested and showed that diosgenin did not dissolve completely in DPBS at pH 5.5 or 7.4, confirming that diosgenin has poor bioavailability.26 However, the diagrams show that diosgenin was successfully released into DPBS at pH 5.5 and 7.4, indicating that the ZnO-NPs increased its solubility. The amounts of diosgenin released were not significantly affected by the pH. The pattern for releasing active compounds bound to ZnO-NPs was the same as seen in several previous studies: the longer the time, the greater the number of compounds released.30,31 However, diosgenin release was more spontaneous in the early time range (before 20 minutes) at pH 7.4 compared to pH 5.5. This difference can be attributed to the increased ionic strength at pH 7.4.54
Conclusions
This research succeeded in preparing ZnO-NPs via HLE-mediated phytofabrication techniques using various solvents, including ethanol, methanol, and distilled water, along with several zinc precursors, such as zinc chloride, zinc acetate, and zinc nitrate. All of the ZnO-NPs have identical characteristics except for the particle sizes. The lowest particle size was obtained with the methanol extract of H. tiliaceus leaves and zinc acetate as the precursor (ZnO-HLMEA), which was then selected for further analysis. Diosgenin was successfully loaded on ZnO-HLMEA with loadings of 79.972% for ZnO-HLMEA-D500 and 39.775% for ZnO-HLMEA-D1000. The higher solubility of diosgenin from ZnO-HLMEA-D500 and ZnO-HLMEA-D1000 compared to free diosgenin was shown by the successful release of diosgenin in less than 100 minutes into DPBS at pH 5.5 and 7.4. The release of diosgenin from ZnO-HLMEA-D500 and ZnO-HLMEA-D1000 was not significantly affected by the pH. Those findings confirm that ZnO-NPs can potentially be delivery vehicles for diosgenin and, conceivably, other water-insoluble drugs. However, it is necessary to find alternative solutions to overcome its cytotoxicity towards normal cells.Methods
Materials
H. tiliaceus leaf powder (HLP) was obtained from Laboratorium Herbal Materia Medica Batu, Batu City, East Java, Indonesia, and had certificate number 074/154A/102.7/2020. Ethanol (Merck), methanol (Merck), zinc chloride [ZnCl2] (Merck), zinc acetate dihydrate [Zn(CH3COO)2·2H2O] (Merck), zinc nitrate hexahydrate [Zn(NO3)2·6H2O] (Merck), potassium hydroxide [KOH] (Merck), diosgenin (Alfa Aesar), Vero normal cells (ATCC), MTT (Sigma Aldrich) and Dulbecco's phosphate buffered saline [DPBS], pH 5.5 and 7.4, were of the analytical grade and used directly as obtained.Preparation of HLE
HLE was prepared via a modified procedure.55 The steps were as follows: 10 g of HLP sifted through a 30 mesh filter was mixed with 100 mL of solvents, ethanol, methanol, or distilled water, each in separate Erlenmeyer flasks. The mixtures were stirred with a rotary shaker at 150 rpm for 24 h and filtered with Whatman paper number 1. The filtrates were recorded as HLEE for the H. tiliaceus leaf ethanol extract, HLME for the H. tiliaceus leaf methanol extract, and HLWE for the H. tiliaceus leaf distilled water extract.Preparation of phytofabricated ZnO-NPs
The ZnO-NPs phytofabrication process used modified procedures.56,57 KOH (0.05 moles) was dissolved in 25 mL of distilled water while heating at 60 °C and stirring. This solution was filled into the burette for further use. First, 0.05 moles of the zinc precursors, including zinc chloride, zinc acetate, or zinc nitrate, in separate Erlenmeyer flasks were dissolved in 50 mL of distilled water while heating at 60 °C and stirring. The KOH solution was slowly dripped into the zinc precursor solution while heated at 60 °C and stirred until the pH reached 12 for the optimum synthetic conditions.58 After a white precipitate was formed, 50 mL of HLE was added to the mixture and stirred with a rotary shaker at 150 rpm for 24 h. The mixture was then