Open Access Article
Open Access ArticleAn antiviral oligomerized linear thiopeptide with a nitrile group from soil-derived Streptomyces sp. CPCC 203702†
Zhe Guo‡
a,
Dewu Zhang‡ *a,
Yujia Wangb,
Jinglin Baiab,
Jun Hua,
Shan Cenb and
Liyan Yu*ab
*a,
Yujia Wangb,
Jinglin Baiab,
Jun Hua,
Shan Cenb and
Liyan Yu*ab
aChina Pharmaceutical Culture Collection, Institute of Medicinal Biotechnology, Chinese Academy of Medical Sciences and Peking Union Medical College, Beijing 100050, People's Republic of China. E-mail: zhangdewuever@163.com; yly@cpcc.ac.cn
bDivision for Medicinal Microorganisms Related Strains, CAMS Collection Center of Pathogenic Microorganisms, Beijing 100050, People's Republic of China
First published on 11th March 2024
Abstract
A new linear thiopeptide, bernitrilecin (1), was isolated from Streptomyces sp. CPCC 203702. Compound 1 is the first example of a nitrile-bearing thiopeptide. Its structure and absolute configuration were elucidated by extensive analysis of spectroscopic data and Marfey's method. The biosynthesis of the nitrile unit for 1 was proposed to be through oxidations, decarboxylation, and dehydration. Compound 1 exhibited significant anti-influenza A virus activity with the IC50 value of 16.7 μM.
Introduction
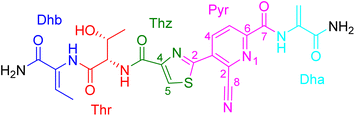
Thiopeptides or thiazolyl peptides are a family of ribosomally synthesized and post-translationally modified peptide (Ripp) natural products, which are characterized by structural features including thiazole and/or oxazole rings with highly modified amino acids. Thiopeptide antibiotics are potent inhibitors of protein synthesis in Gram-positive bacteria, including methicillin-resistant Staphylococcus aureus (MRSA), vancomycin-resistant Enterococcus spp. (VRE), and penicillin-resistant Streptococcus pneumonia (PRSP).1–5 In recent years, more biological activities of thiopeptide antibiotics such as anticancer, antiviral, immunogenic, and anti-malaria activities were reported by medicinal chemists and pharmacologists.5–10 Structurally, most thiopeptides commonly feature a macrocyclic framework (cyclic thiopeptide). According to the size of the macrocycle core, thiopeptides can be divided into four classes: 26-membered, 29-membered, 32-membered, and 35-membered, which were assembled by over ten amino acids. However, there are few reports regarding linear thiopeptides. In particular, oligomerized linear thiopeptide assembled by less than ten amino acids are even rarer.1,5Our previous chemical investigation of Streptomyces sp. CPCC 203702 afforded four new thiopeptides with potent antiviral activities.11 As part of our ongoing search for bioactive thiopeptide from this strain, the extracts of Streptomyces sp. CPCC 203702 were further investigated, leading to the isolation of a new oligomeric linear thiopeptide, bernitrilecin (1) (Fig. 1), which was assembled by seven amino acids. Compound 1 is the first example of a thiopeptide with a nitrile group. In this article, we report the fermentation, isolation, structural elucidation, and biological activity of bernitrilecin (1).
