 Open Access Article
Open Access ArticleGraphene quantum dots for biosensing and bioimaging
P. Abdul Rasheed *ab,
Menon Ankitha
*ab,
Menon Ankitha b,
Vijayamohanan K. Pillai
b,
Vijayamohanan K. Pillai c and
Subbiah Alwarappan
c and
Subbiah Alwarappan *d
*d
aDepartment of Biological Sciences and Engineering, Indian Institute of Technology Palakkad, Palakkad, Kerala 678 557, India. E-mail: abdulrasheed@iitpkd.ac.in
bDepartment of Chemistry, Indian Institute of Technology Palakkad, Palakkad, Kerala 678 557, India
cDepartment of Chemistry, Indian Institute of Science Education and Research, Rami Reddy Nagar, Mangalam, Tirupati, AP 517507, India
dElectrodics & Electrocatalysis Division, CSIR-Central Electrochemical Research Institute, Karaikudi 630003, Tamilnadu, India. E-mail: alwarappan@cecri.res.in
First published on 17th May 2024
Abstract
Graphene Quantum Dots (GQDs) are low dimensional carbon based materials with interesting physical, chemical and biological properties that enable their applications in numerous fields. GQDs possess unique electronic structures that impart special functional attributes such as tunable optical/electrical properties in addition to heteroatom-doping and more importantly a propensity for surface functionalization for applications in biosensing and bioimaging. Herein, we review the recent advancements in the top-down and bottom-up approaches for the synthesis of GQDs. Following this, we present a detailed review of the various surface properties of GQDs and their applications in bioimaging and biosensing. GQDs have been used for fluorescence imaging for visualizing tumours and monitoring the therapeutic responses in addition to magnetic resonance imaging applications. Similarly, the photoluminescence based biosensing applications of GQDs for the detection of hydrogen peroxide, micro RNA, DNA, horse radish peroxidase, heavy metal ions, negatively charged ions, cardiac troponin, etc. are discussed in this review. Finally, we conclude the review with a discussion on future prospects.
1. Introduction
Graphene quantum dots (GQDs) are considered tiny chunks of graphene and are zero-dimensional fragments of bulk carbon materials with properties of both carbon dots and graphene.1 An ideal GQD will have only one atomic layer of carbon atoms although the lateral sizes may be large.2 However, most of the synthesized GQDs have multiple atomic layers with sizes less than 10 nm and also contain functional groups like oxygen and hydrogen.3 GQDs show different unique properties owing to their small size, tunable surface edges, edge effects and quantum confinement effects.4 Since GQDs possess a graphene structure inside the dots, the extraordinary characteristics of graphene are also retained in GQDs.5 Owing to these factors, GQDs possess fascinating optical, electrical, and electrochemical features. Compared to semiconductor QDs, GQDs show better properties in terms of good photoluminescence properties, biocompatibility, high water solubility, easy surface functionalization, high stability, and low toxicity. Hence, GQDs have emerged as a popular material for biosensing and bioimaging applications.6,7The band gap of a material is a fundamental property that determines its electronic behavior. In bulk graphene, there is no intrinsic band gap, making it a zero-band gap semiconductor or a semimetal. However, when graphene is confined to nanoscale dimensions, such as in the form of GQDs, quantum confinement effects come into play, leading to the emergence of a band gap. The band gap of GQDs depends on several factors, with one of the most significant being the size of the GQD itself. Generally, as the size of the GQD decreases, the band gap increases.8,9 This behavior arises from the quantum confinement effect, which effectively restricts the motion of electrons and holes within the GQD. The shape of the GQD and the specific arrangement of atoms at its edges also influence the band gap. Zigzag-edged GQDs tend to have larger band gaps compared to armchair-edged GQDs.10,11 Introducing functional groups or doping the GQD with foreign atoms can also alter the band gap. For example, nitrogen-doped GQDs tend to have smaller band gaps compared to undoped GQDs. The interaction between individual GQDs can also influence their electronic properties. Stacking or aggregating GQDs can lead to interfacial states and affect the effective band gap.12,13 Functionalized GQDs can be integrated into materials, impacting their electronic band structure and properties. This can enhance their mechanical, thermal, and electrical performance, contributing to the development of advanced nanocomposites.14,15 Functionalization with biocompatible molecules or polymers can modify the band gap of GQDs, ensuring they are suitable for applications like drug delivery, imaging, and therapy. This allows for precise control over the electronic properties for specific biomedical tasks.16 Surface functionalization can introduce specific chemical recognition sites on the GQD surface, influencing the band gap and allowing for highly sensitive and selective detection of target molecules. This is vital for applications like environmental monitoring, healthcare diagnostics, and food safety. In summary, surface functionalization provides a powerful means to tailor the band gap of GQDs, enabling their customization for specific applications across various scientific and technological domains. This control over the electronic properties is instrumental in harnessing the full potential of GQDs in diverse fields of research and industry.17,18
2. Synthesis methods
A number of methods have been established for the synthesis of GQDs using either top-down synthesis approaches or bottom-up synthesis approaches.4,19,20 In top-down approaches, GQDs are produced from a larger carbon structure by various important methods such as hydrothermal/solvothermal synthesis,20,21 electrochemical oxidation and exfoliation,22,23 Liquid phase exfoliation,24–26 and oxidative cleavage or oxidation cutting.27 While the bottom-up approach produces GQDs from molecular precursors under specific experimental conditions and the methods comprise chemical synthesis methods,28 pyrolysis,29 microwave assisted chemical routes.30 Fig. 1 shows the schematic diagram representing the top-down and bottom-up approaches for the synthesis of GQDs. It has already been established that the optimum reaction time and an effective precursor are essential to get GQDs having exceptional luminescent properties with a maximum quantum yield and high photo-stability.31 The synthesized GQDs were characterized by typical physiochemical methods to understand their morphology, chemical compositions, structural defects, presence of functional groups, optoelectronic properties, etc.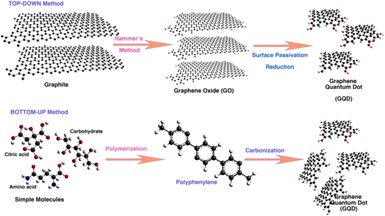 | ||
| Fig. 1 Schematic diagram representing the top-down and bottom-up approaches for the synthesis of GQDs. Reprinted with permission from ref. 6. Copyright @ 2022, MDPI. CCBY 4.0. | ||
2.1. Top-down synthesis methods
A simple facile hydrothermal method has been developed by Tian et al. for the synthesis of GQDs from GO with variable photoluminescence by reacting with hydrogen peroxide.34 Just by varying the time of reaction, the particle size of GQDS could be controlled to lead to variation in photoluminescence. Another method uses one pot hydrothermal reaction between GO and polyethylene glycol (PEG) which results in GQDs with surface passivated by PEG.35 This GQD-PEG showed excellent luminescent properties with high quantum yield (28%) and higher photocurrent generation capability in addition to its upconversion photoluminescence. Li et al. reported a facile hydrothermal approach for cutting preoxidized N-doped graphene into N-doped GQDs which exhibits excellent blue photoluminescence and upconversion photoluminescent properties.36 Similarly, GQDs have been synthesized from a green precursor corn powder37 and rice husk biomass38 as well through hydrothermal methods.
Recently, Facure et al. optimized the concentration, pH and temperature of the synthesis in the hydrothermal synthesis of GQDs to control the structure and luminescence.39 They found that the best synthesis condition was at 2 mg mL−1, pH 8 and 175 °C which produced GQDs with high quantum yield (8.9%) and without multiple steps or use of dopant. By changing the conditions, the amount of specific oxygen functionalities on to the surface of GQDs can be tailored which impacts the luminescent properties.
Solvothermal process is another method for the synthesis of GQDS which uses organic solvents such as dimethyl sulfoxide (DMSO), N,N-dimethylformamide (DMF) or benzene. A solvothermal method for the preparation of GQDs by hydrogen peroxide was introduced by Tian et al.30 They started with expended graphite after heating expandable graphite at 800 °C for 10 s followed by mixing with DMF by ultrasonication for 5 min. Then hydrogen peroxide was added to this reaction mixture and stirred to get homogenous suspension. This was followed by transferring to an autoclave and heating at 170 °C for 5 h to obtain GQDs after vacuum filtration. Shin et al. synthesized GQDs by acid free and oxone oxidant assisted solvothermal synthesis methods for different carbon materials as resources.40 This method uses oxone (potassium monopersulfate) as acid free oxidant and it generates sulfate radicals and hydroxyl radicals which are responsible for the formation of GQDs through a combination of radical oxidation and solvothermal reduction. A solvothermal synthesis method was used to tune the size and surface state of GQDs and was introduced by Qi et al.41 They have made two different series of GQDs with particle sizes of 2.6 to 4.5 nm; one with the same size and different surface chemistry and another with same size with different oxygen degrees. Liu et al. reported a biocompatible nitrogen-doped GQDs (N-GQDs) synthesized by a facile solvothermal method using DMF as a solvent and as a nitrogen source.42 They have used this N-GQDs as efficient two-photon fluorescent probes for cellular and deep-tissue imaging considering its good biocompatibility and photostability.
Li et al. synthesized GQDs by a facile, reliable, direct and reproducible electrochemical approach in which the produced GQDs of uniform size (3–5 nm) exhibited a green luminescence.45 They used phosphate buffer as electrolyte, graphene film as the working electrode, Pt wire as the counter electrode and Ag/AgCl as reference electrodes and the pH dependence of the luminescence was obvious. The GQDs were soluble in aqueous media due to O-containing groups on the surface which also facilitates further functionalization towards various applications. The same group replaced phosphate buffer with acetonitrile containing tetrabutylammonium perchlorate as the electrolyte, resulting in nitrogen doped GQDs (N-GQDs) with oxygen rich functional groups.46 These GQDS were of the size of 2–5 nm with blue luminescence and exhibited electrocatalytic activity towards oxygen reduction reaction (ORR). Shinde et al. electrochemically synthesized GQDs form multiwalled carbon nanotube (MWCNT) by a two-step electrochemical oxidation with propylene carbonate containing LiClO4 as the electrolyte.47 In this method, the sizes of the GQDs can be controlled by changing the temperature and electrolytic process time. This is the first report of an electrochemical approach to synthesize size-tunable GQDs from MWCNTs at room temperature in non-aqueous medium without any capping agents. Considering the intrinsic luminescence behavior, enhanced fluorescence quantum yield and higher photostability, these GQDs are specifically useful for cellular and molecular imaging applications. In another work, GQDs were synthesized by a simple electrochemical unzipping of MWCNTs with good control over size and shape of the GQDs. The synthesized GQDs were modified with PEG to reduce the cytotoxicity and it has been used for bioimaging and drug delivery applications.48 Huang et al. used ammonia solution (weak electrolyte) as electrolyte to prepare controllable and efficient method for the synthesis of GQDs with high quantum yield.49 Here, the circular graphene paper was used as anode, Pt sheet as cathode and ammonia solution acted as nitrogen source. Fig. 2 shows the electrolytic process of graphene paper in ammonia and in NaOH under different reaction times. The electrochemical process operated at 30 V for 2 h to produce GQDs of 3–8 nm size with quantum yield of 28% and better crystallinity. They also found that the other weak electrolytes such as HF and H2S can be used to synthesize other types of GQDs. In another work, Chen et al. reported a facile electrolyzing method to synthesize boron doped GQDs which is used as a fluorescent probe to detect Fe3+ ion levels from water samples.50 Here, a constant potential electrolysis method was used in a two electrode cell in which graphite rod as anode and Pt sheet as cathode.
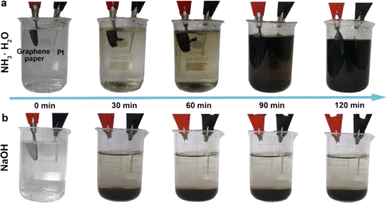 | ||
| Fig. 2 The electrolytic process of graphene paper in (a) ammonia and (b) NaOH under different reaction times. Reprinted with permission from ref. 49. Copyright © 2018, The American Chemical Society. | ||
The GQDs produced by the electrochemical oxidation method show good stability, however, the pretreatment and the purification of GQDs products take a longer time. Additionally, the mass production is very difficult since the product yield is very low.
Lu et al. reported a fast and industrial promising method for large scale synthesis of GQDs through an ultrasonic-assisted liquid-phase exfoliation technique.26 They demonstrated GQDs of different sizes with edge structures and defect distribution can be made by using precursors as low defect nano graphite and defect rich acetylene black respectively. The photoluminescent properties of the synthesized GQDs enable them to be used as a promising fluorescent probe in cell imaging. Liu et al. synthesized pristine GQDs by shaking graphite nanoparticles (GNPs) in ethanol/H2O mixture on a vortex mixer for 1 h followed by centrifugation.53 They used GNPs with 4 nm diameter to synthesize monolayer GQDs with circular shapes and diameter of less than 4 nm. The homogeneity of GQDs in terms of size and shape is improved with blue photoluminescence.
Other than graphene or GO, fullerenes, and carbon fibers can also be used as a precursor to synthesize GQDs. Fullerene was used to synthesize very small GQDs of size 2–3 nm by Pumera et al. by mixing a strong acid and a chemical oxidant (H2SO4 and KMnO4).55 This induces the oxidation, and cage opening of fullerene and then fragmentation process of fullerene to GQDs. The synthesized GQDs remained good aqueous dispersion and exhibited strong luminescence properties. Similarly, carbon fiber was used as precursor for the synthesis of GQDs by H2SO4/HNO3 at high temperature.56 However, this carbon fiber derived GQDs show a narrower size distribution of 1–4 nm and the size of GQDs vary strongly with both reaction temperature and the photoluminescence. Owing to the luminescence stability, biocompatibility, low toxicity, and high solubility, these GQDs can be used as bioimaging and biosensing applications. There are other reports where the GQDs were synthesized from CNTs57 and black carbon58 as well. Abbas et al. introduced a synthesis approach involving oxidative cutting of carbon precursor derived from pyrolysis of biomass waste.59 The GQDs were synthesized in as short time of 30 min and exhibited excellent optical properties and high product yield of ∼84%. Additionally, these GQDs were used for selective and sensitive detection of Fe3+ ions after a simple hydrothermal treatment.
2.2. Bottom-up synthesis methods
Yan et al. controlled the redox potential and bandgap of GQDs by the surface functionalization of GQDs by electron-withdrawing or electron donating groups.9 The main advantage of this wet chemistry route is that it can be used to synthesize a variety of functionalized and well-ordered GQDs. However, synthesizing and purifying larger GQDs with these methods is quite challenging.62 In another work, solution chemistry approach was used to synthesize nitrogen-doped colloidal GQDs with well-defined structures by Li et al. and this material has been used for catalysing the oxygen reduction reaction (ORR).63 They found that N-doping significantly affects the properties of GQDs such as size-dependent electrocatalytic activity for ORR. Similarly, sulfur doped GQDs (S-GQDs) with bright blue emission have been synthesized by a facile one pot hydrothermal treatment.64 They used 1,3,6-trinitropyrene as carbon source and 3-mercaptopropionic acid as sulfur source as well as for carboxyl group modification. Accordingly, S-GQDs were similar to single layer graphene structure of mean size of 2.5 nm. The sulfur doping enhances their electronic and chemical properties, and it can be used as sensing probe for Ag+ ions. The limit of detection of the sensor was found to be 30 nM with a wide linear range of 0.1–130 μM. Zhou et al. reported a simple and effective approach for the synthesis of GQDs by using polycyclic aromatic hydrocarbons (PAHs) as the precursors.65 The obtained GQDs were of sizes between 5–10 nm and exhibited excellent water solubility as well as tunable fluorescence. They have also used these GQDs for bioimaging applications as well as effective sensing for Fe3+ and hydrogen peroxide.