sonicated at 40 kHz for 30 min. To obtain the residue, the mixture was centrifuged. The white precipitate was then dried in an oven at a temperature of 50–60 °C until it reached a constant weight. Nine types of ZnO-NPs were successfully synthesized with this process, including ZnO-HLEEC (ZnO-NPs from the zinc chloride precursor mediated by HLEE), ZnO-HLEEA (ZnO-NPs from the zinc acetate precursor mediated by HLEE), ZnO-HLEEN (ZnO-NPs from the zinc nitrate precursor mediated by HLEE), ZnO-HLMEC (ZnO-NPs from the zinc chloride precursor mediated by HLME), ZnO-HLMEA, ZnO-HLMEN (ZnO-NPs from the zinc nitrate precursor mediated by HLME), ZnO-HLWEC (ZnO-NPs from the zinc chloride precursor mediated by HLWE), ZnO-HLWEA (ZnO-NPs from the zinc acetate precursor mediated by HLEE), and finally ZnO-HLWEN (ZnO-NPs from the zinc nitrate precursor mediated by HLWE). A summary table of ZnO-NPs nomenclature is presented in the ESI file.†Characterization of the ZnO-NPs
XRF (PANalytical, Minipal 4) was used to examine the chemical composition of ZnO-NPs. The crystallinity and shapes of the ZnO-NPs were determined by XRD (PANalytical, X'Pert PRO) with a 2θ range of 10–90° and scan intervals of 0.017°/0.7 seconds. Fourier transform infrared (FTIR) spectrometer (Shimadzu, IR Prestige 21) at 4000–400 cm−1 was used to figure out the functional groups contained in the ZnO-NPs. The surface morphology and particle sizes of the ZnO-NPs were observed with SEM (FEI, Inspect-S50) after coating with Au–Pd. ZnO-HLMEA exhibited the smallest particle sizes, as shown by SEM. It was also investigated with TEM (Hitachi, HT7700) and selected for further processing.Loading of diosgenin on the ZnO-NPs
The diosgenin was loaded on ZnO-NPs as previously described but with modifications.30 The doping process was begun by preparing diosgenin solutions in ethanol with concentrations of 500 μg mL−1 and 1000 μg mL−1. Next, 0.1 g of ZnO-HLMEA was added into two different Erlenmeyer flasks, then mixed with 20 mL of the diosgenin solutions with concentrations of 500 μg mL−1 and 1000 μg mL−1. After that, the mixture was stirred at 100 rpm for 12 h. The products were precipitated by centrifugation at 4000 rpm for 30 min 2 times at a minimum. The obtained ZnO-NPs/diosgenin residue was dried in an oven at 50 °C for 24 h. Two samples of ZnO-NPs/diosgenin were produced by doping with 500 μg mL−1 diosgenin, called ZnO-HLMEA-D500, and 1000 μg mL−1, called ZnO-HLMEA-D1000.Characterization of ZnO-HLMEA-D500 and ZnO-HLMEA-D1000
The functional groups contained in ZnO-HLMEA-D500 and ZnO-HLMEA-D1000 were analyzed by FTIR (Shimadzu, 8400S) over the spectral range 4000–400 cm−1. Thermogravimetric Analyses (TGA) (TA Instruments, TGA 5500) were used to determine the stability and thermal degradation trends for ZnO-HLMEA-D500 and ZnO-HLMEA-D1000 at a maximum temperature of 1000 °C.Cell viability assay
The cell viability assay of normal Vero cells exposed to ZnO-HLMEA-D1000 and diosgenin was performed using the MTT method. The exposed concentrations of ZnO-HLMEA-D1000 and diosgenin ranged from 0 to 200 μg mL−1. After 24 hours of incubation, live cells were counted with the performance of the MTT reagent.Evaluation of diosgenin release
Diosgenin released from ZnO-HLMEA-D500 and ZnO-HLMEA-D1000 was determined by a modified procedure that was previously reported.30 The diosgenin release process was carried out by immersing 0.0500 g of ZnO-HLMEA-D500 and ZnO-HLMEA-D1000 in 250 mL of DPBS solutions with pH 5.5 and 7.4 at 37 °C with gentle stirring. The amount of released diosgenin was determined with a UV-vis spectrophotometer (Thermo Scientific, Genesys 10S UV-vis) at wavelengths of 203 nm for pH 5.5 and 230 nm for pH 7.4. Samples were taken every 15 minutes and stopped when 100% of the diosgenin was released. The diosgenin released (DR) amount was determined with eqn (2).