Results and discussion
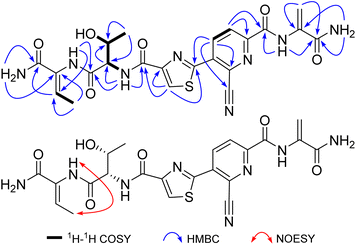
Bernitrilecin (1) was isolated as white amorphous powder. Its molecular formula was determined as C22H22N8O6S by HRESIMS with m/z 527.1451 [M + H]+, requiring 16 degrees of unsaturation. The 13C NMR and DEPT data (Table 1) of 1 revealed the presence of 22 carbons, including thirteen nonprotonated carbons (δC 168.7, 165.8, 164.7, 160.1, 160.1, 160.0, 150.5, 150.3, 134.1, 133.5, 130.5, 128.3, and 116.6), six methine carbons (δC 139.8, 129.0, 128.5, 125.8, 66.8, and 58.8), one methylene carbons (δC 103.4), and two methyl carbons (δC 19.8 and 13.3). The 1H NMR and HSQC data (Table 1) of 1 showed 22 protons, including six sp2 proton (δH 8.86, 8.70, 8.50, 6.57, 6.47, and 5.86). In the HMBC spectrum, the cross peaks from terminal –NH2 (δH 7.16, 7.14) to Dhb-C![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) O (δC 165.8) and Dhb-Cα (δC 130.5), from Dhb-Hβ (δH 6.47) to Dhb-C
O (δC 165.8) and Dhb-Cα (δC 130.5), from Dhb-Hβ (δH 6.47) to Dhb-C![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) O (δC 165.8), Dhb-Cα (δC 130.5), and Dhb-Cγ (δC 13.3) revealed the presence of dehydrobutyrine group. The HMBC correlations from Thr-Hγ (δH 1.19) to Thr-Cα (δC 58.8) and Thr-Cβ (δC 66.8), from Thr-Hα (δH 4.50) to Thr-C
O (δC 165.8), Dhb-Cα (δC 130.5), and Dhb-Cγ (δC 13.3) revealed the presence of dehydrobutyrine group. The HMBC correlations from Thr-Hγ (δH 1.19) to Thr-Cα (δC 58.8) and Thr-Cβ (δC 66.8), from Thr-Hα (δH 4.50) to Thr-C![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) O (δC 168.7), Thr-Cβ (δC 66.8), Thr-Cγ (δC 19.8), and Thz-C
O (δC 168.7), Thr-Cβ (δC 66.8), Thr-Cγ (δC 19.8), and Thz-C![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) O (δC 160.1), along with the 1H-1H COSY correlations of Thr-NH (δH 8.24)/Thr-Hα/(δH 4.50)/Thr-Hβ (δH 4.20)/Thr-Hγ (δH 1.19) indicated the presence of threonine moiety. The HMBC correlations from Thz-H-5 (δH 8.70) to Thz-C
O (δC 160.1), along with the 1H-1H COSY correlations of Thr-NH (δH 8.24)/Thr-Hα/(δH 4.50)/Thr-Hβ (δH 4.20)/Thr-Hγ (δH 1.19) indicated the presence of threonine moiety. The HMBC correlations from Thz-H-5 (δH 8.70) to Thz-C![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) O (δC 160.1), Thz-C-2 (δC 160.1), and Thz-C-4 (δC 150.3) and typical chemical shifts of Thz-C-2 (δC 160.1), Thz-C-4 (δC 150.3), and Thz-C-5 (δC 128.5) for thiazole unit in thiopeptide derivative established the presence of thiazole group. The HMBC cross peaks from Pyr-H-4 (δH 8.86) to Pyr-C-2 (δC 150.5), Pyr-C-3 (δC 128.3), and Thz-C-2 (δC 160.1), from Pyr-H-5 (δH 8.50) to Pyr-C-6 (δC 134.1) and Pyr-C-7 (δC 160.0), together with the 1H-1H COSY correlations for Pyr-H-4 (δH 8.86)/Pyr-H-5 (δH 8.50) determined the presence of 2,3,6-trisubstituted pyridine group. The HMBC correlations from Dha-Hβ (δH 6.57, 5.86) to Dha-C
