Open Access Article
Open Access ArticlePalladium(II), platinum(II), and silver(I) complexes with 3-acetylcoumarin benzoylhydrazone Schiff base: Synthesis, characterization, biomolecular interactions, cytotoxic activity, and computational studies†
Shadia A. Elsayed *,
Islam M. Elnabky
*,
Islam M. Elnabky ,
Mohamed M. Aboelnga
,
Mohamed M. Aboelnga * and
Ahmed M. El-Hendawy
* and
Ahmed M. El-Hendawy
Chemistry Department, Faculty of Science, Damietta University, New Damietta, 34517, Egypt. E-mail: shadia.elsayed@du.edu.eg; mohamed-aboelnga@du.edu.eg
First published on 18th June 2024
Abstract
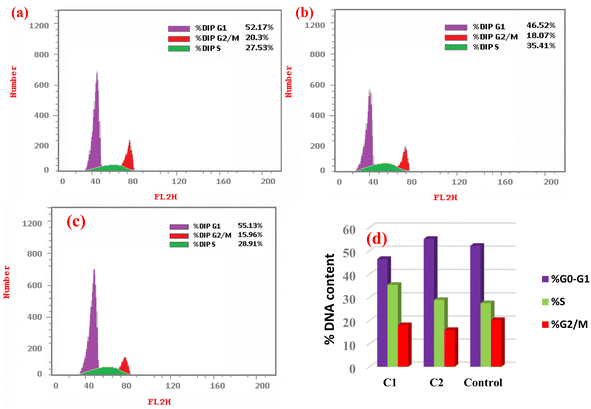
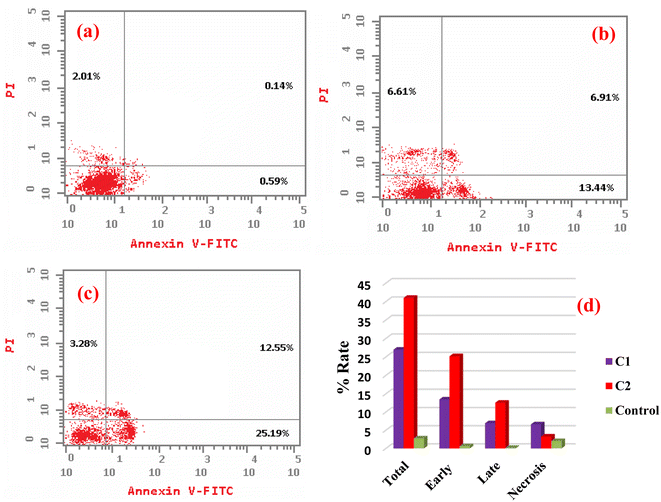
New Pd(II) (C1), Pt(II) (C2), and Ag(I) (C3) complexes derived from 3-acetylcoumarin benzoylhydrazone (HL) Schiff base were synthesized and characterized by FTIR, 1H NMR, UV-visible spectroscopies along with elemental analysis (C, H, N), magnetic, molar conductivity measurements, and DFT calculations. The obtained results suggested that the ligand had different behaviors in the complexes: mono-negative tridentate (C1) and neutral tridentate (C2) as an ONO-donor and neutral bidentate (C3) as an ON-donor. Quantum chemistry calculations were performed to validate the stability of the suggested geometries and indicated that all the complexes possess tetra-coordinated metal ions. The binding affinity of all the compounds toward calf thymus (ctDNA), yeast (tRNA), and bovine serum albumin (BSA) was evaluated by absorption/emission spectral titration studies, which revealed the intercalative binding to ctDNA and tRNA and static binding upon complex formation with BSA. Molecular insights into the binding affinity of the characterized complexes were provided through conducting molecular docking analysis. Moreover, the cytotoxic activity (in vitro) of the compounds was screened against human cancerous cell lines and a non-cancerous lung fibroblast (WI38) one using cis-platin as a reference drug. The IC50 and selective index (SI) values indicated the higher cytotoxic activity of all the metal complexes compared to their parent ligand. Among all the compounds, the complex C2 showed the highest activity. These results confirmed the improvement of the anticancer activity of the ligand by incorporating the metal ions. In addition, flow cytometry results showed that complexes C1 and C2 induced cell cycle arrest at S and G1/S, respectively.
1. Introduction
Coumarins (2H-1-benzopyran-2-one) are a crucial class of oxygen-based heterocyclic compounds of natural origin and can be also found in synthetic medicinally active compounds.1,2 They have been used in flavoring foods and in cosmetic products as a fragrant.3 Coumarin-containing compounds have attracted considerable interest in biological applications including in vitro/in vivo anticancer,4,5 antimicrobial, antioxidant, and anti-inflammatory activities.1 In comparison to free coumarin-based ligands, the anticancer and antimicrobial activities have been enhanced by coordination with metal ions.6–8 Recently, coumarin-derived Schiff bases along with associated metal complexes have been considered to be the most efficient classes of compounds that exhibit a wide range of biological activities.7 It has been found that the C3-position of coumarin is an effective position that can improve the cytotoxic efficacy against several human cell lines.9–11 In particular, 3-acetylcoumarin has demonstrated high reactivity to form remarkably stable Schiff base ligands such as hydrazone (ONO-donor ligands) and the synthesis of their metal complexes.12On the other hand, hydrazone Schiff bases have also attracted considerable scientific attention owing to their unique structural and biological/pharmacological properties.13–15 The keto–enol tautomerism of the hydrazone structure gives them the possibility to coordinate metal ions through azomethine nitrogen, or ketonic or enolic amide oxygen. Additionally, the coordination site from the substituent attached to the hydrazide amine group offers diverse coordination modes.16–20 In fact, a series of hydrazide/hydrazine coumarin-based derivatives have been reported for their potential antitumor activity.21 Moreover, hydrazone-based complexes were reported to improve the binding affinity to DNA, as well as the antitumor activity.22,23 Therefore, the structural improvement of hydrazone/coumarin may lead to an important category of compounds with utility for designing and developing new potential drugs.24,25 Indeed, the literature is rich with numerous coumarin-derived complexes with therapeutic applications due to their anticancer, antimicrobial, and antioxidant effects and as enzyme inhibitors.7,26 For instance, a series of 3-acetylcoumarin hydrazone Schiff base complexes containing nickel(II), cobalt(II), copper(II),27 ruthenium(II), Rh(III), and Ir(III)28 have been reported for their anticancer and antibacterial activities.
Based on the reported biological activities of coumarin, hydrazone Schiff bases, and their metal chelates, and also on a continuation of our research on acetylcoumarin and hydrazone Schiff bases,29–31 we sought to develop novel metal complexes containing Pd(II), Pt(II), and Ag(I) based on 3-acetylcouamrin benzoylhydrazone Schiff base. Moreover, we evaluated their interaction with biomolecules (ctDNA, tRNA, and BSA), and in addition, their in vitro cytotoxic activity against human breast carcinoma (MCF7), cervical carcinoma (HeLa), and normal lung fibroblast (WI38) cell lines. Moreover, the cell death mechanism was elucidated by flow cytometry. Later, DFT calculations were performed to further support the proposed structures while molecular docking was carried out to provide deep insights into their interaction with BSA.
2. Experimental
2.1. Materials and equipment
Details on the materials and physical measurements are presented in the ESI materials (ESI,† Section 1.1).2.2. Synthesis
![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) O)lactone 1721, ν(C
O)lactone 1721, ν(C![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) O)hydrazone1664, ν(C
O)hydrazone1664, ν(C![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) N) 1611, ν(N–N) 963 cm−1. 1H NMR (400 MHz, DMSO-d6, δ, ppm): δ 10.85 (s, 1H, NH), δ 8.26 (s, 1H, H4), δ 7.88 (d, 2H, H6,9), δ 7.67 (t, 2H, H7,8), δ 7.53 (t, 2H, H16,18), δ 7.47 (d, 2H, H15,19), δ 7.41 (t, 1H, H17), δ 2.35 (s, 3H, H12). 13C NMR (100 MHz, DMSO-d6, δ, ppm): C(2) 159.7, C(3) 127.3, C(4) 142.4, C(5) 119.5, C(6) 116.5, C(7) 125.3, C(8) 133.5, C(9) 128.7, C(10) 151.6, C(11) 153.9, C(13) 128.3, C(14) 134.5, C(15,19) 130, C(16, 18) 129.7, C(17) 128.8, C(CH3) 16.7. UV-vis (DMSO, 2.5 × 10−5 M): λmax (nm) (ε, M−1 cm−1): 232 (100
N) 1611, ν(N–N) 963 cm−1. 1H NMR (400 MHz, DMSO-d6, δ, ppm): δ 10.85 (s, 1H, NH), δ 8.26 (s, 1H, H4), δ 7.88 (d, 2H, H6,9), δ 7.67 (t, 2H, H7,8), δ 7.53 (t, 2H, H16,18), δ 7.47 (d, 2H, H15,19), δ 7.41 (t, 1H, H17), δ 2.35 (s, 3H, H12). 13C NMR (100 MHz, DMSO-d6, δ, ppm): C(2) 159.7, C(3) 127.3, C(4) 142.4, C(5) 119.5, C(6) 116.5, C(7) 125.3, C(8) 133.5, C(9) 128.7, C(10) 151.6, C(11) 153.9, C(13) 128.3, C(14) 134.5, C(15,19) 130, C(16, 18) 129.7, C(17) 128.8, C(CH3) 16.7. UV-vis (DMSO, 2.5 × 10−5 M): λmax (nm) (ε, M−1 cm−1): 232 (100![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 000), 336 (90
000), 336 (90![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 000).
