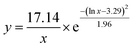Open Access Article
Open Access ArticleSignal amplified colorimetric nucleic acid detection based on autocatalytic hairpin assembly
Yunhua Liu†
*a,
Limin Jin†c,
Jianfei Mao b,
Ru Dengb,
Fengyi Lin
b,
Ru Dengb,
Fengyi Lin b,
Yuxin Chengb,
Min Lib and
Jianyuan Dai
b,
Yuxin Chengb,
Min Lib and
Jianyuan Dai *b
*b
aSchool of Chemical Engineering, Guizhou Institute of Technology, Guiyang 550000, China. E-mail: yhliu@git.edu.cn; Tel: +86-13458610501
bCollege of Chemistry, Sichuan University, Chengdu 610064, China. E-mail: daijy@scu.edu.cn; Tel: +86-18380216833
cInstitute of Biomedical Engineering, Chinese Academy of Medical Sciences, Peking Union Medical College, Tianjin 300192, China
First published on 28th May 2024
Abstract
Herein, a nucleic acid assay based on autocatalytic hairpin assembly (ACHA) was proposed. In this system, two split G-quadruplex sequences were integrated into H1 and H2, respectively. And a DNA strand with the same sequence to target DNA was integrated into the assistant hairpin H3. In the presence of target DNA, the hairpin structure of H1 was opened and catalytic hairpin assembly (CHA) was activated, and then a series of DNA assembly steps based on the toehold-mediated DNA strand displacement were triggered and the product H1–H2 with sticky ends on both sides was formed. On the one side of H1–H2, the split two G-quadruplex sequences were close enough to form the intact G-quadruplex for the signal readout. At the same time, two sticky ends on the other side of H1–H2 hybridized with H3 and a new sticky end with the sequence same to the target DNA was exposed, which can immediately trigger the autocatalytic hairpin assembly reaction, and then the reaction rate of CHA was effectively accelerated and the colorimetric signal was significantly amplified. This ACHA signal amplified strategy has been successfully applied for the rapid and colorimetric nucleic acid detection.
1. Introduction
The detection of nucleic acids plays an important role in biomedical therapy and disease diagnosis.1–4 Polymerase chain reaction (PCR) is the most extensively used assay where the reaction proceeded exponentially to amplify trace amounts of nucleic acid to detectable levels. Nevertheless, PCR has limitations in terms of time consumption, high-precision thermal cycling, and sometimes nonspecificity.4,5 Recently, several isothermal amplification techniques, such as rolling circle amplification,6,7 nuclease-assisted target recycling amplification,3,8 strand displacement amplification,9,10 and loop-mediated isothermal amplification,11,12 have been developed. Compared with PCR, the aforementioned techniques can operate at a constant reaction temperature, which improved the amplification efficiency, but some special conditions such as the use of specific DNA polymerases and the ligation of a padlock probe increased the cost and complexity of the experiment.13,14 Therefore, the requirements of developing room-temperature and enzyme-free techniques for accurate nucleic acid detection were urgent.15 Recently, enzyme-free DNA circuits such as hybridization chain reaction (HCR),16 catalyzed hairpin assembly (CHA),17 and entropy-driven circuit (EDC)18 have been developed, and DNA hybridization has been used to replace the enzyme reaction for signal amplification.16,19,20 However, these new target recycling amplification methods were constrained by relatively long