O (δC 160.1), Thz-C-2 (δC 160.1), and Thz-C-4 (δC 150.3) and typical chemical shifts of Thz-C-2 (δC 160.1), Thz-C-4 (δC 150.3), and Thz-C-5 (δC 128.5) for thiazole unit in thiopeptide derivative established the presence of thiazole group. The HMBC cross peaks from Pyr-H-4 (δH 8.86) to Pyr-C-2 (δC 150.5), Pyr-C-3 (δC 128.3), and Thz-C-2 (δC 160.1), from Pyr-H-5 (δH 8.50) to Pyr-C-6 (δC 134.1) and Pyr-C-7 (δC 160.0), together with the 1H-1H COSY correlations for Pyr-H-4 (δH 8.86)/Pyr-H-5 (δH 8.50) determined the presence of 2,3,6-trisubstituted pyridine group. The HMBC correlations from Dha-Hβ (δH 6.57, 5.86) to Dha-C![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) O (δC 164.7) and Dha-Cα (δC 133.5), from terminal –NH2 (δH 8.20, 7.74) to Dha-C
O (δC 164.7) and Dha-Cα (δC 133.5), from terminal –NH2 (δH 8.20, 7.74) to Dha-C![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) O (δC 164.7) and Dha-Cα (δC 133.5) indicated the presence of dehydroalanine residue. The critical HMBC correlations from Dhb-NH (δH 9.34) to Dhb-Cβ (δC 129.0) and Thr-C
O (δC 164.7) and Dha-Cα (δC 133.5) indicated the presence of dehydroalanine residue. The critical HMBC correlations from Dhb-NH (δH 9.34) to Dhb-Cβ (δC 129.0) and Thr-C![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) O (δC 168.7), from Thr-NH (δH 8.24) to Thr-C
O (δC 168.7), from Thr-NH (δH 8.24) to Thr-C![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) O (δC 168.7), Thr-Cα (δC 58.8), Thr-Cβ (δC 66.8), and Thz-C
O (δC 168.7), Thr-Cα (δC 58.8), Thr-Cβ (δC 66.8), and Thz-C![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) O (δC 160.1), from Dha-NH (δH 10.44) to Dha-C
O (δC 160.1), from Dha-NH (δH 10.44) to Dha-C![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) O (δC 164.7), Dha-Cβ (δC 103.4), and Pyr-C-7 (δC 160.0) established the linkage of each residue. Furthermore, the long-range correlations (four bond) in HMBC spectrum from Pyr-H-4 (δH 8.86) to Pyr-C-8 (δC 116.6), along with the molecular formula and chemical shift of Pyr-C-8 suggested the existence of nitrile group. Based on the above observations, the planar structure of 1 was determined to be an unusual thiopeptide with nitrile moiety. The NOESY correlation (Fig. 2) between methyl protons (δH 1.63) and amide proton (δH 9.34) indicated the Z configuration of methyl-substituted double bond in dehydrobutyrine residue. The configuration of Thr was established as L using Marfey's method (Fig. S1†).11