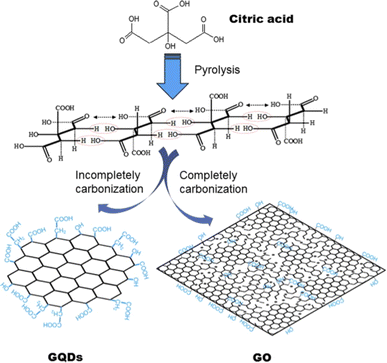 | ||
| Fig. 3 Schematics showing the Pyrolysis technique for producing GQDs and GO by using citric acid as raw material. Reprinted with permission from ref. 66. Copyright © Elsevier, 2012. | ||
In a slightly modified synthesis, Tang et al. made monodispersed GQDs (2.9 to 3.9 nm diameter) by pyrolyzing glucose through combining hydrothermal and microwave techniques.67 The synthesized GQDs displays the shortest emission wavelength (deep ultraviolet (DUV) emission of 4.1 eV) compared to all the solution-based QDs. The same research group made another glucose derived GQDs through pyrolysis by a soft template microwave assisted hydrothermal method.68 They synthesized monodispersed GQDs of diameter below 5 nm with high quantum yield of 15% which is the highest among all other carbon based additive free QDs.
Naik et al. reported a single-step synthesis of GQDs using pyrolysis of citric acid which yield different sized GQDs at different pH.28 Here, the pyrolysis of citric acid was carried out in which hydronium ion formed by the decomposition of the acid and it acts as a catalyst in subsequent decomposition reaction stages. In another work, blue fluorescent nitrogen-doped GQDs (N-GQDs) were synthesized by a hydrothermal method through the pyrolysis of citric acid in presence of urea as the nitrogen source.69 They used the N-GQDs as an efficient drug delivery system after conjugating the anticancer drug, methotrexate.
A single layer GQDs with green photoluminescence has been synthesized by using glucose as a precursor through hydrothermal treatment.70 They dispersed glucose in DI water and heated at 200 °C for 8 h to get single-layered GQDs. This synthesis method needs high temperatures need to disperse the pyrolyzed products in alkaline media.
Zheo et al. reported a microwave assisted method for synthesizing green fluorescent GQDs derived from deoiled asphalt.73 The as prepared GQDs show a size distribution of 1–6 nm, and displays green luminescence as well as high quantum yield up to 14%. In another report, Kumawat et al. synthesized red-luminescent GQDs from ethanol extracts obtained from Mangifera indicia (mango) leaves by one-pot microwave assisted synthesis method.74 The size distribution of the synthesized GQDs ranges from 2 to 8 nm and the fluorescence emission in the near infrared (NIR) region, hence it can be used as NIR responsive fluorescent bioimaging probes.
GQDs with good biocompatibility and capability for sensing and in vivo bioimaging has been synthesized by Campbell et al. using glucosamine–HCl solution as a carbon source and different dopant precursors.75 They have made N-GQDs, sulfur doped GQDs (S-GQDs) and boron/nitrogen doped GQDs (BN-GQDs) by using sulfur thiourea or benezeneboronic acid as doping agents. The solution mixture was exposed to microwave treatment for 40 min and QDs with quantum yield of 15–20% were obtained. These GQDs have great potential in drug delivery, pH-sensing of cancerous environments, and multicolor visible/NIR fluorescence imaging. Table 1 shows the comparison of different methods for the synthesis of GDs with merits and demerits of each methods.
| Synthesis technique | Process | Size range | Quantum yield | Merits | Demerits | Ref. | |
|---|---|---|---|---|---|---|---|
| Top-down synthesis | Hydrothermal/solvothermal synthesis | Carbon material is transformed into GQDs under high temperature and pressure | As small as 1.5 nm | 5–28% | Ecofriendly, non-toxic, inexpensive | Less control over size | 32 and 33 |
| Electrochemical oxidation and exfoliation | Carbon material are oxidatively cleaved into the GQDs under redox voltage | 2–8 nm | Up to 28% | Less time-consuming and does not require strong chemicals, good stability | Product yield is very low | 44, 45 and 48 | |
| Liquid phase exfoliation method | Carbon material is exfoliated into GQDs | Less than 100 nm | 1.8–20% | GQDs of different sizes with edge structures, facile, inexpensive and scalable | Need for organic solvent | 26 and 53 | |
| Oxidative cleavage method | The carbon–carbon bonds of graphene GO or CNT are chemically broken by oxidizers | 1–19 nm | 4.3–23% | Facile and simple method, no post processing steps | Use of strong oxidizers | 57–59 | |
| Bottom-up synthesis | Chemical synthesis | Step wise chemical synthesis | 2.5–5 nm | — | Uniform size distribution, synthesize in large quantities | Chance of aggregation of GQDs | 9 and 61–63 |
| Pyrolysis | Thermal decomposition of raw materials at high temperatures | 1–8 nm | 9–15% | Rapid and one pot synthesis | Reaction occurs at high temperature and need to disperse the pyrolyzed products in alkaline media | 67 and 68 | |
| Microwave assisted chemical routes | Microwave assisted pyrolysis | 1.8–8 nm | 11.7–22.9% | Fast, inexpensive, non-toxic | Poor control over sizes | 73 and 74 | |
3. Properties of GQDs
3.1. Physical properties of GQDs
The nanoparticles that exhibit quantum confinement along all the three axes of direction are termed as quantum dots. Therefore, GQDs are supposed to be such 0-dimensional particles which are chemically composed of graphene. Upon the conversion to a 0D material, if the size of the GQDs are comparatively smaller than the Bohr radius of the exciton it leads to electron redistribution and eventually quantum confinement.76 The presence of sp2 hybridized lattice gives high crystallinity to the carbon structure.77,78 In comparison to normal carbon dots (CDs) the GQDs are superior in its anisotropic character in addition to the layered morphology that owes to a “molecular feature”. On the other hand, CDs are quasi-spherical sizes which possess a typical size less than 10 nm imparting a colloidal character due to less sp2 hybridized carbon atoms in the lattice.78–80 The carbon atoms in the GQDs are arranged in a hexagonal manner with honey comb like crystal lattice. Each carbon is bonded to three other carbon atoms with π electrons delocalized over the sheet in a perpendicular direction. This electron cloud above and below the plane of carbon atoms imparts excellent optical and electrical properties.81The edges of the GQDs are popularly known as armchair and zig zag, as seen in Fig. 4. The former has structures with triple at the ends similar to carbynes while the latter has a pair of electrons along the edges like carbene systems.82–84 The types of edges are one of the factors contributing to the shape of the GQDs. As seen from the Fig. 4, when same type of edges has encountered the angle of formation is 120° giving rise to hexagonal shape. While the combination of these two types of edges makes an angle of 90° at the corners of the GQDs leading to the formation of rectangular structures. There are GQDs with spherical and oval shapes as well depending on their synthetic routes that result from undeveloped corners or borders.84 The edge types affect the electronic, electrochemical and optical properties of GQDs as it effects the shape of the GQDs. Along edge types, their size distribution also contributes for physical and chemical properties. For example, the band gap of GQDs is dependent on their surface states and shapes despite graphene being a material with zero-band gap. Typically, the size of GQDs is inversely proportional to its bandgap.83,85
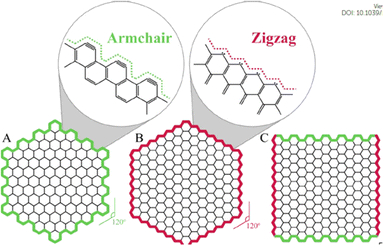 | ||
| Fig. 4 The shapes of graphene quantum dots depending on the type of edge: (A) armchair, (B) zigzag, and (C) hybrid armchair zigzag GQDs. The amplified images in (A) and (B) show structure details of the armchair and zigzag edges, respectively. Reproduced with permission from ref. 82. Copyright © Elsevier 2020. | ||
The structure of GQDs is analyzed using various characteristic techniques. One among the preliminary technique to confirm the formation of GQDs is the UV-vis spectroscopy. Using this, the band corresponding to the π to π* transition of aromatic C![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) C atoms is observed around 230–270 nm. Another should peak around 290–320 nm is also observed owing to the n-π* transition that could arise due to the surface oxidized C
C atoms is observed around 230–270 nm. Another should peak around 290–320 nm is also observed owing to the n-π* transition that could arise due to the surface oxidized C![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) O bonds on GQDs.80,82 The presence of oxygen functionalities can also be investigated through Fourier transform infra-red (FTIR) spectroscopy. Usually, the graphene oxide quantum dotes (GOQDs) are chemically treated using reducing agents to remove the surface oxygen groups that will remove the shoulder peak in the UV-vis spectra and also exhibit visible changes in the IR spectra as well.80 The degree of crystallinity of the synthesized GQDs can be investigated using Raman spectroscopy and X-ray diffraction analysis (XRD) techniques. In Raman spectrum, the characteristic D and G bands appear at 1350 cm−1 and 1590 cm−1 respectively. The ratio of the corresponding band intensities ID/IG ratio determines the extend of crystallinity. A material with lower ratio attributes to the high crystallinity of the lattice.86,87
O bonds on GQDs.80,82 The presence of oxygen functionalities can also be investigated through Fourier transform infra-red (FTIR) spectroscopy. Usually, the graphene oxide quantum dotes (GOQDs) are chemically treated using reducing agents to remove the surface oxygen groups that will remove the shoulder peak in the UV-vis spectra and also exhibit visible changes in the IR spectra as well.80 The degree of crystallinity of the synthesized GQDs can be investigated using Raman spectroscopy and X-ray diffraction analysis (XRD) techniques. In Raman spectrum, the characteristic D and G bands appear at 1350 cm−1 and 1590 cm−1 respectively. The ratio of the corresponding band intensities ID/IG ratio determines the extend of crystallinity. A material with lower ratio attributes to the high crystallinity of the lattice.86,87
From the XRD analysis a peak around 25° corresponds to (002) plane which can undergo shifts in accordance with the surface terminations of GQDs. The d-spacing of the GQDs are reported to be similar to that of graphite approximated to be 0.34 nm which can be further confirmed with the interlayer spacing reflected in the High resolution-transmission electron microscopy (HR-TEM) fringes.88,89 The TEM results also provide an idea of the average size, its distribution and crystallinity of the material.83,87 Atomic force microscopy (AFM) also reveals the shape, size and topography of GQDs. As AFM enables to investigate the number of layers present in GQDs by measuring the height of the sample, it enables us to differentiate between single-layered, few layered and multi layered GQDs.70,86
3.2. Optical properties
The optical properties of GQDs have been thoroughly investigated with the aid of photoluminescence spectroscopy and absorption in UV and visible wavelengths of light.90–92 Interestingly, GQDs exhibit photoluminescence properties which is absent in pristine graphene. Generally, GQDs show an absorption band in the deep UV region and often in UV-Visible regions as seen in Section 3.1. However, with changes in the surface morphology and its nature they exhibit unique absorption and luminescence feature. Peaks with relatively shorter wavelength is attributed to the π to π* transition and the ones with higher wavelengths arises due to less energetic n to π* transition.93,94 Various factors can tune the position of these absorption bands which include its shape, size, solvent, temperature, surface functionalization, physical and chemical bonds.92,95–98 This enables various strategies to be incorporated to tune the optical properties of GQDs with additional advantages of imparting low toxicity and biocompatibility. The alterations in the synthetic route employed leads to the preparation of GQDs with wide range of sizes, shape, edge configurations and many more which eventually led to changes in the absorption peaks in photoluminescence spectra. The superior photostability of GQDs in comparison with other quantum dots makes it a capable for wide range of applications.79,99,100 In GQDs, the phenomenon of photoluminescence is governed by variety of factors viz. extend of conjugation, shape and size, pH, degree of surface functionalization and extend of oxidation.101,102 The intensity of fluorescence is measured using quantum yield (QY) whose values are generally low for GQDs.103Peng et al. studied the effect of tuning the size and its effect on the emission wavelengths in photoluminescence spectra. By controlling the temperature during the synthesis, they prepared GQDs in three different ranges of size; 1–4 nm, 4–8 nm and 7–11 nm. With change in size of the GQDs, the energy gap decreased eventually and exhibited photoluminescence color from blue to green to yellow.101 The photoluminescence emission also depends on the layer dependent size of GQDs. On comparing the single layered and multilayered GQDs synthesized from a carbon black under acidic conditions, the latter showed broader absorption bands than former. The average height of single layered GQDs were 0.5 nm showed yellow photoluminescence emission while multilayered GQDs consisting of 2–6 layers of height 1–3 nm appeared green.104 Tailoring the surface chemistry to tune the optical properties by introducing various reactive sites such as carboxyl (–COOH), epoxy (–C–O–C–), carbonyl (–C![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) O) facilitates the formation of “surface oxidation states”. Tetsuka et al. developed GQDs with amine functionalization of oxidized graphene sheets to yield quantum dots of tunable optical properties.96 The oxidized graphene oxide sheets had epoxy groups which were made to treat with ammonia solution 70–150 °C to form primary amines on the surface. This was followed by annealing at 100 °C to give amine functionalized GQDs that exhibited a wide range of photoluminescence emission under a single excitation wavelength with a quantum yield of 40%. Similarly, Shen et al. prepared GQDs with polyethylene glycol diamine to prepare particles that imparted strong photoluminescence color in blue wavelength regime with a quantum yield of 7.4%.27 The aromatic carbon atoms along the edges of the GQDs also serve as active centres of functionalization. This was studied by Luo et al. where GQDs where modified by aryl groups and its substituted counterparts.105 The photoluminescence emissions of these GQDs were tailored in the range of 418 nm to 447 nm with increase in the quantum by 6 fold attributed due to the resonance between graphene planes and the aryl groups.
O) facilitates the formation of “surface oxidation states”. Tetsuka et al. developed GQDs with amine functionalization of oxidized graphene sheets to yield quantum dots of tunable optical properties.96 The oxidized graphene oxide sheets had epoxy groups which were made to treat with ammonia solution 70–150 °C to form primary amines on the surface. This was followed by annealing at 100 °C to give amine functionalized GQDs that exhibited a wide range of photoluminescence emission under a single excitation wavelength with a quantum yield of 40%. Similarly, Shen et al. prepared GQDs with polyethylene glycol diamine to prepare particles that imparted strong photoluminescence color in blue wavelength regime with a quantum yield of 7.4%.27 The aromatic carbon atoms along the edges of the GQDs also serve as active centres of functionalization. This was studied by Luo et al. where GQDs where modified by aryl groups and its substituted counterparts.105 The photoluminescence emissions of these GQDs were tailored in the range of 418 nm to 447 nm with increase in the quantum by 6 fold attributed due to the resonance between graphene planes and the aryl groups.
The introduction of hetero atoms creates distortions at the electronic and structural level of GQDs.106 Various dopants with tailored size and valency have been explored including doping with nitrogen,107–110 boron,108,111–114 phosphorous,115,116 fluorine,115,117,118 chlorine,119 sulfur120,121 etc. Combining the dopants promotes the coupling effect of heteroatoms and GQDs allowing the synergistic modulation of optical properties.121–124 Xia et al. prepared N, S co-doped GQDs using citric acid as the carbon source and L-cysteine as the dopant. The N, S co-doped GQDS exhibited yellow-green photoluminescence emission (500 nm) while the N, S free GQDs exhibited blue color (417 nm) as depicted in Fig. 5.