response time, inducing high signal-to-noise ratios and non-specific background leakage.21,22 Self-replication is a procedure in which the system could be replicated, causing exponential amplification itself.23–25 In our recent work, a novel strategy known as self-replicating catalyzed hairpin assembly (SRCHA) was developed,26 and the signal amplification rate was significantly accelerated by DNA self-replication. In this work, a DNA strand with the same sequence to target DNA was integrated into the assistant hairpin, which can be exposed in the CHA reaction as the replica to trigger the autocatalytic hairpin assembly (ACHA) reaction. Finally, a rapid and colorimetric assay for DNA detection was successfully developed.2. Experimental section
2.1 Materials and reagents
Trishydroxymethylaminomethane hydrochloride (Tris–HCl), ABTS−, sodium chloride, sodium citrate, magnesium chloride, potassium chloride, sodium hydroxide, hemin, and oligonucleotides were synthesized and purified by Sangon Biotech. Co., Ltd. (Shanghai, China), and their sequences are listed in Table 1. A hemin stock solution was prepared in DMSO and stored darkly at −20 °C. All other chemicals were none need of extra purification. The water (≥18.2 MΩ cm) used in all of the experimental processes was purified by the Millipore filtration system.| Name | Sequences (5′–3′) |
|---|---|
| a The bold sequences in H1 and H2 strands are the two split G-quadruplex sequences, respectively. The bold sequence in H3 is the DNA strand with the same sequence to the target DNA. T: perfectly matched target DNA; DT: deleted target DNA; IT: inserted target DNA, MT: mismatched target DNA. | |
| H1 | AGGAGGCAGACTGATGTTGGGTTAGCTTATCAACATCAGTCTGATAAGCTATTGGGT |
| H2 | AGGGCGGGAGGGTTAGCTTATCAGACTGATGTTGATAAGCTAACCCAACATCAGTAGTAGAGGT |
| H3 | TAGCTTATCAGACTGATGTTGAACCTCTACTCTGCCTCCTTGATAAGCTA |
| T | TAGCTTATCAGACTGATGTTGA |
| DT | TAGCTTATCA_ACTGATGTTGA |
| IT | TAGCTTATCAGTACTGATGTTGA |
| MT | TAGCTTATCACACTGATGTTGA |
2.2 Instruments
Photographs were captured by a Sony DSC-WX150 digital camera. The concentration of DNA was detected by Qubit 2.0 Fluorometer (Thermo Fisher Scientific Inc.). UV-Vis absorption spectra were measured by a U-2900 spectrophotometer (Hitachi Co. Ltd., Japan).2.3 Native PAGE analysis
The different reaction products with a load buffer of ten microliters were loaded into the polyacrylamide gel lane (10%, Acr![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) Bis = 29
Bis = 29![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1). The gel was operated at a constant voltage of 100 V for 90 min in the TBE buffer (1×) at room temperature. It was dyed with ethidium bromide for 40 min to image the location of DNA. Finally, the electrophoretic image of polyacrylamide gel was obtained in visible light by a gel imaging system.
1). The gel was operated at a constant voltage of 100 V for 90 min in the TBE buffer (1×) at room temperature. It was dyed with ethidium bromide for 40 min to image the location of DNA. Finally, the electrophoretic image of polyacrylamide gel was obtained in visible light by a gel imaging system.
2.4 DNA assay
H1, H2, and H3 were prepared in Tris–HCl buffer solution (20 mM, 50 mM KCl, 10 mM MgCl2, pH 7.6), annealed at 95 °C for 5 min, and then slowly cooled to room temperature for 3 h. For DNA assays, 500 nM H1, 500 nM H2, and 100 nM H3 were mixed with different concentrations of target DNA in 100 μL buffer solution. Subsequently, 2.5 μL pf 50 μM hemin, 7.5 μL of 50 mM ABTS2−, and 3 μL of 500 nM H2O2 were added. The resulting solutions were incubated at room temperature for 10 min, and then subjected to visual observation and ultraviolet measurement.3. Results and discussion
