Open Access Article
Open Access ArticleCreative Commons Attribution 3.0 Unported Licence
Unveiling the multifaceted antiproliferative efficacy of Cichorium endivia root extract by dual modulation of apoptotic and inflammatory genes, inducing cell cycle arrest, and targeting COX-2†
Abdullah R. Alzahrania,
Nora Hosny bc,
Doaa I. Mohamedd,
Hebatallah H. Abo Nahase,
Abdulaziz Albogamif,
Tahani Mohamed Ibrahim Al-Hazanig,
Ibrahim Abdel Aziz Ibrahim
bc,
Doaa I. Mohamedd,
Hebatallah H. Abo Nahase,
Abdulaziz Albogamif,
Tahani Mohamed Ibrahim Al-Hazanig,
Ibrahim Abdel Aziz Ibrahim a,
Alaa Hisham Falembana,
Ghazi A. Bamagousa and
Essa M. Saied
a,
Alaa Hisham Falembana,
Ghazi A. Bamagousa and
Essa M. Saied *hi
*hi
aDepartment of Pharmacology and Toxicology, Faculty of Medicine, Umm Al-Qura University, Makkah, Saudi Arabia. E-mail: aralzahrani@uqu.edu.sa; iamustafa@uqu.edu.sa; ahfalemban@uqu.edu.sa; gabamagous@uqu.edu.sa
bMedical Biochemistry and Molecular Biology Department, Faculty of Medicine, Suez Canal University, Ismailia 41522, Egypt. E-mail: Ahmedn@umn.edu
cCenter of Excellence in Molecular and Cellular Medicine, Faculty of Medicine, Suez Canal University, Ismailia, Egypt
dDepartment of Clinical Pharmacology and Therapeutics, Faculty of Medicine, Ain Shams University, Cairo 11566, Egypt. E-mail: doaapharma@med.asu.edu.eg
eZoology Department, Faculty of Science, Port Said University, Port Said 42526, Egypt. E-mail: hebatallah_hassan@science.suez.edu.eg
fBiology Department, Faculty of Science, Al-Baha University, Al Aqiq, Saudi Arabia. E-mail: aalbogami@bu.edu.sa
gBiology Department, College of Science and Humanities, Prince Sattam bin Abdulaziz University, P. O. Box: 83, Al-Kharj, 11940, Saudi Arabia. E-mail: t.alhazani@psau.edu.sa
hChemistry Department, Faculty of Science, Suez Canal University, 41522 Ismailia, Egypt
iInstitute for Chemistry, Humboldt Universität zu Berlin, 12489 Berlin, Germany. E-mail: saiedess@hu-berlin.de
First published on 17th June 2024
Abstract
Chicory (Cichorium endivia L. divaricatum) is a renowned medicinal plant traditionally used for various ailments, yet the pharmacological potential of its roots, particularly in terms of antitumor activity, remains elusive. In the present study, we explore, for the first time, the metabolomic profile of ethanolic extract from Cichorium endivia roots (CIR) and further unveil its antiproliferative potential. The untargeted phytochemical analysis UPLC/T-TOF-MS/MS identified 131 metabolites in the CIR extract, covering acids, amino acids, flavonoids, alkaloids, nucleotides, and carbohydrates. The antiproliferative activity of the CIR extract was tested in 14 cancer cell lines, revealing significant cytotoxicity (IC50: 2.85–29.15 μg mL−1) and a high selectivity index. Among the cells examined, the CIR extract recorded the most potent antiproliferative activity and selectivity toward HepG2 and Panc-1 cells, with an IC50 of 2.85 μg mL−1 and 3.86 μg mL−1, respectively, and SI > 10. Insights into the mode of action of the antiproliferative activity revealed that CIR extract induces cell arrest in the S phase while diminishing cell distribution in the G0/G1 and G2/M phases in HepG-2 and Panc-1 cells. Flow cytometric and RT-PCR analysis revealed that the CIR extract significantly triggers apoptosis and modulates the expression of pro-apoptotic and anti-apoptotic genes. Furthermore, the CIR extract exhibited a pronounced anti-inflammatory activity, as evidenced by down-regulating key cytokines in LPS-induced RAW 264.7 cells and selectively inhibiting the COX-2 enzyme. Finally, the CIR extract showed a robust total antioxidant capacity, together with potent free radicals and metal scavenging properties, highlighting its role in alleviating oxidative stress. Taken together, this study highlights the multifaceted therapeutic potential of CIR extract as a natural-based antitumor supplement.
1 Introduction
Cancer is one of the leading causes of premature death worldwide and its burden is expected to double by 2070 compared to 2020, with an estimated 34 million new cases in 2070.1 Hepatocellular carcinoma (HCC), the primary type of liver cancer, ranks globally as the fourth leading cause of cancer-related death. The burden of liver cancer varies significantly by sex and geographic region due to varying risk factors.2 These risk factors include infections such as hepatitis B virus, hepatitis C virus, and liver flukes in endemic areas, as well as behavioral factors such as alcohol and tobacco use, metabolic factors such as excess body fat, and exposure to aflatoxins.3 As the global community grappled with these increasing cancer concerns, pancreatic cancer emerged as a growing threat. By 2050, it is projected to have an incidence rate of 18.6 cases per 100![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 000, experiencing an annual increase of 1.1%.4 Despite advances in medical science, pancreatic cancer stubbornly held its position as the seventh leading cause of cancer-related mortality.5
000, experiencing an annual increase of 1.1%.4 Despite advances in medical science, pancreatic cancer stubbornly held its position as the seventh leading cause of cancer-related mortality.5
Phytochemical substances, also known as secondary metabolites, are considered to be very promising because of their diverse biological and medicinal applications. They remain a key source of innovation in drug discovery, contributing to understanding essential biological pathways crucial for the drug development process.6–8 Exploring naturally occurring compounds in plants has led to the development of promising anticancer drugs. Cutting-edge technologies such as nanoparticle-based medicines are being used to enhance these drugs by allowing controlled drug release and innovative administration methods.9,10 In recent years, herbal medicines have gained attention in cancer treatment and prevention due to concerns about the toxicity of conventional chemotherapy drugs. Many medicinal plant extracts and phytochemicals show anticancer properties and around 60% of drugs are derived from natural sources, primarily medicinal plants.11 Plant-based phytochemicals exhibit a wide range of mechanisms that collectively contribute to inhibiting cancer development. They achieve this by neutralizing harmful free radicals, impeding the survival and growth of malignant cells, and reducing tumor invasiveness and angiogenesis of tumors.12,13 Furthermore, these metabolites have various and intricate effects on various molecular targets and signal transduction pathways, including membrane receptors, kinases, proteins that activate or suppress tumors, transcription factors, microRNAs, cyclins, and caspases.14 These phytochemicals further induce apoptosis, a programmed cell death process, by activating pro-apoptotic proteins and mitigating anti-apoptotic ones, while simultaneously modulating the expression of genes associated with apoptosis.15 Paclitaxel and resveratrol are the most common plant-based anticancer drugs approved for cancer treatment.16,17 Paclitaxel, originally derived from the Pacific yew tree, is a crucial anticancer drug used in the treatment of breast, ovarian, lung, and pancreatic cancers. It works as a microtubule-stabilizing agent, disrupting cell division, and inhibiting cancer cell proliferation by preventing the breakdown of microtubules. Often administered in combination with other anticancer drugs, paclitaxel has significantly improved cancer treatment outcomes. In the case of pancreatic cancer, it is used in conjunction with drugs such as gemcitabine to interfere with microtubules, disrupting the cell cycle and leading to cell death. This mechanism makes paclitaxel effective even in the challenging context of pancreatic cancer, slowing disease progression and enhancing overall treatment results.18,19 Resveratrol, a natural polyphenol found in various plants, including grapes and red wine, has been extensively investigated for its potential in cancer treatment.20 Resveratrol has been shown to inhibit the growth of various tumor cells, such as those in the breast, colon, prostate, liver, and lung. It also reduces tumor growth and metastasis in mice with highly metastatic lung carcinoma. These effects are attributed to the ability of resveratrol to impede DNA synthesis and inhibit neovascularization, and angiogenesis.21,22 In a mammary cancer model induced by DMBA, resveratrol reduces tumor incidence and multiplicity while extending the latency period. It also suppresses the activation of nuclear factor-κB, a regulator of genes associated with cancer development, including cyclooxygenase-2 and matrix metalloproteinase-9.23
Cichorium endivia, a perennial herb widely found in temperate regions, occupies a significant place in traditional healing systems such as Unani, Ayurveda, and Siddha.24 Different components of the plant have been used as traditional remedies worldwide. However, it should be noted that the root, in particular, harbors a plethora of significant phytochemical compounds.25 It is traditionally applied to address issues related to the renal system, hepatobiliary system, anorexia, and dyspepsia.26,27 Furthermore, beyond its medicinal applications, Cichorium intybus is used as an appetizer and in the treatment of conditions such as jaundice, liver failure, intermittent fever, and various skin disorders.28 Despite its extensive distribution, many of its constituents remain unexplored for their potential pharmacological benefits, and currently there are limited toxicological data available for the Cichorium intybus supplement.29
To thoroughly investigate the varied chemical classes, properties, and the extensive spectrum of metabolite concentrations present in plants, it is imperative to employ a diverse array of analytical techniques. Among the techniques, LC-MS/MS spectrometry has been used successfully and potentially for the exploration of the metabolic profile of various plant-derived extracts.30 In recent years, untargeted metabolomics has become a valuable technique for comprehensive analysis of a wide range of compounds within a single plant extract.31 This approach involves the simultaneous quantification of multiple low molecular weight compounds using techniques such as ultra-performance liquid chromatography (UPLC) coupled with mass spectrometry (MS).32 The application of advanced liquid chromatography tandem mass spectrometry (LC-MS/MS), distinguished by its exceptional resolution, has allowed a thorough exploration of the metabolome in various biological samples, including those from herbal medicine.33 Through the analysis of mass fragmentation (MS/MS) spectra, valuable insights into the structural characteristics of metabolites can be inferred from their unique spectral patterns.34 Metabolomics is extensively applied in studies of plant metabolism to explore the plant system at the molecular level and furnish a characterization of the complete metabolite pool within a plant tissue. This approach yields substantial information about chemical species exhibiting diverse physical properties.35 Untargeted metabolomics, which encompasses the comprehensive and unbiased measurement of all metabolites within a plant extract, provides a broader perspective on the fundamental biochemical processes occurring within that particular plant extract.36 In light of these facts and our keen interest in deciphering new pharmacological potential agents,37–43 we aimed in this study to reveal the phytochemical profile of the ethanolic extract of Cichorium intybus roots (CIR) and to explore its antiproliferative potency in a set of 15 cell lines. In this regard, we employed various analytical and chemical techniques, including UPLC/T-TOF-MS/MS-based analysis, to deep explore the phytochemical profile of the CIR extract. We further screened the cytotoxic activity of the CIR extract in a panel of cancer cells and performed a comprehensive analysis to reveal the mode of action of the antiproliferative activity by assessing programmed cell death, cell cycle arrest, and apoptotic-related genes. Furthermore, due to the fact that cyclooxygenases play a crucial role in inflammatory conditions, including cancer initiation and progress,44,45 we evaluated the antiproliferation activity of CIR extract by evaluating the inflammatory signaling of cyclooxygenase 1 & 2 and a subset of downstream targets such as the IL-1b, IL-6 and TNF-α genes.46 Finally, the antioxidant activity of the CIR extract was explored by evaluating not only the total antioxidant capacity, but also its free radical and metal scavenging activity.
2 Materials and method
2.1. Materials
Analytical grade solvents, including acetonitrile, ethyl acetate, methanol, ethanol, dichloromethane, and isopropanol, were obtained from Thermo-Fisher. Reagents including ammonium hydroxide, MTT, formic acid, DMSO, ammonium acetate, trypan blue dye, and ammonium formate were sourced from Sigma-Aldrich. Cell culture reagents, including fetal bovine serum, DMEM, RPMI-1640, HEPES buffer solution, L-glutamine and gentamycin, were purchased from Lonza (Belgium).2.2. Plant collection and extraction method
Cichorium endivia L. divaricatum, synoun (Cichorium inytubs L.), family Asteraceae is an annual winter plant indigenous to the Mediterranean region. Species were collected in March 2023 from cultivated fields in Egypt, particularly sourced from fields of Trifolium alexandrinum (Egyptian clover or berseem clover). Plant roots were cleansed with distilled water, lyophilized and pulverized using a mixer mill (Retsch MM400, Germany) before the extraction process. Taxonomic identification was conducted according to the methodology outlined by Boulos L. in “Flora of Egypt, vol. 1–4” (1999–2005), published by Al Hadara Publishing in Cairo, Egypt. The samples were deposited in the Alexandria Herbarium for preservation and reference. Five hundred grams of powdered roots were mixed with 70% ethanol (EtOH) or deionized water (240 mL) and were extracted for 3 days on a Kuhner IRC-1-U rotary shaker (Clim-O-Shake system Kuhner IRC-1-U) at 28 °C. The resulting extract mixture was double filtered using filter paper under vacuum and then the collected filtrate was dried using a rotary evaporator to yield the dry extract as waxy oil. The crude extract was finally lyophilized to provide the ethanolic CIR extract as a powder which was kept at −5 °C until utilization.47,482.3. Chemical characterization
![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) ACN (2
ACN (2![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1 ratio, v/v), and subsequently subjected to a vortex mixing, ultrasonication, and centrifugal to provide a transparent supernatant of concentration of 1.0 μg mL−1 concentration. The obtained sample was injected (10 μL) into HPLC/QTOF-MS/MS to be analyzed in positive and negative electrospray ionization (ESI, at +4500/+80 V and 4500/80 V) modes using a C18-RP column with (2.1 × 150 mm, 2.5 μm particle size, Waters, XSelect HSS T3, Milford, MA, USA). The mass spectrometer functioned by conducting a 50 ms survey scan (TOF-MS), followed by automatic selection of the top 20 ions for subsequent fragmentation spectra. The mass range covered during the analysis ranged from 50 to 1100 m/z. Implementing a gradient elution strategy at 0.3 mL min−1, the mobile phase consisted of two components. Part A, a mixture of 5 mM ammonium formate solution in water with 1% (v/v) methanol, adjusted to pH 3.0, and part B, is composed of HPLC grade acetonitrile. The elution gradient started with 10% part B up to 20 minutes, increased to 90% up to 25 minutes, and then reverted to 10% up to 28 minutes. Data were gathered through IDA (AB SCIEX analyst TF 1.7.1). Multivariate data analysis for nontargeted metabolomic analysis was performed comprehensively utilizing MS-DIAL (3.70 Yokohama, Kanagawa 230-0045 software, Japan), along with Peak View software from SCIEX.39,50,51
1 ratio, v/v), and subsequently subjected to a vortex mixing, ultrasonication, and centrifugal to provide a transparent supernatant of concentration of 1.0 μg mL−1 concentration. The obtained sample was injected (10 μL) into HPLC/QTOF-MS/MS to be analyzed in positive and negative electrospray ionization (ESI, at +4500/+80 V and 4500/80 V) modes using a C18-RP column with (2.1 × 150 mm, 2.5 μm particle size, Waters, XSelect HSS T3, Milford, MA, USA). The mass spectrometer functioned by conducting a 50 ms survey scan (TOF-MS), followed by automatic selection of the top 20 ions for subsequent fragmentation spectra. The mass range covered during the analysis ranged from 50 to 1100 m/z. Implementing a gradient elution strategy at 0.3 mL min−1, the mobile phase consisted of two components. Part A, a mixture of 5 mM ammonium formate solution in water with 1% (v/v) methanol, adjusted to pH 3.0, and part B, is composed of HPLC grade acetonitrile. The elution gradient started with 10% part B up to 20 minutes, increased to 90% up to 25 minutes, and then reverted to 10% up to 28 minutes. Data were gathered through IDA (AB SCIEX analyst TF 1.7.1). Multivariate data analysis for nontargeted metabolomic analysis was performed comprehensively utilizing MS-DIAL (3.70 Yokohama, Kanagawa 230-0045 software, Japan), along with Peak View software from SCIEX.39,50,512.4. Assessment of antiproliferative activity
