Open Access Article
Open Access ArticleDegradation of different wastewater by a biological sponge iron system: microbial growth and influencing factors†
Huina Xie *,
Wei Zhao,
Jing Li and
Jie Li
*,
Wei Zhao,
Jing Li and
Jie Li
School of Environmental and Municipal Engineering, Lanzhou Jiaotong University, Lanzhou 730070, China. E-mail: xiehuina@lzjtu.edu.cn
First published on 29th May 2024
Abstract
The bio-ZVI process has undergone widespread development in wastewater treatment in recent years. However, there has been limited examination of the growth and degradation characteristics of functional microorganisms within the system. In the present research, strains were isolated and identified from the bio-ZVI system constructed by sponge iron (encoded as SFe-M). The consistency of operating conditions in treating different wastewater was explored. Three SFe-acclimated microorganisms exhibiting characteristics of degrading organic pollutants and participating in the nitrogen removal process were isolated. The adaptation time of these microorganisms prolonged as the substrate toxicity increased, while the pollutant degradation was related to their metabolic rate in the logarithmic phase. All these functional bacteria exhibited the ability to treat wastewater in a wide pH range (5–8). However, the improper temperature (such as 10 °C and 40 °C) significantly inhibited their growth, and the optimal working temperature was identified as 30 °C. The iron dosage had a significant impact on these function bacteria, ranging from 1 g L−1 to 150 g L−1. It was inferred that the SFe-acclimated microorganisms are capable of resisting the poison of excessive iron, that is, they all have strong adaptability. The results provide compelling evidence for further understanding of the degradation mechanism involved in the bio-ZVI process.
1 Introduction
Zero-valent iron (ZVI) with its high reduction, non-toxicity, cost-effectiveness, and reliability has been widely used in wastewater treatment.1–4 In recent years, the bio-ZVI process was developed by combining ZVI and biological processing for eliminating contaminants from wastewater.5,6 ZVI can degrade contaminants directly. More importantly, during the ZVI biochemical corrosion process, Fe(II) and Fe(III) are released, which can serve as alternative electron donors or acceptors for the growth and reproduction of microorganisms, such as iron-oxidizing bacteria.7,8 Meanwhile, microorganisms can induce the production of active oxygen species while metabolising iron ions.9 Hence, the coupling of ZVI and biological processing can improve the biodegradability of refractory pollutants, including chlorpyrifos, tetracycline, nitrate, and nitrobenzene.Microorganisms are responsible for the degradation of pollutants in biochemical processing. It is important to note that the presence of ZVI or its released ions can alter the microbial structure.10,11 Furthermore, it is worth mentioning that microorganisms are domesticated into obligate bacteria for different types of wastewater. However, studies on the bio-ZVI process have mainly focused on degradation effects,12,13 with little attention paid to the growth characteristics of acclimated microbes. Furthermore, the consistency of operating conditions for acclimated microorganisms was not described in depth.
Sponge iron (SFe) is a special type of ZVI, which mainly contains ZVI and other elements, such as carbon, aluminum, nickel, copper and manganese.14,15 When SFe is submerged in wastewater, countless microscale galvanic cells are formed. What's more, SFe with loose micropores provides a good habitat for the attached microbes. Besides, along with the iron ions releasing, dissolved oxygen (DO) in the wastewater will be consumed. It offers a special “microenvironment” structure of anaerobic-anoxic-aerobic for the symbiosis of microorganisms. Thus a special bio-ZVI process (encoded SFe-M system) was developed by combining SFe and biological processing.16 Previous research has successfully employed the SFe-M system for the treatment of various types of wastewater, yielding promising results.17,18 However, limited attention has been given to the functional microorganisms involved in this process. Here, this study focuses on isolating and identifying the functional microorganisms from different bio-SFe reactors. The degradation characteristics and operating conditions of different wastewaters treated by the acclimated microorganisms were evaluated.