 | (2) |
Data availibility
The data supporting this article have been included as part of the ESI.†Author contributions
OKP: conceptualization, data curation, writing – original draft. LOR: data curation. YK: conceptualization, writing – review & editing. AF: conceptualization, writing – review & editing. RS: data curation. MS: conceptualization, writing – review & editing.Conflicts of interest
The authors have no competing interest to proclaim.Acknowledgements
The authors gratefully acknowledge financial support from the Institut Teknologi Sepuluh Nopember for this work under the project scheme of Penelitian Kerjasama antar Perguruan Tinggi (PAKERTI) 2024 with contract number 1256/PKS/ITS/2024.References
- J. Jiang, J. Pi and J. Cai, Bioinorg. Chem. Appl., 2018, 2018, 1–18 CrossRef.
- A. Aboelnga, H. Salaheldin and A. Elsayed, Egypt. J. Chem., 2023, 67, 555–562 Search PubMed.
- O. K. Putri, H. Syafdhani, H. Holilah, A. Fadlan, Y. Kusumawati, M. Santoso, D. Prasetyoko and H. J. Achmad, Rasayan J. Chem., 2022, 15, 2835–2843 CrossRef CAS.
- B. Boro, J. S. Boruah, C. Devi, A. Alemtoshi, B. Gogoi, P. Bharali, P. V. B. Reddy, D. Chowdhury and P. Kalita, J. Mol. Struct., 2024, 1300, 1–15 CrossRef.
- S. Faisal, H. Jan, S. A. Shah, S. Shah, A. Khan, M. T. Akbar, M. Rizwan, F. Jan, W. Wajidullah, N. Akhtar, A. Khattak and S. Syed, ACS Omega, 2021, 6, 9709–9722 CrossRef CAS.
- S. Keerthana and A. Kumar, Crit. Rev. Toxicol., 2020, 50, 47–71 CrossRef CAS PubMed.
- A. Sharma, R. Nagraik, B. Venkidasamy, A. Khan, K. Dulta, P. Chauhan, D. Kumar and D. Shin, Luminescence, 2023, 38, 1139–1148 CrossRef CAS.
- R. Ananthalakshmi, S. R. X. R. Rathinam and A. M. Sadiq, Res. J. Pharm. Technol., 2021, 14, 2004–2008 Search PubMed.
- K. Dulta, G. K. Ağçeli, P. Chauhan, R. Jasrotia and P. K. Chauhan, J. Inorg. Organomet. Polym., 2021, 31, 180–190 CrossRef CAS.
- A. Es-haghi, M. E. Taghavizadeh Yazdi, M. Sharifalhoseini, M. Baghani, E. Yousefi, A. Rahdar and F. Baino, Biomimetics, 2021, 6, 34 CrossRef CAS PubMed.
- T. A. Ibrahim, T. A. Salman and S. al-Rudha Abbas, Plant Arch., 2021, 21, 329–332 CrossRef.
- A. V. Blinov, M. D. Kachanov, A. A. Gvozdenko, A. A. Nagdalian, A. A. Blinova, Z. A. Rekhman, A. B. Golik, D. S. Vakalov, D. G. Maglakelidze, A. G. Nagapetova, A. D. Pokhilko and I. V. Burkina, Gels, 2023, 9, 57 CrossRef CAS.
- P. Rajiv, S. Rajeshwari and R. Venckatesh, Spectrochim. Acta, Part A, 2012, 112, 384–387 CrossRef PubMed.
- T. Kang, R. Guan, X. Chen, Y. Song, H. Jiang and J. Zhao, Nanoscale Res. Lett., 2013, 8, 1–8 CrossRef.
- M. Husnah, S. Suhartono and Y. S. Ismail, IOP Conf. Ser. Earth Environ. Sci., 2021, 711, 012019 CrossRef.
- M. F. S. Hermandy, M. Z. M. Yusoff, M. S. Yahya and M. R. Awal, Int. J. Emerg. Trends Eng. Res., 2020, 8, 6896–6902 CrossRef.
- A. N. Mustapa, Á. Martin, R. B. Mato and M. J. Cocero, Ind. Crops Prod., 2015, 74, 83–94 CrossRef CAS.