000).[Pd(L)Cl] (C1): Yellow precipitate, yield: 75%, m.p.: 190–192 °C. Elemental analysis for C18H13ClN2O3Pd: calcd (found): C, 48.35 (48.03); H, 2.93 (2.80); N, 6.26 (6.14). FTIR (cm−1): ν(C![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) O)lactone 1620, ν(C–O)new 1221, ν(C
O)lactone 1620, ν(C–O)new 1221, ν(C![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) N) 1589, ν(C
N) 1589, ν(C![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) N)new 1554, ν(N–N) 975, ν(M–O) 575, ν (M–N) 442. 1H NMR (400 MHz, DMSO-d6, δ, ppm): δ 8.53 (s, 1H, H4), δ, 8.02, 7.85 (d, 2H, H6, 9), δ 7.66 (t, 2H, H7,8), δ 7.32 (t, 2H, H16,18), δ 7.42 (d, 2H, H15,19), δ 7.03 (t, 1H, H17), δ 2.66 (s, 3H, H12). UV-vis (DMSO, 2.5 × 10−5 M): λmax (nm) (ε, M−1 cm−1): 276 (127
N)new 1554, ν(N–N) 975, ν(M–O) 575, ν (M–N) 442. 1H NMR (400 MHz, DMSO-d6, δ, ppm): δ 8.53 (s, 1H, H4), δ, 8.02, 7.85 (d, 2H, H6, 9), δ 7.66 (t, 2H, H7,8), δ 7.32 (t, 2H, H16,18), δ 7.42 (d, 2H, H15,19), δ 7.03 (t, 1H, H17), δ 2.66 (s, 3H, H12). UV-vis (DMSO, 2.5 × 10−5 M): λmax (nm) (ε, M−1 cm−1): 276 (127![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 000), 330 (52
000), 330 (52![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 000). Molar conductivity (10−3 M, DMSO), Λm = 2.0 Ω m2 mol−1.
000). Molar conductivity (10−3 M, DMSO), Λm = 2.0 Ω m2 mol−1.
[Pt(HL)Cl]Cl (C2): Orange precipitate, yield: 64%, m.p.: 200–202 °C. Elemental analysis for C18H14Cl2N2O3Pt, calcd (found): 37.78 (37.86); H, 2.47 (2.39); N, 4.89 (4.77). FTIR (KBr, cm−1): ν (NH) 3278, ν(C![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) O)lactone 1724, ν(C
O)lactone 1724, ν(C![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) O)hydrazone 1663, ν(C
O)hydrazone 1663, ν(C![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) N) 1575, ν(N–N) 1021, ν(M–O) 529, ν (M–N) 435. 1H NMR (400 MHz, DMSO-d6, δ, ppm): δ 10.72 (s, 1H, NH), δ 8.84 (s, 1H, H4), δ 8.13 (d, 2H, H6,9), δ 7.89 (t, 2H, H7,8), δ 7.60 (t, 2H, H16,18), δ 7.77 (d, 2H, H15,19), δ 7.50 (t, 1H, H17), δ 2.63 (s, 3H, H12). UV-vis (DMSO, 2.5 × 10−5 M): λmax (nm) (ε, M−1 cm−1): 276 (33
N) 1575, ν(N–N) 1021, ν(M–O) 529, ν (M–N) 435. 1H NMR (400 MHz, DMSO-d6, δ, ppm): δ 10.72 (s, 1H, NH), δ 8.84 (s, 1H, H4), δ 8.13 (d, 2H, H6,9), δ 7.89 (t, 2H, H7,8), δ 7.60 (t, 2H, H16,18), δ 7.77 (d, 2H, H15,19), δ 7.50 (t, 1H, H17), δ 2.63 (s, 3H, H12). UV-vis (DMSO, 2.5 × 10−5 M): λmax (nm) (ε, M−1 cm−1): 276 (33![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 500), 327 (25
500), 327 (25![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 700). Molar conductivity (10−3 M, DMSO), Λm = 65 Ω m2 mol−1.
700). Molar conductivity (10−3 M, DMSO), Λm = 65 Ω m2 mol−1.
![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) O)lactone 1698, ν(C
O)lactone 1698, ν(C![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) O)hydrazone 1664, ν(C
O)hydrazone 1664, ν(C![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) N) 1608, ν(N–N) 971, ν(M–O) 545, ν (M–N) 439. 1H NMR (400 MHz, DMSO-d6, δ, ppm): δ 11.05 (s, 1H, NH), δ 8.29 (s, 1H, H4), δ 7.90 (d, 2H, H6,9), δ 7.68 (t, 2H, H7,8), δ 7.56 (t, 2H, H16,18), δ 7.48 (d, 2H, H15,19), δ 7.42 (t, 1H, H17), δ 2.38 (s, 3H, H12). 13C NMR (100 MHz, DMSO-d6, δ, ppm): C(2) 161.6, C(3) 127.7, C(4) 142.5, C(5) 119.2, C(6) 116.7, C(7) 125.2, C(8) 133.3, C(9) 128.6, C(10) 151.6, C(11) 154.1, C(13), 128.7, C(14) 134.0, C(15,19) 129.7, C(16, 18) 129.5, C(17) 128.6, C(CH3) 16.7. UV-vis (DMSO, 2.5 × 10−5 M): λmax (nm) (ε, M−1 cm−1): 226 (230
N) 1608, ν(N–N) 971, ν(M–O) 545, ν (M–N) 439. 1H NMR (400 MHz, DMSO-d6, δ, ppm): δ 11.05 (s, 1H, NH), δ 8.29 (s, 1H, H4), δ 7.90 (d, 2H, H6,9), δ 7.68 (t, 2H, H7,8), δ 7.56 (t, 2H, H16,18), δ 7.48 (d, 2H, H15,19), δ 7.42 (t, 1H, H17), δ 2.38 (s, 3H, H12). 13C NMR (100 MHz, DMSO-d6, δ, ppm): C(2) 161.6, C(3) 127.7, C(4) 142.5, C(5) 119.2, C(6) 116.7, C(7) 125.2, C(8) 133.3, C(9) 128.6, C(10) 151.6, C(11) 154.1, C(13), 128.7, C(14) 134.0, C(15,19) 129.7, C(16, 18) 129.5, C(17) 128.6, C(CH3) 16.7. UV-vis (DMSO, 2.5 × 10−5 M): λmax (nm) (ε, M−1 cm−1): 226 (230![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 000), 332 (70
000), 332 (70![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 000). Molar conductivity (10−3 M, DMSO), Λm = 53 Ω m2 mol−1.