2 Materials and methods
2.1 Materials and chemicals
Ammonia nitrogen, aniline (AN), and nitrobenzene (NB) were purchased from Shanghai Sinopharm Chemical Reagent Co. Ltd, China. SFe with a diameter of 2–3 mm was provided by Beijing Jinke Composite Materials Co., Ltd (Beijing, China). Expect for 90% elemental iron, it also contains a small amount of manganese, chromium, nickel, magnesium oxide, carbon (0.28%, 0.01%, 0.038%, 0.051%, 3%). SFe was flushed with dilute sulfuric acid to remove the impurities and surface oxide layer, flushed with deionized water several times until the pH of the supernatant was neutral before being dried (378 K). Distilled water was used through all our experiments. Unless otherwise indicated, all chemicals used were analytical reagent grade as received.Seed sludge was collected from the reactors operated under steady-state that the SFe-M method was employed to treat nitrogen-containing (NC), AN, and NB wastewater in our laboratory (Text S1†), encoded as SNC, SAN, and SNB, respectively.
The synthetic NC wastewater was prepared with (NH4)2SO4 (320 mg L−1), K2CO3 (320 mg L−1), NaHCO3 (160 mg L−1), K2HPO4·3H2O (48.64 mg L−1), KH2PO4 (19.2 mg L−1), glucose (2000 mg L−1). The synthetic AN wastewater was prepared with NaHCO3 (80 mg L−1), KH2PO4 (9.6 mg L−1), K2HPO4 (24 mg L−1), K2CO3 (160 mg L−1), AN (600 mg L−1). The synthetic NB wastewater was prepared with Na2HPO4·12H2O (3800 mg L−1), KH2PO4 (1000 mg L−1), NaCl (1000 mg L−1), MgSO4·7H2O (200 mg L−1), NH4Cl (100 mg L−1), NB (300 mg L−1).19 Unless otherwise indicated, all chemicals used were analytical grade.
2.2 Experimental procedures
The growth of bacteria was monitored spectrophoto-metrically in the conical flasks with an effective volume of 150 mL. Specifically, 1 mL of bacterial solution was added to the inorganic salt medium containing NC, AN, and NB, respectively. The reactors were placed in the thermostat oscillator (TS-200B, China) with 140 rpm oscillation frequency at 30 ± 2 °C. The optical density at 600 nm (OD600) and organic matter concentration were measured at a certain interval during the cultivation.
All the reactors in batch experiments were conducted in 500 mL conical flasks. It was replaced by 20 mL of bacterial-sludge mixture and 180 mL of synthetic wastewater. Among them, the conical flask containing the NB wastewater was sealed with butyl rubber stoppers. The reactors were placed in the thermostat oscillator (TS-200B, China) with 140 rpm oscillation frequency at 30 ± 2 °C. Water samples were collected at regular intervals. The initial pH value was 5.0–8.0, the temperature was 10–40 °C, and the SFe dosages in this study were 0–150 g L−1.
2.3 Analytical methods
The analysis of NB was carried out using high-performance liquid chromatography (HPLC, Agilent 1200, USA) equipped with a UV detector. Methanol and water (70![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 30, V/V%) served as the mobile phase pumped at a flow rate of 1.00 mL min−1. The injection volume was 10 μL. AC18 column (150 mm × 4.6 mm × 4 μm) was conducted at a constant temperature of 30 °C to separate the pollutants. AN was quantified using the spectrophotometric method with N-(1-naphthyl) ethylenedia-mine. Samples were filtered with a 0.22 μm filter before measurement. The pH was monitored by a pH meter (pHS-3C, Leici, China). Following the standard methods for water and wastewater examination,23 the determination of chemical oxygen demand (COD) was conducted using the dichromate method. Ammonia nitrogen (NH4+–N) was quantified utilizing Nessler's reagent spectrophotometry. Nitrate nitrogen (NO3−–N) were assessed via ultraviolet spectrophotometry, while nitrite nitrogen (NO2−–N) was measured using N-(1-naphthyl)-ethylenediamine spectrophotometry. Total nitrogen (TN) was determined employing the potassium persulfate oxidation ultraviolet spectrophotometric method. The growth of bacteria was evaluated in terms of absorbance change at 600 nm (OD600).