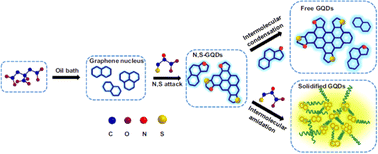 | ||
| Fig. 5 Synthetic strategy for the preparation of N–S co-doped GQDs. Reproduced with permission from ref. 122. Copyright @ Elsevier, 2017. | ||
3.3. Magnetic properties
The presence of electrons in d and f orbitals leads to magnetic properties in many materials, whereas in GQDs their magnetic behavior arises out of their spin polarization along a specific conformation. The electronic states of GQDs are spin-polarized edge states along the zigzag conformations.125 Additionally, the introduction of heteroatoms may also impart magnetic moment within the lattice of GQDs. Theoretical studies convey the importance of geometry for the magnetic nature of GQDs. It states the switching off of the spin polarization occurs if defects are present at the edge sites of the GQDs.126 Experimentally GQDs with low defect were found to be paramagnetic at lower temperatures. The defects at the edge sites do reduce the intrinsic value of magnetic moment.127 However, the incorporation of heteroatoms such as boron114 or fluorine118 exhibited significant magnetism. These materials are ideal and promising in the field of magnetic resonance imaging (MRI). In a typical study by Yuo et al., bare zigzag sites were distorted to pentagon–heptagon structures (ZZ57) after undergoing annealing (as seen in Fig. 6(a)) which changes the geometry along the edge sites and thus suppresses the magnetic property.127 On the other hand, presence of hydroxyl groups along the edges can preserve the magnetic moment even when annealed at higher temperatures as shown in Fig. 6(b).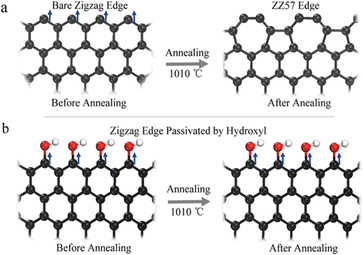 | ||
| Fig. 6 Schematic representation of (a) bare zigzag edge being distorted to edges comprising pentagons and heptagon structures (ZZ57) (b) preserving the edge sites by hydroxyl functionalization of zigzag conformation. Reproduced with permission from ref. 127. Copyright © 2017 Springer Nature Limited. | ||
3.4. Electrical and electrochemical properties
As already seen in the previous sections, the π electron cloud and its delocalization throughout the lattice impart high electrical conductivity to GQDs. These properties of GQDs can be tailored upon surface functionalization. Usually, the preparation of a composite out of GQDs and other materials impairs its inherent conductivity due to a change in its hybridization from sp2 to sp3 carbon atoms.85,128–130 This is evident from one of the works reported by Feng et al. wherein they show the difference in the electronic properties of the material in two different types of modifications.131 One modification is the modification of edges using oxygen containing groups and the other is modifying the basal planes with oxygen functionalities. They observed that the latter showed comparatively poorer conductivity due to the abundant number of sp3 hybrid carbon atoms formed along the basal plane. While such a change in the hybridization only along the edges imparts fewer alterations to the inherent conductivity.Li et al. studied the correlation between the bandgap and electronic structure of GQDs with the aid of computational models.132 The results show that the size of the band gap decreases with the size of GQDs. These values of band gap were found to be smaller for GQDs with zigzag conformation than the ones with armchair edges. The combination of conductivity along with large surface area and active edge sites enables GQDs to acquire good electrochemical activities as well. These are also tailored by changing functional groups along the edges, surfaces or via doping.80,85,128 The small sizes of GQDs help for the small molecules/analytes to interact with GQDs. The key attributes towards the sensing applications are the surface area to volume ratio, active edge sites and fast charge transfer kinetics.85,89 The electrochemical works span over capacitors,133–135 hydrogen evolution reaction (HER),136,137 fuel cells,138,139 ORR140,141 and photovoltaic cells.142,143 The combination of photoluminescence and electrochemical aspects of GQDs helps in the development of electrochemiluminescence sensors144,145 and photo detector devices.146
3.5. Biological properties
Although many carbon-based nanoparticles are toxic due to aggregation, GQDs exhibit great water solubility and offer a non-toxic substitute to nanoparticles composed of metal by removing the fear of toxicity related to metallic residues that cannot be eliminated from human body as GQDs. Examining cytotoxicity is one way to assess biocompatibility. Numerous investigations have demonstrated that GQDs are not harmful, with variations in cytotoxicity brought on by surface changes.147 While GQDs modified with hydroxyl groups have demonstrated substantial cytotoxicity at levels above 100 μg mL−1, NH2, COOH, and CHON(CH3)2 modified GQDs have been proven to exhibit low toxicity at doses up to 200 μg mL−1.148 Before, cellular death and the production of intracellular reactive oxygen species (ROS) were thought to be the mechanisms underlying the cytotoxicity caused by hydroxylated GQDs.149,150 Although research on the in vivo toxicity is still in its infancy, recently studies have amply demonstrated the impact of particular GQDs in (mice) animal models within intravenous injection. The biodistribution and cytotoxicity of carboxyl functionalized GQDs were studied in mouse models; in vivo imaging of the mice and ex vivo organ analyses revealed that the GQDs had started to be eliminated from the body within 12 hours.151 A complete blood count and histopathological analysis of mice treated with GQD and saline revealed no significant differences among both the experimental and the control groups. At high doses of GQDs after 30 days, there were only minor variations in the histological examination of the liver and kidney in both experiments. These in vivo investigations' results that GQDs are not acutely hazardous are encouraging evidence of their biocompatibility.152 The efficacy of these in vivo investigations as complete indications of GQDs' biocompatibility is still under question because, as was shown in in vitro studies, changes to the chemical functions of GQDs might result in appreciable modifications in how they interact with living things.GQDs have been shown to have several notable impacts in cellular settings, including the production of photoinduced ROS and cytotoxicity that follows.153,154 While PDT and other fields can benefit from the potential of GQDs to augment ROS concentration following exposure to incident radiation, it is crucial to address the degree of ROS formation by GQDs upon photoirradiation for practical applications. It was looked at how functional groups, specifically ketone, hydroxyl, and carboxylic groups that contain oxygen, affect the capacity of GQDs to produce ROS. According to the findings, all functional groups that contain oxygen cause an increase in ROS production, with the ketonic group producing the most ROS. While the survival of cells after storage in darkness remained unaffected, a higher rate of ROS production was directly connected to lower cell viability under continuing irradiation. Other studies have shown that no cytotoxicity was seen in cells exposed to GQDs without irradiation. The aforementioned findings show that the ROS produced by irradiating GQDs result in considerable cytotoxicity.155,156
For the biological imaging and biosensing applications of GQDs, the purity of GQDs is very important, which may affect the sensitivity and detection range during the analysis process. The various methods commonly used for the purification and separation of GQDs are size-selective filtration, ultracentrifugation, dialysis–salting out extraction, salting out extraction and acid/base treatment.
Size-selective filtration involves passing the GQD solution through a membrane with specific pore sizes. Since GQDs have a smaller size compared to larger impurities, they can pass through the filter, while larger particles are retained. This technique helps in separating GQDs based on size.157,158 Ultracentrifugation involves spinning the GQD solution at very high speeds. The centrifugal force separates particles based on size and density. Typically, heavier impurities sediment faster than lighter GQDs, enabling their separation.159,160 In dialysis–salting out extraction, GQDs are placed in dialysis bag (1000 Da) in DI water under magnetic stirring. For purification purposes the water is sequentially replaced after every 7 h for required number of days. Thereafter the GQDs are freeze dried and processed for further modifications. Using different pore sizes, GQDs can easily be separated and the effects of size of GQDs on structural and optical parameters can be observed.161,162
The salting out mechanism is based on differences in the dispersibility of the individual GQDs. Usually, ammonium sulfate is used for the salting out process where in, relatively low ammonium sulfate concentrations (10 mM), large, GOs with predominant hydrophobic domains are precipitated. As the salt concentration increases, smaller and less GOs with predominant hydrophobic domains are purified, and eventually graphene quantum dots (GQDs) are isolated. The salting-out process for GOs is simple and scalable, and can be used to achieve large-scale purification.163 Treatment with acids or bases can be used to remove functional groups and impurities from the surface of GQDs. Acid treatment can remove metal impurities, while base treatment can remove functional groups introduced during synthesis. Often, a combination of purification methods is used to achieve higher purity. For example, a combination of size-selective filtration followed by dialysis can effectively remove both size-based impurities and soluble contaminants.164
4. Bioimaging and biosensing applications of GQDs
4.1. Bioimaging applications of GQDs
Fluorescence molecular imaging has evolved as one of the best tools for clinical diagnosis along with therapy management and assessment. A wide variety of fluorescence agents are explored as it allows the sensitive, specific and non-invasive monitoring of physiological events. As discussed in (Section 3.2), GQDs are employed to monitor the therapeutic responses and its distribution in vivo owing to its excellent photostability and versatile photoluminescence properties.165 They are well known for their application as bioimaging agents by visualizing the tumors with targeted drug delivery. These properties in combination with biocompatibility, tunable surface properties and low toxicity makes them promising in the field of bioimaging in the modern era.166,167 The use of bioimaging is crucial as it enables the detailed, isolated observation of biological processes like cellular uptake, targeted delivery, and biodistribution of therapeutics using distinct electromagnetic radiations. GQDs can be employed as optical probes in fluorescence imaging because of their inherent photoluminescence.168 On the other hand, other nanoparticle platforms require extra conjugation of fluorescent dyes. Recent studies show the ability of GQDs to be employed for NIR imaging and magnetic resonance (MR) imaging.165 These unique properties motivated the researchers to use GQDs for cancer cell imaging such as HeLa cells bioimaging,169–171 MG-63 cells,172 MCF-7 cells,173,174 and so on.This fluorescent imaging method uses visible and near-IR radiations to visually examine the distribution of different analyte molecules in animal body cells and tissues.175–177 The versatility with which GQDs can be modified enables them to be employed as the imaging probes for cell nuclei. By solvothermal method, nitrogen doped and chlorine doped amphiphilic GQDs were synthesized. The modified structures showed a highly positive zeta potentials that enabled them to cross the cell membranes.178 This also helped them to directly bind to the histones present inside the cell nuclei. Studies showed that GQDs modified with 4′,6-diamidino-2-phenylindole exhibited targeting of the nuclei without the requirement of any conjugated ligands.179 As they are proved to be minimally cytotoxic these structures are facile candidates as stable fluorescent agents for in vitro imaging. As these zero-dimensional structures are highly soluble in water they enter the cells without the help of additional targeting molecules. Tailoring the photoluminescence can help in combination with other fluorescent probes for analyzing the processes within the cell.
Wang et al. synthesized GQDs that are co-functionalized with amine and sulphur for two-photon microscopy.180 The hydrothermal synthesis was carried by mixing 1,3,6-trinitropyrene with aqueous solution of ammonium sulfide and subjected to a temperature of 200° for 12 hours. The as prepared GQDs showed blue fluorescence. For two-photon confocal imaging, HeLa cells were treated with GQDs and subjected to a laser beam of 800 nm wavelength. Images of the treated cells showed GQD uptake by the cells as well as intense GQD fluorescence, demonstrating the suitability of these GQDs for long-term two-photon fluorescence microscopy. Similarly, GQDs that are doped with nitrogen (N-GQD) were prepared by treating dimethyl formaldehyde and graphene oxide.42 Two-photon-induced fluorescence of N-GQD was systematically investigated and utilized for efficient two-photon cellular and deep-tissue imaging. Fig. 7(a) shows the photoluminescence emission spectra of one-photon fluorescence (OPF) and two-photon fluorescence (TPF) of N-GQDs. The good biocompatibility and extraordinary photostability of N-GQD helps in long-term two-photon-fluorescence imaging of biological tissues. The as synthesized GQDs showed blue-green fluorescence and fluorescence images shows its penetration into the cytoplasm but not into the nuclei of HeLa cells (Fig. 7(b and c)).
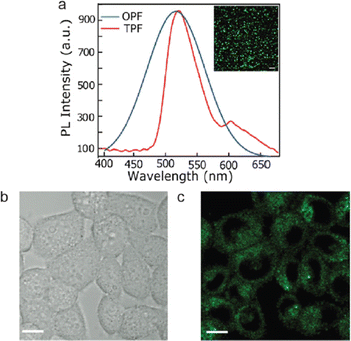 | ||
| Fig. 7 N-GQDs for cell and tissue imaging. (a) Photoluminescence emission spectra of one-photon fluorescence (OPF) and two-photon fluorescence (TPF). (b) Bright field and (c) fluorescence images under 800 nm excitation of HeLa cells (scale bar = 10 μm). Reprinted with permission from ref. 42. Copyright © 2013, American Chemical Society. | ||
Recently, Wang et al. created N–B-GQDs for NIR-II bioimaging, which were GQDs doped with nitrogen and boron (N–B-GQDs).181 These N–B-GQDs were made using a 3-aminophenylboronic acid monohydrate. This was pyrolyzed in one pot at 230 °C in an acetone and hydrogen peroxide solution, producing extremely monodisperse 5 nm-diameter particles containing two or three layers of graphene per particle. The N–B-GQDs displayed NIR-II photoluminescence emission in the 950–1100 nm range when exposed to an 808 nm excitation source. According to the author's hypothesis, the N–B-GQDs' nitrogen and boron doping produced a sizable number of vacancy defects, which caused the photoluminescence emission peak to shift into the NIR-II window. The authors first demonstrated that N–B-GQDs are noncytotoxic in three different cell lines, and then investigated the in vivo photoluminescence abilities of their NIR-II fluorophores by taking a series of photoluminescence images beginning one minute after intravenous tail-vein injection of N–B-GQDs in nude athymic mice over duration of 120 minutes (Fig. 8).
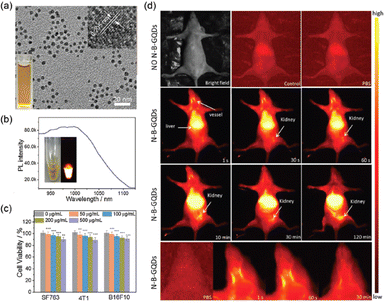 | ||
| Fig. 8 Nitrogen and boron dual-doped GQDs for NIR-II bioimaging. (a) TEM image showing the monodisperse particles (b) photoluminescence spectrum of N–B-GQDs exhibiting NIR-II emission when excited with an 808 nm laser source. The insets display an optical image an a photoluminescence image of N–B-GQDs in aqueous solution. (c) In vitro cytotoxicity study of N–B-GQDs performed by assessing the viability of SF763, 4T1, and B16F10 cells 72 h after incubation with N–B-GQDs. (d) In vivo NIR-II imaging of live mice. Reprinted with permission from ref. 181. Copyright @ Elsevier 2019. | ||
While optical imaging techniques largely use in vitro imaging to demonstrate a fluorescent probe's capabilities, in vivo imaging is challenging due to the need to take into consideration additional factors like in vivo biocompatibility, biotoxicity, and the dynamic metabolism of the organism that are being investigated. The sheer number of studies using GQDs for in vivo applications like cell tracking and nuclear targeting may be a sign of recent developments in GQD research. Using two-photon fluorescence microscopy, GQDs made from graphite were utilized to monitor human adipose-derived stem cells (hADSCs) in a mouse model.182 The two-photon photoluminescence spectra revealed emission wavelengths of 400–520 nm with excitation wavelengths of 680–860 nm with no detectable cytotoxicity. Athymic mice's dorsal area was where the hADSCs were administered after being given GQD treatment. Fluorescence signal was still visible after 24 hours after exposure to 670 nm radiation. In vivo, GQDs showed no substantial toxicity or insolubility in aqueous settings and retained their photoluminescence property without considerable photobleaching. Therefore, rather than using traditional organic dyes, GQD-based systems could be employed as fluorescence probes in cell tracking.
To obtain anatomical images and track physiological processes, MR imaging uses radio frequency signals. Due to its noninvasive nature, high spatial resolution, and practically limitless tissue penetration depth, MR imaging is a popular technique for clinical imaging. The biological traits being studied can be highlighted by contrast agents by either making them brighter (T1 contrast agents) or darker (T2 contrast agents). While safe superparamagnetic iron oxide nanoparticles have enabled advancements in T2 contrast agents, T1 contrast agents are typically restricted to transition metal ion chelates, particularly those based on gadolinium (Gd). However, it is widely recognized that transition metals are hazardous to the body.183 By creating local paramagnetic moments around the dopants, doped graphene nanoparticles with different atoms causes the creation or improvement of magnetic characteristics. B-GQDs, or boron-doped GQDs, were developed using a hydrothermal process. Here, the B-GQDs were assessed as T1 contrast agents both in vitro and in vivo, with the boron-dopants acting as the paramagnetic centers. B-GQDs in aqueous solution had an in vitro longitudinal MR relaxation time that was equivalent to clinical Gd-based contrast agents. Additionally, mice were administered B-GQDs by intravenous injection to test the drug's MR imaging capabilities. When compared to the pre-injection signal, elevated T1 signal intensities were seen in the mice's hearts, stomachs, and kidneys. These developments have demonstrated the development of metal-free, graphene-based MR imaging CAs.184 Fig. 9 shows the in vivo T1-weighted MR images of abdominal cross-sections of mice treated with B-GQDs with dynamic time-resolved MR imaging acquired at various time points after intravenous injection. Table 2 shows a comparison table of bioimaging applications of GQDs in in vitro and in vivo imaging as well as MR imaging.