The cytotoxic activity of the CIR root extract was assessed for a panel of cell lines, including HepG-2 cells (human pancreatic cancer cell line), HCT-116 cells (human colon cancer cell line), MCF-7 cells (human breast cancer cell line), MDA-MB-231 cells (human breast cancer cell line), PC-3 cells (human prostate cancer cell line), A-549 (human lung cancer), A-431 cells (human skin cancer cell line), HeLa cells (human cervical cancer), RD cells (human muscle rhabdomyosarcoma cancer cell line), Panc-1 cells (human pancreatic cancer cell line), CACO2 cells (human intestinal cancer cell line), HEp-2 cells (human larynx cancer), SKOV-3 cells (human ovarian adenocarcinoma), MRC-5 cells (normal human lung fibroblast cells) and SAOS-2 cells (human osteosarcoma). These cell lines were obtained from (the Regional Center for Mycology and Biotechnology at Al-Azhar University). Cells were propagated in specific growth medium and maintained under standard conditions at a temperature of 37 °C in a well-humidified environment with 5% carbon dioxide with regular subculture. Evaluation of antiproliferative activity was carried out following the protocol previously reported.42,43,52 Briefly, 96-well plates were utilized for cell seeding. After a 24 hours incubation, CIR was introduced at varying concentrations. Control wells treated with only DMSO were also included. Following another 24 hours incubation, the evaluation of viable cell numbers was conducted using the MTT assay. In particular, the culture medium was substituted with an MTT reagent and plates were incubated. Subsequently, DMSO was introduced and the optical density was measured at 590 nm with the microplate reader. The relation between surviving cells and CIR concentration was plotted using GraphPad Prism software to obtain the survival curve of each cell line and the IC50 value was derived from a graph.2.5. Microscopic assessment of tumor cell morphology
After treatment of cells with CIR extract at the designated concentration, the medium was drained by inverting the plate and washed with pH 7.2 phosphate buffered saline. Subsequently, cells were immobilized on the plate by exposure to 10% formalin for 15 minutes at room temperature. Cells underwent the staining process using 100 μL of 0.25% crystal violet for a duration of 20 minutes. After the staining process, the excess dye was eliminated, and the plates underwent a deionized water before air drying. Using an inverted microscope (CKX41; Olympus, Japan), images were taken to assess cellular morphology and detect changes compared to control cells. Microscopic examination at 100× magnification facilitated the observation of cytopathic effects or morphological variations.2.6. Evaluation of cell cycle arrest
Cell cycle analysis was performed using the propidium iodide (PI) flow cytometry kit (ab139418, Abcam, USA) by measuring the DNA content using a flow cytometer following the manufacturer's instructions.43,53,54 Briefly, the CIR was dispersed in the cells, centrifugated at 500 g for 5 minutes, washed with 1× PBS, fixed with 66% ethanol on ice and resuspension of the cell pellet in 1× PBS followed by 100% ethanol. The subsequent PI staining procedure involved equilibrating the cells at room temperature, washing with 1× PBS and resuspending the cells in 1× PI + RNase staining solution. After incubation for 1 h in the dark at 37 °C, cells were analyzed using a flow cytometer with an excitation at 493 nm and an emission at 636 nm.2.7. Annexin V analysis for the evaluation of apoptosis
The Annexin V detection method was utilized to assess the apoptosis and necrosis activity of the CIR extract following previous reports.53 Briefly, cells undergo incubation with Annexin V-FITC to stain apoptotic cells, collected by centrifugation and resuspended in 500 μL of 1× binding buffer. Subsequently, 5 μL of Annexin V-FITC and 5 μL of propidium iodide (PI 50 mg mL−1) were added and incubated at room temperature for 5 minutes in the dark. The quantification of Annexin V was subsequently analyzed by flow cytometry through the FITC signal detector, while PI staining was detected via the phycoerythrin emission signal detector (usually FL2).2.8. Evaluation of apoptotic and inflammatory-related gene expression
We evaluated the expression of key genes related to apoptosis and inflammation, including Bcl-2-associated X protein (Bax), B cell lymphoma 2 (BCl2), tumor protein P53 (P53), Caspase-3 (Casp3), cytochrome C (CYC), TNFα, IL1B, and IL6 (Table S1†). This examination was carried out on HepG2 and Panc-1 cells after a treatment period lasting two days with CIR extract at 37 °C compared to staurosporine or untreated control cells. RNA extraction and purification were conducted using the Qiagen RNeasy Mini Kit manufactured by (Hilden, Germany). Subsequently, 1 μg of RNA was used for cDNA synthesis using the cDNA kit (Sigma-Aldrich, product no. 11117831001). Real-time PCR measurements were performed using the Thermo Fisher Quantstudio 5 0.1 ML system and the iScript TM SYBR® Green One-Step RT-PCR Kit was used (Bio-Rad, Hercules, California, United States).55–58 Briefly, 10 ng of cDNA was used and the cycling conditions consisted of an initial heating stage for 5 minutes at 95 °C, followed by 45 cycles of denaturation at 95 °C for 10 seconds, annealing at 60 °C for 30 seconds and extension at 72 °C for 1 minute. Subsequently, the relative gene expression was assessed using the 2−ΔΔCT equation, with the data normalized to the reference gene GAPDH.2.9. Evaluation of antioxidant activity
![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1) mixing copper(II) chloride (10 mM) and neocuproine solution (7.5 mM) with ammonium acetate buffer solution (1 M, pH 7.0). Subsequently, CIR extract at different concentrations (0.2 mL, 0.5 to 1000 μg mL−1) was introduced to the CUPRAC solution and the resulting mixture was incubated for 1 h at room temperature. Finally, CUPRAC activity was evaluated by examining the absorbance by a 450 nm spectrophotometer. In our analysis, EDTA was employed as a control in this assay. The antioxidant activity was then determined and expressed as EDTA equivalents per gram of the CIR extract. The IC50 value was determined from the inhibition concentration curve using the GraphPad program.
1) mixing copper(II) chloride (10 mM) and neocuproine solution (7.5 mM) with ammonium acetate buffer solution (1 M, pH 7.0). Subsequently, CIR extract at different concentrations (0.2 mL, 0.5 to 1000 μg mL−1) was introduced to the CUPRAC solution and the resulting mixture was incubated for 1 h at room temperature. Finally, CUPRAC activity was evaluated by examining the absorbance by a 450 nm spectrophotometer. In our analysis, EDTA was employed as a control in this assay. The antioxidant activity was then determined and expressed as EDTA equivalents per gram of the CIR extract. The IC50 value was determined from the inhibition concentration curve using the GraphPad program.2.10. Evaluation of anti-inflammatory activity
2.11. Molecular modeling study
The molecular coupling study presented was conducted using Molecular Operating Environment (MOE, 2015.10) software to conduct the molecular coupling study of selected secondary metabolites detected in the CIR extract towards the binding pocket of cyclooxygenase 1 & 2 (COX1 & 2) proteins. Toward this end, the crystal structure of the targeted proteins was acquired from the protein database (COX-1: PDB ID 1eqg, COX-2 PDB ID: 4ph9).65,66 The crystal structures were carefully checked and selected on the basis of the nature of the cocrystallized ligand. In our study, we selected crystal structures that cocrystallized with ibuprofen as a reference drug. The acquired crystal structures were initially adjusted by deleting additional chains and water molecules and protonating the protein using the default protocol. The structure of selected secondary metabolites was acquired from the ChemDraw program, and energy minimization and geometry verification were assessed using the MOE program as previously reported.53,54,67–70 The modelling protocol was adjusted for the force field (MMFF94x), the scoring function (London dG), and the placement (Triangle) and subsequently validated by modelling the binding mode of ibuprofen. The binding poses were then evaluated to affirm the original binding mode of ibuprofen to the target proteins, compared to the reported data. The adjusted methodology was then used to explore the binding mode and affinity of the selected secondary metabolites of the CIR extract toward the selected COX proteins. The collected data was analyzed and visualized using the MOE program to provide the binding affinity (kcal mol−1) and binding mode of the examined metabolites.2.12. Statistical analysis
Statistical analysis was performed using GraphPad Prism version 9 software (GraphPad Software, San Diego, CA, USA). Data are presented as mean ± S. D. The Student's t-test was used for the comparison of two groups. The ANOVA test was used for multiple group comparison followed by the Tukey test as a post hoc. The differences between the groups were considered significant when p < 0.05 (*p < 0.05, **p < 0.01, ***p < 0.001 and * ***p < 0.0001).3 Results and discussion
3.1. Phytochemical characterization
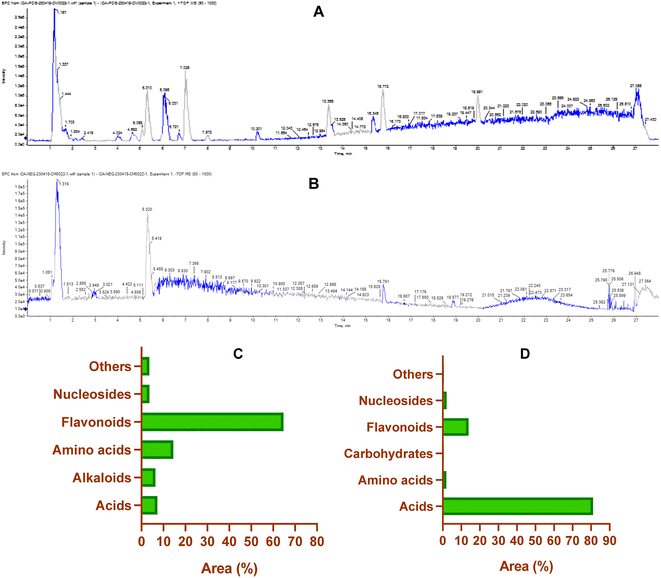
On the other hand, the LCMS/MS analysis of the CIR root extract in negative ionization mode successfully detected 59 metabolites (Table S4†), spanning six diverse classes, namely acids (16 metabolites, 80% peak area), amino acids (7 metabolites, 1.89% peak area), carbohydrates (4 metabolites, 0.5% peak area), flavonoids (15 metabolites, 14% peak area), nucleosides (11 metabolites, 2.1% peak area) and other miscellaneous metabolites (6 metabolites, 0.6% peak area). Within the category of flavonoids, 3 metabolites belonged to flavonoids, while 12 metabolites belonged to flavonoid-O-glycosides. Apigenin emerged as the predominant metabolite in this class, contributing 8.4% of the total peak area, while peonidine-3-O-glucoside chloride exhibited a total peak area of 1.5%. Our findings emphasized that more than 80% of the peak area of the identified metabolites falls within the acid-based class, with 16 distinct metabolites detected. Remarkably, γ-linolenic acid led this category with 58.9% of the total peak area, followed by gluconate (8.2% peak area), D-(−)-quinic acid (5.4% peak area) and D-(+)-malic acid (5.1% peak area). Nucleotide-based metabolites were detected with approximately 2.1% of the total peak area. Uridine, as the most predominant metabolite within this category, represented 0.7% of the total peak area. Furthermore, amino acid metabolites constituted 1.9% of the total detected metabolites in the negative ionization mode. Notably, L-γ-homoisoleucine emerged as the most prevalent amino acid, accounting for 0.9% (peak area). Our analysis further revealed that CIR extract possessed considerable carbohydrate-based metabolites (0.5% peak area, 4 metabolites). Finally, a group of six miscellaneous metabolites collectively account for 0.6% of the peak area. Among these metabolites, sodium deoxycholate was the most abundant, with a detection of 0.25% (peak area).
Our comprehensive profiling of the CIR root extract illuminated the intricate metabolite composition inherent in this extract. The most prominent metabolites (59-1% peak area) identified by untargeted metabolomic analysis of CIR extract are detailed in Table 1 and Fig. 2. Consequently, CIR extract emerges as a promising candidate for natural supplementary interventions addressing a wide array of health issues. Our analysis indicated that the CIR extract possesses high concentrations of acid-based metabolites including γ-linolenic acid (GLA), gluconate, D-(−)-quinic acid, D-(+)-malic acid, and 12-oxo-10,15(Z)-phytodienoic acid. GLA was represented as the most prominent metabolite in this class and as a major constituent of the CIR extract. Several studies showed that GLA has the potential to mitigate conditions associated with chronic inflammation. Furthermore, GLAs showed an acknowledged role in maintaining skin health that extends to applications in skincare formulations. Furthermore, GLA contributes to the support of the immune system and cardiovascular well-being by regulating cholesterol levels.74,75 Gluconate, a naturally carboxylic acid, contributes significantly contributes to cellular metabolism76 and its biological impact extends to various applications, including anti-inflammatory, antioxidant, immunomodulatory, antimicrobial, and antiviral effects.77,78
| Title | RT (min) | Precursor (m/z) | Area | Error (PPM) | Adduct | Reference (m/z) | Formula | Ontology |
|---|---|---|---|---|---|---|---|---|
| Maleic acid | 0.87 | 115.0034 | 637![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 716 716 |
−0.6 | [M − H]− | 115.00368 | C4H4O4 | Dicarboxylic acids |
| D-(+)-Malic acid | 0.88 | 133.0128 | 3![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 334 334![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 704 704 |
6.6 | [M − H]− | 133.01425 | C4H6O5 | β-Hydroxy acids |
| D-(−)-Quinic acid | 0.93 | 191.0543 | 3![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 485 485![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 385 385 |
7.4 | [M − H]− | 191.05611 | C7H12O6 | Quinic acids |
| Gluconate | 0.96 | 195.0495 | 5![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 327 327![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 352 352 |
6 | [M − H]− | 195.05103 | C6H12O7 | Hydroxy acids |
| N,N-Dimethylglycine | 1.053 | 104.1062 | 4![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 379 379![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 100 100 |
3.9 | [M + H]+ | 104.0706 | C4H9NO2 | Amino acid |
| 12-Oxo-10,15(Z)-phytodienoic acid | 1.12 | 293.0635 | 1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 886 886![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 257 257 |
−1.2 | [M + H]+ | 293.21112 | C18H28O3 | Acid |
| Trigonelline | 1.14 | 138.0552 | 1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 573 573![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 969 969 |
−0.1 | [M + H]+ | 138.05496 | C7H7NO2 | Alkaloids |
| DL-5-Hydroxylysine | 1.18 | 163.058 | 673![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 048.7 048.7 |
12.8 | [M + H]+ | 163.10771 | C6H14N2O3 | Amino acid |
| L-Asparagine | 1.29 | 133.0507 | 512![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 949.9 949.9 |
−4.8 | [M + H]+ | 133.06078 | C4H8N2O3 | Amino acid |
| Harmaline | 5.06 | 215.1256 | 1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 403 403![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 891 891 |
0.1 | [M + H]+ | 215.11789 | C13H14N2O | Alkaloids |
| 3′,4′,5,7-Tetrahydroxyflavanone | 5.27 | 289.163 | 7![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 469 469![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 832 832 |
0.8 | [M + H]+ | 289.07068 | C15H12O6 | Flavonoids |
| Isorhamnetin-3-O-glucoside | 6.79 | 479.12 | 1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 024 024![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 418 418 |
0.8 | [M + H]+ | 479.11841 | C22H22O12 | Flavonoids |
| Peonidine-3-O-glucoside chloride | 7.39 | 461.1098 | 1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 011 011![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 160 160 |
−0.8 | [M − 2H]− | 461.1084 | C22H23O11 | Anthocyanidin-3-O-glycosides |
| Naringenin | 7.53 | 273.1644 | 4![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 594 594![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 841 841 |
13.8 | [M + H]+ | 273.07574 | C15H12O5 | Flavonoids |
| Apigenin | 10.40 | 269.0444 | 5![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 452 452![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 233 233 |
3.3 | [M − H]− | 269.04553 | C15H10O5 | Flavones |
| γ-Linolenic acid | 19.07 | 277.2176 | 37![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 943 943![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 284 284 |
−0.9 | [M − H]− | 277.21732 | C18H30O2 | Lineolic acids |
| 3,5,7-Trihydroxy-4′-methoxyflavone | 19.66 | 301.1429 | 3![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 023 023![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 031 031 |
0.6 | [M + H]+ | 301.07068 | C16H12O6 | Flavonoids |