3.1 Principle of the ACHA system
A schematic of this ACHA system for rapid signal amplification and its colorimetric detection of DNA are displayed in Scheme 1. In this system, part of the complementary sequences of H1, H2, and H3 were integrated into hairpins, thereby they remained metastable in the CHA system. Two split G-quadruplex sequences were integrated into H1 and H2, respectively. A DNA strand with the same sequence to target DNA was introduced into the assistant H3. In the presence of target DNA, the hairpin structure of H1 was opened by target DNA (step 1), where the exposed sticky ends will hybridize to the sticky ends of H2 to produce target–H1–H2 as an intermediate (step 2); and then the target DNA will be released by H2 through the strand displacement reaction (step 3) to trigger the next CHA reaction. In the end, the duplex DNA assembly H1–H2 was formed to bring the two split G-quadruplex sequences close enough to form the complete G-quadruplex (recycling I). Successively, this complete G-quadruplex interacted with hemin to produce a hemin/G-quadruplex horseradish peroxidase (HRP)-mimicking DNAzyme and catalyze the H2O2-mediated oxidation of colorless ABTS2− to the green-colored ABTS˙− for signal identification. Therefore, this colorimetric signal readout could be simply identified with the naked eye.27 Moreover, H3 was opened by the new formed sticky ends on the another side of H1–H2 (step 4), and the DNA sequence same to target DNA was exposed, which can be used as the target replica to trigger the ACHA reaction (step 5–6) for the signal amplification (recycling II). In this ACHA system, the formation of numerous target replicas can considerably increase the reaction rate and shorten the detection time. Finally, a rapid and signal amplified colorimetric DNA assay was realized.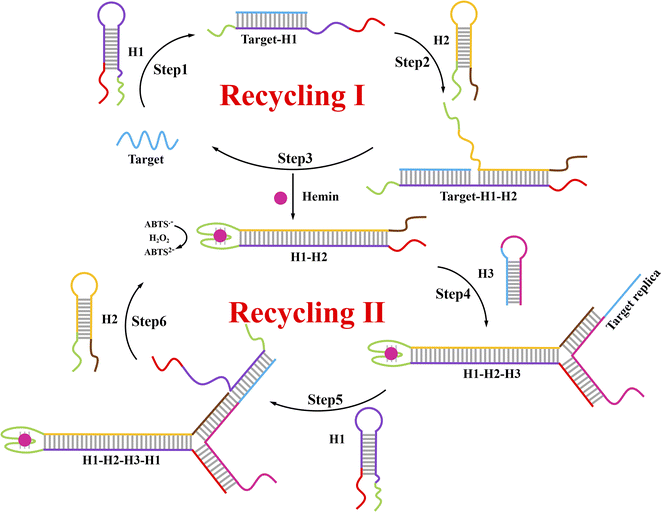 | ||
| Scheme 1 Principle for the rapid signal amplification based on the novel autocatalytic hairpin assembly (ACHA) and its application for DNA detection. | ||
3.2 Feasibility of the ACHA system
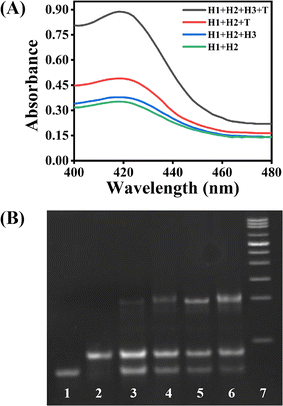
To examine the feasibility of this ACHA system, a non-autocatalytic hairpin assembly (non-ACHA) system without H3 was used as a control. The absorbance intensities of ACHA and non-ACHA systems were measured before and after the addition of 5 nM target DNA (Fig. 1A). In the absence of target DNA, there should be no increase in absorbance intensity. However, because of the uncatalyzed reaction between H1 and H2 caused by the “breathing” of hairpin helix, slight increases of absorbance intensity were observed in both the ACHA and non-ACHA systems. In the presence of target DNA, the absorbance intensity of the ACHA system at 420 nm was considerably higher compared to that of the non-ACHA system. These results confirmed that the reaction rate of the ACHA system has been truly accelerated and the rapid signal amplification was successfully realized. This result was proved by gel electrophoresis. Similar to the absorbance and colorimetric assay, there were more H1–H2 products observed from the ACHA system compared with those from the non-ACHA system (Fig. 1B).3.3 Optimization and analytical performance of the strategy for target DNA detection