30, V/V%) served as the mobile phase pumped at a flow rate of 1.00 mL min−1. The injection volume was 10 μL. AC18 column (150 mm × 4.6 mm × 4 μm) was conducted at a constant temperature of 30 °C to separate the pollutants. AN was quantified using the spectrophotometric method with N-(1-naphthyl) ethylenedia-mine. Samples were filtered with a 0.22 μm filter before measurement. The pH was monitored by a pH meter (pHS-3C, Leici, China). Following the standard methods for water and wastewater examination,23 the determination of chemical oxygen demand (COD) was conducted using the dichromate method. Ammonia nitrogen (NH4+–N) was quantified utilizing Nessler's reagent spectrophotometry. Nitrate nitrogen (NO3−–N) were assessed via ultraviolet spectrophotometry, while nitrite nitrogen (NO2−–N) was measured using N-(1-naphthyl)-ethylenediamine spectrophotometry. Total nitrogen (TN) was determined employing the potassium persulfate oxidation ultraviolet spectrophotometric method. The growth of bacteria was evaluated in terms of absorbance change at 600 nm (OD600).
3 Results and discussion
3.1 Isolation and identification of microorganisms in the SFe-M system
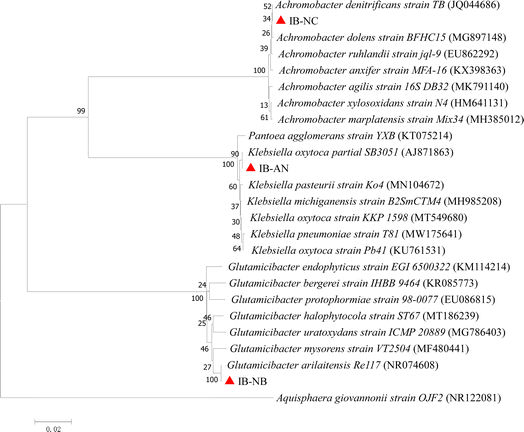
A strain with a strong AN degrading ability was screened from the bacteria liquid of SAN, named IB-AN. IB-AN was flat in shape, metallic luster, radial edges, and a diameter of about 5 mm. Similarly, two strains were isolated from the bacteria liquid of SNC and SNB, named IB-NC and IB-NB respectively. IB-NC was an aerobic denitrification bacteria with a high nitrogen removal performance. IB-NC was spherical in shape, smooth in appearance and approximately 7 mm in diameter. IB-NB with a strong NB degrading ability was round in shape, with neat edges and about 2.5 mm in diameter. In particular, the ferrous oxidation rate of the three strains was above 90% in the physiological and biochemical identification experiments, indicating that these three strains all had iron oxidation function.To establish the identity of IB-NC, IB-AN, and IB-NB more accurately, the 16S rRNA gene sequences were used to construct a phylogenetic tree. The phylogenetic tree was constructed by the neighbor-joining method (Fig. 1). The phylogenetic tree showed that IB-NC was closely related to Achromobacter denitrificans, which showed better NO reduction and sulfamethoxazole degradation efficiency.24,25 Similarly, IB-AN and IB-NB exhibited 99% homology in the 16S rRNA gene sequence with the type strains of Klebsiella oxytoca and Glutamicibacter arilaitensis, respectively. Klebsiella oxytoca, isolated from sediment, has demonstrated proficiency in nitrate removal from industrial wastewater26 and utilize recalcitrant organics as the sole source of nitrogen.27 Glutamicibacter arilaitensis exhibits distinctive traits, including a high number of Fe(III)/siderophore transport systems,28 the ability to catabolize organics while transforming various forms of nitrogen compounds, such as ammonium, hydroxylamine, nitrate, and nitrite.29 In conclusion, the SFe-M system contributed to the evolution of microorganisms into organic contaminants obligate degradation bacteria for different types of wastewater. Specially, these obligate bacteria possess characteristics conducive to the degradation of organic pollutants and participating in the nitrogen removal processes.