| 2′-Deoxycytidine | 19.92 | 228.2302 | 800![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 974 974 |
12 | [M + H]+ | 228.09789 | C9H13N3O4 | Nucleosides |
| trans-Cinnamate | 22.58 | 149.0246 | 1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 220 220![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 626 626 |
−1.9 | [M + H]+ | 149.05971 | C9H8O2 | Acid |
| Acacetin-7-O-rutinoside | 24.03 | 593.2783 | 12![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 237 237![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 822 822 |
0.1 | [M + H]+ | 593.18646 | C28H32O14 | Flavonoids |
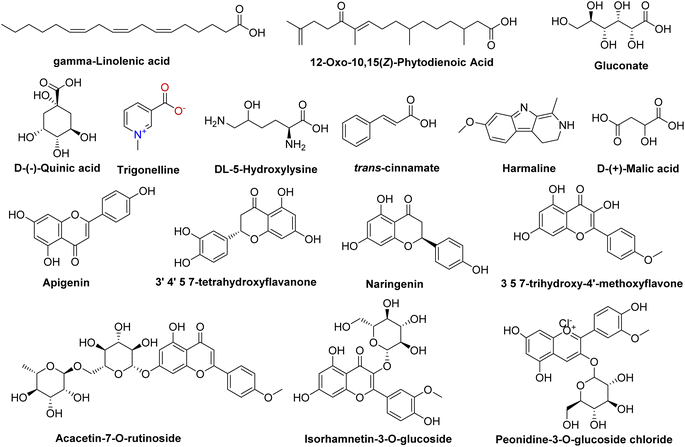 | ||
| Fig. 2 Representative structures of the most prominent metabolites detected in the CIR extract by UPLC/T-TOF-MS/MS analysis in positive and negative ionization modes. | ||
D-(+)-Malic acid exhibits notable biological activities, mainly known for its hepatoprotective effects.79 Additionally, it plays a role in the regulation of acidity in the body and participates in the citric acid cycle, an essential metabolic pathway.80 Additionally, 12-oxo-10,15 (Z)-phytodienoic acid has been involved in the regulation of various physiological processes in plants, including defense responses.81 Finally, trans-cinnamate demonstrated a wide diversity of bioactivity including antioxidant, antimicrobial, and anti-inflammatory potentials, suggesting its potential to alleviate conditions associated with inflammation.82 Flavonoids and flavonoid O-glycoside metabolites have also been detected in substantial concentrations, including acacetin-7-O-rutinoside, 3′,4′,5,7-tetrahydroxy-flavanone, naringenin, apigenin, 3,5,7-trihydroxy-4′-methoxyflavone, peonidine-3-O-glucoside and isorhamnetin-3-O-glucoside. Acacetin-7-O-rutinoside, the second highly detected metabolite, plays a crucial role in various biological activities, including anti-inflammatory, antimicrobial, and anticancer properties.83,84 Acacetin also demonstrates a range of biological activities, emerging as a potential agent for improving insulin sensitivity and regulating blood sugar levels, thus promising in the management of diabetes.85 Similarly, 3′,4′,5,7-tetrahydroxyflavanone, a naturally occurring flavone, is characterized by its anticarcinogenic, anti-inflammatory and antioxidant capabilities, but also offers cardiovascular protection and stimulates the immune response.86,87 The CIR extract also exhibited a notable abundance of naringenin, which encompasses a diverse array of therapeutic potential, including antioxidant, anti-inflammatory, anti-apoptotic, anti-ulcer, anti-osteoporotic, and anti-carcinogenic effects.88,89 Extensive research has empirically supported its pharmacological benefits as a multifaceted agent with hepatoprotective, antiatherogenic, antimutagenic and antimicrobial properties.90,91 Peonidine-3-O-glucoside chloride, a flavonoid glycoside detected in CIR extract, holds promise as antioxidant activity, contributing to cell protection against oxidative stress.92 Apigenin, another detected flavonoid, possesses various bioactive properties, including antibacterial, antiviral, anti-inflammatory, and anticancer activities.93–95 Its versatile pharmacological profile involves the modulation of signaling pathways and cellular processes, making it a promising natural compound for potential therapeutic applications, including rheumatoid arthritis, autoimmune disorders, and neurodegenerative diseases.96 Relatedly, 3,5,7-trihydroxy-4′-methoxyflavone has been recognized for its potential pharmacological applications as an anticancer, anti-inflammatory, and antioxidant.97 LCMS-MS analysis also revealed a considerable abundance of amino acid metabolites including N,N-dimethylglycine, L-asparagine, and 5-hydroxylysine. N,N-Dimethylglycine, commonly denoted as vitamin B16, has versatile therapeutic applications that include the treatment of depression in middle-aged and elderly individuals,98 the facilitation of the human immune system,99 and the mitigation of cholesterol levels within the body.100 Although DL-5-hydroxylysine has been implicated in collagen formation,101 L-asparagine plays diverse roles in protein synthesis, neurotransmitter regulation, and cellular energy metabolism.102,103 Two alkaloids, trigonelline and harmaline, were also identified in the CIR root extract in remarkable abundance. Trigonelline has potential antioxidant, neuroprotective and anti-inflammatory properties and has been implicated in the treatment of diabetes through improved glucose metabolism.104,105 On the other hand, harmaline exhibited substantial neuropharmacological and antiamnesic effects. In addition, studies demonstrated its ability to improve cognition dysfunction associated with neurodegenerative diseases.106,107 Collectively, comprehensive metabolomic analysis underscores the potential of the CIR extract as a valuable source of bioactive compounds with promising applications in the treatment of various health issues. Aligning with our findings, previous investigations revealed that the extract of C. intybus exhibits a set of unique metabolites that include guaianolides (e.g., lactucin, lactucopicrin), polyacetylenes (e.g. putrescine, spermidine) and phenolic compounds (e.g., chlorogenic acid, caffeic acid), flavonoids (e.g., apigenin-7-O-glucoside, kaempferol-7-O-glucoside, quercetin-3-O-glucoside) and sterols (e.g., β-sitosterol, campesterol, stigmasterol). The phytochemical diversity of C. intybus further underscores its potential as a source of bioactive metabolites with miscellaneous therapeutical effects.29 Another investigation of the phenolic metabolites of red C. intybus utilizing high-performance liquid chromatography analysis (HPLC-DAD-ESI-MS/MS) identified malic acid and quinic acid among the free small organic acids, various caffeic acid derivatives, and different isomers of caffeoylquinic acids. Furthermore, various derivatives of kaempferol, which include mono and diglycosides, along with other flavonols such as isorhamnetin, quercetin, and myricetin derivatives, were also characterized based on their MS fragmentation patterns.108
3.2. Assessment of cytotoxic activity
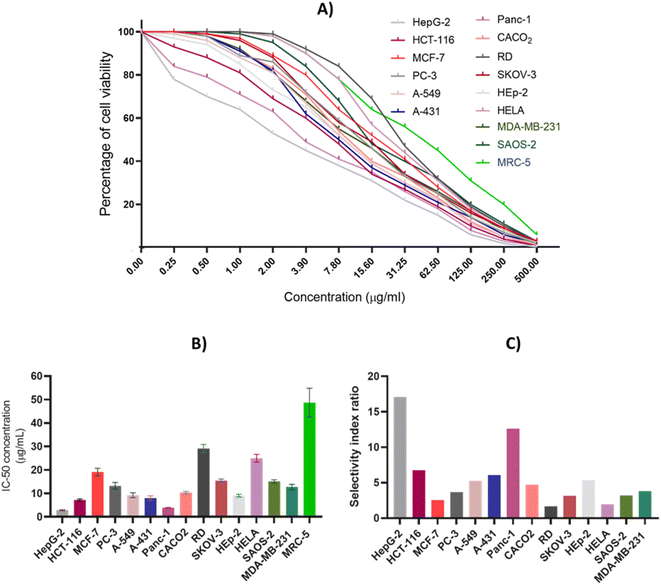
A comprehensive antiproliferative assay was conducted to detect and evaluate the cytotoxic impact of CIR extract against a panel of 15 human cell lines including HepG-2, HCT-116, MCF-7, PC-3, A-549, A-431, Panc-1, CACO2, RD, SKOV-3, HEp-2, HELA, SAOS-2, MDA-MB-231, and MRC-5. The CIR extract was assessed dose-dependently, with concentrations ranging from 0.25 to 500 μg mL−1, and cell viability was examined by MTT assay utilizing a spectrophotometer. As depicted in Fig. 3, our results showed that the CIR extract exhibits a remarkable dose-dependent cytotoxic activity in all cancer cell lines examined with IC50 values 2.85–29.15 μg mL−1. While the extract exhibited moderate antiproliferative activity toward the cell viability of HeLa, MCF-7, RD, SKOV-3, and SAOS-2 cells with IC50 > 15 μg mL−1, it showed considerable cytotoxic activity toward PC-3, A-549, CACO2 and MDA-MB-231 cells with IC50 > 10 μg mL−1. Furthermore, the extract revealed significant antiproliferative activity towards HCT-116, A-431, and HEp-2 cells with IC50 < 10 μg mL−1. Interestingly, among the cancer cell lines examined, the CIR extract demonstrated the most pronounced antiproliferative activity in cellular viability of HepG-2 and Panc-1 cells with IC50 < 5 μg mL−1 (IC50 = 2.85, and 3.86 μg mL−1, respectively). These findings suggest that the extract has promise as a potential antiproliferative with a broad spectrum of antiproliferative activity, particularly for HepG-2 and Panc-1 cells. To examine the effect of the CIR extract on normal cells, we further explored the cytotoxic effect of the extract on normal human fibroblast cells (MRC-5 cell line). In this regard, cells were treated with extract in dose-dependent (0.25–500 μg mL−1) and viability was assessed as previously using the MTT assay. As shown in Fig. 3, the extract showed a low antiproliferative activity towards MRC-5 cells with an IC50 of 48.70 μg mL−1. These findings indicate that the extract has selectivity toward cancer cells with low effect on normal cells, suggesting its antitumor potential. Furthermore, we assessed the selectivity index (SI) of the CIR extract towards the cancer cell lines examined compared to that of the MRC-5 cells. A notable variation was reported among the cancer cell lines tested, with an SI range of 1.7–17.1. The CIR extract showed a moderate selectivity index (SI < 5) toward MCF-7, PC-3, RD, A-549, CACO2, MDA-MB-231, SKOV-3, SAOS-2 and HeLa cells, while it exhibited considerable selectivity (SI > 5) toward HCT-116, A-431, HepG-2, Panc-1and Hep-2 cells, signifying the effect of disparity of the CIR extract on the investigated cell lines. Algin with the observed antiproliferative activity, the CIR extract displayed the highest selectivity index (SI > 10) toward the HepG-2 and Panc-1 cells, indicating the significant selectivity of the CIR extract in those cancer cells. Various studies have explored the phytochemical composition of C. intybus and its therapeutical efficacy.29,108–111 3-O-p-Coumaroyl quinic acid derived from the C. intybus flower extract exhibited antiproliferative activity against PC-3 and undifferentiated noncancerous 3T3L1 fibroblast cells. Furthermore, 5-caffeoylquinic acid substantially inhibited the invasion of nonsmall cell lung cancer cells (H1299).112 The C. intybus leaf extract showed effective cytotoxicity against LNCaP prostate cancer cells, while the root extract showed significant effectiveness against MCF-7 breast cancer cells, C32 amelanotic melanoma cells and ACHN renal adenocarcinoma cells.113 Furthermore, the methanolic extract of C. intybus exhibited a time-dependent reduction in the viability of SKBR3 breast cancer cells, with IC50 values of 800 μg mL−1 at 24 h. In another study, the methanolic extract of C. intybus efficiently inhibited the growth of Jurkat cells (IC50 = 138 μg mL−1) and moderately inhibited the growth of Fen bladder carcinoma cells and HeLa cervical epithelioid carcinoma cells (25% inhibition at 200 μg mL−1).114 Aligned with these facts, our study revealed that the CIR extract exerts a noteworthy inhibitory impact on the viability of specific cancer cells, specifically Hep-G2 and Panc-1. Based on the metabolomic analysis of the extract, the pronounced antiproliferative activity of the extract could be attributed to its unique bioactive content, including γ-linolenic acid which has been documented to produce significant antiproliferative and growth inhibitory effects in the treatment of breast and pancreatic cancer cell lines.115,116 Furthermore, the presence of acacetin-7-O-rutinoside, 3,4,5,7-tetrahydroxyflavanone, which has the potential to serve as anticancer agents with significant antiproliferative capabilities against cancer cells, augments the cytotoxic effect of the extract.117–119 Furthermore, the extract includes abundant amounts of gluconate that has well-characterized cytotoxic effects both in vitro and in vivo.120 Moreover, Murugesan et al. observed the cytotoxic impact of quinic acid derivatives on human glioblastoma cell lines SNB19 and LN229.121 Consequently, the list of bioactive metabolites detected in the CIR extract (Table 1) aligns with the observed antiproliferative potential of the CIR extract against various types of cancer cells.Based on these results, we explored the morphology of HepG-2, Panc-1, and HCT-116 cells after extract treatment (Fig. 3). The extract was applied at different concentrations (1, 4, 20, 100, and 500 μg mL−1) to the cells and the cellular morphology was assessed after a duration of 24 hours under an inverted microscope. As presented in Fig. S1,† the treatment of cells with CIR extract revealed a remarkable pattern of cell loss. Hep-G2, Panc-1, and HCT116 cells, which are polyhedral in shape and typically grow in a tightly connected manner, exhibit normal healthy morphology with a distinct cytoskeleton in the untreated control state. Upon treatment with the CIR extract, the cellular morphology was significantly distorted in a dose-dependent manner. At 4 μg mL−1, some cells exhibited a round shape, and areas of cell loss became apparent. With increasing doses, a significant cytotoxic effect was observed, evident in detached cells with characteristic membrane blebbing at 20 μg mL−1. In the 100–500 μg mL−1 range, cell destruction became more pronounced, with only a few remaining cells attached to the culture dish. These cells displayed distinguishable morphological signs of apoptosis, showing less homogeneity and loss of membrane integrity (Fig. S1†). Our findings showed that the CIR extract has a pronounced cytotoxic effect on HepG-2 and Panc-1 cells. Accordingly, our investigations were directed to explore the underlying mode of action of the observed antiproliferative activity of the extract.
3.3. Assessment of cell cycle arrest
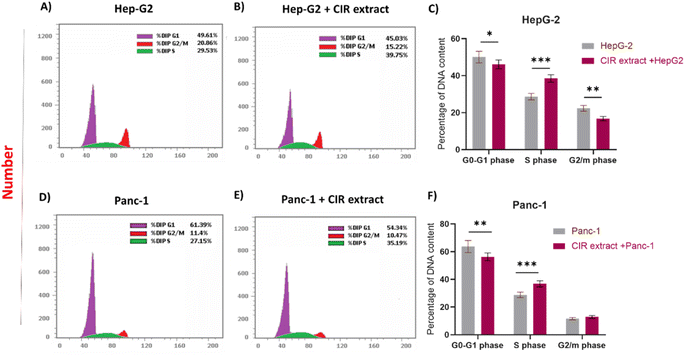
The MTT analysis demonstrated the substantial ability of the CIR extract to significantly reduce the viability of HepG2 and Panc-1 cells. Consequently, we extended our investigation to explore the impact of CIR extract on the cell cycle of HepG-2 and Panc-1 cells (Fig. 4). To assess this, we conducted a flow cytometry analysis to examine the cell cycle phases in the examined cells after administration of the CIR extract at its IC50 value. In the case of HepG-2 cells, treatment with CIR extract (2.85 μg mL−1) resulted in a significant decrease in cell percentage in the G1 phase (approximately 5%), and a similar decrease in the G2/M phase (approximately 5.5%), compared to control cells. In contrast, the percentage of the S phase in the extract-treated group was elevated by approximately 10%, indicating blockage of the cell cycle in the S phase (Fig. 4 ACC). In the context of Panc-1 cells, the application of the CIR extract (3.86 μg mL−1) resulted in a reduction of approximately 7% in cell percentage in phase G1, compared to control cells. Meanwhile, the phase G2/M exhibited a minor decrease, with a decline from 11.4% to 10.47%. On the other hand, the percentage of cells in phase S increased by around 8% in cells treated with CIR, indicating blockage of the cell cycle in phase S (Fig. 4D–F). These findings indicate that the antiproliferative activity of CIR extract could be associated with its ability to induce cell growth arrest in the S phase. Cell cycle dysregulation is the hallmark of cancer development.122 Most human cancers have been reported to originate from disturbances in G1/S cell cycle control.123,124 The G1/S phase transition is carefully regulated by various checkpoints. Disruptions in any of the cell cycle regulators or checkpoints, particularly those that guard the transition of cells from the G1 to the S phase, can be debilitating.124 Such disturbances may account for uncontrolled proliferation, contributing to the initiation and promotion of cancer; alternatively, they may induce cell death in therapeutic strategies targeting these processes.122The use of drugs or bioactive compounds for the therapeutic manipulation of cell growth arrest in the G1–S phase has demonstrated potent antiproliferative activity and apoptotic effects in many human cancer cell lines. Preclinical studies suggest that cells with impaired checkpoint function are more susceptible to anticancer agents.122 Therefore, our findings highlight the impact of CIR extract as an antitumor agent, given its potential to impact crucial cell cycle checkpoints. These findings could be attributed to the presence of specific active metabolites within the CIR extract, such as γ-linolenic acid, acacetin-7-O-rutinoside, harmaline and trigonelline. These metabolites have previously been documented for their ability to halt the cell cycle of tumor cells.125–127 For example, the Jia-Ren Liu group has documented that linoleic acid can block the cell cycle of the gastric adenocarcinoma cell line SGC-7901.128 Furthermore, Liu et al. reported that harmaline has the ability to induce cell cycle arrest and trigger cellular apoptosis through the mitochondrial pathway in SW620 cells, through inhibition of the Akt and ERK signaling pathways.129 In this regard, this study demonstrated a substantial increase in the population of cells arrested in the S phase, as compared to the control cells. With increasing concentrations of harmaline, the population of cells in the S phase increased in a dose-dependent manner, while the population of cells in the G1 phase decreased. These data are consistent with our findings, underscoring that the cell cycle distribution was significantly arrested in the S phase.129 Consequently, it can be inferred that the CIR extract effectively hinders the progression of HepG2 and Panc-1 cells by arresting the cell cycle, thus impeding their replication and proliferation.