000). Molar conductivity (10−3 M, DMSO), Λm = 53 Ω m2 mol−1.2.3. Solution stability
First, 50 μL (1.0 × 10−3 M) of the test compounds (in DMSO) was added to 1950 μL phosphate buffered saline (PBS, pH 7.2) in a UV-visible cuvette, until the final concentration reached 2.5 × 10−5 M. The electronic spectra were measured at set time intervals (0, 20, 40, 60 min, 24, and 48 h).2.4. Biological studies
2.5. Theoretical calculations
3. Results and discussion
3.1. Synthesis
The ligand 3-acetylcomarin benzoylhydrazone Schiff base (HL) and its complexes (C1)–(C3) were prepared and characterized, as shown in the Experimental section (Section 2.2). The analytical data revealed that the molar ratio (M![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) L) of the complexes was 1
L) of the complexes was 1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1 metal-to-ligand except for [Ag(HL)2], which seemed to have a 1
1 metal-to-ligand except for [Ag(HL)2], which seemed to have a 1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 2 ratio. All the complexes were completely soluble in DMF, and DMSO but sparingly soluble in common organic solvents (methanol, ethanol, CH2Cl2, CH3CN, etc.). The molar conductance values of 10−3 M in DMSO suggested the neutrality of the [Pd(L)Cl] complex (C1) (ΛM = 2.0 Ω−1 cm2 mol−1), while the 1
2 ratio. All the complexes were completely soluble in DMF, and DMSO but sparingly soluble in common organic solvents (methanol, ethanol, CH2Cl2, CH3CN, etc.). The molar conductance values of 10−3 M in DMSO suggested the neutrality of the [Pd(L)Cl] complex (C1) (ΛM = 2.0 Ω−1 cm2 mol−1), while the 1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
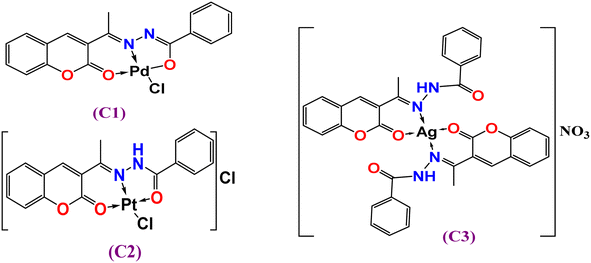
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1 electrolytic nature41 of the other complexes Pt(II) (C2) ((ΛM = 65 Ω−1 cm2 mol−1) and Ag(I) (C3) were in the range of ΛM = 53 Ω−1 cm2 mol−1). The analytical data suggest that the complexes C1–C3 were four coordinates, as indicated in Fig. 1.
1 electrolytic nature41 of the other complexes Pt(II) (C2) ((ΛM = 65 Ω−1 cm2 mol−1) and Ag(I) (C3) were in the range of ΛM = 53 Ω−1 cm2 mol−1). The analytical data suggest that the complexes C1–C3 were four coordinates, as indicated in Fig. 1.
3.2. Characterization
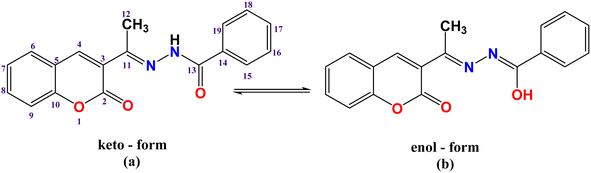
![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) O) of the hydrazone moiety, respectively, suggesting that the ligand displayed an amido keto-form in its solid state (Scheme 1a). In addition, the strong bands that appeared at 1721 cm−1 were attributed to ν(C
O) of the hydrazone moiety, respectively, suggesting that the ligand displayed an amido keto-form in its solid state (Scheme 1a). In addition, the strong bands that appeared at 1721 cm−1 were attributed to ν(C![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) O) of the lactone ring. The strong band at 1611 cm−1 was due to azomethine ν(C
O) of the lactone ring. The strong band at 1611 cm−1 was due to azomethine ν(C![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) N) stretching vibrations.42 Moreover, bands assigned to amide(II), amide(III), and ν(N–N) were observed at 1536, 1273, and 963 cm−1, respectively.43,44
N) stretching vibrations.42 Moreover, bands assigned to amide(II), amide(III), and ν(N–N) were observed at 1536, 1273, and 963 cm−1, respectively.43,44
Generally, the azomethine stretching band ν(C![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) N) was shifted to a lower frequency, suggesting the participation of azomethine-N in coordination with the metal center; however, a diversity of coordination modes was shown by the complexes. For example, in complex [Pd(L)Cl] (C1) (Fig. 1), the HL ligand behaved in a mono-negative tridentate (ONO) manner. This is illustrated by the lower frequency shifts of both the azomethine (C
N) was shifted to a lower frequency, suggesting the participation of azomethine-N in coordination with the metal center; however, a diversity of coordination modes was shown by the complexes. For example, in complex [Pd(L)Cl] (C1) (Fig. 1), the HL ligand behaved in a mono-negative tridentate (ONO) manner. This is illustrated by the lower frequency shifts of both the azomethine (C![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) N) and lactone (C
N) and lactone (C![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) O) groups. Also, there was a disappearance of the ν(NH) and ν(C
O) groups. Also, there was a disappearance of the ν(NH) and ν(C![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) O)hydrazinic through coordination (deprotonation after enolization (Scheme 1b), as supported by the appearance of a new ν(C
O)hydrazinic through coordination (deprotonation after enolization (Scheme 1b), as supported by the appearance of a new ν(C![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) N) group45 near 1589 cm−1, besides the shift of the ν(N–N) band to a higher frequency by 7–58 cm−1. This, in turn, indicated that the bonding occurred through enolate oxygen, accordingly, and new ν(C–O) bands were obtained at 1221 and 1213, respectively. This assumes that the ligand behaves as a mono-negative tridentate using azomethine-N, carbonyl-O, and lactone-O.45,46 In the FTIR spectrum of [Pt(HL)Cl]Cl (C2), the presence of ν(NH) and ν(C
N) group45 near 1589 cm−1, besides the shift of the ν(N–N) band to a higher frequency by 7–58 cm−1. This, in turn, indicated that the bonding occurred through enolate oxygen, accordingly, and new ν(C–O) bands were obtained at 1221 and 1213, respectively. This assumes that the ligand behaves as a mono-negative tridentate using azomethine-N, carbonyl-O, and lactone-O.45,46 In the FTIR spectrum of [Pt(HL)Cl]Cl (C2), the presence of ν(NH) and ν(C![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) O)hydrazinic bands, and the lower frequency shifts of azomethine (C
O)hydrazinic bands, and the lower frequency shifts of azomethine (C![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) N) and lactone (C
N) and lactone (C![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) O) indicate that the coordination took place in a neutral tridentate (ONO) manner. In the FTIR spectrum of [Ag(HL)2]NO3 (C3), the ν(NH) band was shifted to a higher frequency (3277 cm−1), while the ν(HC
O) indicate that the coordination took place in a neutral tridentate (ONO) manner. In the FTIR spectrum of [Ag(HL)2]NO3 (C3), the ν(NH) band was shifted to a higher frequency (3277 cm−1), while the ν(HC![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) N) and lactone-O bands were shifted to lower frequencies; hence the ligand coordinated in a neutral bidentate manner (ON-donor). In addition, the strong absorption near 1365 cm−1 was attributed to the stretching vibration of ν(NO3).47 For all the complexes, new medium bands were observed at 435–449 and 529–575 cm−1, assigned to ν(M–N) and ν(M–O), respectively.47
N) and lactone-O bands were shifted to lower frequencies; hence the ligand coordinated in a neutral bidentate manner (ON-donor). In addition, the strong absorption near 1365 cm−1 was attributed to the stretching vibration of ν(NO3).47 For all the complexes, new medium bands were observed at 435–449 and 529–575 cm−1, assigned to ν(M–N) and ν(M–O), respectively.47
The 13C NMR of HL (Fig. S3a and Table S3†) showed signals at 153.9 and 159.7 and 16.7 ppm assigned to C(11) and C(2) and the methyl carbon CH3 group, respectively similar to an earlier reported ligand.45 These signals were slightly shifted downfield in the spectrum of the Ag(I) complex (Fig. S3b†). This downfield shift by ∼2 ppm indicated the contribution of lactone carbonyl oxygen in the coordination. Moreover, the upfield shift of azomethine-C indicates that the ligand coordinated as an ON-donor through lactone carbonyl oxygen, and azomethine nitrogen. Noteworthily, the 1H and 13C of the silver complex exhibited very little shift in comparison to the parent ligand. This was anticipated due to the coordination of the ligand in its neutral mode and the Ag(I) ion's d10 electronic configuration.