3.2 Characteristics of microbial growth
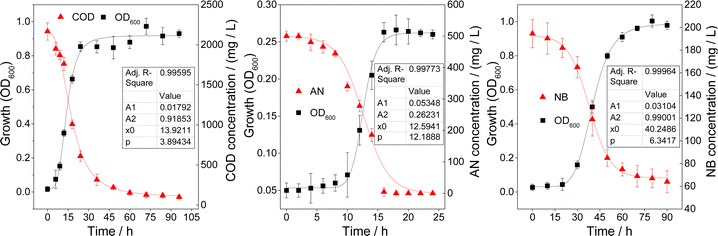
The logistic model is a basic and important microbial growth model that mainly explains the impact of resources (such as food and space), environmental conditions and interspecific competition on the growth rate of microorganisms.30 The logistic model was used to fit the growth curves of IB-AN, IB-NC, and IB-NB, the results are shown in Fig. 2 and Table 1.| Y = A2 + (A1 − A2)/(1 + (X/X0)p) | ||||
|---|---|---|---|---|
| Microbes | A1 | A2 | X0 | p |
| a IB-NC: a strain was screened from sludge in the SFe-M reactor treating nitrogen-containing wastewater. IB-AN: a strain was screened from sludge in the SFe-M reactor treating aniline wastewater. IB-NB: a strain was screened from sludge in the SFe-M reactor treating nitrobenzene wastewater. | ||||
| IB-NC | 0.0179 | 0.9185 | 13.9211 | 3.8943 |
| IB-AN | 0.0535 | 0.2623 | 12.5941 | 12.1888 |
| IB-NB | 0.0310 | 0.9900 | 40.2486 | 6.3417 |
Carbon source has a significant impact on the microbial community and microbial activity.31 Therefore, the growth rate of bacteria is closely related to the carbon source. Fig. 2 showed that the logistic equation had a high fit degree for the growth of IB-NC, IB-AN, and IB-NB, and the regression coefficient R2 was 0.9959, 0.9618, and 0.99932, respectively. The strains had experienced growth stages such as the adaptation phase, logarithmic phase and stable phase. During the adaptation period, high concentrations of pollutants were toxic to microorganisms. Various enzyme systems in the cell adapt to the growth environment. The size of strains increased, but the number did not increase. Glucose is a carbon source that is easily degraded by microorganisms, so IB-NC could adapt to the initial load in 5 h. However, the adaptation period of IB-AN, and IB-NB was longer than 10 h for AN and NB were highly toxic and poorly bioavailable. That is, the order of the adaptation time was IB-NC < IB-AN < IB-NB. It indicated that the adaptation time of SFe-acclimated microorganisms was prolonged as the substrate toxicity increased. Microorganisms multiplied rapidly in the logarithmic growth phase, which accelerated the degradation of organic pollutants. The order of microbial rapid propagation time was IB-NC < IB-AN < IB-NB. Clearly, IB-AN had a relatively faster degradation rate of AN. It was speculated that the degradation rate of organic matters by SFe-acclimated microorganisms in the logarithmic phase was related to their metabolic rate.
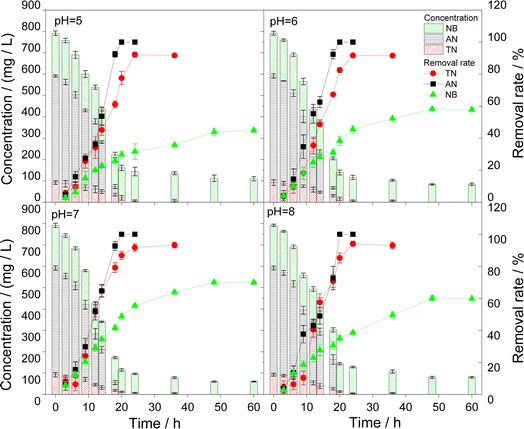
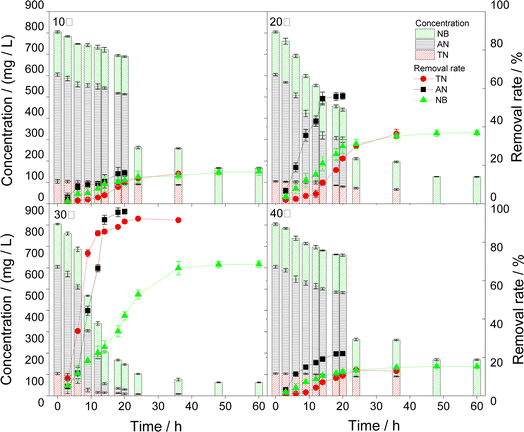
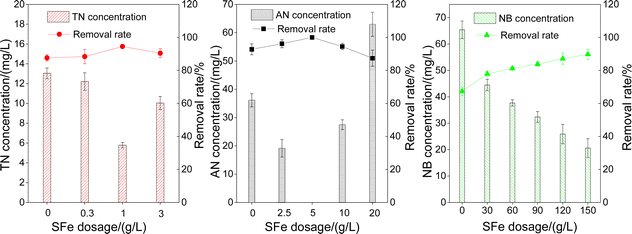
3.3 Influencing factors of pollutants degradation by microorganisms
3.4 Nitrogen compound analysis
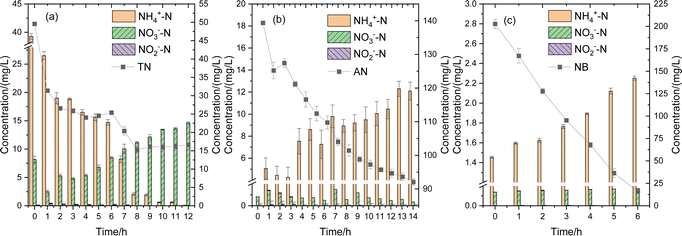