3.4. Evaluation of programmed cell death (apoptosis and necrosis)
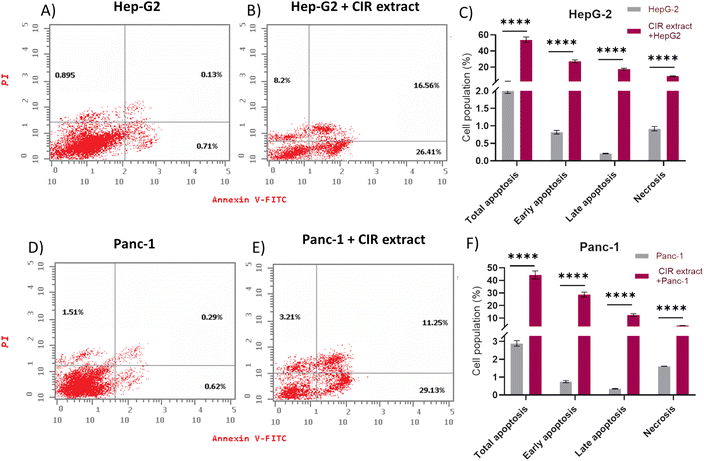
Next, our objective was to explore whether the antiproliferative effect of the CIR extract is associated with its ability to induce programmed cell death in HepG2 and Panc-1 cells. In this regard, a flow cytometry assay was conducted to analyze the effect of CIR extract on apoptosis and necrosis induction. Cell staining with Annexin V-FITC and PI was used to distinguish and quantitatively determine the percentage of apoptotic and necrotic cells. Viable cells are seen in the left lower quadrant (FITC-/PI−), necrotic cells are seen in the left upper quadrant (FITC-/PI+), early apoptotic cells in the right lower quadrant (FITC+/PI−) and late apoptotic cells in the right upper quadrant (FITC+/PI+).130,131 As illustrated in Fig. 5, treatment of HepG-2 cells with CIR extract at the dose of IC50 (2.85 μg mL−1) demonstrated the induction of apoptosis in cells, as indicated by the significant increase in total apoptosis in cells treated with CIR. The percentage of early apoptotic cells stained by Annexin V-FITC was approximately 26%, a 37-fold increase compared to control cells, and the percentage of late apoptotic cells double stained by Annexin V-FITC and PI was around 16%, a 127-fold increase compared to control cells. Consequently, total apoptosis increased from 1.73% for control cells to 51.17% for CIR-treated cells, with an almost 30-fold increase. Similarly, necrosis levels showed an increase from 0.89% in untreated control cells to 8.2% in CIR extract-treated cells, demonstrating a 9-fold increase in regulation (Fig. 5C). According to Panc-1 cells, the proportion of early apoptotic cells increased from around 0.6% in control cells to 29% in CIR treated cells (48 times greater), while the percentage of late apoptotic cells increased from 0.29% in control cells to 11.25% in CIR treated cells (39 times greater). Therefore, total apoptotic levels in cells treated with CIR extract reached 43.59% compared to 2.42% in untreated control cells, demonstrating an increase of 18 times. Additionally, necrosis proportions exhibited an approximately twofold upregulation of 1.51% to 3.21% after treatment with CIR extract (Fig. 5D–F). Taken together, these findings indicate that the antiproliferative activity of CIR extract is associated with its ability to substantially trigger programmed cell death pathways, such as apoptosis and necrosis, in cancer cells and further underscore the pharmacological potential of CIR extract as a natural-based antitumor supplement.3.5. Evaluation of apoptosis-related gene expression
To gain further insight into the apoptotic mode of action of the CIR extract, we extended our investigation to assess the expression of key apoptotic-associated genes in HepG2 and Panc-1 cells. The intrinsic apoptotic pathway is regulated by the Bcl-2 (B cell lymphoma 2) family of proteins. The proteins in this family can be classified by their anti-apoptotic (such as Bcl-2) or pro-apoptotic (such as Bax) actions. Activation of these pathways results in activation of caspases that lead to apoptosis.132 In particular, the pro-apoptotic gene Bax is one of the downstream effectors of p53, which is a main tumor suppressor in multicellular organisms that induces cell cycle arrest, DNA repair, and apoptosis to promote genomic stability and tissue homeostasis.133 A variety of apoptotic stimuli cause mitochondrial cytochrome c release, which in turn induces a series of biochemical reactions that result in caspase activation and subsequent cell death.134 The crosstalk between these genes and their role in inducing and controlling apoptosis is demonstrated in the apoptotic pathway and protein interactions, as indicated by the KEGG and String databases (Fig. 6 and S2†).46,135 Toward this end, we evaluated the expressions of the Casp3, Bax, Bcl-2, P53 and CYC genes after cells were treated with CIR extract at the corresponding IC50 values.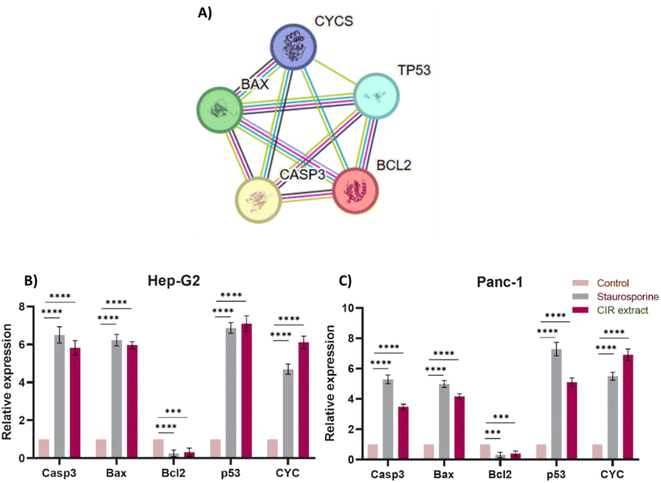 | ||
| Fig. 6 Effect of the CIR extract on the expression of apoptotic genes in Hep-G2 and Panc-1 cells. (A) Protein–protein interaction network for the investigated genes involved in apoptosis. The nodes represent proteins, and the edges denote predicted associations. The colored lines indicate different types of evidence, with red representing fusion evidence, light blue representing database evidence, green representing neighborhood evidence, blue representing co-occurrence evidence, purple representing experimental evidence, black representing co-expression evidence and yellow representing text mining evidence, interaction scores are detailed in supplementary Table S5.† Relative gene expression of the Casp3, Bax, Bcl-2, P53, and CYC genes in Hep-G2 (B) and Panc-1 (C) cells after treatment with CIR extract (at IC50 value) or staurosporine, compared to control cells. Data were expressed as mean ± S. D. in triplicate. Comparisons were made using the ANOVA test and Tukey as a post hoc test. Differences between groups were considered significant when p ≤ 0.05 (*p ≤ 0.05, **p ≤ 0.01, ***p ≤ 0.001, and ****p < 0.0001). | ||
In our investigations, staurosporine, a recognized apoptotic inducer, was used as a control reference drug. As shown in Fig. 6, treatment of HepG2 cells with CIR extract at 2.85 μg mL−1 demonstrated the ability to significantly upregulate the expression of Casp3 (5.8 times), Bax (5.9 times), P53 (7 times) and CYC genes (6 times), compared to control HepG2 cells. However, it considerably decreased the expression of the Bcl2 gene (0.3-fold change), compared to untreated cells. Interestingly, the efficacy of CIR extract was comparable to that of the well-known staurosporine drug which showed the ability to increase the expression of Casp3 (6.5 times change), Bax (6.2 times change), P53 (6.9 times change) and CYC genes (4.7 times change), while it reduced the expression of the Bcl2 gene (0.2 times change). Regarding Panc-1 cells, treatment with CIR extract at 3.86 μg mL−1 demonstrated the ability to substantially increase the expression of the Casp3 (3.4-fold change), Bax (4.2-fold change), P53 (5-fold change) and CYC genes (6.8-fold change), compared to control Panc-1 cells. However, it considerably mitigated the expression of the Bcl2 gene (0.38-fold change), compared to untreated cells. Similarly, comparing the efficacy of CIR extract with that of the well-known staurosporine drug showed the ability to increase the expression of the Casp3 (5.2 times), Bax (4.9 times), P53 (7.3 times) and CYC genes (5.5 times), while reducing the expression of the Bcl2 gene (0.29 times) (Fig. 6). These results further indicate that the antiproliferative activity of CIR extract could be related to its potential ability to modulate the expression of apoptotic genes in cancer cells. The observed apoptotic effect of the CIR extract can be attributed to the presence of several bioactive metabolites, including γ-linolenic acid,136,137 3′,4′,5,7-tetrahydroxyflavanone,138 trans-cinnamate139 and naringenin.140,141 Mechanistically, γ-linolenic acid showed the potential to induce apoptosis in K562/ADM cells by triggering lipid peroxidation and caspase-3.137 The necrotic effect of γ-linolenic acid was also demonstrated in human gliomas where it induced extensive necrosis and cystic degeneration.142 Furthermore, naringenin showed a strong antitumor effect in MDA-MB-231 cells by blocking cells in the G0/G1 phase, altering the mitochondrial-mediated intrinsic pathway responsible for apoptosis and inducing inflammatory reaction.141 Furthermore, the apoptotic effect of naringenin was related to its ability to modulate pro-apoptotic genes including the Bax, P53 and Casp3 genes.143,144 Kanno et al. 2006 showed that naringenin induces necrotic activity in human pro-myeloleukemia HL-60 cells and triggers mitochondrial dysfunctions by depleting intracellular ATP.145 Collectively, the remarkable cytotoxic activity of CIR extract in the cellular growth of cancer cells could be further explained by the unique phytochemical profile of CIR extract, which possesses a set of bioactive metabolites that significantly trigger apoptotic levels in cancer cells. These findings further affirm the antitumor potential of the CIR extract and support CIR as a natural pharmacological supplement for human diseases.
3.6. Evaluation of anti-inflammatory activity
To further explore the pharmacological impacts of CIR extract, we aimed to explore its anti-inflammatory activity by assessing its ability to target the pro-inflammatory enzymes COX-1 & 2 and to mediate the expression of inflammation-related genes (IL-1b, IL-6, and TNF-α genes).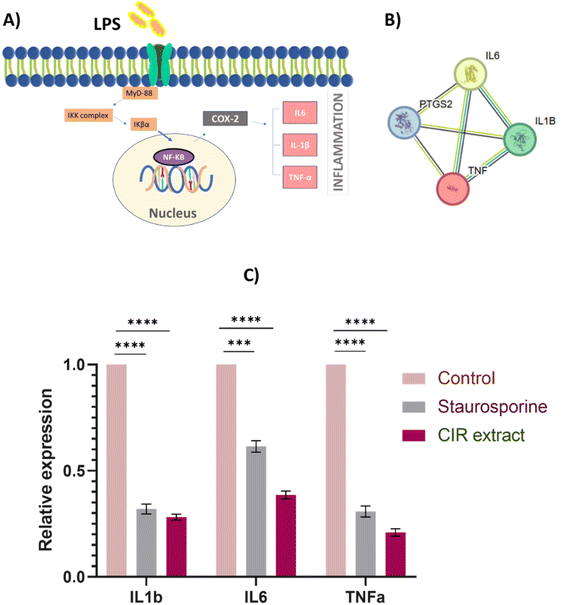 | ||
| Fig. 8 Effect of the CIR extract on the expression of inflammatory genes in LPS-induced RAW 264.7 cells. (A) The proposed KEGG pathway shows IL-1b, IL-6 and TNF-α as selected inflammatory markers associated with the COX2 mediator. (B) Protein–protein interaction network for the investigated genes involved in the inflammatory process. The nodes represent proteins, and edges denote predicted associations. The colored lines indicate different types of evidence, with red representing fusion evidence, light blue representing database evidence, green representing neighborhood evidence, blue indicating co-occurrence evidence, purple representing experimental evidence, black indicating co-expression evidence, and yellow representing text mining evidence, interaction scores are detailed in supplementary Table S6.† STRING analysis135 (C) the relative expression of the genes IL-1b, IL-6 and TNF-α genes in LPS-induced RAW 264.7 cells after treatment with CIR extract (at IC50 value) or staurosporine, compared to control cells. Data were expressed as mean ± S. D. in triplicate. Comparisons were performed using the ANOVA test and Tukey as a post hoc test. Differences between groups were considered significant when p ≤ 0.05 (*p ≤ 0.05, **p ≤ 0.01, ***p ≤ 0.001, and ****p < 0.0001). | ||
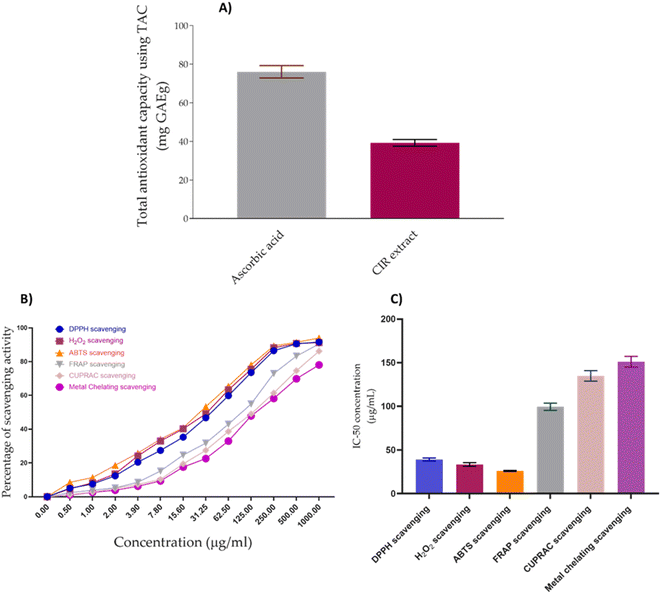
3.7. Assessment of antioxidant activity
Cancer has been linked to ROS-mediated damage to biological macromolecules, which arises from an imbalance between radical generating and radical-scavenging systems.164 In this regard, we have extended our investigations to explore the antioxidant impact of CIR extract. To achieve this aim, we have initially assessed the total antioxidant capacity (TAC) of the CIR extract. Our analysis revealed that the CIR extract has considerable antioxidant capacity with a TAC of 39.21 ± 1.75 mg GAE per g, compared to ascorbic acid (TAC = 73.6 mg GAE per g). Based on these results, we extended our explorations to assess the antioxidant activity of CIR extract by examining its free radical scavenging activity against DPPH, ABTS and H2O2 radicals, but also its metal scavenging activity (FRAP, metal chelating and CUPRAC scavenging assays). In our investigations, ascorbic acid and EDTA functioned as the standard positive control (ESI Fig. S4–S6†). Evaluation of the free radical scavenging activity of CIR extract towards DPPH, ABTS, and H2O2 radicals revealed that it has substantial and dose-dependent scavenging activity, compared to ascorbic acid (Fig. 9).IC50 evaluation indicated that CIR exhibits an inhibitory activity of 39.06 ± 1.78 μg mL−1 toward DPPH radicals, 26.03 ± 0.69 μg mL−1 toward the ABTS radicals, and 33.36 ± 2.09 μg mL−1 toward the H2O2 radicals, compared to ascorbic acid (IC50 = 10.19 ± 0.75 μg mL−1, 10.66 ± 0.89, 14.77 ± 0.69, respectively). These results indicate that the CIR extract exhibits substantial antioxidant activity by scavenging free radicals. Similarly, an evaluation of the metal scavenging activity of the CIR extract using FRAP, metal chelating, and CUPRAC scavenging assays indicated that CIR extract displays considerable antioxidant activity in a concentration-dependent manner (Fig. 9). Our analysis revealed that CIR shows a considerable reducing potential toward copper (Cu2+) and iron (Fe3+) ions, as indicated by CUPRAC and FRAP assays, with an IC50 of 134.82 ± 5.98, and 99.50 ± 4.09 μg mL−1, respectively. Furthermore, the CIR extract exhibited satisfactory metal chelating activity with an IC50 of 151.1 ± 6.14 μg mL−1. These findings indicate that the CIR extract possesses significant reduction power and metal chelating activity, further highlighting the antioxidant potential of the CIR extract. The observed antioxidant capacity of CIR extract could be correlated with the unique content of bioactive metabolites present in the extract, as described in Table 1. Several detected metabolites present with a high prevalence in the CIR extract, including γ-linolenic acid, acacetin-7-O-rutinoside, 3′,4′,5,7-tetrahydroxyflavanone, naringenin, N,N-dimethylglycine, apigenin, and gluconate, are well recognized for their significant antioxidant capacity.165–167 In alignment with our findings, Celep et al. showed that a three-edible fruit extract exhibits a substantial antioxidant capacity, as evidenced by the observed potent free radical and metal scavenging activities.168 Similar to the CIR extract presented, the phytochemical profile of the investigated extract showed an abundance of flavonoids, including acacetin-7-O-rutinoside, naringenin, and apigenin.168