![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) N– and –C
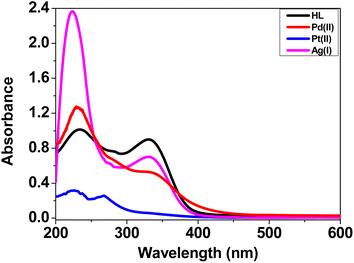
N– and –C![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) O– moieties,53,54 respectively. The electronic spectra of the complexes exhibited a hypsochromic (blue-shift) of these transition bands, along with changes in the absorbance intensity, which were due to the coordination to the center of the metal ions.55,56
O– moieties,53,54 respectively. The electronic spectra of the complexes exhibited a hypsochromic (blue-shift) of these transition bands, along with changes in the absorbance intensity, which were due to the coordination to the center of the metal ions.55,56
![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
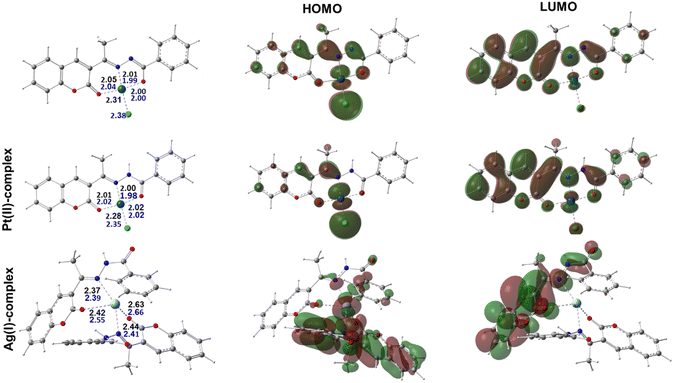
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1 ratio was subtly different with a double ligation from each HL molecule. Its coordination distances were slightly longer than the corresponding ones in Pt(II) and Pd(II) complexes with an average distance of 2.47 Å. Interestingly, the calculated geometries for the three complexes were in accordance with the proposal from the experimental results.
1 ratio was subtly different with a double ligation from each HL molecule. Its coordination distances were slightly longer than the corresponding ones in Pt(II) and Pd(II) complexes with an average distance of 2.47 Å. Interestingly, the calculated geometries for the three complexes were in accordance with the proposal from the experimental results.
Moreover, the most stable geometries for the studied complexes were fully optimized in aqueous medium using a similar functional and basis set combination to further validate their stability in solution. A comparison between the obtained geometries from these later calculations demonstrated there were insignificant changes in both their overall structure and the ligation distances, which agreed well with the experimental observations, Fig. 3. To better explain the chemical reactivity of the three complexes, the energy gap parameter, ΔE, was determined for each complex from the difference between the energy of the highest occupied molecular orbital (EHOMO) and the energy of the lowest unoccupied molecular orbital (ELUMO). In fact, the ΔE value is a stronger procurer for the chemical reactivity of the compounds and the smaller the gap the higher the reactivity of the complex. According to our calculations, the determined energy gap values were found to be 0.0902, 0.0974, and 0.135 au for the Pt(II)-, Pd(II)-, and Ag(I)-containing complexes. Moreover, the softness quantum chemical parameter (σ = 1/ΔE) was calculated and the obtained order for their values was Pt(II) > Pd(II) > Ag(I), with the Pt(II)-containing complex demonstrating the highest value. Therefore, the obtained order of the reactivity from our calculation from both the ΔE and σ values agreed well with the experimental observations.
The HOMO and LUMO were also displayed to better represent the delocalization and localization behavior of the electrons over the characterized complexes, Fig. 3. It was also noted that the HOMO of the Pt(II)- and Pd(II)-containing complexes behaved similarly and mainly localized over both the metal centers and the ligated chloride ions. It is also important to point out that a minor contribution from the ligated atom in the Pd(II) complex in the HOMO was also noticed. For the Ag(I) complex, however, in addition to the localization over the metal center, a significant participation of the aromatic motifs for one of the chelated ligands was observed. Meanwhile, the delocalization of the LUMO also showed some similarity among the three complexes in being distributed over conjugated π-systems of the aromatic rings in the ligand. Overall, this later observation could suggest that the geometry around the metal center together with the conjugated bonds over the aromatic motifs in the ligand are key players in the activity of the studied complexes.
4. Biological applications
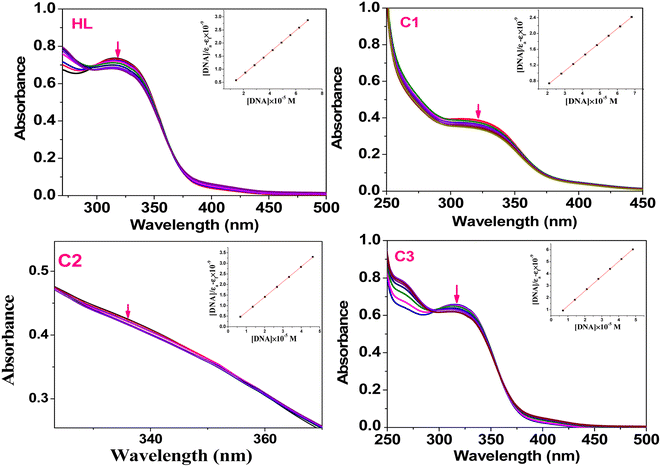
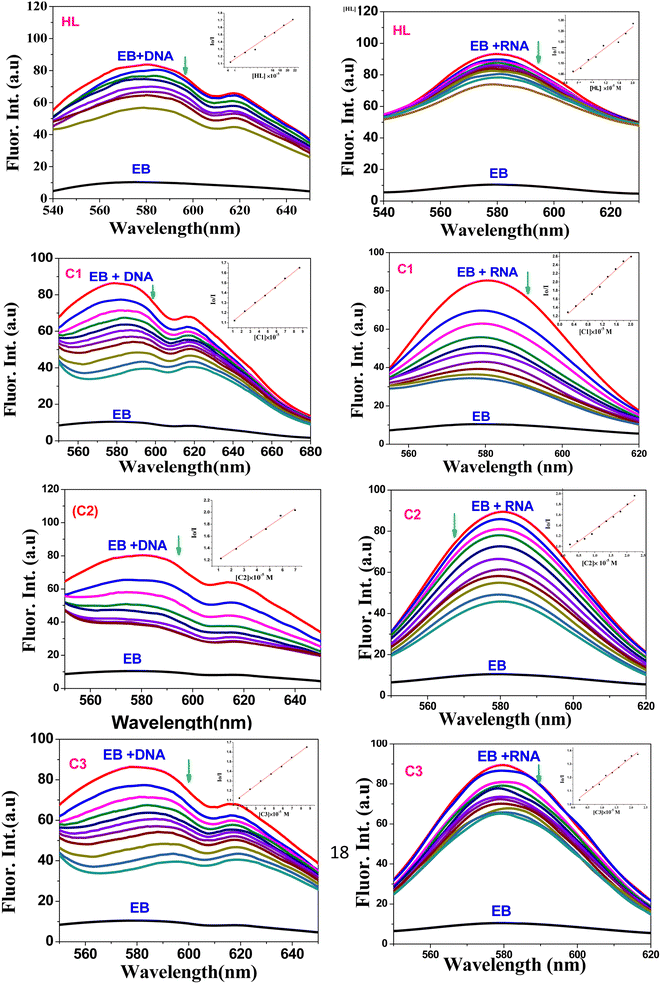
4.1. ctDNA and tRNA interaction studies