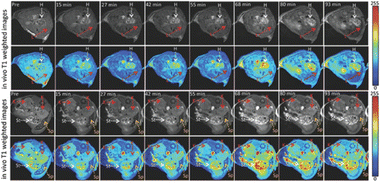 | ||
| Fig. 9 In vivo T1-weighted MR images of abdominal cross-sections of mice treated with boron-doped GQDs with dynamic time-resolved MR imaging acquired at various time points after intravenous injection. The arrows indicate various organs: heart (H), liver (L), kidneys (K), spleen (Sp), and stomach (St). The heart and stomach show the greatest contrast 68 min after administration. Reproduced with permission from.184 Copyright © The Royal Society of Chemistry 2020. | ||
| Material | Size (nm) | PL colour | Excitation (nm) | Quantum yield | PL sensitivity | Ref. |
|---|---|---|---|---|---|---|
| GQD-amine | 7.5 | Green | 500 | 1.5, 4.4 | Excitation | 188 |
| GQD-PEG | 15 | Green | 400 | 18.8 | Excitation | 189 |
| GQD-RGD | 3.7 | Yellow green | 460 | — | Excitation | 190 |
| GQD-HA-HAS | 5 | Green | 430, 540 | 14 | Excitation | 191 |
| GQD-Cyc-HCl | 5 | Green | 400–500 | 58 | Excitation | 192 |
| Edge-functionalized GQDs with N and Cl ligands | 1–5 | Green | 450 | 30 | Excitation | 193 |
| N–B-GQDs | 5 | Green | 368 | 1 | Excitation | 181 |
| SL-BGQD | 4.6 ± 1.4 | Blue | 650 | — | Excitation | 184 |
Other than fluorescent imaging applications, GQDS can be utilized in photoacoustic imaging and radionuclide imaging. In photoacoustic imaging using quantum dots (QDs), the QDs serve as exogenous contrast agents that can be selectively accumulated in target tissues or cells through various targeting strategies. These QDs are typically surface-functionalized to improve their biocompatibility, stability, and targeting specificity. Once accumulated in the target region, the QDs absorb laser light and efficiently convert it into heat, resulting in localized thermoelastic expansion and the generation of photoacoustic signals. These signals are then detected by ultrasound transducers and reconstructed to form high-resolution images of the target tissue or cells. Several advantages make QDs attractive contrast agents for photoacoustic imaging. Their broad absorption spectra, which can be tuned by controlling their size, composition, and surface properties, allow for multi-wavelength imaging and depth penetration into biological tissues. Additionally, their high photostability enables long-term imaging with minimal signal degradation over time. Furthermore, the ability to functionalize QDs with targeting ligands or biomolecules enhances their specificity for imaging specific molecular targets or cellular markers, making them valuable tools for both preclinical research and clinical diagnostics. For e.g. Xuan et al. synthesised N-GQD with photoacoustic properties by a hydrothermal method using citric acid as the carbon source.185 The synthesised N-GQDs exhibited visible light fluorescence and NIR fluorescence with promising photothermal conversion efficiency. After coupling with folic acid, the N-GQDs probe could achieve dual modal imaging and targeted therapy in a single material.
Radionuclide imaging is a powerful medical imaging technique that uses radioactive tracers, or radionuclides, to visualize physiological processes within the body. GQDs have garnered interest as potential contrast agents for radionuclide imaging due to their unique properties, including high surface area, tunable optical properties, and excellent biocompatibility. While GQDs themselves do not emit ionizing radiation, they can be functionalized or conjugated with radionuclides to serve as carriers or delivery platforms for targeted imaging applications.
One approach to radionuclide imaging using GQDs involves the conjugation of GQDs with radionuclide-containing molecules or nanoparticles. For example, GQDs can be functionalized with chelating agents that bind specific radionuclides, such as technetium-99m (99mTc) or indium-111 (111In), which are commonly used in single-photon emission computed tomography (SPECT) imaging.186 The resulting GQD-radionuclide complexes can then be administered intravenously and allowed to accumulate in target tissues or organs, where they emit gamma rays that are detected by gamma cameras and reconstructed into three-dimensional images. Another approach involves labeling GQDs with positron-emitting radionuclides for positron emission tomography (PET) imaging.187 In this method, GQDs are functionalized with chelators or linkers that can bind the positron-emitting radionuclides. After radiolabeling, the GQD-radionuclide complexes are administered to the patient, and the emitted positrons interact with nearby electrons, producing pairs of gamma rays that are detected by PET scanners to create detailed images of metabolic activity or molecular targets within the body.
The use of GQDs in radionuclide imaging offers several advantages. Their high surface area and functionalizability enable the precise control of radionuclide binding and distribution, allowing for targeted imaging of specific tissues or biomarkers. Additionally, the optical properties of GQDs can provide complementary information for multimodal imaging when combined with radionuclide imaging techniques. Moreover, GQDs exhibit excellent biocompatibility and low toxicity, minimizing adverse effects on biological systems. In summary, radionuclide imaging using graphene quantum dots holds great potential for a wide range of biomedical applications, including cancer detection, molecular imaging, and drug delivery monitoring. Continued research and development in this field are expected to further optimize the performance and versatility of GQD-based imaging agents, ultimately improving the diagnosis and treatment of various diseases.
In a nutshell, GQDs may have limited penetration depth in certain biological tissues, which can be a drawback for deep tissue imaging. Making them unsuitable for imaging structures located deep within the body. Moreover, GQDs might not always be highly specific to particular targets or molecules. This could result in non-specific binding or imaging of unintended structures. For efficient in vivo imaging applications, the fluorescence of GQDs should be in the NIR-I or NIR II windows. It was found that GQDs synthesized from biomass-derived materials normally shows the NIR fluorescence.194,195 Similarly, the GQDS doped with boron or/and nitrogen also shows the NIR fluorescence.181 In addition, the reduced graphene oxide-derived GQDs also exhibited the NIR fluorescence particularly for NIR imaging applications196,197
Biocompatibility and toxicity is yet another challenge encountered. Depending on the specific properties and functionalization of GQDs, there may be concerns about their biocompatibility and potential toxicity when used in biological systems. Functionalization allows for the attachment of targeting ligands or molecules, which can increase the specificity of GQDs for particular biomolecules or cellular structures. Functionalization can also introduce biocompatible coatings or molecules on the surface of GQDs, reducing potential toxicity and improving their compatibility with biological systems. Functionalized GQDs can also be used in combination with other imaging agents or techniques, allowing for multimodal imaging approaches that provide complementary information.
Photostability is crucial for fluorescent agents as it ensures that the fluorescence signal remains consistent over time during imaging experiments. In the case of GQDs, their excellent photostability can be attributed to their unique structure and electronic properties. The graphene-like structure of GQDs with sp2 hybridized carbon atoms provides exceptional mechanical and chemical stability, making them resistant to degradation or photobleaching under continuous excitation. The presence of edge states and quantum confinement effects in GQDs contributes to their robust photostability. These features enable efficient energy transfer processes and minimize non-radiative recombination pathways, resulting in prolonged fluorescence lifetimes and minimal loss of fluorescence intensity over time.198 The photostability of GQDs has been demonstrated in various imaging applications, including fluorescence microscopy, bioimaging, and biosensing. Their ability to maintain fluorescence under prolonged illumination makes GQDs suitable for long-term imaging studies, dynamic tracking of biological processes, and real-time monitoring of cellular events. The inherent chemical inertness and structural stability of GQDs contribute to their excellent stability profile. Additionally, surface functionalization strategies can further enhance the stability of GQDs by preventing aggregation, minimizing non-specific interactions, and improving biocompatibility. Moreover, surface functionalization of GQDs with various functional groups, such as hydroxyl, carboxyl, or amine, can further enhance their stability by providing additional protection against aggregation, oxidation, or chemical interactions.199
Biodegradability is an important consideration for fluorescent agents intended for biomedical applications, particularly for in vivo imaging and diagnostic purposes. GQDs exhibit favorable biodegradability characteristics, making them suitable for use in living organisms without causing long-term accumulation or toxicity concerns. The biodegradation of GQDs primarily occurs through enzymatic and metabolic pathways within biological systems. Studies have shown that GQDs can undergo gradual degradation into smaller carbon-based fragments under physiological conditions. These degradation products are typically non-toxic and can be metabolized or excreted from the body through natural clearance mechanisms. The biodegradability of GQDs enhances their biocompatibility and reduces the risk of adverse effects, making them suitable for various biomedical applications, including drug delivery, theranostics, and targeted imaging.200,201 GQDs exhibit excellent stability both in vitro and in vivo, retaining their fluorescence intensity and dispersibility over extended periods of time. This stability is crucial for applications such as bioimaging, drug delivery, and theranostics, where GQDs are utilized for real-time visualization or targeted delivery within biological systems. Overall, the combination of structural integrity, chemical inertness, surface functionalization, and interactions with biomolecules contributes to the promising stability profile of GQDs, making them versatile candidates for a wide range of biomedical applications.
The targeted delivery/imaging of GQDs can be achieved with enhanced specificity and efficiency by employing certain strategies, which paving the way for personalized cancer diagnostics and therapeutics with reduced off-target effects and improved clinical outcomes. The surface of GQDs can be modified with targeting ligands, such as antibodies, peptides, or small molecules, enables specific recognition and binding to molecular markers overexpressed on the surface of cancer cells. These targeted ligands facilitate the selective uptake of GQDs by cancer cells while minimizing interactions with normal cells, enhancing the specificity of imaging and therapy. Han et al. designed in situ automatic DNA assembly reaction and applied it for the simultaneous identification of dual targets using GQDs-based probes.202 As anticancer antibodies are quite expensive, many researchers investigated alternative ligand-based QDs including folic acid (FA), epidermal growth factors, transferrin, and a few aptamers to target cancer cells. Active targeting strategies involve exploiting the enhanced permeability and retention (EPR) effect exhibited by tumors to achieve selective accumulation of GQDs within the tumor microenvironment. This can be achieved by conjugating GQDs with molecules that target tumor vasculature or tumor-specific receptors, enabling preferential accumulation and retention of GQDs in cancerous tissues.
4.2. Biosensing applications of GQDs
The optical or electrochemical characteristics of GQDs can be applied to biosensing in addition to bioimaging. The use of GQDs has enabled isolated visualization of certain cells and tissues of interest and better contrast in MR images, even though both bioimaging and biosensing applications utilize the photoluminescence of GQDs and necessitate the detection of emitted photons.168 GQDs in biosensing systems, on the other hand, serve to identify and signal the presence of biomolecules. As previously mentioned, changing the edge groups' electron structures can change the optical characteristics of GQDs. Different electronic states can be produced by the pair's association when a functional group conjugated to the GQD attaches to the analyte. Analyte detection can then be determined as a change in photoluminescence intensity by changing the electrical structure of the GQD. This can be either the quenching or enhancing the photoluminescence intensity in normal cases. The photoluminescence intensity might change with respect to the pH of the environment and in this case, it can be used as pH sensors. It has been shown that biosensor systems based on GQDs are capable of detecting ions, DNA, and a variety of other compounds. High sensitivity and selectivity are key requirements for biosensing; GQDs' photostability and the photoluminescence-based systems' quick response time position them as a promising biosensing platform.203–205 The promising electrical conductivity of GQDs makes them attractive material for the fabrication of electrochemical sensing platforms in addition to their fascinating properties such as quantum confinement; high surface area and variable bandgap energy.152 The GQDs based biosensors can be broadly classified in to photoluminescence based sensors and electrochemical sensors.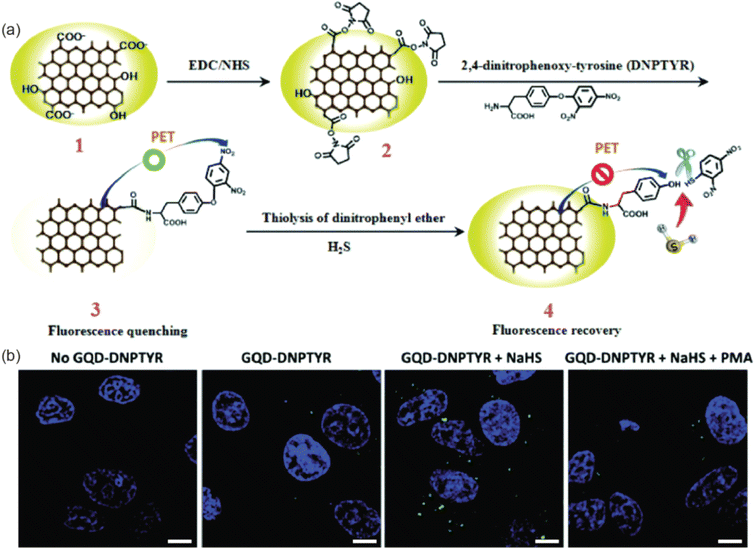 | ||
| Fig. 10 (a) Schematic illustrations of the synthesis and photoluminescence quench mechanisms of GQD–DNPTYR. (b) Confocal images of MCF-7 cells treated with modified GQDs. Scale bar = 10 μm. Reproduced with permission reproduced with permission from ref. 207. Copyright © The Royal Society of Chemistry 2018. | ||
Non-thermal plasma technology has been used as the most suitable methods to synthesize N-GQDs.208 Kurniawan et al. introduced a rational design of N-GQD through microplasma-enabled band-structure engineering for developing pH sensors.209 They have used chitosan biomass-derived N-GQDs for the experiment. The pH sensing capability of N-GQDs are originated from the protonation/deprotonation of –OH group of N-GQDs, which lead to the maximum pH-dependent luminescence peak shift. The change in the luminescence peak shift comes from the broadening or narrowing of bandgap as the pH varies. The developed sensor achieved rapid and label-free pH sensing with a wide range of pH 1.8 to 13.6. The same research group made another pH sensor by synthesising N-GQDs by using chitosan as the sole carbon and nitrogen precursor through atmospheric-pressure microplasmas under ambient conditions.210 The photoluminescence properties of N-GQDs varies with the pH and this can be used to sense pH from 1.78 to 13.56 with two linear ranges from pH 1.25–10.17 and pH 10.17–13.24. The pH sensing property originated from the protonation/deprotonation of N-GQDs due to the presence of N dopants and surface functional groups on N-GQDs.
Tris(hydroxymethyl)aminomethane was utilized to impart amine functionalities on the surface of the N-GQDs based biosensor for detection of 2,4,6-trinitrophenol (TNP). To evaluate the specificity of the N-GQDs for TNP, the amount of photoluminescence quenching in the presence of TNP was contrasted with that in the presence of other metal ions and other aromatic compounds with TNP-like structures. The outcomes demonstrated that considerable photoquenching only happened in the presence of TNP.211 In a different investigation, N-GQDs were employed as colorimetric indicators of glucose and hydrogen peroxide as well as catalysts in the reduction of hydrogen peroxide.212 3,3′,5,5′-Tetramethylbenzidine and horseradish peroxidase (HRP) were used in the comparison of the kinetics of N-GQDs. Comparing the Michaelis–Menten constant (Km), the results showed the higher affinity of N-doped GQDs towards aromatic compounds.
Bioresource-derived colloidal N-GQDs were used as ultrasensitive sensors for detection of cancer and neurotransmitter biomarkers by Chen et al.213 They utilized an atmospheric pressure microplasma system for converting chitosan into N-GQDs. From the photoluminescence measurements at different pH conditions, the N-GQDs were able to detect uric acid, folic acid, epinephrine, and dopamine simultaneously. The developed sensor showed linear relationships between the photoluminescence intensity and the biomarker concentrations with LOD of 123.1, 157.9, 80.5, and 91.3 nM for uric acid, folic acid, epinephrine, and dopamine respectively. Similarly, another photoluminescence-based sensor for the selective detection of folic acid, dopamine and epinephrine by using N-GQDs synthesized from chitosan using atmospheric pressure microplasmas.214 The selective detection is achieved by regulating the pH with the LOD of 81.7, 57.8, and 16.7 nM for folic acid, dopamine and epinephrine respectively. The process of making the N-GQDs is highly scalable as the made stable colloidal N-GQD dispersion with 100 μg mL−1 concentrations in just 1 h and this can be used for at least 100 photoluminescence measurements. The enhanced sensing performance of the sensor can be attributed to the size-dependent π–π stacking attraction between the N-GQDs and the pH-regulated chemical states of the analytes.