3.8. Molecular modeling study
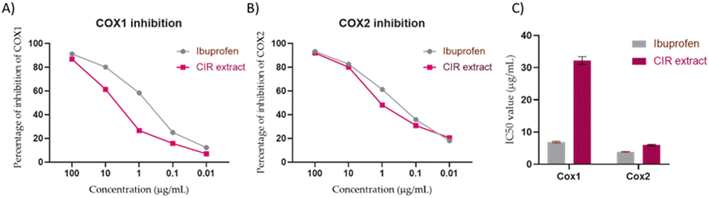
To gain more insight into the anti-inflammatory activity of the CIR extract and its selectivity to cyclooxygenase enzymes (COX-1 & 2), we extended our investigation to explore the binding affinity of the most prominent secondary metabolites detected in the CIR extract to the binding cavity of the COX-1 and COX-2 proteins. In this regard, we conducted extensive molecular coupling studies for a set of 12 secondary metabolites, including acacetin-7-O-rutinoside; 3′,4′,5,7-tetrahydroxy-flavanone; naringenin; 3,5,7-trihydroxy-4′-methoxy-flavone; harmaline; trans-cinnamate; γ-linolenic acid; gluconate; isorhamnetin-3-O-glucoside; 12-oxo-10,15-(Z)-phytodienoic acid; apigenin and peonidine-3-O-glucoside. The crystal structures of COX-1 & 2 were selected and acquired from PDB, and the applied protocol was adjusted to confirm the essential interactions reported for the co-crystallized ligand.| Ligand | Score (kcal mol−1) | Interactions | |
|---|---|---|---|
| Hydrophilic | Hydrophobic | ||
| Ibuprofen | −10.26 | Arg120 (2.91 Å, 3.59 Å, 2.83 Å), Tyr355 (2.78 Å) | Val116, Leu531, Val349, Leu352, Met522, Ala527, Ile523, Tro387, Phe518, Leu359 |
| Acacetin-7-O-rutinoside | −9.35 | Arg120 (2.56 Å), Glu524 (2.62 Å), Arg83 (2.78 Å) | Leu93, Met522, Trp387, Leu384, Val116, Leu531, Val349, Leu115, Leu359, Met113, Ala527, Val119, Phe86, Ile89 |
| 3′,4′,5,7-Tetrahydroxy-flavanone | −8.62 | Ser530 (2.88 Å, 2.67 Å), Arg120 (3.09 Å) | Phe518, Leu359, Val116, Ala527, Trp387, Leu531, Leu352, Ile523, Val349, Met522 |
| Naringenin | −6.18 | Ile523 (3.59 Å), Gly526 (3.19 Å) | Met522, Leu384, Phe381, Ala527, Phe518, Leu531, Trp387, Val116, Val344, Val349, Leu352 |
| 3,5,7-Trihydroxy-4′-methoxy-flavone | −7.04 | Ser530 (2.71 Å), Met522 (2.75 Å) | Leu531, Val349, Val116, Phe518, Ile523, Trp387, Ala527, Leu384, Phe381 |
| Harmaline | −7.61 | Ser530 (3.44 Å), Ala527 (3.94 Å, 3.72 Å) | Met522, Leu531, Leu352, Ile523, Leu359, Val116, Val349, Phe518, Trp387 |
| trans-Cinnamate | −7.83 | Arg120 (3.27 Å) | Leu352, Ile523, Val116, Phe518, Val527, Val349, Leu531 |
| γ-Linolenic acid | −8.96 | Arg120 (3.03 Å, 3.13 Å), Phe209 (3.74 Å) | Met522, Phe205, Leu534, Val228, Ile523, Leu531, Ala527, Val116, Val349, Phe381, Trp387, Val344, Leu352 |
| Gluconate | −6.79 | Ser530 (2.83 Å), Ile523 (3.36 Å) | Val349, Phe518, Met522, Ala527, Trp387, Leu352 |
| Isorhamnetin-3-O-glucoside | −12.17 | Ile89 (3.82 Å), Arg120 (2.64 Å), Tyr355 (2.62 Å) | Pro86, Leu93, Leu359, Val349, Ile523, Val116, Leu531, Ala527, Leu357, Ile112, Leu115, Trp100 |
| 12-Oxo-10,15(Z)-phytodienoic acid | −8.14 | Arg120 (3.03 Å) | Leu531, Ile523, Val116, Met522, Ala527, Phe518, Pro86, Ile89, Leu93, Leu352, Val349, Trp387 |
| Apigenin | −8.47 | Arg120 (3.98 Å), Ser530 (2.6 Å) | Val116, Phe518, Trp387, Met522, Val349, Ile523, Leu352, Leu531, Leu534, Ala527 |
| Peonidine-3-O-glucoside | −9.11 | Arg120 (2.73 Å), Glu524 (2.96 Å), Val116 (3.30 Å) | Ile89, Ile521, Leu93, Pro86, Ala527, Met522, Phe518, Leu352, Met113, Ile345, Val349, Leu354, Leu357 |
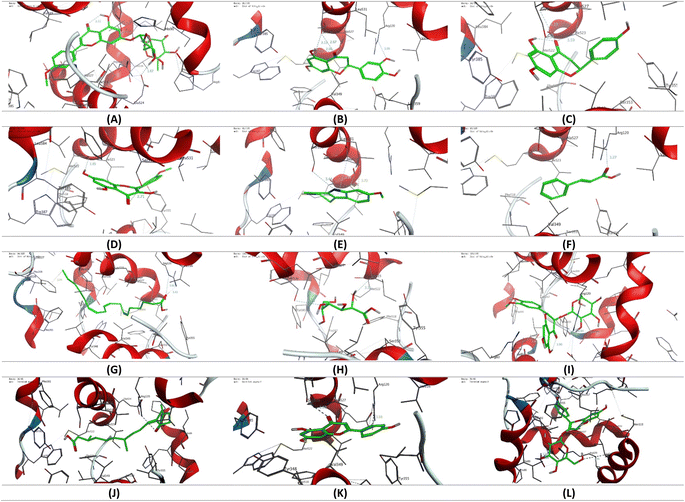 | ||
| Fig. 10 3D representation of the binding mode of selected secondary metabolites identified in the CIR extract to the binding cavity of the COX-1 protein (PDB code: 1eqg). (A) Acacetin-7-O-rutinoside; (B) 3′,4′,5,7-tetrahydroxy-flavanone; (C) naringenin; (D) 3,5,7-trihydroxy-4′-methoxy-flavone; (E) harmaline; (F) trans-cinnamate; (G) γ-linolenic acid; (H) gluconate; (I) isorhamnetin-3-O-glucoside; (J) 12-oxo-10,15(Z)-phytodienoic acid; (K) apigenin; (L) peonidine-3-O-glucoside. | ||
| Ligand | Score (kcal mol−1) | Interactions | |
|---|---|---|---|
| Hydrophilic | Hydrophobic | ||
| Ibuprofen | −11.36 | Arg121 (3.67 Å, 2.97 Å, 3.01 Å), Tyr356 (2.42 Å) | Leu532, Leu360, Ala528, Phe519, Val117, Met523, Val524, Val350, Trp388, Leu353 |
| Acacetin-7-O-rutinoside | −13.51 | Arg121 (2.79 Å, 2.61 Å), Tyr356 (2.45 Å), Arg514 (2.93 Å), Glu525 (2.54 Å, 3.03 Å), Pro86 (3.72 Å), Ser120 (2.79 Å), Lys83 (2.83 Å) | Lys360, Val350, Met523, Phe382, Ala528, Val524, Leu385, Leu353, Phe519, Val89, Leu93, Val117, Trp388, Phe471 |
| 3′,4′,5,7-Tetrahydroxy-flavanone | −9.45 | Met523 (4.25 Å), Glu193 (3.26 Å), His90 (3.03 Å), Val350 (3.27 Å) | Ala528, Leu385, Val524, Trp388, Phe519, Leu353, Ile518, Ala517 |
| Naringenin | −7.88 | Met523 (3.26 Å), Val350 (3.26 Å) | Leu532, Leu353, Val524, Trp388, Leu385, Ala528, Val117 |
| 3,5,7-Trihydroxy-4′-methoxy-flavone | −7.16 | Val350 (3.3 Å), Met523 (4.26 Å) | Try388, Leu353, Ala517, Phe519, Leu385, Val524, Phe382, Ala528 |
| Harmaline | −6.52 | Ala528 (3.59 Å, 3.97 Å) | Phe519, Val117, Leu360, Leu532, Val524, Meth523, Leu353, Val350, Trp388 |
| trans-Cinnamate | −8.39 | Try386 (4.64 Å), Ala528 (3.54 Å), Ser531 (2.98 Å) | Val350, Leu532, Leu353, Val524, Trp388, Leu385 |
| γ-Linolenic acid | −9.11 | Arg121 (2.93 Å) | Val345, Trp388, Val350, Phe206, Phe210, Phe382, Val524, Leu353, Ala528, Leu535, Leu532, Val117, Leu93 |
| Gluconate | −10.26 | Ala528 (3.44 Å), Val524 (3.43 Å), Ser353 (3.5 Å), Ser531 (3.39 Å) | Trp388, Phe519, Met523, Leu385, Phe382, Leu353, Val350 |
| Isorhamnetin-3-O-glucoside | −12.40 | Met523 (4.25 Å), Tyr356 (2.5 Å), Arg121 (2.5 Å, 3.25 Å), His90 (2.74 Å) | Phe382, Phe519, Ala517, Ile518, Leu360, Val117, Val350, Leu353, Leu532, Leu385, Val524, Trp388, Ala528 |
| 12-Oxo-10,15(Z)-phytodienoic acid | −6.94 | Ser120 (2.93 Å, 3.23 Å) | Phe519, Val524, Leu532, Leu353, Val350, Leu360, Val89, Leu93, Val117 |
| Apigenin | −8.70 | Ser531 (2.83 Å), Met523 (4.09 Å), His90 (2.92 Å) | Val350, Ala528, Trp388, Val524, Leu385, Phe519, Leu353, Ala517, Ile518 |
| Peonidine-3-O-glucoside | −11.65 | Arg121 (3.54 Å, 2.88 Å), Ser354 (3.28 Å), Ser531 (2.81 Å), Val524 (3.43 Å) | Phe302, Phe519, Trp388, Ala517, Met523, Val350, Leu532, Leu360, Ala528, Val117 |
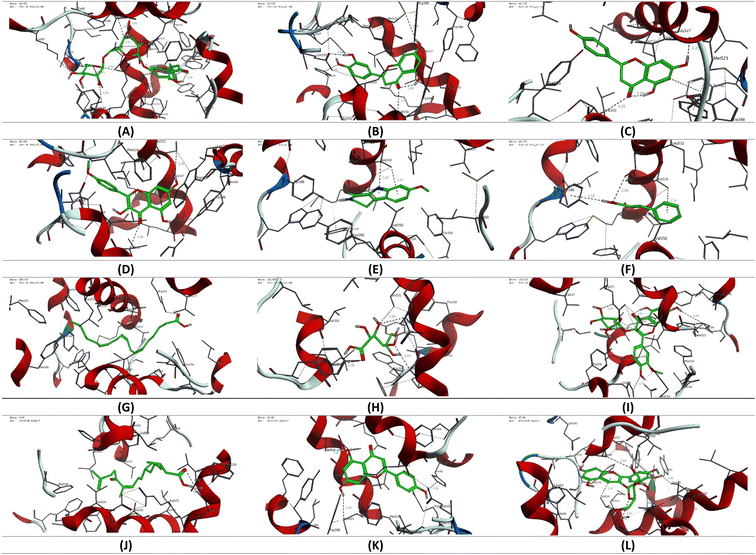 | ||
| Fig. 11 3D representation of the binding mode of selected secondary metabolites identified in the CIR extract toward the binding cavity of the COX-2 protein (PDB code: 4ph9). (A) Acacetin-7-O-rutinoside; (B) 3′,4′,5,7-tetrahydroxy-flavanone; (C) naringenin; (D) 3,5,7-trihydroxy-4′-methoxy-flavone; (E) harmaline; (F) trans-cinnamate; (G) γ-linolenic acid; (H) gluconate; (I) isorhamnetin-3-O-glucoside; (J) 12-oxo-10,15(Z)-phytodienoic acid; (K) apigenin; (L) peonidine-3-O-glucoside. | ||
4 Conclusion
In conclusion, this study bridged the gap in the limited explorations of the pharmacological potential of the CIR extract. The conducted untargeted UPLC/T-TOF-MS/MS metabolite profiling revealed a diverse array of secondary metabolites, including amino acids, flavonoids, alkaloids, nucleotides and carbohydrates, shedding light on the intricate chemical composition of CIR extract. The cytotoxic potential of the CIR extract was investigated in 15 human cell lines, which revealed that the CIR extract has a wide spectrum of antiproliferative activity with a high selectivity index, compared to that of MRC-5 cells. The most pronounced cytotoxic activity and selectivity index of the CIR extract was observed in the Hep-G2 and Panc-1 cells. Exploring the mode of action in Hep-G2 and Panc-1 cells revealed that the CIR extract substantially triggers the arrest of the cell cycle in the S phase, but also early and late apoptosis. These findings were further demonstrated by the notable alternation in the expression of pro-apoptotic and anti-apoptotic genes in both cell lines. Furthermore, our study revealed the antioxidant capacity of CIR extract, which displays a considerable total antioxidant capacity, together with notable free radicals and metal scavenging activities. Insights into anti-inflammatory activity, our results indicated that the CIR extract displays the ability to modulate inflammatory genes (IL-1b, IL-6, TNF-α), but also exhibits a pronounced selectivity towards COX-2 activity, suggesting its significant implications for the treatment of inflammatory and oxidative stress-related conditions. Finally, our in silico coupling studies further demonstrated the anti-inflammatory potential of the CIR extract and unveiled the observed selectivity towards cyclooxygenases 1 & 2. Collectively, these findings underscore the antitumor value of CIR extract as a natural-based therapeutic agent with multifaceted pharmacological benefits, including antioxidant, anti-inflammatory, and antiproliferative activities. Our results indicated that the CIR extract could interfere with key cellular processes including cell cycle, apoptosis, oxidative stress, and inflammatory pathways, which warrants further explorations of its applications in inflammatory conditions and cancer treatment.Data availability
Data supporting the results reported in this manuscript are included in this article. The raw data supporting the conclusions of this article will be made available by the authors without any undue reservation.Author contributions
Conceptualization, A. R. A., N. H., D. I. M., T. M. I. A., I. A. A. I., G. A. B., and E. M. S.; methodology, A. R. A., N. H., D. I. M., H. H. A. N., T. M. I. A., A. A., I. A. A. I., A. H. F., G. A. B., and E. M. S.; software N. H., D. I. M., T. M. I. A., A. A., I. A. A. I., A. H. F., and E. M. S.; validation, A. R. A., H. H. A. N., I. A. A. I., A. H. F., G. A. B., and E. M. S.; formal analysis, A. R. A., N. H., D. I. M., T. M. I. A., G. A. B., and E. M. S.; investigation, D. I. M., A. A., I. A. A. I., A. H. F., G. A. B., and E. M. S.; resources, A. R. A., N. H., D. I. M., T. M. I. A., A. A., and E. M. S.; data curation, A. R. A., N. H., D. I. M., H. H. A. N., T. M. I. A., A. A., I. A. A. I., A. H. F., G. A. B., and E. M. S.; writing—original draft preparation, A. R. A., N. H., D. I. M., H. H. A. N., T. M. I. A., A. A., I. A. A. I., A. H. F., G. A. B., and E. M. S.; writing—review and editing, A. R. A., N. H., D. I. M., A. A., H. H. A. N., T. M. I. A., I. A. A. I., A. H. F., G. A. B., and E. M. S.; visualization, A. R. A, H. H. A. N., A. S. A, I. A. A. I., S. M. Y. M., I. M. A., and I. S.; supervision A. R. A., A. H. F., G. A. B., and E. M. S.; project administration, A. R. A., N. H., D. I. M., T. M. I. A., I. A. A. I., G. A. B., and E. M. S.; funding acquisition, A. R. A., D. I. M., T. M. I. A., and E. M. S. All authors have read and agreed to the published version of the manuscript.Conflicts of interest
The authors declare no conflict of interest.Acknowledgements
The authors extend their appreciation to the Deanship for Research & Innovation, Ministry of Education in Saudi Arabia for funding this research work through the project number: IFP22UQU4360890DSR222.References
- I. Soerjomataram and F. Bray, Planning for tomorrow: global cancer incidence and the role of prevention 2020-2070, Nat. Rev. Clin. Oncol., 2021, 18(10), 663–672 CrossRef PubMed
.