O (δC 164.7), Dha-Cβ (δC 103.4), and Pyr-C-7 (δC 160.0) established the linkage of each residue. Furthermore, the long-range correlations (four bond) in HMBC spectrum from Pyr-H-4 (δH 8.86) to Pyr-C-8 (δC 116.6), along with the molecular formula and chemical shift of Pyr-C-8 suggested the existence of nitrile group. Based on the above observations, the planar structure of 1 was determined to be an unusual thiopeptide with nitrile moiety. The NOESY correlation (Fig. 2) between methyl protons (δH 1.63) and amide proton (δH 9.34) indicated the Z configuration of methyl-substituted double bond in dehydrobutyrine residue. The configuration of Thr was established as L using Marfey's method (Fig. S1†).11
| Unit | No. | δC, type | δH (J in Hz) |
|---|---|---|---|
| Dhb | NH | 9.34, s | |
| CO | 165.8, C | ||
| α | 130.5, C | ||
| β | 129.0, CH | 6.47, q (6.6) | |
| γ | 13.3, CH3 | 1.63, d (6.6) | |
| NH2 | 7.16, s | ||
| 7.14, s | |||
| Thr | NH | 8.24, d (7.2) | |
| OH | 5.52, br s | ||
| CO | 168.7, C | ||
| α | 58.8, CH | 4.50, dd (7.2, 4.2) | |
| β | 66.8, CH | 4.20, m | |
| γ | 19.8, CH3 | 1.19, d (6.6) | |
| Thz | CO | 160.1, C | |
| 2 | 160.1, C | ||
| 4 | 150.3, C | ||
| 5 | 128.5, CH | 8.70, s | |
| Pyr | 2 | 150.5, C | |
| 3 | 128.4, C | ||
| 4 | 139.8, CH | 8.86, d (8.4) | |
| 5 | 125.8, CH | 8.50, d (8.4) | |
| 6 | 134.1, C | ||
| 7 | 160.0, C | ||
| 8 | 116.6, C | ||
| Dha | NH | 10.44, s | |
| CO | 164.8, C | ||
| α | 133.5, C | ||
| β | 103.4, CH2 | 6.57, s | |
| 5.86, s | |||
| NH2 | 8.20, s | ||
| 7.74, s |
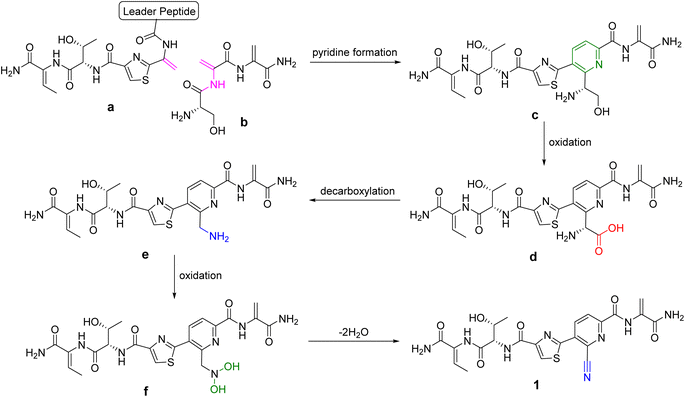
Nitrile-containing natural products produced by microorganisms are relatively rare and the biosynthesis routes to form nitriles are also rarely reported in the literature. To the best of our knowledge, bernitrilecin (1) represents the first example of thiopeptide bearing nitrile group. The biosynthesis of 1 is proposed to be through classical thiopeptide biosynthetic pathway combined with special oxidation reactions (Scheme 1). Two linear peptide residues a and b take part in the pyridine-forming event to produce the linear thiopeptide scaffold (c), and the subsequent oxidation give carboxyl intermediate (d). The intermediate d undergoes decarboxylation to make amino intermediate (e), which can be converted to N,N-dihydroxyamine intermediate (f), and further dehydration lead to the formation of 1 featuring nitrile moiety. We speculate that cytochrome P450 enzyme might play a critical role in the conversion of amino to nitrile, as of examples, similar enzymic catalytic reactions can be found in the biosynthesis of dhurrin and borrelidin.12,13
Compound 1 was evaluated for anti-influenza A virus (IAV) and antibacterial activities.13 Compound 1 displayed significant anti-IAV (H1N1) activities with the IC50 value of 16.7 μM. Compound 1 was also tested for antibacterial activities against Staphylococcus aureus ATCC 29213, Escherichia coli ATCC 25922, Enterococcus faecalis ATCC 29212, Pseudomonas syringae CPCC 101099, and Bacillus cereus CPCC 101254. Compound 1 showed no antimicrobial activity against all pathogenic bacteria with MIC >32 μg mL−1.