Transition metal complexes have a very important function in metallodrug design. They can interact with DNA/RNA via different binding modes: intercalation (insertion), groove binding (major or minor), or electrostatic interaction.60 The intercalative binding is considered the most important one,61 because the molecules are inserted among the base pairs of the DNA double helix, forming π–π stacking interactions that induce elongation and unwinding of the DNA double helix due to the separation of the base pairs to accommodate the drug, and consequently, the DNA binding is responsible for various biological activities; for example, antitumor, antimicrobial, antiviral, anti-inflammatory, and antimalarial properties.62,63 In this section, we evaluated the ctDNA/tRNA binding affinity to the test compounds using both absorption and emission spectroscopies.The presence of the aromatic chromophore existing in the ligand aided the interaction of the compounds with the CT DNA bases via non-covalent π–π* stacking interactions, wherein the π*-orbital of the ligand in the complexes could couple with the π-orbital of the DNA base pairs. The binding constants (Kb) was determined using the Wolfe–Shimer eqn (1):66
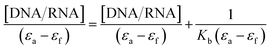 | (1) |
| Compd | λmax | Absorption spectroscopy | Emission spectroscopy (at λem = 580 nm) | ||||||
|---|---|---|---|---|---|---|---|---|---|
| (Kb), × 106 | % H* | (Ksv),×104 | % H | ||||||
| ctDNA | tRNA | ctDNA | tRNA | ctDNA | tRNA | ctDNA | tRNA | ||
| HL | 314 | 1.10 | 1.36 | 8.35 | 7.20 | 0.34 | 1.19 | 32.4 | 26.1 |
| (C1) | 320 | 4.71 | 10.3 | 12.7 | 8.67 | 4.91 | 7.52 | 55.4 | 61.5 |
| (C2) | 329 | 2.58 | 8.96 | 10.4 | 7.60 | 1.42 | 4.51 | 54.2 | 48.8 |
| (C3) | 319 | 1.22 | 1.51 | 6.81 | 6.66 | 0.52 | 1.6 | 50.1 | 26.9 |
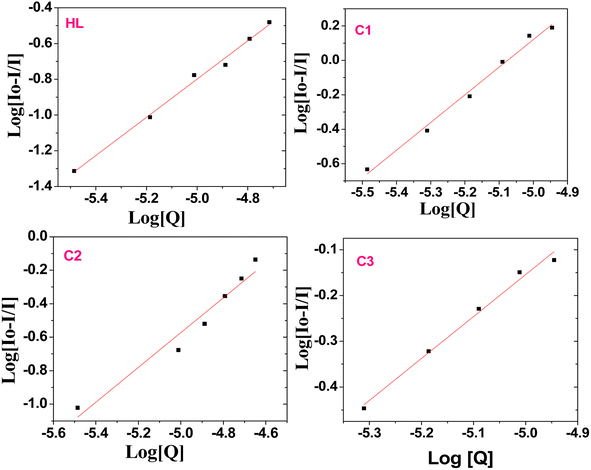
The quenching constant (Ksv) values (Table 1) were determined by the Stern–Volmer eqn (2):67
| Io/I = 1 + KSV[Q] | (2) |
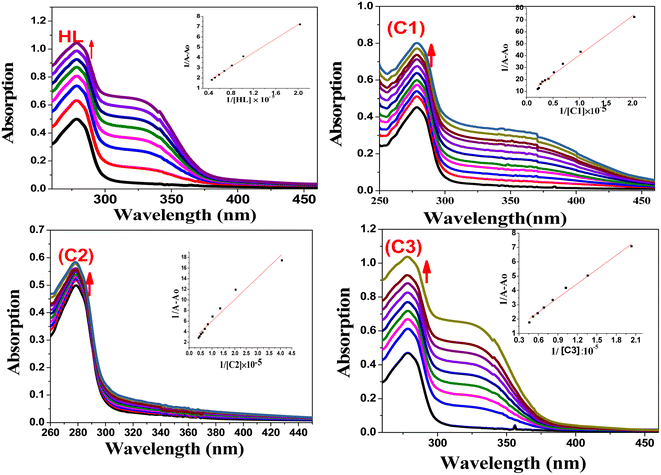
4.2. Bovine serum albumin (BSA) interaction studies
The protein interaction with metal complexes has become an interesting research area, where anticancer drugs are mostly administrated via intravenous injection into the bloodstream, where they are bound to plasma protein.68 The drug–protein binding strength displays a significant role in its accessibility to diffuse from the blood to the target.69 In the following section, we evaluated the binding affinity of BSA protein (human serum albumin analog) by employing absorption and emission measurements.| 1/(Ao − A) = 1/Ao + 1/(K × Ao × CQ) | (3) |
| Absorption spectroscopy λmax = 278 nm | Emission spectroscopy λmax = 345 nm | ||||||
|---|---|---|---|---|---|---|---|
| Compound | (Kb), × 104 | % hyper | % hypo | (Ksv), × 104 | (Kq), × 1012 | (Kb) | n |
| a The observed BSA binding constant (K) values for all the compounds are presented in (2). It was found that the C1 complex exhibited the highest binding strength with BSA in the order of C1 > C2 > C3 > HL. | |||||||
| HL | 1.0 | 39.03 | 27.18 | 2.33 | 2.33 | 8.9 × 103 | 0.93 |
| (C1) | 4.6 | 36.05 | 69.44 | 5.68 | 5.68 | 4.0 × 105 | 1.61 |
| (C2) | 2.5 | 11.90 | 42.63 | 5.39 | 5.39 | 3.9 × 104 | 1.03 |
| (C3) | 1.52 | 40.74 | 35.69 | 4.98 | 4.98 | 2.6 × 104 | 0.92 |
| Kq = Ksv/τo | (4) |
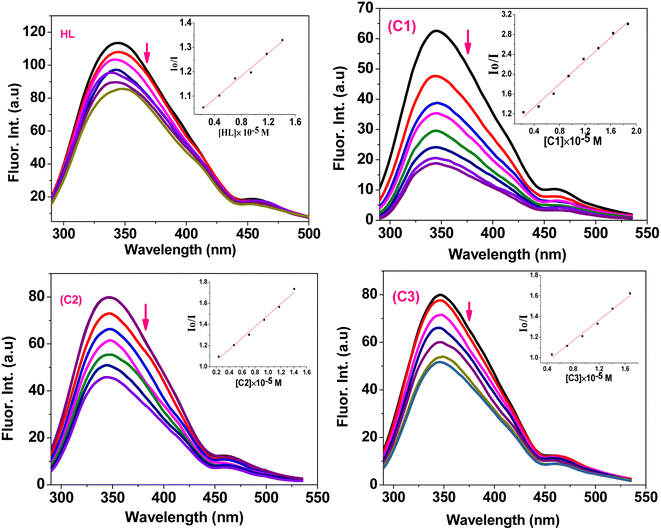 | ||
| Fig. 7 Emission spectra of BSA (50 μM) upon addition of different concentrations of HL and complexes (C1–C3) (0–50 μM) in DMSO/PBS solution. (Inset): plots of Io/I vs. [compound]. | ||
The maximum collision quenching constant (Kq) for biomacromolecules with various quenchers is 1010 M−1 s−1.80 In our study, the resulting Kq values were 0.5–6.1 × 1012 M−1 s−1, which were higher than 1010 M−1 s−1, confirming the static quenching (rather than dynamic) because of the formation of a ground state complex between BSA and the quenchers,81 which confirmed the data obtained from the absorption spectra.
The static binding constant (Kb) and the number of binding sites can be further calculated from the modified Stern–Volmer eqn (5):69,82,83
log[(Io − I)/I] = log![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) Kb + n Kb + n![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) log[Q] log[Q]
| (5) |
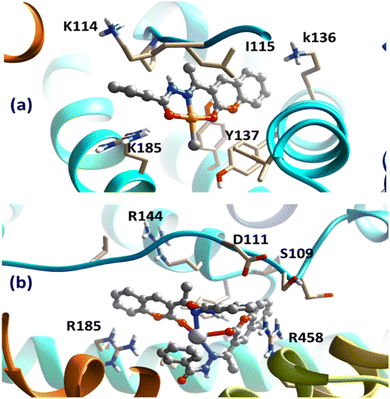 | ||
| Fig. 9 Obtained bound complexes for (a) Pt(II) and (b) Ag(I) complexes with the BSA target. The key amino acids participating in the interactions are highlighted. | ||
Despite binding to distinct locations over the BSA target, both ligands were involved in different types of interaction with the surrounding amino acids of the protein. Most importantly, the cation–π interaction between cationic amino acids, arginine and lysine in particular, with the aromatic domains of the chelating HL were the most dominant interaction. Interestingly, the subtle difference in the obtained binding scores of the two complexes suggested the binding preference of Pt(II)-containing compound over the Ag(I) one, in accordance with the experimental binding results. These obtained findings from the docking analysis further support the validity of the synthesized complexes to act as potential drugs.