- M. Natsuizaka, T. Omura, T. Akaike, Y. Kuwata, K. Yamazaki and T. Sato, et al., Clinical features of hepatocellular carcinoma with extrahepatic metastases, J. Gastroenterol. Hepatol., 2005, 20(11), 1781–1787 CrossRef PubMed
.
- Global Burden of Disease Liver Cancer C, The Burden of Primary Liver Cancer and Underlying Etiologies From 1990 to 2015 at the Global, Regional, and National Level: Results From the Global Burden of Disease Study 2015, JAMA Oncol., 2017, 3(12), 1683–1691 CrossRef PubMed
.
- J. X. Hu, C. F. Zhao, W. B. Chen, Q. C. Liu, Q. W. Li and Y. Y. Lin, et al., Pancreatic cancer: A review of epidemiology, trend, and risk factors, World J. Gastroenterol., 2021, 27(27), 4298–4321 CrossRef PubMed
.
- H. Sung, J. Ferlay, R. L. Siegel, M. Laversanne, I. Soerjomataram and A. Jemal, et al., Global Cancer Statistics 2020: GLOBOCAN Estimates of Incidence and Mortality Worldwide for 36 Cancers in 185 Countries, Ca-Cancer J. Clin., 2021, 71(3), 209–249 CrossRef PubMed
.
- Z. A. Reshi, W. Ahmad, A. S. Lukatkin and S. B. Javed, From Nature to Lab: A Review of Secondary Metabolite Biosynthetic Pathways, Environmental Influences, and In Vitro Approaches, Metabolites, 2023, 13(8), 895 CrossRef CAS PubMed
.
- B. A. Abdel-Wahab, F. Abd El-Kareem H, A. Alzamami, A. Fahmy C, H. Elesawy B and M. Mahmoud M, et al., Novel Exopolysaccharide from Marine Bacillus subtilis with Broad Potential Biological Activities: Insights into Antioxidant, Anti-Inflammatory, Cytotoxicity, and Anti-Alzheimer Activity, Metabolites, 2022, 12(8), 715 CrossRef CAS PubMed
.
- R. A. Dixon and A. J. Dickinson, A century of studying plant secondary metabolism—From “what?” to “where, how, and why?”, Plant Physiol., 2024, kiad596 Search PubMed
.
- C. A. Dehelean, I. Marcovici, C. Soica, M. Mioc, D. Coricovac and S. Iurciuc, et al., Plant-Derived Anticancer Compounds as New Perspectives in Drug Discovery and Alternative Therapy, Molecules, 2021, 26(4), 1109 CrossRef CAS PubMed
.
- A. S. Choudhari, P. C. Mandave, M. Deshpande, P. Ranjekar and O. Prakash, Phytochemicals in Cancer Treatment: From Preclinical Studies to Clinical Practice, Front. Pharmacol, 2020, 10, 1614 CrossRef PubMed
.
- A. G. Atanasov, S. B. Zotchev, V. M. Dirsch, I. E. Orhan, M. Banach and J. M. Rollinger, et al., Natural products in drug discovery: advances and opportunities, Nat. Rev. Drug Discovery, 2021, 20(3), 200–216 CrossRef CAS PubMed
.
- A. Lyubitelev and V. Studitsky, Inhibition of Cancer Development by Natural Plant Polyphenols: Molecular Mechanisms, Int. J. Mol. Sci., 2023, 24(13), 10663 CrossRef CAS PubMed
.
- M. Greenwell and P. K. Rahman, Medicinal Plants: Their Use in Anticancer Treatment, Int. J. Pharm. Sci. Res., 2015, 6(10), 4103–4112 CAS
.
- T. Khan, M. Ali, A. Khan, P. Nisar, S. A. Jan and S. Afridi, et al., Anticancer Plants: A Review of the Active Phytochemicals, Applications in Animal Models, and Regulatory Aspects, Biomolecules, 2019, 10(1), 47 CrossRef PubMed
.
- V. Shrihastini, P. Muthuramalingam, S. Adarshan, M. Sujitha, J.-T. Chen and H. Shin, et al., Plant Derived Bioactive Compounds, Their Anti-Cancer Effects and In Silico Approaches as an Alternative Target Treatment Strategy for Breast Cancer: An Updated Overview, Cancers, 2021, 13(24), 6222 CrossRef CAS PubMed
.
- M. Bensa, I. Vovk and V. Glavnik, Resveratrol Food Supplement Products and the Challenges of Accurate Label Information to Ensure Food Safety for Consumers, Nutrients, 2023, 15(2), 474 CrossRef CAS PubMed
.
- C. Khanna, M. Rosenberg and D. M. Vail, A Review of Paclitaxel and Novel Formulations Including Those Suitable for Use in Dogs, J. Vet. Intern. Med., 2015, 29(4), 1006–1012 CrossRef CAS PubMed
.
- J. Gallego-Jara, G. Lozano-Terol, R. A. Sola-Martínez, M. Cánovas-Díaz and T. de Diego Puente, A Compressive Review about Taxol®: History and Future Challenges, Molecules, 2020, 25(24), 5986 CrossRef CAS PubMed
.
- I. Mutanda, J. Li, F. Xu and Y. Wang, Recent Advances in Metabolic Engineering, Protein Engineering, and Transcriptome-Guided Insights Toward Synthetic Production of Taxol, Front. bioeng. biotechnol., 2021, 9, 632269 CrossRef PubMed
.
- V. Cocetta, V. Quagliariello, F. Fiorica, M. Berretta and M. Montopoli, Resveratrol as Chemosensitizer Agent: State of Art and Future Perspectives, Int. J. Mol. Sci., 2021, 22(4), 2049 CrossRef CAS PubMed
.
- B. Song, W. Wang, X. Tang, R. M. Goh, W. L. Thuya and P. C. Ho, et al., Inhibitory Potential of Resveratrol in Cancer Metastasis: From Biology to Therapy, Cancers, 2023, 15(10), 2758 CrossRef CAS PubMed
.
- L.-X. Zhang, C.-X. Li, M. U. Kakar, M. S. Khan, P.-F. Wu and R. M. Amir, et al., Resveratrol (RV): A pharmacological review and call for further research, Biomed. Pharmacother., 2021, 143, 112164 CrossRef CAS PubMed
.
- S. Banerjee, C. Bueso-Ramos and B. B. Aggarwal, Suppression of 7,12-dimethylbenz(a)anthracene-induced mammary carcinogenesis in rats by resveratrol: role of nuclear factor-kappaB, cyclooxygenase 2, and matrix metalloprotease 9, Cancer Res., 2002, 62(17), 4945–4954 CAS
.
- H. P. Bais and G. A. Ravishankar, Cichorium intybus L – cultivation, processing, utility, value addition and biotechnology, with an emphasis on current status and future prospects, J. Sci. Food Agric., 2001, 81(5), 467–484 CrossRef CAS
.
- I. Süntar, E. Küpeli Akkol, H. Keles, E. Yesilada, S. D. Sarker and T. Baykal, Comparative evaluation of traditional prescriptions from Cichorium intybus L. for wound healing: Stepwise isolation of an active component by in vivo bioassay and its mode of activity, J. Ethnopharmacol., 2012, 143(1), 299–309 CrossRef PubMed
.
- B. Ahmed, T. A. Al-Howiriny and A. B. Siddiqui, Antihepatotoxic activity of seeds of Cichorium intybus, J. Ethnopharmacol., 2003, 87(2–3), 237–240 CrossRef PubMed
.
- G. Gazzani, M. Daglia, A. Papetti and C. Gregotti, In vitro and ex vivo anti- and prooxidant components of Cichorium intybus, J. Pharm. Biomed. Anal., 2000, 23(1), 127–133 CrossRef CAS PubMed
.
- K. Janda, I. Gutowska, M. Geszke-Moritz and K. Jakubczyk, The Common Cichory (Cichorium intybus L.) as a Source of Extracts with Health-Promoting Properties—A Review, Molecules, 2021, 26(6), 1814 CrossRef CAS PubMed
.
- R. A. Street, J. Sidana and G. Prinsloo, Cichorium intybus: Traditional Uses, Phytochemistry, Pharmacology, and Toxicology, Evid. Based Complement. Alternat. Med., 2013, 2013, 579319 Search PubMed
.
- L. He and C. S. Toh, Recent advances in analytical chemistry-a material approach, Anal. Chim. Acta, 2006, 556(1), 1–15 CrossRef CAS PubMed
.
- M. A. Salem, L. Perez de Souza, A. Serag, A. R. Fernie, M. A. Farag and S. M. Ezzat, et al., Metabolomics in the Context of Plant Natural Products Research: From Sample Preparation to Metabolite Analysis, Metabolites, 2020, 10(1), 37 CrossRef CAS PubMed
.
- L. Perez de Souza, S. Alseekh, T. Naake and A. Fernie, Mass Spectrometry-Based Untargeted Plant Metabolomics, Curr. Protoc. Plant Biol., 2019, 4(4), e20100 CrossRef CAS PubMed
.
- S. Guo, S. Qiu, Y. Cai, Z. Wang, Q. Yang and S. Tang, et al., Mass spectrometry-based metabolomics for discovering active ingredients and exploring action mechanism of herbal medicine, Front. Chem., 2023, 11, 1142287 CrossRef CAS PubMed
.
- M. A. Beniddir, K. B. Kang, G. Genta-Jouve, F. Huber, S. Rogers and J. J. J. N. Van Der Hooft, Advances in decomposing complex metabolite mixtures using substructure-and network-based computational metabolomics approaches, Nat. Prod. Rep., 2021, 38(11), 1967–1993 RSC
.
- U. Salam, S. Ullah, Z.-H. Tang, A. A. Elateeq, Y. Khan and J. Khan, et al., Plant Metabolomics: An Overview of the Role of Primary and Secondary Metabolites against Different Environmental Stress Factors, Life, 2023, 13(3), 706 CrossRef CAS PubMed
.
- F. Gevi, G. Fanelli, L. Zolla and S. Rinalducci, Untargeted Metabolomics of Plant Leaf Tissues, Methods Mol. Biol., 2019, 1978, 187–195 CrossRef CAS PubMed
.
- S. M. Khirallah, H. M. M. Ramadan, A. Shawky, S. H. Qahl, R. S. Baty and N. Alqadri, et al., Development of Novel 1,3-Disubstituted-2-Thiohydantoin Analogues with Potent Anti-Inflammatory Activity; In Vitro and In Silico Assessments, Molecules, 2022, 27(19), 6271 CrossRef CAS PubMed
.
- E. M. Saied, S. Diederich and C. Arenz, Facile synthesis of the CERT inhibitor HPA-12 and some novel derivatives, Chem. - Asian J., 2014, 9(8), 2092–2094 CrossRef CAS PubMed
.
- E. A. Ahmed, M. O. El-Derany, A. M. Anwar, E. M. Saied and S. Magdeldin, Metabolomics and Lipidomics Screening Reveal Reprogrammed Signaling Pathways toward Cancer Development in Non-Alcoholic Steatohepatitis, Int. J. Mol. Sci., 2023, 24(1), 210 CrossRef CAS PubMed
.
- E. M. Saied, S. Banhart, S. E. Bürkle, D. Heuer and C. Arenz, A series of ceramide analogs modified at the 1-position with potent activity against the intracellular growth of Chlamydia trachomatis, Future Med. Chem., 2015, 7(15), 1971–1980 CrossRef CAS PubMed
.
- E. M. Saied and C. Arenz, Stereoselective synthesis of novel sphingoid bases utilized for exploring the secrets of Sphinx, Int. J. Mol. Sci., 2021, 22(15), 8171 CrossRef CAS PubMed
.
- E. M. Radwan, E. Abo-Elabass, A. E. Abd El-Baky, H. A. Alshwyeh, R. A. Almaimani and G. Almaimani, et al., Unveiling the antitumor potential of novel N-(substituted-phenyl)-8-methoxycoumarin-3-carboxamides as dual inhibitors of VEGFR2 kinase and cytochrome P450 for targeted treatment of hepatocellular carcinoma, Front. Chem., 2023, 11, 1231030 CrossRef CAS PubMed
.
- M. G. Salem, S. A. Abu El-ata, E. H. Elsayed, S. N. Mali, H. A. Alshwyeh and G. Almaimani, et al., Novel 2-substituted-quinoxaline analogs with potential antiproliferative activity against breast cancer: insights into cell cycle arrest, topoisomerase II, and EGFR activity, RSC Adv., 2023, 13(47), 33080–33095 RSC
.
- S. Zha, V. Yegnasubramanian, W. G. Nelson, W. B. Isaacs and A. M. De Marzo, Cyclooxygenases in cancer: progress and perspective, Cancer Lett., 2004, 215(1), 1–20 CrossRef CAS PubMed
.
- C. Schneider and A. Pozzi, Cyclooxygenases and lipoxygenases in cancer, Cancer Metastasis Rev., 2011, 30, 277–294 CrossRef CAS PubMed
.
- M. Kanehisa and S. Goto, KEGG: kyoto encyclopedia of genes and genomes, Nucleic Acids Res., 2000, 28(1), 27–30 CrossRef CAS PubMed
.
- S. E. Elshaer, G. M. Hamad, E. E. Hafez, H. H. Baghdadi, F. M. El-Demerdash and J. Simal-Gandara, Root extracts of Saussurea costus as prospective detoxifying food additive against sodium nitrite toxicity in male rats, Food Chem. Toxicol., 2022, 113225 CrossRef CAS PubMed
.
- G. Almaimani, A. A. J. Jabbar, I. A. A. Ibrahim, A. R. Alzahrani, G. A. Bamagous and R. A. Almaimani, et al., Anise (Pimpinella anisum L.) attenuates azoxymethane-induced colorectal cancer by antioxidant, anti-inflammatory, and anti-apoptotic pathways in rats, Environ. Sci. Pollut. Res., 2024, 31(3), 4439–4452 CrossRef CAS PubMed
.
- K. M. Radha, S. Puri, A. Pundir, S. P. Bangar and S. Changan, et al., Evaluation of Nutritional, Phytochemical, and Mineral Composition of Selected Medicinal Plants for Therapeutic Uses from Cold Desert of Western Himalaya, Plants, 2021, 10(7), 1429 CrossRef PubMed
.
- H. Tsugawa, T. Cajka, T. Kind, Y. Ma, B. Higgins and K. Ikeda, et al., MS-DIAL: data-independent MS/MS deconvolution for comprehensive metabolome analysis, Nat. Methods, 2015, 12(6), 523–526 CrossRef CAS PubMed
.
- N. M. Fayek, M. A. Farag, A. R. Abdel Monem, M. Y. Moussa, S. M. Abd-Elwahab and N. D. El-Tanbouly, Comparative Metabolite Profiling of Four Citrus Peel Cultivars via Ultra-Performance Liquid Chromatography Coupled with Quadrupole-Time-of-Flight-Mass Spectrometry and Multivariate Data Analyses, J. Chromatogr. Sci., 2019, 57(4), 349–360 CAS
.
- I. A. Aziz Ibrahim, A. R. Alzahrani, I. M. Alanazi, N. Shahzad, I. Shahid and A. H. Falemban, et al., Bioactive compound D-Pinitol-loaded graphene oxide-chitosan-folic acid nanocomposite induced apoptosis in human hepatoma HepG-2 cells, J. Drug Delivery Sci. Technol., 2024, 92, 105282 CrossRef CAS
.
- M. G. Salem, D. M. A. El-Maaty, Y. I. M. El-Deen, B. H. Elesawy, A. E. Askary and A. Saleh, et al., Novel 1,3-Thiazole Analogues with Potent Activity against Breast Cancer: A Design, Synthesis, In Vitro, and In Silico Study, Molecules, 2022, 27(15), 4898 CrossRef CAS PubMed
.
- I. H. El Azab, E. M. Saied, A. A. Osman, A. E. Mehana, H. A. Saad and N. A. A. Elkanzi, Novel N-bridged pyrazole-1-carbothioamides with potential antiproliferative activity: Design, synthesis, in vitro and in silico studies, Future Med. Chem., 2021, 13(20), 1743–1766 CrossRef CAS PubMed
.
- D. I. Mohamed, D. A. E. D. A. El-Waseef, E. S. Nabih, O. A. El-Kharashi, H. F. A. El-Kareem and H. H. A. Nahas, et al., Acetylsalicylic Acid Suppresses Alcoholism-Induced Cognitive Impairment Associated with Atorvastatin Intake by Targeting Cerebral miRNA155 and NLRP3: In Vivo, and In Silico Study, Pharmaceutics, 2022, 14(3), 529 CrossRef CAS PubMed
.
- D. I. Mohamed, S. F. Ezzat, W. M. Elayat, O. A. El-Kharashi, H. F. Abd El-Kareem and H. H. Abo Nahas, et al., Hepatoprotective Role of Carvedilol against Ischemic Hepatitis Associated with Acute Heart Failure via Targeting miRNA-17 and Mitochondrial Dynamics-Related Proteins: An In Vivo and In Silico Study, Pharmaceuticals, 2022, 15(7), 832 CrossRef CAS PubMed
.