Conclusions
In summary, a new linear thiopeptide, bernitrilecin (1) was isolated from Streptomyces sp. CPCC 203702. Compound 1 was unusual oligomeric linear thiopeptide with nitrile group. Compound 1 showed significant anti-IAV activity.Experimental
General experimental procedures
Optical rotations were recorded on an Autopol IV automatic polarimeter. The CD spectra were measured on a JASCO J-815 spectropolarimeter. UV spectra were obtained on a Persee TU-1901 UV-vis spectrometer. 1D and 2D NMR spectra were performed at 600 MHz for 1H NMR and 150 MHz for 13C NMR on Bruker ARX-600 spectrometer. Chemical shifts (δ) are given in ppm, and coupling constants (J) are given in hertz (Hz). ESIMS data were recorded on a Thermo LTQ mass spectrometer. HRESIMS data were measured using a Thermo LTQ Orbitrap XL mass spectrometer. Column chromatography (CC) were carried out with silica gel (200–300 mesh, Qingdao Marine Chemical Inc. Qingdao, PR China). Analytical TLC was carried out on pre-coated silica gel GF254 plates (Qingdao Marine Chemical Industry, Qingdao, China), and spots were visualized under UV light.Biological material
The strain Streptomyces sp. CPCC 203702 was isolated from the soil, collected in Xunhua County, Qinghai Province, China. The strain was identified as a member of the genus Streptomyces on the basis of 16S rRNA gene sequence analysis by China Pharmaceutical Culture Collection. A BLAST search displayed that the sequence was the same (100%) as the sequence of Streptomyces atroolivaceus NRRL ISP-5137(T).Fermentation, extraction and isolation
Strain CPCC 203702 was spread onto slants of ISP2 medium and incubated at 28 °C for 8–10 days. Fresh spores of the strain was inoculated into 500 mL Erlenmeyer flasks containing 100 mL of sterile fermentation medium (glucose 0.5%, malt extract 1.0%, cotton seed meal 1.0%, amylogen 2.0%, yeast extract 0.5%, K2HPO4 0.05%, (NH4)2SO4 0.5%, CaCO3 0.3%, NaCl 0.1%) at 28 °C on a rotary shaker (200 rpm) for 48 hours to prepare the seed culture. Then 2 L of seed culture was transferred into 20 L fermentor containing the same medium and cultured at 28 °C for 4 days.The cultures (20 L) were filtered under reduced pressure to afford the filtrate and mycelia. The mycelia was extracted with EtOAc three times. The EtOAc extract was evaporated under reduced pressure to yield 20 g of residue, which was subjected to silica gel CC eluting with a chloroform–acetone gradient (100![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 0–0
0–0![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 100) to produce twenty-one fractions (Fr.1–Fr.21) on the basis of TLC analysis. Fraction Fr.18 (111 mg) was subjected to Sephadex LH-20 CC to give seven fractions (Fr.18.1–Fr.18.7), fraction Fr.18.4 (10.2 mg) was further isolated by reversed-phase preparative HPLC eluting with CH3CN–H2O (10
100) to produce twenty-one fractions (Fr.1–Fr.21) on the basis of TLC analysis. Fraction Fr.18 (111 mg) was subjected to Sephadex LH-20 CC to give seven fractions (Fr.18.1–Fr.18.7), fraction Fr.18.4 (10.2 mg) was further isolated by reversed-phase preparative HPLC eluting with CH3CN–H2O (10![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 90) at 4 mL min−1 to give 1 (1.1 mg).
90) at 4 mL min−1 to give 1 (1.1 mg).
Bernitrilecin (1): white amorphous powder; [α]25D +85.6 (c 0.03, MeOH); UV (MeOH) λmax: 207, 314; CD (MeOH) Δε (nm): −4.75 (213), +7.69 (245), +1.15 (313); ESIMS m/z 527.2 [M + H]+; HRESIMS m/z 527.1451 [M + H]+ (calcd for C22H23N8O6S, 527.1461); 1H and 13C NMR data, see Table 1.