4.3. In vitro anticancer activity
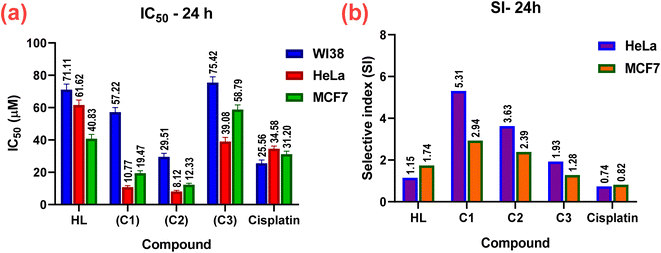
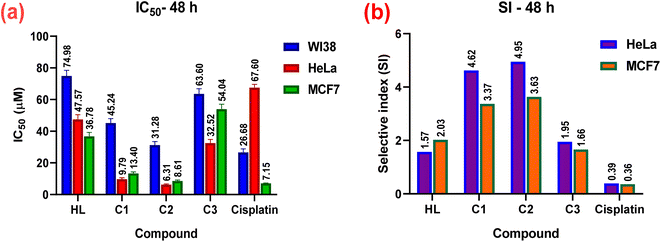
The in vitro cytotoxic effects of HL and its complexes (C1–C3) were evaluated by the colorimetric MTT assay against two human cancer cell lines, namely hormone-dependent breast (MCF7) and cervical (HeLa), in addition to a non-cancerous one, namely the human lung fibroblast (WI38), using cis-platin as a standard drug with exposure times of 24 and 48 h. The results of this study (Table 3, Fig. 10 and 11) showed that all the complexes possessed higher antitumor activity than their parent ligand based on the IC50 and selective index (SI) values. The data revealed that the HeLa cell lines were more sensitive to the complexes than the MCF7 cell lines, as observed from the IC50 and SI values (Table 3). Moreover, all the complexes were found to be more toxic than cis-platin in the HeLa and MCF7 cell lines.| Compound no. | In vitro cytotoxicity IC50 (μM) and selective indices values | ||||
|---|---|---|---|---|---|
| WI38 | HeLa | SI | MCF7 | SI | |
| 24 h | |||||
| HL | 71.11 ± 3.4 | 61.62 ± 3.1 | 1.15 | 40.83 ± 2.6 | 1.74 |
| (C1) | 57.22 ± 2.8 | 10.77 ± 0.9 | 5.31 | 19.47 ± 1.5 | 2.94 |
| (C2) | 29.51 ± 2.2 | 8.12 ± 0.7 | 3.63 | 12.33 ± 0.9 | 2.39 |
| (C3) | 75.42 ± 3.6 | 39.08 ± 2.5 | 1.93 | 58.79 ± 2.9 | 1.28 |
| Std. | 25.56 ± 2.1 | 34.58 ± 1.63 | 0.74 | 31.2 ± 1.8 (ref. 84) | 0.82 |
![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) |
|||||
| 48 h | |||||
| HL | 74.98 ± 3.5 | 47.57 ± 2.9 | 1.57 | 36.78 ± 2.4 | 2.03 |
| (C1) | 45.24 ± 2.7 | 9.79 ± 0.8 | 4.62 | 13.40 ± 0.9 | 3.37 |
| (C2) | 31.28 ± 2.2 | 6.31 ± 0.4 | 4.95 | 8.61 ± 0.6 | 3.63 |
| (C3) | 63.60 ± 3.3 | 32.52 ± 2.3 | 1.95 | 54.04 ± 3.0 | 1.66 |
| Cisplatin | 26.68 ± 2.2 | 67.6 ± 2.0 (ref. 85) | 0.39 | 72.38 ± 0.1 (ref. 86) | 0.36 |
The cytotoxicity and the mechanism of metal complexes mainly depend on their geometry, oxidation state of the metal ion, and redox potential.87–89 According to the IC50 values, the Pt(II) complex (C2) showed the highest cytotoxic activity (i.e., a lower IC50, specifically, IC50 = 8.12 ± 0.7 for HeLa, and 12.33 ± 0.9 for MCF7), which may be due to the cationic nature of the Pt(II)86 rather than the neutral Pd(II) complex, where IC50 = 10.77 ± 0.9 for HeLa and 19.47 ± 1.5 for MCF7. It has been reported that Pd(II) complexes are less stable than Pt(II) ones, because they have a higher aquation rate.90–92 However, the SI values of the Pd(II) complex(C1) (SI = 5.31 for HeLa and 2.94 for MC7) were higher than those of the Pt(II) complex (C2) (SI = 3.63 for HeLa and 2.39 for MCF7), indicating that the Pd(II) complex was more selective and safe for both cell lines. The silver(I) complex had less effect, which may be attributed to the steric hindrance90,93,94 from the introduction of two ligands around the Ag(I) ion (or reduced electrophilicity), which retards the intercalation of the silver(I) complex with DNA (as shown from the binding study, mentioned before), and hence decreased its cytotoxic activity (higher IC50) for both cell lines, as shown in Table 3. It also demonstrated higher selectivity toward both cell lines. Among all the complexes, complex C1 showed the highest selectivity for the tumorous HeLa and MCF7 cells, so, it will be selected in the future for further investigations for better understanding the mechanism of action.
5. Conclusions
In this study, three new metal complexes of Pd(II), Pt(II), and Ag(I) (C1–C3) derived from 3-acetylcoumarin benzoylhydrazone (HL) were prepared and characterized by elemental analysis, spectroscopic techniques, and DFT calculations. The analytical data revealed that the ligand acted as an ONO-donor forming square planar geometries for the Pd(II) and Pt(II) complexes. The HL was coordinated to Pd(II) centers in its deprotonated form, while in the case of Pt(II) it was coordinated in its neutral form. However, in the Ag(I) complex it was coordinated as a neutral ON-donor. The binding affinity of all the compounds toward ctDNA, yeast tRNA, and BSA was assessed by electronic and fluorescence spectra. All the compounds showed the intercalative mode of binding, and the data revealed that the Pd(II) and Pt(II) complexes showed the highest binding strength (Kb) to these molecules. This latter observation was further emphasized by our molecular docking calculations, which provided a higher binding score for the Pt(II) complex than Ag(I). The key interaction responsible for the tight interaction between the targeted BSA and the two complexes was mainly a cation–pi interaction, where the cationic residues arginine and lysine represent the cationic motifs. Furthermore, the cytotoxic activity (in vitro) of the compounds was estimated against a non-cancerous WI38 and cancerous HeLa and MCF7 cell lines using the MTT assay for 24 and 48 h. The data showed that the Pd(II) and Pt(II) complexes had the highest toxicity based on the IC50 and selective indices (SI) values, consistent with the biomolecular interaction studies. Furthermore, the results from flow cytometry demonstrated that complexes C1 and C2 caused cell cycle arrest at S and G1/S, respectively.Abbreviations
| B3LYP | Becke, 3-parameter, Lee–Yang–Parr |
| BSA | Bovine serum albumin |
| ctDNA | Calf thymus deoxyribonucleic acid |
| DFT | Density functional theory |
| DMSO | Dimethylsulfoxide |
| EB | Ethidium bromide |
| FTIR | Fourier-transform infrared |
| HeLa | Cervical carcinoma cell line |
| HOMO | Highest unoccupied molecular orbital |
| Kb | Intrinsic binding constant |
| Ksv | Stern–Volmer constant |
| Kq | Quenching binding constant |
| LANL2DZ | Los Alamos National Laboratory 2 double-zeta |
| LMCT | Ligand-to-metal charge transfer |
| LUMO | Lowest unoccupied molecular orbital |
| MCF7 | Mammary gland breast cancer |
| MGL | Machine graded lumber |
| MTT | 3-(4,5-Dimethylthiazol-2-yl)-2,5-diphenyltetrazoliumbromide |
| NA | Nucleic acid |
| NMR | Nuclear magnetic resonance |
| PBS | Phosphate buffered saline |
| PI | Propidium iodide |
| SI | Selective index |
| tRNA | Transfer ribonucleic acid |
| Tris–HCl | Tris(hydroxymethyl)aminomethane hydrochloride |
| UV-vis | Ultraviolet-visible spectroscopy |
| WI38 | Human lung fibroblast cell line |
| εa | Apparent extinction coefficients |
| εb | Extinction coefficients bound compound |
| εf | Extinction coefficients of the free complex |
Data availability
The data that support the findings of this study are available from the corresponding author upon reasonable request.Author contributions
Shadia A. Elsayed: supervision, conceptualization, formal analysis, software, resources, visualization, methodology, software, validation, writing – original draft. Islam M. Elnabky: software, validation, methodology. Ahmed M. El-Hendawy: supervision, conceptualization, visualization, writing – review & editing. Mohamed M. Aboelnga: supervision, visualization, data curation, software, formal analysis, calculation, writing – original draft.Conflicts of interest
There are no conflicts of interest to declare.Acknowledgements
We acknowledge the Ministry of Higher Education of Egypt for CIQAP (CP3-016-MAN) project, and the Academy of Scientific Research and Technology (ASRT), Egypt, under initiatives of science up capacity building (Grant No. 6389) for research equipment funds.References
- T. Pereira, D. Franco, F. Vitorio and A. Kummerle, Curr. Top. Med. Chem., 2018, 18, 124–148 CrossRef CAS PubMed.
- O. Halter and H. Plenio, Eur. J. Inorg. Chem., 2018, 2018, 2935–2943 CrossRef CAS.
- P. Manojkumar, T. K. Ravi and G. Gopalakrishnan, Acta Pharm., 2009, 59, 159–168 CAS.
- M. P. Sathisha, U. N. Shetti, V. K. Revankar and K. S. R. Pai, Eur. J. Med. Chem., 2008, 43, 2338–2346 CrossRef CAS PubMed.
- M. Mladenović, N. Vuković, S. Sukdolak and S. Solujić, Molecules, 2010, 15, 4294–4308 CrossRef PubMed.
- M. P. Sathisha, U. N. Shetti, V. K. Revankar and K. S. R. Pai, Eur. J. Inorg. Chem., 2008, 43, 2338–2346 CAS.
- S. A. Patil, V. Kandathil, A. Sobha, S. B. Somappa, M. R. Feldman, A. Bugarin and S. A. Patil, Molecules, 2022, 27, 5220 CrossRef CAS PubMed.