- L. S. Kalinichenko, C. Mühle, T. Jia, F. Anderheiden, M. Datz and A. L. Eberle, et al., Neutral sphingomyelinase mediates the co-morbidity trias of alcohol abuse, major depression and bone defects, Mol. Psychiatry, 2021, 26(12), 7403–7416 CrossRef CAS PubMed
.
- D. I. Mohamed, D. A. Abou-Bakr, S. F. Ezzat, H. F. Abd El-Kareem, H. H. Abo Nahas and H. A. Saad, et al., Vitamin d3 prevents the deleterious effects of testicular torsion on testis by targeting mirna-145 and adam17: In silico and in vivo study, Pharmaceuticals, 2021, 14(12), 1222 CrossRef CAS PubMed
.
- M. Valko, D. Leibfritz, J. Moncol, M. T. Cronin, M. Mazur and J. Telser, Free radicals and antioxidants in normal physiological functions and human disease, Int. J. Biochem. Cell Biol., 2007, 39(1), 44–84 CrossRef CAS PubMed
.
- R. J. Ruch, S. J. Cheng and J. E. Klaunig, Prevention of cytotoxicity and inhibition of intercellular communication by antioxidant catechins isolated from Chinese green tea, Carcinogenesis, 1989, 10(6), 1003–1008 CrossRef CAS PubMed
.
- A. Cano, O. Alcaraz, M. Acosta and M. B. Arnao, On-line antioxidant activity determination: comparison of hydrophilic and lipophilic antioxidant activity using the ABTS•+ assay, Redox Rep., 2002, 7(2), 103–109 CrossRef CAS PubMed
.
- N. Chaves, A. Santiago and J. C. Alías, Quantification of the Antioxidant Activity of Plant Extracts: Analysis of Sensitivity and Hierarchization Based on the Method Used, Antioxidants, 2020, 9(1), 76 CrossRef CAS PubMed
.
- R. Apak, K. Güçlü, B. Demirata, M. Ozyürek, S. E. Celik and B. Bektaşoğlu, et al., Comparative evaluation of various total antioxidant capacity assays applied to phenolic compounds with the CUPRAC assay, Molecules, 2007, 12(7), 1496–1547 CrossRef CAS PubMed
.
- P. Prieto, M. Pineda and M. Aguilar, Spectrophotometric Quantitation of Antioxidant Capacity through the Formation of a Phosphomolybdenum Complex: Specific Application to the Determination of Vitamin E, Anal. Biochem., 1999, 269(2), 337–341 CrossRef CAS PubMed
.
- B. J. Orlando, M. J. Lucido and M. G. Malkowski, The structure of ibuprofen bound to cyclooxygenase-2, J. Struct. Biol., 2015, 189(1), 62–66 CrossRef CAS PubMed
.
- B. S. Selinsky, K. Gupta, C. T. Sharkey and P. J. Loll, Structural Analysis of NSAID Binding by Prostaglandin H2 Synthase: Time-Dependent and Time-Independent Inhibitors Elicit Identical Enzyme Conformations, Biochemistry, 2001, 40(17), 5172–5180 CrossRef CAS PubMed
.
- R. D. Healey, E. M. Saied, X. Cong, G. Karsai, L. Gabellier and J. Saint-Paul, et al., Discovery and Mechanism of Action of Small Molecule Inhibitors of Ceramidases, Angew. Chem., Int. Ed., 2022, 61(2), e202109967 CrossRef CAS PubMed
.
- D. Samaha, H. H. Hamdo, X. Cong, F. Schumacher, S. Banhart and Ö. Aglar, et al., Liposomal FRET Assay Identifies Potent Drug-Like Inhibitors of the Ceramide Transport Protein (CERT), Chem.—Eur. J., 2020, 26(70), 16616–16621 CrossRef CAS PubMed
.
- S. M. Khirallah, H. M. M. Ramadan, H. A. A. Aladl, N. O. Ayaz, L. A. F. Kurdi and M. Jaremko, et al., Antidiabetic Potential of Novel 1,3,5-Trisubstituted-2-Thioxoimidazloidin-4-One Analogues: Insights into α-Glucosidase, α-Amylase, and Antioxidant Activities, Pharmaceuticals, 2022, 15(12), 1576 CrossRef CAS PubMed
.
- S. Fathy Elhabal, M. A. El-Nabarawi, N. Abdelaal, M. F. M. Elrefai, S. A. Ghaffar and M. M. Khalifa, et al., Development of canagliflozin nanocrystals sublingual tablets in the presence of sodium caprate permeability enhancer: formulation optimization, characterization, in-vitro, in silico, and in-vivo study, Drug Delivery, 2023, 30(1), 2241665 CrossRef PubMed
.
- M. Kharbach, I. Marmouzi, M. El Jemli, A. Bouklouze and Y. Vander Heyden, Recent advances in untargeted and targeted approaches applied in herbal-extracts and essential-oils fingerprinting-A review, J. Pharm. Biomed. Anal., 2020, 177, 112849 CrossRef CAS PubMed
.
- A. Alonso, S. Marsal and A. Julià, Analytical methods in untargeted metabolomics: state of the art in 2015, Front. Bioeng. Biotechnol., 2015, 3, 23 Search PubMed
.
- A. C. Schrimpe-Rutledge, S. G. Codreanu, S. D. Sherrod and J. A. McLean, Untargeted metabolomics strategies—challenges and emerging directions, J. Am. Soc. Mass Spectrom., 2016, 27(12), 1897–1905 CrossRef CAS PubMed
.
- Y.-Y. Fan and R. S. Chapkin, Importance of Dietary γ-Linolenic Acid in Human Health and Nutrition, J. Nutr., 1998, 128(9), 1411–1414 CrossRef CAS PubMed
.
- F. Wang, M. Miao, B. Chen, R. Wang, B. Sun and D. Ren, et al., Antineoplastic activity of γ-linolenic acid extract from Spirulina platensis on HepG2 cells and its inhibition effect on platelet aggregation, Food Agric. Immunol., 2015, 26(1), 97–108 CrossRef CAS
.
- J. A. Wushensky, T. Youngster, C. M. Mendonca and L. Aristilde, Flux Connections Between Gluconate Pathway, Glycolysis, and Pentose “Phosphate Pathway During Carbohydrate Metabolism in Bacillus megaterium QM B1551, Front. Microbiol., 2018, 9, 2789 CrossRef PubMed
.
- W. Deng, L. Wu, Z. Xiao, Y. Li, Z. Zheng and S. Chen, Structural Characterization and Anti-Inflammatory Activity of Polysaccharides from Tremella fuciformis on Monosodium Urate-Stimulated RAW264.7 Macrophages, Foods, 2023, 12(24), 4398 CrossRef CAS PubMed
.
- H. Claus-Desbonnet, E. Nikly, V. Nalbantova, D. Karcheva-Bahchevanska, S. Ivanova and G. Pierre, et al., Polysaccharides and Their Derivatives as Potential Antiviral Molecules, Viruses, 2022, 14(2), 426 CrossRef CAS PubMed
.
- Y. Wang, N. Zhang, J. Zhou, P. Sun, L. Zhao and F. Zhou, Protective Effects of Several Common Amino Acids, Vitamins, Organic Acids, Flavonoids and Phenolic Acids against Hepatocyte Damage Caused by Alcohol, Foods, 2022, 11(19), 3014 CrossRef CAS PubMed
.
- A. T. Tuncel, T. Ruppert, B.-T. Wang, J. G. Okun, S. Kölker and M. A. Morath, et al., Maleic Acid – but Not Structurally Related Methylmalonic Acid – Interrupts Energy Metabolism by Impaired Calcium Homeostasis, PLoS One, 2015, 10(6), e0128770 CrossRef PubMed
.
- W. Liu and S.-W. Park, 12-oxo-Phytodienoic Acid: A Fuse and/or Switch of Plant Growth and Defense Responses?, Front. Plant Sci., 2021, 12, 724079 CrossRef PubMed
.
- T. V. Pham, H. N. T. Hoang, H. T. Nguyen, H. M. Nguyen, C. T. Huynh and T. Y. Vu, et al., Anti-Inflammatory and Antimicrobial Activities of Compounds Isolated from Distichochlamys benenica, BioMed Res. Int., 2021, 2021, 6624347 CrossRef PubMed
.
- S. Singh, P. Gupta, A. Meena and S. Luqman, Acacetin, a flavone with diverse therapeutic potential in cancer, inflammation, infections and other metabolic disorders, Food Chem. Toxicol., 2020, 145, 111708 CrossRef CAS PubMed
.
- T. P. T. Cushnie and A. J. Lamb, Antimicrobial activity of flavonoids, Int. J. Antimicrob. Agents, 2005, 26(5), 343–356 CrossRef CAS PubMed
.
- R. K. Al-Ishaq, M. Abotaleb, P. Kubatka, K. Kajo and D. Büsselberg, Flavonoids and Their Anti-Diabetic Effects: Cellular Mechanisms and Effects to Improve Blood Sugar Levels, Biomolecules, 2019, 9(9), 430 CrossRef CAS PubMed
.
- E. Amjad, B. Sokouti and S. Asnaashari, A systematic review of anti-cancer roles and mechanisms of kaempferol as a natural compound, Cancer Cell Int., 2022, 22(1), 260 CrossRef CAS PubMed
.
- N. Aziz, M.-Y. Kim and J. Y. Cho, Anti-inflammatory effects of luteolin: A review of in vitro, in vivo, and in silico studies, J. Ethnopharmacol., 2018, 225, 342–358 CrossRef CAS PubMed
.
- W.-S. Li, S.-C. Lin, C.-H. Chu, Y.-K. Chang, X. Zhang and C.-C. Lin, et al., The Gastroprotective Effect of Naringenin against Ethanol-Induced Gastric Ulcers in Mice through Inhibiting Oxidative and Inflammatory Responses, Int. J. Mol. Sci., 2021, 22(21), 11985 CrossRef CAS PubMed
.
- MlF. Manchope, R. Casagrande and W. A. Verri, Naringenin: an analgesic and anti-inflammatory citrus flavanone, Oncotarget, 2017, 8(3), 3766–3767 CrossRef PubMed
.
- A. Duda-Madej, J. Stecko, J. Sobieraj, N. Szymńska and J. Kozłowska, Naringenin and Its Derivatives—Health-Promoting Phytobiotic against Resistant Bacteria and Fungi in Humans, Antibiotics, 2022, 11(11), 1628 CrossRef CAS PubMed
.
- A. G. Veiko, E. Olchowik-Grabarek, S. Sekowski, A. Roszkowska, E. A. Lapshina and I. Dobrzynska, et al., Antimicrobial Activity of Quercetin, Naringenin and Catechin: Flavonoids Inhibit Staphylococcus aureus-Induced Hemolysis and Modify Membranes of Bacteria and Erythrocytes, Molecules, 2023, 28(3), 1252 CrossRef CAS PubMed
.
- N. Tena, J. Martín and A. AnG, State of the Art of Anthocyanins: Antioxidant Activity, Sources, Bioavailability, and Therapeutic Effect in Human Health, Antioxidants, 2020, 9(5), 451 CrossRef CAS PubMed
.
- P. Kashyap, D. Shikha, M. Thakur and A. Aneja, Functionality of apigenin as a potent antioxidant with emphasis on bioavailability, metabolism, action mechanism and in vitro and in vivo studies: A review, J. Food Biochem., 2022, 46(4), e13950 CrossRef CAS PubMed
.
- R. Ginwala, R. Bhavsar, D. G. I. Chigbu, P. Jain and Z. K. Khan, Potential Role of Flavonoids in Treating Chronic Inflammatory Diseases with a Special Focus on the Anti-Inflammatory Activity of Apigenin, Antioxidants, 2019, 8(2), 35 CrossRef PubMed
.
- M. Wang, J. Firrman, L. Liu and K. Yam, A Review on Flavonoid Apigenin: Dietary Intake, ADME, Antimicrobial Effects, and Interactions with Human Gut Microbiota, BioMed Res. Int., 2019, 2019, 7010467 Search PubMed
.
- K. Gaur and H. Y. Siddique, Effect of Apigenin on Neurodegenerative Diseases, CNS Neurol. Disord.: Drug Targets, 2024, 23(4), 468–475 CrossRef CAS PubMed
.
- K. Patel, M. Gadewar, V. Tahilyani and D. K. Patel, A review on pharmacological and analytical aspects of diosmetin: A concise report, Chin. J. Integr. Med., 2013, 19(10), 792–800 CrossRef CAS PubMed
.
- J. Hintikka, T. Tolmunen, A. Tanskanen and H. Viinamäki, High vitamin B12 level and good treatment outcome may be associated
in major depressive disorder, BMC Psychiatry, 2003, 3(1), 17 CrossRef PubMed
.
- C. D. Graber, J. M. Goust, A. D. Glassman, R. Kendall and C. B. Loadholt, Immunomodulating Properties of Dimethylglycine in Humans, J. Infect. Dis., 1981, 143(1), 101–105 CrossRef CAS PubMed
.
- M. Magnusson, T. J. Wang, C. Clish, G. Engström, P. Nilsson and R. E. Gerszten, et al., Dimethylglycine Deficiency and the Development of Diabetes, Diabetes, 2015, 64(8), 3010–3016 CrossRef CAS PubMed
.
- E. A. Popenoe, R. B. Aronson and D. D. Van Slyke, Hydroxylysine Formation from Lysine during Collagen Biosynthesis, Proc. Natl. Acad. Sci. U. S. A., 1966, 55(2), 393–397 CrossRef CAS PubMed
.
- R. Dalangin, A. Kim and R. E. Campbell, The Role of Amino Acids in Neurotransmission and Fluorescent Tools for Their Detection, Int. J. Mol. Sci., 2020, 21(17), 6197 CrossRef CAS PubMed
.
- J. Zhang, J. Fan, S. Venneti, R. Cross Justin, T. Takagi and B. Bhinder, et al., Asparagine Plays a Critical Role in Regulating Cellular Adaptation to Glutamine Depletion, Mol. Cell, 2014, 56(2), 205–218 CrossRef CAS PubMed
.
- A. A. Chowdhury, N. B. Gawali, R. Munshi and A. R. Juvekar, Trigonelline insulates against oxidative stress, proinflammatory cytokines and restores BDNF levels in lipopolysaccharide induced cognitive impairment in adult mice, Metab. Brain Dis., 2018, 33(3), 681–691 CrossRef CAS PubMed
.
- Y. Liang, X. Dai, Y. Cao, X. Wang, J. Lu and L. Xie, et al., The neuroprotective and antidiabetic effects of trigonelline: A review of signaling pathways and molecular mechanisms, Biochimie, 2023, 206, 93–104 CrossRef CAS PubMed
.
- S.-P. Li, Y.-W. Wang, S.-L. Qi, Y.-P. Zhang, G. Deng and W.-Z. Ding, et al., Analogous β-Carboline Alkaloids Harmaline and Harmine Ameliorate Scopolamine-Induced Cognition Dysfunction by Attenuating Acetylcholinesterase Activity, Oxidative Stress, and Inflammation in Mice, Front. Pharmacol, 2018, 9, 346 CrossRef PubMed
.
- J. A. Fuentes and V. G. Longo, An investigation on the central effects of harmine, harmaline and related β-carbolines, Neuropharmacology, 1971, 10(1), 15–23 CrossRef CAS PubMed
.
- C. Carazzone, D. Mascherpa, G. Gazzani and A. Papetti, Identification of phenolic constituents in red chicory salads (Cichorium intybus) by high-performance liquid chromatography with diode array detection and electrospray ionisation tandem mass spectrometry, Food Chem., 2013, 138(2–3), 1062–1071 CrossRef CAS PubMed
.
- M. Bisma, P. Tanveer Bilal, T. Inayatullah and R. Reiaz Ul, Chemo-profiling, Antioxidant Potential and Ionomic Analysis of Cichorium intybus L, Pharmacogn. J., 2017, 9(6), 917–928 CrossRef
.
- A. S. Kandil, F. Abou-Elella and H. A. El Shemy, Cytotoxic profile activities of ethanolic and methanolic extracts of chicory plant (Cichorium intybus L.), J. Radiat. Res. Appl. Sci., 2019, 12(1), 106–111 Search PubMed
.
- A. Epure, A. E. E. Pârvu, L. Vlase, D. Benedec, D. Hanganu and A. M. Gheldiu, et al., Phytochemical Profile, Antioxidant, Cardioprotective and Nephroprotective Activity of Romanian Chicory Extract, Plants, 2020, 10(1), 64 CrossRef PubMed
.
- J. K. In, J. K. Kim, J. S. Oh and D. W. Seo, 5-Caffeoylquinic acid inhibits invasion of non-small cell lung cancer cells through the inactivation of p70S6K and Akt activity: Involvement of p53 in differential regulation of signaling pathways, Int. J. Oncol., 2016, 48(5), 1907–1912 CrossRef CAS PubMed
.