C3 Marfey's analysis
Approximately 0.1 mg of object was hydrolyzed with 0.2 mL of 6 mol L−1 HCl and heated at 90 °C for 12 h. After removing the solvent under reduced pressure, the hydrolysate was dissolved in 50 μL of H2O, and then 50 μL of L-FDAA (1% solution in acetone) and 20 μL of 1 mol L−1 NaHCO3 were added to the mixture. The reaction mixture was heated for 1 h at 40 °C, cooled to room temperature, neutralized with 20 μL of 1 mol L−1 HCl, and then diluted with 500 μL MeCN. The FDAA derivatives were subjected to HPLC-DAD-MS analysis (Agilent Zorbax SB-C3 column, 5 μm, 250 × 4.6 mm, 30 °C, 1 mL min−1, linear gradient elution from 10% to 60% MeCN–H2O (0.1% formic acid) in 50 min, UV detection at 340 nm).Anti-IAV activity assays
Bernitrilecin (1) was evaluated for anti-IAV activity as previously described methods.11Conflicts of interest
There are no conflicts to declare.Acknowledgements
The work was financially supported by the National Natural Science Foundation of China (82073744), CAMS Innovation Fund for Medical Sciences (2021-I2M-1-055), and the National Microbial Resource Center (No. NMRC-2023-3).Notes and references
- X. Shen, M. Mustanfa, Y. Chen, Y. Gao and J. Gao, Med. Chem. Res., 2019, 28, 1063–1098 CrossRef CAS.
- M. C. Bagley, J. W. Dale, E. A. Merritt and X. Xiong, Chem. Rev., 2005, 105, 685–714 CrossRef CAS PubMed.
- A. A. Vinogradov and H. Suga, Cell Chem. Biol., 2020, 27, 1032–1051 CrossRef CAS PubMed.
- A. Rouf and C. Tanyeli, Eur. J. Med. Chem., 2015, 97, 911–927 CrossRef CAS PubMed.
- D. C. K. Chan and L. L. Burrows, J. Antibiot., 2021, 74, 161–175 CrossRef CAS PubMed.
- Y. B. Hsu, M. C. Lan, Y. L. Kuo, C. Y. F. Huang and M. Y. Lan, Invest. New Drugs, 2020, 38, 264–273 CrossRef CAS PubMed.
- A. A. Vinogradow, Y. Zhang, K. Hamada, J. S. Chang, C. Okada, H. Nishimura, N. Terasaka, Y. Goto, K. Ogata, T. Sengoku, H. Onaka and H. Suga, J. Am. Chem. Soc., 2022, 144, 20332–20341 CrossRef PubMed.
- W. Peng, Z. Hong, X. Chen, H. Gao, Z. Dai, J. Zhao, W. Liu, D. Li and K. Deng, Antimicrob. Agents Chemother., 2020, 64, 023288 CrossRef PubMed.
- Y. Wang, W. Xie, J. Humeau, G. Chen, P. Liu, J. Pol, Z. Zhang, O. Kepp and G. Kroemer, J. Immunother. Cancer, 2020, 8, e000462 CrossRef PubMed.
- C. Bailly, Eur. J. Pharmacol., 2022, 914, 174661 CrossRef CAS PubMed.
- D. Zhang, Y. Wang, X. Hu, X. Wang, L. Li, G. Gu, B. Zhang, S. Cen, X. You and L. Yu, Chin. J. Chem., 2021, 39, 3277–3284 CrossRef CAS.
- R. A. Kahn, T. Fahrendorf, B. A. Halkier and B. L. Møller, Arch. Biochem. Biophys., 1999, 363, 9–18 CrossRef CAS PubMed.
- C. Olano, S. J. Moss, A. F. Braña, R. M. Sheridan, V. Math, A. J. Weston, C. Méndez, P. F. Leadlay, B. Wilkinson and J. A. Salas, Mol. Microbiol., 2004, 52, 1745–1756 CrossRef CAS PubMed.
Footnotes |
| † Electronic supplementary information (ESI) available: HRESIMS, UV, ECD, Marfey's analyse, 1D and 2D NMR for 1. See DOI: https://doi.org/10.1039/d4ra01496k |
| ‡ These authors have contributed equally to this work. |
| This journal is © The Royal Society of Chemistry 2024 |