Li et al. developed a sensing platform based on GQDs modified with tyramine (TYR-GQDs).215 This fluorescence-based sensor was employed for sensitive and selective measurement of metabolites with peroxidase activities. The catalysis of H2O2 by TYR-GQDs results in the generation of a hydroxyl radical (˙OH), which causes photoluminescence quenching. Based on the correlation between the photoluminescence intensity of TYRGQDs and the concentration of H2O2, a sensitive turn-off sensing method with a detection limit of 0.32 nM is devised. The proposed ultrasensitive sensing technology based on this idea can be used to measure other metabolites like glucose, L-lactate, xanthine and cholesterol. The schematic representation of the fluorimetric sensor is depicted in Fig. 11.
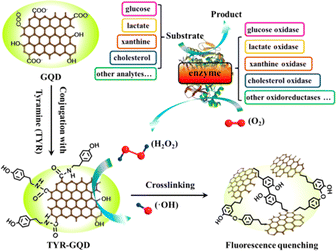 | ||
| Fig. 11 The schematic representation of TYR-GQD based fluorometric sensor for various analytes. Reproduced with permission from ref. 215. Copyright © 2016, American Chemical Society. | ||
The metallic cations and certain anions play a vital role in the regulation of various proteins in the human body and thus its accumulation will cause problematic physiological activities. Thus, the selective and sensitive detection of these ions are important issues in the field of biosensing. Changes in the photoluminescence caused as the result of interaction between the inorganic species with the functionalized GQDs are utilized in the quantitative detection of various ions Pb2+, Cu2+, Hg2+, Fe3+.59,216–221 A functionalized GQDs with 1-butyl-3-methylimidazolium hexafluorophosphate (BMIMPF6, BMIM+) has been used as probe to measure ferric ion (Fe3+) due to the strong binding affinity between the imidazole ring of BMIMPF6, BMIM+ with Fe3+. This interaction causes aggregation of GQDs and induces photoluminescence quenching. The developed probe had a detection limit of 7.22 μM.217 Amine functionalized GQDs have been developed for the detection of Cu2+ level in living cells.216 Literature states that Cu2+ has a higher binding affinity towards nitrogen and oxygen than other transition metals thus interact effectively with the surface of amino functionalized GQDs than other transition-metal ions. This highly selective probe of Cu2+ had a low detection limit of 6.9 nM. A photoluminescence turn-on platform for Pb2+ was proposed by Qian et al. based on aptamer-functionalized GQDs.219 The aptamer modified GQDs were made to electrostatically interact with GO and these interactions due to pi–pi stacking quenched the photoluminescence of the GQDs. This was then recovered by the addition of Pb2+ that facilitated the liberation from GO. With an extremely low LOD of 0.6 nM, Pb2+ was precisely and selectively detected. Similarly, ratiometric photoluminescence probes based on GQDs have gained significant interest with enhanced sensitivity and selectivity. A novel ratiometric photoluminescence probe made of GQDs covalently coupled to rhodamine derivative was developed by Liu et al.221 The developed sensor is based on the fluorescence resonance energy transfer (FRET) process between GQDs (energy donor) and rhodamine derivative. When Hg2+ is added, the photoluminescence emission of GQDs at 500 nm gradually diminishes with the addition of Hg2+ and a new emission peak develops at 585 nm. From 0.6 to 12 μM, a linearity between the Hg2+ concentration and the emission intensity ratio was reported in this work. The use of GQDs for the quantification of negatively charged ions viz. chloride (Cl−), nitrite (NO2−), sulfide (S2−) etc. are also reported in a similar manner in different reports.222–224
Using a similar FRET method, CNTs in place of the quencher might be utilized to quantify DNA by Qian et al.225 Based on FRET between GQDs and CNTs, they built an ultrasensitive nanosensor for DNA detection. Reduced GQDs with strong photoluminescence emission are first produced by reducing with NaBH4. Single-stranded DNA (ssDNA) is then functionalized on the surface of rGQDs to prepare ssDNA-rGQDs. The photoluminescence quenching results from FRET between ssDNA-rGQDs and oxidized CNTs. This happens because of the interactions with ssDNA-rGQDs via electrostatic attraction and stacking interaction. When target DNA (tDNA) is introduced to the system, double-stranded DNA-rGQDs (dsDNA-rGQDs) are liberated along with photoluminescence restoration. The working principle and schematic representation of the developed nano sensor is depicted in Fig. 12. To identify complementary and mismatched nucleic acid sequences, a special photoluminescence “on–off–on” sensing platform was developed with high sensitivity in a linear range of 1.5–133.0 nM.
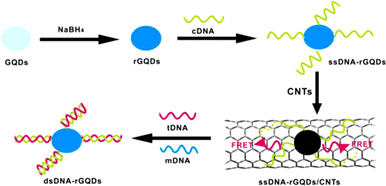 | ||
| Fig. 12 Schematic illustration of nano sensor based on FRET for quantification of DNA developed on GQDs and CNTs. Reproduced with permission from ref. 225. Copyright © 2014 Elsevier B. V. | ||
To analyze micro RNAs (miRNA) quantitatively, FRET between GQDs and pyrene-functionalized molecular beacon probes (py-MBs) has been used.226 Pyrene starts effective FRET from GQDs to fluorescent dyes. The unfolding of hairpin structures and the development of more rigid duplexes structures are both induced when target miRNA is added to the FRET system through the hybridization between the loop structure of py-MBs and the target miRNA. The fluorescent dyes' photoluminescence intensity is decreased as a result of this process, which inhibits the FRET between GQDs and fluorescent dyes. With a detection limit of 100 pM, a linear range of 155 0.1–200 nM was thus made possible with the developed sensor.
In order to measure horseradish peroxidase (HRP), Huang et al. proposed a ratiometric photoluminescence probe using 2,3-diaminophenazine (DAP), the oxidation product of o-phenylenediamine (OPD).227 This was utilized as the specific response signal and amino-functionalized GQDs were used as the reference fluorophore. The catalytic oxidation of OPD by HRP results in a sharp increase in the photoluminescence at 553 nm, whereas the formation of the modified-GQDs-DAP complex simultaneously quenches the photoluminescence of amino functionalized-GQDs at 440 nm. As a result, HRP was precisely measured with a detection limit as low as 0.21 fM. Additionally, this sensing system's colour variation may be used as a visual assay for the blind detection of HRP at extremely low levels.
![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 000 μM.
000 μM.A HRP based biosensor was developed by Muthurasu et al. by using carboxyl functionalized GQDs.229 Initially, carboxyl functionalized GQDs were immobilized onto the GCE which was allowed to conjugate to the amine groups of the enzyme. The modified platform could detect H2O2 using the method of amperometry with a detection limit of 530 nM.
Vasilescu et al. prepared a composite out of GQDs and MoS2 flakes for the first time which was modified on screen printed carbon electrode (SPCE).230 Having known the ability of MoS2 to sense a wide array of analytes, the combination of MoS2 with a band gap of 1.8 eV and that of zero band gap of GQDs led to superior electrocatalytic activity. The composite had better electrocatalytic activity compared to GQDs and MoS2 flakes when taken individually. The MoS2/GQDs/SCPE was further modified with laccase enzyme for the chronoamperometric determination with very low detection limits of 2.4, 0.19, and 0.32 μM for epicatechin, chlorogenic acid and caffeic acid respectively. In order to give an extremely large surface area for antibody immobilisation as electrode modifications, Bhatnagar et al. created an ultrasensitive electrochemical immunosensor for cardiac troponin I (cTnI).231 In this technique, GQDs were covalently attached on an amino functionalized Au screen-printed electrode using EDC (1-ethyl-3-diaminopropyl carbodiimide) NHS (N-hydroxysuccinimide) chemistry. To create an ultra-high surface area for the immobilisation of the capture antibody, poly(amidoamine) and 4-aminothiophenol and the dendrimer were progressively attached to GQDs using EDC coupling. As evaluated by DPV and CV, the sensor exhibited LODs of 25 and 20 fg mL−1, respectively, served as a marker for the recognition of the target protein. Fig. 13 shows the fabrication of electrochemical immunosensor for the determination of cTnI by using Au/GQD/PAMAM nanohybrid electrode.
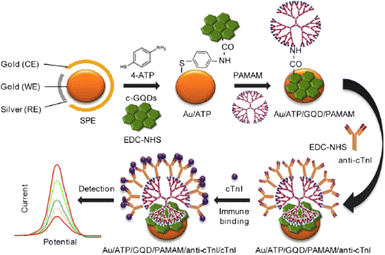 | ||
| Fig. 13 Electrochemical immunosensor constructed for the determination of cTnI involving the use of an Au/GQD/PAMAM nanohybrid electrode. Reproduced with permission from ref. 231. Copyright © 2016 Elsevier. | ||
For the detection of Mycobacterium tuberculosis antigen, Tufa et al. presented an electrochemical sandwich immunosensor using a nanotriplex made of a GQD-coated Fe3O4@Ag core–shell nanostructure (Fe3O4@Ag/GQD) as the GCE modifier and antibody-AuNPs as labels for signal amplification.232 The synthesis of Fe3O4@Ag/GQD NP nanotriplex probe and the inmmunosensor preparation are given in Fig. 12. Due to the distinct functions of the three nanomaterials; Ag improved electrical conductivity, GQDs permitted greater antibody loadings on the electrode, and Fe3O4 increased the surface-to-volume ratio. The sensing platform using the nanotriplex material demonstrated a noticeable synergetic electrochemical performance. The immunosensor was successfully used to analyze spiked human urine samples and displayed a broad linear range with a detection limit of 0.33 ng mL−1 (Fig. 14).
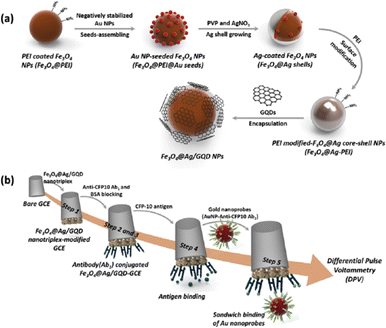 | ||
| Fig. 14 Synthesis of Fe3O4@Ag/GQD NP nanotriplex probe (a) inmmunosensor preparation (b): drop casting onto the GCE (step 1), incubation with CAb (step 2), blocking with BSA (step 3), incubation with the target antigen (step 4), and incubation with the AbD-AuNPs (step 5). Reprinted with permission from ref. 232. Copyright © 2018 Elsevier Ltd. | ||
Table 3 shows the comparison of various luminescence based sensors made of GQDs for the detection of different analytes. The design and development of biosensing platforms with improved performance has been prompted by the growing need for sensitive and selective single or multiplexed determination of pertinent analytes at various molecular levels in matrices of a variety of nature and complexity using simple and rapid protocols. These specific carbon nanomaterials have been utilized in electrochemical biosensing as signal tags, either to replace or carry enzymatic systems or to modify the working electrode surface, always looking for signal amplification. The use of GQDs enables the creation of sensitive, focused, and biocompatible biosensors for the detection of pertinent analytes in biological matrices.
| Material | Analyte | Transduction mechanism | Sensitivity | LOD | Linear range | Ref. |
|---|---|---|---|---|---|---|
| Ligninsulfonate/GQDs | Fe3+ | Fluorescence quenching | 0.004 μM−1 | 0.5 nM | 0.005–500 μM | 233 |
| GQDs from carbon black | Fe3+ | Fluorescence quenching | 0.0099 μM−1 | 0.45 μM | 0–60 μM | 234 |
| GQDs from 3D CVD graphene | Fe3+ | Fluorescence quenching | — | 7.22 μM | 0–80 μM | 217 |
| GQDs from biowaste | Fe3+ | Fluorescence quenching | 2.5 μM | 1–100 μM | 235 | |
| RBD-GQDs | Fe3+ | Fluorescence enhancement | — | 0.02 μM | 0 to 1 μM | 218 |
| B-GQD | Al3+ | Fluorescence enhancement | — | 3.64 μM | 0–100 μM | 236 |
| GQDs-graphene oxide | Pb2+ | Fluorescence recovery | — | 0.6 nM | 9.9–435.0 nM | 219 |
| GDQs/Au NPs | Pb2+ | Fluorescence recovery | — | 16.7 nM | 50 nM to 4 μM | 220 |
| Ethylenediamine functionalized GQDs | Photoluminescence quenching | — | 30 nM | 0.1–50 μM | 206 | |
| afGQDs | Cu2+ | Fluorescence quenching | 6.9 nM | 0 to 100 nM | 216 | |
| GQDs–SR | Hg2+ | Fluorescence enhancement | 23 μM | 0.6 to 12 μM | 221 | |
| N-GQDs | H2O2 | Colorimetric sensing | 5.3 μM | 20–1170 μM | 212 | |
| Glucose | 16 μM | 25 to 375 μM | ||||
| GQDs/py-MBs | microRNA (miR-155) | Fluorescence quenching | — | 100 pM | 0.1 nM to 200 nM | 226 |
| N-GQDs | Tyrosinase | Fluorescence quenching | — | 0.15 U mL−1 | 0.43–3.85 U mL−1 | 237 |
| GQD–DNPTYR | H2S | Fluorescence enhancement | — | 0.89 nM | 2 nM to 1.5 μM | 207 |
| SSGQDs | S2− | Fluorescence enhancement | — | 12.6 nM | 0 to 3.75 μM | 224 |
| Ascorbic acid | — | 12.3 nM | Of 0–12 μM | |||
| GQDs from citric acid | Chlorine | Fluorescence quenching | — | 0.05 μM | 0.05 to 10 μM | 223 |
| N, P-GQDs | Nitrite | Fluorescence quenching | — | 2.5 nM | 5–30 nM | 222 |
| B-GQD | Cytochrome C | Fluorescence quenching | — | 5.9 μg mL−1 | 10–300 μg mL−1 and 300–2000 μg mL−1 | 238 |
| GQDs/MoS2 | Cholesterol | Fluorescence enhancement | — | 35 nM | 0.08–300 μM | 239 |
| Tyramine@GQDs | H2O2 | Fluorescence quenching | 0.32 nM | 1 nM–0.15 μM | 215 | |
| Cholesterol | 1.2 nM | 8 × 10−8 to 1 × 10−5 | ||||
| Lactate | 47 nM | 2 × 10−7 to 2.5 × 10−5 | ||||
| Xanthine | 32 nM | 1 × 10−7 to 1.8 × 10−5 | ||||
| GQDs from graphene oxide | Aniline | Photoluminescence quenching | 11.50 M−1 | — | 0–0.44 M | 240 |
| Amino-functionalized GQDs | HRP enzyme | Fluorescence quenching | 0.21 fM | 2–800 fM | 227 | |
| N-GQDs | TNP | Fluorescence quenching | — | 0.3 μM | 1–60 μM | 211 |
| GQDs/CNT | ssDNA | Fluorescence enhancement | 0.4 nM | 1.5–133.0 nM | 225 | |
| GQDs from graphene sheets | ssDNA-2 | Electrochemical enhancement | — | 100 nM | 200–500 nM | 204 |
| Fe3O4@Ag/GQD | Mtb antigen CFP-10 | Electrochemical immunosensor | — | 0.33 ng mL−1 | 0.005–500 μg mL−1 | 232 |
| GQD-PAMAM | cTnI | Electrochemical immunosensor | — | 20 fg mL−1 | 10−6 to 10−3 ng/6 μL | 231 |
| GQDs/laccase | Epinephrine | Electrochemical enhancement | 2.9 μA mM−1 cm−2 | 83 nM | 1–120 μM | 241 |
| Fe3O4@GQD/multiwalled carbon, nanotubes | Progesterone | Electrochemical enhancement | 16.84 μA μM−1 | 2.18 nM | 0.01–0.5 μM and 0.5–3.0 μM | 242 |
| N-GQD@Ncarbon@Pd | H2O2 | Electrochemical enhancement | 0.59 mA mM−1 cm−2 | 20 nM | 0–1.4 mM | 243 |
| HRP/GQDs | H2O2 | Electrochemical sensing | 0.905 and 7.057 μA mM−1 | 530 nM, 2.16 μM | 100 μM to 1.3 mM, 1.7 to 2.6 mM | 229 |
| GQDs/MoS2 | Caffeic acid | Electrochemical sensing | 17.92 nA μM−1 | 0.32 μM | 0.38–100 μM | 230 |
5. Conclusion and future perspectives
Herein, the top-down and bottom-up approaches employed for the synthesis of graphene quantum dots (GQDs) followed by a detailed review about various surface properties of GQDs has been discussed. Further, the role of dopants on the surface properties were discussed in detail. Following this, the in vivo and in vitro bioimaging applications of GQDs were also discussed. The usefulness of GQDs in optical and electrochemical biosensing were also discussed in detail. GQDs are increasingly employed in the research area of bio-imaging and biosensing despite its issues such as low yield, low QY (<20%) and short wavelength emission. Therefore, there should be intensive futuristic research efforts on the synthetic methodologies that can improve the yield. Further, research should be directed towards synthetic methodology that can yield GDQs with uniform size, geometry and edges which is expected to improve the QY for bio-imaging and biosensing applications. Most of the research articles describe the usefulness of GQDs on fluorescence imaging, very recently there are efforts towards the development of other modes of imaging such as magnetic resonance imaging together with fluorescence and tuning the PL window of GQDs that will be useful in NIR imaging. As a result of GQDs non-invasive and deep tissue penetration depth, it can be employed for in vivo imaging. On the other hand, the safety of GQDs for biological applications are yet to be explored in detail. However, results from initial short term experiments suggests low residual accumulation and low cytotoxicity which is encouraging. The presence of plentiful functionalities and π-orbitals on the surface of GQDs, high percentage of drug loading is possible and thereby it finds potential therapeutic applications. Upon DNA complexation GQDs can also be used for gene therapy. As GQDs being a relatively new material, its applications in bio-imaging and biosensing remains boundless.Author contributions
The manuscript was written through contributions of all authors. All authors have given approval to the final version of the manuscript.Conflicts of interest
The authors declare no conflicts of interests.Acknowledgements
PAR acknowledges Ramalingaswami re-entry fellowship (BT/RLF/Re-entry/75/2020) from the Department of Biotechnology (DBT), Govt. of India. MA acknowledges the PMRF fellowship (31202903) from Govt. of India.References
- V. Kansara, R. Shukla, S. J. S. Flora, P. Bahadur and S. Tiwari, Mater. Today Commun., 2022, 31, 103359 CrossRef CAS.