- S. Balcioğlu, M. Karataş, B. Ates, B. Alici and I. Ozdemir, Bioorg. Med. Chem. Lett., 2020, 30, 126805 CrossRef PubMed.
- C. Ranjan Sahoo, J. Sahoo, M. Mahapatra, D. Lenka, P. Kumar Sahu, B. Dehury, R. Nath Padhy and S. Kumar Paidesetty, Arab. J. Chem., 2021, 14, 102922 CrossRef CAS.
- B. S. Creaven, E. Czeglédi, M. Devereux, É. A. Enyedy, A. Foltyn-Arfa Kia, D. Karcz, A. Kellett, S. McClean, N. V. Nagy, A. Noble, A. Rockenbauer, T. Szabó-Plánka and M. Walsh, Dalton Trans., 2010, 39, 10854–10865 RSC.
- A. Pangal, J. Shaikh and E. Khan, Int. J. Pharmaceut. Sci. Rev. Res., 2017, 42, 161–168 CAS.
- S. Aslkhademi, N. Noshiranzadeh, M. S. Sadjadi, K. Mehrani and N. Farhadyar, Polyhedron, 2019, 160, 115–122 CrossRef CAS.
- R. Kaplánek, M. Havlík, B. Dolenský, J. Rak, P. Džubák, P. Konečný, M. Hajdúch, J. Králová and V. Král, Bioorg. Med. Chem., 2015, 23, 1651–1659 CrossRef PubMed.
- S. Omidi and A. Kakanejadifard, RSC Adv., 2020, 10, 30186–30202 RSC.
- G. Le Goff and J. Ouazzani, Bioorg. Med. Chem., 2014, 22, 6529–6544 CrossRef CAS PubMed.
- Y. Li, Z. Yang, M. Zhou, J. He, X. Wang, Y. Wu and Z. Wang, J. Mol. Struct., 2017, 1130, 818–828 CrossRef CAS.
- P. K. Suganthy, R. N. Prabhu and V. S. Sridevi, Polyhedron, 2015, 88, 57–62 CrossRef CAS.
- R. N. Prabhu and R. Ramesh, J. Organomet. Chem., 2012, 718, 43–51 CrossRef CAS.
- H. H. Monfared, M. Vahedpour, M. M. Yeganeh, M. Ghorbanloo, P. Mayer and C. Janiak, Dalton Trans., 2011, 40, 1286–1294 RSC.
- S. A. Elsayed, I. M. Elnabky, A. di Biase and A. M. El-Hendawy, Appl. Organomet. Chem., 2022, 36, e6481 CrossRef CAS.
- T. Nasr, S. Bondock and M. Youns, Eur. J. Med. Chem., 2014, 76, 539–548 CrossRef CAS PubMed.
- M. Mishra, K. Tiwari, S. Shukla, R. Mishra and V. P. Singh, Spectrochim. Acta, Part A, 2014, 132, 452–464 CrossRef CAS PubMed.
- I. Ali, K. Saleem, D. Wesselinova and A. Haque, Med. Chem. Res., 2013, 22, 1386–1398 CrossRef CAS.
- P. P. Netalkar, A. Kamath, S. P. Netalkar and V. K. Revankar, Spectrochim. Acta, Part A, 2012, 97, 762–770 CrossRef CAS PubMed.
- Y.-h. Li, B.-d. Wang and Z.-y. Yang, Spectrochim. Acta, Part A, 2007, 67, 395–401 CrossRef PubMed.
- Ł. Balewski, S. Szulta, A. Jalińska and A. Kornicka, Front. Chem., 2021, 9, 781779 CrossRef PubMed.
- C. T. G. Retnam, S. V. Rose and B. S. Kumari, J. Mol. Struct., 2023, 1282, 135162 CrossRef CAS.
- L. Dkhar, A. K. Verma, V. Banothu, W. Kaminsky and M. R. Kollipara, Appl. Organomet. Chem., 2022, 36, e6589 CrossRef CAS.
- S. A. Elsayed, H. M. El-Gharabawy, I. S. Butler and F. M. Atlam, Appl. Organomet. Chem., 2020, 34, e5643 CrossRef CAS.
- S. A. Aboafia, S. A. Elsayed, A. K. El-Sayed and A. M. El-Hendawy, J. Mol. Struct., 2018, 1158, 39–50 CrossRef CAS.
- S. A. Elsayed, I. M. Elnabky, A. di Biase and A. M. El-Hendawy, Appl. Organomet. Chem., 2022, 36, e6481 CrossRef CAS.
- M. M. Aboelnga, RSC Adv., 2022, 12, 15543–15554 RSC.
- M. M. Aboelnga and S. Kalyaanamoorthy, ACS Sustainable Chem. Eng., 2022, 48, 15857–15868 CrossRef.
- R. Kaur, M. M. Aboelnga, D. J. Nikkel and S. D. Wetmore, Phys. Chem. Chem. Phys., 2022, 24, 29130–29140 RSC.
- M. M. Aboelnga and J. W. Gauld, J. Phys. Chem. B, 2017, 25, 6163–6174 CrossRef PubMed.
- M. M. Aboelnga, J. J. Hayward and J. W. Gauld, ACS Catal., 2017, 7, 51805193 Search PubMed.
- S. Forli, R. Huey and M. Pique, et al., Nat. Protoc., 2016, 11, 905–919 CrossRef CAS PubMed.
- G. M. Morris, R. Huey, W. Lindstrom, M. F. Sanner, R. K. Belew, D. S. Goodsell and A. J. Olson, J. Comput. Chem., 2009, 16, 2785–2791 CrossRef PubMed.
- S. D. Rossella Castagna, P. Colnago, A. Serafini, E. Parisini and C. Bertarelli, ACS Omega, 2019, 8, 13270–13278 CrossRef PubMed.
- T. D. Goddard, E. F. Pettersen, C. C. Huang, G. S. Couch, D. M. Greenblatt, E. C. Meng and T. E. Ferrin, J. Comput. Chem., 2004, 25(13), 1605–1612 CrossRef PubMed.
- A. Hussain, M. F. AlAjmi, M. T. Rehman, S. Amir, F. M. Husain, A. Alsalme, M. A. Siddiqui, A. A. AlKhedhairy and R. A. Khan, Sci. Rep., 2019, 9, 1–17 CrossRef PubMed.
- M. Carcelli, P. Cozzini, T. Maccagni, C. Pelizzi and L. Righi, Inorg. Chim. Acta, 2000, 303, 238–243 CrossRef CAS.
- F. Hueso-Ureña, N. A. Illán-Cabeza, M. N. Moreno-Carretero, A. L. Peñas-Chamorro and R. Faure, Polyhedron, 2000, 19, 689–693 CrossRef.
- F. B. Tamboura, P. M. Haba, M. Gaye, A. S. Sall, A. H. Barry and T. Jouini, Polyhedron, 2004, 23, 1191–1197 CrossRef.
- R. S. Hunoor, B. R. Patil, D. S. Badiger, R. S. Vadavi, K. B. Gudasi, V. M. Chandrashekhar and I. S. Muchchandi, Spectrochim. Acta, Part A, 2010, 77, 838–844 CrossRef PubMed.
- R. C. Maurya and S. Rajput, J. Mol. Struct., 2007, 833, 133–144 CrossRef CAS.
- K. Nakamoto, Infrared and Raman Spectra of Inorganic and Coordination Compounds, Part B, Applications in Coordination, Organometallic, and BioInorg. Chem., John Wiley & Sons, Inc., 6th edn, 2009 Search PubMed.
- M. Ghorbanloo, R. Bikas and G. Małecki, Inorg. Chim. Acta, 2016, 445, 8–16 CrossRef CAS.
- V. Kamat, A. Kotian, A. Nevrekar, K. Naik, D. Kokare and V. K. Revankar, Inorg. Chim. Acta, 2017, 466, 625–631 CrossRef CAS.
- G. S. Kurdekar, S. M. Puttanagouda, N. V. Kulkarni, S. Budagumpi and V. K. Revankar, Med. Chem. Res., 2011, 20, 421–429 CrossRef CAS.
- R. S. Hunoor, B. R. Patil, D. S. Badiger, R. S. Vadavi, K. B. Gudasi, V. Chandrashekhar and I. Muchchandi, Spectrochim. Acta, Part A, 2010, 77, 838–844 CrossRef PubMed.
- I. L. Paiva, G. S. G. de Carvalho, A. D. da Silva, P. P. Corbi, F. R. G. Bergamini, A. L. B. Formiga, R. Diniz, W. R. do Carmo, C. Q. F. Leite, F. R. Pavan and A. Cuin, Polyhedron, 2013, 62, 104–109 CrossRef CAS.