Since all three strains exhibited nitrogen removal capabilities, the variations in nitrogen compounds during pollutant degradation were monitored, as depicted in Fig. 6. In Fig. 6(a), the removal of conventional nitrogen-containing wastewater by IB-NC was illustrated. It was obvious that with the degradation of ammonia nitrogen, the concentration of nitrate nitrogen initially increases before decreasing. This suggested that the denitrification process of IB-NC was inhibited after 6 h, leading to nitrate accumulation. TN reached a stable removal rate after 8 h. In Fig. 6(b), the removal of AN wastewater by IB-AN was shown. As AN degrades, NH4+–N, NO3−–N, and NO2−–N were detected in the reactor. Amino group on the benzene ring is a primary contributor to AN's high toxicity. The concentration of NH4+–N increased, which indicating that the nitrogen element in AN could be released as the ammonia ions during the degradation of AN.44 To a certain extent, it indicated the effective transformation and degradation of AN. In Fig. 6(c), the removal of NB wastewater by IB-NB was presented. It was evident that NH4+–N concentration increases as NB degrades. Presumably, NB was converted to phenyl hydroxylamine and diaminophenol with NH4+–N release, indicating that partial reduction of NB may be the primary pathway in this system.453.5 Discussion on mechanism
A proposed model illustrates the enhanced mechanism of the SFe-M system (Fig. 7). This system entails a symbiotic relationship between sponge iron and microorganisms, including micro-electrolysis, Fe/O2 chemical reactions, iron microbial and chemical corrosion. A significant amount of Fe(II) and H2O2 was detected at the beginning of the reaction as the SFe continuously corroded.19,44,45 It initiated the Fenton reaction between Fe(II) and H2O2, resulting in the sustained generation of hydroxyl free radicals and hydroxyl free radicals exhibited strong reactivity towards the toxic functional groups of pollutants. It's imperative to recognize that SFe contributes to the evolution of microorganisms into obligate degradation bacteria for different types of wastewater. These functional microorganisms play a pivotal role in metabolizing pollutants, thereby facilitating internal enhancement and removal. It is interesting that all these functional bacteria exhibit efficient wastewater treatment capabilities across a broad pH range (5–8). However, a significant difference in iron dosage was observed, ranging from 1 g L−1 to 150 g L−1. The internal degradation mechanism of the bio-ZVI process was further revealed, shedding light on its intricate workings.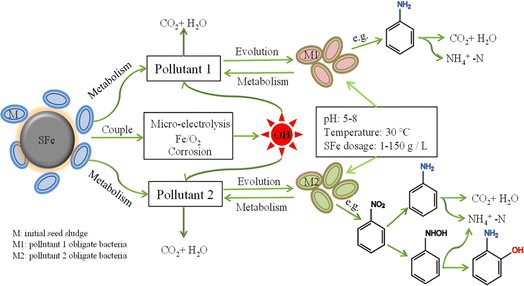 | ||
| Fig. 7 Mechanism of microbial evolution during the degradation of pollutants by biological sponge iron system. | ||
In summary, SFe-M system, as a novel wastewater treatment technology, exhibits promising applications. In fact, in our early exploration, the SFe-M system was applied to treat acrylic fiber wastewater and fine chemical wastewater. Results demonstrated that the SFe-M system achieved the removal rates of 60% for COD and 90% for NH4+–N in acrylic fiber wastewater. Additionally, compared with the control group, SFe had a good effect on stimulating the growth of bacteria.46 In the treatment of fine chemical wastewater, the SFe-M system exhibited impressive removal efficiencies, with COD, TN, and TP removal rates reaching 31.83%, 23.67%, and 65.17%, respectively. Following biochemical treatment, effluent COD, TN, and TP levels met regulatory standards.47 However, our current focus on refractory wastewater remains in the experimental research phase. As noted by Xu, further exploration is required to transition the bio-ZVI process to practical implementation