- F. Jiang, D. Chen, R. Li, Y. Wang, G. Zhang, S. Li, J. Zheng, N. Huang, Y. Gu, C. Wang and C. Shu, Nanoscale, 2013, 5, 1137–1142 RSC.
- N. Nesakumar, S. Srinivasan and S. Alwarappan, Microchim. Acta, 2022, 189, 258 CrossRef CAS PubMed.
- T. D. Thangadurai, N. Manjubaashini, D. Nataraj, V. Gomes and Y. I. Lee, J. Mater. Sci. Eng. B, 2022, 278, 115633 CrossRef CAS.
- A. Ghaffarkhah, E. Hosseini, M. Kamkar, A. A. Sehat, S. Dordanihaghighi, A. Allahbakhsh, C. van der Kuur and M. Arjmand, Small, 2022, 18, 2102683 CrossRef CAS PubMed.
- S. M. Mousavi, S. A. Hashemi, M. Yari Kalashgrani, D. Kurniawan, A. Gholami, V. Rahmanian, N. Omidifar and W.-H. Chiang, Biosensors, 2022, 12, 461 CrossRef CAS PubMed.
- M. Farshbaf, S. Davaran, F. Rahimi, N. Annabi, R. Salehi and A. Akbarzadeh, Artif. Cells, Nanomed., Biotechnol., 2018, 46, 1331–1348 CrossRef CAS PubMed.
- Z. Ji, E. Dervishi, S. K. Doorn and M. Sykora, J. Phys. Chem. Lett., 2019, 10, 953–959 CrossRef CAS PubMed.
- X. Yan, B. Li, X. Cui, Q. Wei, K. Tajima and L.-s. Li, J. Phys. Chem. Lett., 2011, 2, 1119–1124 CrossRef CAS PubMed.
- L. Kittiratanawasin and S. Hannongbua, Integr. Ferroelectr., 2016, 175, 211–219 CrossRef CAS.
- S. Kim, S. W. Hwang, M.-K. Kim, D. Y. Shin, D. H. Shin, C. O. Kim, S. B. Yang, J. H. Park, E. Hwang and S.-H. Choi, ACS Nano, 2012, 6, 8203–8208 CrossRef CAS PubMed.
- M. Zhu, Q. Liu, W. Chen, Y. Yin, L. Ge, H. Li and K. Wang, ACS Appl. Mater. Interfaces, 2017, 9, 38832–38841 CrossRef CAS PubMed.
- Y. Dai, H. Long, X. Wang, Y. Wang, Q. Gu, W. Jiang, Y. Wang, C. Li, T. H. Zeng and Y. Sun, Part. Part. Syst. Charact., 2014, 31, 597–604 CrossRef CAS.
- K. Geethalakshmi, T. Y. Ng and R. Crespo-Otero, J. Mater. Chem. C, 2016, 4, 8429–8438 RSC.
- R. Sekiya, Y. Uemura, H. Naito, K. Naka and T. Haino, Chem.–Eur. J., 2016, 22, 8198–8206 CrossRef CAS PubMed.
- K. L. Schroeder, R. V. Goreham and T. Nann, Pharm. Res., 2016, 33, 2337–2357 CrossRef CAS.
- T. A. Tabish, H. Hayat, A. Abbas and R. J. Narayan, Biosensors, 2022, 12, 77 CrossRef CAS PubMed.
- F. Faridbod and A. L. Sanati, Curr. Anal. Chem., 2019, 15, 103–123 CrossRef CAS.
- Y. Wang and A. Hu, J. Mater. Chem. C, 2014, 2, 6921–6939 RSC.
- M. Xu, Z. Li, X. Zhu, N. Hu, H. Wei, Z. Yang and Y. Zhang, Nano Biomed. Eng., 2013, 5, 65–71 CAS.
- M. Zhao, Appl. Sci., 2018, 8, 1303 CrossRef.
- H. Kalita, V. S. Palaparthy, M. S. Baghini and M. Aslam, Carbon, 2020, 165, 9–17 CrossRef CAS.
- S. Sarkar, D. Gandla, Y. Venkatesh, P. R. Bangal, S. Gosh, Y. Yang and S. Misra, Phys. Chem. Chem. Phys., 2016, 18, 21278–21287 RSC.
- S. Wang, Z.-G. Chen, I. Cole and Q. Li, Carbon, 2015, 82, 304–313 CrossRef CAS.
- L. Zdrazil, R. Zahradnicek, R. Mohan, P. Sedlacek, L. Nejdl, V. Schmiedova, J. Pospisil, M. Horak, M. Weiter, O. Zmeskal and J. Hubalek, J. Lumin., 2018, 204, 203–208 CrossRef CAS.
- L. Lu, Y. Zhu, C. Shi and Y. T. Pei, Carbon, 2016, 109, 373–383 CrossRef CAS.
- J. Shen, Y. Zhu, C. Chen, X. Yang and C. Li, Chem. Commun., 2011, 47, 2580–2582 RSC.
- J. P. Naik, P. Sutradhar and M. Saha, J. Nanostruct. Chem., 2017, 7, 85–89 CrossRef CAS.
- R. Liu, D. Wu, X. Feng and K. Müllen, J. Am. Chem. Soc., 2011, 133, 15221–15223 CrossRef CAS PubMed.
- R. Tian, S. Zhong, J. Wu, W. Jiang, Y. Shen, W. Jiang and T. Wang, Opt. Mater., 2016, 60, 204–208 CrossRef CAS.
- T. Ogi, H. Iwasaki, K. Aishima, F. Iskandar, W.-N. Wang, K. Takimiya and K. Okuyama, RSC Adv., 2014, 4, 55709–55715 RSC.
- D. Pan, J. Zhang, Z. Li and M. Wu, Adv. Mater., 2010, 22, 734–738 CrossRef CAS PubMed.
- D. Pan, L. Guo, J. Zhang, C. Xi, Q. Xue, H. Huang, J. Li, Z. Zhang, W. Yu, Z. Chen, Z. Li and M. Wu, J. Mater. Chem., 2012, 22, 3314–3318 RSC.
- R. Tian, S. Zhong, J. Wu, W. Jiang and T. Wang, RSC Adv., 2016, 6, 40422–40426 RSC.
- J. Shen, Y. Zhu, X. Yang, J. Zong, J. Zhang and C. Li, New J. Chem., 2012, 36, 97–101 RSC.
- R. G. Acres, A. V. Ellis, J. Alvino, C. E. Lenahan, D. A. Khodakov, G. F. Metha and G. G. Andersson, J. Phys. Chem. C, 2012, 116, 6289–6297 CrossRef CAS.
- H. Teymourinia, M. Salavati-Niasari, O. Amiri and H. Safardoust-Hojaghan, J. Mol. Liq., 2017, 242, 447–455 CrossRef CAS.
- Z. Wang, J. Yu, X. Zhang, N. Li, B. Liu, Y. Li, Y. Wang, W. Wang, Y. Li, L. Zhang, S. Dissanayake, S. L. Suib and L. Sun, ACS Appl. Mater. Interfaces, 2016, 8, 1434–1439 CrossRef CAS PubMed.
- M. H. M. Facure, R. Schneider, L. A. Mercante and D. S. Correa, Mater. Today Chem., 2022, 23, 100755 CrossRef CAS.
- Y. Shin, J. Park, D. Hyun, J. Yang, J.-H. Lee, J.-H. Kim and H. Lee, Nanoscale, 2015, 7, 5633–5637 RSC.
- B. P. Qi, X. Zhang, B. B. Shang, D. Xiang and S. Zhang, J. Nanoparticle Res., 2018, 20, 1–9 CrossRef CAS.
- Q. Liu, B. Guo, Z. Rao, B. Zhang and J. R. Gong, Nano Lett., 2013, 13, 2436–2441 CrossRef CAS PubMed.
- W. Chen, G. Lv, W. Hu, D. Li, S. Chen and Z. Dai, Nanotechnol. Rev., 2018, 7, 157–185 CrossRef CAS.
- H. Lu, W. Li, H. Dong and M. Wei, Small, 2019, 15, 1902136 CrossRef PubMed.
- Y. Li, Y. Hu, Y. Zhao, G. Shi, L. Deng, Y. Hou and L. Qu, Adv. Mater., 2011, 23, 776–780 CrossRef CAS PubMed.
- Y. Li, Y. Zhao, H. Cheng, Y. Hu, G. Shi, L. Dai and L. Qu, J. Am. Chem. Soc., 2012, 134, 15–18 CrossRef CAS PubMed.
- D. B. Shinde and V. K. Pillai, Chem.–Eur. J., 2012, 18, 12522–12528 CrossRef CAS PubMed.
- A. Chandra, S. Deshpande, D. B. Shinde, V. K. Pillai and N. Singh, ACS Macro Lett., 2014, 3, 1064–1068 CrossRef CAS PubMed.
- H. Huang, S. Yang, Q. Li, Y. Yang, G. Wang, X. You, B. Mao, H. Wang, Y. Ma, P. He, Z. Liu, G. Ding and X. Xie, Langmuir, 2018, 34, 250–258 CrossRef CAS.
- L. Chen, C. Wu, P. Du, X. Feng, P. Wu and C. Cai, Talanta, 2017, 164, 100–109 CrossRef CAS PubMed.
- M. Xu, W. Zhang, Z. Yang, F. Yu, Y. Ma, N. Hu, D. He, Q. Liang, Y. Su and Y. Zhang, Nanoscale, 2015, 7, 10527–10534 RSC.
- L. Zdrazil, R. Zahradnicek, R. Mohan, P. Sedlacek, L. Nejdl, V. Schmiedova, J. Pospisil, M. Horak, M. Weiter and O. Zmeskal, J. Lumin., 2018, 204, 203–208 CrossRef CAS.
- F. Liu, M.-H. Jang, H. D. Ha, J.-H. Kim, Y.-H. Cho and T. S. Seo, Adv. Mater., 2013, 25, 3657–3662 CrossRef CAS PubMed.
- C. Zhou, W. Jiang and B. K. Via, Colloids Surf., B, 2014, 118, 72–76 CrossRef CAS PubMed.
- C. K. Chua, Z. Sofer, P. Šimek, O. Jankovský, K. Klímová, S. Bakardjieva, Š. Hrdličková Kučková and M. Pumera, ACS Nano, 2015, 9, 2548–2555 CrossRef CAS.
- J. Peng, W. Gao, B. K. Gupta, Z. Liu, R. Romero-Aburto, L. Ge, L. Song, L. B. Alemany, X. Zhan, G. Gao, S. A. Vithayathil, B. A. Kaipparettu, A. A. Marti, T. Hayashi, J.-J. Zhu and P. M. Ajayan, Nano Lett., 2012, 12, 844–849 CrossRef CAS.
- W. Chen, F. Li, C. Wu and T. Guo, Appl. Phys. Lett., 2014, 104, 063109 CrossRef.
- Q. Lu, C. Wu, D. Liu, H. Wang, W. Su, H. Li, Y. Zhang and S. Yao, Green Chem., 2017, 19, 900–904 RSC.
- A. Abbas, T. A. Tabish, S. J. Bull, T. M. Lim and A. N. Phan, Sci. Rep., 2020, 10, 21262 CrossRef CAS PubMed.
- X. Yan, X. Cui, B. Li and L.-s. Li, Nano Lett., 2010, 10, 1869–1873 CrossRef CAS PubMed.
- X. Yan, B. Li and L.-s. Li, Acc. Chem. Res., 2013, 46, 2254–2262 CrossRef CAS PubMed.
- L.-s. Li and X. Yan, J. Phys. Chem. Lett., 2010, 1, 2572–2576 CrossRef CAS.
- Q. Li, S. Zhang, L. Dai and L.-s. Li, J. Am. Chem. Soc., 2012, 134, 18932–18935 CrossRef CAS PubMed.
- S. Bian, C. Shen, Y. Qian, J. Liu, F. Xi and X. Dong, Sens. Actuators, B, 2017, 242, 231–237 CrossRef CAS.
- L. Zhou, J. Geng and B. Liu, Part. Part. Syst. Charact., 2013, 30, 1086–1092 CrossRef CAS.
- Y. Dong, J. Shao, C. Chen, H. Li, R. Wang, Y. Chi, X. Lin and G. Chen, Carbon, 2012, 50, 4738–4743 CrossRef CAS.
- L. Tang, R. Ji, X. Cao, J. Lin, H. Jiang, X. Li, K. S. Teng, C. M. Luk, S. Zeng, J. Hao and S. P. Lau, ACS Nano, 2012, 6, 5102–5110 CrossRef CAS PubMed.
- L. Tang, R. Ji, X. Li, K. S. Teng and S. P. Lau, Part. Part. Syst. Charact., 2013, 30, 523–531 CrossRef CAS.
- F. Khodadadei, S. Safarian and N. Ghanbari, Mater. Sci. Eng., C, 2017, 79, 280–285 CrossRef CAS PubMed.
- A. Bayat and E. Saievar-Iranizad, J. Lumin., 2017, 192, 180–183 CrossRef CAS.
- S. A. Prabhu, V. Kavithayeni, R. Suganthy and K. Geetha, Carbon Letters, 2021, 31, 1–12 CrossRef.
- C. Zhang, Y. Cui, L. Song, X. Liu and Z. Hu, Talanta, 2016, 150, 54–60 CrossRef CAS PubMed.
- P. Zhao, C. Li and M. Yang, J. Dispersion Sci. Technol., 2017, 38, 769–774 CrossRef CAS.
- M. K. Kumawat, M. Thakur, R. B. Gurung and R. Srivastava, ACS Sustain. Chem. Eng., 2017, 5, 1382–1391 CrossRef CAS.
- E. Campbell, M. T. Hasan, R. Gonzalez Rodriguez, G. R. Akkaraju and A. V. Naumov, ACS Biomater. Sci. Eng., 2019, 5, 4671–4682 CrossRef CAS PubMed.
- R. P. Choudhary, S. Shukla, K. Vaibhav, P. B. Pawar and S. Saxena, Mater. Res. Express, 2015, 2, 095024 CrossRef.
- T. K. Henna and K. Pramod, Mater. Sci. Eng., C, 2020, 110, 110651 CrossRef CAS PubMed.
- E. Haque, J. Kim, V. Malgras, K. R. Reddy, A. C. Ward, J. You, Y. Bando, M. S. A. Hossain and Y. Yamauchi, Small Methods, 2018, 2, 1800050 CrossRef.