- L. M. Fostiak, I. García, J. K. Swearingen, E. Bermejo, A. Castiñeiras and D. X. West, Polyhedron, 2003, 22, 83–92 CrossRef CAS.
- N. K. Ngan, K. M. Lo and C. S. R. Wong, Polyhedron, 2012, 33, 235–251 CrossRef CAS.
- S. A. Elsayed, E. E. Saleh, M. M. Aboelnga and E. A. Toson, J. Inorg. Biochem., 2023, 241, 112132 CrossRef CAS PubMed.
- A. M. Ismail, S. A. El Sayed, I. S. Butler and S. I. Mostafa, J. Mol. Struct., 2020, 1200, 127088 CrossRef CAS.
- M. Muralisankar, J. Haribabu, N. S. Bhuvanesh, R. Karvembu and A. Sreekanth, Inorg. Chim. Acta, 2016, 449, 82–95 CrossRef CAS.
- G. Ayyannan, M. Mohanraj, M. Gopiraman, R. Uthayamalar, G. Raja, N. Bhuvanesh, R. Nandhakumar and C. Jayabalakrishnan, Inorg. Chim. Acta, 2020, 119868 CrossRef CAS.
- V. Censi, A. B. Caballero, M. Perez-Hernandez, V. Soto-Cerrato, L. Korrodi-Gregorio, R. Perez-Tomas, M. M. Dell'Anna, P. Mastrorilli and P. Gamez, J. Inorg. Biochem., 2019, 198, 110749 CrossRef CAS PubMed.
- S. Delaney, M. Pascaly, P. K. Bhattacharya, K. Han and J. K. Barton, Inorg. Chem., 2002, 41, 1966–1974 CrossRef CAS PubMed.
- K. L. Bergeron, E. L. Murphy, O. Majofodun, L. D. Muñoz, J. C. Williams and K. H. Almeida, Mutat. Res. Genet. Toxicol. Environ. Mutagen, 2009, 673, 141–148 CrossRef CAS PubMed.
- K. Sudeepa, N. Narsimha, B. Aparna, S. Sreekanth, A. V. Aparna, M. Ravi, J. Mohmed and C. S. Devi, J. Chem. Sci., 2018, 130, 52 CrossRef.
- Y. Jung and S. J. Lippard, Chem. Rev., 2007, 107, 1387–1407 CrossRef CAS PubMed.
- H.-K. Liu and P. J. Sadler, Acc. Chem. Res., 2011, 44, 349–359 CrossRef CAS PubMed.
- E. Movahedi, A. R. Rezvani and H. Razmazma, Int. J. Biol. Macromol., 2019, 126, 1244–1254 CrossRef CAS PubMed.
- A. Wolfe, G. H. Shimer Jr and T. Meehan, Biochemistry, 1987, 26, 6392–6396 CrossRef CAS PubMed.
- Y. Sun, S. Bi, D. Song, C. Qiao, D. Mu and H. Zhang, Sens. Actuators, B, 2008, 129, 799–810 CrossRef CAS PubMed.
- M. V. Babak, S. M. Meier, A. A. Legin, M. S. Adib Razavi, A. Roller, M. A. Jakupec, B. K. Keppler and C. G. Hartinger, Chem.–Eur. J., 2013, 19, 4308–4318 CrossRef CAS PubMed.
- V. D. Suryawanshi, L. S. Walekar, A. H. Gore, P. V. Anbhule and G. B. Kolekar, J. Biol. Chem., 2016, 6, 56–63 Search PubMed.
- S. Kathiresan, S. Mugesh, M. Murugan, F. Ahamed and J. Annaraj, RSC Adv., 2016, 6, 1810–1825 RSC.
- M. Idowu, E. Lamprecht and T. Nyokong, J. Photochem. Photobiol. Chem., 2008, 198, 7–12 CrossRef CAS.
- K. Sakthikumar, R. V. Solomon and J. D. Raja, RSC Adv., 2019, 9, 14220–14241 RSC.
- M. Anjomshoa, S. J. Fatemi, M. Torkzadeh-Mahani and H. Hadadzadeh, Spectrochim. Acta, Part A, 2014, 127, 511–520 CrossRef CAS PubMed.
- D. Xu, X. Wang, D. Fei and L. Ding, Nucleos Nucleot., 2010, 29, 854–866 CrossRef CAS PubMed.
- A. Ray, B. K. Seth, U. Pal and S. Basu, Spectrochim. Acta, Part A, 2012, 92, 164–174 CrossRef CAS PubMed.
- J. Toneatto and G. A. Argüello, J. Inorg. Biochem., 2011, 105, 645–651 CrossRef CAS PubMed.
- M. H. Chowdhury, K. Aslan, S. N. Malyn, J. R. Lakowicz and C. D. Geddes, J. Fluoresc., 2006, 16, 295–299 CrossRef CAS PubMed.
- J. R. Lakowicz, Principles of Fluorescence Spectroscopy, Springer science & business media, 2013 Search PubMed.
- J. R. Lakowicz and G. Weber, Biochem., 1973, 12, 4161–4170 CrossRef CAS PubMed.
- W. R. Ware, J. Phys. Chem., 1962, 66, 455–458 CrossRef CAS.
- X. Zhang, S. Li, L. Yang and C. Fan, Spectrochim. Acta, Part A, 2007, 68, 763–770 CrossRef PubMed.
- E. Froehlich, J. Mandeville, C. Jennings, R. Sedaghat-Herati and H. Tajmir-Riahi, J. Phys. Chem. B, 2009, 113, 6986–6993 CrossRef CAS PubMed.
- N. Wang, L. Ye, F. Yan and R. Xu, Int. J. Pharm., 2008, 351, 55–60 CrossRef CAS PubMed.
- R. A. Khan, I. I. BinSharfan, S. S. Alterary, H. Alsaeedi, F. A. Qais, A. AlFawaz, A. D. Hadi and A. Alsalme, Appl. Organomet. Chem., 2022, 36, e6550 CrossRef CAS.
- P. Wiji Prasetyaningrum, A. Bahtiar and H. Hayun, Sci. Pharm., 2018, 86, 25 CrossRef PubMed.
- S. A. Elsayed, H. E. Badr, A. di Biase and A. M. El-Hendawy, J. Inorg. Biochem., 2021, 223, 111549 CrossRef CAS PubMed.
- S. Roy, S. Saha, R. Majumdar, R. R. Dighe and A. R. Chakravarty, Polyhedron, 2010, 29, 3251–3256 CrossRef CAS.
- D. A. Guk, O. O. Krasnovskaya and E. K. Beloglazkina, Russ. Chem. Rev., 2021, 90, 1566 CrossRef.
- H. Y. Khan, M. O. Ansari, G. Shadab, S. Tabassum and F. Arjmand, Bioorg. Chem., 2019, 88, 102963 CrossRef CAS PubMed.
- M. Marloye, G. Berger, M. Gelbcke and F. Dufrasne, Future Med. Chem., 2016, 8, 2263–2286 CrossRef CAS PubMed.
- Y. Wang, J. Hu, Y. Cai, S. Xu, B. Weng, K. Peng, X. Wei, T. Wei, H. Zhou, X. Li and G. Liang, J. Med. Chem., 2013, 56, 9601–9611 CrossRef CAS PubMed.
- K. A. Abu-Safieh, A. S. Abu-Surrah, H. D. Tabba, H. A. AlMasri, R. M. Bawadi, F. M. Boudjelal and L. H. Tahtamouni, J. Chem., 2016, 2016, 508724 Search PubMed.
- S. D. Brown, K. D. Trotter, O. B. Sutcliffe, J. A. Plumb, B. Waddell, N. E. B. Briggs and N. J. Wheate, Dalton Trans., 2012, 41, 11330–11339 RSC.
- S. K. Liew, S. Malagobadan, N. M. Arshad and N. H. Nagoor, Biomolecules, 2020, 10, 138 CrossRef CAS PubMed.
- C.-R. Sihn, E.-J. Suh, K.-H. Lee, T.-Y. Kim and S. H. Kim, Cancer Lett., 2003, 201, 203–210 CrossRef CAS PubMed.
- I. Vermes, C. Haanen and C. Reutelingsperger, J. Immunol. Methods, 2000, 243, 167–190 CrossRef CAS PubMed.
Footnote |
| † Electronic supplementary information (ESI) available. See DOI: https://doi.org/10.1039/d4ra02738h |
| This journal is © The Royal Society of Chemistry 2024 |