on a commercial scale, particularly in addressing effluents with multiple contaminants.48 In addition, the practical application of the bio-ZVI system is constrained by various factors, including the passivation of ZVI, initial inhibition of iron to microorganisms, and the way of iron dosing. These limitations have spurred the development of enhanced treatment methods such as electrochemical methods, activated carbon, and bacteria-mediated approaches. Ultimately, the bio-ZVI process holds significant potential for widespread adoption. Ultimately, the bio-ZVI process holds significant potential for widespread adoption.
4 Conclusions
Microorganisms play an important role in biochemical processing. Three high-efficiency SFe-acclimated strains were isolated from the SFe-M system for treating NC, AN, and NB wastewater. These strains exhibited a remarkable 99% homology in the 16S rRNA gene sequence to the type strain of Achromobacter denitrificans, Klebsiella oxytoca and Glutamicibacter arilaitensis, respectively. Notably, all three strains demonstrated proficiency in degrading organic pollutants and participating in the nitrogen removal process. The growth curves denoted that the adaptation time followed the order IB-NC < IB-AN < IB-NB, indicating that the adaptation time was prolonged as the substrate toxicity increased. But the logarithmic phase depended on each strain's individual metabolic rate. The SFe-acclimated microorganisms displayed uniform temperature requirements when treating wastewater separately, and the optimal temperature was 30 °C. Conversely, they exhibited less sensitivity to initial pH values. The optimal SFe dosage of IB-NC, IB-AN, and IB-NB was 1 g L−1, 5 g L−1, and 150 g L−1, which proved that they have better adaptive capacity to iron dosage in the complex SFe-M system.Author contributions
Huina Xie: conceptualization, writing – reviewing and editing, supervision, project administration, funding. Wei Zhao: investigation, data curation, funding. Jing Li: investigation, data curation. Jie Li: methodology, supervision, validation, formal analysis.Conflicts of interest
There are no conflicts to declare.Acknowledgements
This work was supported by the Gansu Province Higher Education Innovation Fund Project (2022A-039), Science and technology project of China Railway Qinghai-Tibet Group Co., Ltd (QZ2022-Q09), Gansu Youth Science and Technology Fund (23JRRA888), Tianyou Youth Talent Lift Program of Lanzhou Jiaotong University, and National Natural Science Foundation of China (51768032).Notes and references
- Q. Dou, L. Zhang, S. La, S. Hao, W. Guo, Q. Sun, Y. Wan, Y. Peng, X. Wang and J. Yang, Bioresour. Technol., 2022, 356, 127317 CrossRef CAS PubMed.
- M. Minella, S. Bertinetti, K. Hann, C. Minero and D. Vione, Environ. Res., 2019, 179, 108750 CrossRef CAS PubMed.
- L. Xu, Y. Yang, J. Su, C. He, J. Shi, H. Yan and H. Wei, Environ. Res., 2023, 219, 115163 CrossRef CAS PubMed.
- J. Wang and J. Tang, Chemosphere, 2021, 276, 130177 CrossRef CAS PubMed.
- Z. Xu, W. Wang, Y. Liu, Y. Zhao, X. Zhang and Y. Ban, Bioresour. Technol., 2023, 386, 129516 CrossRef CAS PubMed.
- G. Yo, P. Wang, J. Hou, C. Wang, Y. Xu, L. Mia, B. Lv, Y. Yang and F. Zhang, Crit. Rev. Environ. Sci. Technol., 2017, 47, 877–907 CrossRef.