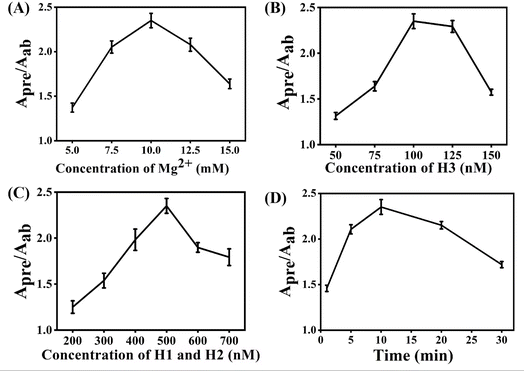
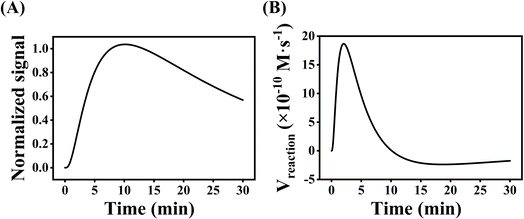
Several experimental conditions, such as concentration of Mg2+ and hairpin probes, and reaction time have been optimized to realize the highest sensing performance of ACHA system (Fig. 2). Obviously, the Apre/Aab (where Apre and Aab are the absorbance intensities of the ACHA system in the presence and absence of target DNA, respectively) value increased with increasing concentrations of the Mg2+, H1, H2, and H3, and reaction time, and reached the peak at 10 mM Mg2+, 100 nM H3, 500 nM H1, and H2, and reaction time of 10 min. Then, the Apre/Aab value decreased with further increase of their concentrations due to the background increasement. According to the above results, these best optimum conditions were adopted in the subsequent experiments.Reaction rate (Vreaction) of the ACHA system in the presence of 5 nM target DNA was calculated as per the reported literature.28 Firstly, the ratios of Apre/Aab that vary with reaction time were normalized to the interval [0,1] (Fig. 3A). Subsequently, the log-normal formula was utilized to obtain a smooth fit of the normalized results (Fig. 3B), which is
Therefore, Vreaction at each time point can be calculated by calculating the slope of tangent, where the maximal reaction rate Vmax is 18.67 × 10−10 M s−1.
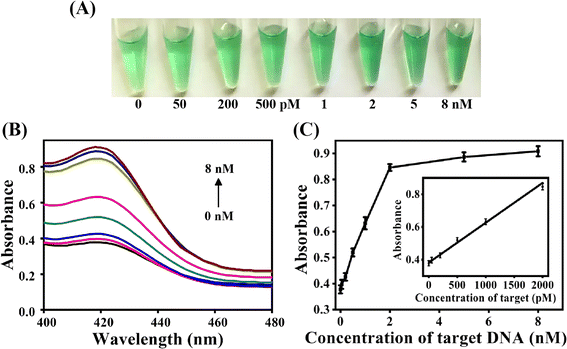
Fig. 4A demonstrated the color change of the ACHA system at different concentration of target DNA. The solution color gradually turned green with the increase of target DNA concentration, and the color change can be distinguished by the naked eyes when the target concentration was as low as 200 pM. UV-Vis data demonstrated that the absorbance at 420 nm increased with increase in the target DNA concentration (Fig. 4B). The absorbance intensity at 420 nm was adopted to detect the target concentration (Fig. 4C). A good linearity between Apre/Aab and the target concentration from 0.05 nM to 2 nM was obtained, and the detection limit was determined to be 16 pM based on the 3σ calculation (S/N = 3) of the blank tests (n = 11), which was comparable with previously reported signal amplification DNA assays but with significant short detection time (Table 2).29–33
| Strategy | Signal readout | Detection limit | Detection time | Ref. |
|---|---|---|---|---|
| Hybridization chain reaction (HCR) | Colorimetry | 50 pM | 60 min | 31 |
| Hairpin-free DNA amplification strategy | Fluorescence | 5 pM | 60 min | 32 |
| Catalyzed hairpin assembly (CHA) | Fluorescence | 10 pM | 120 min | 30 |
| Catalyzed hairpin assembly (CHA) | Fluorescence | 19 pM | 90 min | 29 |
| Catalyzed hairpin assembly (CHA) | Colorimetry | 9 pM | 90 min | 19 |
| Catalyzed hairpin assembly (CHA) | Colorimetry | 0.1 pM | 60 min | 33 |
| Autocatalytic hairpin assembly (ACHA) | Colorimetry | 16 pM | 10 min | This work |
3.4 Selectivity of this ACHA system
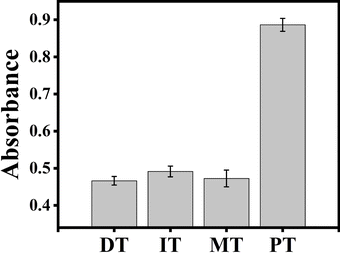
To investigate the selectivity of the proposed ACHA system, four different target DNAs, deleted, inserted, mismatched, and perfectly matched target DNAs were tested under the same experimental conditions. As shown in Fig. 5, the deleted, inserted, and mismatched target DNAs only produced low absorbance, while the perfectly matched DNA showed obvious enhancement of absorbance intensity, indicating that the rapid signal amplification was triggered by the perfectly matched DNA.3.5 Detection of target DNA in human serum samples