- K. M. S. U. Imam, Y. Xie, Y. Liu, F. Wang and F. Xin, Cytotoxicity of Cichorium intybus L. metabolites (Review), Oncol. Rep., 2019, 42(6), 2196–2212 CAS
.
- M. Esmaeilbeig, S. A. Kouhpayeh and Z. Amirghofran, An Investigation of the Growth Inhibitory Capacity of Several Medicinal Plants From Iran on Tumor Cell Lines, Iran. J. Cancer Prev., 2015, 8(5), e4032 Search PubMed
.
- J. Menendez, M. del Mar Barbacid, S. Montero, E. Sevilla, E. Escrich and M. Solanas, et al., Effects of gamma-linolenic acid and oleic acid on paclitaxel cytotoxicity in human breast cancer cells, Eur. J. Cancer, 2001, 37(3), 402–413 CrossRef CAS PubMed
.
- P. Whitehouse, A. Cooper and C. Johnson, Synergistic activity of gamma-linolenic acid and cytotoxic drugs against pancreatic adenocarcinoma cell lines, Pancreatology, 2003, 3(5), 367–374 CrossRef CAS PubMed
.
- S. K. El-Desouky, L. F. Ibrahim, S. A. Kawashty, M. A. El-Ansari, Y. S. Kim and H.-S. Chong, et al., Phytochemical constituents and biological activities of Origanum syriacum, Z. Naturforsch., B: J. Chem. Sci., 2009, 64(4), 447–451 CrossRef CAS
.
- I. Zakaria, N. Ahmat, F. M. Jaafar and A. Widyawaruyanti, Flavonoids with antiplasmodial and cytotoxic activities of Macaranga triloba, Fitoterapia, 2012, 83(5), 968–972 CrossRef CAS PubMed
.
- S. P. Kumar, K. Birundha, K. Kaveri and K. R. Devi, Antioxidant studies of chitosan nanoparticles containing naringenin and their cytotoxicity effects in lung cancer cells, Int. J. Biol. Macromol., 2015, 78, 87–95 CrossRef CAS PubMed
.
- J. X. Liu, J. Werner, T. Kirsch, J. D. Zuckerman and M. S. Virk, Cytotoxicity evaluation of chlorhexidine gluconate on human fibroblasts, myoblasts, and osteoblasts, J. Bone Jt. Infect., 2018, 3(4), 165–172 CrossRef PubMed
.
- A. Murugesan, S. Holmstedt, K. C. Brown, A. Koivuporras, A. S. Macedo and N. Nguyen, et al., Design and synthesis of novel quinic acid derivatives: in vitro cytotoxicity and anticancer effect on glioblastoma, Future Med. Chem., 2020, 12(21), 1891–1910 CrossRef CAS PubMed
.
- Z. A. Stewart, M. D. Westfall and J. A. Pietenpol, Cell-cycle dysregulation and anticancer therapy, Trends Pharmacol. Sci., 2003, 24(3), 139–145 CrossRef CAS PubMed
.
- C. J. Sherr, Cancer cell cycles, Science, 1996, 274(5293), 1672–1677 CrossRef CAS PubMed
.
- Y. Lee, N. F. Lahens, S. Zhang, J. Bedont, J. M. Field and A. Sehgal, G1/S cell cycle regulators mediate effects of circadian dysregulation on tumor growth and provide targets for timed anticancer treatment, PLoS Biol., 2019, 17(4), e3000228 CrossRef CAS PubMed
.
- W. G. Jiang, R. P. Bryce, D. F. Horrobin and R. E. Mansel, gamma-Linolenic acid blocks cell cycle progression by regulating phosphorylation of p27kip1 and p57kip2 and their interactions with other cycle regulators in cancer cells, Int. J. Oncol., 1998, 13(3), 611–618 CAS
.
- Y. Wang, C. Wang, C. Jiang, H. Zeng and X. He, Novel mechanism of harmaline on inducing G2/M cell cycle arrest and apoptosis by up-regulating Fas/FasL in SGC-7901 cells, Sci. Rep., 2015, 5(1), 18613 CrossRef CAS PubMed
.
- A. Sathiyaseelan, K. Saravanakumar, J. Jayalakshmi, M. Gopi, A. Shajahan and K. Barathikannan, et al., Trigonelline-loaded chitosan nanoparticles prompted antitumor activity on glioma cells and biocompatibility with pheochromocytoma cells, Int. J. Biol. Macromol., 2020, 163, 36–43 CrossRef CAS PubMed
.
- J. R. Liu, B. X. Li, B. Q. Chen, X. H. Han, Y. B. Xue and Y. M. Yang, et al., Effect of cis-9, trans-11-conjugated linoleic acid on cell cycle of gastric adenocarcinoma cell line (SGC-7901), World J. Gastroenterol., 2002, 8(2), 224–229 CrossRef CAS PubMed
.
- J. Liu, Q. Li, Z. Liu, L. Lin, X. Zhang and M. Cao, et al., Harmine induces cell cycle arrest and mitochondrial pathway-mediated cellular apoptosis in SW620 cells via inhibition of the Akt and ERK signaling pathways, Oncol. Rep., 2016, 35(6), 3363–3370 CrossRef CAS PubMed
.
- R. Raina, S. Pramodh, N. Rais, S. Haque, J. Shafarin and K. Bajbouj, et al., Luteolin inhibits proliferation, triggers apoptosis and modulates Akt/mTOR and MAP kinase pathways in HeLa cells, Oncol. Lett., 2021, 21, 192 CrossRef CAS PubMed
.
- L. C. Crowley, B. J. Marfell, A. P. Scott and N. J. Waterhouse, Quantitation of Apoptosis and Necrosis by Annexin V Binding, Propidium Iodide Uptake, and Flow Cytometry, Cold Spring Harb. Protoc., 2016, 2016(11), pdb.prot087288 CrossRef PubMed
.
- R. Carpenter and M. F. Brady, BAX Gene, StatPearls, Treasure Island (FL) ineligible companies, Disclosure: Mark Brady declares no relevant financial relationships with ineligible companies, StatPearls Publishing Copyright © 2023, StatPearls Publishing LLC, 2023 Search PubMed
.
- P. De Feudis, S. Vignati, C. Rossi, T. Mincioni, R. Giavazzi and M. D'Incalci, et al., Driving p53 response to Bax activation greatly enhances sensitivity to taxol by inducing massive apoptosis, Neoplasia, 2000, 2(3), 202–207 CrossRef CAS PubMed
.
- X. Jiang and X. Wang, Cytochrome C-mediated apoptosis, Annu. Rev. Biochem., 2004, 73(1), 87–106 CrossRef CAS PubMed
.
- D. Szklarczyk, A. L. Gable, K. C. Nastou, D. Lyon, R. Kirsch and S. Pyysalo, et al., The STRING database in 2021: customizable protein-protein networks, and functional characterization of user-uploaded gene/measurement sets, Nucleic Acids Res., 2021, 49(D1), D605 CrossRef CAS PubMed
.
- H. Ge, X. Kong, L. Shi, L. Hou, Z. Liu and P. Li, Gamma-linolenic acid induces apoptosis and lipid peroxidation in human chronic myelogenous leukemia K562 cells, Cell Biol. Int., 2009, 33(3), 402–410 CrossRef CAS PubMed
.
- X. Kong, H. Ge, L. Hou, L. Shi and Z. Liu, Induction of apoptosis in K562/ADM cells by gamma-linolenic acid involves lipid peroxidation and activation of caspase-3, Chem.-Biol. Interact., 2006, 162(2), 140–148 CrossRef CAS PubMed
.
- H. W.-C. Leung, C.-J. Lin, M.-J. Hour, W.-H. Yang, M.-Y. Wang and H.-Z. Lee, Kaempferol induces apoptosis in human lung non-small carcinoma cells accompanied by an induction of antioxidant enzymes, Food Chem. Toxicol., 2007, 45(10), 2005–2013 CrossRef CAS PubMed
.
- H. Kostrhunova, J. Zajac, L. Markova, V. Brabec and J. Kasparkova, A Multi-action PtIV Conjugate with Oleate and Cinnamate Ligands Targets Human Epithelial Growth Factor Receptor HER2 in Aggressive Breast Cancer Cells, Angew. Chem., 2020, 132(47), 21343–21348 CrossRef
.
- D. Arul and P. Subramanian, Naringenin (citrus flavonone) induces growth inhibition, cell cycle arrest and apoptosis in human hepatocellular carcinoma cells, Pathol. Oncol. Res., 2013, 19, 763–770 CrossRef CAS PubMed
.
- Z. Zhao, G. Jin, Y. Ge and Z. Guo, Naringenin inhibits migration of breast cancer cells via inflammatory and apoptosis cell signaling pathways, Inflammopharmacology, 2019, 27, 1021–1036 CrossRef CAS PubMed
.
- U. Das, V. Prasad and D. R. Reddy, Local application of γ-linolenic acid in the treatment of human gliomas, Cancer Lett., 1995, 94(2), 147–155 CrossRef CAS PubMed
.
- A. Rauf, M. A. Shariati, M. Imran, K. Bashir, S. A. Khan and S. Mitra, et al., Comprehensive review on naringenin and naringin polyphenols as a potent anticancer agent, Environ. Sci. Pollut. Res., 2022, 29(21), 31025–31041 CrossRef CAS PubMed
.
- J. Stabrauskiene, D. M. Kopustinskiene, R. Lazauskas and J. Bernatoniene, Naringin and Naringenin: Their Mechanisms of Action and the Potential Anticancer Activities, Biomedicines, 2022, 10(7), 1686 CrossRef CAS PubMed
.
- S.-i. Kanno, A. Tomizawa, T. Ohtake, K. Koiwai, M. Ujibe and M. Ishikawa, Naringenin-induced apoptosis via activation of NF-κB and necrosis involving the loss of ATP in human promyeloleukemia HL-60 cells, Toxicol. Lett., 2006, 166(2), 131–139 CrossRef CAS PubMed
.
- J. Hong, T. J. Smith, C.-T. Ho, D. A. August and C. S. Yang, Effects of purified green and black tea polyphenols on cyclooxygenase-and lipoxygenase-dependent metabolism of arachidonic acid in human colon mucosa and colon tumor tissues, Biochem. Pharmacol., 2001, 62(9), 1175–1183 CrossRef CAS PubMed
.
- C. Selvam and S. M. Jachak, A cyclooxygenase (COX) inhibitory biflavonoid from the seeds of
Semecarpus anacardium, J. Ethnopharmacol., 2004, 95(2), 209–212 CrossRef CAS PubMed
.
- R. Gautam and S. M. Jachak, Recent developments in anti-inflammatory natural products, Med. Res. Rev., 2009, 29(5), 767–820 CrossRef CAS PubMed
.
- S. Khan, E. M. Shin, R. J. Choi, Y. H. Jung, J. Kim and A. Tosun, et al., Suppression of LPS-induced inflammatory and NF-κB responses by anomalin in RAW 264.7 macrophages, J. Cell. Biochem., 2011, 112(8), 2179–2188 CrossRef CAS PubMed
.
- P. Arulselvan, W. S. Tan, S. Gothai, K. Muniandy, S. Fakurazi and N. M. Esa, et al., Anti-Inflammatory Potential of Ethyl Acetate Fraction of Moringa oleifera in Downregulating the NF-κB Signaling Pathway in Lipopolysaccharide-Stimulated Macrophages, Molecules, 2016, 21(11), 1452 CrossRef PubMed
.
- S. Ōmura, Y. Asami and A. Crump, Staurosporine: New lease of life for parent compound of today's novel and highly successful anti-cancer drugs, J. Antibiot., 2018, 71(8), 688–701 CrossRef PubMed
.
- M. M. Silva, E. K. Iriguchi, C. A. L. Kassuya, M. C. Vieira, M. A. Foglio and J. E. Carvalho, et al., Schinus terebinthifolius: phenolic constituents and in vitro antioxidant, antiproliferative and in vivo anti-inflammatory activities, Rev. Bras. Farmacogn., 2017, 27, 445–452 CrossRef
.
- F. Conforti, S. Sosa, M. Marrelli, F. Menichini, G. A. Statti and D. Uzunov, et al., In vivo anti-inflammatory and in vitro antioxidant activities of Mediterranean dietary plants, J. Ethnopharmacol., 2008, 116(1), 144–151 CrossRef CAS PubMed
.
- T. Ernawati, T. Yuliani, M. Minarti and I. D. Dewijanti, Anti-inflammatory Activities of Methyl trans-Cinnamate derivatives on Carrageenan-Induced Paw Edema in Male Sprague-Dawley Rats, International Summit on Science Technology and Humanity, 2019, p-ISSN: 2477-3328, e-ISSN:2615-1588 Search PubMed
.
- X. Liu, M. Li, S. Tan, C. Wang, S. Fan and C. Huang, Harmine is an inflammatory inhibitor through the suppression of NF-κB signaling, Biochem. Biophys. Res. Commun., 2017, 489(3), 332–338 CrossRef CAS PubMed
.
- R. Kapoor and Y.-S. Huang, Gamma Linolenic Acid: An Antiinflammatory Omega-6 Fatty Acid, Curr. Pharm. Biotechnol., 2006, 7(6), 531–534 CAS
.
- S.-J. Tsai, C.-S. Huang, M.-C. Mong, W.-Y. Kam, H.-Y. Huang and M.-C. Yin, Anti-inflammatory and antifibrotic effects of naringenin in diabetic mice, J. Agric. Food Chem., 2012, 60(1), 514–521 CrossRef CAS PubMed
.
- B. R. Lawson, S. M. Belkowski, J. F. Whitesides, P. Davis and J. W. Lawson, Immunomodulation of murine collagen-induced arthritis by N, N-dimethylglycine and a preparation of Perna canaliculus, BMC Complementary Altern. Med., 2007, 7(1), 1–9 CrossRef PubMed
.
- E. A. Miles, T. Banerjee, M. M. Dooper, L. M'Rabet, Y. M. Graus and P. C. Calder, The influence of different combinations of γ-linolenic acid, stearidonic acid and EPA on immune function in healthy young male subjects, Br. J. Nutr., 2004, 91(6), 893–903 CrossRef CAS PubMed
.
- V. S. Pelly, A. Moeini, L. M. Roelofsen, E. Bonavita, C. R. Bell and C. Hutton, et al., Anti-Inflammatory Drugs Remodel the Tumor Immune Environment to Enhance Immune Checkpoint Blockade Efficacy, Cancer Discovery, 2021, 11(10), 2602–2619 CrossRef CAS PubMed
.
- A. Mantovani, P. Allavena, A. Sica and F. Balkwill, Cancer-related inflammation, Nature, 2008, 454, 436–444 CrossRef CAS PubMed
.
- J. Dulak and A. Józkowicz, Anti-angiogenic and anti-inflammatory effects of statins: relevance to anti-cancer therapy, Curr. Cancer Drug Targets, 2005, 5(8), 579–594 CrossRef CAS PubMed
.
- E. R. Rayburn, S. J. Ezell and R. Zhang, Anti-inflammatory agents for cancer therapy, Mol. Cell. Pharmacol., 2009, 1(1), 29 CrossRef CAS PubMed
.
- D. Taubert, T. Breitenbach, A. Lazar, P. Censarek, S. Harlfinger and R. Berkels, et al., Reaction rate constants of superoxide scavenging by plant antioxidants, Free Radicals Biol. Med., 2003, 35(12), 1599–1607 CrossRef CAS PubMed
.
- J. Nelson, S. DeMichele, E. Pacht and A. Wennberg, Effect of enteral feeding with eicosapentaenoic acid, gamma-linolenic acid, and antioxidants on antioxidant status in patients with acute respiratory distress syndrome, J. Parenter. Enteral Nutr., 2003, 27(2), 98–104 CrossRef CAS PubMed
.
- N. H. Zaidun, Z. C. Thent and A. Abd Latiff, Combating oxidative stress disorders with citrus flavonoid: Naringenin, Life Sci., 2018, 208, 111–122 CrossRef CAS PubMed
.
- J. P. V. Singh, K. Selvendiran, S. M. Banu, R. Padmavathi and D. Sakthisekaran, Protective role of Apigenin on the status of lipid peroxidation and antioxidant defense against hepatocarcinogenesis in Wistar albino rats, Phytomedicine, 2004, 11(4), 309–314 CrossRef CAS PubMed
.
- E. Celep, A. Aydın and E. Yesilada, A comparative study on the in vitro antioxidant potentials of three edible fruits: Cornelian cherry, Japanese persimmon and cherry laurel, Food Chem. Toxicol., 2012, 50(9), 3329–3335 CrossRef CAS PubMed
.
Footnote |
| † Electronic supplementary information (ESI) available. See DOI: https://doi.org/10.1039/d4ra02131b |
| This journal is © The Royal Society of Chemistry 2024 |