- S. Wang, A. Zhou, J. Zhang, Z. Liu, J. Zheng, X. Zhao and X. Yu, RSC Adv., 2019, 9, 1176–1186 RSC.
- J. Wang, H. Fang, H. Jia, G. Yang, F. Gao and W. Liu, Environ. Sci. Pollut. Res., 2018, 25, 749–757 CrossRef CAS PubMed.
- X. Wang and T. Waite, Water Res., 2010, 44, 3511–3521 CrossRef CAS PubMed.
- J. Xu, X. Liu, J. Huang, M. Huang, T. Wang, S. Bao, W. Tang and T. Fang, RSC Adv., 2020, 10, 23212–23220 RSC.
- W. Xu, H. Zhao, H. Cao, Y. Zhang, Y. Sheng, T. Li, S. Zhou and H. Li, Bioresour. Technol., 2019, 300, 122667 CrossRef PubMed.
- T. Guo, Y. Ji, J. Zhao, H. Horn and J. Li, Water Res., 2020, 186, 116331 CrossRef CAS PubMed.
- C. Wang, Y. Xu, J. Hou, P. Wang, F. Zhang, Q. Zhou and G. You, Sci. Total Environ., 2019, 689, 1044–1053 CrossRef CAS PubMed.
- Y. Ju, X. Liu, Z. Li, J. Kang, X. Wang, Y. Zhang, J. Fang and D. D. Dionysiou, J. Hazard. Mater., 2015, 283, 469–479 CrossRef CAS PubMed.
- X. Hou, L. Chu, Y. Wang, X. Song, Y. Liu, D. Li and X. Zhao, Bioresour. Technol., 2023, 384, 129270 CrossRef CAS PubMed.
- H. Xie, J. Li, Y. Wang, W. Zhao, L. Zhang and J. Li, Process Biochem., 2021, 101, 230–236 CrossRef CAS.
- Y. Wang, J. Li, S. Zhai, Z. Wei and J. Feng, Water Sci. Technol., 2015, 72, 1257–1265 CrossRef CAS PubMed.
- Q. Jin, Z. Hu, Z. Jin, L. Qiu, W. Zhong and Z. Pa, Bioresour. Technol., 2012, 117, 148–154 CrossRef CAS PubMed.
- J. Li, Y. Wang, H. Xie, W. Zhao, L. Zhang and J. Li, Bioprocess Biosyst. Eng., 2022, 45, 117–130 CrossRef CAS PubMed.
- G. Fu, L. Zhao, L. K. Huangshen and J. Wu, Bioresour. Technol., 2019, 290, 121725 CrossRef CAS PubMed.
- D. Li, X. Liang, Y. Jin, C. Wu and R. Zhou, Appl. Biochem. Biotechnol., 2019, 188, 540–554 CrossRef CAS PubMed.
- Y. Zheng, B. Mu, P. Wang, Y. Wang and J. Li, China Environ. Sci., 2018, 38, 2535–2541 CAS.
- China NEPA, Water and Wastewater Monitoring Methods, Chinese Environmental Science Publishing House, Beijing, China, 4th edn, 2012 Search PubMed.
- C. Chen, Y. Wang, H. Liu, Y. Chen, J. Yao, J. Chen, D. Hrynsphanb and S. Tatsianab, Chemosphere, 2020, 253, 126739 CrossRef CAS PubMed.
- P. Y. Nguyen, A. F. Silva, A. C. Reis, O. C. Nunes, A. M. Rodrigues, J. E. Rodrigues, V. V. Cardoso, M. J. Benoliel, M. A. M. Reis, A. Oehmen and G. Carvalho, Sci. Total Environ., 2019, 648, 44–55 CrossRef CAS PubMed.
- P. Guadalupe, O. José-Miguel, S. B. Luis, C. Victor and L. R. Juan, Biotechnol. Bioeng., 1998, 58, 510–514 CrossRef.
- C. Kao, K. Chen, J. Liu, S. Chou and S. Chen, Appl. Microbiol. Biotechnol., 2006, 71, 228–233 CrossRef CAS PubMed.