To investigate the practicality of the ACHA system in real biological samples, targets with different concentrations (100, 200, and 400 pM) were added to 100-fold diluted human serum samples, respectively. The results are shown in Table 3. The recoveries of target DNA ranged from 96.75% to 102.8%, and RSDs ranged from 3.4 to 4.7%. The results showed that the rapid and colorimetric DNA assay possesses the ability for the real biological samples detection.| Sample | Added (pM) | Founded (pM) | Recovery (%) | RSD (%) |
|---|---|---|---|---|
| Serum | 100.0 | 102.8 | 102.8 | 4.1 |
| 200.0 | 193.5 | 96.75 | 3.4 | |
| 400.0 | 392.6 | 98.15 | 4.7 |
4. Conclusion
In this work, a novel ACHA system was constructed by introducing a DNA strand with the same sequence to target DNA into the assistant hairpin, and a rapid and colorimetric signal amplified DNA assay was successfully developed. Benefiting from the highly efficient autocatalysis in the ACHA system, our proposed assay exhibits high sensitivity towards target DNA with a short detection time and has been successfully applied for the determination of DNA in the serum samples with satisfying results, exhibiting a wide application prospect in the biomedical field.Ethical statement
All experiments were performed in accordance with the guidelines (approval number 20170201) that was granted on 20 August 2019. All animal procedures were performed in accordance with the Guidelines for Care and Use of Laboratory Animals of Sichuan University and experiments were approved by the Animal Ethics Committee of Sichuan University.Data availability
All the data used in the manuscript are available in the tables and figures. The datasets generated during and/or analyzed during the current study are available from the corresponding author on reasonable request.Conflicts of interest
The authors declare no competing financial interests.Acknowledgements
This work was supported by the Cooperative Project of Chengdu Xiaojia Technology Co., Ltd (No. 22H1111), Cooperative Project of Analysis and Testing Center of Sichuan Academy of Agricultural Science (No. 20H1090, 0020303410001), the Science and Technology Planning Project of Guizhou Province (No. Qiankehezhicheng [2020]1Y167).References
- X. Cao, C. Chen and Q. Zhu, Talanta, 2023, 253, 123977 CrossRef CAS PubMed.
- M.-S. Kim, G. Stybayeva, J. Y. Lee, A. Revzin and D. J. Segal, Nucleic Acids Res., 2011, 39, e29 CrossRef CAS PubMed.
- X. Zuo, F. Xia, Y. Xiao and K. W. Plaxco, J. Am. Chem. Soc., 2010, 132, 1816–1818 CrossRef CAS PubMed.
- A. R. Connolly and M. Trau, Angew. Chem., Int. Ed., 2010, 49, 2720–2723 CrossRef CAS PubMed.
- J. Zhou, Q.-x. Wang and C.-y. Zhang, J. Am. Chem. Soc., 2013, 135, 2056–2059 CrossRef CAS PubMed.
- P. M. Lizardi, X. Huang, Z. Zhu, P. Bray-Ward, D. C. Thomas and D. C. Ward, Nat. Genet., 1998, 19, 225–232 CrossRef CAS PubMed.
- W. Zhao, M. M. Ali, M. A. Brook and Y. Li, Angew. Chem., Int. Ed., 2008, 47, 6330–6337 CrossRef CAS PubMed.
- W. Xu, X. Xue, T. Li, H. Zeng and X. Liu, Angew. Chem., Int. Ed., 2009, 48, 6849–6852 CrossRef CAS PubMed.
- G. T. Walker, M. C. Little, J. G. Nadeau and D. D. Shank, Proc. Natl. Acad. Sci. U.S.A., 1992, 89, 392–396 CrossRef CAS PubMed.
- Q. Guo, X. Yang, K. Wang, W. Tan, W. Li, H. Tang and H. Li, Nucleic Acids Res., 2009, 37, e20 CrossRef PubMed.