- M. Li, T. Chen, J. J. Gooding and J. Liu, ACS Sens., 2019, 4, 1732–1748 CrossRef CAS PubMed.
- R. Xie, Z. Wang, W. Zhou, Y. Liu, L. Fan, Y. Li and X. Li, Anal. Methods, 2016, 8, 4001–4016 RSC.
- T. Ohta, A. Bostwick, T. Seyller, K. Horn and E. Rotenberg, Science, 2006, 313, 951–954 CrossRef CAS PubMed.
- M. H. Facure, R. Schneider, L. A. Mercante and D. S. Correa, Environ. Sci.: Nano, 2020, 7, 3710–3734 RSC.
- L. Li, G. Wu, G. Yang, J. Peng, J. Zhao and J.-J. Zhu, Nanoscale, 2013, 5, 4015–4039 RSC.
- M. WooáLee and J. SangáSuh, RSC Adv., 2015, 5, 67669–67675 RSC.
- Y. Yan, J. Gong, J. Chen, Z. Zeng, W. Huang, K. Pu, J. Liu and P. Chen, Adv. Mater., 2019, 31, 1808283 CrossRef PubMed.
- S. Kundu and V. K. Pillai, Physical Sciences Reviews, 2019, 5, 20190013 Search PubMed.
- M. O. Valappil, V. K. Pillai and S. Alwarappan, Appl. Mater. Today, 2017, 9, 350–371 CrossRef.
- Z. Zhang, C. Fang, X. Bing and Y. Lei, Materials, 2018, 11, 512 CrossRef PubMed.
- S. Gupta, T. Smith, A. Banaszak and J. Boeckl, Nanomaterials, 2017, 7, 301 CrossRef PubMed.
- D. Pan, J. Zhang, Z. Li and M. Wu, Adv. Mater., 2010, 22, 734–738 CrossRef CAS PubMed.
- L. Tang, R. Ji, X. Li, G. Bai, C. P. Liu, J. Hao, J. Lin, H. Jiang, K. S. Teng, Z. Yang and S. P. Lau, ACS Nano, 2014, 8, 6312–6320 CrossRef CAS PubMed.
- S. Zhu, J. Zhang, X. Liu, B. Li, X. Wang, S. Tang, Q. Meng, Y. Li, C. Shi, R. Hu and B. Yang, RSC Adv., 2012, 2, 2717–2720 RSC.
- L. Li, G. Wu, G. Yang, J. Peng, J. Zhao and J.-J. Zhu, Nanoscale, 2013, 5, 4015–4039 RSC.
- Z. Luo, Y. Lu, L. A. Somers and A. T. C. Johnson, J. Am. Chem. Soc., 2009, 131, 898–899 CrossRef CAS.
- S. Kim, S. W. Hwang, M.-K. Kim, D. Y. Shin, D. H. Shin, C. O. Kim, S. B. Yang, J. H. Park, E. Hwang, S.-H. Choi, G. Ko, S. Sim, C. Sone, H. J. Choi, S. Bae and B. H. Hong, ACS Nano, 2012, 6, 8203–8208 CrossRef CAS PubMed.
- H. Tetsuka, R. Asahi, A. Nagoya, K. Okamoto, I. Tajima, R. Ohta and A. Okamoto, Adv. Mater., 2012, 24, 5333–5338 CrossRef CAS.
- S. Zhu, J. Zhang, C. Qiao, S. Tang, Y. Li, W. Yuan, B. Li, L. Tian, F. Liu, R. Hu, H. Gao, H. Wei, H. Zhang, H. Sun and B. Yang, Chem. Commun., 2011, 47, 6858–6860 RSC.
- S. H. Jin, D. H. Kim, G. H. Jun, S. H. Hong and S. Jeon, ACS Nano, 2013, 7, 1239–1245 CrossRef CAS.
- D. Pan, L. Guo, J. Zhang, C. Xi, Q. Xue, H. Huang, J. Li, Z. Zhang, W. Yu and Z. Chen, J. Mater. Chem., 2012, 22, 3314–3318 RSC.
- S. Zhu, J. Shao, Y. Song, X. Zhao, J. Du, L. Wang, H. Wang, K. Zhang, J. Zhang and B. Yang, Nanoscale, 2015, 7, 7927–7933 RSC.
- J. Peng, W. Gao, B. K. Gupta, Z. Liu, R. Romero-Aburto, L. Ge, L. Song, L. B. Alemany, X. Zhan and G. Gao, Nano Lett., 2012, 12, 844–849 CrossRef CAS.
- J. Valenta, Nanosci. Methods, 2014, 3, 11–27 CrossRef CAS.
- Z. Zhang, J. Zhang, N. Chen and L. Qu, Energy Environmental Science, 2012, 5, 8869–8890 RSC.
- Y. Dong, C. Chen, X. Zheng, L. Gao, Z. Cui, H. Yang, C. Guo, Y. Chi and C. M. Li, J. Mater. Chem., 2012, 22, 8764–8766 RSC.
- P. Luo, Z. Ji, C. Li and G. Shi, Nanoscale, 2013, 5, 7361–7367 RSC.
- R. Suryawanshi, R. Kurrey, S. Sahu and K. K. Ghosh, RSC Adv., 2023, 13, 701–719 RSC.
- C. T. Cao, S.-W. Kim, H. J. Kim, R. Purbia, S. H. Kim, D. Kim, K. J. Choi, H. Park and J. M. Baik, Nano Energy, 2022, 96, 107117 CrossRef CAS.
- Y.-X. Wang, M. Rinawati, W.-H. Huang, Y.-S. Cheng, P.-H. Lin, K.-J. Chen, L.-Y. Chang, K.-C. Ho, W.-N. Su and M.-H. Yeh, Carbon, 2022, 186, 406–415 CrossRef CAS.
- H. L. Tran, V. D. Dang, N. K. Dega, S.-M. Lu, Y.-F. Huang and R.-a. Doong, Sens. Actuators, B, 2022, 368, 132233 CrossRef CAS.
- R. V. Khose, P. Bangde, M. P. Bondarde, P. S. Dhumal, M. A. Bhakare, G. Chakraborty, A. K. Ray, P. Dandekar and S. Some, Spectrochim. Acta, Part A, 2022, 266, 120453 CrossRef CAS PubMed.
- O. Karaman, N. Özcan, C. Karaman, B. B. Yola, N. Atar and M. L. Yola, Mater. Today Chem., 2022, 23, 100666 CrossRef CAS.
- R. Kaimal, P. N. Mansukhlal, B. Aljafari, S. Anandan and M. Ashokkumar, Ultrason. Sonochem., 2022, 83, 105921 CrossRef CAS PubMed.
- X. Liu, J. Deng, J. Li, J. Dong, H. Liu, J. Zhao, X. Luo, D. Huo and C. Hou, Sens. Actuators, B, 2023, 376, 132989 CrossRef CAS.
- H. Wang, M. R. Revia, K. Wang, H. Wang, R. Revia, Q. Mu, G. Lin, C. Yen and M. Zhang, Nanoscale Horiz., 2020, 5, 573–579 RSC.
- H. Sun, H. Ji, E. Ju, Y. Guan, J. Ren and X. Qu, Chem.–Eur. J., 2015, 21, 3791–3797 CrossRef CAS PubMed.
- A. Al-Azmi and S. Keshipour, J. Colloid Interface Sci., 2022, 612, 701–709 CrossRef CAS PubMed.
- W. Zuo, L. Tang, J. Xiang, R. Ji, L. Luo, L. Rogée and S. Ping Lau, Appl. Phys. Lett., 2017, 110, 221901 CrossRef.
- D. D. Chronopoulos, A. Bakandritsos, M. Pykal, R. Zbořil and M. Otyepka, Appl. Mater. Today, 2017, 9, 60–70 CrossRef.
- X. Wang, G. Sun, P. Routh, D.-H. Kim, W. Huang and P. Chen, Chem. Soc. Rev., 2014, 43, 7067–7098 RSC.
- X. Qin, Z. Zhan, R. Zhang, K. Chu, Z. Whitworth and Z. Ding, Nanoscale, 2023, 15, 3864–3871 RSC.
- J. Peng, Z. Zhao, M. Zheng, B. Su, X. Chen and X. Chen, Sens. Actuators, B, 2020, 304, 127383 CrossRef CAS.
- C. Xia, X. Hai, X.-W. Chen and J.-H. Wang, Talanta, 2017, 168, 269–278 CrossRef CAS PubMed.
- S. e. Özönder, C. Ünlü, C. Güleryüz and L. Trabzon, ACS Omega, 2023, 8, 2112–2118 CrossRef.
- M. Chatterjee, P. Nath, S. Kadian, A. Kumar, V. Kumar, P. Roy, G. Manik and S. Satapathi, Sci. Rep., 2022, 12, 9061 CrossRef CAS PubMed.
- T. Enoki and K. Takai, Solid State Commun., 2009, 149, 1144–1150 CrossRef CAS.
- J. Jiang, W. Lu and J. Bernholc, Phys. Rev. Lett., 2008, 101, 246803 CrossRef CAS.
- Y. Sun, Y. Zheng, H. Pan, J. Chen, W. Zhang, L. Fu, K. Zhang, N. Tang and Y. Du, npj Quantum Mater., 2017, 2, 5 CrossRef.
- X. T. Zheng, A. Ananthanarayanan, K. Q. Luo and P. Chen, Small, 2015, 11, 1620–1636 CrossRef CAS.
- P. Tian, L. Tang, K. Teng and S. Lau, Mater. Today Chem., 2018, 10, 221–258 CrossRef CAS.
- H. Fei, R. Ye, G. Ye, Y. Gong, Z. Peng, X. Fan, E. L. Samuel, P. M. Ajayan and J. M. Tour, ACS Nano, 2014, 8, 10837–10843 CrossRef CAS.
- J. Feng, H. Dong, L. Yu and L. Dong, J. Mater. Chem. C, 2017, 5, 5984–5993 RSC.
- Y. Li, H. Shu, S. Wang and J. Wang, J. Phys. Chem. C, 2015, 119, 4983–4989 CrossRef CAS.
- B. Shen, J. Lang, R. Guo, X. Zhang and X. Yan, ACS Appl. Mater. Interfaces, 2015, 7, 25378–25389 CrossRef CAS PubMed.
- W. W. Liu, Y. Q. Feng, X. B. Yan, J. T. Chen and Q. J. Xue, Adv. Funct. Mater., 2013, 23, 4111–4122 CrossRef CAS.
- H. Zhu, L. Li, M. Shi, P. Xiao, Y. Liu and X. Yan, Chem. Eng. J., 2022, 437, 135301 CrossRef CAS.
- B. S. Reghunath, S. Rajasekaran, S. D. Kr, D. Pinheiro and J. R. J. Uc, Int. J. Hydrogen Energy, 2023, 48, 2906–2919 CrossRef CAS.
- J. Gong, Z. Zhang, S. Xi, W. Wang, J. Lu and P. Chen, Chem. Eng. J., 2023, 451, 138951 CrossRef CAS.
- Y. Yan, Y. Hou, Z. Yu, L. Tu, S. Qin, D. Lan, S. Chen, J. Sun and S. Wang, Chemosphere, 2022, 286, 131908 CrossRef CAS PubMed.
- N. Shaari, S. K. Kamarudin and R. Bahru, Int. J. Energy Res., 2021, 45, 1396–1424 CrossRef CAS.
- D. Geng, Y. Huang, S. Yuan, Y. Jiang, H. Ren, S. Zhang, Z. Liu, J. Feng, T. Wei and Z. Fan, Small, 2023, 2207227, DOI:10.1002/smll.202207227.
- M. Favaro, L. Ferrighi, G. Fazio, L. Colazzo, C. Di Valentin, C. Durante, F. Sedona, A. Gennaro, S. Agnoli and G. Granozzi, ACS Catal., 2015, 5, 129–144 CrossRef CAS.
- M. Dutta, S. Sarkar, T. Ghosh and D. Basak, J. Phys. Chem. C, 2012, 116, 20127–20131 CrossRef CAS.
- S. Mahalingam, A. Manap, R. Rabeya, K. S. Lau, C. H. Chia, H. Abdullah, N. Amin and P. Chelvanathan, Electrochim. Acta, 2023, 439, 141667 CrossRef CAS.
- M. Kaur, M. Kaur and V. K. Sharma, Adv. Colloid Interface Sci., 2018, 259, 44–64 CrossRef CAS PubMed.
- S. Chen, X. Chen, T. Xia and Q. Ma, Biosens. Bioelectron., 2016, 85, 903–908 CrossRef CAS PubMed.
- S. Gupta, J. Walden, A. Banaszak and S. B. Carrizosa, MRS Adv., 2018, 3, 817–824 CrossRef CAS.
- S. Naahidi, M. Jafari, F. Edalat, K. Raymond, A. Khademhosseini and P. Chen, J. Controlled Release, 2013, 166, 182–194 CrossRef CAS PubMed.
- X. Yuan, Z. Liu, Z. Guo, Y. Ji, M. Jin and X. Wang, Nanoscale Res. Lett., 2014, 9, 1–9 CrossRef PubMed.
- Y. Xie, B. Wan, Y. Yang, X. Cui, Y. Xin and L.-H. Guo, J. Environ. Sci., 2019, 77, 198–209 CrossRef CAS PubMed.
- X. Tian, B. Xiao, A. Wu, L. Yu, J. Zhou, J. Wang, N. Wang, H. Guan and Z. Shang, Toxicol. Res., 2016, 5, 1639–1648 CrossRef CAS PubMed.
- M. Nurunnabi, Z. Khatun, K. M. Huh, S. Y. Park, D. Y. Lee, K. J. Cho and Y.-k. Lee, ACS Nano, 2013, 7, 6858–6867 CrossRef CAS PubMed.
- T. A. Tabish, C. J. Scotton, D. C. J Ferguson, L. Lin, A. v. der Veen, S. Lowry, M. Ali, F. Jabeen, M. Ali and P. G. Winyard, Nanomedicine, 2018, 13, 1923–1937 CrossRef CAS PubMed.
- Z. M. Markovic, B. Z. Ristic, K. M. Arsikin, D. G. Klisic, L. M. Harhaji-Trajkovic, B. M. Todorovic-Markovic, D. P. Kepic, T. K. Kravic-Stevovic, S. P. Jovanovic and M. M. Milenkovic, Biomaterials, 2012, 33, 7084–7092 CrossRef CAS PubMed.
- B. Z. Ristic, M. M. Milenkovic, I. R. Dakic, B. M. Todorovic-Markovic, M. S. Milosavljevic, M. D. Budimir, V. G. Paunovic, M. D. Dramicanin, Z. M. Markovic and V. S. Trajkovic, Biomaterials, 2014, 35, 4428–4435 CrossRef CAS PubMed.
- Y. Zhou, H. Sun, F. Wang, J. Ren and X. Qu, Chem. Commun., 2017, 53, 10588–10591 RSC.
- D. Wang, L. Zhu, J.-F. Chen and L. Dai, Nanoscale, 2015, 7, 9894–9901 RSC.
- S.-G. Yim, Y. J. Kim, Y.-E. Kang, B. K. Moon, E. S. Jung and S. Y. Yang, Nanomaterials, 2018, 8, 959 CrossRef PubMed.
- H.-H. Cho, H. Yang, D. J. Kang and B. J. Kim, ACS Appl. Mater. Interfaces, 2015, 7, 8615–8621 CrossRef CAS PubMed.
- L. Feng, X.-Y. Tang, Y.-X. Zhong, Y.-W. Liu, X.-H. Song, S.-L. Deng, S.-Y. Xie, J.-W. Yan and L.-S. Zheng, Nanoscale, 2014, 6, 12635–12643 RSC.
- C.-Y. Poon, Q. Li, J. Zhang, Z. Li, C. Dong, A. W.-M. Lee, W.-H. Chan and H.-W. Li, Anal. Chim. Acta, 2016, 917, 64–70 CrossRef CAS PubMed.
- S. Bak, D. Kim and H. Lee, Curr. Appl. Phys., 2016, 16, 1192–1201 CrossRef.
- E. Hwang, H. M. Hwang, Y. Shin, Y. Yoon, H. Lee, J. Yang, S. Bak and H. Lee, Sci. Rep., 2016, 6, 39448 CrossRef CAS PubMed.