- M. Singh, A. Jha, A. Kumar, N. Hettiarachchy, A. K. Rai and D. Sharma, J. Food Sci. Technol., 2014, 51, 2070–2077 CrossRef CAS PubMed.
- P. R. Verma, F. Khan and S. Banerjee, Inorg. Nano-Met. Chem., 2021, 51, 427–433 CrossRef CAS.
- S. Sharma, K. Kumar, N. Thakur, S. Chauhan and M. S. Chauhan, J. Mater. Sci., 2020, 43, 1–20 Search PubMed.
- S. Fakhari, M. Jamzad and H. K. Fard, Green Chem. Lett. Rev., 2019, 12, 19–24 CrossRef CAS.
- R. Pachaiappan, S. Rajendran, G. Ramalingam, D.-V. N. Vo, P. M. Priya and M. Soto-Moscoso, Chem. Eng. Technol., 2021, 44, 551–558 CrossRef CAS.
- N. Babayevska, Ł. Przysiecka, I. Iatsunskyi, G. Nowaczyk, M. Jarek, E. Janiszewska and S. Jurga, Sci. Rep., 2022, 12, 1–13 CrossRef.
- P. Semwal, S. Painuli, T. Abu-Izneid, A. Rauf, A. Sharma, S. D. Daştan, M. Kumar, M. M. Alshehri, Y. Taheri, R. Das, S. Mitra, T. B. Emran, J. Sharifi-Rad, D. Calina and W. C. Cho, Oxid. Med. Cell. Longev., 2022, 2022, 1–17 CrossRef.
- K. Moodley, J. Rarey and D. Ramjugernath, J. Chem. Thermodyn., 2017, 106, 199–207 CrossRef CAS.
- F.-C. Wu and J.-G. Jiang, Food Funct., 2019, 10, 7022–7036 RSC.
- T.-H. T. Nguyen, N.-T. Trinh, H. N. Tran, H. T. Tran, P. Q. Le, D.-N. Ngo, H. Tran-Van, T. V. Vo, L. B. Vong and Y. Nagasaki, J. Contr. Release, 2021, 331, 515–524 CrossRef CAS.
- G. F. Paciotti, J. Zhao, S. Cao, P. J. Brodie, L. Tamarkin, M. Huhta, L. D. Myer, J. Friedman and D. G. I. Kingston, Bioconjugate Chem., 2016, 27, 2646–2657 CrossRef CAS PubMed.
- M. Akbarian, S. Mahjoub, S. M. Elahi, E. Zabihi and H. Tashakkorian, Colloids Surf., B, 2020, 186, 1–18 CrossRef PubMed.
- P. Sathishkumar, Z. Li, R. Govindan, R. Jayakumar, C. Wang and F. Gu, Appl. Surf. Sci., 2021, 536, 147741 CrossRef CAS.
- Q. Wang, S. Mei, P. Manivel, H. Ma and X. Chen, Curr. Res. Nutr. Food Sci., 2022, 5, 868–877 CAS.
- Z. M. AL-Asady, A. H. AL-Hamdani and M. A. Hussein, AIP Conf. Proc., 2020, 2213, 020061 CrossRef CAS.
- M. F. Al-Ajmi, A. Hussain, E. Alsharaeh, F. Ahmed, S. Amir, M. S. Anwar, M. A. Siddiqui, A. A. Al-Khedhairy and B. H. Koo, Adv. Mater. Sci., 2018, 10, 349–355 CrossRef CAS.
- J. Arumugam, S. Thambidurai, S. Suresh, M. Selvapandiyan, M. Kandasamy, N. Pugazhenthiran, S. Karthick Kumar, T. Muneeswaran and F. Quero, Chem. Phys. Lett., 2021, 783, 139040 CrossRef CAS.
- G. Sharmila, M. Thirumarimurugan and C. Muthukumaran, Microchem. J., 2019, 145, 578–587 CrossRef CAS.
- A. S. Bhosale, K. K. Abitkar, P. S. Sadalage, K. D. Pawar and K. M. Garadkar, J. Mater. Sci. Mater. Electron., 2021, 32, 20510–20524 CrossRef CAS.