- T. Notomi, H. Okayama, H. Masubuchi, T. Yonekawa, K. Watanabe, N. Amino and T. Hase, Nucleic Acids Res., 2000, 28, e63 CrossRef CAS PubMed.
- N. Tomita, Y. Mori, H. Kanda and T. Notomi, Nat. Protoc., 2008, 3, 877–882 CrossRef CAS PubMed.
- Y. S. Jiang, S. Bhadra, B. Li and A. D. Ellington, Angew. Chem., Int. Ed., 2014, 53, 1845–1848 CrossRef CAS PubMed.
- Y. Guo, J. Wu and H. Ju, Chem. Sci., 2015, 6, 4318–4323 RSC.
- X. Xia, H. Yang, J. Cao, J. Zhang, Q. He and R. Deng, Trends Anal. Chem., 2022, 153, 116641 CrossRef CAS.
- R. M. Dirks and N. A. Pierce, Proc. Natl. Acad. Sci. U.S.A., 2004, 101, 15275–15278 CrossRef CAS PubMed.
- P. Yin, H. M. T. Choi, C. R. Calvert and N. A. Pierce, Nature, 2008, 451, 318–322 CrossRef CAS PubMed.
- D. Y. Zhang, A. J. Turberfield, B. Yurke and E. Winfree, Science, 2007, 318, 1121–1125 CrossRef CAS PubMed.
- H. He, J. Dai, Z. Duan, Y. Meng, C. Zhou, Y. Long, B. Zheng, J. Du, Y. Guo and D. Xiao, Biosens. Bioelectron., 2016, 86, 985–989 CrossRef CAS PubMed.
- F. Huang, M. You, D. Han, X. Xiong, H. Liang and W. Tan, J. Am. Chem. Soc., 2013, 135, 7967–7973 CrossRef CAS PubMed.
- A. R. Connolly and M. Trau, Nat. Protoc., 2011, 6, 772–778 CrossRef CAS PubMed.
- K. Hsieh, A. S. Patterson, B. S. Ferguson, K. W. Plaxco and H. T. Soh, Angew. Chem., Int. Ed., 2012, 51, 4896–4900 CrossRef CAS PubMed.
- T. Wang, R. Sha, R. Dreyfus, M. E. Leunissen, C. Maass, D. J. Pine, P. M. Chaikin and N. C. Seeman, Nature, 2011, 478, 225–228 CrossRef CAS PubMed.
- J. Kim, J. Lee, S. Hamada, S. Murata and S. Ha Park, Nat. Nanotechnol., 2015, 10, 528–533 CrossRef CAS PubMed.
- S. Li, L. Zhu, S. Lin and W. Xu, Biosens. Bioelectron., 2023, 220, 114922 CrossRef CAS PubMed.
- J. Dai, H. He, Z. Duan, Y. Guo and D. Xiao, Anal. Chem., 2017, 89, 11971–11975 CrossRef CAS PubMed.
- Y. Xiao, V. Pavlov, T. Niazov, A. Dishon, M. Kotler and I. Willner, J. Am. Chem. Soc., 2004, 126, 7430–7431 CrossRef CAS PubMed.
- L. Wu, G. A. Wang and F. Li, J. Am. Chem. Soc., 2024, 146, 6516–6521 CrossRef CAS PubMed.
- Z. Duan, Z. Li, J. Dai, H. He and D. Xiao, Talanta, 2017, 164, 34–38 CrossRef CAS PubMed.
- J. Huang, X. Su and Z. Li, Anal. Chem., 2012, 84, 5939–5943 CrossRef CAS PubMed.
- C.-H. Lu, F. Wang and I. Willner, J. Am. Chem. Soc., 2012, 134, 10651–10658 CrossRef CAS PubMed.
- Y. Lv, L. Cui, R. Peng, Z. Zhao, L. Qiu, H. Chen, C. Jin, X.-B. Zhang and W. Tan, Anal. Chem., 2015, 87, 11714–11720 CrossRef CAS PubMed.
- W. Yun, J. Jiang, D. Cai, P. Zhao, J. Liao and G. Sang, Biosens. Bioelectron., 2016, 77, 421–427 CrossRef CAS PubMed.
Footnote |
| † These authors contributed equally to this work. |
| This journal is © The Royal Society of Chemistry 2024 |