- S. Ryu, B. Lee, S. Hong, S. Jin, S. Park, S. H. Hong and H. Lee, Carbon, 2013, 63, 45–53 CrossRef CAS.
- M. J. Sweetman, S. M. Hickey, D. A. Brooks, J. D. Hayball and S. E. Plush, Adv. Funct. Mater., 2019, 29, 1808740 CrossRef.
- M. R. Younis, G. He, J. Lin and P. Huang, Front. Chem., 2020, 8, 424 CrossRef CAS PubMed.
- K. P. Loh, Q. Bao, G. Eda and M. Chhowalla, Nat. Chem., 2010, 2, 1015–1024 CrossRef CAS PubMed.
- A. Rakovich and T. Rakovich, J. Mater. Chem. B, 2018, 6, 2690–2712 RSC.
- S. Chung, R. A. Revia and M. Zhang, Adv. Mater., 2021, 33, 1904362 CrossRef CAS PubMed.
- S. Nsanzamahoro, W.-F. Wang, Y. Zhang, C.-B. Wang, Y. Iradukunda, Y.-P. Shi and J.-L. Yang, Sens. Actuators, B, 2023, 381, 133478 CrossRef CAS.
- B. Lee, G. A. Stokes, A. Valimukhametova, S. Nguyen, R. Gonzalez-Rodriguez, A. Bhaloo, J. Coffer and A. Naumov, Nanomaterials, 2023, 13, 805 CrossRef CAS PubMed.
- S. Asghari and M. Mahmoudifard, J. Biomed. Mater. Res., Part B, 2023, 111, 1121–1132 CrossRef CAS PubMed.
- H. Lu, W. Li, H. Dong and M. Wei, Small, 2019, 15, 1902136 CrossRef PubMed.
- H. Yan, Q. Wang, J. Wang, W. Shang, Z. Xiong, L. Zhao, X. Sun, J. Tian, F. Kang and S. H. Yun, Adv. Mater., 2023, 2210809, DOI:10.1002/adma.202210809.
- E. S. Zadeh, N. Ghanbari, Z. Salehi, S. Derakhti, G. Amoabediny, M. Akbari and M. A. Tokmedash, Mater. Chem. Phys., 2023, 127336, DOI:10.1016/j.matchemphys.2023.127336.
- J. Lin, Y. Huang and P. Huang, Adv. Drug Delivery Rev., 2018, 105, 242–254 CrossRef PubMed.
- W. R. Algar, M. Massey, K. Rees, R. Higgins, K. D. Krause, G. H. Darwish, W. J. Peveler, Z. Xiao, H.-Y. Tsai and R. Gupta, Chem. Rev., 2021, 121, 9243–9358 CrossRef CAS PubMed.
- S. Sarkar, H. Lee, H. G. Ryu, S. Singha, Y. M. Lee, Y. J. Reo, Y. W. Jun, K. H. Kim, W. J. Kim and K. H. Ahn, ACS Sens., 2020, 6, 148–155 CrossRef PubMed.
- L. Wang, B. Wu, W. Li, S. Wang, Z. Li, M. Li, D. Pan and M. Wu, Adv. Biosyst., 2018, 2, 1700191 CrossRef.
- C. Zhao, X. Song, Y. Liu, Y. Fu, L. Ye, N. Wang, F. Wang, L. Li, M. Mohammadniaei, M. Zhang, Q. Zhang and J. Liu, J. Nanobiotechnol., 2020, 18, 142 CrossRef CAS PubMed.
- L. Yin, J. Zhou, W. Li, J. Zhang and L. Wang, RSC Adv., 2019, 9, 9301–9307 RSC.
- H. Wang, Q. Mu, K. Wang, R. A. Revia, C. Yen, X. Gu, B. Tian, J. Liu and M. Zhang, Appl. Mater. Today, 2019, 14, 108–117 CrossRef PubMed.
- H. Yoon, M. Park, J. Kim, T. G. Novak, S. Lee and S. Jeon, Chem. Phys. Rev., 2021, 2, 031303 CrossRef.
- S. Ponrartana, M. M. Moore, S. S. Chan, T. Victoria, J. R. Dillman and G. B. Chavhan, Pediatr. Radiol., 2021, 51, 736–747 CrossRef PubMed.
- H. Wang, R. Revia, Q. Mu, G. Lin, C. Yen and M. Zhang, Nanoscale Horiz., 2020, 5, 573–579 RSC.
- Y. Xuan, R. Y. Zhang, X. S. Zhang, J. An, K. Cheng, C. Li, X. L. Hou and Y. D. Zhao, Nanotechnology, 2018, 29, 355101 CrossRef PubMed.
- R. Ganji Arjenaki, G. Samieepour, S. E. Sadat Ebrahimi, M. Pirali Hamedani, M. Saffari, M. Seyedhamzeh, A. N. Kamali, A. Najdian and M. Shafiee Ardestani, Arabian J. Chem., 2024, 17, 105518 CrossRef CAS.
- X. Sun, W. Cai and X. Chen, Acc. Chem. Res., 2015, 48, 286–294 CrossRef CAS PubMed.
- F. Jiang, D. Chen, R. Li, Y. Wang, G. Zhang, S. Li, J. Zheng, N. Huang, Y. Gu and C. Wang, Nanoscale, 2013, 5, 1137–1142 RSC.
- Z. Wang, J. Xia, C. Zhou, B. Via, Y. Xia, F. Zhang, Y. Li, L. Xia and J. Tang, Colloids Surf., B, 2013, 112, 192–196 CrossRef CAS PubMed.
- J. Dong, K. Wang, L. Sun, B. Sun, M. Yang, H. Chen, Y. Wang, J. Sun and L. Dong, Sens. Actuators, B, 2018, 256, 616–623 CrossRef CAS.
- P. Nigam, S. Waghmode, M. Louis, S. Wangnoo, P. Chavan and D. Sarkar, J. Mater. Chem. B, 2014, 2, 3190–3195 RSC.
- M. Thakur, A. Mewada, S. Pandey, M. Bhori, K. Singh, M. Sharon and M. Sharon, Mater. Sci. Eng., C, 2016, 67, 468–477 CrossRef CAS PubMed.
- L. Wang, B. Wu, W. Li, S. Wang, Z. Li, M. Li, D. Pan and M. Wu, Adv. Biosyst., 2018, 2, 1700191 CrossRef.
- S. Reagen, Y. Wu, X. Liu, R. Shahni, J. Bogenschuetz, X. Wu, Q. R. Chu, N. Oncel, J. Zhang, X. Hou, C. Combs, A. Vasquez and J. X. Zhao, ACS Appl. Mater. Interfaces, 2021, 13, 43952–43962 CrossRef CAS PubMed.
- X. Wu, F. Tian, W. Wang, J. Chen, M. Wu and J. X. Zhao, J. Mater. Chem. C, 2013, 1, 4676–4684 RSC.
- M. T. Hasan, B. H. Lee, C.-W. Lin, A. McDonald-Boyer, R. Gonzalez-Rodriguez, S. Vasireddy, U. Tsedev, J. Coffer, A. M. Belcher and A. V. Naumov, 2D Materials, 2021, 8, 035013 CrossRef CAS.
- B. H. Lee, R. L. McKinney, M. T. Hasan and A. V. Naumov, Materials, 2021, 14, 616 CrossRef CAS PubMed.
- T. Kulahava, N. Belko, M. Parkhats, A. Bahdanava, S. Lepeshkevich, V. Chizhevsky and D. Mogilevtsev, J. Photochem. Photobiol., B, 2023, 248, 112800 CrossRef CAS PubMed.
- Z. Qian, J. Ma, X. Shan, L. Shao, J. Zhou, J. Chen and H. Feng, RSC Adv., 2013, 3, 14571–14579 RSC.
- C. Martín, G. Jun, R. Schurhammer, G. Reina, P. Chen, A. Bianco and C. Ménard-Moyon, Small, 2019, 15, 1905405 CrossRef PubMed.
- K. Bhattacharya, S. P. Mukherjee, A. Gallud, S. C. Burkert, S. Bistarelli, S. Bellucci, M. Bottini, A. Star and B. Fadeel, Nanomed. Nanotechnol. Biol. Med., 2016, 12, 333–351 CrossRef CAS PubMed.
- B. Han, L. Sha, X. Yu, M. Yang, Y. Cao and J. Zhao, Biosens. Bioelectron., 2021, 176, 112913 CrossRef CAS PubMed.
- Q. Xiang, J. Huang, H. Huang, W. Mao and Z. Ye, RSC Adv., 2018, 8, 1820–1825 RSC.
- J. Zhao, G. Chen, L. Zhu and G. Li, Electrochem. Commun., 2011, 13, 31–33 CrossRef CAS.
- V. Vinoth, R. Kaimal, M. Selvamani, R. Michael, N. Pugazhenthiran, R. V. Mangalaraja, H. Valdés and S. Anandan, J. Electroanal. Chem., 2023, 934, 117302 CrossRef CAS.
- A. Xu, P. He, T. Huang, J. Li, X. Hu, P. Xiang, D. Chen, S. Yang, G. Wang and G. Ding, Synth. Met., 2018, 244, 106–112 CrossRef CAS.
- N. Li, A. Than, J. Chen, F. Xi, J. Liu and P. Chen, Biomater. Sci., 2018, 6, 779–784 RSC.
- D. Kurniawan, J. Mathew, M. R. Rahardja, H.-P. Pham, P.-C. Wong, N. V. Rao, K. Ostrikov and W.-H. Chiang, Small, 2023, 19, 2206813 CrossRef CAS PubMed.
- D. Kurniawan, B. A. Anjali, O. Setiawan, K. K. Ostrikov, Y. G. Chung and W.-H. Chiang, ACS Appl. Mater. Interfaces, 2022, 14, 1670–1683 CrossRef CAS PubMed.
- D. Kurniawan and W.-H. Chiang, Carbon, 2020, 167, 675–684 CrossRef CAS.
- L. Lin, M. Rong, S. Lu, X. Song, Y. Zhong, J. Yan, Y. Wang and X. Chen, Nanoscale, 2015, 7, 1872–1878 RSC.
- L. Lin, X. Song, Y. Chen, M. Rong, T. Zhao, Y. Wang, Y. Jiang and X. Chen, Anal. Chim. Acta, 2015, 869, 89–95 CrossRef CAS PubMed.
- Y.-Y. Chen, D. Kurniawan, S. M. Mousavi, P. V. Fedotov, E. D. Obraztsova and W.-H. Chiang, J. Mater. Chem. B, 2022, 10, 9654–9661 RSC.
- D. Kurniawan, R.-C. Jhang, K. K. Ostrikov and W.-H. Chiang, ACS Appl. Mater. Interfaces, 2021, 13, 34572–34583 CrossRef CAS PubMed.
- N. Li, A. Than, X. Wang, S. Xu, L. Sun, H. Duan, C. Xu and P. Chen, ACS Nano, 2016, 10, 3622–3629 CrossRef CAS PubMed.
- H. Sun, N. Gao, L. Wu, J. Ren, W. Wei and X. Qu, Chem.–Eur. J., 2013, 19, 13362–13368 CrossRef CAS PubMed.
- A. Ananthanarayanan, X. Wang, P. Routh, B. Sana, S. Lim, D. H. Kim, K. H. Lim, J. Li and P. Chen, Adv. Funct. Mater., 2014, 24, 3021–3026 CrossRef CAS.
- R. Guo, S. Zhou, Y. Li, X. Li, L. Fan and N. H. Voelcker, ACS Appl. Mater. Interfaces, 2015, 7, 23958–23966 CrossRef CAS PubMed.
- Z. S. Qian, X. Y. Shan, L. J. Chai, J. R. Chen and H. Feng, Biosens. Bioelectron., 2015, 68, 225–231 CrossRef CAS PubMed.
- X. Niu, Y. Zhong, R. Chen, F. Wang, Y. Liu and D. Luo, Sens. Actuators, B, 2018, 255, 1577–1581 CrossRef CAS.
- M. Liu, T. Liu, Y. Li, H. Xu, B. Zheng, D. Wang, J. Du and D. Xiao, Talanta, 2015, 143, 442–449 CrossRef CAS PubMed.
- R. Liu, J. Zhao, Z. Huang, L. Zhang, M. Zou, B. Shi and S. Zhao, Sens. Actuators, B, 2017, 240, 604–612 CrossRef CAS.
- Y. Dong, G. Li, N. Zhou, R. Wang, Y. Chi and G. Chen, Biosens. Bioelectron., 2012, 84, 8378–8382 CAS.
- W. Na, Z. Qu, X. Chen and X. Su, Sens. Actuators, B, 2018, 256, 48–54 CrossRef CAS.
- Z. S. Qian, X. Y. Shan, L. J. Chai, J. J. Ma, J. R. Chen and H. Feng, Biosens. Bioelectron., 2014, 60, 64–70 CrossRef CAS PubMed.
- H. Zhang, Y. Wang, D. Zhao, D. Zeng, J. Xia, A. Aldalbahi, C. Wang, L. San, C. Fan and X. J. A. a. m. Zuo, ACS Appl. Mater. Interfaces, 2015, 7, 16152–16156 CrossRef CAS PubMed.
- S. Huang, L. Wang, C. Huang, W. Su and Q. Xiao, Sens. Actuators, B, 2016, 234, 255–263 CrossRef CAS.
- Y.-C. Chen, W.-H. Chiang, D. Kurniawan, P.-C. Yeh, K.-i. Otake and C.-W. Kung, ACS Appl. Mater. Interfaces, 2019, 11, 35319–35326 CrossRef CAS PubMed.
- A. Muthurasu and V. Ganesh, Appl. Biochem. Biotechnol., 2014, 174, 945–959 CrossRef CAS PubMed.
- I. Vasilescu, S. A. Eremia, M. Kusko, A. Radoi, E. Vasile and G.-L. Radu, Biosens. Bioelectron., 2016, 75, 232–237 CrossRef CAS PubMed.
- D. Bhatnagar, I. Kaur and A. Kumar, Int. J. Biol. Macromol., 2017, 95, 505–510 CrossRef CAS PubMed.
- L. T. Tufa, S. Oh, J. Kim, K.-J. Jeong, T. J. Park, H.-J. Kim and J. Lee, Electrochim. Acta, 2018, 290, 369–377 CrossRef CAS.
- L. Xu, W. Mao, J. Huang, S. Li, K. Huang, M. Li, J. Xia and Q. Chen, Sens. Actuators, B, 2016, 230, 54–60 CrossRef CAS.
- X. Zhu, Z. Zhang, Z. Xue, C. Huang, Y. Shan, C. Liu, X. Qin, W. Yang, X. Chen and T. Wang, Biosens. Bioelectron., 2017, 89, 12054–12058 CAS.
- A. Abbas, T. A. Tabish, S. J. Bull, T. M. Lim and A. N. Phan, Sci. Rep., 2020, 10, 21262 CrossRef CAS PubMed.
- Z. Fan, Y. Li, X. Li, L. Fan, S. Zhou, D. Fang and S. Yang, Carbon, 2014, 70, 149–156 CrossRef CAS.
- Z. Qu, W. Na, X. Liu, H. Liu and X. Su, Anal. Chim. Acta, 2018, 997, 52–59 CrossRef CAS PubMed.
- S. Ge, J. He, C. Ma, J. Liu, F. Xi and X. Dong, Talanta, 2019, 199, 581–589 CrossRef CAS PubMed.
- J. Hassanzadeh and A. Khataee, Talanta, 2018, 178, 992–1000 CrossRef CAS PubMed.
- T. Ghosh, S. Chatterjee and E. Prasad, J. Phys. Chem. A, 2015, 119, 11783–11790 CrossRef CAS PubMed.
- S. Baluta, A. Lesiak and J. Cabaj, Electroanalysis, 2018, 30, 1781–1790 CrossRef.
- M. Arvand and S. Hemmati, Sens. Actuators, B, 2017, 238, 346–356 CrossRef CAS.
- J. Xi, C. Xie, Y. Zhang, L. Wang, J. Xiao, X. Duan, J. Ren, F. Xiao and S. Wang, ACS Appl. Mater. Interfaces, 2016, 8, 22563–22573 CrossRef CAS PubMed.
| This journal is © The Royal Society of Chemistry 2024 |