- C. Monnet, V. Loux, J. F. Gibrat, E. Spinnler, V. Barbe, B. Vacherie, F. Gavory, E. Gourbeyre, P. Siguier, M. Chandler, R. Elleuch, F. Irlinger and T. Vallaeys, PLoS One, 2010, 5, e15489 CrossRef PubMed.
- T. He, D. Xie, J. Ni, Z. Li and Z. Li, Sci. Total Environ., 2020, 701, 134730 CrossRef CAS PubMed.
- Q. Zhou, S. Takenaka, S. Murakami, P. Seesuriyachan, A. Kuntiya and K. Aoki, J. Biosci. Bioeng., 2007, 103, 185–191 CrossRef CAS PubMed.
- A. Mielcarek, J. Rodziewicz, W. Janczukowicz and J. Struk-Sokołowska, Sci. Total Environ., 2020, 720, 137377 CrossRef CAS PubMed.
- J. C. Aciego Pietri and P. C. Brookes, Soil Biol. Biochem., 2009, 41, 1396–1405 CrossRef CAS.
- T. He and Z. Li, Desalin. Water Treat., 2015, 57, 19181–19189 CrossRef.
- S. Deng, D. Li, X. Yang, W. Xing, J. Li and Q. Zhang, Bioresour. Technol., 2016, 219, 677–686 CrossRef CAS PubMed.
- B. Maria, J. P. M. Carlos, F. Milton, P. M. S. N. Maria Graça, F. F. Maria Amparo and A. Adelaide, Sci. Total Environ., 2023, 860, 160427 CrossRef PubMed.
- A. Konstantinos, Ł. Mateusz, G. A. Antonio, M. Antonios, Z. Wen, N. G. Hariklia and V. S. Ioannis, Chem. Eng. J., 2020, 393, 124739 CrossRef.
- J. Ma, M. Xie, N. Zhao, Y. Wang, Q. Lin, Y. Zhu, Y. Chao, Z. Ni and R. Qiu, Sci. Total Environ., 2023, 878, 162720 CrossRef CAS PubMed.
- Y. Sun, J. Li, T. Huang and X. Guan, Water Res., 2016, 100, 277–295 CrossRef CAS PubMed.
- Z. Ai, S. Zheng, D. Liu, S. Wang, H. Wang, W. Huang, Z. Lei, Z. Zhang, F. Yang and W. Huang, Chemosphere, 2022, 306, 135544 CrossRef CAS PubMed.
- B. Wang, L. Lu, Y. Zhang, K. Fang, D. An and H. Li, Sci. Total Environ., 2020, 753, 142073 CrossRef PubMed.
- J. Wang and Z. Sun, J. Hazard. Mater., 2020, 392, 122293 CrossRef CAS PubMed.
- B. Yang, J. Wang, J. Wang, H. Xu, X. Song, Y. Wang, F. Li, Y. Liu and J. Bai, Bioresour. Technol., 2018, 250, 374–381 CrossRef CAS PubMed.
- A. V. Vahatalo, Y. Xiao and K. Salonen, Water Res., 2021, 188, 116483 CrossRef CAS PubMed.
- H. Xie, J. Li, Y. Wang, W. Zhao, L. Zhang and J. Li, Process Biochem., 2021, 101, 230–236 CrossRef CAS.
- K. Guo, W. Li, Y. Wang, Y. Zheng, B. Ji, F. Mao, H. Xie and J. Li, Process Biochem., 2021, 108, 146–152 CrossRef CAS.
- H. Xie, Y. Wang, J. Li, W. Zhao and B. Ji, China Environ. Sci., 2018, 38, 3406–3412 CAS.
- W. Xu, D. Wei, J. Li, F. Zhang and W. Zhou, Pearl River, 2020, 41, 91–97 Search PubMed.
- Y. Xu, C. Wang, J. Hou, P. Wang, G. You, L. Miao, B. Lv, Y. Yang and F. Zhang, Rev. Environ. Sci. Biotechnol., 2017, 16, 667–693 CrossRef.
Footnote |
| † Electronic supplementary information (ESI) available. See DOI: https://doi.org/10.1039/d4ra02696a |
| This journal is © The Royal Society of Chemistry 2024 |