- D. Borah, M. K. Baruah, P. P. Saikia, K. K. Senapoty, M. Barua and R. Singha, J. Mater. Environ. Sci., 2016, 7, 331–336 CAS.
- W. Muhammad, N. Ullah, M. Haroon and B. H. Abbasi, RSC Adv., 2019, 9, 29541–29548 RSC.
- M. Sundrarajan, S. Ambika and K. Bharathi, Adv. Powder Technol., 2015, 26, 1294–1299 CrossRef CAS.
- F. Rashid, I. Pervaiz, H. Malik, Z. Kanwal, M. Rafique and S. S. A. Gillani, Comb. Chem. High Throughput Screen., 2022, 25, 1200–1206 CrossRef CAS PubMed.
- P. Sharma, M. Urfan, R. Anand, M. Sangral, H. R. Hakla, S. Sharma, R. Das, S. Pal and M. Bhagat, Physiol. Mol. Biol. Plants, 2022, 28, 363–381 CrossRef CAS.
- J. N. Hasnidawani, H. N. Azlina, H. Norita, N. N. Bonnia, S. Ratim and E. S. Ali, Procedia Chem., 2016, 19, 211–216 CrossRef CAS.
- B. Rabha, K. K. Bharadwaj, D. Baishya, T. Sarkar, H. A. Edinur and S. Pati, Polymers, 2021, 13, 1–17 Search PubMed.
- S. Ghosh, P. More, A. Derle, R. Kitture, T. Kale, M. Gorain, A. Avasthi, P. Markad, G. C. Kundu, S. Kale, D. D. Dhavale, J. Bellare and B. A. Chopade, J. Nanosci. Nanotechnol., 2015, 15, 9464–9472 CrossRef CAS PubMed.
- P. Kumar, V. Pillay and Y. E. Choonara, Sci. Rep., 2021, 11, 3104 CrossRef CAS.
- G. Murtaza, R. Ahmad, M. S. Rashid, M. Hassan, A. Hussnain, M. A. Khan, M. E. Ul Haq, M. A. Shafique and S. Riaz, Curr. Appl. Phys., 2014, 14, 176–181 CrossRef.
- N. Gong, Y. Wang, B. Zhang, D. Yang, G. Du and Y. Lu, Steroids, 2019, 143, 18–24 CrossRef CAS PubMed.
- K. Nithya and S. Kalyanasundharam, OpenNano, 2019, 4, 100024 CrossRef.
- M. Bata and S. Rahayua, Curr. Bioact. Compd., 2017, 13, 157–164 CrossRef CAS.
- F. Chen, L. Fu, L. Feng, C. Liu and B. Ren, Russ. J. Phys. Chem., 2013, 87, 1611–1614 CrossRef CAS.
- Ö. S. Aslantürk, IntechOpen, 2018, 1, 1–18 Search PubMed.
- L. Tolosa, M. T. Donato and M. J. Gómez-Lechón, Methods Mol. Biol., 2015, 1250, 333–348 CrossRef CAS PubMed.
- H. Maheswaran, S. Djearamane, A. C. T. A. Dhanapal and L. S. Wong, Heliyon, 2024, 10, e31316 CrossRef CAS PubMed.
- S. Pavlukhina, Y. Lu, A. Patimetha, M. Libera and S. Sukhishvili, Biomacromolecules, 2010, 11, 3448–3456 CAS.
- O. K. Putri, L. O. Rahayu, G. F. Hadiwibowo and Y. Kusumawati, E3S Web Conf., 2023, 374, 00025 CrossRef CAS.
- M. A. M. Moazzen, S. M. Borghei and F. Taleshi, Appl. Nanosci., 2013, 3, 295–302 CrossRef.
- E. K. Droepenu, E. A. Asare, B. S. Wee, R. B. Wahi, F. Ayertey and M. O. Kyene, Beni-Suef Univ. J. Basic Appl. Sci., 2021, 10, 1–10 Search PubMed.
- F. H. Abdullah, N. H. H. Abu Bakar and M. Abu Bakar, Optik, 2020, 206, 164279 CrossRef CAS.
Footnote |
| † Electronic supplementary information (ESI) available. See DOI: https://doi.org/10.1039/d4ra03249g |
| This journal is © The Royal Society of Chemistry 2024 |